Significance
We describe here the application of genetic synthetic lethal screening to the mammalian central nervous system. The principle of synthetic lethality is that factors that are dispensable in a healthy cell are rendered essential in a diseased cell; these factors thus define the pathways responsible for increased cellular vulnerability in that disease. Our synthetic lethality screening in mouse models of Huntington’s disease (HD) reveals that a glutathione peroxidase, Gpx6, can regulate the emergence of HD model symptoms in mice.
Keywords: Huntington’s disease, synthetic lethality, striatum, glutathione peroxidase, pooled screening
Abstract
Huntington’s disease, the most common inherited neurodegenerative disease, is characterized by a dramatic loss of deep-layer cortical and striatal neurons, as well as morbidity in midlife. Human genetic studies led to the identification of the causative gene, huntingtin. Recent genomic advances have also led to the identification of hundreds of potential interacting partners for huntingtin protein and many hypotheses as to the molecular mechanisms whereby mutant huntingtin leads to cellular dysfunction and death. However, the multitude of possible interacting partners and cellular pathways affected by mutant huntingtin has complicated efforts to understand the etiology of this disease, and to date no curative therapeutic exists. To address the general problem of identifying the disease-phenotype contributing genes from a large number of correlative studies, here we develop a synthetic lethal screening methodology for the mammalian central nervous system, called SLIC, for synthetic lethal in the central nervous system. Applying SLIC to the study of Huntington’s disease, we identify the age-regulated glutathione peroxidase 6 (Gpx6) gene as a modulator of mutant huntingtin toxicity and show that overexpression of Gpx6 can dramatically alleviate both behavioral and molecular phenotypes associated with a mouse model of Huntington’s disease. SLIC can, in principle, be used in the study of any neurodegenerative disease for which a mouse model exists, promising to reveal modulators of neurodegenerative disease in an unbiased fashion, akin to screens in simpler model organisms.
All of the major neurodegenerative diseases display characteristic nerve-cell (neuronal) vulnerability patterns, as well as an increased prevalence with advanced age. Due to the latter, it has been reasoned that knowledge of the normal aging-associated gene expression changes in vulnerable neuronal populations would reveal pathways that intersect with disease-associated molecular mechanisms. Numerous studies have examined age-related gene expression changes in human and rodent brain, revealing altered expression of genes involved in many cellular processes, including synaptic plasticity, mitochondrial function, proteasomal function, antioxidant responses, as well as DNA damage repair (1–4). However, these studies have focused almost exclusively on gene expression changes averaged over entire brain regions without resolving the multiple distinct neuronal cell types within these regions. To study normal age-associated molecular pathways in individual neurodegenerative disease-relevant cell types in situ, we used the translating ribosome affinity purification (TRAP) methodology (5, 6) to create cell-type–specific molecular profiles of cell populations during normal mouse brain aging. Due to their vulnerability in Huntington’s disease (HD), we profiled dopaminoceptive neurons in the striatum and cortex, as defined by cells that express the Drd1a or Drd2 dopamine receptors at 6 wk and 2 y of age (all research involving vertebrate animals was approved by the MIT institutional review board). Each cell type displayed a unique pattern of gene expression changes associated with aging that were consistent across technical replicates and across mice (Datasets S1–S4 and Fig. 1). Only five genes, including two predicted pseudogenes, displayed a statistically significant altered expression with aging in all cell types (Tnnt2, Gm5425, Rnd3, Pisd, and Pisd-ps3), indicating that there is not a common aging program across the cell types studied, but rather that even closely related cell types show distinct molecular changes during normal aging. Pathways analysis of age-regulated genes revealed several molecular pathways altered by aging in each cell type (Datasets S5–S8). In Drd2-expressing striatal neurons, which displayed the greatest number of altered gene pathways during aging, “glutathione-mediated detoxification” and “glutathione redox reactions” were among the top gene pathways altered with age (including the genes Gsta3, Gsta4, Gstm1, Gstm6, Gpx1, Gpx2, and Gpx6). Oxidative damage has long been linked to aging (7). Given that oxidative damage to DNA, proteins, and lipids has been reported to increase with age in the brain (8–10), the increases to glutathione-dependent enzymes that we report here likely reflect a homeostatic neuronal response to increased oxidative damage in this cell population.
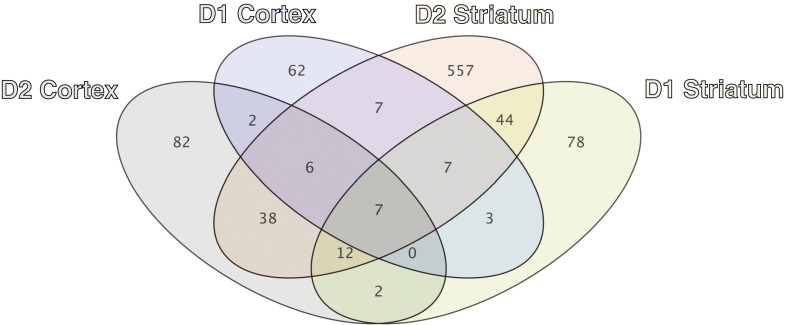
Fig. 1.
Gene expression changes associated with normal aging in cortical and striatal dopaminoceptive cell types. Venn diagram showing the number and overlap of statistically significant gene expression changes in dopamine receptor 1a (D1)- or dopamine receptor 2 (D2)-expressing cortical or striatal neurons, based on a comparison of mice aged 6 wk of age versus 2 y of age. Statistically significant changes are defined as genes displaying ≥1.2-fold change and a Benjamini–Hochberg adjusted P value from Welch’s t test of ≤0.05.
Although these experiments identified a promising set of age-regulated candidate genes that may be relevant to the age-associated progression of Huntington’s disease, they did not single out any one factor as contributing above others. In addition, the large aggregate number of changes observed in these cell types prevented us from inspecting the effects of each gene expression change on Huntington’s disease progression with traditional means such as mouse knockout or overexpression studies. To address this problem, we sought to develop a genetic screening platform that could be used in the mammalian nervous system in a fashion similar to the way that genetic screens are used in simpler model organisms such as Caenorhabditis elegans and Drosophila melanogaster. We term this methodology SLIC (for synthetic lethal in the CNS). SLIC is composed of four parts: a short hairpin RNA (shRNA) library, stereotaxic intracranial injection, mouse models of disease, and DNA sequencing (Fig. 2). The principle of synthetic lethality is that factors that are dispensable in a healthy cell are rendered essential in a diseased cell; these factors thus define the pathways responsible for increased cellular vulnerability in that disease. To identify synthetic lethal interactions, SLIC uses a pooled screen approach that was pioneered in yeast (11) and that has been adapted to identify tumor suppressors and oncogenes in mammalian cancer models (12–15). In SLIC, each neuron in a defined brain region receives a single perturbation, in this case a lentivirus encoding an shRNA, allowing many perturbations to be tested simultaneously in a single mouse, as opposed to one mouse being used as a screening vehicle for a single perturbation as in traditional knockout or knockdown experiments. Based on test injections into the mouse striatum, we calculate that we can transduce up to 2.8 × 105 striatal cells per mouse (SI Appendix, Fig. S1) and that over 80% of viral-transduced cells are neurons (SI Appendix, Fig. S2).
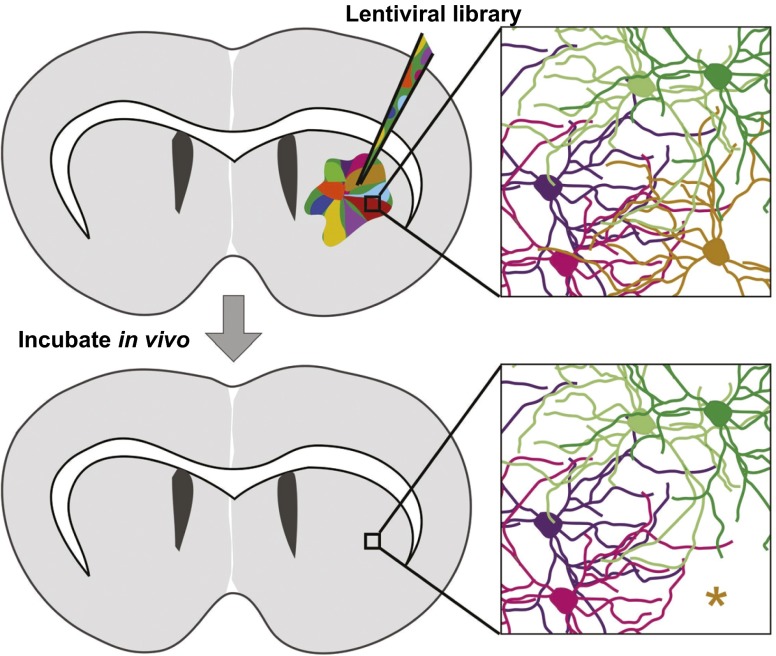
Fig. 2.
Synthetic lethal in the CNS (SLIC) screening. (Top) Lentiviral knockdown libraries are injected into the striatum, such that each neuron or glial cell receives a distinct element (schematized by different colors). Lentivirus integrates into the cell’s genome and expresses either a cDNA or shRNA. (Bottom) After incubation in vivo, cells that have received a synthetic lethal hit will die (*) and the representation of these library elements will be lost (an event that can be revealed by sequencing of all of the lentiviruses still present in the brain). As neurons in adult animals do not proliferate, the change in abundance detected by sequencing is due to cell loss. When injections are performed in a paired fashion, comparing disease model mice to wild-type littermates, genes that cause synthetic lethality only in combination with a disease-causing mutation can be identified.
For our first SLIC screen, we focused on Huntington’s disease, as it is an ideal test case for a genetic screen: the disease is monogenic, affects defined cell populations in an age-dependent manner, and several mouse models have been created that display minimal cell loss. This latter feature is particularly advantageous to our screening scheme, as synthetic lethal screens require a mild phenotype around which to screen for an enhanced phenotype. We performed a SLIC screen in the R6/2 Huntington’s disease model line (16), seeking genes that, when knocked down, would enhance mutant huntingtin toxicity. We used 95 shRNAs in our screen (Dataset S9), including 1 positive control shRNA, 7 negative control shRNAs, and 87 shRNAs targeting 28 candidate genes. The 28 candidates included 19 genes identified in our cell-type–specific aging study, six genes previously linked to Huntington’s disease, and three randomly chosen genes (see Dataset S9 for details). Viral pools were injected bilaterally into mouse striata of disease model and control littermates at 6 wk of age. Genomic DNA was then harvested at a control time point (2 d postinjection, once the lentivirus has integrated into the cellular DNA but before significant target protein knockdown), and two experimental time points at which we expected to observe synthetic lethality (4 and 6 wk postinjection). Finally, shRNA-coding sequences were amplified and sequenced to assess the relative abundance of the 95 shRNAs in the pool.
To validate our SLIC approach, we first focused on the positive and negative control shRNAs included in our screen. Comparison of viral shRNA representation in the wild-type control (nonmodel) mouse striatal samples at 4 wk versus 2 d revealed that the positive control lentivirus, carrying an shRNA targeting the Psmd2 gene product (a proteasomal subunit, depletion of which is expected to lead to cell death), was greatly reduced in representation, whereas negative controls, which have no expected target in the mouse genome, were not reduced in representation (Fig. 3A). We next asked which shRNAs were lost to a higher degree from R6/2 Huntington’s disease model mice versus control littermates at the 4 and 6 wk experimental time points. This comparison revealed that all shRNAs targeting Gpx6, a glutathione peroxidase that by homology is predicted to detoxify H2O2 to water, demonstrated synthetic lethality with mutant huntingtin. Using a gene-scoring scheme that incorporates the synthetic lethality ranks of the top two shRNAs for each gene, Gpx6 was the top-ranking gene at both the 4 and 6 wk time points (Fig. 3 B and C; Datasets S10–S12; SI Appendix). We also observed several shRNAs that were depleted in wild-type cells, and thus presumably are deleterious even in healthy cells. Importantly, these shRNAs were lost to an equal or lesser extent in R6/2 brains, demonstrating that the Gpx6 interaction is specific and not a consequence of cells in the disease model showing increased vulnerability to a broad spectrum of potential cell stressors (Fig. 3B).
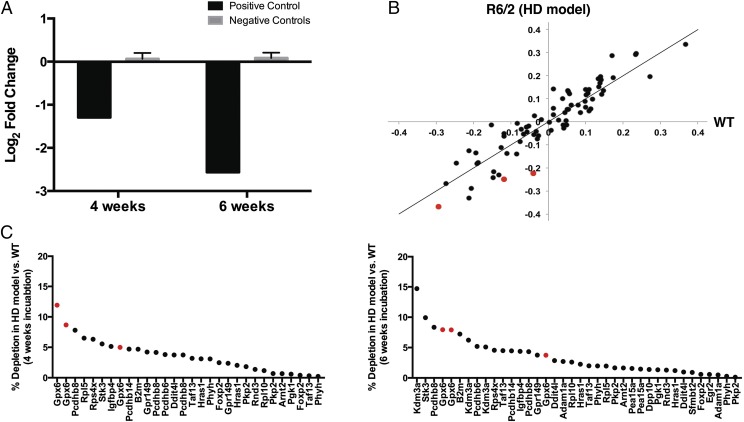
Fig. 3.
SLIC screening in mouse models of Huntington’s disease. (A) Changes in control shRNA representation, as determined by sequencing, in the striatum of wild-type animals at 4 and 6 wk after injection compared with the control 2-d time point. A negative number reflects loss versus the control time point. Error bars reflect ±SD across shRNAs. (B) shRNA representation at the first SLIC HD time point. The log2 fold changes in shRNA representation at 4 wk compared with the control 2-d time point for the HD model (R6/2, y axis) are plotted versus the log2 fold changes at the same two time points for wild-type controls (WT, x axis). The positive control targeting the Psmd2 gene product is not plotted for the purposes of scaling. Diagonal line represents equal representation for visual reference (x = y). Genes causing synthetic lethality are expected to be offset to the right of the diagonal. Gpx6-targeting shRNAs are denoted in red. (C) SLIC results for shRNAs showing synthetic lethality in the HD model. The y axes show the percentage of depletion of the indicated shRNAs in the HD model compared with wild-type controls, where “depletion” is defined as “the log2 fold change in representation between early (2 d) and late (4 wk, Left; 6 wk, Right) time points in each genotype” and “% depletion” is defined as “the difference in depletion in the disease model relative to wild-type controls {[(WT-R6/2)/WT] × 100}.” Sample values were averaged across four replicates. Multiple annotations of the same target gene on the x axis represent distinct shRNAs targeting these genes. Gpx6-targeting shRNAs are denoted in red.
Because little is known about Gpx6 function and expression, we next assessed its distribution across brain region and age. We found Gpx6 to be highly expressed in the olfactory bulb, striatum, and frontal cerebral cortex (SI Appendix, Fig. S3) and, confirming our TRAP results, observed that Gpx6 expression increases with age (SI Appendix, Fig. S4). Because our screen identified Gpx6 as a gene that, when reduced, enhances mutant huntingtin toxicity, we next asked whether overexpression of this gene product would have a therapeutic effect on phenotype progression in a Huntington’s disease mouse model. We overexpressed Gpx6 in the striatum of the R6/2 model and congenic wild-type control mice via adeno-associated viral (AAV) transduction starting at 6 wk of age. Two weeks after viral injection, we observed a dramatic rescue of open-field and rota-rod motor behavior in R6/2 mice, but no effect on motor behavior in wild-type control mice (Fig. 4A and SI Appendix, Fig. S6). Finally, analysis of a molecular marker of Huntington’s disease progression, loss of DARPP-32 striatal expression (17), revealed that Gpx6 overexpression also increases DARPP-32 expression in the R6/2 model (Fig. 4B). Thus, Gpx6 overexpression protects against both behavioral and molecular phenotypes associated with disease progression in this mouse model.
Fig. 4.
(A) Rescue of open-field motor behavior in Huntington’s disease model mice overexpressing Gpx6. Huntington’s disease model mice (R6/2) or wild-type (WT) congenic controls were injected in the striatum bilaterally with Gpx6 or control (EGFP-L10a construct expressing; EGFP-L10a is most prominently observed in the nucleolus) AAV9 virus at 6 wk of age. After 2 wk of recovery, motor function was assessed by open field assay. Average performance is plotted ±SEM for each data point, reflecting total distance in centimeters traveled during a 1-h interval (R6/2 + Gpx6, n = 10; R6/2+ control, n = 10; WT + Gpx6, n = 12; WT + control, n = 11). R6/2 + Gpx6 vs. R6/2 + control, P value = 0.0165; WT + Gpx6 vs. WT + control, P value = 0.7826 (no significance). (B) Increased DARPP-32 expression in Huntington’s disease model mice overexpressing Gpx6. Huntington’s disease model mice (R6/2) or wild-type (WT) congenic controls were unilaterally injected with control (EGFP-L10a construct; left hemisphere) or Gpx6 overexpressing (right hemisphere) AAV9 virus at 6 wk of age. After 2 wk of recovery, mice were killed and brain tissue was processed for indirect immunofluorescent staining. (Top) Representative images of R6/2 mice injected with Gpx6 and control AAV9. (Bottom) Quantitation of images (mean pixel intensity across imaging field) from equivalent points in the dorsal striatum; P value = 0.0026. No significant difference between control and Gpx6-injected hemispheres was observed in wild-type congenic controls (SI Appendix, Fig. S5). A.U. signifies arbitrary fluorescence units.
Our data demonstrate that our SLIC methodology can be used to identify genes that display synthetic lethality in combination with a disease-associated mutation. This screen tested 95 shRNAs using just a single mouse brain hemisphere per replicate, demonstrating its power and scaleability. In principle, both genome-wide overexpression and knockdown screens can be conducted in the mammalian central nervous system using SLIC. Our screen used constitutively expressing lentiviruses to target all striatal cells, but increased cellular specificity could be achieved through use of conditional systems such as the TVA/EnvA system (18). The gene identified in our screen as enhancing the toxicity of mutant huntingtin when knocked down, Gpx6, is particularly appealing given the abundant literature linking oxidative stress to Huntington’s disease pathophysiology (19), a study reporting that mice deficient in cellular glutathione peroxidase show enhanced vulnerability to a chemical model of Huntington’s disease (20), a recent study identifying glutathione peroxidase activity as a suppressor of mutant Huntingtin toxicity in yeast (21), and a study reporting Gpx6 protein levels to be altered in the striatum of human Huntington’s disease patients (22). Furthermore, blood–brain barrier-penetrant, orally active glutathione peroxidase-mimicking hydrogen peroxide scavengers have been identified (23, 24). Given that Gpx6 interacts genetically with the mutant huntingtin, and that we find that it is up-regulated with age, our screen suggests that increases to reactive oxygen species (hydrogen peroxide in particular) contribute to the enhancement of mutant huntingtin toxicity that is seen with advancing age.
Supplementary Material
Acknowledgments
We acknowledge members of the M.H. laboratory for discussions and feedback. This work was supported by National Institute of Neurological Disorders and Stroke Award R01NS085880 (to M.H.), a William N. and Bernice E. Bumpus Foundation Early Career Investigator Innovation Award (to M.H.), a JPB Foundation award (to M.H.), and a European Molecular Biology Organization long-term fellowship awarded to R. Shema.
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
Data deposition: Microarray data have been deposited at the National Center for Biotechnology Information Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE64140).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1417231112/-/DCSupplemental.
References
- 1.Xu X, et al. Gene expression atlas of the mouse central nervous system: Impact and interactions of age, energy intake and gender. Genome Biol. 2007;8(11):R234. doi: 10.1186/gb-2007-8-11-r234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lee CK, Weindruch R, Prolla TA. Gene-expression profile of the ageing brain in mice. Nat Genet. 2000;25(3):294–297. doi: 10.1038/77046. [DOI] [PubMed] [Google Scholar]
- 3.Zahn JM, et al. AGEMAP: A gene expression database for aging in mice. PLoS Genet. 2007;3(11):e201. doi: 10.1371/journal.pgen.0030201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lu T, et al. Gene regulation and DNA damage in the ageing human brain. Nature. 2004;429(6994):883–891. doi: 10.1038/nature02661. [DOI] [PubMed] [Google Scholar]
- 5.Heiman M, et al. A translational profiling approach for the molecular characterization of CNS cell types. Cell. 2008;135(4):738–748. doi: 10.1016/j.cell.2008.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Doyle JP, et al. Application of a translational profiling approach for the comparative analysis of CNS cell types. Cell. 2008;135(4):749–762. doi: 10.1016/j.cell.2008.10.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Harman D. Aging: A theory based on free radical and radiation chemistry. J Gerontol. 1956;11(3):298–300. doi: 10.1093/geronj/11.3.298. [DOI] [PubMed] [Google Scholar]
- 8.Mecocci P, et al. Oxidative damage to mitochondrial DNA shows marked age-dependent increases in human brain. Ann Neurol. 1993;34(4):609–616. doi: 10.1002/ana.410340416. [DOI] [PubMed] [Google Scholar]
- 9.Dei R, et al. Lipid peroxidation and advanced glycation end products in the brain in normal aging and in Alzheimer’s disease. Acta Neuropathol. 2002;104(2):113–122. doi: 10.1007/s00401-002-0523-y. [DOI] [PubMed] [Google Scholar]
- 10.Smith CD, et al. Excess brain protein oxidation and enzyme dysfunction in normal aging and in Alzheimer disease. Proc Natl Acad Sci USA. 1991;88(23):10540–10543. doi: 10.1073/pnas.88.23.10540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shoemaker DD, Lashkari DA, Morris D, Mittmann M, Davis RW. Quantitative phenotypic analysis of yeast deletion mutants using a highly parallel molecular bar-coding strategy. Nat Genet. 1996;14(4):450–456. doi: 10.1038/ng1296-450. [DOI] [PubMed] [Google Scholar]
- 12.Berns K, et al. A large-scale RNAi screen in human cells identifies new components of the p53 pathway. Nature. 2004;428(6981):431–437. doi: 10.1038/nature02371. [DOI] [PubMed] [Google Scholar]
- 13.Zender L, et al. An oncogenomics-based in vivo RNAi screen identifies tumor suppressors in liver cancer. Cell. 2008;135(5):852–864. doi: 10.1016/j.cell.2008.09.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Possemato R, et al. Functional genomics reveal that the serine synthesis pathway is essential in breast cancer. Nature. 2011;476(7360):346–350. doi: 10.1038/nature10350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Beronja S, et al. RNAi screens in mice identify physiological regulators of oncogenic growth. Nature. 2013;501(7466):185–190. doi: 10.1038/nature12464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mangiarini L, et al. Exon 1 of the HD gene with an expanded CAG repeat is sufficient to cause a progressive neurological phenotype in transgenic mice. Cell. 1996;87(3):493–506. doi: 10.1016/s0092-8674(00)81369-0. [DOI] [PubMed] [Google Scholar]
- 17.Bibb JA, et al. Severe deficiencies in dopamine signaling in presymptomatic Huntington’s disease mice. Proc Natl Acad Sci USA. 2000;97(12):6809–6814. doi: 10.1073/pnas.120166397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Beier KT, Samson ME, Matsuda T, Cepko CL. Conditional expression of the TVA receptor allows clonal analysis of descendants from Cre-expressing progenitor cells. Dev Biol. 2011;353(2):309–320. doi: 10.1016/j.ydbio.2011.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Browne SE, Ferrante RJ, Beal MF. Oxidative stress in Huntington’s disease. Brain Pathol. 1999;9(1):147–163. doi: 10.1111/j.1750-3639.1999.tb00216.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Klivenyi P, et al. Mice deficient in cellular glutathione peroxidase show increased vulnerability to malonate, 3-nitropropionic acid, and 1-methyl-4-phenyl-1,2,5,6-tetrahydropyridine. J Neurosci. 2000;20(1):1–7. doi: 10.1523/JNEUROSCI.20-01-00001.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mason RP, et al. Glutathione peroxidase activity is neuroprotective in models of Huntington’s disease. Nat Genet. 2013;45(10):1249–1254. doi: 10.1038/ng.2732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sorolla MA, et al. Proteomic and oxidative stress analysis in human brain samples of Huntington disease. Free Radic Biol Med. 2008;45(5):667–678. doi: 10.1016/j.freeradbiomed.2008.05.014. [DOI] [PubMed] [Google Scholar]
- 23.Imai H, Masayasu H, Dewar D, Graham DI, Macrae IM. Ebselen protects both gray and white matter in a rodent model of focal cerebral ischemia. Stroke. 2001;32(9):2149–2154. doi: 10.1161/hs0901.095725. [DOI] [PubMed] [Google Scholar]
- 24.Day BJ. Catalase and glutathione peroxidase mimics. Biochem Pharmacol. 2009;77(3):285–296. doi: 10.1016/j.bcp.2008.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.