Abstract
Currently, the best scenario for earliest forms of life is based on RNA molecules as they have the proven ability to catalyze enzymatic reactions and harbor genetic information. Evolutionary principles valid today become apparent in such models already. Furthermore, many features of eukaryotic genome architecture might have their origins in an RNA or RNA/protein (RNP) world, including the onset of a further transition, when DNA replaced RNA as the genetic bookkeeper of the cell. Chromosome maintenance, splicing, and regulatory function via RNA may be deeply rooted in the RNA/RNP worlds. Mostly in eukaryotes, conversion from RNA to DNA is still ongoing, which greatly impacts the plasticity of extant genomes. Raw material for novel genes encoding protein or RNA, or parts of genes including regulatory elements that selection can act on, continues to enter the evolutionary lottery.
Many features of eukaryotic genomes (e.g., the modular rearrangement of genes) may have their origins in an RNA or RNA/protein world.
Everything has been said already, but not yet by everyone.
—Karl Valentin
Sturgeon's Revelation: Ninety percent of science fiction is crud, but then, ninety percent of everything is crud.
—Theodore Sturgeon
They think that intelligence is about noticing things that are relevant (detecting patterns); in a complex world, intelligence consists in ignoring things that are irrelevant (avoiding false patterns).
—Nassim Nicholas Taleb (Taleb 2010)
Of all extant cellular macromolecules, RNA is the most ancient, persisting as much as 4 × 109 years in our planet’s life-forms. The ability to combine genotype with phenotype such as catalytic activity (Noller and Chaires 1972; Kruger et al. 1982; Guerrier-Takada et al. 1983; Noller et al. 1992) leveled a major hurdle in understanding the origin of life. The salient discoveries eliminated the virtually impossible prerequisite for two to three different classes of macromolecules to converge as an evolving unit. At the same time, RNA provides a required continuity in the path of evolution (Yarus 2011) during various genetic takeovers or evolutionary transitions (Cairns-Smith 1982; Szathmáry and Smith 1995). In a remarkably insightful article dating back half a century, Alex Rich foresaw much of what now is becoming mainstream, for example, that RNA was ancestral to protein and DNA (Rich 1962). This landmark publication received little attention over the years; even early proponents of an RNA world did not refer to this article (Woese 1967; Crick 1968; Orgel 1968; Gilbert 1986), although at least one of the investigators must have had knowledge about the article, as it was cited in a different context concerning the stereochemical possibility of six distinct base pairs (Crick 1968). The origin of the DNA genome from RNA and that “DNA may be regarded as a derivative molecule which has evolved in the form that it only carries out part of the primitive nucleic acid function” is another correct prediction (Rich 1962). Furthermore, the investigator presaged mechanisms such as antisense RNA control of gene expression, short interfering RNAs (siRNAs), and perhaps microRNAs (miRNAs): “If both strands are active, then the DNA would produce two RNA strands which are complementary to each other. Only one of these might be active in protein synthesis, and the other strand might be a component of the control or regulatory signal” (Rich 1962).
In this article, I shall present the rise and persistence of RNA from the dawn of an RNA world and discuss current evolutionary principles already apparent in an RNA world. In comparison to Archaea and Bacteria, the eukaryotic genome is a better vantage point, as archaeal and bacterial genomes are more derived and, thus, lost many of the RNA signatures that eukaryotes still show. It is likely that eukaryotic DNA genomes not only kept much more of their RNA/RNP world heritage than previously anticipated, but also continue to evolve novel RNAs in various functional roles.
WHEN DOES LIFE BEGIN?
An excellent treatise of possible scenarios leading to and continuing in an RNA world to the last universal common ancestor (LUCA) is available (Atkins et al. 2011). Can the beginning of life be defined along the transitions from physicochemical to biological reactions? Like almost everything in biology, clear boundaries are difficult to demarcate and thus the definition of the first life-form rather occupies a bandwidth on a continuum. One of several possible thresholds to consider would be the fortuitous generation of one or two molecules that could replicate themselves or each other. Should the threshold be set at the transition when the molecules involved could change during replication and the variants are subjected to selection—the initial Darwinian ancestor (IDA) (Szathmáry 2006; Yarus 2011)? The first self-replicating macromolecules must not necessarily have been RNA. Derivatives of RNA, especially with altered backbones, have been suggested as predecessors of RNA owing to more favorable chemistries/stabilities for spontaneous generation and persistence of oligomerization at the presumed planetary conditions (Joyce et al. 1987; Schöning et al. 2000; Zhang et al. 2005; Powner et al. 2009; Robertson and Joyce 2012; Neveu et al. 2013).
An interesting question is, if it is that “simple,” why did life not evolve multiple times? There are a number of explanations. The early environment of the planet with conditions favoring the necessary chemical reactions differed from the more temperate conditions now (Robertson and Joyce 2012). Perhaps life did evolve before LUCA numerous times independently, but the descendants of LUCA are the only survivors. Perhaps primitive forms of life, for example, in the form of IDAs, still do arise, but we are not aware of them, in part, because we have not searched for such simple and different life-forms. Another reason is that a nascent form of life would easily be outcompeted by the established ones, as the latter had a great chronological advantage adapting to current conditions. New forms of life might have a chance only if their metabolism is sufficiently different and, thus, not useful prey to LUCA-related life-forms or if they happened to evolve in an unoccupied niche so as not to succumb to immediate predation by the fitter “incumbents.”1
RETRACING THE PATH
In any event, by applying in vitro synthesis and selection procedures (Ellington and Szostak 1990; Tuerk and Gold 1990; Gold et al. 2012), several laboratories are making great strides toward generating RNA molecules with the ability to self-replicate (Doudna and Szostak 1989; Johnston et al. 2001; Zaher and Unrau 2007; Lincoln and Joyce 2009; Shechner et al. 2009; Wochner et al. 2011; Attwater et al. 2013; Mast et al. 2013), although these RNA polymerase ribozymes still fail to completely self-replicate (Deamer 2005). Cooperation of two or more RNA enzymes in hypercycles (Eigen and Schuster 1977) may be a solution to this problem (Vaidya et al. 2012) (see also below).
Once a self-replicating ribozyme (mono- or multimeric) arose with the further potential to evolve into a replicator not restricted to only self-copy but to copy other RNA templates as well, a prerequisite for a metabolically self-sufficient RNA conglomerate, further challenges are apparent. First, the replicator indiscriminately copying any “junk RNA” in the mix hardly would be able to persist. Second, if further RNA molecules would arise by copying with errors—just like in extant organisms, new genes still arise by duplication and variation—to eventually take over metabolic functions other than replication (e.g., activated compounds, including nucleotides), these associations would be fleeting, at best, because of diffusion.
SEQUESTRA, AMPLIFICA, DIVIDE ET IMPERA!
A compartmentalization of cellular constituents in droplets, as suggested by Carl Woese (1979), stabilized by simple fatty acids (Szostak et al. 2001) was an early evolutionary transition (Maynard Smith and Szathmáry 1995) crucial for the continuation of life. Recent experiments showed that such bilayered partitions were sufficiently permeable for uptake of small molecules from the environment and copying nucleic acids in such vesicles is possible (Sacerdote and Szostak 2005; Mansy et al. 2008; Adamala and Szostak 2013). This sequestration was a prerequisite for cellular life and evolution as we know it today, with many evolutionary principles in place already (Brosius 2003c). Vesicles could grow along with their RNA contents, divide, fuse while shuffling their contents and divide again, in other words, performing sexual acts. Another “forecast” of the mechanisms generating genomic diversity would be recombination not only between cells, but also between different RNA molecules, a mechanism that actually had been observed in a two-component ribozyme system (Lincoln and Joyce 2009). The origin of viruses could date to this early stage of cellular evolution as well. RNA molecules could, perhaps protected by a lipid envelope, move from cell to cell, blurring the line between infection and horizontal transfer.
MOST EVOLUTIONARY PRINCIPLES ARE AT LEAST AS ANCIENT AS THE RNA WORLD
Lessons from the RNA world apply remarkably well to extant organisms and their genomes. In a primitive RNA cell, conflict and cooperation, selfishness and altruism had to coexist and establish a fine balance. Importantly, the success of individual ribozymes also depended to a large degree on functional interactions with other cellular RNAs (today: gene products), namely, the (genetic) background of the protocell (Brosius 2003c). In contemporary biology and medicine, considerations of interactions of various alleles within the genomic background, as well as individual variability of gene expression levels including complete gene depletions, are beginning to gain wider acceptance (Williams and Nesse 1991; Sibilia and Wagner 1995; Brosius 2003b; Ganten and Nesse 2012; Nesse et al. 2012).
Balances between selfishness and cooperation had to evolve early; this is recently documented by in vitro selection research that revealed in an experiment to generate Tetrahymena group I ribozymes with improved DNA cleavage capability that one of the two RNA aptamers, itself being catalytically inactive, participated in a productive intermolecular interaction with an active ribozyme evolving in parallel, thus, ensuring the survival of both RNAs in the nucleic acid population (Hanczyc and Dorit 1998). Other studies also show that parasites can cause the evolution of further complexity (Takeuchi and Hogeweg 2008). In the case of the bacterially derived AzoΔ ribozyme, a clonal preparation showed no activity toward the phosphorothioate substrate. Presumably, this sequence alone fails to show a functional fold, but could form an active complex in an intermolecular partnership with other RNA molecules (Hayden et al. 2011). Recently, it was shown that cooperative cycles of replication involving three or four participating RNAs have a selective advantage over selfish replication cycles (Vaidya et al. 2012), as formulated earlier in the hypercycle principle of natural self-organization (Eigen and Schuster 1978) and placed into context with extant organisms (Brosius 2003c). The participation of several small RNAs also has the potential to significantly increase the complexity of ribozymes.
CHROMOSOMAL RNA
The dispensation of essential RNA molecules between daughter cells was initially stochastic, probably facilitated by the availability of sufficient copies of each RNA species in the parent cell, such that each daughter would have a reasonable chance to end up with at least one complete set of RNAs. If initially a few different RNAs were strung together as “mini RNA chromosomes,” the advantage would be achievement of a more balanced distribution of RNAs. Perhaps they were distributed and replicated as such, with some of them cut up into functional RNAs, conceivably the birth of RNA processing. Processing signals might have been placed, in part, between the fragments corresponding to mature RNAs, presumably the origin of intergenic or even intronic space (Brosius 2003c). With the advent of templated protein biosynthesis, a major evolutionary transition included co-option of existing functional RNAs as well as longer “chromosomal RNAs” by RNA cutting and pasting to stitch together messenger RNAs (mRNAs) with open and increasingly longer reading frames, which might suggest a very early origin of RNA splicing in a ribonucleoprotein world (RNP world) (Reanney 1974; Darnell and Doolittle 1986). Alternative splicing and other rearrangements would be one of the mechanisms to enhance variation for generating translation products out of a limited repertoire of functional RNAs also doubling as templates for translation (Brosius 2001).
RNA SIGNATURES WRITTEN ALL OVER EXTANT DNA GENOMES
Linear arrangement of RNA genes, as well as the transition to the RNP world evolving an ancestor of the reverse transcriptase enzyme, happened to constitute a useful precondition for a next major evolutionary transition, namely, the conversion of RNA to DNA, the latter merely serving as bookkeeper (Darnell and Doolittle 1986; Gould 2002). For this and other reasons, the central dogma of biology could be revisited or supplemented by a different graphic account, both in appreciation of the major significance of RNA in the cell and the chronological order of the major transitions (Fig. 1) (Maizels and Weiner 1987; Brosius 2003a; Cech 2012). Remarkably, the process of reverse transcription of any RNA (Brosius 1999b) and more or less random integration into DNA genomes still persists and contributes much to their landscapes in a number of ways. The modular arrangement of functional with nonfunctional DNA and its plasticity in extant eukaryotic genomes might be a vestige of early evolutionary transitions (Brosius 2009).
Figure 1.
Alternative to the central dogma of molecular biology. The left part depicts the original central dogma of molecular biology with several adjustments incorporated, for example, the discovery of reverse transcription (thin upward arrow) (Crick 1958, 1970). The grouping on the right better reflects the evolutionary transitions and primacy of RNA. RNA could be replicated directly (semicircular arrow), albeit in extant organisms (e.g., plants, viruses) only with the catalytic activity of protein (RNA-dependent RNA polymerase). In this scheme, catalysis is indicated by (RNA) and or (protein) in parentheses. The major significance of RNAs for peptidyl transferase activity during translation (Noller 2012) is represented by the larger font for (RNA) compared to (Protein). Execution includes structural, catalytic, and regulatory tasks in the cell. The evolutionary developments underscore Stephen Jay Gould’s view that DNA merely is the agent of bookkeeping (Gould 2002). RNAs used to be bookkeepers as well, but remained agents of causality (Brosius 2005a).
Reverse transcriptase not only played an important role in generating the DNA genome but continues to be essential for chromosome maintenance via the action of telomerase. The extant enzyme synthesizes telomeres that serve as protective caps of chromosome ends, thus counteracting their shortening during replication cycles. Telomerase is a complex between RNA and protein in which the RNA serves as template and protein as reverse transcriptase (Greider and Blackburn 1989; Blackburn et al. 2006; Blackburn and Collins 2011). Group II introns, when transcribed as RNA are self-splicing with the activity residing on the RNA at high salt conditions in vitro (van der Veen et al. 1986). For in vivo activity and integration at new genomic sites in a process termed reverse splicing, a reverse transcriptase encoded in the intron is necessary (Lambowitz and Zimmerly 2011). The ribozyme has been proposed to be ancestral to non-LTR (long terminal repeat) retroposons as well as the spliceosome attributable to similarities of their reverse transcriptase and some small nuclear RNA (snRNA) components of the spliceosome, including aspects of the splice mechanism itself, respectively (Xiong and Eickbush 1990; Guthrie 1991; Sharp 1991; Fica et al. 2013). The rapid spread of introns in eukaryotes has been ascribed to group II introns after they were imported by endosymbionts (Cavalier-Smith 1991), perhaps necessitating the separation of slow mRNA production in the nucleus from the fast translation in the cytoplasm, one of the hallmarks of the eukaryotic cell (Martin and Koonin 2006).2
Importantly, the process of continuously converting RNA to DNA and its genomic integration has the potential to grossly inflate genomes with neutrally evolving material and, in conjunction with larger deletion of segments by recombination, leads to a high turnover rate of sequences on an evolutionary time scale, once more calling into question the ENCODE claim that ∼80% of the human genome is functional (Doolittle 2013; Graur et al. 2013; Niu and Jiang 2013). Generally, specific retroposons3 become active in certain lineages, and reverse transcripts from one or several master gene transcripts, such as LINEs (long interspersed elements), autonomous as they harbor gene encoding the retroposition machinery such as reverse transcriptase and nonautonomous SINEs (short interspersed elements), such as Alu or B1 elements. Nonautonomous retroposons rely on the machinery of the autonomous retroposons. Most retrocopies are not active as they are transcriptionally silent because of the lack of internal (e.g., truncation of LINEs) or external promoters in SINEs. Intact and transcribed master genes can spawn, over a few tens or hundreds of million years, more than one million copies, as is the case for Alu elements (Weiner 2006). With a few exceptions (see below), there is no selective pressure on such elements and, accordingly, they deviate over time from the consensus sequence. The human genome has been estimated to contain ∼43% retroposons, a relatively small amount of DNA transposons (∼3%), and as little as 5% conserved sequences (Lindblad-Toh et al. 2011; Ward and Kellis 2012) that potentially have function (Fig. 2).4 As argued before, conversion of RNA to DNA is an ancient process and it has been suggested that most DNA in the human genome has been derived by a virtually unabated bombardment of retroposons (Brosius 1999a). Over time, these elements are blurring into oblivion as randomized sequences by incessant changes, such as point mutations and small indels. Recently, it was confirmed by more sensitive computational strategies involving P-clouds, that almost 70% of the human genome may harbor repetitive elements (de Koning et al. 2011). Extrapolating back to or forward from the origin of RNA → DNA genome transition, there is no reason why this figure, except for the generally lesser contributions of DNA transposons, should not approach almost 100% (Brosius 1999a). The fact that essential sequences are interspersed with nonessential sequences as landing pads for transposed elements lessening their detrimental impact genome-wide could be chalked up to the ENCODE project (Bernstein et al. 2012), as well as adherents to intelligent design, creationists, and the like as “functional,” which would cleanse our species from the blemish of living with ∼80%–90% junk DNA in our genomes (Doolittle 2013; Graur et al. 2013; Niu and Jiang 2013).
Figure 2.
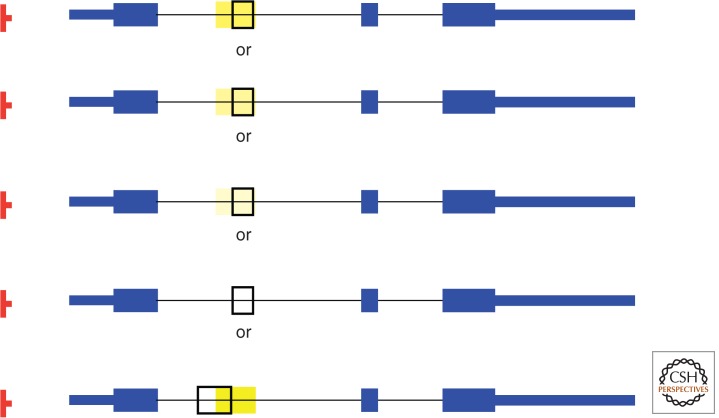
Exaptation of a new gene module at any stage of transposed element (TE) deterioration. In this example, a SINE, such as Alu, B1, identifier (ID), or mammalian-wide interspersed repeat (MIR), is retroposed into the first intron of a gene (introns depicted by thin black lines and the SINE as wide yellow bar). The gene harbors three exons (in blue). Open reading frames (ORFs) are shown as wide bars, whereas the 5′-UTR (untranslated region) and 3′-UTR are shown as narrower blue bars, in the terminal exons, respectively. The transcription promoter is depicted in red. The four top representations indicate the gradual decay of the SINE element by continuous mutation fading from yellow to white; thus, blending into the other anonymous sequence of the intron. At any of those stages, part of the TE can be exapted as novel exon (framed in black). Sometimes part of a discernible TE and adjacent anonymous intron can be exapted as novel gene module (bottom).
YESTERDAY’S JUNK COULD BECOME TOMORROW’S NOVEL GENE MODULE, IF ONLY TEMPORARILY SO
The lack of purifying selection concerning nonharmful TEs, including mRNA-derived retrocopies, should not divert from the fact that, occasionally, such sequences can be exapted as genetic novelties (Brosius 1991; Brosius and Gould 1992). Gene duplication as a means of generating novel genes has been realized for a long time (Haldane 1933; Muller 1935; Bridges 1936; Lewis 1951; Stephens 1951; Nei 1969). Gradual change from a duplicated gene, via those encoding isoforms, up to the acquisition of novel functions including subfunctionalization (Lynch and Force 2000) is now well documented (Roth et al. 2007; Kaessmann 2010; Chen et al. 2013). Gene amplification can occur via segmental duplication (Bailey and Eichler 2006) or retroposition (Brosius 1999a; Babushok et al. 2007) with a lower “success rate” for the latter. Retroposition of a copy of the mature mRNA requires the fortuitous presence of a promoter element upstream, which, as a potential benefit, could immediately alter regulation of the retrogene in comparison to the parent gene (Brosius 1999b). Genes, chiefly those encoded by endogenous retroviruses, were independently exapted or domesticated numerous times into novel functions, some of them are meanwhile essential for procreation or survival (Volff 2006, 2009). Also, DNA copies of nonretroviral RNA genomes, or parts thereof, can be integrated into genomes (Koonin 2010). In fungi, exaptations of such genes were reported (Taylor and Bruenn 2009), underscoring once more the notion that any RNA can be a template for the retroposition machinery (Brosius 1999b).
Can novel genes arise de novo from previously gene-free neutrally evolving genome regions? Despite some false positives (Monte et al. 1997; Kriegs et al. 2005), recruitment of entire protein-coding genes out of neutrally evolving sequences does occur (Long et al. 2003, 2013; Heinen et al. 2009; Kaessmann 2010; Carvunis et al. 2012; Murphy and McLysaght 2012; Neme and Tautz 2013). Recently, it has been emphasized that many long non-protein-coding RNAs originated from TEs (Kapusta et al. 2013). This RNA class is discussed in more detail below. In any event, it is much more common that novel gene modules are being added to existing genes. Such co-opted or exapted modules can be derived from inter- or intragenic space and constitute novel protein-coding exons and regulatory regions such as promoters and enhancers (Brosius 2005b, 2009; Baertsch et al. 2008; Rebollo et al. 2012).5 A prominent case is the exonization of parts of Alu elements, usually as alternatively spliced exons (Makalowski et al. 1994; Nekrutenko and Li 2001; Sorek et al. 2002; Lev-Maor et al. 2003; Krull et al. 2005; Shen et al. 2011). Many such events are, over evolutionary time, not stable. That is, they persist for a certain time if they are not or only slightly detrimental, especially when the novel splice product only constitutes a fraction of the functional RNA. Should, over time, the novel splice variant happen to become beneficial, it will be under purifying selection and, by point mutations in and around splice signals, its ratio, in comparison to the canonical splice form, might change in its favor. In a phylogenetic study on primates, it has been shown that such exons derived from Alu elements are lost at a high rate in the trial and error mode, typical for the evolution of novelties (Krull et al. 2005). A similar study involving all mammals and the older MIRs revealed that many of the events were already fixed and currently are under negative selection. The study also shows, in the case of a relatively recent exonization of an ancient MIR element, that exaptation can occur at any stage of TE decay (Fig. 3), and consequently also with any nondescript randomized DNA (Krull et al. 2007). Recently, a mechanism was described by which a snoRNA (small nucleolar RNA) extended its RNA-coding region by alternative processing of its 3′ end by introducing a single-point mutation near the site important for processing, generating both the canonical snoRNA and an extended variant (Mo et al. 2013). This event, detected in a cluster of rat Snord115 snoRNA genes in the imprinted Prader-Willi syndrome locus, but not in mouse, must be relatively recent. The additional gene product, L-Snord115, has most likely not yet acquired a function. The odds are that this snoRNA variant will not survive another dozen or so million years.
Figure 3.
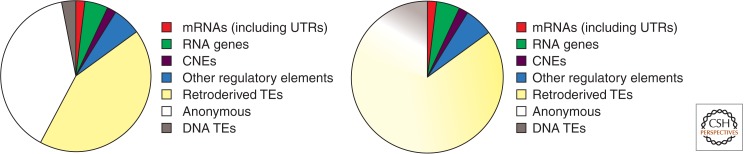
The functional versus nonfunctional human genome. The pies represent the various segments of the genome applying more conservative estimates. mRNAs (∼1.3% accounting for ORFs) including their 5′- and 3′-UTRs (∼0.8%) occupy slightly >2% of the human genome. All other RNAs (non-protein-coding RNAs) are estimated to cover about 5% (green), maximally 10%. This figure is under debate. Likewise, there is a conservative estimate for regulatory elements of ∼8%. Other sources argue up to 20% (Bernstein et al. 2012; Shen et al. 2012). Because conserved non-protein-coding DNA elements (CNEs, ∼2%, purple) often encode cis-regulatory elements (Hiller et al. 2012), the other regulatory elements are shown as 6% only (blue). TEs derived from RNA intermediates contribute ∼43% (yellow) and DNA transposons only 3% (brown). The remainder (39%) corresponds to anonymous, scrambled sequences (white). The estimated 26% introns (Bernstein et al. 2012) occupy mostly the yellow and white segments; intronic regulatory elements and encoded RNAs (e.g., snoRNAs, miRNAs) are accounted for in the respective segments. The pie on the right is identical, except that the retroposons (yellow) and, to a lesser extent, the DNA transposons (brown), blend into the anonymous sequence (white) to reflect TE ancestry of the nondescript sequences as well.
ALL WIRED UP ON REWIRING
Examples for TE contributions to gene expression by providing promoter or enhancer modules have been known for some time and are ample (Jordan et al. 2003; van de Lagemaat et al. 2003; Medstrand et al. 2005). Interestingly, TEs that were conserved at unusually high levels over hundreds of millions of years were reported to act as enhancers (Bejerano et al. 2006; Santangelo et al. 2007; Sasaki et al. 2008; Lindblad-Toh et al. 2011; Lowe and Haussler 2012). Recently, a number of publications proposed that the spreading of copies from active TE classes can lead to rapid rewiring, affecting hundreds of genes whose expression is being altered as a consequence, and is caused by the action of additional transcription factors binding to those enhancers (Wang et al. 2007; Bourque et al. 2008; Feschotte 2008; Xie et al. 2010; Lynch et al. 2011; Rebollo et al. 2012). This might even involve Alu elements in the form of Alu-derived miRNAs or non-miRNAs (Du et al. 2013; Hoffman et al. 2013; Liang and Yeh 2013; Mandal et al. 2013; Spengler et al. 2014).
Nevertheless, caution should be applied for the following reasons:
(1) Four ultraconserved elements were deleted in mice. Although there is no indication that these elements show immediate ancestry to TEs, the lesson equally applies to TE-derived enhancers. In this study, enhancer elements adjacent to genes whose protein-coding exons, when deleted, showed clear phenotypes in mice were chosen for deletion and the elements also were known to function as enhancers. In all four cases, the mice were not only viable and fertile, but also failed to reveal any obvious phenotype among the many parameters tested in the laboratory (Ahituv et al. 2007). Experimentally, it appears to be much easier to obtain a positive expression result with reporter constructs than a phenotype by enhancer ablation (Nelson and Wardle 2013).
(2) Should a newly acquired or activated TE class with “ready-to-use” enhancer activity populate a genome in high numbers, it might not smoothly rewire expression of a set of genes in a functionally viable manner, but simply wreak havoc in a genome by juxtaposition of new enhancers to resident genes. Such eruptions of TEs would be expected to have at least as many detrimental effects than beneficial ones. A less radical scenario would be the following: TEs carry sequences resembling transcription factor binding sites (TFBS) or other regulatory regions that pending minor mutations have the potential to become functional.
(3) Enhancers usually harbor clusters of homotypic or heterotypic TFBS and therefore it is less likely that most TEs are instantly functional. Enhancers usually act in a modular fashion and rather than changing expression patterns, they add a cell-type/tissue or a developmental window to existing expression patterns. After a period of testing (just as in the case of exonization; see above), this or other modules might get lost (adding or exchanging a few mains, yes; rewiring, hardly). For a recent comprehensive assessment of these problems, see de Souza et al. (2013).
TE FUNCTIONS: TO THE MOON!
TEs are insertional mutagens and often enough, integration near or into functional modules of the genome is disadvantageous and, for example, can cause disease (Chen et al. 2005; Callinan and Batzer 2006; Iskow et al. 2010; Hancks and Kazazian 2012). Because of affordable high-throughput sequencing technologies, the search for somatic de novo insertions of TEs becomes feasible. Numerous such events could be detected in tumor tissues and cells (Miki et al. 1992; Lee et al. 2012), some of which might be causal. Somatic integration even was proposed to contribute to neuropsychiatric disease, such as schizophrenia (Bundo et al. 2014). Apart from being detrimental or neutral, a minority of events has the potential to turn out beneficial.
TEs of a certain class might harbor not only TFBS, but also numerous other functional sequences in their consensus sequences, and, perhaps, mainly because these elements are defined by their designation, proposals with highly diverse functions are being published, even involving TEs with very narrow phylogenetic distributions (Allen et al. 2004; Espinoza et al. 2004, 2007; Lunyak et al. 2007; Mariner et al. 2008; Gong and Maquat 2011; Yakovchuk et al. 2011; Carrieri et al. 2012; Jady et al. 2012; Holdt et al. 2013; Ponicsan et al. 2013; Wang et al. 2013). Somatic LINE element integrations even have been implied in the development of the brain (Muotri et al. 2005; Coufal et al. 2009; Faulkner et al. 2009; Singer et al. 2010; Baillie et al. 2011; Upton et al. 2011; Perrat et al. 2013; Reilly et al. 2013). Either TEs are chock full of regulatory motifs and control elements, which can be argued in case of promoters, for example, LTRs (Feuchter and Mager 1990), or it is the fact that TEs are defined and designated nucleic acid sequences (nuons) (Brosius and Gould 1992) and, therefore, receive more attention instead of randomized and anonymous sequences in attempts to investigate their functions. Perhaps one or the other of these exhilarating findings will share the fate of ID repetitive elements and TEs of the SINE class, in a development that unfolded about three decades ago. A lot of excitement was generated by reports that ID elements regulate brain-specific gene expression (Milner et al. 1984; Sutcliffe et al. 1984a,b; McKinnon et al. 1986). Unfortunately, these claims did not stand the test of time (Owens et al. 1985; Sapienza and St-Jacques 1986; Goldman et al. 2014).6 For sure, any seemingly insignificant novelty could have far-reaching consequences for future lineages (Martignetti and Brosius 1993; Kapitonov and Jurka 2005), but in their infancy, the functional significance, if not potential of novelties, is not easy to assess and often might be transitory. And what is true for regulatory elements and protein-coding genes and their exonic modules should also apply to functions of non-protein-coding RNAs.
FUNCTIONAL RNA: ALIVE AND KICKING
Despite the growing recognition of RNA’s functional significance and versatility, as well as its preeminence in the evolution of life, up to ∼15 years ago, most scholars in the life sciences still deemed RNA molecules as fossils or remnants from bygone eras. The most complex cytoplasmic RNA class, messengers between the genetic information on DNA and ribosomes, organelles in which structural and functional macromolecules (proteins) of a cell are being assembled, did not generate much excitement any longer after the genetic code had been cracked (Nirenberg et al. 1965; Söll et al. 1965).
A minority of investigators sensed that the previously known and rather abundant nonprotein-coding RNAs, such as transfer and ribosomal RNAs, were only the tip of the iceberg (Prestayko and Busch 1968; Zieve and Penman 1976; Lerner et al. 1980; Brown and Fournier 1984; Lee et al. 1985; Erdmann and Wolters 1987; Mattick 1994; Brosius 1996) and that even novel functional RNAs could evolve (DeChiara and Brosius 1987; Brosius 1991). The floodgates began to open when sequencing of copy DNA generated from non-mRNA fractions revealed a plethora of novel RNAs and RNA classes including miRNAs (Filipowicz 2000; Huttenhofer et al. 2001; Lagos-Quintana et al. 2001; Couzin 2002). The development of deep and ultradeep sequencing methods for cellular, organ-specific, and whole transcriptomes of organisms greatly accelerated the deluge of data (Wang et al. 2009). The past 15 years have surprised most of the scientific community with the incessant discovery of thousands of novel RNAs, some extending RNA species from already known classes such as snoRNAs, some establishing novel RNA classes (e.g., miRNAs and siRNAs), and some unclassified (Fire et al. 1998; Ambros 2001; Lagos-Quintana et al. 2001; Lau et al. 2001; Moss 2001; Ruvkun 2001; Couzin 2002; Carninci et al. 2005; Derrien et al. 2012; Djebali et al. 2012; Kapranov et al. 2012; Ross et al. 2014). Even extracellular RNAs are receiving renewed attention (Benner 1988; Wu et al. 2002; Dunoyer et al. 2007; Leslie 2013a). Major RNA classes are summarized in Table 1.7
Table 1.
RNA classification
| RNA class | Approximate size (nt) | Function |
|---|---|---|
| Tiny RNAs, <50 nt | ||
| microRNA, miRNA | 21–24 | Gene regulation, for example, fine-tuning at the translational level |
| Short interfering RNA, siRNA | 20–25 | RNA interference, defense |
| PIWI-interacting RNA, piRNA | 26–31 | Epigenetic and posttranscriptional gene silencing of TEs |
| Small RNAs, ∼50–500 nt | ||
| Extracellular RNA, exRNA | Wide range | Intercellular communication |
| Small nuclear RNA (snRNA) | ||
| Spliceosomal RNAs, U1, U2, U4, U5, etc. RNAs | 100–200 | Splicing of mRNA out of primary transcripts |
| U7 RNA | 50 | 3′ processing of histone mRNA precursors |
| Ribonuclease P RNA, RNase P RNA or H1 RNA | 340 | tRNA processing |
| 7SK RNA | 330 | Regulation of transcription |
| Y RNA | 90–100 | DNA replication |
| Telomerase RNA component, TERC | 450 | Maintenance of telomeres |
| Small nucleolar RNA, snoRNA | 60–300 | Guide RNAs for nucleotide modification |
| C/D box snoRNA | 2′ O-methylation of the ribose moiety of rRNA | |
| H/ACA box snoRNA | rRNA pseudouridinylation | |
| Cajal body–specific RNA, scaRNA | Composite C/D and H/ACA guide RNA, modification of snRNAs | |
| U3, U8, U14, U17, and U22 snoRNAs | Regulation of preribosomal RNA (pre-rRNA) folding, mediation of correct nucleolytic processing (maturation) | |
| “Orphan” snoRNAs | Target of modification (if any) and function unknown, examples are SNORD115 and SNORD116 in the PWS (Prader-Willi syndrome) locus | |
| Small cytoplasmic RNA, scRNA | ||
| 5S and 5.8S small ribosomal RNAs | 120, 150 | Translation |
| Transfer RNAs, tRNAs | 73–94 | Adapter molecules in translation |
| Signal recognition particle RNA, 7SL RNA or SRP RNA | 300 | Targets protein to the endoplasmic reticulum |
| Vault RNAs | 90–100 | Components of the vault organelle, function not clear yet |
| Ribonuclease MRP, mitochondrial RNA processing RNA | 290 | Mitochondrial DNA replication and rRNA processing in the nucleus |
| BC1 RNA | 150 | Neuronal cytoplasmic RNA (some expression in testes) in soma, but also transported to dendrites, possibly involved in regulation of translation; phylogenetically restricted to rodents, originated from tRNA retroposition |
| BC200 RNA | 200 | Neuronal cytoplasmic RNA (some expression in testes) in soma, but also transported to dendrites, possibly involved in regulation of translation; phylogenetically restricted to anthropoid primates; not homologous to BC1 RNA, originated from a monomeric Alu element |
| Long RNAs, >500 nt | ||
| 18S and 28S large ribosomal RNA, rRNA | 1900, 5000 | Translation |
| Messenger RNAs, mRNAs | Various | Templates for protein biosynthesis |
| Long intervening non-protein-coding RNAs, lincRNAs | Various | Various suggested functions, not many are certain; regulation of gene expression; gene/chromosome silencing/imprinting by interaction with chromatin |
| Enhancer RNA, eRNA | >1000 | |
| Multiexonic poly(A)+ RNA, meRNA | Various | |
| Long antisense RNAs, asRNA, or aRNA or natural antisense RNA, NAS RNA | Various | Possibly regulation of gene expression |
| Pseudogene transcripts | Various | Decoy or sink for sequestration of RNA or protein |
| Circular RNA | Decoy or sink for sequestration of RNA or protein | |
It is difficult to classify RNAs because they are so diverse. This table attempts to categorize according to size; other groupings take their subcellular locations or functions into account. In any event, the distinctions are blurry and apply only to one species or lineage. In yeast, for example, the U-type snRNAs are significantly larger than in vertebrates. More recently, an arbitrary line between large and small RNAs is being drawn at 200 nt, the latter including tiny RNAs, such as miRNAs. The three size classes here reflect the historical division between small and large RNA at ∼500 nt to accommodate SRP RNA, 7SK RNA, or mitochondrial RNA processing (MRP) RNA that are well >200 nt in length, but have been known since their discoveries as small RNAs. Large or long RNAs are defined as being in the size range of mRNAs, sometimes even displaying mRNA-like attributes (e.g., processing, polyadenylation), but devoid of a functional open reading frame. Initially, miRNAs were designated as tiny RNAs to distinguish them from conventional small RNAs (Ruvkun 2001). This tripartite categorization is kept in the table. There is a flurry of attempts to identify additional RNA classes. Their functions are mostly unknown and the RNAs might correspond to spurious or aborted transcription or other by-products of gene expression (see the text). These include promoter-associated short and long RNA (PASR, PALR), termini-associated RNA (TASR), promoter upstream RNA (PROMPT) transcription initiated RNA (tiRNA), transcription start site antisense RNA (TSSa), antisense termini-associated short RNA (aTASR), retrotransposon derived RNA, satellite region RNA, telomere repeat region antisense RNA (TERRA), etc. For further information and original references see Saxena and Carninci (2011a). PIWI, P-element-induced wimpy testis.
It appears that the mining of novel tiny and small RNAs is reaching a point of deceleration (Saxena and Carninci 2011b) and, as a result, long RNAs are receiving more attention (Jacquier 2009; Ponting et al. 2009; Carninci 2010; Derrien et al. 2012; Hu et al. 2012; St Laurent et al. 2012; Cloutier et al. 2013; Di Ruscio et al. 2013; Geisler and Coller 2013; Kung et al. 2013; Orom and Shiekhattar 2013; Sabin et al. 2013; Ulitsky and Bartel 2013; Fatica and Bozzoni 2014; Yang et al. 2014). Oligonucleotide array sequence analysis, conventional RNA sequencing, and RNA-sequencing technologies (FANTOM) (Carninci et al. 2005; Cheng et al. 2005; Birney et al. 2007; Mercer et al. 2012), even in combination with biocomputation, taking phylogenetic considerations and/or the potential for forming secondary structures into account, mostly come up with at least ten thousand long non-protein-coding RNA candidates (Washietl et al. 2007; Managadze et al. 2013; Necsulea et al. 2014; Nielsen et al. 2014).
DID RNA ENTER BUBBLE TERRITORY?
The percentage of the human genome that is being transcribed appears to be approaching the maximum asymptotically. A simple explanation is the increased coverage with ultradeep sequencing methods and coverage of any cell type including tumors, many developmental stages, as well as (tumor) cell lines numerous times over (Carninci 2009; Ritz et al. 2011; Haas et al. 2012; Ho et al. 2012; Shah et al. 2012; An et al. 2013; Eswaran et al. 2013; Hu et al. 2013; Schonberg et al. 2013; Yoshihara et al. 2013). With all this overabundance, there is a debate raging between those that claim almost any identified transcript and snippet of RNA to be functional (Mattick 2003; Lee et al. 2009; Carninci 2010; Kishore et al. 2010; Clark et al. 2011; Bernstein et al. 2012; Kapranov and St Laurent 2012; Khayrullina et al. 2012; Gebetsberger and Polacek 2013; Mattick and Dinger 2013) and more conservative voices (Brosius 2005c; Huttenhofer et al. 2005; Robinson 2010; van Bakel et al. 2010; Graur et al. 2013). The writer concurs with the argumentation for spurious and stochastic transcripts (Kowalczyk et al. 2012; Jensen et al. 2013; Mudge et al. 2013), especially as many transcription promoters are bidirectional (Seila et al. 2008; Neil et al. 2009; Xu et al. 2009; Wei et al. 2011; Uesaka et al. 2014). Even the yeast Saccharomyces cerevisiae featuring a compact genome abounds with transcriptional noise and erroneous initiation of transcription by RNA polymerase II (Struhl 2007). Furthermore, some transcription might be involved in gene regulation without the transcribed RNA being functional (Tisseur et al. 2011). Examples include regulation of transcription by upstream promoters, previously known as promoter occlusion or transcriptional interference (Adhya and Gottesman 1982; Cullen et al. 1984). In yeast, for example, it has been shown that the SER3 gene is repressed by the act of transcription from the upstream SRG1 gene (Martens et al. 2004). In contrast, the PHO5 gene is activated by intergenic transcription in the opposite orientation presumably influencing chromatin remodeling (Uhler et al. 2007). In other words, the act of transcription exerts the function whereas the RNA is merely a by-product, and this mechanism regulates gene expression in multicellular eukaryotes as well (Kornienko et al. 2013). Another relatively passive role for some RNAs could be the following: Many proteins can bind RNA and proteins can associate with each other. Perhaps an RNA molecule could have the sole function to broker the interaction of two or more proteins that otherwise would not be able to interact with each other. However, if the proteins bind directly or even via other RNA binding proteins to the RNA, close proximity might facilitate functional or structural interactions. In addition, such an RNP complex might even serve as a shuttle into subcellular domains or environments that one or the other component, for lack of the appropriate signals, would not be able to reach by itself (Brosius 2005b).
It has been argued that many of the newly discovered long RNAs represent 3′-UTRs extending beyond the annotated 3′-ends of the mRNAs by using alternative distal poly(A) addition signals. If there are large introns, 5′-UTRs with alternative upstream promoters also can be located far from the ORF. For that reason, some investigators took measures to stay clear of transcripts that are located in a certain proximity to annotated protein-coding genes (Managadze et al. 2013). Nevertheless, splicing does occur in 3′-UTRs as well (Bicknell et al. 2012; Camacho-Vanegas et al. 2012; Zhernakova et al. 2013) and, hence, the corresponding exons could map to corresponding protein-coding genes at a much greater distance to the ORFs. Furthermore, apart from spurious initiation of transcription anywhere in the genome, a certain level of readthrough into gene distal regions could account for a significant portion of these long RNA candidates (Finta and Zaphiropoulos 2000; Akiva et al. 2006; Parra et al. 2006; Richard and Manley 2009).
RNAs overlapping with annotated genes or transcribed in antisense orientation, albeit showing regulatory potential, are generally being avoided for now as their structures, regulation, and functions are more difficult to investigate (Ulitsky and Bartel 2013). Hence, the focus has narrowed to investigating long intergenic noncoding RNAs (lincRNAs).8 Another point is being overlooked frequently. For the most part, the arbitrary cutoff for an open reading frame is at 100 amino acids in length. If humans were not pentadactyls, but hexa- or tetradactyls, the cutoff would probably have been chosen at 144 or 64 amino acids, respectively. There are numerous mRNAs that have even shorter ORFs, namely, those encoding peptides (Rudd et al. 1998; Sousa et al. 2001; Frith et al. 2006a; Kastenmayer et al. 2006; Savard et al. 2006; Galindo et al. 2007; Ghabrial 2007; Hanada et al. 2007; Hashimoto et al. 2008; Magny et al. 2013). Perhaps a significant fraction of mRNA-like long intervening non-protein-coding RNAs are mRNAs after all, encoding peptides and short proteins. The question on how many of the non-protein-coding RNAs are associated with polysomes for productive translation is currently under debate (Ingolia et al. 2011; Guttman et al. 2013; Ingolia et al. 2013; Ulitsky and Bartel 2013; van Heesch et al. 2014).
NOVEL RNAs OUT OF THE BLUE
De novo arisen long (intergenic or better intervening) non-protein-coding RNA genes out of neutrally evolving DNA including transposed elements also are receiving increased attention (Guttman et al. 2009; Marques and Ponting 2009; Esteller 2011; Hadjiargyrou and Delihas 2013; Kapusta et al. 2013; Managadze et al. 2013; Ulitsky and Bartel 2013). Their abundance as a class is not surprising, however, because parts of TEs, especially LINEs and lone LTRs, can harbor active transcription promoters. As a consequence, a large proportion of long intervening non-protein-coding RNAs show sequence similarities to TEs.
Most investigators agree that among the tens of thousands of long RNA candidates, there will be hundreds if not thousands of bona fide functional RNAs. One article estimates the number to be ∼10% of all suggested candidates (Ulitsky and Bartel 2013). In addition, quite a few of these spurious and currently nonfunctional RNAs might one day be exapted into novel functions and, as discussed above, as protein-coding exons or regulatory regions. The majority will be rendered inactive again and a few, if beneficial, will eventually persist under selective pressure (Brosius 2005c; Siepel 2009; Polev 2012). In an analogy to Alex Rich’s predictions with respect to an RNA world and the persisting significance of RNA, Henry Harris envisaged the potential of spurious transcripts as raw material for evolution about half a century ago: “Only a small proportion of the RNA made in the nucleus of animal and higher plant cells serves as a template for the synthesis of protein. … Most of the nuclear RNA, however, is made on parts of the DNA, which do not contain information for the synthesis of specific proteins. … It plays no role in the synthesis of a cell protein, but serves as a background on which mutation and selection may operate to produce new templates for protein synthesis” (Harris 1965, 2013). This also applies to new genes encoding functional RNAs.
The process of new genes arising out of nongenic regions might not appear as extraordinary if one considers the presumably more trivial reverse process, namely, formation and gradual decay of pseudogenes. A single-point mutation or small deletion can turn an mRNA into a non-protein-coding RNA. Further mutations can silence its transcription and, after enough time, the remnants of a gene cannot be recognized as such any longer.
COMPETITIVE RNAs
Interestingly, transcribed pseudogenes, including those that arose via retroposition, reportedly can be functional as RNAs or mRNAs encoding truncated proteins (Balakirev and Ayala 2003; Duret et al. 2006; Frith et al. 2006b; Pink et al. 2011; Wen et al. 2012; Sen and Ghosh 2013). For example, retropseudogene PTENP1 derived from tumor suppressor gene PTEN is transcribed. Like the canonical mRNA expressed from the parent gene, it harbors binding sites for several miRNAs in what used to be the 3′-UTR. These miRNA target sites are competing for the corresponding miRNAs, thus, ameliorating their inhibitory effect on the intact mRNA, resulting in higher levels of the tumor suppressor protein PTEN (Poliseno et al. 2010). Even two isoforms of an antisense RNA generated from the PTENP1 pseudogene have been reported to be involved in gene regulation (Johnsson et al. 2013). In addition, a plethora of naturally occurring circular RNAs have been discovered, which may regulate gene expression by sponging up complementary RNAs, such as miRNAs (Salzman et al. 2012; Hansen et al. 2013; Memczak et al. 2013; Tay et al. 2014). RNAs not only can be decoys or sinks for other RNAs but also for proteins. A long RNA processing product located between two snoRNAs has been reported to act as a sink for Fox2 splicing factor and, as a consequence, alter the relative abundance of alternative splice products of a number of genes (Yin et al. 2012).
THE (NUCLEIC) ACID TEST FOR FUNCTION
As one of the early advocates of a vibrant RNA componentry in extant organisms (DeChiara and Brosius 1987; Brosius 1991, 1996; Petherick 2008), the writer never doubted that the number of functional non-protein-coding RNAs could easily match those for mRNAs in line with more conservative estimates being in the 3% range (Clamp et al. 2007; Church et al. 2009; Cabili et al. 2011; Managadze et al. 2013).9 Considering genes encoding RNAs smaller than long RNAs as well and leaving generous space for potentially functional RNAs overlapping other genes in sense or antisense orientation, the figure might top 5%. It does not matter whether the numbers will end up even at 10% of the total genome, they will not be in the range of 75% (Djebali et al. 2012). There is a tremendous task ahead of us to determine which of the detected transcripts are bona fide RNAs, and what their individual functions and contributions to the cell are. Phylogenetic conservation helps, but its absence is not a perfect criterion for lack of function. Not many investigators would deny the functional significance of the UTRs of mRNAs. These regions, flanking ORFs, can encode other, for example, regulatory functions such as structures to modulate turnover or translation efficiencies, including sequence complementarities to regulatory miRNAs. Yet, for the most part, these sequences show much less phylogenetic conservation in comparison to ORFs. This agrees with findings about Xist, an ∼17-kb-long non-protein-coding RNA in humans that, at best, is conserved only in small patches between mammals (Duret et al. 2006). Inactivation of Xist RNA expression on the paternal X chromosome leads to early embryonic death in a mouse knockout model (Marahrens et al. 1997). Even the best test for functional significance, gene inactivation in knockout model systems, does not always provide immediate and simple answers. In yeast, very few knockouts of snoRNAs showed a clear phenotype as was the case for U14 (Li et al. 1990), but not, for example, snR189 (Thompson et al. 1988), or only over time or under certain conditions with respect to other snoRNAs (Badis et al. 2003; King et al. 2003; Esguerra et al. 2008). Bacterial genomes like those of Escherichia coli do not have much space to waste and everything without a selective advantage would not remain in the genome for long. In agreement, the knockout of the 4.5S RNA gene had been shown to be essential (Brown and Fournier 1984). Surprisingly, ablation of the gene encoding 6S RNA with a wide phylogenetic distribution and structural conservation in bacteria (Barrick et al. 2005) had no effect on immediate viability (Lee et al. 1985). Later, 6S RNA was shown to regulate transcription and enhance long-term survival (Wassarman and Storz 2000; Trotochaud and Wassarman 2004).
In mammalian genomes, not every inactivation of a protein-coding gene results in a discernible phenotype. The same is to be expected from genes encoding RNA. Snora35 is located within the second intron (almost 100 kb in length) of the serotonin 2c receptor gene (Htr2c) and is highly conserved between placental mammals. As a consequence, the brain-specific Snora35 (earlier termed MBI-36 snoRNA) is cotranscribed with and processed out of the primary Htr2c heterogeneous nuclear RNA (hnRNA), for example, in the choroid plexus (Cavaille et al. 2000). Snora35 gene-depleted mice do not appear to display a phenotype different from their wild-type siblings, thus far (BV Skryabin and J Brosius, unpubl.). In another example of an RNA knockout, it took several years of work to tease out a reduced exploratory behavior in mice when small cytoplasmic BC1 RNA, phylogenetically restricted to rodents and expressed in neurons where it is delivered to dendritic processes, was deleted (Tiedge et al. 1991; Martignetti and Brosius 1993; Lewejohann et al. 2004). In a further mouse snoRNA knockout, the entire cluster encoding Snord116 RNA isoforms (earlier termed MBII-85 snoRNA) was deleted from a locus associated with Prader-Willi syndrome, a neurodevelopmental disorder in humans. Two independent studies revealed that the mice showed failure to thrive and growth retardation, but not all symptoms described for humans (Skryabin et al. 2007; Ding et al. 2008). In many multicellular organisms, snoRNAs are cotranscribed with introns of protein-coding genes or non-protein-coding genes. Snord116 is paternally imprinted and expressed in the brain. The host RNA is processed into a set of abundant long non-protein-coding RNAs by splicing out the introns (Runte et al. 2001). Attempts to compensate for the loss of Snord116 with constructs coexpressing some copies of the respective snoRNA in introns of a different host transgene independently failed thus far (Ding et al. 2008; BV Skryabin and J Brosius, unpubl.). The following could explain the findings: (1) The number of Snord116 RNA isoforms from the original transcription unit is not sufficient in the transgenic construct; (2) spatial, temporal expression and/or its levels are not appropriate; and (3) truncation of the long host hnRNA-derived, processed non-protein-coding RNA is the underlying cause for the mouse phenotype and, by extension, human disease.
An interesting case involves the Hotair long RNA that had been shown ex vivo to regulate HoxD genes (Rinn et al. 2007); deletion of the HoxC cluster harboring the Hotair gene was devoid of a phenotype (Schorderet and Duboule 2011), whereas targeted disruption of the Hotair locus alone led to homeotic transformation of the spine and malformation of metacarpal and carpal bones (Li et al. 2013).
A great stride forward has been made through a comprehensive gene knockout project involving 18 large RNA candidates (Sauvageau et al. 2013). The corresponding genes were selected not to overlap with any other gene or gene module, and their coding regions were replaced with the lacZ and neoR marker genes. At the current level of investigation, five of the 18 mouse lines revealed a phenotype. In three lines peri- and postnatal lethality was reported, two of which had some survivors, but they had growth defects. For the two remaining lines with a phenotype, growth defects were also reported. The most lethal knockout involved Fendrr, a gene with six exons encoding a nuclear RNA of about 2.4 kb in length. An independent knockout only involving the first exon by inserting a transcriptional stop cassette was lethal as well, albeit showing different organ defects (Grote et al. 2013). The <30%, but >10% “success” rate of 18 knockouts could mean a number of things, for example, that the RNA gene candidates were carefully selected in favor of potential functionality and phenotypic consequences (Sauvageau et al. 2013). Still, two-thirds of the gene ablations did not show a noticeable impact under the laboratory conditions tested, but, nevertheless, could be beneficial for the long-term survival of mice in natural, ever-changing environments or that some of the RNAs have no function per se. Also, it remains valid that, in exceptional cases, such transitory “protogenes” could become exaptations encoding novel functional RNAs, even proteins, or predominantly disappear again into the noise of the neutrally evolving genomic mass. In any event, major points in an article written to balance some of the extreme views are confirmed: “[t]here is no doubt that a number of these non-protein-coding RNAs have important regulatory functions in the cell” and “… aberrant transcripts or processing products embody evolutionary potential and provide novel RNAs that natural selection can act on” (Brosius 2005c).
CONCLUDING REMARKS
All scenarios for the beginning of life are far from perfect. The most plausible hypothesis is based on RNA as a primordial (auto)catalytic macromolecule, leading over a variety of transitions (Cairns-Smith 1982; Szathmáry and Smith 1995), of which one of the first was acquisition of a simple lipid enclosure (Szostak et al. 2001; Mansy et al. 2008) to extant forms of life. In the RNA world, pheno- and genotype were united in the same macromolecule. This union began to separate during the major transition to the RNP world with a “division of labor” between RNA and protein. Even before, linkage of RNA molecules, confinement (of sets of RNAs), and discrimination, on one hand, versus escape (reminiscent of viruses and horizontal gene transfer) and more expansive exchange by reshuffling of the RNA componentry (sex), are apparent. This resembles a tug of war, constantly in need of balancing the forces and not allowing for a victory of either side. Up to this day, at the cellular, organismal, and societal level, the predicament of discrimination and exchange as well as the quandary of selfishness and cooperation remains an essence of evolution and life.10
Perhaps the modular arrangement of genes and chromosomal DNA has its origins in the RNA or RNP worlds as well, reflecting the hypothetical structure of several linked RNA molecules. This also would mean that RNA processing and trimming had very early origins. Components of the extant telomerase as well as group II introns, ancestral to the spliceosome and non-LTR retroposons, might date back as far as the RNP or RNA worlds. The continued assault of extant eukaryotic genomes with retroposons (somewhat countered and alleviated by deletions via recombination) clearly had its origin in the transition period when RNA was replaced by DNA for the task of storing and replicating genetic material. This process still can shape genomes quite drastically in relatively short evolutionary time frames. Importantly, out of this sea of change with islands of rather conserved and fixed gene modules, new modules can be generated fortuitously and “tested” for added or modified function by the forces of selection. Most disappear again with a few eventually exapted as novel gene modules and, occasionally, even as novel genes encoding protein or functional RNA.
After a long lag phase on the sidelines, functional RNA currently is in the spotlight of biology, even medicine, as RNomics shows promise to detect additional disease genes, greatly develop the diagnostic toolbox, and revolutionize therapeutic possibilities (Esteller 2011; Cech 2012; Erdmann and Barciszewski 2012; Gold et al. 2012). However, the development from only two decades ago when the mere mention of RNA generally exposed grant proposals to monkey hammering resulting in poorer scores and the current situation in which a feeding frenzy of RNA discovery fueled by ultradeep RNA-sequencing technologies endorses almost any detected transcript or degradation product as functional RNA borders on the grotesque. Clearly, the pendulum has swung to the other extreme.11 In any event, the renewed interest in RNA and advances in understanding its evolution and biology will continue to fascinate us. RNA, this ancient macromolecule, will grant us exciting new insights into the works of life past, present, and possibly future.
An afterthought worthy of note is the dichotomy with respect to life’s fragility considering individuals, even species versus the resilience of life as a whole, over an ∼4-billion-year timescale.
As mentioned above, some of the intron gain could date back as far as the RNA world, or at least back to LUCA, because type II introns are present in bacteria (Ferat and Michel 1993) and in the more derived bacterial and archaeal genomes, introns could have been lost (Poole et al. 1999). Massive intron loss would be reminiscent of a relatively recent purge of most introns in the yeast S. cerevisiae via retroposition (Fink 1987).
A general term for retroposed sequences as well as DNA transposons is “transposable” elements (TEs). However, because following retroposition most elements are not able to transpose any longer attributable to truncations, lack of transcription, etc., the writer prefers “transposed” elements.
See discussion points below that absence or low levels of conservation must not necessarily rule out function and vice versa.
Even modules that reduce or destroy the activity of a targeted gene and/or its product can be beneficial to the host. Xmrk is an epidermal growth factor receptor-related oncogene in certain Xiphophorus fish hybrids and, when overexpressed, leads to melanoma. Insertion of an autonomous non-LTR retrotransposon disrupted and deactivated Xmrk. As a consequence, the individuals harboring this insertion do not develop tumors (Schartl et al. 1999).
Ironically, the presence of a brain-specific RNA (BC1 RNA) with similarity to the consensus sequence of ID elements had been noticed in early publications, but dismissed as by-product of a functional act of transcription (Sutcliffe et al. 1984a). This activity by RNA polymerase III had been suggested to open the chromatin structure of brain-specific genes to allow transcription by RNA polymerase II (Sutcliffe et al. 1984b). It turned out, however, that BC1 RNA is encoded by a single active gene, is a master gene for the ID repetitive SINEs, and is functional (DeChiara and Brosius 1987; Martignetti and Brosius 1993; Kim et al. 1994; Wang et al. 2002; Lewejohann et al. 2004; Iacoangeli and Tiedge 2013). This perfectly reflects the “Zeitgeist” of the era and is in stark contrast to the current situation in which almost anything that features a ribogroup is being considered functional (see below).
Detailed descriptions of the RNA classes and their (potential) functions can readily be found in reviews and the vast original literature cited therein, for example, Prasanth and Spector 2007; Costa 2010; Aalto and Pasquinelli 2012; Djebali et al. 2012; Kapranov et al. 2012; Dieci et al. 2013; Gomes et al. 2013; Leslie 2013b.
This is a double blunder in the troubled RNA nomenclature. First, there hardly is a bona fide noncoding RNA (except, of course, for the nonfunctional noise) and most RNAs carry a code (Trifonov 1989; Barbieri 2003), and, second, if the locus encodes an RNA, it is a gene itself and not something intergenic. At least one laboratory has begun to address these macromolecules as long “intervening” non-[protein]-coding RNAs (Ulitsky and Bartel 2013).
ORFs amount to ∼1.3% in the mouse and human genomes and an additional ∼0.8% serve as 5′- and 3′-UTRs at their extremities (International Human Genome Sequencing Consortium 2004; Church et al. 2009; Managadze et al. 2013). Accordingly, all sequences that end up in mature mRNAs cover somewhat >2% of their respective genomes. Strictly speaking, mRNAs are chimera between templates for translation and non-protein-coding RNAs and many long (intervening) non-protein-coding RNAs are quite similar, except for the lack of (long) ORFs.
“Without Contraries is no progression. Attraction and Repulsion […] are necessary to […] existence.” The original William Blake quote, not shortened by the writer is given as: “Without Contraries is no progression. Attraction and Repulsion, Reason and Energy, Love and Hate, are necessary to Human existence” (Blake 1975).
See zmbe.uni-muenster.de/institutes/iep/iepcartoon.htm for a cartoon.
Editors: Patrick J. Keeling and Eugene V. Koonin
Additional Perspectives on The Origin and Evolution of Eukaryotes available at www.cshperspectives.org
REFERENCES
- Aalto AP, Pasquinelli AE 2012. Small non-coding RNAs mount a silent revolution in gene expression. Curr Opin Cell Biol 24: 333–340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adamala K, Szostak JW 2013. Nonenzymatic template-directed RNA synthesis inside model protocells. Science 342: 1098–1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adhya S, Gottesman M 1982. Promoter occlusion: Transcription through a promoter may inhibit its activity. Cell 29: 939–944. [DOI] [PubMed] [Google Scholar]
- Ahituv N, Zhu Y, Visel A, Holt A, Afzal V, Pennacchio LA, Rubin EM 2007. Deletion of ultraconserved elements yields viable mice. PLoS Biol 5: e234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akiva P, Toporik A, Edelheit S, Peretz Y, Diber A, Shemesh R, Novik A, Sorek R 2006. Transcription-mediated gene fusion in the human genome. Genome Res 16: 30–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen TA, Von Kaenel S, Goodrich JA, Kugel JF 2004. The SINE-encoded mouse B2 RNA represses mRNA transcription in response to heat shock. Nat Struct Mol Biol 11: 816–821. [DOI] [PubMed] [Google Scholar]
- Ambros V 2001. microRNAs: Tiny regulators with great potential. Cell 107: 823–826. [DOI] [PubMed] [Google Scholar]
- An J, Lai J, Lehman ML, Nelson CC 2013. miRDeep*: An integrated application tool for miRNA identification from RNA sequencing data. Nucleic Acids Res 41: 727–737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atkins JF, Gesteland RF, Cech TR 2011. RNA Worlds: From life’s origins to diversity in gene regulation. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- Attwater J, Wochner A, Holliger P 2013. In-ice evolution of RNA polymerase ribozyme activity. Nat Chem 5: 1011–1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babushok DV, Ostertag EM, Kazazian HH Jr 2007. Current topics in genome evolution: Molecular mechanisms of new gene formation. Cell Mol Life Sci 64: 542–554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badis G, Fromont-Racine M, Jacquier A 2003. A snoRNA that guides the two most conserved pseudouridine modifications within rRNA confers a growth advantage in yeast. RNA 9: 771–779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baertsch R, Diekhans M, Kent WJ, Haussler D, Brosius J 2008. Retrocopy contributions to the evolution of the human genome. BMC Genomics 9: 466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey JA, Eichler EE 2006. Primate segmental duplications: Crucibles of evolution, diversity and disease. Nat Rev Genet 7: 552–564. [DOI] [PubMed] [Google Scholar]
- Baillie JK, Barnett MW, Upton KR, Gerhardt DJ, Richmond TA, De Sapio F, Brennan PM, Rizzu P, Smith S, Fell M, et al. 2011. Somatic retrotransposition alters the genetic landscape of the human brain. Nature 479: 534–537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balakirev ES, Ayala FJ 2003. Pseudogenes: Are they “junk” or functional DNA? Annu Rev Genet 37: 123–151. [DOI] [PubMed] [Google Scholar]
- Barbieri M 2003. The organic codes: An introduction to semantic biology. Cambridge University Press, Cambridge. [Google Scholar]
- Barrick JE, Sudarsan N, Weinberg Z, Ruzzo WL, Breaker RR 2005. 6S RNA is a widespread regulator of eubacterial RNA polymerase that resembles an open promoter. RNA 11: 774–784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bejerano G, Lowe CB, Ahituv N, King B, Siepel A, Salama SR, Rubin EM, Kent WJ, Haussler D 2006. A distal enhancer and an ultraconserved exon are derived from a novel retroposon. Nature 441: 87–90. [DOI] [PubMed] [Google Scholar]
- Benner SA 1988. Extracellular “communicator RNA.” FEBS Lett 233: 225–228. [DOI] [PubMed] [Google Scholar]
- Bernstein BE, Birney E, Dunham I, Green ED, Gunter C, Snyder M 2012. An integrated encyclopedia of DNA elements in the human genome. Nature 489: 57–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bicknell AA, Cenik C, Chua HN, Roth FP, Moore MJ 2012. Introns in UTRs: Why we should stop ignoring them. BioEssays 34: 1025–1034. [DOI] [PubMed] [Google Scholar]
- Birney E, Stamatoyannopoulos JA, Dutta A, Guigo R, Gingeras TR, Margulies EH, Weng Z, Snyder M, Dermitzakis ET, Thurman RE, et al. 2007. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature 447: 799–816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackburn EH, Collins K 2011. Telomerase: An RNP enzyme synthesizes DNA. Cold Spring Harb Perspect Biol 3: a003558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackburn EH, Greider CW, Szostak JW 2006. Telomeres and telomerase: The path from maize, Tetrahymena and yeast to human cancer and aging. Nat Med 12: 1133–1138. [DOI] [PubMed] [Google Scholar]
- Blake W 1975. The marriage of heaven and hell. Oxford University Press, Oxford (first published in 1790). [Google Scholar]
- Bourque G, Leong B, Vega VB, Chen X, Lee YL, Srinivasan KG, Chew JL, Ruan Y, Wei CL, Ng HH, et al. 2008. Evolution of the mammalian transcription factor binding repertoire via transposable elements. Genome Res 18: 1752–1762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bridges CB 1936. The bar “gene” a duplication. Science 83: 210–211. [DOI] [PubMed] [Google Scholar]
- Brosius J 1991. Retroposons—Seeds of evolution. Science 251: 753. [DOI] [PubMed] [Google Scholar]
- Brosius J 1996. More haemophilus and Mycoplasma genes. Science 271: 1302–1303. [PubMed] [Google Scholar]
- Brosius J 1999a. Genomes were forged by massive bombardments with retroelements and retrosequences. Genetica 107: 209–238. [PubMed] [Google Scholar]
- Brosius J 1999b. RNAs from all categories generate retrosequences that may be exapted as novel genes or regulatory elements. Gene 238: 115–134. [DOI] [PubMed] [Google Scholar]
- Brosius J 2001. tRNAs in the spotlight during protein biosynthesis. Trends Biochem Sci 26: 653–656. [DOI] [PubMed] [Google Scholar]
- Brosius J 2003a. The contribution of RNAs and retroposition to evolutionary novelties. Genetica 118: 99–116. [PubMed] [Google Scholar]
- Brosius J 2003b. From Eden to a hell of uniformity? Directed evolution in humans. BioEssays 25: 815–821. [DOI] [PubMed] [Google Scholar]
- Brosius J 2003c. Gene duplication and other evolutionary strategies: From the RNA world to the future. J Struct Funct Genomics 3: 1–17. [DOI] [PubMed] [Google Scholar]
- Brosius J 2005a. Disparity, causation, adaptation, exaptation, and contingency at the genome level. Paleobiology 31: 1–16. [Google Scholar]
- Brosius J 2005b. Echoes from the past—Are we still in an RNP world? Cytogenet Genome Res 110: 8–24. [DOI] [PubMed] [Google Scholar]
- Brosius J 2005c. Waste not, want not—Transcript excess in multicellular eukaryotes. Trends Genet 21: 287–288. [DOI] [PubMed] [Google Scholar]
- Brosius J 2009. The fragmented gene. Ann NY Acad Sci 1178: 186–193. [DOI] [PubMed] [Google Scholar]
- Brosius J, Gould SJ 1992. On “genomenclature”: A comprehensive (and respectful) taxonomy for pseudogenes and other “junk DNA.” Proc Natl Acad Sci 89: 10706–10710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown S, Fournier MJ 1984. The 4.5 S RNA gene of Escherichia coli is essential for cell growth. J Mol Biol 178: 533–550. [DOI] [PubMed] [Google Scholar]
- Bundo M, Toyoshima M, Okada Y, Akamatsu W, Ueda J, Nemoto-Miyauchi T, Sunaga F, Toritsuka M, Ikawa D, Kakita A, et al. 2014. Increased l1 retrotransposition in the neuronal genome in schizophrenia. Neuron 81: 306–313. [DOI] [PubMed] [Google Scholar]
- Cabili MN, Trapnell C, Goff L, Koziol M, Tazon-Vega B, Regev A, Rinn JL 2011. Integrative annotation of human large intergenic noncoding RNAs reveals global properties and specific subclasses. Genes Dev 25: 1915–1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cairns-Smith AG 1982. Genetic takeover and the mineral origins of life. Cambridge University Press, Cambridge. [Google Scholar]
- Camacho-Vanegas O, Camacho SC, Till J, Miranda-Lorenzo I, Terzo E, Ramirez MC, Schramm V, Cordovano G, Watts G, Mehta S, et al. 2012. Primate genome gain and loss: A bone dysplasia, muscular dystrophy, and bone cancer syndrome resulting from mutated retroviral-derived MTAP transcripts. Am J Hum Genet 90: 614–627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carninci P 2009. Is sequencing enlightenment ending the dark age of the transcriptome? Nat Methods 6: 711–713. [DOI] [PubMed] [Google Scholar]
- Carninci P 2010. RNA dust: Where are the genes? DNA Res 17: 51–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carninci P, Kasukawa T, Katayama S, Gough J, Frith MC, Maeda N, Oyama R, Ravasi T, Lenhard B, Wells C, et al. 2005. The transcriptional landscape of the mammalian genome. Science 309: 1559–1563. [DOI] [PubMed] [Google Scholar]
- Carrieri C, Cimatti L, Biagioli M, Beugnet A, Zucchelli S, Fedele S, Pesce E, Ferrer I, Collavin L, Santoro C, et al. 2012. Long non-coding antisense RNA controls Uchl1 translation through an embedded SINEB2 repeat. Nature 491: 454–457. [DOI] [PubMed] [Google Scholar]
- Carvunis AR, Rolland T, Wapinski I, Calderwood MA, Yildirim MA, Simonis N, Charloteaux B, Hidalgo CA, Barbette J, Santhanam B, et al. 2012. Proto-genes and de novo gene birth. Nature 487: 370–374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavaille J, Buiting K, Kiefmann M, Lalande M, Brannan CI, Horsthemke B, Bachellerie JP, Brosius J, Huttenhofer A 2000. Identification of brain-specific and imprinted small nucleolar RNA genes exhibiting an unusual genomic organization. Proc Natl Acad Sci 97: 14311–14316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavalier-Smith T 1991. Intron phylogeny: A new hypothesis. Trends Genet 7: 145–148. [PubMed] [Google Scholar]
- Cech TR 2012. The RNA worlds in context. Cold Spring Harb Perspect Biol 4: a006742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S, Krinsky BH, Long M 2013. New genes as drivers of phenotypic evolution. Nat Rev Genet 14: 645–660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng J, Kapranov P, Drenkow J, Dike S, Brubaker S, Patel S, Long J, Stern D, Tammana H, Helt G, et al. 2005. Transcriptional maps of 10 human chromosomes at 5-nucleotide resolution. Science 308: 1149–1154. [DOI] [PubMed] [Google Scholar]
- Church DM, Goodstadt L, Hillier LW, Zody MC, Goldstein S, She X, Bult CJ, Agarwala R, Cherry JL, DiCuccio M, et al. 2009. Lineage-specific biology revealed by a finished genome assembly of the mouse. PLoS Biol 7: e1000112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clamp M, Fry B, Kamal M, Xie X, Cuff J, Lin MF, Kellis M, Lindblad-Toh K, Lander ES 2007. Distinguishing protein-coding and noncoding genes in the human genome. Proc Natl Acad Sci 104: 19428–19433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark MB, Amaral PP, Schlesinger FJ, Dinger ME, Taft RJ, Rinn JL, Ponting CP, Stadler PF, Morris KV, Morillon A, et al. 2011. The reality of pervasive transcription. PLoS Biol 9: e1000625; discussion e1001102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cloutier SC, Wang S, Ma WK, Petell CJ, Tran EJ 2013. Long noncoding RNAs promote transcriptional poising of inducible genes. PLoS Biol 11: e1001715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa FF 2010. Non-coding RNAs: Meet thy masters. BioEssays 32: 599–608. [DOI] [PubMed] [Google Scholar]
- Coufal NG, Garcia-Perez JL, Peng GE, Yeo GW, Mu Y, Lovci MT, Morell M, O’Shea KS, Moran JV, Gage FH 2009. L1 retrotransposition in human neural progenitor cells. Nature 460: 1127–1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couzin J 2002. Breakthrough of the year. Small RNAs make big splash. Science 298: 2296–2297. [DOI] [PubMed] [Google Scholar]
- Crick FH 1958. On protein synthesis. Symp Soc Exp Biol 12: 138–163. [PubMed] [Google Scholar]
- Crick FH 1968. The origin of the genetic code. J Mol Biol 38: 367–379. [DOI] [PubMed] [Google Scholar]
- Crick F 1970. Central dogma of molecular biology. Nature 227: 561–563. [DOI] [PubMed] [Google Scholar]
- Cullen BR, Lomedico PT, Ju G 1984. Transcriptional interference in avian retroviruses—Implications for the promoter insertion model of leukaemogenesis. Nature 307: 241–245. [DOI] [PubMed] [Google Scholar]
- Darnell JE, Doolittle WF 1986. Speculations on the early course of evolution. Proc Natl Acad Sci 83: 1271–1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deamer D 2005. A giant step towards artificial life? Trends Biotechnol 23: 336–338. [DOI] [PubMed] [Google Scholar]
- DeChiara TM, Brosius J 1987. Neural BC1 RNA: cDNA clones reveal nonrepetitive sequence content. Proc Natl Acad Sci 84: 2624–2628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Koning AP, Gu W, Castoe TA, Batzer MA, Pollock DD 2011. Repetitive elements may comprise over two-thirds of the human genome. PLoS Genet 7: e1002384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derrien T, Johnson R, Bussotti G, Tanzer A, Djebali S, Tilgner H, Guernec G, Martin D, Merkel A, Knowles DG, et al. 2012. The GENCODE v7 catalog of human long noncoding RNAs: Analysis of their gene structure, evolution, and expression. Genome Res 22: 1775–1789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Souza FS, Franchini LF, Rubinstein M. 2013. Exaptation of transposable elements into novel cis-regulatory elements: Is the evidence always strong? Mol Biol Evol 30: 1239–1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dieci G, Conti A, Pagano A, Carnevali D 2013. Identification of RNA polymerase III-transcribed genes in eukaryotic genomes. Biochim Biophys Acta 1829: 296–305. [DOI] [PubMed] [Google Scholar]
- Ding F, Li HH, Zhang S, Solomon NM, Camper SA, Cohen P, Francke U 2008. SnoRNA Snord116 (Pwcr1/MBII-85) deletion causes growth deficiency and hyperphagia in mice. PloS ONE 3: e1709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Ruscio A, Ebralidze AK, Benoukraf T, Amabile G, Goff LA, Terragni J, Figueroa ME, De Figueiredo Pontes LL, Alberich-Jorda M, Zhang P, et al. 2013. DNMT1-interacting RNAs block gene-specific DNA methylation. Nature 503: 371–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djebali S, Davis CA, Merkel A, Dobin A, Lassmann T, Mortazavi A, Tanzer A, Lagarde J, Lin W, Schlesinger F, et al. 2012. Landscape of transcription in human cells. Nature 489: 101–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doolittle WF 2013. Is junk DNA bunk? A critique of ENCODE. Proc Natl Acad Sci 110: 5294–5300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doudna JA, Szostak JW 1989. RNA-catalysed synthesis of complementary-strand RNA. Nature 339: 519–522. [DOI] [PubMed] [Google Scholar]
- Du ZQ, Yang CX, Rothschild MF, Ross JW 2013. Novel microRNA families expanded in the human genome. BMC Genomics 14: 98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunoyer P, Himber C, Ruiz-Ferrer V, Alioua A, Voinnet O 2007. Intra- and intercellular RNA interference in Arabidopsis thaliana requires components of the microRNA and heterochromatic silencing pathways. Nat Genet 39: 848–856. [DOI] [PubMed] [Google Scholar]
- Duret L, Chureau C, Samain S, Weissenbach J, Avner P 2006. The Xist RNA gene evolved in eutherians by pseudogenization of a protein-coding gene. Science 312: 1653–1655. [DOI] [PubMed] [Google Scholar]
- Eigen M, Schuster P 1977. The hypercycle. A principle of natural self-organization: Part A. Emergence of the hypercycle. Naturwissenschaften 64: 541–565. [DOI] [PubMed] [Google Scholar]
- Eigen M, Schuster P 1978. The hypercycle. A principle of natural self-organization: Part C. The realistic hypercycle. Naturwissenschaften 65: 341–369. [DOI] [PubMed] [Google Scholar]
- Ellington AD, Szostak JW 1990. In vitro selection of RNA molecules that bind specific ligands. Nature 346: 818–822. [DOI] [PubMed] [Google Scholar]
- Ellison CE, Bachtrog D 2013. Dosage compensation via transposable element mediated rewiring of a regulatory network. Science 342: 846–850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erdmann VA, Barciszewski J 2012. From nucleic acids sequences to molecular medicine. Springer, Berlin. [Google Scholar]
- Erdmann VA, Wolters J 1987. The Berlin RNA Databank. Protein Seq Data Anal 1: 127. [PubMed] [Google Scholar]
- Esguerra J, Warringer J, Blomberg A 2008. Functional importance of individual rRNA 2′-O-ribose methylations revealed by high-resolution phenotyping. RNA 14: 649–656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espinoza CA, Allen TA, Hieb AR, Kugel JF, Goodrich JA 2004. B2 RNA binds directly to RNA polymerase II to repress transcript synthesis. Nat Struct Mol Biol 11: 822–829. [DOI] [PubMed] [Google Scholar]
- Espinoza CA, Goodrich JA, Kugel JF 2007. Characterization of the structure, function, and mechanism of B2 RNA, an ncRNA repressor of RNA polymerase II transcription. RNA 13: 583–596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esteller M 2011. Non-coding RNAs in human disease. Nat Rev Genet 12: 861–874. [DOI] [PubMed] [Google Scholar]
- Eswaran J, Horvath A, Godbole S, Reddy SD, Mudvari P, Ohshiro K, Cyanam D, Nair S, Fuqua SA, Polyak K, et al. 2013. RNA sequencing of cancer reveals novel splicing alterations. Sci Rep 3: 1689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fatica A, Bozzoni I 2014. Long non-coding RNAs: New players in cell differentiation and development. Nat Rev Genet 15: 7–21. [DOI] [PubMed] [Google Scholar]
- Faulkner GJ, Kimura Y, Daub CO, Wani S, Plessy C, Irvine KM, Schroder K, Cloonan N, Steptoe AL, Lassmann T, et al. 2009. The regulated retrotransposon transcriptome of mammalian cells. Nat Genet 41: 563–571. [DOI] [PubMed] [Google Scholar]
- Ferat JL, Michel F 1993. Group II self-splicing introns in bacteria. Nature 364: 358–361. [DOI] [PubMed] [Google Scholar]
- Feschotte C 2008. Transposable elements and the evolution of regulatory networks. Nat Rev Genet 9: 397–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feuchter A, Mager D 1990. Functional heterogeneity of a large family of human LTR-like promoters and enhancers. Nucleic Acids Res 18: 1261–1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fica SM, Tuttle N, Novak T, Li NS, Lu J, Koodathingal P, Dai Q, Staley JP, Piccirilli JA 2013. RNA catalyses nuclear pre-mRNA splicing. Nature 503: 229–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filipowicz W 2000. Imprinted expression of small nucleolar RNAs in brain: Time for RNomics. Proc Nat Acad Sci 97: 14035–14037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fink GR 1987. Pseudogenes in yeast? Cell 49: 5–6. [DOI] [PubMed] [Google Scholar]
- Finta C, Zaphiropoulos PG 2000. The human cytochrome P450 3A locus. Gene evolution by capture of downstream exons. Gene 260: 13–23. [DOI] [PubMed] [Google Scholar]
- Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC 1998. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature 391: 806–811. [DOI] [PubMed] [Google Scholar]
- Frith MC, Forrest AR, Nourbakhsh E, Pang KC, Kai C, Kawai J, Carninci P, Hayashizaki Y, Bailey TL, Grimmond SM 2006a. The abundance of short proteins in the mammalian proteome. PLoS Genet 2: e52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frith MC, Wilming LG, Forrest A, Kawaji H, Tan SL, Wahlestedt C, Bajic VB, Kai C, Kawai J, Carninci P, et al. 2006b. Pseudo-messenger RNA: Phantoms of the transcriptome. PLoS Genet 2: e23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galindo MI, Pueyo JI, Fouix S, Bishop SA, Couso JP 2007. Peptides encoded by short ORFs control development and define a new eukaryotic gene family. PLoS Biol 5: e106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganten D, Nesse R 2012. The evolution of evolutionary molecular medicine: Genomics are transforming evolutionary biology into a science with new importance for modern medicine. J Mol Med (Berl) 90: 467–470. [DOI] [PubMed] [Google Scholar]
- Gebetsberger J, Polacek N 2013. Slicing tRNAs to boost functional ncRNA diversity. RNA Biol 10: 1798–1806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geisler S, Coller J 2013. RNA in unexpected places: Long non-coding RNA functions in diverse cellular contexts. Nat Rev Mol Cell Biol 14: 699–712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghabrial A 2007. Coding RNAs: Separating the grain from the chaff. Nat Cell Biol 9: 617–619. [DOI] [PubMed] [Google Scholar]
- Gilbert W 1986. Origin of life—The RNA world. Nature 319: 618. [Google Scholar]
- Gold L, Janjic N, Jarvis T, Schneider D, Walker JJ, Wilcox SK, Zichi D 2012. Aptamers and the RNA world, past and present. Cold Spring Harb Perspect Biol 4: a003582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldman A, Capoano CA, Gonzalez-Lopez E, Geisinger A 2014. Identifier (ID) elements are not preferentially located to brain-specific genes: High ID element representation in other tissue-specific- and housekeeping genes of the rat. Gene 533: 72–77. [DOI] [PubMed] [Google Scholar]
- Gomes AQ, Nolasco S, Soares H 2013. Non-coding RNAs: Multi-tasking molecules in the cell. Int J Mol Sci 14: 16010–16039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong C, Maquat LE 2011. lncRNAs transactivate STAU1-mediated mRNA decay by duplexing with 3′ UTRs via Alu elements. Nature 470: 284–288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gould SJ 2002. The structure of evolutionary theory. Belknap, Harvard University Press, Cambridge, MA. [Google Scholar]
- Graur D, Zheng Y, Price N, Azevedo RB, Zufall RA, Elhaik E 2013. On the immortality of television sets: “Function” in the human genome according to the evolution-free gospel of ENCODE. Genome Biol Evol 5: 578–590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greider CW, Blackburn EH 1989. A telomeric sequence in the RNA of Tetrahymena telomerase required for telomere repeat synthesis. Nature 337: 331–337. [DOI] [PubMed] [Google Scholar]
- Grote P, Wittler L, Hendrix D, Koch F, Wahrisch S, Beisaw A, Macura K, Blass G, Kellis M, Werber M, et al. 2013. The tissue-specific lncRNA Fendrr is an essential regulator of heart and body wall development in the mouse. Dev Cell 24: 206–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerrier-Takada C, Gardiner K, Marsh T, Pace N, Altman S 1983. The RNA moiety of ribonuclease P is the catalytic subunit of the enzyme. Cell 35: 849–857. [DOI] [PubMed] [Google Scholar]
- Guthrie C 1991. Messenger RNA splicing in yeast: Clues to why the spliceosome is a ribonucleoprotein. Science 253: 157–163. [DOI] [PubMed] [Google Scholar]
- Guttman M, Amit I, Garber M, French C, Lin MF, Feldser D, Huarte M, Zuk O, Carey BW, Cassady JP, et al. 2009. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature 458: 223–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guttman M, Russell P, Ingolia NT, Weissman JS, Lander ES 2013. Ribosome profiling provides evidence that large noncoding RNAs do not encode proteins. Cell 154: 240–251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas BJ, Chin M, Nusbaum C, Birren BW, Livny J 2012. How deep is deep enough for RNA-Seq profiling of bacterial transcriptomes? BMC Genomics 13: 734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadjiargyrou M, Delihas N 2013. The intertwining of transposable elements and non-coding RNAs. Int J Mol Sci 14: 13307–13328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haldane JBS 1933. The part played by recurrent mutation in evolution. Am Nat 67: 5–9. [Google Scholar]
- Hanada K, Zhang X, Borevitz JO, Li WH, Shiu SH 2007. A large number of novel coding small open reading frames in the intergenic regions of the Arabidopsis thaliana genome are transcribed and/or under purifying selection. Genome Res 17: 632–640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanczyc MM, Dorit RL 1998. Experimental evolution of complexity: In vitro emergence of intermolecular ribozyme interactions. RNA 4: 268–275. [PMC free article] [PubMed] [Google Scholar]
- Hansen TB, Kjems J, Damgaard CK 2013. Circular RNA and miR-7 in cancer. Cancer Res 73: 5609–5612. [DOI] [PubMed] [Google Scholar]
- Harris H 1965. The short-lived RNA in the cell nucleus and its possible role in evolution. In Evolving genes and proteins (ed. Bryson V, Vogel HJ), pp. 469–500 Academic, New York. [Google Scholar]
- Harris H 2013. History: Non-coding RNA foreseen 48 years ago. Nature 497: 188. [DOI] [PubMed] [Google Scholar]
- Hashimoto Y, Kondo T, Kageyama Y 2008. Lilliputians get into the limelight: Novel class of small peptide genes in morphogenesis. Dev Growth Differ 50: S269–276. [DOI] [PubMed] [Google Scholar]
- Hayden EJ, Ferrada E, Wagner A 2011. Cryptic genetic variation promotes rapid evolutionary adaptation in an RNA enzyme. Nature 474: 92–95. [DOI] [PubMed] [Google Scholar]
- Heinen TJ, Staubach F, Haming D, Tautz D 2009. Emergence of a new gene from an intergenic region. Curr Biol 19: 1527–1531. [DOI] [PubMed] [Google Scholar]
- Hiller M, Schaar BT, Bejerano G 2012. Hundreds of conserved non-coding genomic regions are independently lost in mammals. Nucleic Acids Res 40: 11463–11476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho DW, Yang ZF, Yi K, Lam CT, Ng MN, Yu WC, Lau J, Wan T, Wang X, Yan Z, et al. 2012. Gene expression profiling of liver cancer stem cells by RNA-sequencing. PloS ONE 7: e37159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman Y, Dahary D, Bublik DR, Oren M, Pilpel Y 2013. The majority of endogenous microRNA targets within Alu elements avoid the microRNA machinery. Bioinformatics 29: 894–902. [DOI] [PubMed] [Google Scholar]
- Holdt LM, Hoffmann S, Sass K, Langenberger D, Scholz M, Krohn K, Finstermeier K, Stahringer A, Wilfert W, Beutner F, et al. 2013. Alu elements in ANRIL non-coding RNA at chromosome 9p21 modulate atherogenic cell functions through trans-regulation of gene networks. PLoS Genet 9: e1003588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu W, Alvarez-Dominguez JR, Lodish HF 2012. Regulation of mammalian cell differentiation by long non-coding RNAs. EMBO Rep 13: 971–983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y, Huang Y, Du Y, Orellana CF, Singh D, Johnson AR, Monroy A, Kuan PF, Hammond SM, Makowski L, et al. 2013. DiffSplice: The genome-wide detection of differential splicing events with RNA-seq. Nucleic Acids Res 41: e39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huttenhofer A, Kiefmann M, Meier-Ewert S, O’Brien J, Lehrach H, Bachellerie JP, Brosius J 2001. RNomics: An experimental approach that identifies 201 candidates for novel, small, non-messenger RNAs in mouse. EMBO J 20: 2943–2953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huttenhofer A, Schattner P, Polacek N 2005. Non-coding RNAs: Hope or hype? Trends Genet 21: 289–297. [DOI] [PubMed] [Google Scholar]
- Iacoangeli A, Tiedge H 2013. Translational control at the synapse: Role of RNA regulators. Trends Biochem Sci 38: 47–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingolia NT, Lareau LF, Weissman JS 2011. Ribosome profiling of mouse embryonic stem cells reveals the complexity and dynamics of mammalian proteomes. Cell 147: 789–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingolia NT, Brar GA, Rouskin S, McGeachy AM, Weissman JS 2013. Genome-wide annotation and quantitation of translation by ribosome profiling. Current protocols in molecular biology (ed. Ausubel FM, et al. ), Chap. 4, Unit 4 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- International Human Genome Sequencing Consortium. 2004. Finishing the euchromatic sequence of the human genome. Nature 431: 931–945. [DOI] [PubMed] [Google Scholar]
- Jacquier A 2009. The complex eukaryotic transcriptome: Unexpected pervasive transcription and novel small RNAs. Nat Rev Genet 10: 833–844. [DOI] [PubMed] [Google Scholar]
- Jady BE, Ketele A, Kiss T 2012. Human intron-encoded Alu RNAs are processed and packaged into Wdr79-associated nucleoplasmic box H/ACA RNPs. Genes Dev 26: 1897–1910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen TH, Jacquier A, Libri D 2013. Dealing with pervasive transcription. Mol Cell 52: 473–484. [DOI] [PubMed] [Google Scholar]
- Johnsson P, Ackley A, Vidarsdottir L, Lui WO, Corcoran M, Grander D, Morris KV 2013. A pseudogene long-noncoding-RNA network regulates PTEN transcription and translation in human cells. Nat Struct Mol Biol 20: 440–446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston WK, Unrau PJ, Lawrence MS, Glasner ME, Bartel DP 2001. RNA-catalyzed RNA polymerization: Accurate and general RNA-templated primer extension. Science 292: 1319–1325. [DOI] [PubMed] [Google Scholar]
- Jordan IK, Rogozin IB, Glazko GV, Koonin EV 2003. Origin of a substantial fraction of human regulatory sequences from transposable elements. Trends Genet 19: 68–72. [DOI] [PubMed] [Google Scholar]
- Joyce GF, Schwartz AW, Miller SL, Orgel LE 1987. The case for an ancestral genetic system involving simple analogues of the nucleotides. Proc Natl Acad Sci 84: 4398–4402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaessmann H 2010. Origins, evolution, and phenotypic impact of new genes. Genome Res 20: 1313–1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapitonov VV, Jurka J 2005. RAG1 core and V(D)J recombination signal sequences were derived from Transib transposons. PLoS Biol 3: e181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapranov P, St Laurent G 2012. Dark matter RNA: Existence, function, and controversy. Front Genet 3: 60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapranov P, Ozsolak F, Milos PM 2012. Profiling of short RNAs using Helicos single-molecule sequencing. Methods Mol Biol 822: 219–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapusta A, Kronenberg Z, Lynch VJ, Zhuo X, Ramsay L, Bourque G, Yandell M, Feschotte C 2013. Transposable elements are major contributors to the origin, diversification, and regulation of vertebrate long noncoding RNAs. PLoS Genet 9: e1003470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kastenmayer JP, Ni L, Chu A, Kitchen LE, Au WC, Yang H, Carter CD, Wheeler D, Davis RW, Boeke JD, et al. 2006. Functional genomics of genes with small open reading frames (sORFs) in S. cerevisiae. Genome Res 16: 365–373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khayrullina GA, Raabe CA, Hoe CH, Becker K, Reinhardt R, Tang TH, Rozhdestvensky TS, Kopylov AM 2012. Transcription analysis and small non-protein coding RNAs associated with bacterial ribosomal protein operons. Curr Med Chem 19: 5187–5198. [DOI] [PubMed] [Google Scholar]
- Kim J, Martignetti JA, Shen MR, Brosius J, Deininger P 1994. Rodent BC1 RNA gene as a master gene for ID element amplification. Proc Natl Acad Sci 91: 3607–3611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King TH, Liu B, McCully RR, Fournier MJ 2003. Ribosome structure and activity are altered in cells lacking snoRNPs that form pseudouridines in the peptidyl transferase center. Mol Cell 11: 425–435. [DOI] [PubMed] [Google Scholar]
- Kishore S, Khanna A, Zhang Z, Hui J, Balwierz PJ, Stefan M, Beach C, Nicholls RD, Zavolan M, Stamm S 2010. The snoRNA MBII-52 (SNORD 115) is processed into smaller RNAs and regulates alternative splicing. Hum Mol Genet 19: 1153–1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koonin EV 2010. Taming of the shrewd: Novel eukaryotic genes from RNA viruses. BMC Biol 8: 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornienko AE, Guenzl PM, Barlow DP, Pauler FM 2013. Gene regulation by the act of long non-coding RNA transcription. BMC Biol 11: 59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kowalczyk MS, Higgs DR, Gingeras TR 2012. Molecular biology: RNA discrimination. Nature 482: 310–311. [DOI] [PubMed] [Google Scholar]
- Kriegs JO, Schmitz J, Makalowski W, Brosius J 2005. Does the AD7c-NTP locus encode a protein? Biochim Biophys Acta 1727: 1–4. [DOI] [PubMed] [Google Scholar]
- Kruger K, Grabowski PJ, Zaug AJ, Sands J, Gottschling DE, Cech TR 1982. Self-splicing RNA: Autoexcision and autocyclization of the ribosomal RNA intervening sequence of Tetrahymena. Cell 31: 147–157. [DOI] [PubMed] [Google Scholar]
- Krull M, Brosius J, Schmitz J 2005. Alu-SINE exonization: En route to protein-coding function. Mol Biol Evol 22: 1702–1711. [DOI] [PubMed] [Google Scholar]
- Krull M, Petrusma M, Makalowski W, Brosius J, Schmitz J 2007. Functional persistence of exonized mammalian-wide interspersed repeat elements (MIRs). Genome Res 17: 1139–1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kung JT, Colognori D, Lee JT 2013. Long noncoding RNAs: Past, present, and future. Genetics 193: 651–669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagos-Quintana M, Rauhut R, Lendeckel W, Tuschl T 2001. Identification of novel genes coding for small expressed RNAs. Science 294: 853–858. [DOI] [PubMed] [Google Scholar]
- Lambowitz AM, Zimmerly S 2011. Group II introns: Mobile ribozymes that invade DNA. Cold Spring Harb Perspect Biol 3: a003616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau NC, Lim LP, Weinstein EG, Bartel DP 2001. An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegans. Science 294: 858–862. [DOI] [PubMed] [Google Scholar]
- Lee CA, Fournier MJ, Beckwith J 1985. Escherichia coli 6S RNA is not essential for growth or protein secretion. J Bacteriol 161: 1156–1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YS, Shibata Y, Malhotra A, Dutta A 2009. A novel class of small RNAs: tRNA-derived RNA fragments (tRFs). Genes Dev 23: 2639–2649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lerner MR, Boyle JA, Mount SM, Wolin SL, Steitz JA 1980. Are snRNPs involved in splicing? Nature 283: 220–224. [DOI] [PubMed] [Google Scholar]
- Leslie M 2013a. Cell Biology. NIH effort gambles on mysterious extracellular RNAs. Science 341: 947. [DOI] [PubMed] [Google Scholar]
- Leslie M 2013b. Cell biology. The immune system's compact genomic counterpart. Science 339: 25–27. [DOI] [PubMed] [Google Scholar]
- Lev-Maor G, Sorek R, Shomron N, Ast G 2003. The birth of an alternatively spliced exon: 3′ splice-site selection in Alu exons. Science 300: 1288–1291. [DOI] [PubMed] [Google Scholar]
- Lewejohann L, Skryabin BV, Sachser N, Prehn C, Heiduschka P, Thanos S, Jordan U, Dell’Omo G, Vyssotski AL, Pleskacheva MG, et al. 2004. Role of a neuronal small non-messenger RNA: Behavioural alterations in BC1 RNA-deleted mice. Behav Brain Res 154: 273–289. [DOI] [PubMed] [Google Scholar]
- Lewis EB 1951. Pseudoallelism and gene evolution. Cold Spring Harb Symp Quant Biol 16: 159–174. [DOI] [PubMed] [Google Scholar]
- Li HD, Zagorski J, Fournier MJ 1990. Depletion of U14 small nuclear RNA (snR128) disrupts production of 18S rRNA in Saccharomyces cerevisiae. Mol Cell Biol 10: 1145–1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Liu B, Wapinski OL, Tsai MC, Qu K, Zhang J, Carlson JC, Lin M, Fang F, Gupta RA, et al. 2013. Targeted disruption of Hotair leads to homeotic transformation and gene derepression. Cell Rep 5: 3–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang KH, Yeh CT 2013. A gene expression restriction network mediated by sense and antisense Alu sequences located on protein-coding messenger RNAs. BMC Genomics 14: 325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lincoln TA, Joyce GF 2009. Self-sustained replication of an RNA enzyme. Science 323: 1229–1232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindblad-Toh K, Garber M, Zuk O, Lin MF, Parker BJ, Washietl S, Kheradpour P, Ernst J, Jordan G, Mauceli E, et al. 2011. A high-resolution map of human evolutionary constraint using 29 mammals. Nature 478: 476–482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long M, Betran E, Thornton K, Wang W 2003. The origin of new genes: Glimpses from the young and old. Nat Rev Genet 4: 865–875. [DOI] [PubMed] [Google Scholar]
- Long M, VanKuren NW, Chen S, Vibranowski MD 2013. New gene evolution: Little did we know. Annu Rev Genet 47: 307–333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowe CB, Haussler D 2012. 29 mammalian genomes reveal novel exaptations of mobile elements for likely regulatory functions in the human genome. PloS ONE 7: e43128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lunyak VV, Prefontaine GG, Nunez E, Cramer T, Ju BG, Ohgi KA, Hutt K, Roy R, Garcia-Diaz A, Zhu X, et al. 2007. Developmentally regulated activation of a SINE B2 repeat as a domain boundary in organogenesis. Science 317: 248–251. [DOI] [PubMed] [Google Scholar]
- Lynch M, Force A 2000. The probability of duplicate gene preservation by subfunctionalization. Genetics 154: 459–473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch VJ, Leclerc RD, May G, Wagner GP 2011. Transposon-mediated rewiring of gene regulatory networks contributed to the evolution of pregnancy in mammals. Nat Genet 43: 1154–1159. [DOI] [PubMed] [Google Scholar]
- Magny EG, Pueyo JI, Pearl FM, Cespedes MA, Niven JE, Bishop SA, Couso JP 2013. Conserved regulation of cardiac calcium uptake by peptides encoded in small open reading frames. Science 341: 1116–1120. [DOI] [PubMed] [Google Scholar]
- Maizels N, Weiner AM 1987. Peptide-specific ribosomes, genomic tags, and the origin of the genetic code. Cold Spring Harb Symp Quant Biol 52: 743–749. [DOI] [PubMed] [Google Scholar]
- Makalowski W, Mitchell GA, Labuda D 1994. Alu sequences in the coding regions of mRNA: A source of protein variability. Trends Genet 10: 188–193. [DOI] [PubMed] [Google Scholar]
- Managadze D, Lobkovsky AE, Wolf YI, Shabalina SA, Rogozin IB, Koonin EV 2013. The vast, conserved mammalian lincRNome. PLoS Comput Biol 9: e1002917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandal AK, Pandey R, Jha V, Mukerji M 2013. Transcriptome-wide expansion of non-coding regulatory switches: Evidence from co-occurrence of Alu exonization, antisense and editing. Nucleic Acids Res 41: 2121–2137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansy SS, Schrum JP, Krishnamurthy M, Tobe S, Treco DA, Szostak JW 2008. Template-directed synthesis of a genetic polymer in a model protocell. Nature 454: 122–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marahrens Y, Panning B, Dausman J, Strauss W, Jaenisch R 1997. Xist-deficient mice are defective in dosage compensation but not spermatogenesis. Genes Dev 11: 156–166. [DOI] [PubMed] [Google Scholar]
- Mariner PD, Walters RD, Espinoza CA, Drullinger LF, Wagner SD, Kugel JF, Goodrich JA 2008. Human Alu RNA is a modular transacting repressor of mRNA transcription during heat shock. Mol Cell 29: 499–509. [DOI] [PubMed] [Google Scholar]
- Marques AC, Ponting CP 2009. Catalogues of mammalian long noncoding RNAs: Modest conservation and incompleteness. Genome Biol 10: R124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martens JA, Laprade L, Winston F 2004. Intergenic transcription is required to repress the Saccharomyces cerevisiae SER3 gene. Nature 429: 571–574. [DOI] [PubMed] [Google Scholar]
- Martignetti JA, Brosius J 1993. Neural BC1 RNA as an evolutionary marker: Guinea pig remains a rodent. Proc Natl Acad Sci 90: 9698–9702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin W, Koonin EV 2006. Introns and the origin of nucleus-cytosol compartmentalization. Nature 440: 41–45. [DOI] [PubMed] [Google Scholar]
- Mast CB, Schink S, Gerland U, Braun D 2013. Escalation of polymerization in a thermal gradient. Proc Natl Acad Sci 110: 8030–8035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattick JS 1994. Introns: Evolution and function. Curr Opin Genet Dev 4: 823–831. [DOI] [PubMed] [Google Scholar]
- Mattick JS 2003. Challenging the dogma: The hidden layer of non-protein-coding RNAs in complex organisms. BioEssays 25: 930–939. [DOI] [PubMed] [Google Scholar]
- Mattick JS, Dinger ME 2013. The extent of functionality in the human genome. HUGO J 7: 2. [Google Scholar]
- Maynard Smith J, Szathmáry E 1995. The major transitions in evolution. Oxford University Press, Oxford. [Google Scholar]
- McKinnon RD, Shinnick TM, Sutcliffe JG 1986. The neuronal identifier element is a cis-acting positive regulator of gene expression. Proc Natl Acad Sci 83: 3751–3755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medstrand P, van de Lagemaat LN, Dunn CA, Landry JR, Svenback D, Mager DL 2005. Impact of transposable elements on the evolution of mammalian gene regulation. Cytogenet Genome Res 110: 342–352. [DOI] [PubMed] [Google Scholar]
- Memczak S, Jens M, Elefsinioti A, Torti F, Krueger J, Rybak A, Maier L, Mackowiak SD, Gregersen LH, Munschauer M, et al. 2013. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature 495: 333–338. [DOI] [PubMed] [Google Scholar]
- Mercer TR, Gerhardt DJ, Dinger ME, Crawford J, Trapnell C, Jeddeloh JA, Mattick JS, Rinn JL 2012. Targeted RNA sequencing reveals the deep complexity of the human transcriptome. Nat Biotechnol 30: 99–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milner RJ, Bloom FE, Lai C, Lerner RA, Sutcliffe JG 1984. Brain-specific genes have identifier sequences in their introns. Proc Natl Acad Sci 81: 713–717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mo D, Raabe CA, Reinhardt R, Brosius J, Rozhdestvensky TS 2013. Alternative processing as evolutionary mechanism for the origin of novel non-protein coding RNAs. Genome Biol Evol 5: 2061–2071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monte SM, Ghanbari K, Frey WH, Beheshti I, Averback P, Hauser SL, Ghanbari HA, Wands JR 1997. Characterization of the AD7C-NTP cDNA expression in Alzheimer’s disease and measurement of a 41-kD protein in cerebrospinal fluid. J Clin Invest 100: 3093–3104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moss EG 2001. RNA interference: It’s a small RNA world. Curr Biol 11: R772–R775. [DOI] [PubMed] [Google Scholar]
- Mudge JM, Frankish A, Harrow J 2013. Functional transcriptomics in the post-ENCODE era. Genome Res 23: 1961–1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller HJ 1935. The origination of chromatin deficiencies as minute deletions subject to insertion elsewhere. Genetics 17: 237–252. [Google Scholar]
- Muotri AR, Chu VT, Marchetto MC, Deng W, Moran JV, Gage FH 2005. Somatic mosaicism in neuronal precursor cells mediated by L1 retrotransposition. Nature 435: 903–910. [DOI] [PubMed] [Google Scholar]
- Murphy DN, McLysaght A 2012. De novo origin of protein-coding genes in murine rodents. PloS ONE 7: e48650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Necsulea A, Soumillon M, Warnefors M, Liechti A, Daish T, Zeller U, Baker JC, Grutzner F, Kaessmann H 2014. The evolution of lncRNA repertoires and expression patterns in tetrapods. Nature 505: 635–640. [DOI] [PubMed] [Google Scholar]
- Nei M 1969. Gene duplication and nucleotide substitution in evolution. Nature 221: 40–42. [DOI] [PubMed] [Google Scholar]
- Neil H, Malabat C, d’Aubenton-Carafa Y, Xu Z, Steinmetz LM, Jacquier A 2009. Widespread bidirectional promoters are the major source of cryptic transcripts in yeast. Nature 457: 1038–1042. [DOI] [PubMed] [Google Scholar]
- Nekrutenko A, Li WH 2001. Transposable elements are found in a large number of human protein-coding genes. Trends Genet 17: 619–621. [DOI] [PubMed] [Google Scholar]
- Nelson AC, Wardle FC 2013. Conserved non-coding elements and cis regulation: Actions speak louder than words. Development 140: 1385–1395. [DOI] [PubMed] [Google Scholar]
- Neme R, Tautz D 2013. Phylogenetic patterns of emergence of new genes support a model of frequent de novo evolution. BMC Genomics 14: 117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nesse RM, Ganten D, Gregory TR, Omenn GS 2012. Evolutionary molecular medicine. J Mol Med (Berl) 90: 509–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neveu M, Kim HJ, Benner SA 2013. The “strong” RNA world hypothesis: Fifty years old. Astrobiology 13: 391–403. [DOI] [PubMed] [Google Scholar]
- Nielsen MM, Tehler D, Vang S, Sudzina F, Hedegaard J, Nordentoft I, Orntoft TF, Lund AH, Pedersen JS 2014. Identification of expressed and conserved human noncoding RNAs. RNA 20: 236–251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nirenberg M, Leder P, Bernfield M, Brimacombe R, Trupin J, Rottman F, O’Neal C 1965. RNA codewords and protein synthesis: VII. On the general nature of the RNA code. Proc Natl Acad Sci 53: 1161–1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu DK, Jiang L 2013. Can ENCODE tell us how much junk DNA we carry in our genome? Biochem Biophys Res Commun 430: 1340–1343. [DOI] [PubMed] [Google Scholar]
- Noller HF 2012. Evolution of protein synthesis from an RNA world. Cold Spring Harb Perspect Biol 4: a003681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noller HF, Chaires JB 1972. Functional modification of 16S ribosomal RNA by kethoxal. Proc Natl Acad Sci 69: 3115–3118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noller HF, Hoffarth V, Zimniak L 1992. Unusual resistance of peptidyl transferase to protein extraction procedures. Science 256: 1416–1419. [DOI] [PubMed] [Google Scholar]
- Orgel LE 1968. Evolution of the genetic apparatus. J Mol Biol 38: 381–393. [DOI] [PubMed] [Google Scholar]
- Orom UA, Shiekhattar R 2013. Long noncoding RNAs usher in a new era in the biology of enhancers. Cell 154: 1190–1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owens GP, Chaudhari N, Hahn WE 1985. Brain “identifier sequence” is not restricted to brain: Similar abundance in nuclear RNA of other organs. Science 229: 1263–1265. [DOI] [PubMed] [Google Scholar]
- Parra G, Reymond A, Dabbouseh N, Dermitzakis ET, Castelo R, Thomson TM, Antonarakis SE, Guigo R 2006. Tandem chimerism as a means to increase protein complexity in the human genome. Genome Res 16: 37–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrat PN, DasGupta S, Wang J, Theurkauf W, Weng Z, Rosbash M, Waddell S 2013. Transposition-driven genomic heterogeneity in the Drosophila brain. Science 340: 91–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petherick A 2008. Genetics: The production line. Nature 454: 1042–1045. [DOI] [PubMed] [Google Scholar]
- Pink RC, Wicks K, Caley DP, Punch EK, Jacobs L, Carter DR 2011. Pseudogenes: Pseudo-functional or key regulators in health and disease? RNA 17: 792–798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polev D 2012. Transcriptional noise as a driver of gene evolution. J Theor Biol 293: 27–33. [DOI] [PubMed] [Google Scholar]
- Poliseno L, Salmena L, Zhang J, Carver B, Haveman WJ, Pandolfi PP 2010. A coding-independent function of gene and pseudogene mRNAs regulates tumour biology. Nature 465: 1033–1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponicsan SL, Houel S, Old WM, Ahn NG, Goodrich JA, Kugel JF 2013. The non-coding B2 RNA binds to the DNA cleft and active-site region of RNA polymerase II. J Mol Biol 425: 3625–3638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponting CP, Oliver PL, Reik W 2009. Evolution and functions of long noncoding RNAs. Cell 136: 629–641. [DOI] [PubMed] [Google Scholar]
- Poole A, Jeffares D, Penny D 1999. Early evolution: Prokaryotes, the new kids on the block. BioEssays 21: 880–889. [DOI] [PubMed] [Google Scholar]
- Powner MW, Gerland B, Sutherland JD 2009. Synthesis of activated pyrimidine ribonucleotides in prebiotically plausible conditions. Nature 459: 239–242. [DOI] [PubMed] [Google Scholar]
- Prasanth KV, Spector DL 2007. Eukaryotic regulatory RNAs: An answer to the “genome complexity” conundrum. Genes Dev 21: 11–42. [DOI] [PubMed] [Google Scholar]
- Prestayko AW, Busch H 1968. Low molecular weight RNA of the chromatin fraction from Novikoff hepatoma and rat liver nuclei. Biochim Biophys Acta 169: 327–337. [DOI] [PubMed] [Google Scholar]
- Reanney DC 1974. On the origin of prokaryotes. J Theor Biol 48: 243–251. [DOI] [PubMed] [Google Scholar]
- Rebollo R, Romanish MT, Mager DL 2012. Transposable elements: An abundant and natural source of regulatory sequences for host genes. Annu Rev Genet 46: 21–42. [DOI] [PubMed] [Google Scholar]
- Reilly MT, Faulkner GJ, Dubnau J, Ponomarev I, Gage FH 2013. The role of transposable elements in health and diseases of the central nervous system. J Neurosci 33: 17577–17586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rich A 1962. On the problems of evolution and biochemical information transfer. In Horizons in biochemistry (ed. Kasha M, Pullman B), pp. 103–126 Academic, New York. [Google Scholar]
- Richard P, Manley JL 2009. Transcription termination by nuclear RNA polymerases. Genes Dev 23: 1247–1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinn JL, Kertesz M, Wang JK, Squazzo SL, Xu X, Brugmann SA, Goodnough LH, Helms JA, Farnham PJ, Segal E, et al. 2007. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell 129: 1311–1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritz K, van Schaik BD, Jakobs ME, Aronica E, Tijssen MA, van Kampen AH, Baas F 2011. Looking ultra deep: Short identical sequences and transcriptional slippage. Genomics 98: 90–95. [DOI] [PubMed] [Google Scholar]
- Robertson MP, Joyce GF 2012. The origins of the RNA world. Cold Spring Harb Perspect Biol 4: a003608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson R 2010. Dark matter transcripts: Sound and fury, signifying nothing? PLoS Biol 8: e1000370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross RJ, Weiner MM, Lin H 2014. PIWI proteins and PIWI-interacting RNAs in the soma. Nature 505: 353–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth C, Rastogi S, Arvestad L, Dittmar K, Light S, Ekman D, Liberles DA 2007. Evolution after gene duplication: Models, mechanisms, sequences, systems, and organisms. J Exp Zool B Mol Dev Evol 308: 58–73. [DOI] [PubMed] [Google Scholar]
- Rudd KE, Humphery-Smith I, Wasinger VC, Bairoch A 1998. Low molecular weight proteins: A challenge for post-genomic research. Electrophoresis 19: 536–544. [DOI] [PubMed] [Google Scholar]
- Runte M, Huttenhofer A, Gross S, Kiefmann M, Horsthemke B, Buiting K 2001. The IC-SNURF-SNRPN transcript serves as a host for multiple small nucleolar RNA species and as an antisense RNA for UBE3A. Hum Mol Genet 10: 2687–2700. [DOI] [PubMed] [Google Scholar]
- Ruvkun G 2001. Molecular biology. Glimpses of a tiny RNA world. Science 294: 797–799. [DOI] [PubMed] [Google Scholar]
- Sabin LR, Delas MJ, Hannon GJ 2013. Dogma derailed: The many influences of RNA on the genome. Mol Cell 49: 783–794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sacerdote MG, Szostak JW 2005. Semipermeable lipid bilayers exhibit diastereoselectivity favoring ribose. Proc Natl Acad Sci 102: 6004–6008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salzman J, Gawad C, Wang PL, Lacayo N, Brown PO 2012. Circular RNAs are the predominant transcript isoform from hundreds of human genes in diverse cell types. PloS ONE 7: e30733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santangelo AM, de Souza FS, Franchini LF, Bumaschny VF, Low MJ, Rubinstein M 2007. Ancient exaptation of a CORE-SINE retroposon into a highly conserved mammalian neuronal enhancer of the proopiomelanocortin gene. PLoS Genet 3: 1813–1826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sapienza C, St-Jacques B 1986. “Brain- specific” transcription and evolution of the identifier sequence. Nature 319: 418–420. [DOI] [PubMed] [Google Scholar]
- Sasaki T, Nishihara H, Hirakawa M, Fujimura K, Tanaka M, Kokubo N, Kimura-Yoshida C, Matsuo I, Sumiyama K, Saitou N, et al. 2008. Possible involvement of SINEs in mammalian-specific brain formation. Proc Natl Acad Sci 105: 4220–4225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauvageau M, Goff LA, Lodato S, Bonev B, Groff AF, Gerhardinger C, Sanchez-Gomez DB, Hacisuleyman E, Li E, Spence M, et al. 2013. Multiple knockout mouse models reveal lincRNAs are required for life and brain development. eLife 2: e01749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savard J, Marques-Souza H, Aranda M, Tautz D 2006. A segmentation gene in tribolium produces a polycistronic mRNA that codes for multiple conserved peptides. Cell 126: 559–569. [DOI] [PubMed] [Google Scholar]
- Saxena A, Carninci P 2011a. Long non-coding RNA modifies chromatin: Epigenetic silencing by long non-coding RNAs. BioEssays 33: 830–839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saxena A, Carninci P 2011b. Whole transcriptome analysis: What are we still missing? Wiley Interdiscip Rev Syst Bio Med 3: 527–543. [DOI] [PubMed] [Google Scholar]
- Schartl M, Hornung U, Gutbrod H, Volff JN, Wittbrodt J 1999. Melanoma loss-of-function mutants in Xiphophorus caused by Xmrk-oncogene deletion and gene disruption by a transposable element. Genetics 153: 1385–1394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schonberg DL, Bao S, Rich JN 2013. Genomics informs glioblastoma biology. Nat Genet 45: 1105–1107. [DOI] [PubMed] [Google Scholar]
- Schöning K, Scholz P, Guntha S, Wu X, Krishnamurthy R, Eschenmoser A 2000. Chemical etiology of nucleic acid structure: The α-threofuranosyl-(3′→2′) oligonucleotide system. Science 290: 1347–1351. [DOI] [PubMed] [Google Scholar]
- Schorderet P, Duboule D 2011. Structural and functional differences in the long non-coding RNA hotair in mouse and human. PLoS Genet 7: e1002071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seila AC, Calabrese JM, Levine SS, Yeo GW, Rahl PB, Flynn RA, Young RA, Sharp PA 2008. Divergent transcription from active promoters. Science 322: 1849–1851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sen K, Ghosh TC 2013. Pseudogenes and their composers: Delving in the “debris” of human genome. Brief Funct Genomics 12: 536–547. [DOI] [PubMed] [Google Scholar]
- Shah SP, Roth A, Goya R, Oloumi A, Ha G, Zhao Y, Turashvili G, Ding J, Tse K, Haffari G, et al. 2012. The clonal and mutational evolution spectrum of primary triple-negative breast cancers. Nature 486: 395–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp PA 1991. Five easy pieces. Science 254: 663. [DOI] [PubMed] [Google Scholar]
- Shechner DM, Grant RA, Bagby SC, Koldobskaya Y, Piccirilli JA, Bartel DP 2009. Crystal structure of the catalytic core of an RNA-polymerase ribozyme. Science 326: 1271–1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen S, Lin L, Cai JJ, Jiang P, Kenkel EJ, Stroik MR, Sato S, Davidson BL, Xing Y 2011. Widespread establishment and regulatory impact of Alu exons in human genes. Proc Natl Acad Sci 108: 2837–2842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Y, Yue F, McCleary DF, Ye Z, Edsall L, Kuan S, Wagner U, Dixon J, Lee L, Lobanenkov VV, et al. 2012. A map of the cis-regulatory sequences in the mouse genome. Nature 488: 116–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sibilia M, Wagner EF 1995. Strain-dependent epithelial defects in mice lacking the EGF receptor. Science 269: 234–238. [DOI] [PubMed] [Google Scholar]
- Siepel A 2009. Darwinian alchemy: Human genes from noncoding DNA. Genome Res 19: 1693–1695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singer T, McConnell MJ, Marchetto MC, Coufal NG, Gage FH 2010. LINE-1 retrotransposons: Mediators of somatic variation in neuronal genomes? Trends Neurosci 33: 345–354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skryabin BV, Gubar LV, Seeger B, Pfeiffer J, Handel S, Robeck T, Karpova E, Rozhdestvensky TS, Brosius J 2007. Deletion of the MBII-85 snoRNA gene cluster in mice results in postnatal growth retardation. PLoS Genet 3: e235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Söll D, Ohtsuka E, Jones DS, Lohrmann R, Hayatsu H, Nishimura S, Khorana HG 1965. Studies on polynucleotides, XLIX. Stimulation of the binding of aminoacyl-sRNA’s to ribosomes by ribotrinucleotides and a survey of codon assignments for 20 amino acids. Proc Natl Acad Sci 54: 1378–1385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorek R, Ast G, Graur D 2002. Alu-containing exons are alternatively spliced. Genome Res 12: 1060–1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sousa C, Johansson C, Charon C, Manyani H, Sautter C, Kondorosi A, Crespi M 2001. Translational and structural requirements of the early nodulin gene enod40, a short-open reading frame-containing RNA, for elicitation of a cell-specific growth response in the alfalfa root cortex. Mol Cell Biol 21: 354–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spengler RM, Oakley CK, Davidson BL 2014. Functional microRNAs and target sites are created by lineage-specific transposition. Hum Mol Genet 23: 1783–1793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens SG 1951. Possible significances of duplication in evolution. Adv Genet 4: 247–265. [DOI] [PubMed] [Google Scholar]
- St Laurent G, Shtokalo D, Tackett MR, Yang Z, Eremina T, Wahlestedt C, Urcuqui-Inchima S, Seilheimer B, McCaffrey TA, Kapranov P 2012. Intronic RNAs constitute the major fraction of the non-coding RNA in mammalian cells. BMC Genomics 13: 504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Struhl K 2007. Transcriptional noise and the fidelity of initiation by RNA polymerase II. Nat Struct Mol Biol 14: 103–105. [DOI] [PubMed] [Google Scholar]
- Sutcliffe JG, Milner RJ, Gottesfeld JM, Lerner RA 1984a. Identifier sequences are transcribed specifically in brain. Nature 308: 237–241. [DOI] [PubMed] [Google Scholar]
- Sutcliffe JG, Milner RJ, Gottesfeld JM, Reynolds W 1984b. Control of neuronal gene expression. Science 225: 1308–1315. [DOI] [PubMed] [Google Scholar]
- Szathmáry E 2006. The origin of replicators and reproducers. Philos Trans R Soc Lond B Biol Sci 361: 1761–1776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szathmáry E, Smith JM 1995. The major evolutionary transitions. Nature 374: 227–232. [DOI] [PubMed] [Google Scholar]
- Szostak JW, Bartel DP, Luisi PL 2001. Synthesizing life. Nature 409: 387–390. [DOI] [PubMed] [Google Scholar]
- Takeuchi N, Hogeweg P 2008. Evolution of complexity in RNA-like replicator systems. Biol Direct 3: 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taleb NN 2010. The bed of Procrustes: Philosophical and practical aphorisms. Random House, New York. [Google Scholar]
- Tay Y, Rinn J, Pandolfi PP 2014. The multilayered complexity of ceRNA crosstalk and competition. Nature 505: 344–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor DJ, Bruenn J 2009. The evolution of novel fungal genes from non-retroviral RNA viruses. BMC Biol 7: 88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JR, Zagorski J, Woolford JL, Fournier MJ 1988. Sequence and genetic analysis of a dispensible 189 nucleotide snRNA from Saccharomyces cerevisiae. Nucleic Acids Res 16: 5587–5601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiedge H, Fremeau RT Jr, Weinstock PH, Arancio O, Brosius J 1991. Dendritic location of neural BC1 RNA. Proc Natl Acad Sci 88: 2093–2097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tisseur M, Kwapisz M, Morillon A 2011. Pervasive transcription—Lessons from yeast. Biochimie 93: 1889–1896. [DOI] [PubMed] [Google Scholar]
- Trifonov EN 1989. The multiple codes of nucleotide sequences. Bull Math Biol 51: 417–432. [DOI] [PubMed] [Google Scholar]
- Trotochaud AE, Wassarman KM 2004. 6S RNA function enhances long-term cell survival. J Bacteriol 186: 4978–4985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuerk C, Gold L 1990. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science 249: 505–510. [DOI] [PubMed] [Google Scholar]
- Uesaka M, Nishimura O, Go Y, Nakashima K, Agata K, Imamura T 2014. Bidirectional promoters are the major source of gene activation-associated non-coding RNAs in mammals. BMC Genomics 15: 35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhler JP, Hertel C, Svejstrup JQ 2007. A role for noncoding transcription in activation of the yeast PHO5 gene. Proc Natl Acad Sci 104: 8011–8016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulitsky I, Bartel DP 2013. lincRNAs: Genomics, evolution, and mechanisms. Cell 154: 26–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Upton KR, Baillie JK, Faulkner GJ 2011. Is somatic retrotransposition a parasitic or symbiotic phenomenon? Mob Genet Elements 1: 279–282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaidya N, Manapat ML, Chen IA, Xulvi-Brunet R, Hayden EJ, Lehman N 2012. Spontaneous network formation among cooperative RNA replicators. Nature 491: 72–77. [DOI] [PubMed] [Google Scholar]
- van Bakel H, Nislow C, Blencowe BJ, Hughes TR 2010. Most “dark matter” transcripts are associated with known genes. PLoS Biol 8: e1000371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van de Lagemaat LN, Landry JR, Mager DL, Medstrand P 2003. Transposable elements in mammals promote regulatory variation and diversification of genes with specialized functions. Trends Genet 19: 530–536. [DOI] [PubMed] [Google Scholar]
- van der Veen R, Arnberg AC, van der Horst G, Bonen L, Tabak HF, Grivell LA 1986. Excised group II introns in yeast mitochondria are lariats and can be formed by self-splicing in vitro. Cell 44: 225–234. [DOI] [PubMed] [Google Scholar]
- van Heesch S, van Iterson M, Jacobi J, Boymans S, Essers PB, de Bruijn E, Hao W, Macinnes AW, Cuppen E, Simonis M. 2014. Extensive localization of long noncoding RNAs to the cytosol and mono- and polyribosomal complexes. Genome Biol 15: R6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volff JN 2006. Turning junk into gold: Domestication of transposable elements and the creation of new genes in eukaryotes. BioEssays 28: 913–922. [DOI] [PubMed] [Google Scholar]
- Volff JN 2009. Cellular genes derived from Gypsy/Ty3 retrotransposons in mammalian genomes. Ann NY Acad Sci 1178: 233–243. [DOI] [PubMed] [Google Scholar]
- Wang H, Iacoangeli A, Popp S, Muslimov IA, Imataka H, Sonenberg N, Lomakin IB, Tiedge H 2002. Dendritic BC1 RNA: Functional role in regulation of translation initiation. J Neurosci 22: 10232–10241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T, Zeng J, Lowe CB, Sellers RG, Salama SR, Yang M, Burgess SM, Brachmann RK, Haussler D 2007. Species-specific endogenous retroviruses shape the transcriptional network of the human tumor suppressor protein p53 Proc Natl Acad Sci 104: 18613–18618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Gerstein M, Snyder M 2009. RNA-Seq: A revolutionary tool for transcriptomics. Nat Rev Genet 10: 57–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Gong C, Maquat LE 2013. Control of myogenesis by rodent SINE-containing lncRNAs. Genes Dev 27: 793–804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward LD, Kellis M 2012. Evidence of abundant purifying selection in humans for recently acquired regulatory functions. Science 337: 1675–1678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Washietl S, Pedersen JS, Korbel JO, Stocsits C, Gruber AR, Hackermuller J, Hertel J, Lindemeyer M, Reiche K, Tanzer A, et al. 2007. Structured RNAs in the ENCODE selected regions of the human genome. Genome Res 17: 852–864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wassarman KM, Storz G 2000. 6S RNA regulates E. coli RNA polymerase activity. Cell 101: 613–623. [DOI] [PubMed] [Google Scholar]
- Watanabe Y, Yamamoto M 1994. S. pombe mei2+ encodes an RNA-binding protein essential for premeiotic DNA synthesis and meiosis I, which cooperates with a novel RNA species meiRNA. Cell 78: 487–498. [DOI] [PubMed] [Google Scholar]
- Wei W, Pelechano V, Jarvelin AI, Steinmetz LM 2011. Functional consequences of bidirectional promoters. Trends Genet 27: 267–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiner AM 2006. SINEs and LINEs: Troublemakers, saboteurs, benefactors, ancestors. In The RNA world (ed. Gesteland RF, Cech TR, Atkins JF), pp. 507–533 Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- Wen YZ, Zheng LL, Qu LH, Ayala FJ, Lun ZR 2012. Pseudogenes are not pseudo any more. RNA Biol 9: 27–32. [DOI] [PubMed] [Google Scholar]
- Williams GC, Nesse RM 1991. The dawn of Darwinian medicine. Q Rev Biol 66: 1–22. [DOI] [PubMed] [Google Scholar]
- Wochner A, Attwater J, Coulson A, Holliger P 2011. Ribozyme-catalyzed transcription of an active ribozyme. Science 332: 209–212. [DOI] [PubMed] [Google Scholar]
- Woese CR 1967. The genetic code: The molecular basis for genetic expression. Harper and Row, New York. [Google Scholar]
- Woese CR 1979. A proposal concerning the origin of life on the planet earth. J Mol Evol 13: 95–101. [DOI] [PubMed] [Google Scholar]
- Wu X, Weigel D, Wigge PA 2002. Signaling in plants by intercellular RNA and protein movement. Genes Dev 16: 151–158. [DOI] [PubMed] [Google Scholar]
- Xie D, Chen CC, Ptaszek LM, Xiao S, Cao X, Fang F, Ng HH, Lewin HA, Cowan C, Zhong S 2010. Rewirable gene regulatory networks in the preimplantation embryonic development of three mammalian species. Genome Res 20: 804–815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong Y, Eickbush TH 1990. Origin and evolution of retroelements based upon their reverse transcriptase sequences. EMBO J 9: 3353–3362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Z, Wei W, Gagneur J, Perocchi F, Clauder-Munster S, Camblong J, Guffanti E, Stutz F, Huber W, Steinmetz LM 2009. Bidirectional promoters generate pervasive transcription in yeast. Nature 457: 1033–1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yakovchuk P, Goodrich JA, Kugel JF 2011. B2 RNA represses TFIIH phosphorylation of RNA polymerase II. Transcription 2: 45–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L, Froberg JE, Lee JT 2014. Long noncoding RNAs: Fresh perspectives into the RNA world. Trends Biochem Sci 39: 35–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yarus M 2011. Getting past the RNA world: The initial Darwinian ancestor. Cold Spring Harb Perspect Biol 3: a003590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin QF, Yang L, Zhang Y, Xiang JF, Wu YW, Carmichael GG, Chen LL 2012. Long noncoding RNAs with snoRNA ends. Mol Cell 48: 219–230. [DOI] [PubMed] [Google Scholar]
- Yoshihara K, Shahmoradgoli M, Martinez E, Vegesna R, Kim H, Torres-Garcia W, Trevino V, Shen H, Laird PW, Levine DA, et al. 2013. Inferring tumour purity and stromal and immune cell admixture from expression data. Nature Comm 4: 2612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaher HS, Unrau PJ 2007. Selection of an improved RNA polymerase ribozyme with superior extension and fidelity. RNA 13: 1017–1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Peritz A, Meggers E 2005. A simple glycol nucleic acid. J Am Chem Soc 127: 4174–4175. [DOI] [PubMed] [Google Scholar]
- Zhernakova DV, de Klerk E, Westra HJ, Mastrokolias A, Amini S, Ariyurek Y, Jansen R, Penninx BW, Hottenga JJ, Willemsen G, et al. 2013. DeepSAGE reveals genetic variants associated with alternative polyadenylation and expression of coding and non-coding transcripts. PLoS Genet 9: e1003594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zieve G, Penman S 1976. Small RNA species of the HeLa cell: Metabolism and subcellular localization. Cell 8: 19–31. [DOI] [PubMed] [Google Scholar]