Abstract
Tightly controlled epithelial and endothelial barriers are a prerequisite for life as these barriers separate multicellular organisms from their environment and serve as first lines of defense. Barriers between neighboring epithelial cells are formed by multiple intercellular junctions including the ‘apical junctional complex—AJC’ with tight junctions (TJ), adherens junctions (AJ), and desmosomes. TJ consist of tetraspan transmembrane proteins like occludin, various claudins that directly control paracellular permeability, and the ‘Junctional Adhesion Molecules’ (JAMs). For establishing tight barriers TJ are essential but at the same time have to allow also selective permeability. For this, TJ need to be tightly regulated and controlled. This is organized by a variety of adaptor molecules, i.e., protein kinases, phosphatases and GTPases, which in turn are regulated and fine-tuned involving microRNAs (miRNAs). In this review we summarize available data on the role and targeting of miRNAs in the maintenance of epithelial and/or endothelial barriers.
Keywords: epithelial and endothelial barriers, miRNA, regulation of barrier functions, tight junctions
Abbreviations: AJ, Adherens junctions; AJC, apical junctional complex; CDS, coding sequence; IBD, inflammatory bowel disease; JAM, junctional adhesion molecule; miRNA, microRNA; ROS, reactive oxygen species; TEM, transmission electron microscopy; TJ, tight junction
Introduction
Tight (TJ) and adherens (AJ) junctions form a decisive part of the ‘Apical Junctional Complex – AJC’ and regulate the paracellular permeability of epithelial layers across the apical/basolateral axis. Different groups of proteins are required to assemble the tight junction complex: transmembrane proteins like occludin, proteins of the claudin family and junctional adhesion molecules (JAM). These proteins are linked with cytosolic regulatory proteins as well as scaffolding and cytoskeletal proteins.1-4 Interactions of barrier components have to be strictly coordinated and tightly controlled to maintain their function in homeostasis of epithelial—or endothelial—barriers including their paracellular permeability. There is increasing evidence on how this sophisticated regulation might be orchestrated by microRNAs (miRNAs), small regulatory RNAs that post-transcriptionally affect most of the assembly steps and synthesis processes of junctional complex proteins.
In this review we aim to summarize current knowledge about regulatory effects of miRNAs involved in the adaptation of tight junctions i.e., modulating epithelial barrier functions in response to environmental challenges such as inflammation or disease. Our main focus is directed toward miRNA involved in TJ of intestinal epithelial barriers, as the gut is constantly challenged by foreign antigens and is by far the largest immunological organ. In addition, we will briefly touch the endothelial blood brain barrier as toxin or drug passage across this barrier is of particular pharmaceutical interest.
Tight Junctions and Adherens Junctions
Tight (TJ) and adherens junctions (AJ) are the major protein complexes of the Apical Junctional Complex (AJC).5,6 The AJC complex tightly connects the polarized epithelial cells in the intestinal mucosa and maintains the homeostasis of the intestinal barrier.7 The AJC represents the key structure for maintaining intestinal barrier functions.8,9 Tight junctions are found in all vertebrate epithelia and represent specialized multi-protein complexes localized at the apical side of lateral membranes of polarized epithelial cells.10 Since their discovery in 1963 more than 50 TJ-associated proteins have been identified.1 Tight junctions allow selective permeability for molecules on the basis of charge and size restrictions.11-13 It also became apparent that TJ are highly dynamic and may vary in different tissues giving rise to distinct barrier properties.1,2 However, despite ever increasing numbers of TJ-associated components, the detailed mechanism(s) of how these components work together to form a controlled selectively permeable barrier has remained largely enigmatic.1,3
Trans-membrane proteins such as occludin, claudins, junctional adhesion molecules (JAMs) constitute the main protein complexes of tight junctions.8,10,11 With the exception of JAMs, all trans-membrane proteins of tight junctions are tetraspanins consisting of 4 trans-membrane domains, which form 2 extracellular loops and one intracellular loop. Adaptor proteins such as ZO-1, -2 and -3 are linked to the cytosolic C-terminus of the trans-membrane proteins. The adaptor proteins interact also with many other proteins and are anchored to the actin cytoskeleton. Interactions between the different tight junction proteins and the cytoskeleton are essential for the normal assembly and maintenance of tight junction and hence also epithelial barrier integrity.
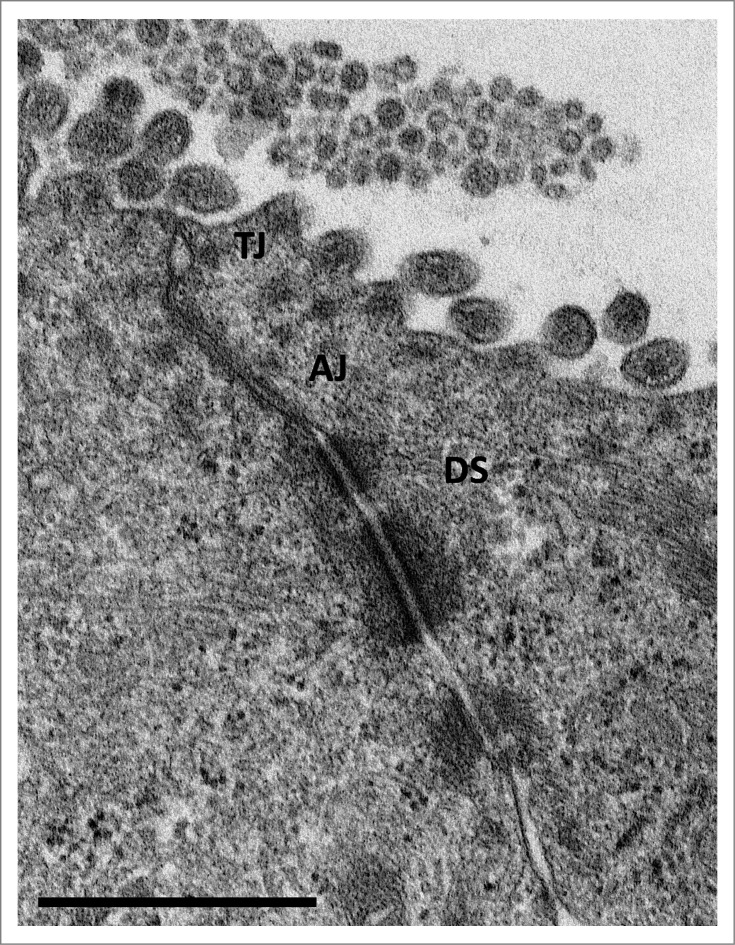
Examination by electron microscopy of the apical junctional complex has revealed a distinct ultrastructure. At the apical side of lateral membranes of polarized epithelial cells transmission electron microscope (TEM) pictures exhibited electron-dense regions consisting of regularly punctates representing the cellular epithelial barrier (Fig. 1). According to freeze fracture electron microscopy tight junctions exist of an anastomosing strands of beads.13
Figure 1.
Transmission electron microscopy (TEM) image of an 'apical junctional complex' of polarized T84 human colorectal carcinoma epithelial cells Depicted are the ‘tight junctions’ (TJ), directly beneath the microvilli, ‘adherens junctions’ (AJ), and ‘desmosomes’ (DS) below the ‘apical junctional complex’. Scale bar = 1 μm (courtesy of Lilo Greune, Institute of Infectiology – Center for Molecular Biology of Inflammation (ZMBE), University of Münster).
Tight junctions in different epithelia are challenged by a variety of factors like toxins, pathogens or inflammation potentially leading to tissue damage.14-16 At the same time these barriers are positively affected by probiotics and/or molecules like growth factors or anti-inflammatory cytokines, which stabilize the barrier function.17-21 The disruption as well as the assembly of tight junctions are regulated by several signaling cascades, involving protein kinases, protein phosphatases and G-proteins.22-25
The second major constituents of AJC are adherens junctions (AJ). AJ are characteristic for anchoring cells to cytoplasmatic actin filaments via a tightly controlled network of adaptor proteins. Cadherins, the transmembrane spanning proteins of 2 opposing cell membranes of epithelial cells (E-cadherins) or endothelial cells (VE-cadherins) are involved in homotypical trans-interactions thereby mediating cell-cell adhesion. Interactions of cadherins to several catenins provide the linkage to the actin cytoskeleton.26
Impact of Tight Junction Regulation
The interactions of tight junction proteins are dynamically regulated by several regulatory mechanisms (see Fig. 2). This results in selectively permeable barriers that are distinct in different tissues. Various extracellular factors such as inflammatory cytokines, reactive oxygen species (ROS) or microbial pathogens and their products disrupt epithelial tight junctions by activating multiple intracellular signaling pathways. In particular, molecules involved in signaling like protein kinases are involved in disruption or assembly of tight junctions. The c-Src-dependent tyrosine phosphorylation of different AJC proteins disrupts barrier functions and intestinal and renal epithelia.23,24
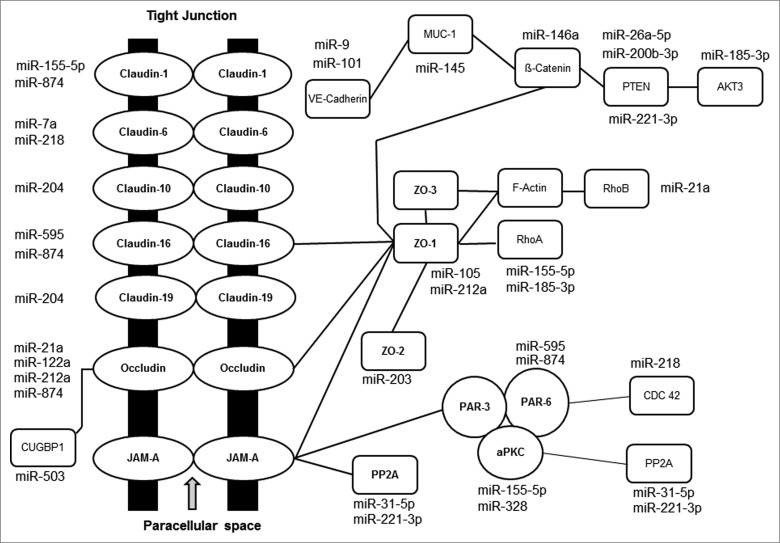
Figure 2.
Schematic representation of currently known TJ pathway genes affected by miRNAs (modified from Veltman et al. 201293).
Work from our and other laboratories showed that PKC isoforms such as PKCλ, PKCζ and PKCη are involved in the assembly of tight junctions,25,27: e.g., the activation of PKCζ effects the availability of occludin in TJ. Other protein phosphatases like PP2A and PP1 are able to effect TJ stability indirectly by dephosphorylating components of Par3, Par6, CDC42 polarity complex, which is essential for the integrity on TJ.28-30
Bacterial lipopolysaccharides (LPS) induce tight junction disruption via NF-kB and the induction of the Toll-like receptor 4 (TLR4) and LPS-binding protein (LBP) pathways. A specific knockdown of TLR4 or LBP significantly attenuates LPS-induced tight junction disruption.31
For several gastrointestinal inflammatory diseases, including Crohn's disease, ulcerative colitis, celiac disease, as well as many diarrheal syndromes induced by pathogenic microbes, the disturbance of TJ functions leads to a disruption of the intestinal barrier. In this way an increased penetration of microbial and other antigens is facilitated, resulting in inflammatory responses.23,32 Correspondingly, restoring or even enhancing the intestinal TJ barrier limits the development of intestinal inflammation and facilitates a more rapid resolution of the inflammatory disease.33,34 However, the role of transmembrane TJ protein depletion in disorders involving increased intestinal permeability remains largely undefined. A marked decrease of occludin levels in inflammatory bowel disease has been reported in several clinical studies.35-37 How occludin depletion causes an increase of intestinal permeability is currently under investigation.
Tumor necrosis factor α (TNF-α) represents one of the central mediators of gastrointestinal inflammation.38,39 Expression levels of TNF-α are significantly elevated in patients with inflammatory conditions, like Crohn's disease, ulcerative colitis, or celiac disease. An effective induction of remission by anti-TNF-α therapy has been achieved in patients with severe active Crohn's disease and ulcerative colitis39,40 as well as refractory celiac disease.41 This suggests that the pro-inflammatory activity of TNF-α may contribute to an increase in intestinal TJ permeability.42-45 TNF-α induces a breakdown of barrier functions, as shown in vitro by employing the Caco-2 monolayer model and in vivo in a mouse intestinal model.45
MicroRNA Biogenesis
MicroRNAs (miRNAs) represent a large family of short, single-stranded, non-coding RNAs (ncRNAs) of about 19–25 nucleotides in length. The human genome encodes for probably more than 2570 miRNAs (mirBase 20).46,47 Different sets of miRNAs are found in different cell types and tissues.48 MiRNAs are synthesized by a defined biogenesis pathway.49,50 The relevant gene regulation and mode of action has been reviewed extensively.46,51-53 The basic principle of gene regulation depends on the final, partial hybridization of one miRNA strand (‘guide strand’) within the RISC complex to partially complementary sequences, localized mostly within the 3′-UTR of target mRNAs (imperfect base-pairing). This leads to gene silencing by triggering mRNA deadenylation and degradation or—as a second option—translation inhibition. However, it is important to emphasize that several other mechanisms of the regulation of gene expression by miRNAs have also been described.54 Since their discovery in 1993 miRNAs have emerged as a new class of gene regulators that modulate and control the activities of thousands of mRNAs and in recent years the number of protein coding genes found to be regulated involving miRNA has been increasing steadily.55 Actually, it has been estimated that at least 60% of all protein-encoding genes are regulated by miRNAs.46,56 MiRNAs are well conserved in eukaryotic organisms and are supposed to be evolutionarily ancient components of gene regulation.57 Hence, it is not surprising that miRNA-deregulation effects many physiological and/or pathological processes58,59 particularly when one takes into account that miRNA are usually able to target different mRNA, and vice versa that a certain mRNA is supposed be targeted by multiple miRNAs.60 Moreover, it has been discovered recently, that miRNA-binding sites might not only be localized within the 3´-UTR46 but also within the coding sequence (CDS)61 or the 5´-UTR of a transcript.62 The genome-wide analyses of miRNA-binding sites, performed by Darnell and others indicate that a substantial number of miRNAs interact with these alternative binding sides.63,64 Therefore, this implies that the number of miRNA target sequences might have been even underestimated up to now.
Nevertheless, the prevailing consent concerning miRNA-mediated regulatory mechanisms is gene silencing by transcript decay due to mRNA deadenylation. However, miRNA-based gene regulation following this mechanism may operate in certain cells differently depending on the locus of miRNA-mRNA interactions and also the physiological state of the cell.
For the identification of miRNAs and the elucidation of potential miRNA targets several algorithms have been developed including TargetScan (http://www.targetscan.org/), MicroCosm Targets (http://www.ebi.ac.uk/enright-srv/microcosm/htdocs/targets/v5/), DIANA LAB (http:// http://diana.cslab.ece.ntua.gr/), miRSearch (https://www.exiqon.com/ mirsearch) or GeneGlobe (http://http://www.qiagen.com/products/genes%20and%20 pathways/) just to mention a few.
MiRNA and Human Disease
As miRNAs are involved in the regulation of many essential physiological processes in eukaryotic cells, dysregulation and aberrant expression of miRNAs has been associated with numerous disease states65 Consequently, miRNA-based diagnostics and therapies are under investigation.66 Here, only a few examples of miRNA involvement in pathologic conditions will be mentioned.
Role of miRNA in Cancer
Chronic lymphocytic leukemia was the first human disease known to be associated with miRNA deregulation.67 Many miRNAs correlated with different types of cancer68 are also referred to as ‘oncomirs’ e.g., miRNA-21 which is linked with different types of cancer such as glioblastoma.69 In human breast cancer cells the expression of claudin-6, as a TJ protein, was shown to be regulated by miR-7 and miR-218.70 Recent studies concerning 5 members of the microRNA-200 family (miR-200a, miR-200b, miR-200c) as well as miR-141 and miR-429 also revealed their regulatory effects during tumor progression of breast cancer.71 Screening assays for regulated miRNAs connected to early detection of colorectal cancer have been developed and are currently undergoing clinical trials. Our own experiments revealed that a sufficient selectivity and specificity can be achieved from less than 100 μl of blood plasma samples. This is due to the fact that cell-free, circulating miRNAs are highly stable in body fluids. This indicates that certain miRNAs might be used in diagnostics to assist clinical decision-making or the monitoring and interpretation of different disease treatment regimes.
Beyond that, just to indicate the scope of miRNA dependent regulation, the impact of miRNAs during heart development in general has been proven by inhibiting miRNA maturation in mouse models and for miR-155, miR-221 and miR-222 a pivotal role for the development of obesity during of the differentiation of stem cell progenitors into adipocytes have been shown.72,73 Furthermore let-7 inhibition might be used obesity and type 2 diabetes treatment.
MiRNA and Barrier Function
MiRNAs are involved in nearly every developmental and physiological process and play decisive roles in the differentiation, cell migration, architecture, and barrier function in intestinal epithelial cells. In recent years the important role of miRNAs in protein expression in the small intestine has been firmly established and it has become increasingly clear that expression patterns of intestinal miRNAs are altered in intestinal diseases. In different mouse models the impact of miRNAs on the impairment of epithelial barrier function was shown e.g., by Mckenna et al.74 In a typical Dicer1-deficient mouse model the intestinal barrier function is impaired leading to spontaneous intestinal inflammation. This is due to the loss of mmu-miR-192, which is normally highly expressed in the intestinal mucosa. Furthermore, in this mutant the transcription factors Regα and Regß as well as Relmß, which play a crucial role in inflammation and infection susceptibility are up-regulated and represent targets for miR-23a and miR-23b. The “recycling perfusion in vivo mouse model” by Ye et al. is discussed in the occludin section.75 In addition, it has been found that the intestinal miRNA signature is also influenced by the presence of microbiota.76,77 During bacterial infections miRNA expression is altered and plays an important role in the onset and progression of intestinal disease (for review: Staedel and Darfeuille, 2013).78 Recently, intestinal barrier dysfunction due to altered miRNA expression has been reported also in HIV and SIV infections.79 Furthermore, miRNAs have been identified as important factors in the host's response against microbial insults.
Inflammatory bowel diseases (IBD) represent a group of chronic, idiopathic, relapsing, and remitting immune disorders of the gastrointestinal tract in genetically susceptible individuals who are exposed to environmental risk factors.80,81 In these diseases disturbances of the intestinal barrier are major factors in aggravating and perpetuating disease pathology. Thus, restoring barrier functions greatly helps to induce remission. MiRNAs have been found to regulate tight junctions in intestinal epithelial cells and in this way also affect intestinal barrier functions.82 Interestingly, clinical studies demonstrated that in IBD including Crohn's disease83 and ulcerative colitis84 miRNA expression patterns are abnormal. From our own work as well as others we know that this differences are also reflected in mouse models which to a certain degree simulate the human diseases. In the more UC like dextran sulfate sodium (DSS) colitis model (miR-155) or in the more CD like T cell transfer model (mir-10a) different miRNAs, indicating the different origin of the disease, are upregulated during the course of the inflammation.85-88 Wu et al. were able to show that different sets of miRNAs also represent the degree of inflammation in ulcerative colitis (UC) and Crohn´s disease (CD).83,84 For example, they were able to characterize miR-192 as up-regulated and miR-16 as down-regulated in patients with active UC. MiR-16 has been shown to be upregulated during inactive UC and is therefore inversely correlated with the status of the disease. For active CD miR-23b has been revealed to be up-regulated and on the other hand miR-19b as downregulated. Pathophysiologically both diseases are correlated with diarrhea and the according breakdown of the barrier function. However, these miRNAs have not yet been linked to the exact target molecules during the course of disease.
Occludin
Recently, it could be shown that miR-21 is upregulated in chronic UC patients.84 MiR-21 induces the degradation of ras homolog gene family member B mRNA, leading to the depletion of occludin with the resulting impairment of tight junctions. Increased levels of miR-21 levels in inflamed tissues were also shown by Takagi et al.89 revealing its important role in pathogenesis of IBD. UC patients revealed an increase in miR-21 levels in serum samples, this was also supported by the presence of miRNAs in peripheral blood cell as it was shown by Paraskevi et al.90 A similar increase in miR-21 levels was also reported in patients with pediatric CD.91 After transfection with miR-21 mimics Caco-2 cells suffer the loss of tight junction proteins and the according structural changes.83 These results indicate that circulating miRNAs such as miR-21 and others might have the potential to be used as biomarkers in clinical applications, which is also supported by our own recent results (Pott et al.; 2014; personal communication). The expression of occludin in Caco-2 cells as well as in mouse intestinal epithelial cells is TNF-α dependent as had been shown by Ye et al.75 TNF-α is increased in patients under inflammatory conditions (anti-TNF-α therapy) which leads to the expression of miR-122a. By binding to the non-coding region of occludin mRNA, miR-122a induces specific mRNA degradation and thus occludin depletion, leading to an increase in intestinal permeability. Therefore, miRNA-122a plays a crucial role in TJ formation and barrier function during intestinal inflammation.
Zonula occludens proteins ZO-1 and ZO-2
The up-regulation of miR-212 expression after alcohol abuse was reported to induce the disruption of TJ by inhibition of ZO-1 translation.92 Work from our own laboratory used miRNA profiling in T84 (human colorectal carcinoma cells) monolayers and employing specific miRNA inhibitors showed that miR-203, miR-483-3p, and miR-595 affect the expression of several adapter molecules of the tight junctional complexes as monitored by expression of associated proteins and transepithelial resistance (TER) in epithelial cellular barrier models (Table 1). Although ZO-2 and PKCζ were found to be affected, interestingly, a modulation of ZO-1 could not be observed.93 Further putative activities of these miRNAs were not investigated in these studies.
Table 1.
miRNAs quoted in this review, affecting the expression of apical junctional complex proteins
| miRNA | miRNA - target | Reference |
|---|---|---|
| miR-7a | Claudin-6 | Li et al. 201270 |
| miR-9 | Aquaporin 3, Claudin-14 | Zhi et al. 201494 |
| miR-21a | Occludin, RhoB | Yang et al. 201383 |
| miR-105 | VE-cadherin, Tight junction protein 1 | Zhou et al. 2014122 |
| miR-122a | Occludin, Dicer1 | Ye et al. 201175 |
| miR-145 | Protein phosphatase 2 regulatory subunit, Tight junction protein 1, Mucin1 | Ma et al. 2010117 |
| miR-146a | ß-Catenin, Protein Kinase C | Hwang et al. 2012121 |
| miR-155-5p | Claudin-1, RhoA, ß-Catenin, Protein Kinase C | Tili et al. 2007104 O’Connell RM et al. 2007105 |
| miR-200 | Pten | Gregory et al. 200771 |
| miR-203 | Tight junction protein 2, Claudin1 | Veltman et al. 201293 |
| miR-212 | Occludin, Tight junction protein 1 | Tang et al. 200892 |
| miR-218 | Claudin-2/6, CDC-42 | Li et al. 201270 |
| miR-221 | Protein phosphatase 2 regulatory subunit, Pten | Romao et al. 201173 |
| miR-223 | Occludin, Protein Kinase C, Tiam1 | Redell J. 2012122 |
| miR-328 | Tiam-1, Claudin-19 | Arora et al. 2011120 |
| miR-483-3p | Protein Kinase-α, | Zang Y-W et al. 2012114 |
| miR-503 | CUG-binding protein 1, Occludin | Yang et al. 2014123 |
| miR-595 | Cell-polarity protein-6 | Veltman et al. 201293 |
| miR-874 | Cell-polarity protein-6 | Veltman et al. 201293 |
Detailed information concerning microRNA sequences and validated or further potential target mRNA molecules can be obtained from `miRBase´or `TargetScan´ databases.
Claudins
McKenna et al. demonstrated that claudin-4 and claudin-7 are not expressed in the apical membrane of intestinal epithelial cells of Dicer 1-deficient mice, which resulted in impaired intestinal barrier functions, which could not be correlated yet to a certain miRNA.74 In contrast Zhi et al. were able to show that claudin-1 is indirectly affected by miR-874 during intestinal dysfunction after ischemic injury, which might result as a complication following intestinal disease or abdominal surgery. It was found that the level of miR-874 was inversely related to the level of aquaporin 3 (AQP3) expression which enhances intestinal permeability by down regulating claudin-1 and occludin.94
Looking at the retinal pigment epithelium another indirect effect by miR-204 on the upregulation of claudin-10 and -19 has been shown.95 The direct target for miR-204 is the TGFß-R2 which in its active form reduces the claudin expression via transcriptionfactor SNAIL2. Furthermore, the dysregulation of TJ is a basic phenomenon, e.g.,: Claudin-1 expression is down-regulated by an increase of miR-155 in ovarian cancer cells.96
Rho-GTPases
Different members of the Rho GTPase family play significant cellular roles e.g. in organizing the actin cytoskeleton.97 RhoB, a Ras GTPase was identified as an additional target for miR-21, which has been already identified as targeting the mRNA of occludin. Unlike RhoA and RhoC, RhoB acts as a tumor suppressor and affects cell cycle, angiogenesis, and apoptosis. On the cellular level it also influences actin organization, cell migration, and cell adhesion.98 In colorectal cancer cell lines,97 hepatocellular carcinoma cell lines,99 and human umbilical vein endothelial cells (HUVECs),100 RhoB is regulated by miR-21 via binding to 3′-UTR. Furthermore, reducing RhoB expression by siRNA treatment resulted in a decrease in TER and a destabilization of the junctional complex emphasizing the role of RhoB in the AJC. RhoA induces and regulates the assembly of the AJC. It has been shown that down-regulation of RhoA leads to disruption of junctional complexes101,102 by interfering with the synthesis of tight junction proteins.103 The mRNA of RhoA contains 3 miRNA binding sites in the 3′-UTR. One of these sites is specifically bound by miR-155.104 MiR-155 plays a pivotal role in the systemic inflammatory response.105 In addition, miR-155 interferes with the activation of Toll-like receptor (TLR) pathway in monocytic cell after LPS-induction.106 Inflammatory cytokines, such as TNF-α and interferons are able to induce miR-155 expression.104,107 The increase of TNF-α in inflammatory bowel disease, which can be induced by bacterial infection leads to the downregulation of ZO-1 and E-cadherin expression and the destabilization of the AJC.108 This further emphasizes a key role of miR-155 for the regulation of barrier function in intestinal epithelial cells.
Currently validated miRNAs and their targeted proteins modulating epithelial and/or endothelial barrier integrity are summarized in Table 1.
MiRNA and Blood Brain Barrier
The complex blood brain barrier (BBB) ensures the barrier functions between the vascular system and the brain and consists of brain microvascular endothelial cells, astroglia and pericytes. The tight junctions and adherens junctions109 between these cells restrict the passage of cells, bioactive molecules, including most therapeutics across this barrier.110,111 Impairment of the homeostasis of these cellular junctions leads to barrier disruption. Experiments in several mouse models revealed the importance of Claudin-based tight junctions for the selectivity of endothelial barrier function. The knock-out of claudin-1 or -5 in different murine models was found to be lethal during embryogenesis.112,113 In esophageal carcinoma cells the displacement of claudin-7 induces the loss of E-cadherin expression and therefore the destabilization of tight junctions.114
VE-cadherin, the endothelial cell-specific AJ and transmembrane protein, is a key regulator of endothelial apical junctional complexes and of endothelial barrier integrity.115 VE-cadherin interferes with a variety of signaling molecules, which coordinate endothelial TJ organization and permeability113 and is targeted by different miRNAs that modulate the transcriptional repression directly and also indirectly. MiR-9 was described to target VE-cadherin, inducing the reduction of ß-catenin, which enhances invasion as well as increased tumor angiogenesis.
The HI-virus is able to cross the BBB and to infect brain macrophages/microglia. The HIV-1 Tat protein induces miRNA-32. This miRNA regulates the expression level of TRAF3, which itself enables the infection of microglial cells.113 This is also achieved by the downregulation of VE-cadherin, due to the upregulation of miR-101 by Tat C protein. Knockdown experiments for miR-101 revealed that the degree of claudin-5 expression is depending on VE-cadherin level.
TJ are also destabilized by metastatic brain tumors. These enable circulating tumor cells to enter the brain, this indicates that the integrity of the BBB is an important barrier for melanoma cells. It has been shown that the disruption of barrier function is correlated with a reduced Claudin-5 and ZO-1 availability, which can be monitored by a loss in transendothelial electrical resistance (TER).116,117 Recently, these findings have been supported by Zhou et al.118 who found that miR-105 which is secreted by metastatic breast cancer cells targets the TJ protein ZO-1. In this way, miR-105 secreted and transferred via exosomes disrupts TJ and facilitates metastatic migration.
Brain metastasis has been linked to the expression of miR-145 and miR-328.119,120 The overexpression of miR-328 has been shown to interfere with the level of protein kinase C α (PKCa) which is one of the TJ regulatory proteins. On the contrary other studies were able to show that the overexpression of miR-146 leads to an increased β-catenin level which suppresses brain invasion by migrating cells.121
Furthermore brain injury induces a variety of signaling pathways, which effect blood-brain barrier permeability. MiR-223 levels seem to be upregulated in cerebral microvasculature where it targets TIAM1 thereby affecting barrier integrity. It has been reported by John Redell in 2012 that blocking miR-223 leads to an improvement in barrier function.122
Summary and Perspective
Cellular junctions such as tight and adherens junctions play a crucial role in regulating paracellular permeability in vertebrate epithelia and endothelia. In order to fulfill this task effectively the assembly and maintenance of junctional complexes need to be properly controlled and regulated. TJ and AJ exist as a set of tetraspins, transmembrane proteins (occluding, claudins) and JAMs, accompanied by many regulatory and adapter proteins like ZO-1, -2 and -3, or kinases and phosphatases. During disease or infection the function of epithelial or endothelial cellular barriers can be severely disturbed. A cellular response mechanism to counteract an impairment of barrier function or to fine-tune and adapt the paracellular permeability are microRNAs, which are able to modulate the AJC proteins post-transcriptionally.
To demonstrate the very complex regulation of TJ proteins, Yang et al.123 reviewed very recently the cooperative and post-transcriptional effects of RNA-binding proteins (RBPs) and miRNAs on mRNA stability coding for TJ proteins in the gastrointestinal mucosa.
The extent of gene regulation byMiRNAs will become more complex as new miRNA binding sites are likely to be discovered.46,124 It is foreseeable that there will be many more regulatory interactions to be identified that are affecting the synthesis of junctional proteins, the proper assembly of the complexes and the fine-tuning of immunological responses. Due to the intrinsic ‘musketeer strategy’ (‘one for all and all for one’) in miRNA-mRNA interactions, the ensuing degeneration of regulatory pathways involving miRNAs will present a formidable task to distill and validate the determining interactions with target mRNAs. Defining certain interactions and regulations of miRNAs with junctional proteins will hopefully also pave the way for potential therapeutic applications that would either reinforce cellular barriers to prevent tissue damage, induced by pathogenic microbes or inflammatory reactions, or would on the contrary provide possibilities to temporarily disrupt or impair barrier functions to facilitate drug delivery.
Acknowledgments
We like to thank L. Greune (Institute of Infectiology – ZMBE, Münster) for excellent electron microscopy.
Funding
Our work has been supported in part by grants from the Deutsche Forschungsgemeinschaft (DFG: GRK1409, SCHM770/15-1 and SFB1009 B03), a fellowship of the Medical Faculty Münster (to H.S.; FKZ: 0012), and the Cells-in-Motion Cluster of Excellence (EXC1003 – CiM), University of Münster, Germany.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
References
- 1.Marchiando AM, Graham WV, Turner JR. Epithelial barriers in homeostasis and disease. Annu Rev Pathol Mech Dis 2010; 5:119–44; PMID: ; http://dx.doi.org/ 10.1146/annurev.pathol.4.110807.092135 [DOI] [PubMed] [Google Scholar]
- 2.Turner HL, Turner JR. Good fences make good neighbors. Gut Microbes 2010; 1:22–29; PMID: ; www.landesbioscience.com/journals/gutmicrobes/article/II427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shen L, Weber CR, Raleigh DR, Yu D, Turner JR. Tight junction pore and leak pathways: a dynamic duo. Annu Rev Physiol 2011; 73:283–309; PMID: ; http://dx.doi.org/ 10.1146/annurev-physiol-012110-142150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Peterson LW, Artis D. Intestinal epithelial cells: regulators of barrier function and immune homeostasis. Nature R Immunol 2014; 14:141–53; PMID: ; http://dx.doi.org/ 10.1038/nri3608 [DOI] [PubMed] [Google Scholar]
- 5.Groschwitz KR, Hogan SP. Intestinal barrier function: molecular regulation and disease pathogenesis. J Allergy Clin Immunol 2009; 124:3–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tsukita S, Yamazaki Y, Katsuno T, Tamura A, Tsukita S. Tight junction-based epithelial microenvironment and cell proliferation. Oncogene 2008; 27:6930–38; PMID: ; http://dx.doi.org/ 10.1038/onc.2008.344 [DOI] [PubMed] [Google Scholar]
- 7.Förster C. Tight junctions and the modulation of barrier function in disease. Histochem Cell Biol 2008; 130:55–70; PMID: ; http://dx.doi.org/ 10.1007/s00418-008-0424-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Anderson JM, Van Itallie CM. Physiology and function of the tight junction. Cold Spring Harb Perspect Biol 2009; 1:a002584; PMID: ; http://dx.doi.org/ 10.1101/cshperspect.a002584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Catalioto RM, Maggi CA, Giuliani S. Intestinal epithelial barrier dysfunction in disease and possible therapeutical interventions. Curr Med Chem 2011; 18:398–426; PMID: ; http://dx.doi.org/ 10.2174/092986711794839179 [DOI] [PubMed] [Google Scholar]
- 10.Mitic LL, Van Itallie CM, Anderson JM. Molecular physiology and pathophysiology of tight junctions I. Tight junction structure and function: lessons from mutant animals and proteins. Am J Physiol Gastrointest Liver Physiol 2000; 275(24):18407–17; PMID: ; http://dx.doi.org/ 10.1074/jbc.M001530200 [DOI] [PubMed] [Google Scholar]
- 11.Van Itallie CM, Anderson JM. Claudins and epithelial paracellular transport. Annu Rev Physiol 2006; 68:403–29; PMID: ; http://dx.doi.org/ 10.1146/annurev.physiol.68.040104.131404 [DOI] [PubMed] [Google Scholar]
- 12.Mitic LL, Anderson JM. Molecular architecture of tight junctions. Annu Rev Physiol 1998; 60:121–42; PMID: ; http://dx.doi.org/ 10.1146/annurev.physiol.60.1.121 [DOI] [PubMed] [Google Scholar]
- 13.Madara JL. Maintenance of the macromolecular barrier at cell extrusion sites in intestinal epithelium: physiological rearrangement of tight junctions. J Membr Biol 1990; 116:177–84; PMID: [DOI] [PubMed] [Google Scholar]
- 14.Ulluwishewa D, Anderson RC, McNabb WC, Moughan PJ, Wells JM, Roy NC. Regulation of tight junction permeability by intestinal bacteria and dietary components. J Nutr 2011; 141:769–76; PMID: ; http://dx.doi.org/ 10.3945/jn.110.135657 [DOI] [PubMed] [Google Scholar]
- 15.Utech M, Mennigen R, Bruewer M. Endocytosis and recycling of tight junction proteins in inflammation. J Biomed Biotechnol 2010; 2010:484987; PMID: ; http://dx.doi.org/ 10.1155/2010/484987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Edelblum KL, Turner JR. The tight junction in inflammatory disease: communication breakdown. Curr Opin Pharmacol 2009; 9:715–20; PMID: ; http://dx.doi.org/ 10.1016/j.coph.2009.06.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Amasheh M, Andres S, Amasheh S, Fromm M, Schulzke JD. Barrier effects of nutritional factors. Ann N Y Acad Sci 2009; 1165:267–73; PMID: ; http://dx.doi.org/ 10.1111/j.1749-6632.2009.04063.x [DOI] [PubMed] [Google Scholar]
- 18.Seth A, Basuroy S, Sheth P, Rao RK. L-Glutamine ameliorates acetaldehyde-induced increase in paracellular permeability in Caco-2 cell monolayer. Am J Physiol Gastrointest Liver Physiol 2004; 287:G510–7; PMID: ; http://dx.doi.org/ 10.1152/ajpgi.00058.2004 [DOI] [PubMed] [Google Scholar]
- 19.Ahrne S, Hagslatt ML. Effect of lactobacilli on paracellular permeability in the gut. Nutrients 2011; 3:104–17; PMID: ; http://dx.doi.org/ 10.3390/nu3010104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Basuroy S, Seth A, Elias B, Naren AP, Rao R. MAPK interacts with occludin and mediates EGF-induced prevention of tight junction disruption by hydrogen peroxide. Biochem J 2006; 393:69–77; PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Seth A, Yan F, Polk DB, Rao RK. Probiotics ameliorate the hydrogen peroxide-induced epithelial barrier disruption by a PKC- and MAP kinase-dependent mechanism. Am J Physiol Gastrointest Liver Physiol 2008; 294:G1060–9; PMID: ; http://dx.doi.org/ 10.1152/ajpgi.00202.2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dörfel MJ, Huber O. Modulation of tight junction structure and function by kinases and phosphatases targeting occludin. J Biomed Biotechnol 2012; 2012:807356; PMID: ; http://dx.doi.org/ 10.1155/2012/807356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Elias BC, Suzuki T, Seth A, Giorgianni F, Kale G, Shen L, Turner JR, Naren A, Desiderio DM, Rao R. Phosphorylation of Tyr- 398 and Tyr-402 in occludin prevents its interaction with ZO-1 and destabilizes its assembly at the tight junctions. J Biol Chem 2009; 284:1559–69; PMID: ; http://dx.doi.org/ 10.1074/jbc.M804783200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rao R. Occludin phosphorylation in regulation of epithelial tight junctions. Ann N Y Acad Sci 2009; 1165:62–8; PMID: ; http://dx.doi.org/ 10.1111/j.1749-6632.2009.04054.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Suzuki T, Elias BC, Seth A, Shen L, Turner JR, Giorgianni F, Desiderio D, Guntaka R, Rao R. PKCeta regulates occludin phosphorylation and epithelial tight junction integrity. Proc Natl Acad Sci U S A 2009; 106:61–6; PMID: ; http://dx.doi.org/ 10.1073/pnas.0802741106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Meng W, Takeichi M. Adherens junction: molecular architecture and regulation. Cold Spring Harb Perspect Biol 2009; 1:a002899; http://dx.doi.org/ 10.1101/cshperspect.a002899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zyrek AA, Cichon C, Helms S, Enders C, Sonnenborn U, Schmidt MA. Molecular mechanisms underlying the probiotic effects of Escherichia coli Nissle 1917 involve ZO-2 and PKCzeta redistribution resulting in tight junction and epithelial barrier repair. Cell Microbiol 2007; 9(3):804–16; PMID: ; http://dx.doi.org/ 10.1111/j.1462-5822.2006.00836 [DOI] [PubMed] [Google Scholar]
- 28.Seth A, Sheth P, Elias BC, Rao R. Protein phosphatases 2A and 1 interact with occluding and negatively regulate the assembly of tight junctions in the CACO-2 cell monolayer. J Biol Chem 2007; 282:11487–98; PMID: ; http://dx.doi.org/ 10.1074/jbc.M610597200 [DOI] [PubMed] [Google Scholar]
- 29.Sheth P, Samak G, Shull JA, Seth A, Rao R. Protein phosphatase 2A plays a role in hydrogen peroxide-induced disruption of tight junctions in Caco-2 cell monolayers. Biochem J 2009; 421:59–70; PMID: http://dx.doi.org/ 10.1042/BJ20081951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nunbhakdi-Craig V, Machleidt T, Ogris E, Bellotto D, White CL, 3 rd, Sontag E. Protein phosphatase 2A associates with and regulates atypical PKC and the epithelial tight junction complex. J Cell Biol 2002; 158:967–78; PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Turner JR. Intestinal mucosal barrier function in health and disease. Nat Rev Immunol 2009; 9:799–809; PMID: ; http://dx.doi.org/ 10.1038/nri2653 [DOI] [PubMed] [Google Scholar]
- 32.Arrieta MC, Madsen K, Doyle J, Meddings J. Reducing small intestinal permeability attenuates colitis in the IL10 gene-deficient mouse. Gut 2009; 58:41–8; PMID: ; http://dx.doi.org/ 10.1136/gut.2008.150888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wyatt J, Vogelsang H, Hubl W, Waldhoer T, Lochs H. Intestinal permeability and the prediction of relapse in Crohn's disease. Lancet 1993; 341:1437–9; PMID: [DOI] [PubMed] [Google Scholar]
- 34.Arnott ID, Kingstone K, Ghosh S. Abnormal intestinal permeability predicts relapse in inactive Crohn disease. Scand J Gastroenterol 2000; 35:1163–9; PMID: [DOI] [PubMed] [Google Scholar]
- 35.Ciccocioppo R, Finamore A, Ara C, Di Sabatino A, Mengheri E, Corazza GR. Altered expression, localization, and phosphorylation of epithelial junctional proteins in celiac disease. Am J Clin Pathol 2006; 125:502–11; PMID: ; http://dx.doi.org/ 10.1309/DTYRA91G8R0KTM8M [DOI] [PubMed] [Google Scholar]
- 36.Gassler N, Rohr C, Schneider A, Kartenbeck J, Bach A, Obermuller N, Otto HF, Autschbach F. Inflammatory bowel disease is associated with changes of enterocytic junctions. Am J Physiol Gastrointest Liver Physiol 2001; 281:G216–28; PMID: [DOI] [PubMed] [Google Scholar]
- 37.Zeissig S, Burgel N, Gunzel D, Richter J, Mankertz J, Wahnschaffe U, Kroesen AJ, Zeitz M, Fromm M, Schulzke JD. Changes in expression and distribution of claudin 2, 5 and 8 lead to discontinuous tight junctions and barrier dysfunction in active Crohn's disease. Gut 2007; 56:61–72; PMID: ; http://dx.doi.org/ 10.1136/gut.2006.094375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Targan SR, Hanauer SB, van Deventer SJ, Mayer L, Present DH, Braakman T, DeWoody KL, Schaible TF, Rutgeerts PJ. A short-term study of chimeric monoclonal antibody cA2 to tumor necrosis factor alpha for Crohn's disease. Crohn's Disease cA2 Study Group. N Engl J Med 1997; 337:1029–35; PMID: [DOI] [PubMed] [Google Scholar]
- 39.Van Deventer SJ. Tumour necrosis factor and Crohn's disease. Gut 1997; 40: 443–8; PMID: ; http://dx.doi.org/ 10.1136/gut.40.4.443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rutgeerts P, Sandborn WJ, Feagan BG, Reinisch W, Olson A, Johanns J, Travers S, Rachmilewitz D, Hanauer SB, Lichtenstein GR, et al. . Infliximab for induction and maintenance therapy for ulcerative colitis. N Engl J Med 2005; 353:2462–76; PMID: ; http://dx.doi.org/ 10.1056/NEJMoa050516 [DOI] [PubMed] [Google Scholar]
- 41.Gillett HR, Arnott ID, McIntyre M, Campbell S, Dahele A, Priest M, Jackson R, Ghosh S. Successful infliximab treatment for steroid-refractory celiac disease: a case report. Gastroenterology 2002; 122:800–5; PMID: ; http://dx.doi.org/ 10.1053/gast.2002.31874 [DOI] [PubMed] [Google Scholar]
- 42.Blair SA, Kane SV, Clayburgh DR, Turner JR. Epithelial myosin light chain kinase expression and activity are upregulated in inflammatory bowel disease. Lab Invest 2006; 86:191–201; PMID: ; http://dx.doi.org/ 10.1038/labinvest.3700373 [DOI] [PubMed] [Google Scholar]
- 43.Ma TY, Boivin MA, Ye D, Pedram A, Said HM. Mechanism of TNF-alpha modulation of Caco-2 intestinal epithelial tight junction barrier: role of myosin light-chain kinase protein expression. Am J Physiol Gastrointest Liver Physiol 2005; 288:G422–30; PMID: ; http://dx.doi.org/ 10.1152/ajpgi.00412.2004 [DOI] [PubMed] [Google Scholar]
- 44.Ma TY, Iwamoto GK, Hoa NT, Akotia V, Pedram A, Boivin MA, Said HM. TNF-alpha induced increase in intestinal epithelial tight junction permeability requires NF-kappa B activation. Am J Physiol Gastrointest Liver Physiol 2004; 286:G367–76; PMID: ; http://dx.doi.org/ 10.1152/ajpgi.00173.2003 [DOI] [PubMed] [Google Scholar]
- 45.Schwarz BT, Wang F, Shen L, Clayburgh DR, Su L, Wang Y, Fu YX, Turner JR. LIGHT signals directly to intestinal epithelia to cause barrier dysfunction via cytoskeletal and endocytic mechanisms. Gastroenterology 2007; 132:2383–94; PMID: ; http://dx.doi.org/ 10.1053/j.gastro.2007.02.052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell 2009; 136:215–233; PMID: ; http://dx.doi.org/ 10.1016/j.cell.2009.01.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. http://www.mirbase.org/index.shtml miRBase Release 20: June 2013. [Google Scholar]
- 48.Lagos-Quintana M, Rauhut R, Yalcin A, Meyer J, Lendeckel W, Tuschl T. Identification of tissue-specific microRNAs from mouse. Curr Biol 2002; 12(9):735–9. PMID: ; http://dx.doi.org/ 10.1016/S0960-9822(02)00809-6 [DOI] [PubMed] [Google Scholar]
- 49.Krol J, Loedige I, Filipowicz W. The widespread regulation of microRNA biogenesis, function and decay. Nat Rev Genet 2010; 11:597–610; PMID: ; http://dx.doi.org/ 10.1038/nrg2843 [DOI] [PubMed] [Google Scholar]
- 50.Libri V, Miesen P, van Rij RP, Buck AH. Regulation of microRNA biogenesis and turnover by animals and their viruses. Cell Mol Life Sci 2013; 70:3525–44; PMID: ; http://dx.doi.org/ 10.1007/s00018-012-1257-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lee HJ. Exceptional stories of microRNAs. Exp Biol Med (Maywood) 2013; 238(4):339–43; PMID: ; http://dx.doi.org/ 10.1258/ebm.2012.012251 [DOI] [PubMed] [Google Scholar]
- 52.Rácz Z1, Kaucsár T, Hamar P. The huge world of small RNAs: regulating networks of microRNAs (review). Acta Physiol Hung. 2011; 98(3):243–51; PMID: ; http://dx.doi.org/ 10.1556/APhysiol.98.2011.3.1 [DOI] [PubMed] [Google Scholar]
- 53.Shruti K, Shrey K, Vibha R. Micro RNAs: tiny sequences with enormous potential. Biochem Biophys Res Commun. 2011; 407(3):445–9; PMID: ; http://dx.doi.org/ 10.1016/j.bbrc.2011.03.058 [DOI] [PubMed] [Google Scholar]
- 54.Stroynowska-Czerwinska A, Fiszer A, Krzyzosiak WJ. The panorama of miRNA-mediated mechanisms in mammalian cells. Cell Mol Life Sci 2014; 71(12):2253–70; PMID: ; http://dx.doi.org/ 10.1007/s00018-013-1551-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 1993; 75(5):843–54; PMID: ; http://dx.doi.org/ 10.1016/0092-8674(93)90529-Y [DOI] [PubMed] [Google Scholar]
- 56.Friedman RC, Farh KK-H, Burge CB, Bartel DP. Most mammalian mRNAs are conserved targets of microRNAs. Genom Res 2009; 19:92–105; PMID: ; http://dx.doi.org/ 10.1101/gr.082701.108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lee CT, Risom T, Strauss WM. Evolutionary conservation of microRNA regulatory circuits: an examination of microRNA gene complexity and conserved microRNA-target interactions through metazoan phylogeny. DNA Cell Biol 2007; 26(4):209–18; PMID: ; http://dx.doi.org/ 10.1089/dna.2006.0545 [DOI] [PubMed] [Google Scholar]
- 58.Mraz M., Pospisilova S.MicroRNAs in chronic lymphocytic leukemia: from causality to associations and back. Expert Rev Hematol 2012; 5(6):579–81; PMID: ; http://dx.doi.org/ 10.1586/ehm.12.54 [DOI] [PubMed] [Google Scholar]
- 59.Jiang Q, Wang Y, Hao Y, Juan L, Teng M, Zhang X, Li M, Wang G, Liu Y. miR2Disease: a manually curated database for microRNA deregulation in human disease. Nucleic Acids Res 2009; 37(Database issue):D98–104; PMC 2686559; PMID: ; http://dx.doi.org/ 10.1093/nar/gkn714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Rajewsky N. microRNA target predictions in animals. Nat Genet 2006; 38(6):S8–13; PMID: ; http://dx.doi.org/ 10.1038/ng1798 [DOI] [PubMed] [Google Scholar]
- 61.Duursma AM, Kedde M, Schrier M, le Sage C, Agami R. miR-148 targets human DNMT3b protein coding region. RNA 2008; 14:872–77; PMID: ; http://dx.doi.org/ 10.1261/rna.972008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Forman JJ, Legesse-Miller A, Coller H. A search for conserved sequences in coding regions reveals that the let-7 microRNA targets Dicer within its coding sequence. Proc Natl Acad Sci U S A 2008; 105:14879–84. PMID: ; http://dx.doi.org/ 10.1073/pnas.0803230105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Chi SW, Zang JB, Mele A, Darnell RB. Argonaute HITSCLIP decodes microRNA-mRNA interaction maps. Nature 2009; 460:479–86; PMID: ; http://dx.doi.org/ 10.1038/nature 08170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hafner M, Landthaler M, Burger L, Khorshid M, Hausser J, Berninger P, Rothballer A, Ascano M, Jr, Jungkamp AC, Munschauer M, et al. Transcriptomewide identification of RNA-binding protein and microRNA target sites by PAR-CLIP. Cell 2010; 141:129–41; PMID: ; http://dx.doi.org/ 10.1016/j.cell.2010.03.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Trang P, Weidhaas JB, Slack FJ. MicroRNAs as potential cancer therapeutics. Oncogene 2008; 27(Suppl 2):S52–7; PMID: ; http://dx.doi.org/ 10.1038/onc.2009.353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hydbring P, Badalian-Very G. Clinical applications of microRNAs. Version 3. F1000Res 2013; 2:136. PMID: ; http://dx.doi.org/ 10.12688/f1000research.2-136.v3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Mraz M, Pospisilova S. MicroRNAs in chronic lymphocytic leukemia: from causality to associations and back. Exp Rev Hematol 2012; 5(6):579–81; PMID: ; http://dx.doi.org/ 10.1586/ehm.12.54 [DOI] [PubMed] [Google Scholar]
- 68.Mraz M, Pospisilova S, Malinova K, Slapak I, Mayer J. MicroRNAs in chronic lymphocytic leukemia pathogenesis and disease subtypes. Leuk Lymphoma 2009; 50(3):506–9; PMID: ; http://dx.doi.org/ 10.1080/10428190902763517 [DOI] [PubMed] [Google Scholar]
- 69.Møller HG, Rasmussen AP, Andersen HH, Johnsen KB, Henriksen M, Duroux M. A systematic review of microRNA in glioblastoma multiforme: Micro-modulators in the mesenchymal mode of migration and invasion. Mol Neurobiol 201347(1):131–144; PMCID:PMC3538124; PMID: ; http://dx.doi.org/ 10.1080/10428190902763517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Li Q, Zhu F, Chen P. MiR-7 and miR-218 epigenetically control tumor suppressor genes RASSF1A and Claudin-6 by targeting HoxB3 in breast cancer. Biochem Biophys Res Commun 2012; 424:28–33; PMID: ; http://dx.doi.org/ 10.1016/j.bbrc.2012.06.028 [DOI] [PubMed] [Google Scholar]
- 71.Gregory PA, Bert AG, Paterson EL, Barry SC, Tsykin A, Farshid G, Vadas MA, Khew- Goodall Y, Goodall GJ. The miR-200 family and miR-205 regulate epithelial to mesenchymal transition by targeting ZEB1 and SIP1. Nat Cell Biol 2008; 10(5):593–601; PMID: ; http://dx.doi.org/ 10.1038/ncb1722 [DOI] [PubMed] [Google Scholar]
- 72.Chen JF, Murchison EP, Tang R, Callis TE, Tatsuguchi M, Deng Z, Rojas M, Hammond SM, Schneider MD, Selzman CH, et al. . Targeted deletion of dicer in the heart leads to dilated cardiomyopathy and heart failure. Proc Natl Acad Sci U S A 2008; 105(6):2111–6; PMID: ; http://dx.doi.org/ 10.1073/pnas.0710228105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Romao JM, Jin W, Dodson MV, Hausman GJ, Moore SS, Guan LL. MicroRNA regulation in mammalian adipogenesis. Exp Biol Med 2011; 236(9):997–1004. PMID: ; http://dx.doi.org/ 10.1258/ebm.2011.011101 [DOI] [PubMed] [Google Scholar]
- 74.McKenna LB, Schug J, Vourekas A, McKenna JB, Bramswig NC, Friedman JR, Kaestner KH. MicroRNAs control intestinal epithelial differentiation, architecture, and barrier function. Gastroenterology 2010; 139:1654–64; PMID: ; http://dx.doi.org/ 10.1053/j.gastro.2010.07.040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ye D, Guo S, Al-Sadi R, Ma TY. MicroRNA regulation of intestinal epithelial tight junction permeability. Gastroenterology 2011; 141:1323–33; PMID: ; http://dx.doi.org/ 10.1053/j.gastro.2011.07.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Dalmasso G, Nguyen HTT, Yan Y, Laroui H, Charania MA, Ayyadurai S, Sitaraman SV, Merlin D. Microbiota modulate host gene expression via microRNAs. PLoS One 2011; 6(4):e19293; PMID: ; http://dx.doi.org/ 10.1371/journal.pone.0019293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Singh N, Shirdel EA, Waldron L, Zhang R-H, Jurisica I, Comelli EM. The murine caecal microRNA signature depends on the presence of the endogeneous microbiota. Int J Biol Sci 2012; 8:171–86; PMID: ; http://dx.doi.org/ 10.7150/ijbs.8.171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Staedel C, Darfeuille F. MicroRNAs and bacterial infections. Cell Microbiol 2013; 15:1496–507; PMID: ; http://dx.doi.org/ 10.1111/cmi.12159 [DOI] [PubMed] [Google Scholar]
- 79.Gaulke CA, Porter M, Han Y-H, Sankaran-Walters S, Grishina I, George MD, Dang AT, Ding SW, Jiang G, Korf I, et al. . Intestinal epithelial barrier disruption through altered mucosal microRNA expression in HIV and SIV infections. J Virol 2014; 88(11):6268–80; PMID: ; http://dx.doi.org/ 10.1128/JVI.00097-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Loftus EV, Jr. Clinical epidemiology of inflammatory bowel disease: incidence, prevalence, and environmental influences. Gastroenterology 2004; 126:1504–17; PMID: ; http://dx.doi.org/ 10.1053/j.gastro.2004.01.063 [DOI] [PubMed] [Google Scholar]
- 81.Pekow JR, Kwon JH. MicroRNAs in inflammatory bowel disease. Inflamm Bowel Dis 2012; 18:187–93; PMID: ; http://dx.doi.org/ 10.1002/ibd.21691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Yang Y, Ma Y, Shi C, Chen H, Zhang H, Chen N, Zhang P, Wang F, Yang J, Yang J, et al. . Overexpression of miR-21 in patients with ulcerative colitis impairs intestinal epithelial barrier function through targeting the Rho GTPase RhoB. Biochem Biophys Res Commun 2013; 434:746–52; PMID: ; http://dx.doi.org/ 10.1016/j.bbrc.2013.03.122 [DOI] [PubMed] [Google Scholar]
- 83.Wu F, Zhang S, Dassopoulos T, Harris ML, Bayless TM. Meltzer SJ, Brant SR, Kwon JH. Identification of microRNAs associated with ileal and colonic Crohn's disease. Inflamm Bowel Dis 2010; 16:1729–38; PMID: ; http://dx.doi.org/ 10.1002/ibd.21267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Wu F, Zikusoka M, Trindade A, Dassopoulos T, Harris ML, Bayless TM, Meltzer SJ, Brant SR, Kwon JH. MicroRNAs are differentially expressed in ulcerative colitis and alter expression of macrophage inflammatory peptide-2 alpha. Gastroenterology 2008: 135:1624–35; PMID: ; http://dx.doi.org/ 10.1053/j.gastro.2008.07.068 [DOI] [PubMed] [Google Scholar]
- 85.Cichon C, Veltman K, Hummel S, Schmidt MA. Mirnas contribute to the probiotic effect of E. coli nissle 1917 on host cells offering new perspectives for inflammatory bowel disease treatment. Gastroenterology 2012; 142(5):S392–3 http://dx.doi.org/ 10.1016/S0016-5085(12)61489-9 [DOI] [Google Scholar]
- 86.Jeker LT, Zhou X, Gershberg K, de Kouchkovsky D, Morar MM, Stadthagen G, Lund AH, Bluestone JA. MicroRNA 10a marks regulatory T cells. PLoS One 2012;7(5):e36684; PMID: ; http://dx.doi.org/ 10.1371/journal.pone.0036684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Pott F, Cichon C, Sabharwal H, Schmidt MA, Bettenworth D. MicroRNA mir-up1 serves as a biomarker for the course of inflammation in experimental colitis as well as in crohns disease. Gastroenterology 2013; 144(5):S420; http://dx.doi.org/ 10.1016/S0016-5085(13)61545-0 [DOI] [Google Scholar]
- 88.Singh UP, Murphy AE, Enos RT, Shamran HA, Singh NP, Guan H, Hegde VL, Fan D, Price RL, Taub DD, et al. . miR-155 deficiency protects mice from experimental colitis by reducing Th1/Th17 responses. Immunology 2014; 31; PMID: ; http://dx.doi.org/ 10.1111/imm.12328 [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Takagi T, Naito Y, Mizushima K, Hirata I, Yagi N, Tomatsuri N, Ando T, Oyamada Y, Isozaki Y, Hongo H, et al. . Increased expression of microRNA in the inflamed colonic mucosa of patients with active ulcerative colitis. J Gastroenterol Hepatol 2010; 25(Suppl. 1)S129–33; PMID: ; http://dx.doi.org/ 10.1111/j.1440-1746.2009.06216.x [DOI] [PubMed] [Google Scholar]
- 90.Paraskevi A, Theodoropoulos G, Papaconstantinou I, Mantzaris G, Nikiteas N, Gazouli M. Circulating microRNA in inflammatory bowel disease. J Crohns Colitis 2012; 6:900–4; PMID: ; http://dx.doi.org/ 10.1016/j.crohns.2012.02.006 [DOI] [PubMed] [Google Scholar]
- 91.Zahm AM, Thayu M, Hand NJ, Horner A, Leonard MB, Friedman JR. Circulating microRNA is a biomarker of pediatric crohn disease. J Pediatr Gastroenterol Nutr 2011; 53:26–33; PMID: ; http://dx.doi.org/ 10.1097/MPG.0b013e31822200cc [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Tang Y, Banan A, Forsyth CB, Fields JZ, Lau CK, Zhang LJ, Keshavarzian A.Effect of alcohol on miR-212 expression in intestinal epithelial cells and its potential role in alcoholic liver disease. Alcohol Clin Exp Res 2008; 32:355–64; PMID: ; http://dx.doi.org/ 10.1111/j.1530-0277.2009.00946.x [DOI] [PubMed] [Google Scholar]
- 93.Veltman K, Hummel S, Cichon C, Sonnenborn U, Schmidt MA. Identification of specific miRNAs targeting proteins of the apical junctional complex that simulate the probiotic effect of E. coli Nissle 1917 on T84 epithelial cells. Intern J Biochem Cell Biol 2012; 44(2):341–349; PMID: ; http://dx.doi.org/ 10.1016/j.biocel.2011.11.006 [DOI] [PubMed] [Google Scholar]
- 94.Zhi X, Tao J, Li Z, Jiang B, Feng J, Yang L, Xu H, Xu Z. MiR-874 promotes intestinal barrier dysfunction through targeting AQP3 following intestinal ischemic injury. FEBS Lett 2014; 588(5):757–63; PMID: ; http://dx.doi.org/ 10.1016/j.febslet.2014.01.022 [DOI] [PubMed] [Google Scholar]
- 95.Wang FE, Zhang C, Maminishkis A, Dong L, Zhi C, Li R, Zhao J, Majerciak V, Gaur AB, Chen S, et al. . MicroRNA-204/211 alters epithelial physiology. FASEB J 2010; 24(5):1552–71; PMID: ; http://dx.doi.org/ 10.1096/fj.08-125856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Qin W, Ren Q, Liu T, Huang Y, Wang J. MicroRNA-155 is a novel suppressor of ovarian cancer-initiating cells that targets CLDN1. FEBS Lett 2013 May 2;587(9):1434–9; PMID: ; http://dx.doi.org/ 10.1016/j.febslet.2013.03.023 [DOI] [PubMed] [Google Scholar]
- 97.Liu M, Tang Q, Qiu M, Lang N, Li M, Zheng Y, Bi F. MiR-21 targets the tumor suppressor RhoB and regulates proliferation, invasion and apoptosis in colorectal cancer cells. FEBS Lett 2011; 585:2998–3005; PMID: ; http://dx.doi.org/ 10.1016/j.febslet.2011.08.014 [DOI] [PubMed] [Google Scholar]
- 98.Vega FM, Colomba A, Reymond N, Thomas M, Ridley AJ. RhoB regulates cell migration through altered focal adhesion dynamics. Open Biol 2012; 2:120076; PMID: ; http://dx.doi.org/ 10.1098/rsob.120076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Connolly EC, Van Doorslaer K, Rogler LE, Rogler CE. Overexpression of miR-21 promotes an in vitro metastatic phenotype by targeting the tumor suppressor RHOB. Mol Cancer Res 2010; 8:691–700; PMID: ; http://dx.doi.org/ 10.1158/1541-7786.MCR-09-0465 [DOI] [PubMed] [Google Scholar]
- 100.Sabatel C, Malvaux L, Bovy N, Deroanne C, Lambert V, Gonzalez ML, Colige A, Rakic JM, Noel A, Martial JA, et al. . MicroRNA-21 exhibits antiangiogenic function by targeting RhoB expression in endothelial cells. PLoS One 2011; 6:e16979; PMID: ; http://dx.doi.org/ 10.1371/journal.pone.0016979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Ozdamar B, Bose R, Barrios-Rodiles M, Wang HR, Zhang Y, Wrana JL. Regulation of the polarity protein Par6 by TGF-beta receptors controls epithelial cell plasticity. Science 2005; 307:1603–9; PMID: [DOI] [PubMed] [Google Scholar]
- 102.Wang HR, Zhang Y, Ozdamar B, Ogunjimi AA, Alexandrova E, Thomsen GH, Wrana JL. Regulation of cell polarity and protrusion formation by targeting RhoA for degradation. Science 2003; 302:1775–9; PMID: [DOI] [PubMed] [Google Scholar]
- 103.Walsh SV, Hopkins AM, Chen J, Narumiya S, Parkos CA, Nusrat A. Rho kinase regulates tight junction function and is necessary for tight junction assembly in polarized intestinal epithelia. Gastroenterology 2001; 121:566–79; PMID: [DOI] [PubMed] [Google Scholar]
- 104.Tili E, Michaille JJ, Cimino A, Costinean S, Dumitru CD, Adair B, Fabbri M, Alder H, Liu CG, Calin GA, et al. . Modulation of miR-155 and miR-125b levels following lipo- polysaccharide/TNF-alpha stimulation and their possible roles in regulating the response to endotoxin shock. J Immunol 2007; 179:5082–9; PMID: ; http://dx.doi.org/ 10.4049/jimmunol.179.8.5082 [DOI] [PubMed] [Google Scholar]
- 105.O’Connell RM, Taganov KD, Boldin MP, Cheng G, Baltimore D. MicroRNA-155 is induced during the macrophage inflammatory response. Proc Natl Acad Sci U S A 2007; 104:1604–9; PMID: ; http://dx.doi.org/ 10.1073/pnas.0610731104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Sheedy FJ, O’Neill LA. Adding fuel to fire: microRNAs as a new class of mediators of inflammation. Ann Rheum Dis 2008; 67(Suppl 3):iii50-5; PMID: ; http://dx.doi.org/ 10.1136/ard.2008.100289 [DOI] [PubMed] [Google Scholar]
- 107.Imaizumi T, Tanaka H, Tajima A, Yokono Y, Matsumiya T, Yoshida H, Tsuruga K, Aizawa-Yashiro T, Hayakari R, Inoue I, et al. . IFN-γ and TNF-α synergistically induce microRNA-155 which regulates TAB2/IP-10 expression in human mesangial cells. Am J Nephrol 2010; 32:462–8; PMID: ; http://dx.doi.org/ 10.1159/000321365 [DOI] [PubMed] [Google Scholar]
- 108.Sasaki M, Sitaraman SV, Babbin BA, Gerner-Smidt P, Ribot EM, Garrett N, Alpern JA, Akyildiz A, Theiss AL, Nusrat A, et al. . Invasive Escherichia coli are a feature of Crohn's disease. Lab Invest 2007; 87:1042–54; PMID: ; http://dx.doi.org/ 10.1038/labinvest.3700661 [DOI] [PubMed] [Google Scholar]
- 109.Dejana E, Orsenigo F, Lampugnani. The role of adherens junctions and VE cadherin in the control of vascular permeability. J Cell Sci 2008; 121:2115–22; PMID: ; http://dx.doi.org/ 10.1242/jcs.017897 [DOI] [PubMed] [Google Scholar]
- 110.Petty MA, Lo EH. Junctional complexes of the blood-brain barrier: Permeability changes in neuroinflammation. Prog Neurobiol 2002; 68:311–23; PMID: ; http://dx.doi.org/ 10.1016/S0301-0082(02)00128-4 [DOI] [PubMed] [Google Scholar]
- 111.Löscher W, Potschka H. Role of drug efflux transporters in the brain for drug disposition and treatment of brain diseases. Prog Neurobiol 2005 May; 76(1):22–76. PMID: ; http://dx.doi.org/ 10.1016/j.pneurobio.2005.04.006 [DOI] [PubMed] [Google Scholar]
- 112.Andl CD. The misregulation of cell adhesion components during tumorigenesis: overview and commentary. J Oncol 20102010:pii: 174715; PMID: ; http://dx.doi.org/ 10.1155/2010/174715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Mishra R, Singh SK. HIV-1 Tat C modulates expression of miRNA-101 to suppress VE- Cadherin in human brain microvascular endothelial cells. J Neurosci 2013; 33(14):5992–6000; PMID: ; http://dx.doi.org/ 10.1523/JNEUROSCI.4796-12.2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Zang Y-W, Gu X-D, Xiang J-B, Chen Z-Y. Brain metastases from colorectal cancer: microenvironment and molecular mechanisms. Int J Mol Sci 2012; 13(12):15784–800; PMID: ; http://dx.doi.org/ 10.3390/ijms131215784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Ma L, Young J, Prabhala H, Pan E, Mestdagh P, Muth D, Teruya-Feldstein J, Reinhardt F, Onder TT, Valastyan S, et al. . MiR-9, a MYC/MYCN- activated microRNA, regulates E-cadherin and cancer metastasis. Nat Cell Biol 2010; 12(3):247–56; PMID: ; http://dx.doi.org/ 10.1038/ncb2024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Fazakas C, Wilhelm I, Nagyoszi P, Farkas AE, Haskó J, Molnár J, Bauer H, Bauer HC, Ayaydin F, Dung NT, et al. . Transmigration of melanoma cells through the blood-brain barrier: role of endothelial tight junctions and melanoma-released serine proteases. PLoS ONE 2011; 6:e20758; PMID: ; http://dx.doi.org/ 10.1371/journal.pone.0020758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Wilhelm I, Fazakas C, Krizbai IA. In vitro models of the blood-brain barrier. Acta Neurobiol Exp (Wars) 2011; 71:113–28; PMID: [DOI] [PubMed] [Google Scholar]
- 118.Zhou W, Fong MY, Min Y, Somlo G, Liu L, Palomares MR, Yu Y, Chow A, O'Connor ST, Chin AR, et al. . Cancer-secreted miR-105 destroys vascular endothelial barriers to promote metastasis. Cancer Cell 2014; 25(4):501–15; PMID: ; http://dx.doi.org/ 10.1016/j.ccr.2014.03.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Lu Y, Govindan R, Wang L, Liu PY, Goodgame B, Wen W, Sezhiyan A, Pfeifer J, Li YF, Hua X, et al. . MicroRNA profiling and prediction of recurrence/relapse- free survival in stage I lung cancer. Carcinogenesis 2012; 33:1046–54; PMID: ; http://dx.doi.org/ 10.1093/carcin/bgs100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Arora S, Ranade AR, Tran NL, Nasser S, Sridhar S, Korn RL, Ross JT, Dhruv H, Foss KM, Sibenaller Z, et al. . MicroRNA-328 is associated with (non-small) cell lung cancer (NSCLC) brain metastasis and mediates NSCLC migration. Int J Cancer 2011; 129:2621–31; PMID: ; http://dx.doi.org/ 10.1002/ijc.25939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Hwang SJ, Seol HJ, Park YM, Kim KH, Gorospe M, Nam DH, Kim HH. MicroRNA-146a suppresses metastatic activity in brain metastasis. Mol Cells 2012; 34(3):329–34. PMID: ; PMCID: PMC3887840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. NIH grant to Redell JB: NIH #R21NS078596-01; MiR-223 regulation of blood brain barrier function after traumatic brain injury 2012; University of Texas health Science Center Houston; http://projectreporter.nih.gov/project_info_description.cfm?projectnumber=1R21NS078596-01 [Google Scholar]
- 123.Yang H, Rao JN, Wang JY. Posttranscriptional regulation of intestinal epithelial tight junction barrier by RNA-binding proteins and microRNAs. Tissue Barriers 2014; 2:e28320; PMID: ; http://dx.doi.org/ 10.4161/tisb.28320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell 2005; 120:15–20; PMID: ; http://dx.doi.org/ 10.1016/j.cell.2004.12.035 [DOI] [PubMed] [Google Scholar]