Abstract
In cancer, epigenetic proteins are intensely studied targets for therapeutic drug discovery, showing great promise. These proteins include the chromatin-modifying enzymes that “write” and “erase” histone posttranslational modifications (PTM), and those that “read” these marks through binding modules. In an effort to find a compound that could disrupt the protein–protein interactions between a PTM and reader, JQ1 has proven to be a first-in-class, drug-like inhibitor of the “bromodomain and extraterminal domain” epigenetic readers (BETs), which recognize histone lysine acetylation marks. JQ1 has facilitated the mechanistic study and therapeutic application in cancer of this kind of epigenetic inhibition. By using this chemical probe, we have discovered that the bromodomain inhibitors (BETi) have compelling activity in preclinical models of multiple myeloma and acute myeloid leukemia. In particular, BETi down-regulates the MYC, IL-7R, and E2F transcriptional programs. We are continuously integrating the transcriptional consequences of BETi with changes in the epigenomic landscapes of cancer cells to elucidate the mechanisms underlying response to BETi using chemical and genetic perturbations.
BET proteins recognize histone lysine acetylation marks and influence gene expression. The inhibition of BET proteins (e.g., using JQ1) down-regulates transcriptional programs that are typically oncogenic (e.g., MYC).
In cancer, epigenetic proteins are intensely studied targets for drug discovery owing to the general view that it is not just the DNA sequence that is altered in epigenetics-based diseases. Studies to date have indeed shown therapeutic promise; inhibitors of DNA methyltransferases (DNMTs) and histone deacetylases (HDACs), for example, have shown substantial clinical efficacy leading to regulatory approval for use in hematologic malignancies. These successes have stimulated broad-based efforts to develop other inhibitors of chromatin-modifying enzymes, so-called epigenetic “writers” and “erasers.” Perhaps owing to the perception that it is difficult to interfere with protein–protein interactions, chromatin-binding modules or epigenetic “readers” have received comparatively little attention. Motivated by this challenge, we developed inhibitors of the BET (for bromodomain and extraterminal domain) epigenetic readers, using a tried and tested thienodiazepine core, which forms the core of a number of pharmaceutical drugs. Compounds identified showing potential were used as chemical probes to study the mechanistic details and its therapeutic application.
Readers are often regarded as functional “effector” proteins that can recognize particular posttranslational modifications (e.g., methylation or acetylation marks) on histone proteins or DNA, placed by “writers” as part of signaling-based transcriptional pathways (Fig. 1). Historically the bromodomain stands as the first well-characterized epigenetic reader, which recognizes the acetylated lysine side chain on histones. The binding affinity between a bromodomain protein reader and the histone tail is low—in the µm range. We designed and synthesized a thienodiazepine-based small molecule called JQ1, which shows excellent inhibition against the BET subfamily with low nanomolar binding potency, especially targeting the BET protein, BRD4. Typically, drug design seeks to achieve a high binding affinity, in the range of 0.1–10 nm with its target. The compound was selected by combining small-molecule library screening and structural information provided by crystallography. The cocrystal structure of JQ1 and the first bromodomain of BRD4 showed excellent structure complementarity between the small molecule and protein, explaining the high binding affinity. JQ1 also possessed good cell permeability, which allowed us to study the extent and effect of bromodomain inhibition in a BRD4-dependent cancer: the NUT-midline carcinoma, which arises from a BRD4-NUT translocation. The effect of the compound was to induce differentiation in patient-derived cancer cell lines in less than 24 hours after treatment. It also showed excellent efficacy in a murine xenograft model without obvious toxicity. Given the reasonable pharmacokinetic properties JQ1 was even entitled to be used in a human patient–derived xenograft model (Filippakopoulos et al. 2010). This prototype drug was further optimized to a preclinic candidate for BRD4-dependent cancers (M Mckewon, K Shaw, and J Qi, in prep.).
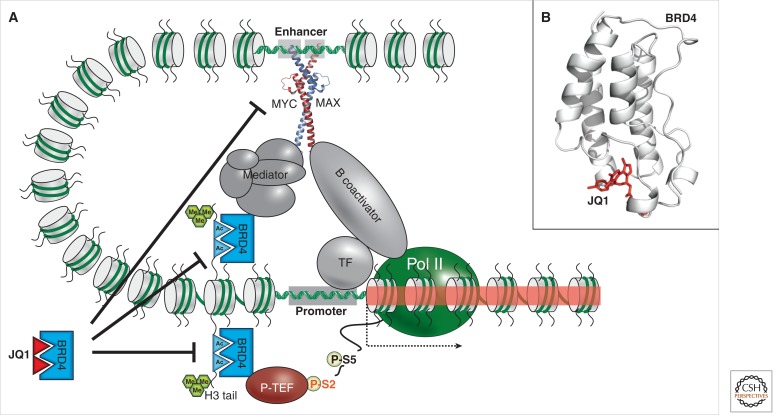
Figure 1.
Model for JQ1 small-molecule inhibition of the BRD4 bromodomain in cancer. (A) The aberrant transcriptional activation of MYC target genes (indicated as red shaded regions of chromatin) is a common feature of many cancers. Transcriptional activation requires the bromodomain “reading” function of BRD4, which recognizes acetylation marks (“Ac”-labeled cyan triangles) on histone H3 tails at promoter-proximal target sequences. Acetyl-bound BRD4 interacts with both the MYC-MAX complex bound to enhancer sequences (via a mediator complex) and the PTEFb phosphorylase required for the release of RNA polymerase II (Pol II) during transcriptional elongation. The competitive binding of the JQ1 bromodomain inhibitor (red triangle) to BRD4 not only reduces transcription of the MYC gene (top inhibition arrow) but also its target genes by abrogating recruitment of enhancer complexes and PTEFb (middle and bottom inhibition arrows), possibly via chromatin looping. The active chromatin mark histone H3K4me3 is illustrated as triple green hexagons. (B) Crystal structure of the human BRD4 protein in complex with JQ1 (red).
With a chemical probe in hand, the role of bromodomain epigenetic readers could be examined in transcriptional regulation. More recent studies showed that BET inhibitors (BETi) have compelling activity in preclinical models of multiple myeloma (Delmore et al. 2011) and acute myeloid leukemia (Zuber et al. 2011; Ott et al. 2012). Specifically, BETi down-regulates typically oncogenic MYC, IL7R, and E2F transcriptional programs. These observations have shown that BET bromodomain inhibition provides an efficient pathway to strategically target certain malignancies and other diseases that can be characterized by the pathologic activation of c-Myc.
The inhibitory effects of JQ1 on the testis-specific BRD member, BRDT, which is essential for chromatin remodeling during spermatogenesis, leads to a complete, but reversible contraceptive effect. As with BRD4, JQ1 occupies the BRDT acetyllysine binding pocket, preventing recognition of acetylated histone H4. In mice, this reduces the seminiferous tubule area, testis size, and spermatozoa number and motility without affecting hormone levels. JQ1-treated males show the inhibitory effects of JQ1 at the spermatocyte and round spermatid stages, in particular. These data establish a rationale for the development of a new contraceptive that can cross the blood–testis boundary and inhibit bromodomain activity during spermatogenesis, providing a lead compound targeting the male germ cell for contraception (Matzuk et al. 2012).
The discovery of the small-molecule inhibitor of bromodomains, JQ1, has shed light on the mechanisms underlying the response to BETi at a transcriptional level in its epigenomic context. Studies involving JQ1 have provided a clear “proof of principle” that other reader domains, both within and beyond the BET subfamily of bromodomains, may be attractive drug targets in years to come as more structural insights are learned from each class of chromatin-associated readers.
Footnotes
Editors: C. David Allis, Marie-Laure Caparros, Thomas Jenuwein, and Danny Reinberg
Additional Perspectives on Epigenetics available at www.cshperspectives.org
REFERENCES
- Delmore JE, Issa GC, Lemieux ME, Rahl PB, Shi JW, Jacobs HM, Kastritis E, Gilpatrick T, Paranal RM, Qi J, et al. 2011. BET bromodomain inhibition as a therapeutic strategy to target c-Myc. Cell 146: 904–917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filippakopoulos P, Qi J, Picaud S, Shen Y, Smith WB, Fedorov O, Morse EM, Keates T, Hickman TT, Felletar I, et al. 2010. Selective inhibition of BET bromodomains. Nature 468: 1067–1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matzuk MM, McKeown MR, Filippakopoulos P, Li Q, Ma L, Agno JE, Lemieux ME, Picaud S, Yu RN, Qi J, et al. 2012. Small-molecule inhibition of BRDT for male contraception. Cell 150: 673–684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ott CJ, Kopp N, Bird L, Paranal RM, Qi J, Bowman T, Rodig SJ, Kung AL, Bradner JE, Weinstock DM 2012. BET bromodomain inhibition targets both c-Myc and IL7R in high-risk acute lymphoblastic leukemia. Blood 120: 2843–2852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuber J, Shi J, Wang E, Rappaport AR, Herrmann H, Sison EA, Magoon D, Qi J, Blatt K, Wunderlich M, et al. 2011. RNAi screen identifies BRD4 as a therapeutic target in acute myeloid leukaemia. Nature 478: 524–528. [DOI] [PMC free article] [PubMed] [Google Scholar]