Significance
The question as to how the proton motive force in mitochondria is distributed among the proteins that require a proton gradient for their work is one of the central unresolved questions in mitochondrial physiology and important for the mechanistic insight in the function of mitochondrial proteins. Our results suggest that the local separation of the proteins on the inner mitochondrial membrane makes it impossible for uncoupling protein 4 (UCP4) to uncouple phosphorylation from proton pumping. Nonetheless, UCP4 should be well able to shortcut excessive transmembrane proton gradients to thereby regulate reactive oxygen species production. It explains how the proton transporter may fulfill that function without being a real UCP like UCP1.
Keywords: mitochondrial membrane proteins, proton diffusion, direct stochastic optical reconstruction microscopy, uncoupling, reactive oxygen species
Abstract
Because different proteins compete for the proton gradient across the inner mitochondrial membrane, an efficient mechanism is required for allocation of associated chemical potential to the distinct demands, such as ATP production, thermogenesis, regulation of reactive oxygen species (ROS), etc. Here, we used the superresolution technique dSTORM (direct stochastic optical reconstruction microscopy) to visualize several mitochondrial proteins in primary mouse neurons and test the hypothesis that uncoupling protein 4 (UCP4) and F0F1-ATP synthase are spatially separated to eliminate competition for the proton motive force. We found that UCP4, F0F1-ATP synthase, and the mitochondrial marker voltage-dependent anion channel (VDAC) have various expression levels in different mitochondria, supporting the hypothesis of mitochondrial heterogeneity. Our experimental results further revealed that UCP4 is preferentially localized in close vicinity to VDAC, presumably at the inner boundary membrane, whereas F0F1-ATP synthase is more centrally located at the cristae membrane. The data suggest that UCP4 cannot compete for protons because of its spatial separation from both the proton pumps and the ATP synthase. Thus, mitochondrial morphology precludes UCP4 from acting as an uncoupler of oxidative phosphorylation but is consistent with the view that UCP4 may dissipate the excessive proton gradient, which is usually associated with ROS production.
Mitochondria are involved in a wide range of cell functions, including fatty acid oxidation, calcium homeostasis, apoptosis, reactive oxygen species (ROS) signaling, and above all, production of ATP (1, 2). In neurons, these organelles are transported along neuronal processes to provide energy for areas of high energy demand, such as synapses (3). To support their functions, mitochondria exhibit a complex morphology consisting of separate and functionally distinct outer mitochondrial membrane (OMM) and inner mitochondrial membrane (IMM). The latter is structurally organized into two domains: an inner boundary membrane (IBM) and a cristae membrane (CM) (4). The current hypotheses imply that the morphology/topology of the IMM is tightly related to biochemical function, the energy state, and the pathophysiological state of mitochondria (5). Whereas the OMM contains porins [e.g., voltage-dependent anion channel (VDAC)], which mediate its permeability to molecules up to 10 kDa, the IMM topology is highly complex. It is comprised of different transport proteins, the ATP synthase (complex V), and complexes I, III, and IV of the electron transport chain, which are responsible for generating the proton motive force (pmf); pmf represents the driving force for not only ATP synthesis, but also other protein-mediated transport activities (for example, phosphate, pyruvate, and glutamate transport). Uncoupling protein 1 (UCP1; thermogenin), a member of the UCP subfamily, is known to dissipate the inner membrane proton gradient for heat production. One of the widely discussed functions for UCP4—another member of the same subfamily that is localized in neurons and neurosensory cells (6–9)—is the regulation of ROS by decreasing the pmf (10, 11). Although there is no unambiguous evidence revealing the exact UCP4 function, it was shown that UCP4 transports protons similar to UCP1 (12). It is, therefore, assumed that UCP4 and other UCPs possibly compete for protons with other proton-consuming proteins, including ATP synthase, but this phenomenon has not yet been studied in detail (13).
Knowledge about exact protein localization at the mitochondrial inner membrane is of utmost importance for understanding the mechanisms behind the allocation of electrochemical potential to various demands, such as ATP production, thermogenesis, ROS regulation, etc. Because of resolution limitations, current data about IMM protein topography are scarce. By implementing immuno-EM, EM tomography, and live cell fluorescence microscopy, it was found that IBM and CM have different protein compositions (14–17). Few studies using superresolution microscopy have investigated nanoscale protein distribution, mainly focusing on respiratory chain proteins (18). In particular, there was strong evidence obtained in yeast, fibroblast-like COS cell line, and heart and liver mitochondria that ATP synthase and complexes I, III, and IV are mainly localized on the CM (14, 15, 19, 20). No data are available on the exact localization of UCPs along the IMM.
In this study, we test the hypothesis that proton gradient-consuming proteins UCP4 and F0F1-ATP synthase are spatially separated within and/or between individual neuronal mitochondria. Therefore, we performed a two-color analysis of pairwise fluorescence-labeled mitochondrial proteins UCP4, VDAC, and F0F1-ATP synthase at 30 nm spatial resolution using superresolution imaging by direct stochastic optical reconstruction microscopy (dSTORM).
Results and Discussion
Coexistence of Mitochondria with Specialized Protein Expression Levels.
We tested the hypothesis that proteins competing for the proton gradient may be located within different neuronal mitochondria. Therefore, we determined the colocalization of UCP4 with the respiratory chain protein ATP synthase using dual-color dSTORM. As a control, we used the VDAC, which is supposed to be abundant in the OMM of each mitochondrion (21). Both UCP4 and ATP synthase were stained with a specific primary antibody and secondary antibodies conjugated to Alexa 488 and Alexa 647, respectively.
To ensure that protein colocalization was correctly detected and rule out fluorophore-specific errors, we first used two differently colored secondary antibodies to label the same protein of interest; in this case, virtually all mitochondria should have been stained in both color channels. Indeed, the overlays obtained for UCP4 and ATP synthase yielded predominantly yellow patches, indicating that nearly all mitochondria were double positive (Fig. S1 A and B). To quantify the degree of colocalization, we counted the number of localizations per mitochondrion in each of two color channels; mitochondria were identified with a cluster analysis (details in Materials and Methods). In the correlation plots (Fig. S1 C and D), the data clearly grouped along the principal diagonal (), indicating a strong correlation between the two color channels. To facilitate comparison between the plots, we defined three sectors (I–III) specified by the polar angle φ: sectors I and III included mitochondria stained predominantly in the green or red channel, respectively, and sector II contained double-positive mitochondria. For both UCP4 and ATP synthase, sector II contained ≥94% of all points, revealing the high degree of protein colocalization.
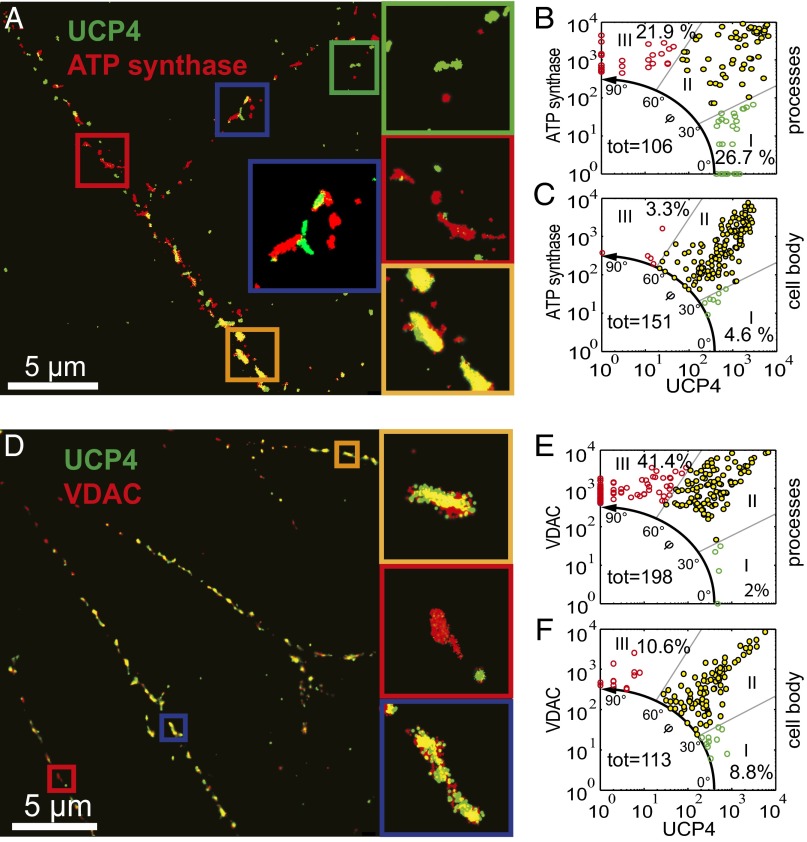
Next, we determined the colocalization of the UCP4 with ATP synthase and VDAC in processes of neurons. Immediately, the images looked different: a high degree of single-positive mitochondria was found for protein pairs UCP4/ATP synthase (Fig. 1 A and B) and UCP4/VDAC (Fig. 1 D and E). Quantitative analysis revealed only 51.4% double-positive mitochondria in case of UCP4/ATP synthase (Fig. 1B, sector II) and about 56% for UCP4/VDAC (Fig. 1E, sector II). Assuming that VDAC is present in every mitochondrion, 41.4% of all mitochondria were observed to be lacking UCP4 (Fig. 1E, sector III).
Fig. 1.
Dual-color dSTORM images of F0F1-ATP synthase, VDAC, and UCP4. Superresolution images are shown pairwise for (A–C) UCP4 (stained with Alexa 488; green) and ATP synthase (stained with Alexa 647; red) and (D–F) UCP4 and VDAC (stained with Alexa 647; red). The different colored boxes represent magnified views. For each protein pair, at least three different images were further analyzed for the number of detected localizations per mitochondrion and shown as scatter plots for UCP/ATP synthase [(B) neuronal processes and (C) neuronal body] and UCP4/VDAC [(E) neuronal processes and (F) neuronal body]. The plots are segmented in three different sectors (I–III) covering angles of 0° to 30°, 30° to 60°, and 60° to 90°, respectively. The colored dots represent clusters of dominating protein: UCP4 (green dots), ATP synthase (red dots in B and C), and VDAC (red dots in E and F). Yellow dots indicate clusters with similar amounts of localizations within the clusters (sector II). Percentages refer to the fractions of detected localizations in the respective sectors, and tot specifies the total amount of mitochondria analyzed.
The results clearly support the hypothesis that UCP4 and ATP synthase are separated at the intermitochondrial level in neuronal processes. Only one-half of all observed mitochondria contained both proteins at similar levels, and the other one-half was enriched in either UCP4 or ATP synthase (21.9% or 26.7%, respectively) (Fig. 1C, sectors I and III), implying that single neuronal mitochondria perform specialized functions. This finding confirms the functional heterogeneity of mitochondria, which has been proposed for various cell types (22–25). The preferential presence of UCP4 or ATP synthase in mitochondria can be rationalized by different energy demands in various neuronal areas. Energy demand is especially high in synapses, which require mitochondria containing higher levels of ATP synthase (26). The prevalence of highly energized mitochondria was observed in the periphery of several cells, including cortical neurons (23). UCP4 may be predominantly located in the neuronal cell body (8), where it would dissipate the excessive proton gradient. This location would also better fit UCP4’s function in neuronal metabolism (7, 27). Indeed, performing two-color dSTORM experiments in the neuronal cell body, we found nearly all mitochondria to be double positive for UCP4/ATP synthase (92.1%) (Fig. 1C) and UCP4/VDAC (80.6%) (Fig. 1F). It seems that the functional maturation of mitochondria in neuronal cells occurs either during transport or at their final destination but not at the site of their biogenesis.
Spatial Arrangement of UCP4 and ATP Synthase at the IMM.
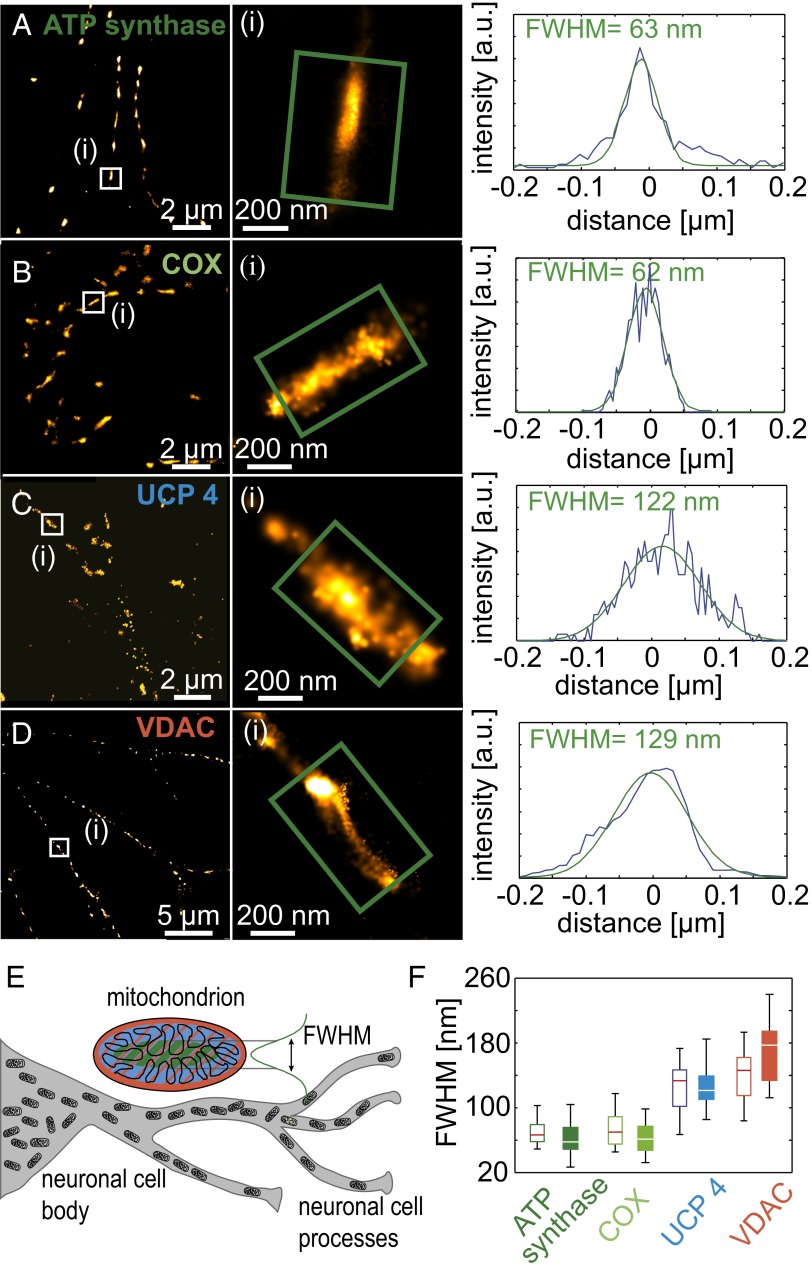
To evaluate the localization of UCP4 relative to other mitochondrial proteins in the same mitochondrion using dSTORM, we chose candidates known to be located at the outer (VDAC) and inner [ATP synthase and cytochrome c oxidase (COX)] mitochondrial membranes. All proteins were stained with the Alexa 647-tagged secondary antibody (Fig. 2 A–D). The intensity line plots (Fig. 2 A, Right, B, Right, C, Right, and D, Right) show average cross-sections along the minor mitochondrial axis for the magnified mitochondria as examples. The signal distributions were fitted with Gaussian functions. The full width at half-maximum (FWHM) describes the average position of the protein within single mitochondria; small values indicate preferential localization at the CM, whereas larger values indicate that proteins localize in the IBM or OMM (Fig. 2E). For every protein, at least 50 mitochondria were analyzed; median and upper and lower quartiles are shown as whisker–box plots for the FWHM of the distributions (Fig. 2F). As expected, VDAC, localized at the outer membrane, showed the broadest distribution, with a median of 144.9 nm in neuronal processes and 173.8 nm in neuronal cell body. Interestingly, we observed similarly broad distributions of 132.3 and 119.2 nm for UCP4 (in processes and cell body, respectively), whereas ATP synthase (65.7 and 57.9 nm, respectively) and COX (69.3 and 60.7 nm, respectively) were distributed significantly closer to the center of the mitochondrion and therefore, were assigned to the CM. Although the expression level of proteins differed between neuronal processes and the neuronal cell body (Fig. 1 B, C, E, and F), their distribution within mitochondria was not affected by the location of mitochondria within the cell (Fig. 2F). The results of these experiments confirm the localization of the respiratory chain proteins in close proximity at the CM; they further imply that mitochondrial proteins, which compete for the proton gradient, populate spatially separated areas within the mitochondria.
Fig. 2.
Spatial arrangement of F0F1-ATP synthase, COX, UCP4, and VDAC within single mitochondria. Superresolution images of (A) ATP synthase, (B) COX, (C) UCP4, and (D) VDAC stained with Alexa 647 and recorded with dSTORM are shown. For each protein, one example (white boxes in A–D) is shown as a magnified view in Center (green boxes in A–D). The areas in the green boxes were fitted with Gaussian intensity distributions and presented as cross-section plots with the obtained FWHM for each protein. (E) The scheme shows the different neuronal regions (gray) that were analyzed. In the magnified view of a mitochondrion, the colored ellipses represent CM (green), IBM (violet), and OMM (red). (F) Whisker–box plots represent the median and lower and upper quartile of the obtained widths (FWHM) for the different protein distributions; each box contains data of at least 50 mitochondria for neuronal processes (white boxes) and neuronal cell bodies (colored boxes).
The separation of UCP4 and ATP synthase at the IMM may be an important condition for their functional decoupling so that no direct competition for the proton gradient would occur between these proteins. Mitchell’s theory (28) assumes that the small compartments at both sides of the IMM ensure fast pH equilibration of their volumes; consequently, under equilibrium conditions, the electrochemical potential would be identical along the entire inner membrane surface. Thus, despite their different localization inside the mitochondria, all proteins would experience the same pmf. However, an increasing amount of evidence implies that protons do not easily equilibrate between the membrane surface and the bulk because of the presence of an energy barrier that has recently been quantified (29, 30). It suggests that proton uptake (or release) transpires at the membrane surface (31) and does not occur from the bulk of the mitochondrial matrix or the bulk of the intermembrane space. As a result of both the membrane proteins’ activity and proton surface to bulk release, the local surface proton concentration varies along the membrane (32, 33). It implies that proteins colocalized with the proton pumps on the CM (e.g., ATP synthase) (34, 35) will experience the highest pmf, whereas the pmf is much lower for proteins localized at the IBM (e.g., UCP4).
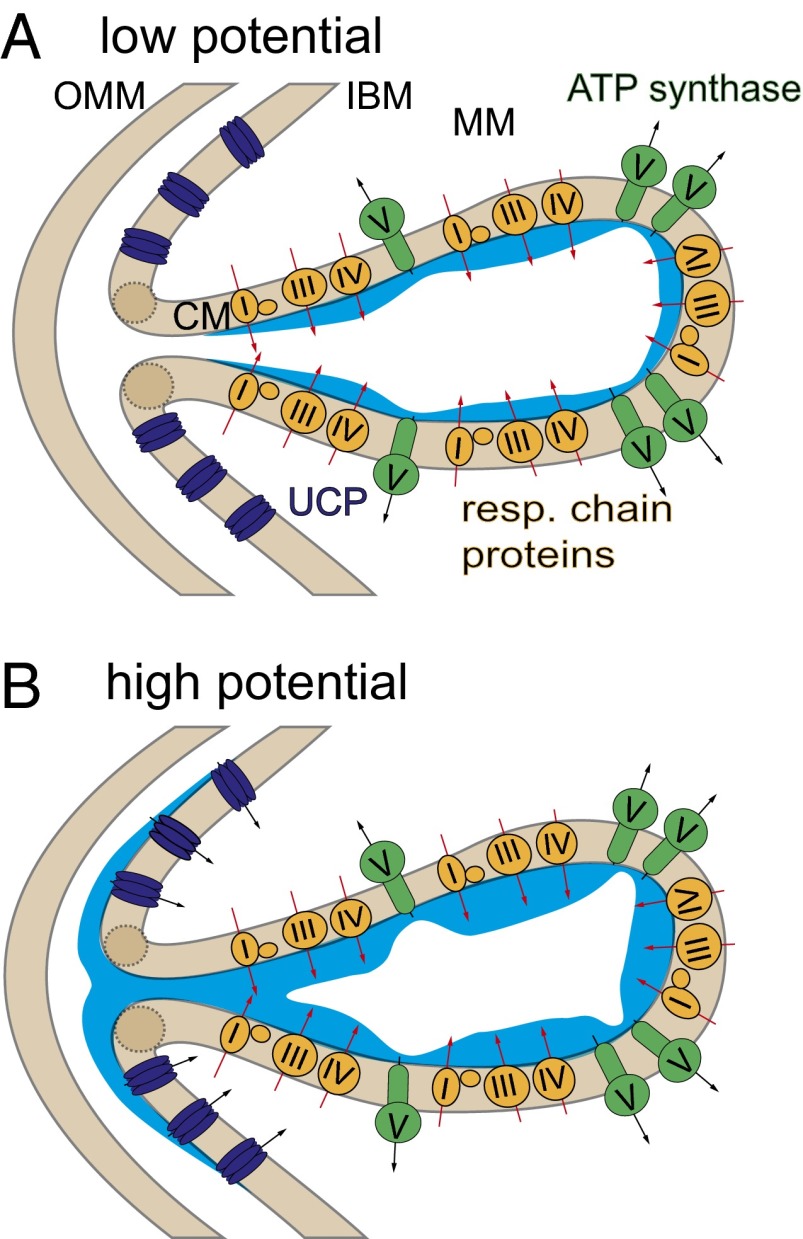
Fig. 3 visualizes the concept of mild uncoupling, originally proposed by Skulachev (36), for neuronal mitochondria. Because UCP4 can only locally decrease proton gradients, it will not be capable of hampering ATP synthesis at working potentials, because the uncoupling occurs at a remote position in respect to the location of the CM (Fig. 3A). The maximal transmembrane proton gradient is limited by fast lateral proton diffusion from the cristae to UCP4, which acts as a proton sink (Fig. 3B). Because UCP4 activity determines the maximal proton gradient at the cristae, it may be regarded as a regulator of ROS production. It is consistent with the hypothesis that UCPs will only lower the mitochondrial membrane potential if it exceeds a certain threshold (37).
Fig. 3.
Scheme of local proton gradients on the IMM at (A) low and (B) high potentials. Although the respiratory chain proteins (yellow) localized at the CM build up the proton gradient, ATP synthase (green), also found at the CM, is its main consumer. UCP4, localized at the IBM, depletes the proton gradient along the membrane only if it becomes excessive. Areas marked in blue indicate local proton concentration. MM, mitochondrial matrix.
Materials and Methods
Primary Neuronal Cell Culture.
The neurons were prepared as previously described (38). In brief, brains were removed from embryonic OFI/SPF mouse (E14). The mesencephali were carefully isolated and cut in small pieces in 0.1 M PBS. Afterward, the neuronal tissue was triturated and homogenized with the help of Pasteur pipettes in DMEM supplemented with heat-inactivated FCS (10% vol/vol), 25 mM Hepes buffer, 2 mM glutamine, 30 mM glucose, 10 U/mL penicillin, and 10 g/mL streptomycin. The dissociated neurons were resuspended, seeded into six-well plates at a density of 750 × 103 cells/mL (3 mL per well) on poly-l-lysine–coated coverslips (0.1 mg/mL in PBS for 1 h), and cultured at 37 °C in a CO2 incubator [5% (vol/vol); relative humidity > 80%].
After 7 d in vitro, cells were fixed in prewarmed 4% paraformaldehyde at room temperature (RT) for 10 min and washed three times in 0.1 M PBS. The autofluorescence of paraformaldehyde was quenched with 50 mM ammonium chloride in 0.1 M PBS for 10 min at RT. After washing them three times with 0.1 M PBS, the cells were permeabilized with 0.5% Triton X-100 and blocked with normal goat serum (Vectashield; Vector Laboratories) for 60 min at RT. Subsequently, the cells were labeled overnight at 4 °C with appropriate primary antibodies (rabbit anti-UCP4, 1:500; mouse anti-ATP synthase subunit-β, 1:1,000, 21351; Invitrogen; mouse anti-COXIV, 1:1,000, 14744; Abcam; mouse anti-VDAC, 1:1,000, 14734; Abcam). The antibodies against F0F1-ATP synthase and UCP4 have previously been shown to be suitable for dSTORM (39) and specific for immunochemistry (9), respectively. After washing the cells, they were incubated with corresponding secondary antibodies in dilution at 1:1,000 (Alexa Fluor 488-, Alexa Fluor 647-, and Alexa Fluor 633-conjugated goat anti-mouse IgG; A-21236, A-11001, A-11008, and A-21244; Invitrogen). The high degree of colocalization shown in Fig. S1 (Results and Discussion) indirectly confirmed the specificity of the secondary antibody labeling. Samples were measured using a blinking buffer containing 50 mM cysteamine, 40 μg/mL catalase, 10% (vol/vol) glucose, and 0.5 μg/mL glucose oxidase in PBS adjusted to pH 7.4 to optimize the ratio of bright vs. dark fluorophores and maximize the number of photons per fluorophore (40).
Superresolution Microscopy.
To obtain superresolution images, we used dSTORM (41). Measurements were performed using a custom-built single-molecule microscope. A 405-nm laser (100 mW; Coherent), a 488-nm laser (200 mW; Coherent), and a 642-nm laser (200 mW; Coherent) were coupled through a single-mode fiber (QiOptics) into a Polytrope and Yanus (TILL Photonics) mounted on an inverted Zeiss Axiovert 200 microscope. The beam was then focused onto the back-focal plane of a high numerical aperture objective (α–Plan-Apochromat 100×/1.46; Zeiss) for highly inclined illumination (illumination intensity ∼ 1 kW/cm2) (42). Emission light was filtered using appropriate filter sets for Alexa 488 and Alexa 647. The emission light was imaged with a back-illuminated iXon DU 897 EMCCD camera (Andor Technology Ltd.), which was water-cooled to −80 °C. The 488- and 642-nm excitation lasers were switched between subsequent frames for dual-color dSTORM imaging, whereas the power of the 405-nm activation laser was subsequently increased over the time course of the measurement to guarantee a constant number of localizations per frame. The lateral drift was typically smaller than 50 nm/h, and drift correction was implemented at the level of the analysis software (see below). Analyzing the widths of the individual signals showed no significant defocusing of the images during the measurements, and therefore, it was not corrected.
Localization Analysis.
The data were analyzed as previously described (43). Briefly, smoothing, nonmaximum suppression, and thresholding revealed the possible locations of single fluorophore molecules. Selected regions of interest were fitted by a pixelated Gaussian function and a homogeneous photon background with a maximum likelihood estimator for Poisson distributed data using a freely available fast GPU (graphics processing unit) fitting routine (44) on a GeForce GT 550 Ti (Nvidia).
We typically acquired 15,000 frames for the reconstruction and data analysis per channel. Lateral drift was corrected based on the imaged features. For drift corrections, subblocks of 3,000 frames, on average, were used to reconstruct one superresolution image. The displacements between the reconstructed image blocks were determined by image correlation, and the maximum was obtained by fitting with elliptical Gaussian function. The displacements corresponding to each time point were averaged using a robust estimator that was interpolated by a spline and used to correct the position of each localization. We estimated that the residual errors for the corrected positions were about 5 nm.
For Alexa 488, 1,100 photons were detected, on average, resulting in an average localization precision of 14 nm (45). For Alexa 647, an average of 2,085 photons were detected, yielding slightly improved average localization precision of 11.2 nm. Only localizations with a precision accuracy better than 30 nm were considered (typically 400,000–700,000 localizations per dSTORM image) for visualization and data analysis.
To reduce overcounting artifacts because of repeated observations of the same fluorophore, we collated multiple localizations from consecutive frames (interrupted by no more than two dark frames) into a single localization if they had a mutual distance < 90 nm; collating removed 56% of the detected localizations. In addition, we only accepted localizations with more than 15 neighbors within a 60-nm search radius, because they most likely corresponded to intact mitochondria; this procedure further reduced the number of localizations to about 28%.
The localization data were rendered using the Thomson blurring (46). Briefly, each localization is represented as a 2D Gaussian function with a width according to the precision of the respective localization determined from the fitted number of photons and background (45). All analysis software was written in MATLAB. All quantitative analyses were done using the list of localization coordinates and not the processed images; the latter are only presented for visualization.
Cluster Analysis.
We applied a cluster finding algorithm to identify mitochondria in both color channels. The reconstructed image was blurred with a Gaussian function with an SD of 100 nm, and a water-shedding algorithm was applied to segment the individual clusters. Furthermore, a linear regression model was fitted to the elliptical clouds of localizations to determine the lengths of their major and minor axes. To remove spherical vesicles from the analysis, the identified clusters were filtered using the Pearson coefficient-ρ in the ranges from −1 to −0.3 and from 0.3 to 1; in addition, only clusters with more than 400 localizations were taken into account. Together with the filters described above, ∼20% of all detected localizations were finally included for analysis.
Three different approaches were tested for determining clusters’ sizes. First, a binning of the data along the minor or major axis resulted in a histogram that we fitted with a Gaussian function to estimate the size by FWHM; this strategy was used for all quantifications shown in Fig. 2. Second, after rotational alignment of the clusters based on the linear regression model, SDs were calculated along the minor and major axes to quantify the mitochondrial dimensions. Third, cluster lengths and widths were calculated using the positions where the Gaussian blurred image decreased below 25% of the maximum. The three approaches showed no significant difference in the relative spatial distributions for the different analyzed proteins.
Comparison of Protein Numbers at Mitochondrial Membranes.
In photoswitching microscopy, the number of obtained single-molecule localizations usually exceeds the number of colocalized protein copies, partly because of repeated appearances of the same chromophore (47) during the recorded stream of images and partly because of a priori unknown degrees of labeling (48). In general, the total number of single-molecule localizations resulting from N copies of a protein is given by , with Lsm being the number of counts per single-protein molecule at the applied labeling conditions, and being the degrees of saturation of primary and secondary antibody labeling normalized to the maximally achievable labeling, respectively, and being the accessibility of the protein to antibody labeling (48). Although it is possible to determine Lsm, , and , the unknown accessibility factor renders calculation of the protein copy number N difficult. We, thus, performed here a comparative analysis, which yields the copy number ratios instead of absolute numbers. We first ensured that reducing the concentrations of primary and secondary antibodies led to similar numbers of detected fluorophores, indicating saturation of the labeling (39) (i.e., and ). In the data analysis, we corrected for multiple detections of the same fluorophore within one burst: localizations occurring in consecutive frames within a circle of 100 nm in diameter were treated as one count.
In the comparative analysis, the angle φ in the correlation plots specifies the count number ratio , with Lx and Ly being the obtained single-molecule localizations for the compared compounds plotted on the x and y axes, respectively. Assuming similar accessibility of each mitochondrion for the different antibodies () and considering saturating binding conditions, we get . In our experiments, we set the imaging conditions such that the two color channels yielded similar amounts of single-molecule localizations (Fig. S1, in which most data points are located in sector II at an angle ), yielding . Taken together, within the experimental errors, provides a valid estimate of the copy number ratios of the compared proteins.
Supplementary Material
Acknowledgments
We thank Barbara Kranner (Institute for Medical Biochemistry, University of Veterinary Medicine) for the preparation of neuronal cells. We also thank Peter Pohl for the valuable discussion and Q. Beatty for excellent editorial assistance. This work was partly supported by Austrian Research Fund Grants F3519-B20 (to G.J.S.) and P25123-B20 (to E.E.P.). E.K. was supported by a Long-Term Fellowship from Federation of European Biochemical Societies (FEBS).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1415261112/-/DCSupplemental.
References
- 1.Nunnari J, Suomalainen A. Mitochondria: In sickness and in health. Cell. 2012;148(6):1145–1159. doi: 10.1016/j.cell.2012.02.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dröse S, Brandt U. Molecular mechanisms of superoxide production by the mitochondrial respiratory chain. Adv Exp Med Biol. 2012;748:145–169. doi: 10.1007/978-1-4614-3573-0_6. [DOI] [PubMed] [Google Scholar]
- 3.Chang DTW, Reynolds IJ. Mitochondrial trafficking and morphology in healthy and injured neurons. Prog Neurobiol. 2006;80(5):241–268. doi: 10.1016/j.pneurobio.2006.09.003. [DOI] [PubMed] [Google Scholar]
- 4.Neupert W. SnapShot: Mitochondrial architecture. Cell. 2012;149(3):722–722.e1. doi: 10.1016/j.cell.2012.04.010. [DOI] [PubMed] [Google Scholar]
- 5.Mannella CA, Lederer WJ, Jafri MS. The connection between inner membrane topology and mitochondrial function. J Mol Cell Cardiol. 2013;62:51–57. doi: 10.1016/j.yjmcc.2013.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mao W, et al. UCP4, a novel brain-specific mitochondrial protein that reduces membrane potential in mammalian cells. FEBS Lett. 1999;443(3):326–330. doi: 10.1016/s0014-5793(98)01713-x. [DOI] [PubMed] [Google Scholar]
- 7.Liu D, et al. Mitochondrial UCP4 mediates an adaptive shift in energy metabolism and increases the resistance of neurons to metabolic and oxidative stress. Neuromolecular Med. 2006;8(3):389–414. doi: 10.1385/NMM:8:3:389. [DOI] [PubMed] [Google Scholar]
- 8.Smorodchenko A, et al. Comparative analysis of uncoupling protein 4 distribution in various tissues under physiological conditions and during development. Biochim Biophys Acta. 2009;1788(10):2309–2319. doi: 10.1016/j.bbamem.2009.07.018. [DOI] [PubMed] [Google Scholar]
- 9.Smorodchenko A, Rupprecht A, Fuchs J, Gross J, Pohl EE. Role of mitochondrial uncoupling protein 4 in rat inner ear. Mol Cell Neurosci. 2011;47(4):244–253. doi: 10.1016/j.mcn.2011.03.002. [DOI] [PubMed] [Google Scholar]
- 10.Krauss S, Zhang CY, Lowell BB. The mitochondrial uncoupling-protein homologues. Nat Rev Mol Cell Biol. 2005;6(3):248–261. doi: 10.1038/nrm1592. [DOI] [PubMed] [Google Scholar]
- 11.Ramsden DB, et al. Human neuronal uncoupling proteins 4 and 5 (UCP4 and UCP5): Structural properties, regulation, and physiological role in protection against oxidative stress and mitochondrial dysfunction. Brain Behav. 2012;2(4):468–478. doi: 10.1002/brb3.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hoang T, Smith MD, Jelokhani-Niaraki M. Toward understanding the mechanism of ion transport activity of neuronal uncoupling proteins UCP2, UCP4, and UCP5. Biochemistry. 2012;51(19):4004–4014. doi: 10.1021/bi3003378. [DOI] [PubMed] [Google Scholar]
- 13.Nicholls DG, Budd SL. Mitochondria and neuronal survival. Physiol Rev. 2000;80(1):315–360. doi: 10.1152/physrev.2000.80.1.315. [DOI] [PubMed] [Google Scholar]
- 14.Vogel F, Bornhövd C, Neupert W, Reichert AS. Dynamic subcompartmentalization of the mitochondrial inner membrane. J Cell Biol. 2006;175(2):237–247. doi: 10.1083/jcb.200605138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wurm CA, Jakobs S. Differential protein distributions define two sub-compartments of the mitochondrial inner membrane in yeast. FEBS Lett. 2006;580(24):5628–5634. doi: 10.1016/j.febslet.2006.09.012. [DOI] [PubMed] [Google Scholar]
- 16.Suppanz IE, Wurm CA, Wenzel D, Jakobs S. The m-AAA protease processes cytochrome c peroxidase preferentially at the inner boundary membrane of mitochondria. Mol Biol Cell. 2009;20(2):572–580. doi: 10.1091/mbc.E07-11-1112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stoldt S, et al. The inner-mitochondrial distribution of Oxa1 depends on the growth conditions and on the availability of substrates. Mol Biol Cell. 2012;23(12):2292–2301. doi: 10.1091/mbc.E11-06-0538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jakobs S, Wurm CA. Super-resolution microscopy of mitochondria. Curr Opin Chem Biol. 2014;20:9–15. doi: 10.1016/j.cbpa.2014.03.019. [DOI] [PubMed] [Google Scholar]
- 19.Gilkerson RW, Selker JM, Capaldi RA. The cristal membrane of mitochondria is the principal site of oxidative phosphorylation. FEBS Lett. 2003;546(2-3):355–358. doi: 10.1016/s0014-5793(03)00633-1. [DOI] [PubMed] [Google Scholar]
- 20.Davies KM, et al. Macromolecular organization of ATP synthase and complex I in whole mitochondria. Proc Natl Acad Sci USA. 2011;108(34):14121–14126. doi: 10.1073/pnas.1103621108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Colombini M, Mannella CA. VDAC, the early days. Biochim Biophys Acta. 2012;1818(6):1438–1443. doi: 10.1016/j.bbamem.2011.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kuznetsov AV, Margreiter R. Heterogeneity of mitochondria and mitochondrial function within cells as another level of mitochondrial complexity. Int J Mol Sci. 2009;10(4):1911–1929. doi: 10.3390/ijms10041911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Collins TJ, Berridge MJ, Lipp P, Bootman MD. Mitochondria are morphologically and functionally heterogeneous within cells. EMBO J. 2002;21(7):1616–1627. doi: 10.1093/emboj/21.7.1616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Breckwoldt MO, et al. Multiparametric optical analysis of mitochondrial redox signals during neuronal physiology and pathology in vivo. Nat Med. 2014;20(5):555–560. doi: 10.1038/nm.3520. [DOI] [PubMed] [Google Scholar]
- 25.Waagepetersen HS, et al. Cellular mitochondrial heterogeneity in cultured astrocytes as demonstrated by immunogold labeling of alpha-ketoglutarate dehydrogenase. Glia. 2006;53(2):225–231. doi: 10.1002/glia.20276. [DOI] [PubMed] [Google Scholar]
- 26.Mironov SL. Complexity of mitochondrial dynamics in neurons and its control by ADP produced during synaptic activity. Int J Biochem Cell Biol. 2009;41(10):2005–2014. doi: 10.1016/j.biocel.2009.04.009. [DOI] [PubMed] [Google Scholar]
- 27.Rupprecht A, et al. Uncoupling protein 2 and 4 expression pattern during stem cell differentiation provides new insight into their putative function. PLoS ONE. 2014;9(2):e88474. doi: 10.1371/journal.pone.0088474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mitchell P. Coupling of phosphorylation to electron and hydrogen transfer by a chemi-osmotic type of mechanism. Nature. 1961;191:144–148. doi: 10.1038/191144a0. [DOI] [PubMed] [Google Scholar]
- 29.Springer A, Hagen V, Cherepanov DA, Antonenko YN, Pohl P. Protons migrate along interfacial water without significant contributions from jumps between ionizable groups on the membrane surface. Proc Natl Acad Sci USA. 2011;108(35):14461–14466. doi: 10.1073/pnas.1107476108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang C, et al. Water at hydrophobic interfaces delays proton surface-to-bulk transfer and provides a pathway for lateral proton diffusion. Proc Natl Acad Sci USA. 2012;109(25):9744–9749. doi: 10.1073/pnas.1121227109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ojemyr LN, Lee HJ, Gennis RB, Brzezinski P. Functional interactions between membrane-bound transporters and membranes. Proc Natl Acad Sci USA. 2010;107(36):15763–15767. doi: 10.1073/pnas.1006109107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rieger B, Junge W, Busch KB. Lateral pH gradient between OXPHOS complex IV and F(0)F(1) ATP-synthase in folded mitochondrial membranes. Nat Commun. 2014;5:3103. doi: 10.1038/ncomms4103. [DOI] [PubMed] [Google Scholar]
- 33.Song DH, et al. Biophysical significance of the inner mitochondrial membrane structure on the electrochemical potential of mitochondria. Phys Rev E Stat Nonlin Soft Matter Phys. 2013;88(6):062723. doi: 10.1103/PhysRevE.88.062723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Strauss M, Hofhaus G, Schröder RR, Kühlbrandt W. Dimer ribbons of ATP synthase shape the inner mitochondrial membrane. EMBO J. 2008;27(7):1154–1160. doi: 10.1038/emboj.2008.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wilkens V, Kohl W, Busch K. Restricted diffusion of OXPHOS complexes in dynamic mitochondria delays their exchange between cristae and engenders a transitory mosaic distribution. J Cell Sci. 2013;126(Pt 1):103–116. doi: 10.1242/jcs.108852. [DOI] [PubMed] [Google Scholar]
- 36.Skulachev VP. Role of uncoupled and non-coupled oxidations in maintenance of safely low levels of oxygen and its one-electron reductants. Q Rev Biophys. 1996;29(2):169–202. doi: 10.1017/s0033583500005795. [DOI] [PubMed] [Google Scholar]
- 37.Rupprecht A, et al. Role of the transmembrane potential in the membrane proton leak. Biophys J. 2010;98(8):1503–1511. doi: 10.1016/j.bpj.2009.12.4301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Moldzio R, et al. Protective effects of resveratrol on glutamate-induced damages in murine brain cultures. J Neural Transm. 2013;120(9):1271–1280. doi: 10.1007/s00702-013-1000-6. [DOI] [PubMed] [Google Scholar]
- 39.van de Linde S, Sauer M, Heilemann M. Subdiffraction-resolution fluorescence imaging of proteins in the mitochondrial inner membrane with photoswitchable fluorophores. J Struct Biol. 2008;164(3):250–254. doi: 10.1016/j.jsb.2008.08.002. [DOI] [PubMed] [Google Scholar]
- 40.Vogelsang J, et al. Make them blink: Probes for super-resolution microscopy. ChemPhysChem. 2010;11(12):2475–2490. doi: 10.1002/cphc.201000189. [DOI] [PubMed] [Google Scholar]
- 41.Heilemann M, et al. Subdiffraction-resolution fluorescence imaging with conventional fluorescent probes. Angew Chem Int Ed Engl. 2008;47(33):6172–6176. doi: 10.1002/anie.200802376. [DOI] [PubMed] [Google Scholar]
- 42.Tokunaga M, Imamoto N, Sakata-Sogawa K. Highly inclined thin illumination enables clear single-molecule imaging in cells. Nat Methods. 2008;5(2):159–161. doi: 10.1038/nmeth1171. [DOI] [PubMed] [Google Scholar]
- 43.Schoen I, Ries J, Klotzsch E, Ewers H, Vogel V. Binding-activated localization microscopy of DNA structures. Nano Lett. 2011;11(9):4008–4011. doi: 10.1021/nl2025954. [DOI] [PubMed] [Google Scholar]
- 44.Smith CS, Joseph N, Rieger B, Lidke KA. Fast, single-molecule localization that achieves theoretically minimum uncertainty. Nat Methods. 2010;7(5):373–375. doi: 10.1038/nmeth.1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Thompson RE, Larson DR, Webb WW. Precise nanometer localization analysis for individual fluorescent probes. Biophys J. 2002;82(5):2775–2783. doi: 10.1016/S0006-3495(02)75618-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Endesfelder U, et al. Multiscale spatial organization of RNA polymerase in Escherichia coli. Biophys J. 2013;105(1):172–181. doi: 10.1016/j.bpj.2013.05.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Annibale P, Vanni S, Scarselli M, Rothlisberger U, Radenovic A. Quantitative photo activated localization microscopy: Unraveling the effects of photoblinking. PLoS ONE. 2011;6(7):e22678. doi: 10.1371/journal.pone.0022678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ehmann N, et al. Quantitative super-resolution imaging of Bruchpilot distinguishes active zone states. Nat Commun. 2014;5:4650. doi: 10.1038/ncomms5650. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.