Abstract
Members of the claudin family of tight junction proteins are critical for establishing epithelial barriers and for the regulation of paracellular transport. To understand their roles during kidney development, we first performed RT-PCR analyses and determined that 23 claudin family members were expressed in embryonic day (E) 13.5 mouse kidneys. Based on their developmental expression and phenotypes in mouse models, we hypothesized that 3 claudin members could affect nephron formation during kidney development. Using whole mount in situ hybridization and immunohistochemistry, we demonstrated that Claudin-7 (Cldn7) was expressed in the nephric duct, the emerging ureteric bud, and in tubules derived from ureteric bud branching morphogenesis. In contrast, Claudin-16 (Cldn16) and Claudin-19 (Cldn19) were expressed at later stages of kidney development in immature renal tubules that become the Loop of Henle. To determine if a loss of these claudins would perturb kidney development, we examined newborn kidneys from mutant mouse models lacking Cldn7 or Cldn16. In both models, we noted no evidence for any congenital renal malformation and quantification of nephron number did not reveal a decrease in nephron number when compared to wildtype littermates. In summary, Cldn7, Cldn16, and Cldn19 are expressed in different epithelial lineages during kidney development. Mice lacking Cldn7 or Cldn16 do not have defects in de novo nephron formation, and this suggests that these claudins primarily function to regulate paracellular transport in the mature nephron.
Keywords: Loop of Henle, nephrogenesis, renal branching morphogenesis, tubule differentiation
Abbreviations: E, embryonic day; P, postnatal; FHHNC, familial hypomagnesaemia with hypercalciuria and nephrocalcinosis; nd, nephric duct; ub, ureteric bud; MET, mesenchymal to epithelial transition
Introduction
Members of the claudin family of integral membrane proteins are components of tight junctions: they connect adjacent epithelial cells to one another in a narrow band just beneath the apical surface to create an occluding seal.1-2 Claudin proteins have well-established roles in renal, gastrointestinal, and lung physiology based on their abilities to regulate paracellular transport. However, many claudins are expressed during embryogenesis, suggesting that these proteins may also contribute to organ formation, including kidney development, through direct effects on cell adhesion, proliferation, and lumen formation.3
During mammalian kidney development, 2 primordial tissues, the ureteric bud and the metanephric mesenchyme, undergo epithelial morphogenesis to form the final metanephric kidney.4 The kidneys and the ureters arise from 2 epithelial tubes that extend along the length of the embryo, the mesonephric ducts. An epithelial swelling emerges from the mesonephric duct at embryonic day (E) 10.5 during mouse embryonic development and is known as the ureteric bud. The ureteric bud invades the adjacent undifferentiated mesenchyme and induces the formation of the metanephric mesenchyme. Reciprocal signaling between the ureteric bud and the metanephric mesenchyme induces the ureteric bud to elongate and bifurcate in a process known as branching morphogenesis that ultimately gives rise to the collecting duct system of the adult kidney. The process of ureteric bud branching morphogenesis is critical for kidney development: each ureteric bud tip induces the adjacent metanephric mesenchyme to undergo mesenchymal-to-epithelial transition and this determines the final number of nephrons formed in utero. As part of mesenchymal-to-epithelial transition, a number of intermediate structures form including mesenchymal condensates, comma-shaped structures, S-shaped structures, and early and late tubules that eventually give rise to all of the nephron segments except the collecting duct.
Data from microarray, SAGE expression profiling, RT-PCR and immunohistochemical analyses of embryonic kidneys have shown that a number of claudins are expressed during kidney development in the mouse and in the rat.5-12 Gene expression microarray analysis of mRNA isolated from ureteric bud and metanephric mesenchyme of embryonic rats demonstrated that Claudin-3 and -9 mRNA transcripts were strongly upregulated in ureteric bud when compared to metanephric mesenchyme by a factor of 12.5 and 19.1 fold, respectively.7 Similarly, Schwab et al. observed that Claudin-3, -4, -7, and -8 were strongly upregulated in E11.5 mouse ureteric bud when compared to metanephric mesenchyme.6 Ohta demonstrated that a number of claudins are expressed in the developing mouse kidney and showed that Claudin-3, -4, and -8 appear to be expressed in collecting duct tubules that are derived from the ureteric bud.9
The presence of claudins prior to the formation of a urinary filtrate suggested that they could have specific roles during kidney development not related to paracellular transport. In this study, we confirmed by RT-PCR and sequencing that 23 members of the claudin family are expressed in the E13.5 mouse kidney. We then focused our analysis on Claudin-7, -16 and -19 to determine if they have a role during kidney development. Claudin-7 is highly expressed in the aldosterone-sensitive segments of the distal nephron13 and when inactivated in the mouse, the mice die at postnatal day 12 from severe dehydration from urinary salt wasting such that a coincident defect in nephron formation could be missed if not considered.14 Mutations in CLDN16 and 19 have been associated with a rare autosomal recessive syndrome known as familial hypomagnesaemia with hypercalciuria and nephrocalcinosis (FHHNC) characterized by renal calcium and magnesium wasting that can lead to chronic renal failure in childhood or adolescence.15-17 The early onset of chronic renal failure in childhood suggests that these patients might have a defect in nephron formation during kidney development in addition to possible loss of nephrons secondary to renal calcium deposition. Here, we report a detailed expression analysis of Cldn7, Cldn16, and Cldn19 during mouse kidney development and have examined mouse models lacking either Cldn7 or Cldn16 to determine if there is evidence for a defect in kidney development.
Materials and Methods
Animal care. Timed-pregnant CD1 and non-pregnant adult mice (Charles River Laboratories, St. Constant, QC) were euthanized using CO2 inhalation followed by cervical dislocation at embryonic day (E) 9.5, 11.0, 11.5, 12.5, 13.5, 16.5, postnatal day (P) 1, or adulthood to retrieve embryonic, newborn, or adult kidneys as previously described.11 Newborn pups were euthanized using decapitation. Whole embryos or metanephric kidneys were dissected in PBS using an M5A stereomicroscope (Wild Leitz, Willowdale, ON). All mice used in these studies were housed in temperature-controlled animal facilities in individual ventilated cages with ad libitum access to food and water. Light cycles for all mice were maintained as 12 hours of light and 12 hours of darkness. The mouse studies were performed in accordance with the rules and regulations of the Canadian Council of Animal Care (AUP #4120) and the Office of Laboratory Animal Welfare in the US. Department of Health and Human Services (AUP#172b).
Cldn7 +/− heterozygous breeders on a C57BL/6J background were used to derive P1 litters. The pups were euthanized and genotyped as described.14 Transgenic Cldn16+/− knockdown mice were derived using a lentiviral construct that contained the pFUGW vector in which the snRNP U6 promoter drives overexpression of an siRNA directed to Cldn16 and the ubiquitin-C promoter drives expression of green fluorescent protein (GFP).18 Transgenic Cldn16+/− knockdown males on a C57BL/6J background were crossed to C57BL/6J female to derive P1 litters. All pups were euthanized, and transgenic progeny were identified by direct observation of GFP fluorescence using a hand-held GFP flashlight as previously reported.18
RT-PCR. Ten E11.5, 12.5, or 13.5 mouse kidneys were pooled and RNA was extracted from 3 different pools using the RNeasy kit (Qiagen). For E16.5 and P1 kidneys, 3 kidneys were pooled for RNA extraction. RNA was treated with RNase-free DNase (Ambion) to degrade any traces of DNA. RT-PCR was performed using the One Step RT-PCR Kit (Qiagen). 2 negative controls were included: in one there was no RNA template, and in the other, the reverse transcriptase enzyme was inactivated. β−actin was used as a positive control for the quality of the RNA used in these reactions. Gene specific primers designed to amplify individual claudin family members are shown in Table 1. RT-PCR amplicons were sequenced to confirm their identity.
Table 1.
Claudin Primer Sequences used in RT-PCR
| Gene | Size | Name of primer | Sequence (5′ to 3′) |
|---|---|---|---|
| Claudin1 | 636 bp | mCldn1-`F | CCATGGCCAACGCGGGGCTGC |
| mCldn1-R | TCACACATAGTCTTTCCCAC | ||
| Claudin2 | 693 bp | mCldn2-F | CCATGGCCTCCCTTGGCGTCC |
| mCldn2-R | CACACATACCCAGTCAGGCTG | ||
| Claudin3 | 657 bp | mCldn3-F | CATATGTCCATGGGCTGG |
| mCldn3-R | TCAGACGTAGTCCTTGCGGT | ||
| Claudin4 | 633 bp | mCldn4-F | CCATGGCGTCTATGGGACTAC |
| mCldn4-R | TTACACATAGTTGCTGGCGG | ||
| Claudin5 | 420 bp | mCldn5-F | CCATGGGGTCTGCAGCGTTGG |
| mCldn5-R | GGCGAACCAGCAGAGCGGCAC | ||
| Claudin6 | 660 bp | mCldn6-F | CCATGGCCTCTACTGGTCTGC |
| mCldn6-R | TCACACATAATTCTTGGTGG | ||
| Claudin7 | 689 bp | mCldn7-F | CCATGGCCAACTCGGGCCTGCAAC |
| mCldn7-R | TCACACGTATTCCTTGGAGG | ||
| Claudin8 | 678 bp | mCldn8-F | CCATGGCAACCTACGCTCTTC |
| mCldn8-R | CTACACATACTGACTTTTGG | ||
| Claudin9 | 654 bp | mCldn9-F | CCATGGCTTCCACTGGCCTTG |
| mCldn9-R | TCACACATAGTCCCTCTTATC | ||
| Claudin10 | 689 bp | mCldn10a-F | CCATGTCCAGGGCACAGATCT |
| mCldn10b-F | CCATGGGTAGCACGGCCTTGG | ||
| 696 bp | mCldn10-R | TTAGACATAGGCATTTTTATC | |
| Claudin11 | 624 bp | mCldn11-F | CCATGGTAGCCACTTGCCTTC |
| mCldn11-R | TTAGACATGGGCACTCTTG | ||
| Claudin12 | 735 bp | mCldn12-F | CCATGGGCTGCCGAGATGTCC |
| mCldn12-R | TTAAGTGCTGTGTGAGACTAC | ||
| Claudin13 | 636 bp | mCldn13-F | CCATGGTCGTCAGCAAACAAG |
| mCldn13-R | TCAAACATCTAAGGTATCG | ||
| Claudin14 | 720 bp | mCldn14-F | CCATGGCCAGCACAGCGGTCC |
| mCldn14-R | TCACACGTAGTCATTCAGCC | ||
| Claudin15 | 684 bp | mCldn15-F | CATATGTCGGTAGCTGTGGAGACC |
| mCldn15-R | CTACACGTATGCGTTTTTGC | ||
| Claudin16 | 708 bp | mCldn16-F | CATATGAAGGATCTTCTTCAGTACG |
| mCldn16-R | TTATACTCTGGTGTCTACAGC | ||
| Claudin17 | 675 bp | mCldn17-F | CCATGGCTTTTTATCCCTTAC |
| mCldn17-R | TTAGACGTAGCTGGTGGAAG | ||
| Claudin18 | 150 bp | mCldn18-F | TTCATCATCTCCGGCATCTG |
| mCldn18-R | CAAAGGTGTACCTGGTCTGA | ||
| Claudin19 | 675 bp | mCldn19-F | CCATGGCCAACTCGGGCCTCCA |
| mCldn19-R | TTACACACCCAGGGGGCCCT | ||
| Claudin20 | 660 bp | mCldn20-F | ATGGCCTCGGCAGGTCTCC |
| mCldn20-R | TTACATGTAATCCTTCAAGTTATG | ||
| Claudin22 | 663 bp | mCldn22-F | CCATGGGCTTAGTCTTCCGAAC |
| mCldn22-R | TTAGATTTCTGGATTGGCT | ||
| Claudin23 | 891 bp | mCldn23-F | CATATGCGGACGCCGGTGGTGATGACG |
| mCldn23-R | CTACAGGTCGGAGTCACAGG | ||
| Claudin24 | 663 bp | mCldn24-F | CCATGGCTTTCATCTTCAGAACG |
| mCldn24-R | TTACACTTTAGGCTGTACAGTTCC | ||
| β-actin | 336 bp | mβ-actin-F | CACAGCTGAGAGGGAAATC |
| mβ-actin-R | TCAGCAATGCCTGGGTAC |
Whole mount in situ hybridization. In situ hybridization was performed on whole E9.5 and 11.0 mouse embryos and on dissected E12.5, 13.5, 16.5, and P1 mouse kidneys as described.19 Kidneys from P1 mice were bisected prior to fixation and in situ hybridization analysis. All samples were fixed overnight in 4% PFA in PBS at 4°C. cDNA clones for Cldn7, Cldn16, Cldn19, and Ret were generated by RT-PCR amplification of the coding sequences using E15 mouse kidney total RNA as a template. The primers sequences were (5′ to 3′): Cldn7 forward CCATGGGCCTGGAGATCACCG, Cldn7 reverse TCAGACGTAGTCCTTGCGGT, Cldn16 forward CATATGAAGGATCTTCTTCAGTACG, Cldn16 reverse TTATACTCTGGTGTCTACAGC, Cldn19 forward CCATGGCCAACTCGGGCCTCCA, Cldn19 reverse TTACACACCCAGGGGGCCCT and Ret9 forward GATTACAAGGATGACGAC and reverse TGAGGTAGACGGTGAGCA. Amplified DNA fragments were sequenced to confirm their identity and then subcloned into vectors. To generate antisense riboprobes for in situ hybridization analysis, PCRII-TOPO vector (Invitrogen, Camarillo, CA) encoding mouse Cldn7 was linearized using BamHI while pSC-A vectors (Strataclone, Agilent Technologies, North Torrey Pines Road La Jolla, CA) encoding mouse Cldn16 and Cldn19 were linearized with Sma1, and the mouse Ret vector was linearized with BamHI respectively. Digoxigenin (DIG)-labeled UTP probes were generated in vitro using SP6 RNA polymerase for Cldn7 and T3 RNA polymerase for Cldn16, Cldn19 and Ret. Anti-DIG antibody conjugated to alkaline phosphatase was used to detect duplexes of DIG-labeled antisense riboprobe hybridized to claudin mRNA sequences. Treated samples were developed using nitro blue tetrazolium chloride, 5-bromo-4-chloro-3-indolyl phosphatase substrate in NTMT. All kidneys were photographed and E16.5 and P1 kidneys were processed for cryosectionning. 15 μm frozen sections from E16.5 and P1 mouse kidneys were counterstained with eosin and coverslipped with glycerol-gelatin aqueous mounting medium (Sigma-Aldrich) for imaging, while earlier time points were imaged as whole mounts.
Immunohistochemistry and immunofluorescence. E16.5, P1, and P28 kidneys from CD1 mice were dissected, fixed in 6:3:1 (ethanol:water:formaldehyde) and processed for paraffin sectioning or cryosectioned for immunofluorescent studies. Mouse anti-Calbindin-D-28K (C9848, Sigma-Aldrich), rabbit anti-Claudin 7 (E10590, Spring Biosciences), rabbit anti-Tamm Horsfall glycoprotein/Uromodulin (BT-590, Biomedical Technologies Inc.), and rabbit anti-Claudin-16 (34–5400, Invitrogen) were used at a 1:200 dilution. Rabbit anti-Claudin-19 antibody was generated by Dr. J. Hou20 and used at a 1:200 dilution. Hematoxylin QS (Vector Laboratories) was used as a counterstain. Color development was performed using the Vectastain ABC kit (Vector Laboratories). For the immunofluorescent studies, mouse anti-ZO-1 (339100, Invitrogen), mouse anti-Occludin (331500, Invitrogen), rabbit anti-Claudin-16 (34-5400, Invitrogen), and rabbit anti-Claudin-19 (Dr. J. Hou) were used, each at a dilution of 1:50. Alexa Fluor 555 goat anti-mouse (A21127, Invitrogen) and Alexa Fluor 488 goat anti-rabbit ( A11008, Invitrogen) were used as secondary antibodies at a concentration of 1:100.
Nephron number assessment. P1 kidneys were dissected from pups derived from Cldn7+/− heterozygous matings and transgenic Cldn16+/− crosses, fixed in formalin, dehydrated and then paraffin-embedded for sectioning. Total nephron counts were obtained by counting every tenth section as we have done previously. 21 Each kidney was sectioned in its entirety and serial sections of 7 μm thickness were obtained and stained with hematoxylin and eosin. The sections were analyzed using Image J (v1.36b). Individual glomeruli were identified by the presence of a glomerular tuft with an adjacent Bowman's capsule and were counted using the point selection tool and the auto measure function. Every tenth section was counted, therefore the total nephron number for each kidney was calculated by multiplying this value by 10. The Student's t-test was used for statistical analysis of the data.
Results
Multiple claudins are expressed during mouse kidney development
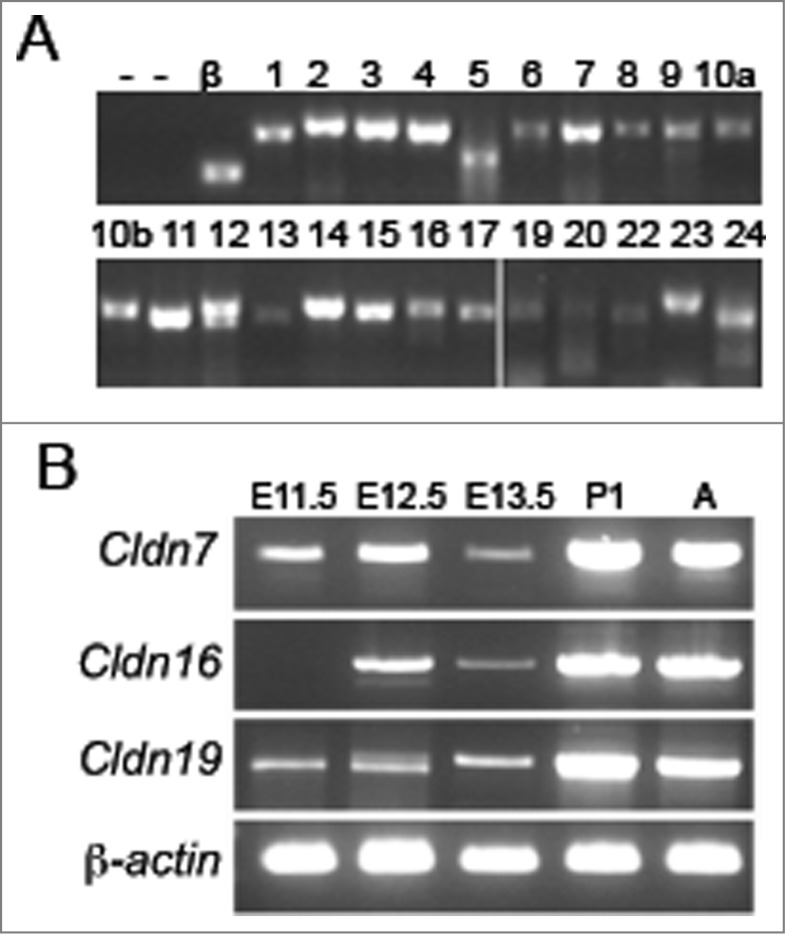
To determine which claudin family members are expressed during kidney development, we performed RT-PCR analyses using RNA that was extracted from a pool of mouse embryonic kidneys at E13.5, a time point at which both ureteric bud branching morphogenesis and nephrogenesis are well underway. The identity of all PCR products was confirmed by Sanger sequencing. At E13.5, 23 mouse Claudin family members were expressed, including Cldn7, Cldn16, and Cldn19 (Fig. 1A). Claudin-18 was the only member that was not amplified at this time point. A more detailed expression analysis was performed for Cldn7, Cldn16, and Cldn19 to track their temporal expression in the embryonic and postnatal kidney. The analysis revealed that Cldn7 and Cldn19 are detected at E11.5, while Cldn16 was not detected until E12.5 (Fig. 1B).
Figure 1.
Temporal Expression Analysis of Claudins during Mouse Kidney Development. (A) RT-PCR analysis of all claudin family members in E13.5 mouse kidneys. β-actin (β) was used as a positive control and RT-PCR reactions without the addition of RNA (lane 1) or with inactivated reverse transcriptase (‘—’; lanes 1 and 2) served as negative controls. The claudin family member is listed above each lane. Cldn18 was not amplified (data not shown). All RT-PCR amplicons were sequenced to confirm identity. (B) RT-PCR analysis of Cldn7, Cldn16, and Cldn19 at different stages of kidney development. β-actin was amplified as an internal control. Cldn7 and Cldn19 mRNA transcripts are expressed from embryonic day (E) 11.5 onwards, whereas Cldn16 mRNA transcripts are expressed from E12.5 onwards.
Claudin-7, -16, and -19 are expressed in specific lineages during kidney development
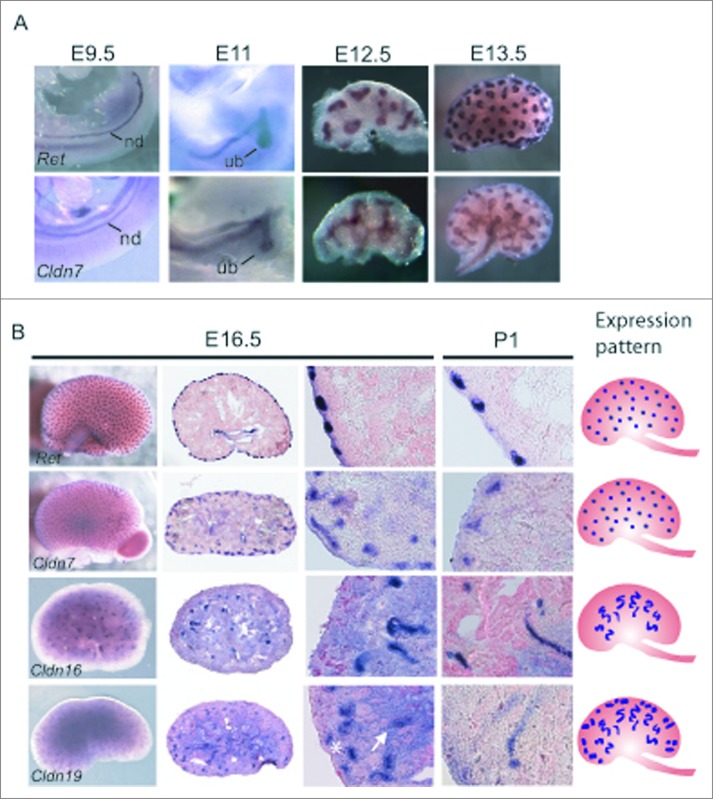
We examined the spatial expression patterns of Cldn7, Cldn16, and Cldn19 using in situ hybridization at critical stages of kidney development including E9.5 when the nephric duct is well-delineated, E11 when the ureteric bud first emerges from the nephric duct and invades the metanephric mesenchyme and branches, and E12.5 and E13.5, when there is more extensive ureteric bud branching morphogenesis and early nephron precursors are detected. We also examined E16.5 when the glomerulus becomes vascularised and nephron segments begin to differentiate22 and postnatal day 1 to complete the developmental expression analysis.
Cldn7 was highly expressed within the nephric duct, the emerging ureteric bud, the ureteric bud trunks and the tips, but by P1, it was mostly detected in the ureteric bud tips (Fig. 2A,B). Whereas Ret was limited to the tips of the ureteric bud branches from E12.5-16.5, Cldn7 was expressed more broadly at the tips as well as in the adjacent ureteric bud trunks. At P1, the expression patterns of Ret and Cldn7 looked more similar with both highly concentrated in the ureteric bud tips.
Figure 2.
Spatial Expression Analysis of Cldn7, Cldn16, and Cldn19 mRNA Transcripts during Mouse Kidney Development. (A-B) Whole mount in situ hybridization was performed on whole embryos at embryonic day (E) 9.5 and E11.0, and on whole kidneys at E12.5, E13.5, E16.5, and P1 for Cldn7, Cldn16, Cldn19, and Ret. E16.5 and P1 kidneys were cryosectioned at 15 μm thickness and counterstained with eosin. Cldn7 expression is observed in the nephric duct (nd) at E9.5 and in the ureteric bud (ub) at E11.0. At E11.0, E12.5, E13.5 and E16.5 Cldn7 is seen in the ureteric bud trunks and the tips. At P1, Cldn7 is detected mostly in ureteric bud tips, similar to Ret expression at the same stage. Cldn16 and Cldn19 transcripts are first detected at E16.5. Cldn16 is predominantly expressed in late tubular structures, while Cldn19 is expressed in both early and late tubular structures that will eventually become the Loop of Henle. On the far right schematic patterns are shown indicating a ureteric bud tip pattern (Cldn7 and Ret), a late tubule pattern (Cldn16), and a combined early and late tubule pattern (Cldn19) as described.23
While Cldn16 and Cldn19 were detected by RT-PCR beginning at E12.5 and E11.5 respectively (Fig. 1B), whole mount in situ hybridization failed to detect any signal for either transcript before E16.5 (data not shown), suggesting that their expression is low. At E16.5, both Cldn16 and Cldn19 mRNA transcripts were expressed in late tubular structures, one of 8 discernible patterns recently used to define gene expression profiles during kidney development (Fig. 2B).23 While both transcripts demonstrated this pattern, Cldn19 transcripts were detected more broadly and were also apparent in early tubular structures. These early and late tubular structures give rise to the Loop of Henle that progresses from an anlage, to a primitive loop, and then to an immature loop prior to its final differentiation.24-25
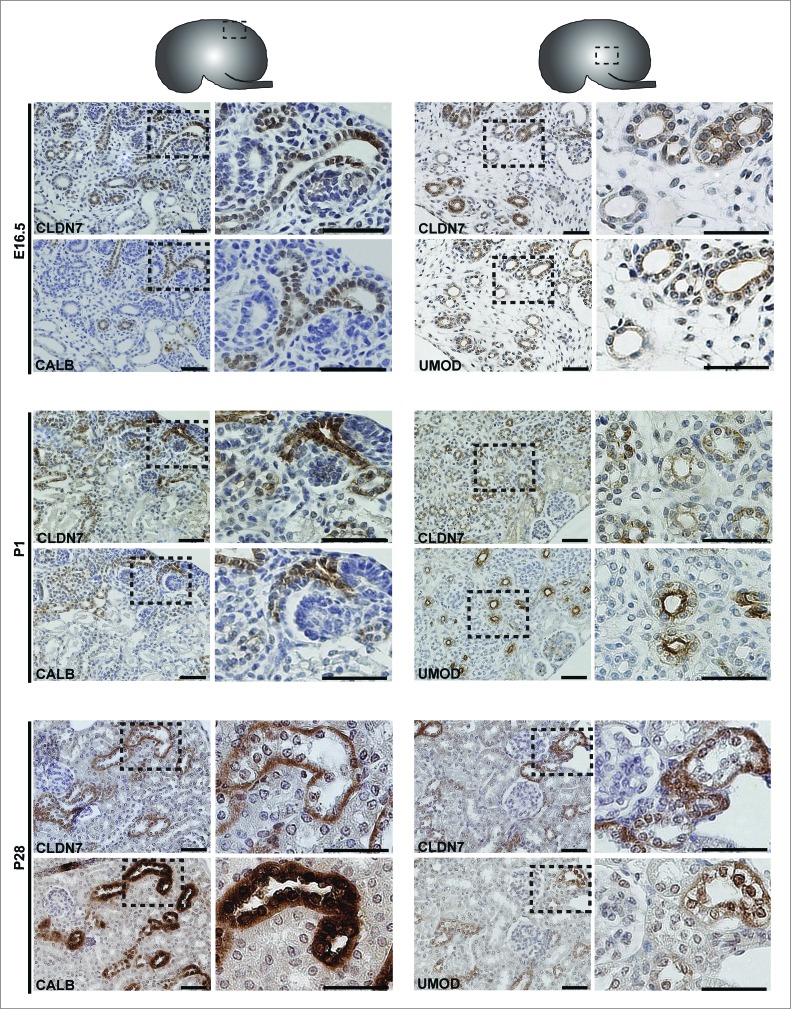
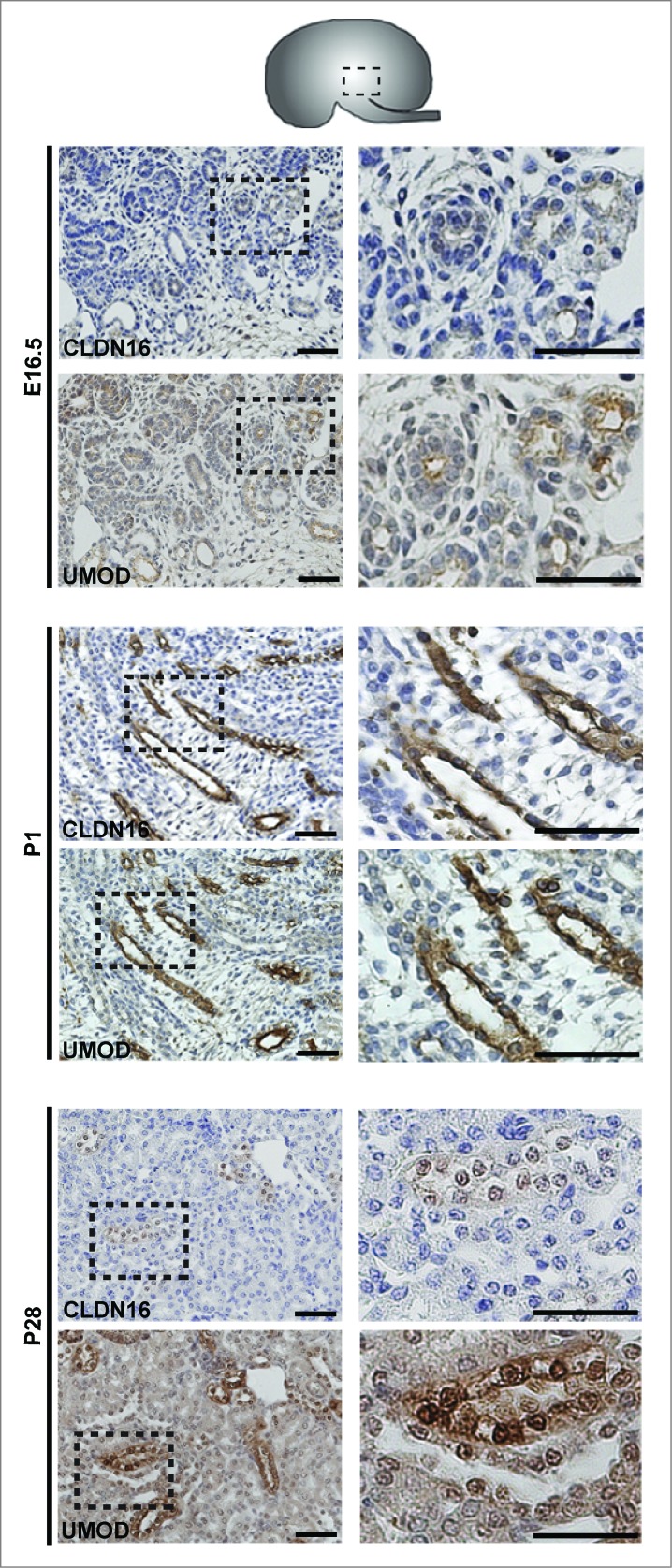
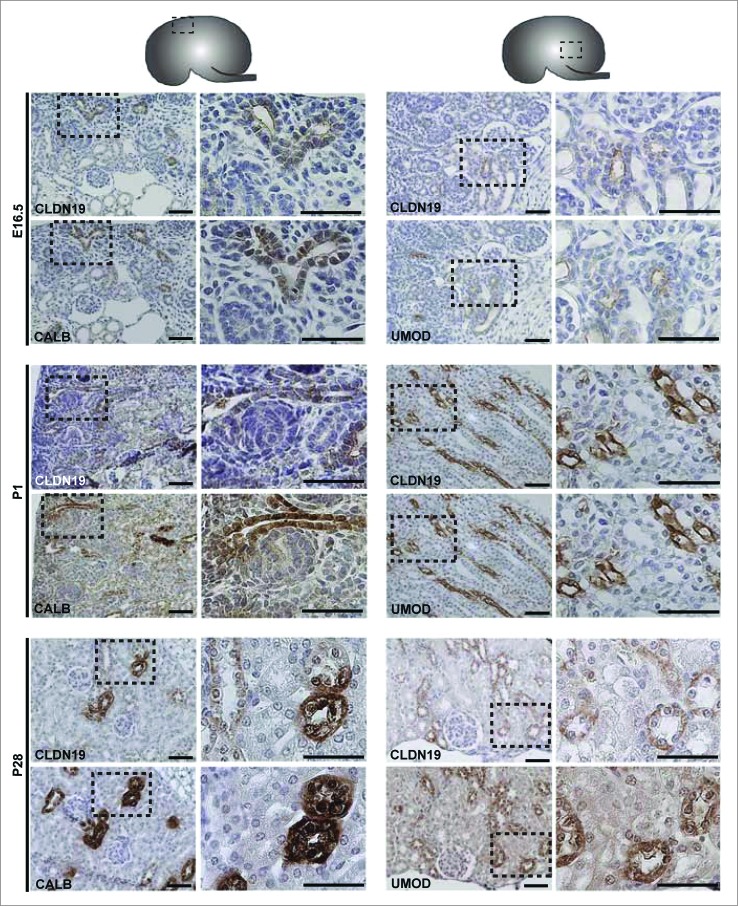
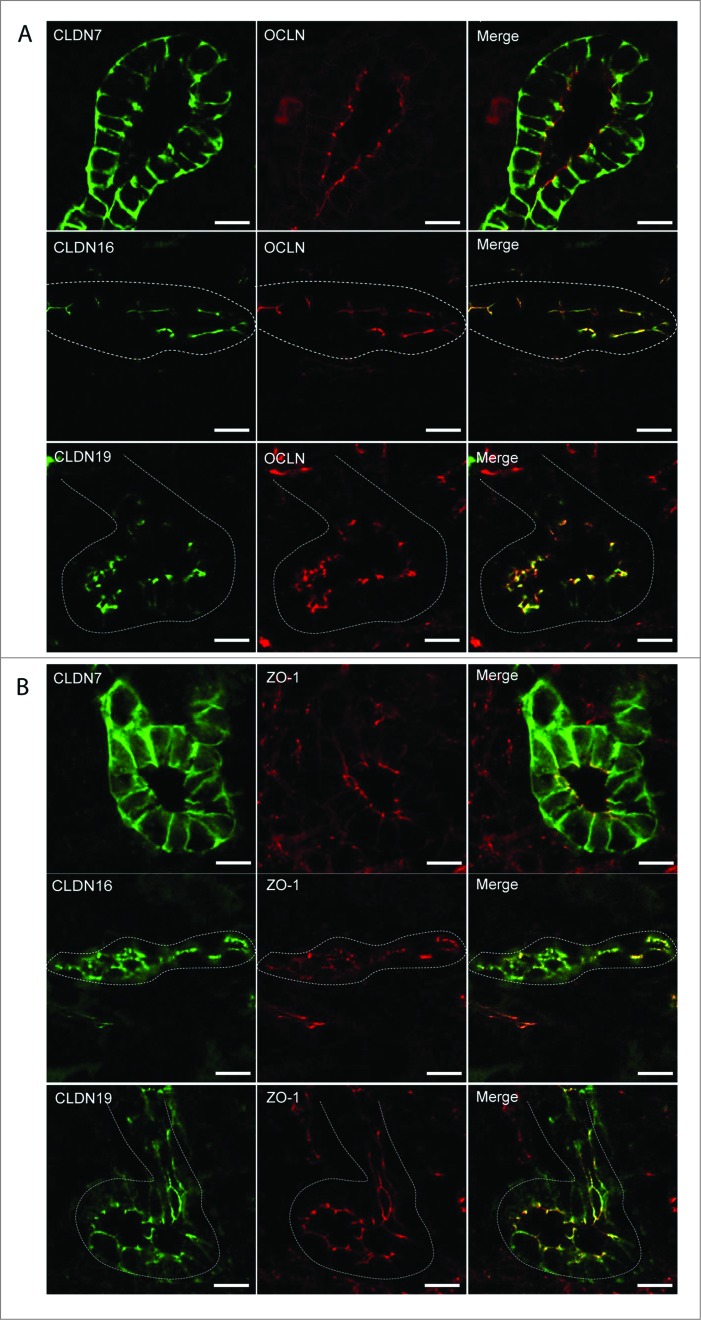
Protein expression was examined by performing immunohistochemistry on paraffin-embedded sections and immunofluorescence on cryosections (Figs. 3–6). At E16.5 and at P1, CLDN7 was detected in uroepithelial cells in ureteric bud trunks and tips and appeared to be expressed both apically and basolaterally (Figs. 3 and 6). Similar expression was noted at P28, but here CLDN7 was predominantly detected in distal tubular segments based on its overlapping expression with Calbindin-D-28K and with Uromodulin.24 Calbindin-D-28K protein is expressed by the distal convoluted tubule, the connecting tubule and the collecting duct, while Tamm Horsfall or Uromodulin protein is expressed by the thin and thick ascending limbs of the Loop of Henle.24 CLDN16 and 19 were strongly expressed in both the primitive and the mature Loop of Henle at E16, P1 and P28 and showed overlapping expression with Uromodulin that also demarcates the Loop of Henle24 (Figs. 4 and 5). CLDN19 showed more broad expression at all stages examined and overlapped with both Calbindin-D-28K and Uromodulin (Fig. 5), while CLDN16 did not show overlapping expression with Calbindin-D-28K (data not shown). By immunofluorescence, it was evident that all 3 claudins co-localize with both ZO-1 and with Occludin, confirming their expression in tight junctions (Fig. 6).
Figure 3.
Protein Expression of Claudin-7 during Mouse Kidney Development. Immunohistochemistry was performed on paraffin-embedded sections taken from mouse kidneys at embryonic day (E) 16.5, postnatal day (P) 1 and P28 as shown. Claudin-7 is expressed in ureteric bud trunks and tips at E16.5 and in distal tubular segments as well at P1 and P28, based on its overlapping expression with Calbindin-D-28K and with Uromodulin. A higher magnification of the boxed regions are shown to the right of each image. Scale bar = 50 μm.
Figure 4.
Protein Expression of Claudin-16 during Mouse Kidney Development. Immunohistochemistry was performed on paraffin-embedded sections taken from mouse kidneys at E16.5, P1 and P28 as shown. Claudin-16 is expressed in primitive and mature Loop of Henle tubules as indicated by its overlapping expression with Uromodulin at all stages. A higher magnification of the boxed regions are shown to the right of each image. Scale bar = 50 μm.
Figure 5.
Protein Expression of Claudin-19 during Mouse Kidney Development. Immunohistochemistry was performed on paraffin-embedded sections taken from mouse kidneys at E16.5, P1 and P28 as shown. Claudin-19 is expressed in primitive and mature Loop of Henle tubules as indicated by its overlapping expression with Uromodulin at all stages. Claudin-19 shows broader expression than Claudin-16 and also overlaps with Calbindin-D-28K. A higher magnification of the boxed regions are shown to the right of each image and show regions of overlap with Uromodulin or Calbindin-D-28K. Scale bar = 50 μm.
Figure 6.
Claudin-7, 16, and -19 Co-Localize with Tight Junction Markers. Immunofluorescence was performed on cryosections taken from mouse P1 kidneys using antibodies to Claudin-7, -16, and -19 and co-localization was performed using antibodies to 2 tight junction markers, Occludin (A), and ZO-1 (B). Claudin-7 is expressed apically and basolaterally in ureteric bud tubules and is co-expressed with both ZO-1 and Occludin. Claudin-16 is expressed apically in Loop of Henle tubules and is co-expressed with ZO-1 and Occludin. Claudin-19 is expressed apically in Loop of Henle tubules and is co-expressed with ZO-1 and Occludin. Dotted white lines outline tubule segments. Scale bar = 10 μm.
Nephron number assessment in mouse models lacking Claudin-7 and Claudin-16
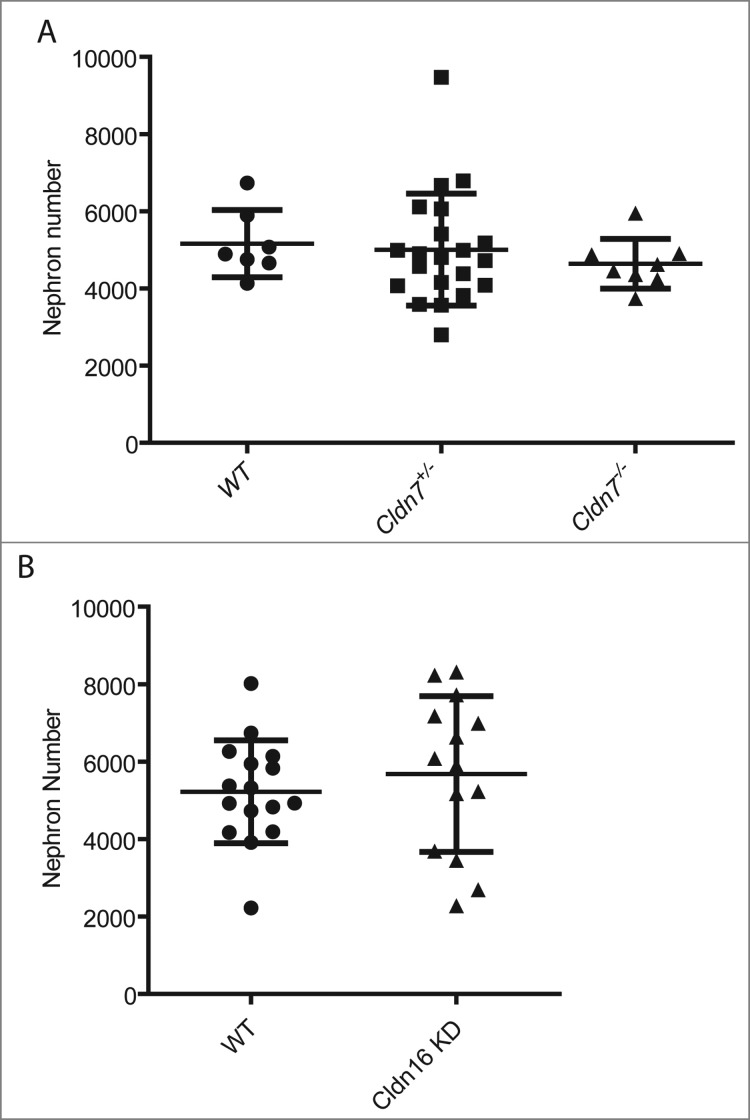
Based on their expression patterns during kidney development, we examined mutant mouse models lacking Claudin-7 and Claudin-16 to determine if these mice had defects in nephron formation. Crosses were established between Cldn7+/− breeders14 and kidneys from the newborn pup offspring were sectioned for histology and total nephron number assessment. Kidney sections from all of the pups including the homozygous and heterozygous mutants appeared grossly normal with no defects noted in the extent of ureteric bud branching morphogenesis or in the organization of the cortex and the medulla. There was no significant difference in nephron number noted in kidneys when comparing wildtype to heterozygous or to homozygous null pups (Fig. 7A, mean nephron number +/− SD; wt: 5160 ± 872 vs. heterozygous: 5099 ± 1312 vs. null: 4648 ± 656, n = 7 kidneys/group P > 0.05).
Figure 7.
Nephron Number in Mouse Models with Mutations in Claudin-7 and Claudin-16. P1 kidneys were paraffin-embedded and sectioned and stained with hematoxylin and eosin for nephron number counts. (A) Total nephron number for the right kidney for each pup was calculated from 6 litters and mean values +/− SD are shown. Kidneys from Cldn7 −/− pups do not have fewer nephrons (n = 11 kidneys; 4648 ± 656 nephrons/kidney) compared to kidneys from wildtype pups (n = 7; 5160 ± 872 nephrons/kidney, p = 0.174) or when compared to kidneys from heterozygous pups (n = 27 kidneys; 5099 ± 1312 nephrons/kidney, p = 0.287). (B) Total nephron number for both kidneys for each pup was calculated from 1 litter and mean values +/− SD are shown. Kidneys from Cldn-16 +/− knockdown pups do not have fewer nephrons (n = 14; 5682 ± 2011 nephrons/kidney) compared to kidneys from wildtype pups (n = 16 kidneys; 5220 ± 1328 nephrons/kidney, p = 0.458).
A similar analysis was performed on transgenic Cldn16+/− knockdown newborn pups that were derived using RNAi18 to determine if they exhibited any evidence of a renal malformation. Kidney sections from wildtype and transgenic Cldn16 knockdown pups appeared grossly normal. Assessment of nephron number revealed no differences when comparing wildtype and transgenic knockdown pups (Fig. 7B, mean nephron number +/− SD; wt: 5220 ± 1328 vs. knockdown: 5682 ± 2011, n = 8 kidneys/group P > 0.05).
Discussion
Our data demonstrate that 23 mouse claudin family members are expressed during embryonic kidney development and that Claudin-7, -16, and -19 have specific and distinct expression patterns. Despite Claudin-7's expression within the ureteric bud lineage and Claudin-16's expression within renal tubules, we noted that neither of the mouse models lacking these claudins showed evidence of renal maldevelopment nor did they demonstrate a deficit in nephron number. We therefore speculate that other claudin family members can compensate for a lack of Claudin-7 and Claudin-16 during kidney development.
During the emergence of the ureteric bud and the subsequent steps of branching morphogenesis, individual cells undergo cell shape changes to elicit the formation of buds and branch points, although the forces that lead to these changes are not well-characterized.26 The purse-string hypothesis suggests that constriction of actin filaments on one side of an epithelial cell will change its shape from columnar to triangular or wedge-shaped,27-28 and this has been proposed as a mechanism by which buds and branch points might form during ureteric bud branching morphogenesis.10 The claudin family of proteins are expressed within epithelial tight junction complexes and are sufficient for the formation of tight junction strands when ectopically expressed in fibroblasts.29 The ureteric bud and its derivatives have tight junctions based on electron microscopic images.11 We therefore speculate that claudin proteins that are expressed in the ureteric bud lineage, including CLDN3, CLDN4, CLDN7, CLDN8, and CLDN9,10,11 may influence cell shape change during ureteric bud branching morphogenesis via their C-terminal tails that interact with PDZ domain adaptor proteins, like ZO-1 and MUPP1, to link tight junctions to the actin cytoskeleton.3
Claudin-16 and Claudin-19 are expressed at later stages of renal development in tubular segments that arise when the metanephric mesenchyme undergoes mesenchymal-to-epithelial transition. Their expression patterns resemble that of Pou3f3 (Brn1)25 and Uromodulin24 such that taken together they define a molecular network that specifies the Loop of Henle. Claudin-19 has a broader expression pattern than Claudin-16 showing overlapping expression with both Calbindin-D-28K and Uromodulin, suggesting that in addition to its expression in the Loop of Henle, it is expressed within the distal convoluted tubule and the connecting tubule. A similar expression pattern for Claudin-19 has been shown by Lee et al. using a different antibody raised to amino acids 23–39.30
Our analysis of Claudin-7 and Claudin-16 deficient mouse models suggests that these 2 members are not essential for kidney development. These studies do not eliminate the possibility that they participate in the development and differentiation of the nephrons and the collecting duct network, however other claudin family members may be able to compensate for their absence. Remarkably, many of the single claudin loss-of-function mouse models exhibit very subtle physiological phenotypes that are more restricted than their expression domains.3 In fact, very few claudin mutations lead to abnormal renal physiology, in spite of the fact that most claudins are expressed within the adult nephron.13,31,32 This strongly suggests that claudins may function in combination with other claudins to exert their effects on paracellular transport. There is evidence that claudins can both heteromerize and homomerize within tight junctions to influence paracellular transport,33 but thus far a deeper understanding of how this influences cell and tissue function remains elusive.
So the question remains, why are so many claudins expressed in the embryonic kidney? While it is possible that their only function is to establish the paracellular barrier, we hypothesize that they are required for MET and branching morphogenesis during nephron differentiation and establishment of the collecting duct network. We propose that the absence of a renal developmental phenotype following removal of a single claudin is due to functional redundancy. Therefore, future studies will need to consider knockdown of multiple claudins in combination to begin to elucidate their roles during renal development.
Disclosure of Potential Conflicts of Interest
There were no conflicts of interest to disclose.
Funding
This work was supported by an operating grant from the Kidney Foundation of Canada (IRG and AKR). IRG holds a salary award from the Fonds de la Recherche en Santé du Québec. AKR and IRG are members of the Research Institute of the McGill University Health Center, which is supported in part by the FRSQ.
References
- 1.Krause G, Winkler L, Mueller SL, Haseloff R F, Piontek J, Blasig I E. Structure and function of claudins. Biochim Biophys Acta 2008; 1778:631; PMID:; http://dx.doi.org/ 10.1016/j.bbamem.2007.10.018 [DOI] [PubMed] [Google Scholar]
- 2.Lal-Nag M, Morin PJ. The claudins. Genome Biol 2009; 10:235; PMID:; http://dx.doi.org/ 10.1186/gb-2009-10-8-235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gupta IR, Ryan AK. Claudins: unlocking the code to tight junction function during embryogenesis and in disease. Clin Genet 2010; 77:314; PMID:; http://dx.doi.org/ 10.1111/j.1399-0004.2010.01397.x [DOI] [PubMed] [Google Scholar]
- 4.Costantini F. Genetic controls and cellular behaviors in branching morphogenesis of the renal collecting system. Wiley Interdiscip Rev Dev Biol 2012; 1:693; PMID:; http://dx.doi.org/ 10.1002/wdev.52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schmidt-Ott KM, Yang J, Chen X, Wang H, Paragas N, Mori K, Li J Y, Lu B, Costantini F, Schiffer M, et al. Novel regulators of kidney development from the tips of the ureteric bud. J Am Soc Nephrol 2005; 16:1993; PMID:; http://dx.doi.org/ 10.1681/ASN.2004121127 [DOI] [PubMed] [Google Scholar]
- 6.Schwab K, Patterson L, Aronow B, Luckas R, Liang H-C, Potter S. A catalogue of gene expression in the developing kidney. Kidney Int 2003; 64:1588; PMID:; http://dx.doi.org/ 10.1046/j.1523-1755.2003.00276.x [DOI] [PubMed] [Google Scholar]
- 7.Stuart RO, Bush KT, Nigam SK. Changes in gene expression patterns in the ureteric bud and metanephric mesenchyme in models of kidney development. Kidney Int 2003; 64:1997; PMID:; http://dx.doi.org/ 10.1046/j.1523-1755.2003.00383.x [DOI] [PubMed] [Google Scholar]
- 8.Davies JA, Little MH, Aronow B, Armstrong J, Brennan J, Lloyd-MacGilp S, Armit C, Harding S, Piu X, Roochun Y, et al. : Access and use of the GUDMAP database of genitourinary development. Methods Mol Biol 2012; 886:185; PMID:; http://dx.doi.org/ 10.1007/978-1-61779-851-1_17 [DOI] [PubMed] [Google Scholar]
- 9.Ohta H, Adachi H, Inaba M. Developmental changes in the expression of tight junction protein claudins in murine metanephroi and embryonic kidneys. J Vet Med Sci 2006; 68:149; PMID:; http://dx.doi.org/ 10.1292/jvms.68.149 [DOI] [PubMed] [Google Scholar]
- 10.Meyer TN, Schwesinger C, Bush KT, Stuart RO, Rose DW, Shah MM, Vaughn DA, Steer DL, Nigam SK. Spatiotemporal regulation of morphogenetic molecules during in vitro branching of the isolated ureteric bud: toward a model of branching through budding in the developing kidney. Dev Biol 2004; 275:44; PMID:; http://dx.doi.org/ 10.1016/j.ydbio.2004.07.022 [DOI] [PubMed] [Google Scholar]
- 11.Haddad N, El Andalousi J, Khairallah H, Yu M, Ryan AK, Gupta IR. The tight junction protein claudin-3 shows conserved expression in the nephric duct and ureteric bud and promotes tubulogenesis in vitro. Am J Physiol Renal Physiol 2011; 301:F1057; PMID:; http://dx.doi.org/ 10.1152/ajprenal.00497.2010 [DOI] [PubMed] [Google Scholar]
- 12.Siddiqui AS, Khattra J, Delaney AD, Zhao Y, Astell C, Asano J, Babakaiff R, Barber S, Beland J, Bohacec S, et al. A mouse atlas of gene expression: large-scale digital gene-expression profiles from precisely defined developing C57BL/6J mouse tissues and cells. Proc Natl Acad Sci U S A 2005; 102:18485; PMID:; http://dx.doi.org/ 10.1073/pnas.0509455102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li WY, Huey CL, Yu AS. Expression of claudin-7 and -8 along the mouse nephron. Am J Physiol Renal Physiol 2004; 286:F1063; PMID:; http://dx.doi.org/ 10.1152/ajprenal.00384.2003 [DOI] [PubMed] [Google Scholar]
- 14.Tatum R, Zhang Y, Salleng K, Lu Z, Lin JJ, Lu Q, Jeansonne BG, Ding L, Chen YH. Renal salt wasting and chronic dehydration in claudin-7-deficient mice. Am J Physiol Renal Physiol 2010; 298:F24; PMID:; http://dx.doi.org/ 10.1152/ajprenal.00450.2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Simon DB, Lu Y, Choate KA, Velazquez H, Al-Sabban E, Praga M, Casari G, Bettinelli A, Colussi G, Rodriguez-Soriano J, et al. Paracellin-1, a renal tight junction protein required for paracellular Mg2+ resorption. Science 1999; 285:103; PMID:; http://dx.doi.org/ 10.1126/science.285.5424.103 [DOI] [PubMed] [Google Scholar]
- 16.Haisch L, Konrad M. Impaired paracellular ion transport in the loop of Henle causes familial hypomagnesemia with hypercalciuria and nephrocalcinosis. Ann N Y Acad Sci 2012; 1258:177; PMID:; http://dx.doi.org/ 10.1111/j.1749-6632.2012.06544.x [DOI] [PubMed] [Google Scholar]
- 17.Konrad M, Schaller A, Seelow D, Pandey AV, Waldegger S, Lesslauer A, Vitzthum H, Suzuki Y, Luk JM, Becker C, et al. Mutations in the tight-junction gene claudin 19 (CLDN19) are associated with renal magnesium wasting, renal failure, and severe ocular involvement. Am J Hum Genet 2006; 79:949; PMID:; http://dx.doi.org/ 10.1086/508617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hou J, Shan Q, Wang T, Gomes A S, Yan Q, Paul D L, Bleich M, Goodenough DA. Transgenic RNAi depletion of claudin-16 and the renal handling of magnesium. J Biol Chem 2007; 282:17114; PMID:; http://dx.doi.org/ 10.1074/jbc.M700632200 [DOI] [PubMed] [Google Scholar]
- 19.Acloque H, Wilkinson DG, Nieto MA. In situ hybridization analysis of chick embryos in whole-mount and tissue sections. Methods Cell Biol 2008; 87:169; PMID:; http://dx.doi.org/ 10.1016/S0091-679X(08)00209-4 [DOI] [PubMed] [Google Scholar]
- 20.Gong Y, Renigunta V, Himmerkus N, Zhang J, Renigunta A, Bleich M, Hou J. Claudin-14 regulates renal Ca(+)(+) transport in response to CaSR signalling via a novel microRNA pathway. EMBO J 2012; 31:1999; PMID:; http://dx.doi.org/ 10.1038/emboj.2012.49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Murawski IJ, Maina RW, Gupta IR. The relationship between nephron number, kidney size and body weight in two inbred mouse strains. Organogenesis 2010; 6:189; PMID:; http://dx.doi.org/ 10.4161/org.6.3.12125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Thiagarajan RD, Georgas KM, Rumballe BA, Lesieur E, Chiu HS, Taylor D, Tang DT, Grimmond SM, Little MH. Identification of anchor genes during kidney development defines ontological relationships, molecular subcompartments and regulatory pathways. PLoS One 2011; 6:e17286; PMID:; http://dx.doi.org/ 10.1371/journal.pone.0017286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yu J, Valerius MT, Duah M, Staser K, Hansard JK, Guo JJ, McMahon J, Vaughan J, Faria D, Georgas K, et al. Identification of molecular compartments and genetic circuitry in the developing mammalian kidney. Development 2012; 139:1863; PMID:; http://dx.doi.org/ 10.1242/dev.074005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Georgas K, Rumballe B, Wilkinson L, Chiu H S, Lesieur E, Gilbert T, Little M H. Use of dual section mRNA in situ hybridisation/immunohistochemistry to clarify gene expression patterns during the early stages of nephron development in the embryo and in the mature nephron of the adult mouse kidney. Histochem Cell Biol 2008; 130:927; PMID:; http://dx.doi.org/ 10.1007/s00418-008-0454-3 [DOI] [PubMed] [Google Scholar]
- 25.Nakai S, Sugitani Y, Sato H, Ito S, Miura Y, Ogawa M, Nishi M, Jishage K, Minowa O, Noda T. Crucial roles of Brn1 in distal tubule formation and function in mouse kidney. Development 2003; 130:4751; PMID:; http://dx.doi.org/ 10.1242/dev.00666 [DOI] [PubMed] [Google Scholar]
- 26.Costantini F, Kopan R. Patterning a complex organ: branching morphogenesis and nephron segmentation in kidney development. Dev Cell 2010; 18:698; PMID:; http://dx.doi.org/ 10.1016/j.devcel.2010.04.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nagele RG, Bush KT, Kosciuk MC, Hunter ET, Steinberg AB, Lee HY. Intrinsic and extrinsic factors collaborate to generate driving forces for neural tube formation in the chick: a study using morphometry and computerized three-dimensional reconstruction. Brain Res Dev Brain Res 1989; 50:101; PMID:; http://dx.doi.org/ 10.1016/0165-3806(89)90129-6 [DOI] [PubMed] [Google Scholar]
- 28.Baker PC, Schroeder TE. Cytoplasmic filaments and morphogenetic movement in the amphibian neural tube. Dev Biol 1967; 15:432; PMID:; http://dx.doi.org/ 10.1016/0012-1606(67)90036-X [DOI] [PubMed] [Google Scholar]
- 29.Furuse M, Sasaki H, Fujimoto K, Tsukita S. A single gene product, claudin-1 or -2, reconstitutes tight junction strands and recruits occludin in fibroblasts. J Cell Biol 1998; 143:391; PMID:; http://dx.doi.org/ 10.1083/jcb.143.2.391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lee NP, Tong MK, Leung PP, Chan VW, Leung S, Tam PC, Chan KW, Lee KF, Yeung WS, Luk JM. Kidney claudin-19: localization in distal tubules and collecting ducts and dysregulation in polycystic renal disease. FEBS Lett 2006; 580:923; PMID:; http://dx.doi.org/ 10.1016/j.febslet.2006.01.019 [DOI] [PubMed] [Google Scholar]
- 31.Kiuchi-Saishin Y, Gotoh S, Furuse M, Takasuga A, Tano Y, Tsukita S. Differential expression patterns of claudins, tight junction membrane proteins, in mouse nephron segments. J Am Soc Nephrol 2002; 13:875; PMID: [DOI] [PubMed] [Google Scholar]
- 32.Reyes JL, Lamas M, Martin D, del Carmen Namorado M, Islas S, Luna J, Tauc M, Gonzalez-Mariscal L. The renal segmental distribution of claudins changes with development. Kidney Int 2002; 62:476; PMID:; http://dx.doi.org/ 10.1046/j.1523-1755.2002.00479.x [DOI] [PubMed] [Google Scholar]
- 33.Angelow S, Ahlstrom R, Yu AS. Biology of claudins. Am J Physiol Renal Physiol 2008; 295:F867; PMID:; http://dx.doi.org/ 10.1152/ajprenal.90264.2008 [DOI] [PMC free article] [PubMed] [Google Scholar]