Abstract
Cardiogenesis is one of the earliest and most important steps during human development and is orchestrated by discrete families of heart progenitors, which build distinct regions of the fetal heart. For the past decade, a lineage map for the distinct subsets of progenitors that generate the embryonic mammalian heart has begun to lay a foundation for the development of new strategies for rebuilding the adult heart after injury, an unmet clinical need for the vast majority of patients with end-stage heart failure who are not heart transplant recipients. The studies also have implications for the root causes of congenital heart disease, which affects 1 in 50 live births, the most prevalent malformations in children. Although much of this insight has been generated in murine models, it is becoming increasingly clear that there can be important divergence with principles and pathways for human cardiogenesis, as well as for regenerative pathways. The development of human stem cell models, coupled with recent advances in genome editing with RNA-guided endonucleases, offers a new approach for the primary study of human cardiogenesis. In addition, application of the technology to the in vivo setting in large animal models, including nonhuman primates, has opened the door to genome-edited large animal models of adult and congenital heart disease, as well as a detailed mechanistic dissection of the more diverse and complex set of progenitor families and pathways, which guide human cardiogenesis. Implications of this new technology for a new generation of human-based, genetically tractable systems are discussed, along with potential therapeutic applications.
Advances in human stem cell models and genome-editing technologies (e.g., the CRISPR-Cas9 system) are ushering in a new era of genetically engineered models of human cardiogenesis.
A DECADE OF ADVANCES IN MURINE CARDIOGENESIS
Over the past decade, major advances in our understanding of conserved principles and pathways for cardiogenesis have been made through genetically engineered mouse models of cardiogenesis. The assumption has been that the major underpinnings of such a conserved and essential organ system would have parallels in human organogenesis. The approach has been largely based on gene knockout approaches, as well as lineage tracing via the conditional irreversible tagging of specific putative heart progenitor cells with a Cre recombinase, capitalizing on markers identified in model systems. For example, a key role for canonical Wnt signaling has been established for mesodermal formation and subsequent heart development via a plasmid containing a multimerized TCF/LEF-binding site, driving expression of H2B-GFP, which was pronuclear injected into one-cell-stage embryo to generate a strain of mice that would report Wnt/β-catenin signaling activity (Ferrer-Vaquer et al. 2010). Analysis of this reporter mouse suggests that canonical Wnt signaling is important for heart development because of its strong expression in the primitive streak region, the precursor region of the heart.
To isolate the first heart field and second heart field (FHF and SHF, respectively) progenitors, a 3.97-kb enhancer fragment from Mef2c followed by dsRed DNA insert was introduced into the pronucleus to generate the SHF-dsRed mice (Domian et al. 2009). After that, timed matings were performed between SHF-dsRed males and Nkx2.5-eGFP females. This two-colored fluorescent reporter system can be used to isolate FHF and SHF progenitors from developing mouse embryos and embryonic stem cells (ESCs) (Domian et al. 2009). In addition, by precise knockin of a fluorescent reporter into a specific gene locus of interest, regulation of this gene can be studied in vivo. Similarly, by knockin of a Cre recombinase into a gene of interest, lineage tracing of this specific gene can be studied. For instance, to study the contribution of isl1-expression cells to the development of the murine heart, an isl1-cre mouse was crossed with a CAG-nlacZ indicator mouse (Cai et al. 2003). In progeny of this cross, Cre-mediated excision brings the nlacZ gene under the control of the ubiquitously expressed CAG-lacZ transgene, enabling the fate of isl1-expressing cells to be followed by staining for β-galactosidase activity, even when transcription from the endogenous isl1 locus has been repressed. This analysis reveals that isl1-expressing cells make a substantial contribution to the embryonic heart, comprising a majority of cells in the outflow tract, right ventricle, and atria, and also contribute to specific regions of the left ventricle.
To answer whether isl1+ cardiac progenitors exist in the murine postnatal heart, tamoxifen-inducible Cre was precisely inserted into isl1 locus. This inducible technology enables selective marking of isl1+ progenitor cell population, including its progeny, at a defined time, and purification to relative homogeneity. Endogenous isl1+ cardiac progenitors, when coculture with neonatal myocytes, display highly efficient conversion to a mature cardiac phenotype with stable expression of myocytic markers, intact Ca2+ cycling, and the generation of action potentials (Laugwitz et al. 2005). The genetic fate-mapping studies have also been used to document that isl1+ precursors from the SHF can generate each of the three diverse cardiovascular cell types in vivo: endothelial cells (ECs), smooth muscle cells, and cardiac muscle cells (Moretti et al. 2006). Using ESCs for in vitro studies, the isl1+/Nkx2.5+/flk1+ cardiovascular progenitors have been clonally isolated and further differentiated. These isl1+ cardiovascular progenitors can give rise to cells of all three lineages in the heart (Moretti et al. 2006). Although SHF isl1+ progenitors have been characterized in detail, comprehensive studies on the FHF have been hampered by the lack of exclusive markers. Lineage-traced Hcn4+ (hyperpolarization-activated cyclic nucleotide-gated channel 4) cells delineate FHF-derived structures in the heart and primarily contribute to cardiomyogenic cell lineages. In conclusion, a primary purpose of the FHF is to generate cardiac muscle and support the contractile activity of the primitive heart tube, whereas SHF-derived progenitors contribute to heart cell lineage diversification (Später et al. 2013).
Although these murine model systems have provided new insights into cardiogenesis, it is becoming increasingly clear that murine models may not have complete fidelity to the more complex, diverse, and scalable system of human cardiogenesis, which occurs over several months as opposed to a few days. In addition, it is well known that the physiology of the rodent cardiovascular system is not a faithful phenocopy of human cardiovascular and metabolic physiology and disease, for example, coronary artery disease and cholesterol metabolism. In these cases, larger animal model systems are needed, particularly nonhuman primates (NHPs), which represent one of the most important animal models for studying human disease with regard to developing therapeutic agents before FDA approval. Previous attempts to generate transgenic NHPs have relied on viral methods, which randomly integrated into the NHP genome in uncontrolled numbers (Sasaki et al. 2009; Niu et al. 2010). The low rates of a conventional homologous recombination strategy make such a method unfeasible in NHP genome editing because monkeys reproduce slowly and in limited numbers. Recent advances with new classes of engineered nucleases, zinc-finger nucleases (ZFNs), transcription activation-like effector nucleases (TALENs), and RNA-guided Cas9 nucleases, have been shown to be able to introduce double-strand breaks (DSBs) at specific sites in the genome. Without a template, these DSBs will be rejoined with errors following the nonhomologous joining mechanism, which basically disrupts gene expression and knocks out the gene of interest. The first cases of successful gene knockouts in rhesus and cynomolgus monkeys have been shown recently. TALEN plasmids targeting an X-linked, Rett syndrome gene, methyl-CpG-binding protein 2 (MECP2) were microinjected into rhesus and cynomolgus zygotes, which subsequently leads to MECP2 mutations in monkeys with no detected offtarget mutagenesis (Liu et al. 2014). Similarly, nuclease Cas9 messenger RNA (mRNA) and guide RNA can also be introduced into one-cell-stage embryos to produce precise gene knockout cynomolgus monkeys (Niu et al. 2014). This review will highlight these recent advances in NHP model systems, as well as the application of the new technology to human stem cell model systems. Implications for future directions in understanding human cardiogenesis, and the related translational importance to new directions in cardiovascular regenerative therapeutics and risk factors for congenital heart disease will also be discussed.
HUMAN PLURIPOTENT STEM CELLS AND CARDIOGENESIS
In 1998, James A. Thomson derived the first human embryonic stem cell (hESC) lines from normal blastocyst-stage embryos (Thomson 1998), which provide unique opportunities to study heart development through in vitro cardiovascular differentiation of these cells. Because these hESCs can be propagated indefinitely while still retaining the capacity to differentiate into all somatic cell types, they are also a potentially inexhaustible supply of human cells for cell therapy. One of the practical challenges for therapeutic application of these cells would be the risk of immune response from allogeneic recipients. In 2007, human-induced pluripotent stem cells (iPSCs) have been generated from individual somatic cells by overexpression of key stem cell–related transcription factors (Takahashi et al. 2007; Yu et al. 2007). Human iPSCs can be generated from patient-specific sources and, thus, can serve as models to study the role of specific genetic backgrounds and monogenic diseases on human cardiogenesis in vitro, with recent advances in driving pluripotent human stem cells into specific heart cell progenitor lineages and their downstream differentiated progeny. Accordingly, human pluripotent stem cells (hPSCs), including hESCs and iPSCs, can serve as important in vitro models for studying human development if we can establish efficient and robust cardiovascular differentiation protocols. In addition, disease modeling with hPSCs and rescuing of these disease-causing mutations would be also possible if robust genome editing in these cells would be feasible.
IN VITRO DIFFERENTIATION OF hPSCs INTO CARDIOMYOCYTES
The most successful in vitro differentiation protocols in ESC model systems have recapitulated the in vivo regulatory pathways that control the establishment of the corresponding lineage in the early embryo. Over the past decade, substantial advances have been made in generating cardiomyocytes from hPSCs by providing developmental cues, such as Activin A/Nodal, BMP, Wnt, and vascular endothelial growth factor (VEGF) signaling pathways, during differentiation. Cardiac differentiation of hPSCs is widely performed in two platforms: the formation of aggregate embryoid bodies (EBs) and culturing hPSCs as a monolayer.
Human cardiomyocytes were first isolated from differentiating EBs in media-containing fetal bovine serum. The purity of the resulting cardiomyocytes in the differentiated cell populations is ∼1%. Insulin-free media, addition of prostaglandin I2, and mitogen-associated protein (MAP) kinase inhibitor SB203580 have been shown to enhance EB cardiac differentiation (Xu et al. 2008). Control of EB size via physical microwell cultivation also promotes cardiac differentiation (Bauwens et al. 2008; Azarin et al. 2012). Proliferation of hPSC-derived cardiomyocytes is mediated via the growth factors insulin-like growth factor 1 (IGF1) and 2 (IGF2) through an IGF/PI3-kinase/Akt signaling pathway (McDevitt et al. 2005). In addition, hPSC-derived cardiomyocytes have appropriate functional responses to substrate stiffness and pharmaceutical agents (Hazeltine et al. 2012). Because of the low efficiency of the differentiation, as well as the use of serum and batch-to-batch variation, a new serum-free, growth-factor-dependent EB technique was developed. The addition of growth factors, including Activin A, BMP4, FGF-2, VEGF, and DKK-1, can enhance cardiomyocyte differentiation in EBs. However, this protocol is not universally applicable across different hPSC lines because it requires monitoring of KDR/c-kit (Yang et al. 2008) or KDR/PDGFRα (Kattman et al. 2011) expression to temporally apply growth factors at optimal concentrations to induce effective cardiac development.
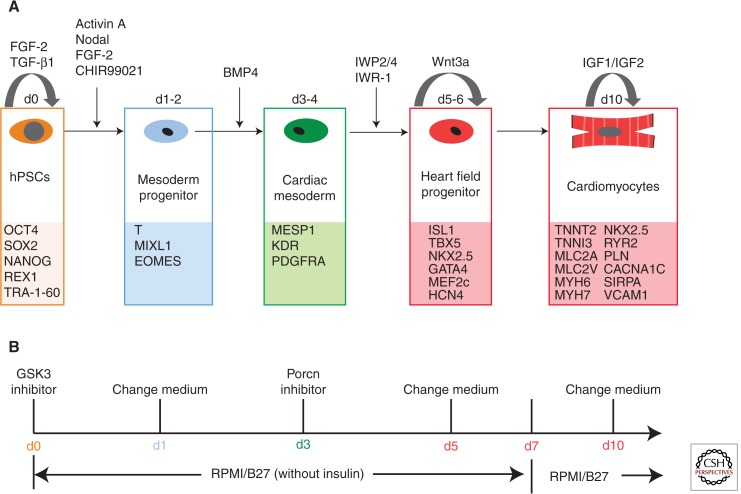
The EB method is time consuming and results in low cardiomyocyte yield and purity partially because of the EB size heterogeneity, which has led to the development of a monolayer-based method. hPSCs were first singularized and replated on Matrigel or defined matrix-coated Petri dishes as a monolayer. When confluent, sequential addition of Activin A and BMP4 to defined RPMI/B27 media is performed (Laflamme et al. 2007). This monolayer method has been reported to be more efficient than EB-based methods, generating >30% cardiomyocytes in the H7 hESC line (Laflamme et al. 2007). Supplement B27 contains a substantial amount of insulin, which greatly inhibits cardiomyocyte differentiation in many hPSC lines (Lian et al. 2013b). One of the improvements for this original protocol is the exclusion of insulin by using B27-minus insulin supplement. Another improvement for this protocol is the inclusion of Matrigel overlay (Zhang et al. 2012) to cells that are subsequently treated with Activin A, BMP4, and FGF-2 in RPMI/B27− insulin media for inducing robust cardiomyocyte differentiation in multiple hPSC lines, known as the matrix-sandwich method (Zhang et al. 2012). Five major cell types are involved in the hPSC cardiac differentiation process: pluripotent stem cells, mesoderm cells, cardiac mesoderm cells, cardiac progenitors, and cardiomyocytes. A summary of the hPSC cardiac differentiation process is listed in Figure 1A.
Figure 1.
Summary of the hPSC cardiac differentiation process.(A) Schematic approaches and growth factors/small molecules involved in hPSC cardiac differentiation. The five major cell types during hPSC cardiomyogenesis are shown: pluripotent stem cells, mesoderm progenitors, cardiac mesoderm cells, heart field progenitors, and cardiomyocytes. (B) Cardiac differentiation GiWi protocol is shown. hPSCs were first treated with a Gsk3 inhibitor to promote mesoderm differentiation and, then, on day 3 of differentiation, cells were treated with a Porcn inhibitor to block Wnt ligand function and, thus, inhibit Wnt signaling for enhancing cardiac differentiation.
These growth-factor-based protocols relied on optimization of the concentration of growth factors and their exposure time (when and for how long) to allow the efficient cardiac differentiation for different hPSC lines. Therefore, it is not suitable for large-scale production of cardiomyocytes because of the high cost of growth factors and optimization cost for individual lines. Instead, small molecules are generally much more stable and cheaper than growth factors. Recently, a robust protocol (Fig. 1B), known as the GiWi method, for generating cardiomyocytes from multiple hPSCs has been shown by exclusively using two small molecules (Lian et al. 2012, 2013a): a glycogen synthase kinase 3 (Gsk3) inhibitor and a Porcn inhibitor (Wnt ligands modification inhibitor). A Gsk3 inhibitor is first used to activate canonical Wnt signaling and promote mesoderm cell formation from hPSCs. At this early stage, knockdown of β-catenin by small hairpin RNA (shRNA) completely blocks cardiomycyte generation, indicating that β-catenin is essential for mesoderm differentiation. Three days after administration of the Gsk3 inhibitor, another inhibitor, IWP2, IWP4, or Wnt-C59 (three commonly used inhibitors for Porcn), was added to inhibit Porcupine, to block the functional modification of Wnt ligands, and thus prevent them from secretion. The inhibition of Wnt ligand processing enhances the cardiac differentiation from mesoderm cells. Consistent with the small molecule experiment, shRNA of β-catenin knockdown at this late stage greatly promotes cardiac muscle differentiation. Spontaneously, contracting cardiomyocytes can be routinely generated with the high purity (80%–95% positive cardiomyocytes) in 7–10 days in many hPSC lines without further individual optimization.
IN VITRO DIFFERENTIATION OF hPSCs INTO ECs
Similar to cardiomyocyte differentiation, two main approaches are used in the endothelial differentiation of hPSCs: differentiation of EBs in suspension and the two-dimensional monolayer differentiation.
The first hESC-derived ECs were isolated from 13-day-old EBs via fluorescence-activated cell sorting using EC-specific antibody CD31 in 2002 (Levenberg et al. 2002). These purified CD31+ cells are able to differentiate and form tube-like structures when cultured on Matrigel in vitro. In addition, when transplanted into severe combined immunodeficiency (SCID) mice, the cells appeared to form microvessels, containing mouse blood cells. However, the efficiency of endothelial differentiation in EBs is usually very low (typically <5% CD31+ cells). Another approach of differentiation involves culturing hESCs as a monolayer and differentiating them by coculturing with various feeder cell layers (Kaufman et al. 2001; Choi et al. 2009a; Hill et al. 2010). For instance, up to 20% of CD34+ cells are present by using coculture of hESCs with OP9 bone marrow stromal cells in 10 days (Vodyanik et al. 2005). Similar to hESCs, human iPSCs are also capable of differentiation into ECs with ∼15% efficiency in a differentiation pattern very similar to that of hESCs (Choi et al. 2009b).
More recently, a defined differentiation condition without feeder cells or serum was developed for efficient EC differentiation, which showed that administration of BMP4 and a GSK-3β inhibitor in an early phase, and treatment with VEGF-A and inhibition of the Notch signaling pathway in a later phase, led to efficient differentiation of hPSCs to the endothelial lineage within 6 days (Sahara et al. 2014). Defined growth factors, Activin A, BMP-4, stem cell factor (SCF), VEGF, fibroblast growth factor 2 (FGF-2) and -8 (FGF-8) were used to enhance EC differentiation (Wang et al. 2007; Costa et al. 2013; Orlova et al. 2014a,b). Besides growth factors and signaling pathway modulators, O2 levels during endothelial cell differentiation are also involved in lineage specification decisions and promote efficient differentiation of hESCs to functional ECs (Prado-Lopez et al. 2010). Functional assessment of hPSC-derived ECs has been shown in vitro and in vivo. To validate EC function in vitro, uptake of acetylated low-density lipoprotein (Ac-LDL), formation of EC networks in Matrigel assays, migration in trans-well assays, and production of nitric oxide (NO) have all been used to test the functionality of the resulting VE-cadherin+/CD31+ cells. In an in vivo assay, after transplantation into SCID mice, the purified hESC-derived ECs contributed to functional blood vessels that were incorporated into local tissue vascular networks of the animals for up to 150 days (Wang et al. 2007).
The signaling pathways controlling the differentiation of ECs and the maintenance of these cells might be different. Several studies have tried to identify specific developmental pathways that are sufficient to support the maintenance of functional ECs in vitro. By using an endothelial cell-specific vascular endothelial (VE)-cadherin promoter driving green fluorescent protein (GFP) to screen for factors that promoter vascular commitment, inhibition of transforming growth factor β (TGF-β) at late stages of differentiation increases GFP+ cells tenfold. Furthermore, TGF-β inhibition also maintains the proliferation and vascular identity of purified ECs. Expansion and maintenance of hESC-derived ECs by TGF-β inhibition is Id1 dependent (James et al. 2010).
The identification of precise paracrine signals that drive the cardiac-endothelial cell-fate decision of multipotent heart progenitors are critical steps toward unlocking their regenerative therapeutic potential. By comparing angiocrine factors expressed by the human outflow tract ECs and noncardiac ECs, VEGF-A was identified as the most abundantly expressed factor. In vitro assays documented that VEGF-A could drive endothelial specification of hPSC-derived Isl1+ progenitors in a VEGF-receptor-dependent manner. In addition, overexpression of VEGF-A via modified RNA transfection promotes not only the endothelial specification, but also engraftment, proliferation, and survival of the human Isl1+ progenitors in vivo (Lui et al. 2013).
In summary, all of these efforts provide a better understanding of the mechanics for differentiation and long-term maintenance of ECs from hPSCs and create a promising future for scalable clinical applications.
GENOME EDITING OF hPSCs
Although hPSCs can be used to delineate cell fate signals, this ultimately requires a faithful and stable genetic reporter line to track gene expression during long-term differentiation. In 2002, the first knockin hESC line was reported using conventional homologous recombinant technology for Oct4 gene (Zwaka and Thomson 2003). The cassettes containing internal ribosomal entry site (IRES) sequence, GFP, the same IRES, and the gene neo (encoding neomycin resistance) were inserted into the locus of Oct4 gene, which permits the isolation of pluripotent stem cells and monitors the beginning of the differentiation process. The efficiency of knockin using the conventional homologous recombinant technology in hESCs is low. After electroporation with the linearized targeting vector for Oct4, 103 drug-resistant clones were obtained, 28 of which (27%) were positive for homologous recombination. To improve the efficiency of knockin reporters in hESCs, a recombinant adeno-associated viral (rAAV)-based gene-targeting method was developed. To test the efficiency of this approach, GFP was inserted into the Sox2 locus in H9 hESCs. Among the drug-resistant clones, surprisingly, >70% of the clones were found to carry the GFP-neo cassette in the Sox2 locus (Brafman et al. 2013), showing the high efficiency of this knockin approach.
For genes related with heart development, GFP was knocked in to the early mesoderm gene Mixl1 and cardiac mesoderm gene Nkx2.5 in hESCs by the conventional homologous recombination approach. Heart muscle cells are derived from primitive streak cells, which were marked by expression of Mixl1 gene. The established Mixl1-GFP knockin hESC line will be very useful for studying both heart and blood development (Davis et al. 2008). To study the cardiac progenitor and cardiomyocyte differentiation, Nkx2.5-GFP knockin cell line (Elliott et al. 2011) was generated for monitoring the cardiac differentiation process. These GFP reporter cell lines provide a real-time analysis of the expression of gene of interest. Similar to the transgenic studies in mice, to study the fate mapping of the gene of interest, Bu et al. (2009) designed an approach for knockin a Cre recombinase into human Isl1 locus, and then randomly integrated a flox red fluorescent protein (RFP) reporter for irreversibly labeling the isl1 lineage cells. This is also the first knockin reporter in hPSC using the Cre-loxp system. This analysis showed that human isl1 progenitors can lead to cardiomyocyte, smooth muscle cell, and endothelial cell lineages (Bu et al. 2009).
GENOME-WIDE SCREENING VIA HAPLOID PLURIPOTENT STEM CELLS
These previous studies address the genotype and phenotype relationship by studying a single gene. However, recent advances in human stem cell biology and genome editing now present a strategy to identify the network of all genes related with certain phenotype through a genome-scale screening method. Traditionally, two approaches are applied for genetic screens aiming to determine gene function, forward, and reverse genetics screening.
The fact that mammals are diploid sets a barrier to rapidly understanding the function of noncoding and coding genes in the genome (Lian and Chien 2013). Conventional gene-trap techniques are high-throughput approaches, which are used to introduce insertional mutations across the mammalian genome. For example, the piggyBac transposon-based genome-wide library of insertionally mutated murine ES cells were reported (Wang et al. 2009). However, for recessive phenotypes, the possibility that gene-trapping randomly generates loss-of-function mutations in both alleles is extremely low, which makes this technique not feasible for recessive phenotype screening. The recent development of haploid embryonic stem cells (haESCs) from both mice and monkeys now makes it possible to set up genome-wide screening efforts to systematically identify entire networks of genes that drive specific differentiation events, and early steps of murine and primate organogenesis, in general, and cardiogenesis, in particular.
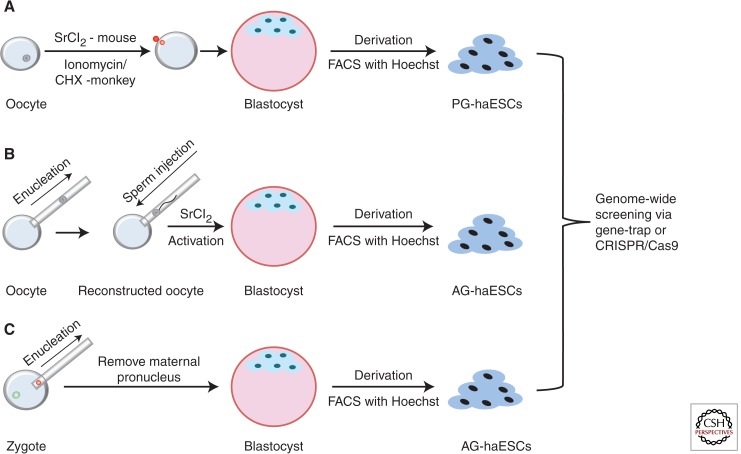
Two types of haESCs have been recently reported: parthenogenetic (PG) and androgenetic (AG) haESCs. For the generation of murine PG-haESCs, metaphase II oocytes were activated with strontium chloride (SrCl2) (Elling et al. 2011; Leeb and Wutz 2011) and further cultivated to the blastocyst stage. With the help of the fluorescence-activated cell-sorting (FACS) technique of Hoechst staining, PG-haESCs can be derived (Fig. 2A). One example for forward genetic screening using these cells is that the PG-haESCs can be used to assay for ricin toxicity (Elling et al. 2011). Ricin is a toxin being used as a bioweapon. Because ricin is highly toxic to mouse ESCs, a lethal dose of ricin was used to challenge the mutagenized haploid cell library cells. The growth of ricin-resistant ESC colonies from the mutagenized haploid cells, indicating that the genes mutated in these colonies are involved in ricin toxicity. These clones were then pooled and deep sequenced to determine the integration sites and mutant genes, resulting in the discovery of Gpr107 as a novel molecule essential for ricin-induced cell death. These results open the possibility of combining the power of a haploid genome with the pluripotency of ESCs to uncover fundamental biological endpoints, including cell fate signals, in defined cell types at a genomic scale.
Figure 2.
The scheme of PG and AG haESC derivation. (A) For the generation of PG-haESCs, metaphase II oocytes were activated with either SrCl2 for mice or ionomycin/cycloheximide for monkeys and further cultivated to the blastocyst stage. With the help of Hoechst FACS technique, PG-haESCs can be derived. (B) For the generation of AG-haESCs, metaphase II oocytes were enucleated followed by sperm injection. In addition, the reconstructed oocytes were activated with SrCl2 for mice and further developed to the blastocyst stage in vitro. AG-haESCs can be derived by several rounds of Hoechst FACS based on DNA content. The derivation of NHP AG-haESCs has not been reported yet. (C) For the generation of rat AG-haESCs, the maternal pronucleus in fertilized zygote was enucleated. The zygotes containing a male pronucleus were transferred into E0.5 pseudopregnant female rats, which were killed at E4.5 to collect the AG haploid morulas and blastocysts. AG-haESCs can be derived by several rounds of Hoechst FACS based on DNA contents.
In another screening approach, mouse PG-haESCs have been used for a forward genetic screening strategy designed to identify the network of genes responsible for dissolving the pluripotency program and enable fate transition. To produce a library of mutagenized PG-haESCs, a piggyBac transposon was used as a vehicle for delivering a gene-trap cassette. These cells were then cultured in highly permissive differentiation conditions for 2 weeks, and the clones associated with persistent self-renewal were manually picked and studied. Genetic exploration of the exit from self-renewal yielded multiple mutants in the Fgf/Erk and Gsk3/Tcf3 modules known to drive differentiation, as well as factors not previously identified, such as Zfp706 and Pum1 (Leeb et al. 2014).
For the generation of murine AG-haESCs, metaphase II oocytes were enucleated followed by sperm injection. In addition, the reconstructed oocytes were activated with SrCl2 and further developed to the blastocyst stage in vitro (Li et al. 2012; Yang et al. 2012). Murine AG-haESCs can be derived by several rounds of Hoechst FACS based on DNA contents (Fig. 2B). These AG-haESCs partially retain paternal imprints, express pluripotency markers, and contribute to various tissues, including the germline, on injection into diploid blastocysts. Remarkably, the AG-haESCs can be used like sperm cells to produce viable and fertile progenies after intracytoplasmic injection into mature oocytes. Viable transgenic mice can also be produced if genetically engineered AG-haESCs cells were used. These findings show the developmental pluripotency of AG haploid cell lines and provide a new tool to quickly produce genetic models for recessive traits. Furthermore, gene targeting via homologous recombination is feasible in the AG-haESCs, which permits the use of knockin AG-haESCs as potential fluorescent reporters for genome-wide screening studies.
This approach to generating haploid cell lines has also been extended to rat cell model systems. To generate rat AG-haESCs, the maternal pronucleus was removed from rat zygotes to generate AG embryos. The AG-haESC lines were then derived from these AG embryos via several rounds of FACS purification based on DNA content (Fig. 2C). Using this model, homozygous mutations can be introduced by both large-scale gene trapping and precise gene targeting. Similar to murine AG-haESCs, rat AG-haESCs also can produce fertile rats after intracytoplasmic injection into oocytes and are able to transmit genetic modifications to offspring.
Finally, these advances in the generation of haESCs lines has allowed the derivation of monkey PG-haESCs (Yang et al. 2013a), providing an ideal tool for genetic analyses in primates. Metaphase II monkey oocytes were first activated with ionomycin, briefly followed by cycloheximide treatment (Fig. 2A). These activated oocytes could develop into blastocysts in vitro, and monkey PG-haESCs can be derived by culturing the inner cell mass in a standard monkey ES cell culture system and using a FACS technique based on DNA content. Importantly, genetic screening is now clearly feasible in monkey PG-haESCs (Yang et al. 2013a). These results show that genome-wide screening in NHP is feasible by using monkey PG-haESCs.
Given these recent advances in haploid cell lines from mice, rats, and monkeys, the extension of these studies to human haESCs is of clear value. Unfortunately, the derivation of human PG- and AG-haESCs has not yet been reported, making this haploid pluripotent cell screening approach unfeasible in human systems.
GENOME-WIDE SCREENING VIA CRISPR-CAS9 FOR DIPLOID PLURIPOTENT STEM CELLS
CRISPR/Cas9 Technology Introduction
Over the past two decades, several genome-editing technologies have been developed, including conventional homology recombination (HR) (Gordon et al. 1980), ZFNs (Kim et al. 1996), TALENs (Hockemeyer et al. 2011), and the RNA-guided CRISPR-Cas nuclease system (Cong et al. 2013; Mali et al. 2013).
The clustered regularly interspaced short palindromic repeats (CRISPR) and CRISPR-associated (Cas) system was first discovered in bacteria and archaea (Bhaya et al. 2011; Terns and Terns 2011; Wiedenheft et al. 2012). CRISPRs are often associated with Cas genes that code for proteins related to CRISPRs. The CRISPR-Cas system is a prokaryotic immune system that confers resistance to foreign genetic elements, such as plasmids and phages, and provides a form of acquired immunity by incorporating fragments of invading phage and plasmid DNA into CRISPR loci and using the corresponding CRISPR RNAs (crRNAs) to guide the degradation of the homologous sequences (Jinek et al. 2012). There are three types of CRISPR-Cas systems (Makarova et al. 2011). The types I and III CRISPR systems assemble the Cas proteins complexed with crRNAs to mediate the recognition and subsequent degradation of target nucleic acids (Jinek et al. 2012). The type II system works in a different manner (Deltcheva et al. 2011; Jinek et al. 2012), consisting of three minimal components: a specificity-determining crRNA, an auxiliary trans-activating crRNA (tracrRNA), and the Cas nuclease Cas9. The crRNA encodes the protospacer sequence and hybridizes with the tracrRNA to form a crRNA:tracrRNA duplex. Nuclease Cas9 contains domains homologous to both HNH and RuvC endonucleases (Jinek et al. 2012; Nishimasu et al. 2014). Sequence recognition and, subsequently, cleavage by the crRNA-tracrRNA-Cas9 complex requires both a complementary sequence to the 20 nucleotide guide sequence in crRNAs, as well as the presence of an appropriate protospacer adjacent motif (PAM) sequence immediately downstream from the guide sequence. Different Cas9 proteins may have different PAM requirements (Mojica et al. 2009). The Cas9 protein from Streptococcus pyogenes (Sp), Neisseria meningitidis (NM), Streptococcus thermophilus (ST), and Treponema denticola (TD) requires a 5′-NGG-3′, 5′-NNNNGATT-3′, 5′-NNAGAA-3′, and 5′-NAAAAC-3′ PAM sequence, respectively.
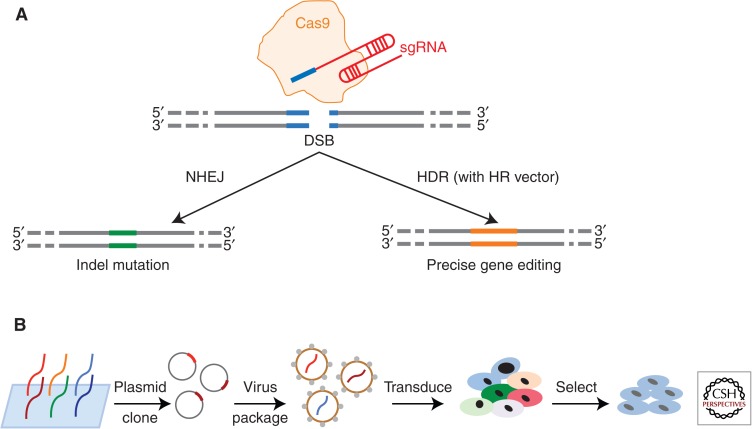
To further simplify the three-component type II CRISPR-Cas system, a single chimeric guide RNA (sgRNA) that mimics the natural crRNA-tracrRNA hybrid has been generated by fusing crRNA and tracrRNA (Jinek et al. 2012). In this two-component system, the SpCas9 protein, along with the sgRNA, could be programmed to cleave virtually any sequence preceding a 5′-NGG-3′ PAM sequence in mammalian cells (Fig. 3A). About 190,000 specific sgRNA target sequences, targeting ∼40.5% exons of genes in the human genome, have been be identified (Mali et al. 2013). The binding of sgRNA/Cas9 complex to the target genome sequence activates the HNH and RuvC domains in Cas9 and creates DSBs in the genomic DNA. A DSB can be repaired through one of two general repair pathways: the nonhomologous end joining (NHEJ) DNA repair pathway or the homology-directed repair (HDR) pathway. In the absence of a suitable repair template, the DSB is repaired by the NHEJ DNA repair pathway. During NHEJ repair, insertions or deletions usually occur as a small number of nucleotides are either randomly inserted or deleted at the DSB site, which causes an “indel” (insertion and/or deletion) mutation (Fig. 3A). These mutations may alter the open reading frame of the gene or introduce a premature stop codon. Any of these outcomes of the NHEJ repair pathway will disrupt the target gene, achieving the goal of disrupting the function of a gene (gene knockout).
Figure 3.
RNA-guided genome editing with engineered type II CRISPR/Cas9 system and its application in genome-scale screening assays. (A) DSBs induced by nuclease Cas9 (yellow) and its sgRNA (blue and red) can be repaired in one of two ways. In the NHEJ pathway, the ends of a DSB are processed by endogenous DNA repair machinery and rejoined, which can result in random indel mutations at the site of junction. Alternatively, a repair template can be supplied to leverage the HDR pathway, which allows precise editing. (B) Genome-wide functional screening can be facilitated by mass synthesis and delivery of guide RNA libraries in viral form. The resulting mutant cell libraries will be selected under certain desired pressures. Last, the final cell populations will be sequenced to identify the sgRNAs and corresponding genome elements.
The newly developed CRISPR-Cas9 nuclease system has several advantages. For instance, its efficiency is higher than HR, which makes it feasible to modify human cells; and it is also considerably easier to design guide RNA constructs than ZFNs and TALENs, making it suitable to perform genome-scale genes knockout screening. The gene knockout capabilities of the CRISPR-Cas9 system have been successfully shown in many model organisms, such as mice (Wang et al. 2013; Wu et al. 2013; Yang et al. 2013b), rats (Hu et al. 2013; Li et al. 2013; Ma et al. 2014), pigs (Hai et al. 2014), and even monkeys (Niu et al. 2014). The CRISPR-Cas9 system can be used to correct a genetic disease in mouse via HDR based on an exogenously supplied oligonucleotide or the endogenous Wilms’ tumor (WT) allele by coinjection into zygotes of Cas9 mRNA and an sgRNA targeting the mutant allele (Wu et al. 2013). Furthermore, delivery of components of the CRISPR-Cas9 system by hydrodynamic injection resulted in correction of a disease mutation and phenotype in the live cell in adult mice (Yin et al. 2014).
CRISPR-Cas9 Genome-Wide Screening Approach
The heart is a complex organ system composed of a diverse set of muscle and nonmuscle cells. A central problem in developmental biology is to identify the network of genes underlying the formation, migration, and assembly of these cells into the heart muscle tissue, the pacemaker and conduction system, and the coronary vasculature. Previously, genome-wide RNA interference (RNAi) loss-of-function screenings have provided a wealth of information in diverse model systems (Berns et al. 2004; Boutros et al. 2004; Carette et al. 2009; Jiang et al. 2009; Rad et al. 2010). Recent advances in CRISPR/Cas9 genome-editing technology and the cardiac differentiation method are now allowing genome-wide loss-of-function screenings in human cells.
CRISPR-Cas9 technology has now been implemented in large-scale genetic screening (Koike-Yusa et al. 2014; Shalem et al. 2014; Wang et al. 2014; Zhou et al. 2014), in a manner analogous to previous RNAi-based screenings. CRISPR-Cas9 screening can be performed in five steps: (1) Design sgRNAs libraries at genome scale by means of bioinformatics, (2) generate a pool of sgRNA-expressing lentivirus constructs by array synthesis and library cloning, (3) produce lentivirus particles and transfer these particles into the cells of interest and generate knockout cell libraries, (4) apply positive or negative selections to the mutant library, and (5) backtrack the mutations by deep-sequencing the sgRNAs integrated in the genome. The mutations that pass the selection will eventually enrich in the population after selection; thus, the mutation-causing sgRNAs will stand out as a hit in response to the screening (Fig. 3B).
There are several improvements of CRISPR-Cas9 over conventional RNAi-based screens. First, CRISPR-Cas9 inactivates genes at the DNA level, whereas RNAi acts at the mRNA level, resulting in an incomplete level of protein depletion. Second, CRISPR-Cas9 can allow the functional interrogation of nontranscribed elements, such as promoters, enhancers, silencers, as well as the intergenic regions, which are inaccessible by the means of RNAi. Third, mutations caused by the CRISPR-Cas9 system are permanently fixed in the genome, independent of the state of the cells, which make it possible to verify mutations at endpoint of a transition process, such as differentiation, whereas RNAi knockdown is only effective while the target gene is actively being transcribed. At the same time, there are also some additional issues that help CRISPR-Cas9 to reach its full potential, including off-target effects and varying knockout efficiencies of different genes. Nevertheless, taken together, CRISPR-Cas9-screening technology has provided a powerful tool for genome-scale functional genetic interrogation.
Future Application to Human Cardiogenesis
The convergence of hPSC technology, efficient differentiation protocols to generate specific human heart progenitor cell lineages, as well as their downstream differentiated progeny, and major advances in genome-editing technologies is ushering in a new era of genetically engineered models of human cardiogenesis. CRISPR-Cas9 knockout-screening technology together with the highly efficient and robust cardiac differentiation technology (Lian et al. 2012, 2013a) have now supported the feasibility of performing genome-wide genetic interrogation of the important genes for human cardiogenesis at the level of specific heart progenitor subsets. Differentiation of hPSCs into the cardiovascular lineages is a multistep process that involves initial epithelial to mesenchymal transition, mesoderm induction and specification, cardiac specification and differentiation, and functional maturation (Laflamme and Murry 2011), and it should be possible to identify the individual intermediate cells via recent advances in single-cell RNA sequencing (RNA-seq). With combinatorial cell surface or other markers, it should be possible to identify the genes that contribute to decisions at the branch points of key intermediate cell types in the human heart cell fate map through CRISPR-Cas9 screenings. Gene knockout NHP model systems, along with other large animal model systems, have already been generated and will ultimately allow direct in vivo interrogation of the physiological phenotype, which has the optimal fidelity of the complex human cardiovascular phenotypes of interest. In short, a new renaissance in large animal and human cardiovascular developmental biology and physiology appears to be on the horizon.
Footnotes
Editors: Margaret Buckingham, Christine L. Mummery, and Kenneth R. Chien
Additional Perspectives on The Biology of Heart Disease available at www.perspectivesinmedicine.org
REFERENCES
- Azarin SM, Lian X, Larson EA, Popelka HM, de Pablo JJ, Palecek SP 2012. Modulation of Wnt/β-catenin signaling in human embryonic stem cells using a 3-D microwell array. Biomaterials 33: 2041–2049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauwens CL, Peerani R, Niebruegge S, Woodhouse KA, Kumacheva E, Husain M, Zandstra PW 2008. Control of human embryonic stem cell colony and aggregate size heterogeneity influences differentiation trajectories. Stem Cells 26: 2300–2310. [DOI] [PubMed] [Google Scholar]
- Berns K, Hijmans EM, Mullenders J, Brummelkamp TR, Velds A, Heimerikx M, Kerkhoven RM, Madiredjo M, Nijkamp W, Weigelt B, et al. 2004. A large-scale RNAi screen in human cells identifies new components of the p53 pathway. Nature 428: 431–437. [DOI] [PubMed] [Google Scholar]
- Bhaya D, Davison M, Barrangou R 2011. CRISPR-Cas systems in bacteria and archaea: Versatile small RNAs for adaptive defense and regulation. Annu Rev Genet 45: 273–297. [DOI] [PubMed] [Google Scholar]
- Boutros M, Kiger AA, Armknecht S, Kerr K, Hild M, Koch B, Haas SA, Paro R, Perrimon N 2004. Genome-wide RNAi analysis of growth and viability in Drosophila cells. Science 303: 832–835. [DOI] [PubMed] [Google Scholar]
- Brafman DA, Moya N, Allen-Soltero S, Fellner T, Robinson M, McMillen ZL, Gaasterland T, Willert K 2013. Analysis of SOX2-expressing cell populations derived from human pluripotent stem cells. Stem Cell Rep 1: 464–478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bu L, Jiang X, Martin-Puig S, Caron L, Zhu S, Shao Y, Roberts DJ, Huang PL, Domian IJ, Chien KR 2009. Human ISL1 heart progenitors generate diverse multipotent cardiovascular cell lineages. Nature 460: 113–117. [DOI] [PubMed] [Google Scholar]
- Cai C-L, Liang X, Shi Y, Chu P-H, Pfaff SL, Chen J, Evans S 2003. Isl1 identifies a cardiac progenitor population that proliferates prior to differentiation and contributes a majority of cells to the heart. Dev Cell 5: 877–889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carette JE, Guimaraes CP, Varadarajan M, Park AS, Wuethrich I, Godarova A, Kotecki M, Cochran BH, Spooner E, Ploegh HL, et al. 2009. Haploid genetic screens in human cells identify host factors used by pathogens. Science 326: 1231–1235. [DOI] [PubMed] [Google Scholar]
- Choi K-D, Yu J, Smuga-Otto K, Salvagiotto G, Rehrauer W, Vodyanik M, Thomson J, Slukvin I 2009a. Hematopoietic and endothelial differentiation of human induced pluripotent stem cells. Stem Cells 27: 559–567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi K-D, Yu J, Smuga-Otto K, Salvagiotto G, Rehrauer W, Vodyanik M, Thomson J, Slukvin I 2009b. Hematopoietic and endothelial differentiation of human induced pluripotent stem cells. Stem Cells 27: 559–567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, Hsu PD, Wu X, Jiang W, Marraffini LA, et al. 2013. Multiplex genome engineering using CRISPR/Cas systems. Science 339: 819–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa M, Sourris K, Lim SM, Yu QC, Hirst CE, Parkington HC, Jokubaitis VJ, Dear AE, Liu HB, Micallef SJ, et al. 2013. Derivation of endothelial cells from human embryonic stem cells in fully defined medium enables identification of lysophosphatidic acid and platelet activating factor as regulators of eNOS localization. Stem Cell Res 10: 103–117. [DOI] [PubMed] [Google Scholar]
- Davis RP, Ng ES, Costa M, Mossman AK, Sourris K, Elefanty AG, Stanley EG 2008. Targeting a GFP reporter gene to the MIXL1 locus of human embryonic stem cells identifies human primitive streak-like cells and enables isolation of primitive hematopoietic precursors. Blood 111: 1876–1884. [DOI] [PubMed] [Google Scholar]
- Deltcheva E, Chylinski K, Sharma CM, Gonzales K, Chao Y, Pirzada ZA, Eckert MR, Vogel J, Charpentier E 2011. CRISPR RNA maturation by trans-encoded small RNA and host factor RNase III. Nature 471: 602–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domian IJ, Chiravuri M, van der Meer P, Feinberg AW, Shi X, Shao Y, Wu SM, Parker KK, Chien KR 2009. Generation of functional ventricular heart muscle from mouse ventricular progenitor cells. Science 326: 426–429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elling U, Taubenschmid J, Wirnsberger G, O’Malley R, Demers S-P, Vanhaelen Q, Shukalyuk AI, Schmauss G, Schramek D, Schnuetgen F, et al. 2011. Forward and reverse genetics through derivation of haploid mouse embryonic stem cells. Cell Stem Cell 9: 563–574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elliott DA, Braam SR, Koutsis K, Ng ES, Jenny R, Lagerqvist EL, Biben C, Hatzistavrou T, Hirst CE, Yu QC, et al. 2011. NKX2-5(eGFP/w) hESCs for isolation of human cardiac progenitors and cardiomyocytes. Nat Methods 8: 1037–1040. [DOI] [PubMed] [Google Scholar]
- Ferrer-Vaquer A, Piliszek A, Tian G, Aho RJ, Dufort D, Hadjantonakis A-K 2010. A sensitive and bright single-cell resolution live imaging reporter of Wnt/β-catenin signaling in the mouse. BMC Dev Biol 10: 121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon JW, Scangos GA, Plotkin DJ, Barbosa JA, Ruddle FH 1980. Genetic transformation of mouse embryos by microinjection of purified DNA. Proc Natl Acad Sci 77: 7380–7384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hai T, Teng F, Guo R, Li W, Zhou Q 2014. One-step generation of knockout pigs by zygote injection of CRISPR/Cas system. Cell Res 24: 372–375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hazeltine LB, Simmons CS, Salick MR, Lian X, Badur MG, Han W, Delgado SM, Wakatsuki T, Crone WC, Pruitt BL, et al. 2012. Effects of substrate mechanics on contractility of cardiomyocytes generated from human pluripotent stem cells. Int J Cell Biol 2012: 508294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill KL, Obrtlikova P, Alvarez DF, King JA, Keirstead SA, Allred JR, Kaufman DS 2010. Human embryonic stem cell-derived vascular progenitor cells capable of endothelial and smooth muscle cell function. Exp Hematol 38: 246–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hockemeyer D, Wang H, Kiani S, Lai CS, Gao Q, Cassady JP, Cost GJ, Zhang L, Santiago Y, Miller JC, et al. 2011. Genetic engineering of human pluripotent cells using TALE nucleases. Nat Biotechnol 29: 731–734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu X, Chang N, Wang X, Zhou F, Zhou X, Zhu X, Xiong J-W 2013. Heritable gene-targeting with gRNA/Cas9 in rats. Cell Res 23: 1322–1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- James D, Nam H, Seandel M, Nolan D, Janovitz T, Tomishima M, Studer L, Lee G, Lyden D, Benezra R, et al. 2010. Expansion and maintenance of human embryonic stem cell-derived endothelial cells by TGF-β inhibition is Id1 dependent. Nat Biotechnol 28: 161–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang D, Zhao L, Clapham DE 2009. Genome-wide RNAi screen identifies Letm1 as a mitochondrial Ca2+/H+ antiporter. Science 326: 144–147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E 2012. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 337: 816–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kattman SJ, Witty AD, Gagliardi M, Dubois NC, Niapour M, Hotta A, Ellis J, Keller G 2011. Stage-specific optimization of activin/nodal and BMP signaling promotes cardiac differentiation of mouse and human pluripotent stem cell lines. Cell Stem Cell 8: 228–240. [DOI] [PubMed] [Google Scholar]
- Kaufman DS, Hanson ET, Lewis RL, Auerbach R, Thomson JA 2001. Hematopoietic colony-forming cells derived from human embryonic stem cells. Proc Natl Acad Sci 98: 10716–10721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim YG, Cha J, Chandrasegaran S 1996. Hybrid restriction enzymes: Zinc finger fusions to Fok I cleavage domain. Proc Natl Acad Sci 93: 1156–1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koike-Yusa H, Li Y, Tan E-P, Velasco-Herrera MDC, Yusa K 2014. Genome-wide recessive genetic screening in mammalian cells with a lentiviral CRISPR-guide RNA library. Nat Biotechnol 32: 267–273. [DOI] [PubMed] [Google Scholar]
- Laflamme MA, Murry CE 2011. Heart regeneration. Nature 473: 326–335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laflamme MA, Chen KY, Naumova AV, Muskheli V, Fugate JA, Dupras SK, Reinecke H, Xu C, Hassanipour M, Police S, et al. 2007. Cardiomyocytes derived from human embryonic stem cells in pro-survival factors enhance function of infarcted rat hearts. Nat Biotechnol 25: 1015–1024. [DOI] [PubMed] [Google Scholar]
- Laugwitz K-L, Moretti A, Lam J, Gruber P, Chen Y, Woodard S, Lin L-Z, Cai C-L, Lu MM, Reth M, et al. 2005. Postnatal isl1+ cardioblasts enter fully differentiated cardiomyocyte lineages. Nature 433: 647–653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leeb M, Wutz A 2011. Derivation of haploid embryonic stem cells from mouse embryos. Nature 479: 131–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leeb M, Dietmann S, Paramor M, Niwa H, Smith A 2014. Genetic exploration of the exit from self-renewal using haploid embryonic stem cells. Cell Stem Cell 14: 385–393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levenberg S, Golub JS, Amit M, Itskovitz-Eldor J, Langer R 2002. Endothelial cells derived from human embryonic stem cells. Proc Natl Acad Sci 99: 4391–4396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Shuai L, Wan H, Dong M, Wang M, Sang L, Feng C, Luo G-Z, Li T, Li X, et al. 2012. Androgenetic haploid embryonic stem cells produce live transgenic mice. Nature 490: 407–411. [DOI] [PubMed] [Google Scholar]
- Li D, Qiu Z, Shao Y, Chen Y, Guan Y, Liu M, Li Y, Gao N, Wang L, Lu X, et al. 2013. Heritable gene targeting in the mouse and rat using a CRISPR-Cas system. Nat Biotechnol 31: 681–683. [DOI] [PubMed] [Google Scholar]
- Lian X, Chien KR 2013. The years of the monkey. Cell Res 23: 1161–1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lian X, Hsiao C, Wilson G, Zhu K, Hazeltine LB, Azarin SM, Raval KK, Zhang J, Kamp TJ, Palecek SP 2012. Robust cardiomyocyte differentiation from human pluripotent stem cells via temporal modulation of canonical Wnt signaling. Proc Natl Acad Sci 109: E1848–E1857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lian X, Zhang J, Azarin SM, Zhu K, Hazeltine LB, Bao X, Hsiao C, Kamp TJ, Palecek SP 2013a. Directed cardiomyocyte differentiation from human pluripotent stem cells by modulating Wnt/β-catenin signaling under fully defined conditions. Nat Protoc 8: 162–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lian X, Zhang J, Zhu K, Kamp TJ, Palecek SP 2013b. Insulin inhibits cardiac mesoderm, not mesendoderm, formation during cardiac differentiation of human pluripotent stem cells and modulation of canonical Wnt signaling can rescue this inhibition. Stem Cells 31: 447–457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Chen Y, Niu Y, Zhang K, Kang Y, Ge W, Liu X, Zhao E, Wang C, Lin S, et al. 2014. TALEN-mediated gene mutagenesis in rhesus and cynomolgus monkeys. Cell Stem Cell 14: 323–328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lui KO, Zangi L, Silva EA, Bu L, Sahara M, Li RA, Mooney DJ, Chien KR 2013. Driving vascular endothelial cell fate of human multipotent Isl1+ heart progenitors with VEGF modified mRNA. Cell Res 23: 1172–1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Y, Zhang X, Shen B, Lu Y, Chen W, Ma J, Bai L, Huang X, Zhang L 2014. Generating rats with conditional alleles using CRISPR/Cas9. Cell Res 24: 122–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarova KS, Haft DH, Barrangou R, Brouns SJJ, Charpentier E, Horvath P, Moineau S, Mojica FJM, Wolf YI, Yakunin AF, et al. 2011. Evolution and classification of the CRISPR-Cas systems. Nat Rev Microbiol 9: 467–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mali P, Yang L, Esvelt KM, Aach J, Guell M, DiCarlo JE, Norville JE, Church GM 2013. RNA-guided human genome engineering via Cas9. Science 339: 823–826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDevitt TC, Laflamme MA, Murry CE 2005. Proliferation of cardiomyocytes derived from human embryonic stem cells is mediated via the IGF/PI3-kinase/Akt signaling pathway. J Mol Cell Cardiol 39: 865–873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mojica FJM, Díez-Villaseñor C, García-Martínez J, Almendros C 2009. Short motif sequences determine the targets of the prokaryotic CRISPR defence system. Microbiology 155: 733–740. [DOI] [PubMed] [Google Scholar]
- Moretti A, Caron L, Nakano A, Lam JT, Bernshausen A, Chen Y, Qyang Y, Bu L, Sasaki M, Martin-Puig S, et al. 2006. Multipotent embryonic isl1+ progenitor cells lead to cardiac, smooth muscle, and endothelial cell diversification. Cell 127: 1151–1165. [DOI] [PubMed] [Google Scholar]
- Nishimasu H, Ran FA, Hsu PD, Konermann S, Shehata SI, Dohmae N, Ishitani R, Zhang F, Nureki O 2014. Crystal structure of Cas9 in complex with guide RNA and target DNA. Cell 156: 935–949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu Y, Yu Y, Bernat A, Yang S, He X, Guo X, Chen D, Chen Y, Ji S, Si W, et al. 2010. Transgenic rhesus monkeys produced by gene transfer into early-cleavage-stage embryos using a simian immunodeficiency virus-based vector. Proc Natl Acad Sci 107: 17663–17667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu Y, Shen B, Cui Y, Chen Y, Wang J, Wang L, Kang Y, Zhao X, Si W, Li W, et al. 2014. Generation of gene-modified cynomolgus monkey via Cas9/RNA-mediated gene targeting in one-cell embryos. Cell 156: 836–843. [DOI] [PubMed] [Google Scholar]
- Orlova VV, Drabsch Y, Freund C, Petrus-Reurer S, van den Hil FE, Muenthaisong S, Ten Dijke P, Mummery CL 2014a. Functionality of endothelial cells and pericytes from human pluripotent stem cells demonstrated in cultured vascular plexus and zebrafish xenografts. Arterioscler Thromb Vasc Biol 34: 177–186. [DOI] [PubMed] [Google Scholar]
- Orlova VV, van den Hil FE, Petrus-Reurer S, Drabsch Y, ten Dijke P, Mummery CL 2014b. Generation, expansion and functional analysis of endothelial cells and pericytes derived from human pluripotent stem cells. Nat Protoc 9: 1514–1531. [DOI] [PubMed] [Google Scholar]
- Prado-Lopez S, Conesa A, Armiñán A, Martínez-Losa M, Escobedo-Lucea C, Gandia C, Tarazona S, Melguizo D, Blesa D, Montaner D, et al. 2010. Hypoxia promotes efficient differentiation of human embryonic stem cells to functional endothelium. Stem Cells 28: 407–418. [DOI] [PubMed] [Google Scholar]
- Rad R, Rad L, Wang W, Cadinanos J, Vassiliou G, Rice S, Campos LS, Yusa K, Banerjee R, Li MA, et al. 2010. PiggyBac transposon mutagenesis: A tool for cancer gene discovery in mice. Science 330: 1104–1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahara M, Hansson EM, Wernet O, Lui KO, Später D, Chien KR 2014. Manipulation of a VEGF-Notch signaling circuit drives formation of functional vascular endothelial progenitors from human pluripotent stem cells. Cell Res 24: 820–841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki E, Suemizu H, Shimada A, Hanazawa K, Oiwa R, Kamioka M, Tomioka I, Sotomaru Y, Hirakawa R, Eto T, et al. 2009. Generation of transgenic non-human primates with germline transmission. Nature 459: 523–527. [DOI] [PubMed] [Google Scholar]
- Shalem O, Sanjana NE, Hartenian E, Shi X, Scott DA, Mikkelsen TS, Heckl D, Ebert BL, Root DE, Doench JG, et al. 2014. Genome-scale CRISPR-Cas9 knockout screening in human cells. Science 343: 84–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Später D, Abramczuk MK, Buac K, Zangi L, Stachel MW, Clarke J, Sahara M, Ludwig A, Chien KR 2013. A HCN4+ cardiomyogenic progenitor derived from the first heart field and human pluripotent stem cells. Nat Cell Biol 15: 1098–1106. [DOI] [PubMed] [Google Scholar]
- Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, Yamanaka S 2007. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell 131: 861–872. [DOI] [PubMed] [Google Scholar]
- Terns MP, Terns RM 2011. CRISPR-based adaptive immune systems. Curr Opin Microbiol 14: 321–327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomson JA 1998. Embryonic stem cell lines derived from human blastocysts. Science 282: 1145–1147. [DOI] [PubMed] [Google Scholar]
- Vodyanik MA, Bork JA, Thomson JA, Slukvin II 2005. Human embryonic stem cell-derived CD34+ cells: Efficient production in the coculture with OP9 stromal cells and analysis of lymphohematopoietic potential. Blood 105: 617–626. [DOI] [PubMed] [Google Scholar]
- Wang ZZ, Au P, Chen T, Shao Y, Daheron LM, Bai H, Arzigian M, Fukumura D, Jain RK, Scadden DT 2007. Endothelial cells derived from human embryonic stem cells form durable blood vessels in vivo. Nat Biotechnol 25: 317–318. [DOI] [PubMed] [Google Scholar]
- Wang W, Bradley A, Huang Y 2009. A piggyBac transposon-based genome-wide library of insertionally mutated Blm-deficient murine ES cells. Genome Res 19: 667–673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Yang H, Shivalila CS, Dawlaty MM, Cheng AW, Zhang F, Jaenisch R 2013. One-step generation of mice carrying mutations in multiple genes by CRISPR/Cas-mediated genome engineering. Cell 153: 910–918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T, Wei JJ, Sabatini DM, Lander ES 2014. Genetic screens in human cells using the CRISPR-Cas9 system. Science 343: 80–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiedenheft B, Sternberg SH, Doudna JA 2012. RNA-guided genetic silencing systems in bacteria and archaea. Nature 482: 331–338. [DOI] [PubMed] [Google Scholar]
- Wu Y, Liang D, Wang Y, Bai M, Tang W, Bao S, Yan Z, Li D, Li J 2013. Correction of a genetic disease in mouse via use of CRISPR-Cas9. Cell Stem Cell 13: 659–662. [DOI] [PubMed] [Google Scholar]
- Xu XQ, Graichen R, Soo SY, Balakrishnan T, Rahmat SNB, Sieh S, Tham SC, Freund C, Moore J, Mummery C, et al. 2008. Chemically defined medium supporting cardiomyocyte differentiation of human embryonic stem cells. Differentiation 76: 958–970. [DOI] [PubMed] [Google Scholar]
- Yang L, Soonpaa MH, Adler ED, Roepke TK, Kattman SJ, Kennedy M, Henckaerts E, Bonham K, Abbott GW, Linden RM, et al. 2008. Human cardiovascular progenitor cells develop from a KDR+ embryonic-stem-cell-derived population. Nature 453: 524–528. [DOI] [PubMed] [Google Scholar]
- Yang H, Shi L, Wang B-A, Liang D, Zhong C, Liu W, Nie Y, Liu J, Zhao J, Gao X, et al. 2012. Generation of genetically modified mice by oocyte injection of androgenetic haploid embryonic stem cells. Cell 149: 605–617. [DOI] [PubMed] [Google Scholar]
- Yang H, Liu Z, Ma Y, Zhong C, Yin Q, Zhou C, Shi L, Cai Y, Zhao H, Wang H, et al. 2013a. Generation of haploid embryonic stem cells from Macaca fascicularis monkey parthenotes. Cell Res 23: 1187–1200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H, Wang H, Shivalila CS, Cheng AW, Shi L, Jaenisch R 2013b. One-step generation of mice carrying reporter and conditional alleles by CRISPR/Cas-mediated genome engineering. Cell 154: 1370–1379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin H, Xue W, Chen S, Bogorad RL, Benedetti E, Grompe M, Koteliansky V, Sharp PA, Jacks T, Anderson DG 2014. Genome editing with Cas9 in adult mice corrects a disease mutation and phenotype. Nat Biotechnol 32: 551–553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J, Vodyanik MA, Smuga-Otto K, Antosiewicz-Bourget J, Frane JL, Tian S, Nie J, Jonsdottir GA, Ruotti V, Stewart R, et al. 2007. Induced pluripotent stem cell lines derived from human somatic cells. Science 318: 1917–1920. [DOI] [PubMed] [Google Scholar]
- Zhang J, Klos M, Wilson GF, Herman AM, Lian X, Raval KK, Barron MR, Hou L, Soerens AG, Yu J, et al. 2012. Extracellular matrix promotes highly efficient cardiac differentiation of human pluripotent stem cells: The matrix sandwich method. Circ Res 111: 1125–1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, Zhu S, Cai C, Yuan P, Li C, Huang Y, Wei W 2014. High-throughput screening of a CRISPR/Cas9 library for functional genomics in human cells. Nature 509: 487–491. [DOI] [PubMed] [Google Scholar]
- Zwaka TP, Thomson JA 2003. Homologous recombination in human embryonic stem cells. Nat Biotechnol 21: 319–321. [DOI] [PubMed] [Google Scholar]