Abstract
Homologous DNA pairing and strand exchange are at the core of homologous recombination. These reactions are promoted by a DNA-strand-exchange protein assembled into a nucleoprotein filament comprising the DNA-pairing protein, ATP, and single-stranded DNA. The catalytic activity of this molecular machine depends on control of its dynamic instability by accessory factors. Here we discuss proteins known as recombination mediators that facilitate formation and functional activation of the DNA-strand-exchange protein filament. Although the basics of homologous pairing and DNA-strand exchange are highly conserved in evolution, differences in mediator function are required to cope with differences in how single-stranded DNA is packaged by the single-stranded DNA-binding protein in different species, and the biochemical details of how the different DNA-strand-exchange proteins nucleate and extend into a nucleoprotein filament. The set of (potential) mediator proteins has apparently expanded greatly in evolution, raising interesting questions about the need for additional control and coordination of homologous recombination in more complex organisms.
At the core of homologous recombination is a DNA-strand-exchange protein filament. Positive recombination mediator proteins (e.g., Rad52 or BRCA2 in eukaryotes) facilitate its formation and functional activation.
Homologous recombination, the exchange of base pairing between homologous DNA molecules, is a central process for life (Morrical 2014). Not only does it create genetic diversity that sustains a population, it is essential at the cellular level for proper replication and maintenance of genomes (Wyman and Kanaar 2006). Given this central role in DNA metabolism, it is not surprising that the core reaction of homologous recombination is highly conserved among all kingdoms of life. The fundamental unit of this reaction is a DNA-strand-exchange protein, a small ATP-binding protein of ∼40 kDa. The DNA-strand-exchange protomers assemble head-to-tail in a right-handed helical filament around single-stranded (ss) DNA. This molecular assembly, programmed by the sequence of the bases of the bound DNA strand, recognizes homology in a double-stranded (ds) partner DNA molecule. Through protein-mediated manipulation of DNA structure and disassembly of protein–protein and protein–DNA interactions, exchange of base-pairing partners is achieved (Wyman 2011; Jasin and Rothstein 2013).
It is essential that the DNA-strand-exchange reaction at the core of homologous recombination is regulated such that it is actively applied in specific and changing conditions. Inappropriate DNA rearrangements also need to be avoided where they would be disastrous rather than beneficial. Thus, there need to be tipping points at which the reaction can either be driven forward or reversed, an important feature required to attain quality control (Kanaar et al. 2008). To achieve intricate levels of regulation, reaction choreographers have evolved, which are often referred to as positive or negative recombination mediators or effectors (Daley et al. 2014). Interestingly, although the fundamentals of the DNA-strand-exchange reaction are the same for bacteriophages, bacteria, archaea, and eukaryotes (Maher et al. 2011; White 2011), it appears that the set of proteins that influences homologous recombination has expanded significantly during evolution of more complex life forms (Fig. 1). In this review, we focus on positive recombination mediators of the highly conserved DNA-strand-exchange protein proteins UvsX (bacteriophages), RecA (bacteria), and RAD51 (eukaryotes).
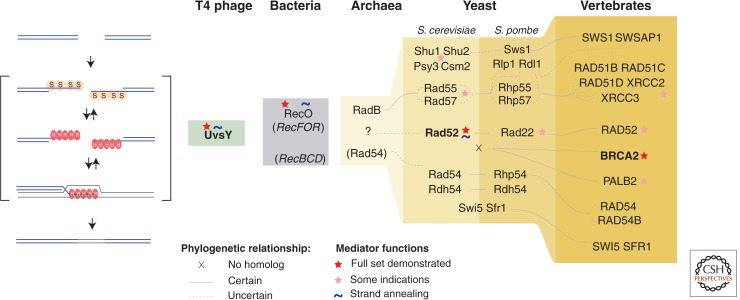
Figure 1.
The figure shows the increasing evolutionary complexity within the group of homologous recombination accessory factors that contribute to the formation and stability of the DNA-strand-exchange protein nucleoprotein filament in the key model organisms. Main homologous recombination steps (resection [Symington 2014], coating with ssDNA-binding protein [S], loading of the DNA-strand-exchange protein [R], and strand invasion) are shown schematically. Phylogenetic relationships between homologous proteins are indicated with solid lines in cases of well-supported orthology or broken lines when the exact evolutionary relationship is uncertain; x indicates no close homolog in a fully sequenced genome. Because phage, bacterial, and archaeal homologous recombination accessory proteins do not show detectably sequence similarity and have likely evolved independently, they are displayed as separate domains. Proteins that most closely meet the “mediator” definition criteria are indicated in bold. ∼ indicates the ability to promote annealing of protein-coated ssDNA, shared by UvsY, RecO, and RAD52, which is a remarkable example of convergent evolution emphasizing the universal usefulness of this biochemical activity.
At the core of homologous recombination is a DNA-strand-exchange protein–ATP–ssDNA nucleoprotein filament (Wyman 2011). The positive recombination mediator proteins facilitate formation and functional activation of this molecular machinery. Specifically, in the original definition, recombination mediator proteins are described to facilitate loading DNA-strand-exchange proteins onto ssDNA that is coated by ssDNA-binding proteins (Beernink and Morrical 1999). Mediators influence the competition for ssDNA binding in favor of the filament forming DNA-strand-exchange protein. Mediators also influence the preferential binding of DNA-strand-exchange proteins to ssDNA in favor of the much more abundant dsDNA in cells. Although in biochemical assays the order of addition of protein can be controlled by the experimenter, with variable results for efficiency of assay outcome, in vivo exposed ssDNA will be rapidly and effectively bound by abundant, high-affinity ssDNA-binding proteins (Dickey et al. 2013). These proteins need to be prevented from binding or replaced by the DNA-strand-exchange protein to initiate homologous recombination. However, in part because of the semantic ambiguity of the term and the large number of factors affecting homologous recombination that still lack a clearly established biochemical function, the term mediator is often applied more generally in the literature. We prefer the term “homologous recombination accessory proteins” for this broader class of proteins, and will focus our discussion here on those mediators that fit the original definition when possible. A model for mediator-assisted sequential handover of ssDNA from the ssDNA-binding protein to the DNA-strand-exchange protein gradually emerged from studies of the slimmed down recombination systems, especially phage T4. The properties attributed to mediators include: ssDNA binding, interaction with the ssDNA-binding protein, interaction with the DNA-strand-exchange protein, and “filament stabilization” by affecting ATPase activity of the nucleoprotein filament (Liu et al. 2011a). This last feature, an effect on nucleotide exchange or ATPase activity by the nucleoprotein filament, became the template used to search for candidate mediators in higher organisms. Typically, the potential mediator proteins were then tested for the specific interactions and functions needed to promote DNA-strand-exchange protein filament formation and activation.
HOW THE ORIGINAL MEDIATOR PROTEIN WORKS AND VARIATIONS ON THE THEME
The UvsY Protein from Bacteriophage T4
The founding member of the class of recombination mediators that replace the ssDNA-binding protein by the DNA-strand-exchange protein is UvsY from bacteriophage T4 (Harris and Griffith 1989; Kodadek et al. 1989; Yonesaki and Minagawa 1989; Beernink and Morrical 1999; Liu et al. 2011a; Maher et al. 2011). The phage T4 recombination system can be considered an archetype in that it includes relatively few components: the UvsX DNA-strand-exchange protein, the gp32 ssDNA-binding protein, and the UvsY recombination mediator. UvsX alone can catalyze DNA-strand-exchange in biochemical assays (Yonesaki and Minagawa 1985), as can all other DNA-strand-exchange proteins, but requires UvsY in vivo. The in vivo requirement for UvsY is explained by the competition between UvsX and gp32 for binding ssDNA. The biochemical activity of UvsY is to load UvsX on gp32-coated ssDNA. This results in the formation of UvsX nucleoprotein filaments active in homology search and strand exchange. In principle, this exchange requires modifying either the ssDNA-binding protein or ssDNA structure so that their interaction is weakened or disfavored. Recent work indicates that UvsY does this by binding ssDNA in a conformation that is incompatible with ssDNA-binding protein–ssDNA complex formation (Xu et al. 2010). Ironically, this picture emerged from an initial observation in single molecule force spectroscopy experiments in which UvsY appeared to exhibit behavior opposite to what was expected. In those experiments, UvsY bound less well to ssDNA than to dsDNA. However, the assay involved holding individual DNA molecules in an optical tweezer setup, in which force can be applied to the DNA molecules. UvsY bound better to dsDNA than ssDNA when the DNAs were held with sufficient force to keep the strands extended. Further analysis indicated that this behavior results from the arrangement of ssDNA bound to UvsY, wrapped around the protein. UvsY binding is disfavored when the ssDNA is held in an extended conformation (Pant et al. 2008). The arrangement of DNA wrapped around UvsY is supported by structural data showing that the protein forms a hexamer in solution and that the bound ssDNA is associated with multiple subunits (Xu et al. 2010). The gp32–ssDNA complex is described as relatively stiff with ssDNA neither wrapped nor kinked in the complex. Thus, ssDNA wrapped around UvsY is constrained in a conformation incompatible with binding to gp32. The arrangement of the mediator in a multimeric ring with ssDNA wrapped around it is shared by eukaryotic mediator Rad52 (see below) (Singleton et al. 2002). The intriguing question of how conformational transitions in DNA-protein and protein–protein interactions cause the exchange of gp32 for UvsX on ssDNA remains to be elucidated.
Different Single-Stranded DNA-Binding Proteins Demand Different Mechanisms of Mediation
Interestingly, different strategies for replacing ssDNA-binding proteins for DNA-strand-exchange proteins are effective depending on the arrangement of ssDNA and the ssDNA binding sites in the protein–ssDNA complex. The T4 gp32 protein apparently holds ssDNA, without wrapping or kinking it, in a stiff filament. Thus, as described above, a mediator acting as an ssDNA chaperone to enforce a curved conformation denies the ssDNA-binding protein of a suitable substrate. Escherichia coli presents a different situation in which the homotetrameric SSB binds ssDNA to identical sites on all four subunits wrapped like a seam of a baseball around the surface. In this case, straightening out ssDNA would disfavor SSB binding. The ssDNA in the RecA nucleoprotein filament is straight, stiff, and stretched, all conformations incompatible with SSB binding (Bianco et al. 1998). Once nucleated, the highly cooperative nature of RecA filament formation on ssDNA could disfavor SSB binding to the downstream DNA.
Escherichia coli RecFOR and RecBCD
The recombination mediators in E. coli are the RecOR and RecFOR complexes (Madiraju et al. 1988, 1992; Umezu et al. 1993; Liu et al. 2011a). The two-component complex of RecOR facilitates RecA filament formation on SSB covered ssDNA. The RecFOR complex specifically loads RecA at ss-dsDNA junctions, presumably during ssDNA gap repair or reactions involving ssDNA-tailed substrates (Morimatsu and Kowalczykowski 2003; Handa et al. 2009). Additional details and insight have come from following RecA filament formation on single DNA molecules. Once started, RecA filament formation is quite cooperative and results in exerting force and torque on ssDNA that can displace bound SSB (van der Heijden et al. 2005). However, SSB displacement by polymerizing RecA required an already nucleated filament (Joo et al. 2006). Elegant single molecule imaging and manipulation experiments have recently shown that RecOR or RecFOR specifically influence RecA nucleation on SSB-bound ssDNA after which filament growth, which is also stimulated by RecOR, is able to displace additional SSB (Bell et al. 2012). Although a recent study suggests that RecOR may bind ssDNA inside a ring-like structure, sterically hindering SSB access (Radzimanowski et al. 2013), how E. coli mediators negatively influence SSB–DNA interactions or deliver RecA have yet to be worked out in detail.
Alternatively, assembly of DNA-strand-exchange protein into nucleoprotein filaments during DNA break repair (Mehta and Haber 2014) can bypass the need to displace ssDNA-binding proteins by coupling DNA end processing (Symington 2014) into ssDNA directly to DNA-strand-exchange protein loading (Anderson and Kowalczykowski 1997). This mechanism is exemplified by the action of E. coli RecBCD. This molecular machine combines several functions including unwinding dsDNA, degrading one strand and loading RecA onto the newly exposed ssDNA, before it is available for SSB. The detailed mechanism of coupling DNA unwinding and strand degradation have become clear, aided by some stunning single molecule analysis (Dillingham and Kowalczykowski 2008) but the way RecA is delivered and loaded is still unknown.
Packaging of Single-Stranded DNA in Eukaryotes
The eukaryotic ssDNA-binding protein, RPA, is a heterotrimer including four ssDNA-binding sites with different affinities (Wold 1997). In addition, the DNA-binding domains are attached to each other and to the complex by flexible linker domains and are not held in a fixed orientation relative to each other. Many eukaryotic repair proteins interact with RPA. These interactions can alter the arrangement of the ssDNA-binding domains or fix them in a specific conformation relative to each other. Clues on how mediators may weaken interactions between DNA and the ssDNA-binding protein come from the structure studies of the RPA–ssDNA complex. The overall arrangement of RPA with and without bound DNA has been determined (Fan and Pavletich 2012; Brosey et al. 2013). This work revealed that an initial complex with 10 nucleotides of DNA bound results in a fixed architecture of the two engaged ssDNA-binding domains. In the absence of bound ssDNA, these domains are associated with the core by a flexible, unstructured linker and not ordered relative to each other. Additional ssDNA binding up to 30 nucleotides engaging all four DNA-binding domains resulted in a compact protein, indicating that DNA binding induces order and restricts conformational freedom of RPA (Fan and Pavletich 2012). Mediator proteins could favor DNA-strand-exchange protein binding by modifying RPA so that it would not effectively interact with ssDNA. RPA interaction partners are proposed to bind the various RPA flexible linkers and hold the ssDNA-binding domains apart or in an arrangement preventing cooperative high-affinity DNA binding (Fan and Pavletich 2012; Brosey et al. 2013). Mediators favoring DNA-strand-exchange protein filament formation would act as RPA protein chaperones or modulators that affect the affinity of the ssDNA-binding protein for ssDNA by altering protein conformation so that high-affinity binding to ssDNA is not likely. A mediator such as Rad52 that binds both RPA and Rad51 may couple the functions of affecting the affinity of RPA and delivering Rad51 to ssDNA (see below). Alternatively, RAD51 may be delivered and loaded by a separate protein/function. The mechanistic and structural details of promoting DNA-strand-exchange protein filament formation remain to be revealed.
EUKARYOTES: SAME MEDIATOR, DIFFERENT FUNCTION; DIFFERENT MEDIATOR, SAME FUNCTION
Eukaryotes encode a distinct and larger set of candidate HR mediator proteins that evolved independently, and at the sequence level show no detectable similarity to the bacterial and phage proteins we discussed in the previous section. This lack of sequence homology makes the mechanistic similarities between the eukaryotic and bacterial mediators all the more remarkable. HR accessory apparatus of eukaryotes is, however, phylogenetically related to some archaeal proteins.
Eukaryotic genes can be traced to different origins. Common to all domains of life, specific to eukarya, shared with bacteria, shared with archaea. This patchwork is described by a well-supported model postulating that the eukaryotic cell arose from an archaeal ancestor bearing a bacterial endosymbiont (van der Giezen and Tovar 2005; Makarova and Koonin 2013; Williams et al. 2013). The latter gave rise to the mitochondrion and, after migration of genes to the nucleus, to the subset of eukaryotic proteins that have “operational” functions (metabolism, biosynthesis, and membranes) and are more closely related to bacterial proteins. The former gave rise to the smaller “information processing” subset, including proteins involved in DNA replication, repair, transcription, and translation (Rivera et al. 1998; Esser 2004; Rivera and Lake 2004).
Eukaryotic HR machinery is clearly more similar to archaeal rather than bacterial counterpart (Seitz et al. 2001; White 2011). The much simpler archaeal HR system may hold valuable clues for understanding the features of eukaryotic HR that are not present in bacteria. The two major eukaryotic mediator proteins that we will discuss in detail—yeast RAD52 and vertebrate BRCA2—have no known archaeal homologs. However, the big group of putative mediators including RAD51 paralogs and the Shu complex subunits appear to have archaeal roots (Fig. 1).
RAD52
RAD52 has a pivotal mediator function in some organisms but appear less critical in others (Mortensen et al. 2009; Lok and Powell 2012). In budding yeast, Rad52 is genetically similar to UvsY of T4 in that defects in either the DNA-strand-exchange protein (UvsX or Rad51) or the mediator (UvsY or Rad52) have similar strong defects in recombination (Cunningham and Berger 1977). In Saccharomyces cerevisiae, Rad52 is the clearly the central player in homologous recombination, having a dual role as a mediator and as a DNA-strand-annealing protein (Krogh and Symington 2004). The mediator function of Rad52 is well established and fits very well with the paradigm established in the phage (UvsY) and bacterial (RecFOR) recombination systems. Rad52 removes the inhibitory effect of RPA on strand exchange reactions in vitro (Sung 1997a; New et al. 1998), and binds DNA (Mortensen et al. 1996), binds Rad51 (Shen et al. 1996), binds RPA (Hays et al. 1998; Shinohara et al. 1998), targets Rad51 to ssDNA (Song and Sung 2000), and facilitates RPA displacement by Rad51 (Sugiyama and Kowalczykowski 2002).
Vertebrate RAD52 has a number of characteristics in common with S. cerevisiae Rad52, but there are also intriguing differences. Human RAD52 interacts with human RAD51 (Shen et al. 1996) and human RPA (Park et al. 1996), which, along with the yeast studies, made it an ideal candidate for the mediator role. However, although some stimulatory activity was reported on joint molecule formation by RAD51 in the absence or RPA (Benson et al. 1998), DNA-strand exchange by human RAD51 was not affected by RAD52 when RPA was present (Jensen et al. 2010). This is consistent with no or very mild effects of RAD52 deficiency in various DNA repair assays in vertebrate cells (Rijkers et al. 1998; Yamaguchi-Iwai et al. 1998).
Interestingly, inactivation of RAD52 is synthetically lethal in chicken DT40 cells deficient for the XRCC3 protein, a candidate mediator from the RAD51 paralog group; (see below) (Fujimori et al. 2001). Similar synthetic lethal phenotypes were detected for RAD52 in human cells with the genuine mediator BRCA2 (Feng et al. 2011), and with the two proteins it forms a complex with: BRCA1 and PALB2 (Lok et al. 2013). It was suggested that in the absence of the (potential) mediators, RAD52 provides a backup pathway. Strikingly, however, no lethality or even increased DNA damage sensitivity was observed in DT40 cells when BRCA2 and RAD52 were simultaneously inactivated (Qing et al. 2011). The discrepancy between the results obtained in human and chicken DT40 cells remains to be resolved and is all the more intriguing because the triple-mutant rad52/xrcc3/brca2-null DT40 cells are viable, although not yet extensively characterized for their homologous recombination associated phenotypes.
BRCA2
Mutations in the human BRCA2 gene strongly predispose to breast and several other types of cancer (Wooster et al. 1995). DNA damage sensitivity of BRCA2-deficient cells (Connor et al. 1997; Sharan et al. 1997) and physical interaction between BRCA2 and RAD51 (Sharan et al. 1997; Katagiri et al. 1998) suggested that BRCA2 is involved in HR, which was confirmed by numerous studies (Davies et al. 2001; Moynahan et al. 2001b). BRCA2 protein is 3418 amino acids long, contains DNA- and protein-binding domains, and interacts with a number of other proteins. The most notable of the interactors are BRCA1 and PALB2, as deficiencies in these proteins manifest similarly to the BRCA2 deficiency (Chen et al. 1999; Moynahan et al. 1999, 2001a; Xia et al. 2006). Moreover, PALB2 was shown to stabilize RAD51 binding to ssDNA in the presence of RPA and promote D-loop formation, and thus satisfies some of the mediator criteria (Buisson et al. 2010; Dray et al. 2010).
Although BRCA2 has long been seen as a prime candidate for a mediator role in vertebrates based on genetic and cytological studies, and in vitro experiments using orthologs and isolated domains (Thompson 2012; Jensen 2013), direct demonstration was delayed by the challenges associated with the purification of a functional full-length BRCA2. After this hurdle was overcome, it was confirmed that BRCA2 binds RAD51 primarily through a set of eight BRC repeats in the middle part of the protein with a maximum stoichiometry of ∼1:6 (Jensen et al. 2010; Liu et al. 2010), a feature that might be important for nucleation of the RAD51 nucleoprotein filament. Furthermore, the biochemical experiments showed that BRCA2 can facilitate RAD51-catalyzed strand exchange by promoting RAD51 binding to RPA-coated ssDNA, imparting on RAD51 the preference for ssDNA over dsDNA binding, and greatly reducing the ATPase activity of ssDNA-bound RAD51 (Jensen et al. 2010; Liu et al. 2010; Thorslund et al. 2010). However, the data did not support a role in localizing filament nucleation to a ssDNA/dsDNA junction as observed for Ustilago maydis ortholog Brh2 and the E. coli RecFOR complex (Yang et al. 2005; Morimatsu et al. 2012). BRCA2 stimulated RAD51 strand exchange when E. coli SSB was used in place of RPA, and no BRCA2–RPA interaction could be observed, indicating that the interaction with the ssDNA-binding protein is not required for its mediator function, which also contrasts with Brh2 and RecFOR. Further biochemical characterization of BRCA2 and its mutants should reveal the importance of the BRC repeats, the roles of the subdomains within the carboxy-terminal DNA-binding domain of BRCA and the role of other BRCA2-interacting proteins (Holloman 2011).
ADDITIONAL CANDIDATES: HAVING SOME BUT NOT ALL MEDIATOR FUNCTIONS
The overview of eukaryotic homologous recombination mediators would be incomplete without the discussion of some of the accessory proteins for which bona fide mediator function has not (yet) been shown. Excluding the true mediators (Rad52 in budding yeast and BRCA2 in vertebrates), the group of homologous recombination accessory proteins with a possible mediator role includes the Rad51 paralogs (2–5 members), Swi5–Sfr1 complex and the Shu/PCSS complex (2–4 members; Fig. 1).
The RAD51 Paralogs
In addition to the RAD51 DNA-strand-exchange protein and its meiosis-specific sibling DMC1, eukaryotic genomes encode a variable number of proteins with low but readily detectable sequence similarities to RAD51, which are referred to as RAD51 paralogs (Kans and Mortimer 1991; Lovett 1994; Schild et al. 2000; Lin et al. 2006). The best characterized of these are budding yeast proteins, Rad55 and Rad57, and vertebrate proteins, RAD51B, RAD51C, RAD51D, XRCC2, and XRCC3. Deficiency in any of these proteins results in a homologous recombination phenotype, which is nearly as severe as RAD51 loss. The paralogs form complexes with each other: Rad55–Rad57 (Sung 1997b) in S. cerevisiae and RAD51B–C–D–X2 and RAD51C–X3 in human cells (Masson et al. 2001; Liu et al. 2002; Wiese et al. 2002).
Despite the large body of literature unequivocally demonstrating a critical requirement of the paralogs for homologous recombination in yeast and vertebrate cells (Game and Mortimer 1974; Saeki et al. 1980; Lovett and Mortimer 1987; Liu et al. 1998; Takata et al. 2001; Thacker 2005), most probably in a close conjunction with the nucleoprotein filament, their exact role remains elusive. This is, in part, because interpretation of cytological and genetic data is complicated by the additional roles attributed to the paralog complexes that have no clear link to nucleoprotein filament stability (Thacker 2005), such as control of centrosome integrity (Cappelli et al. 2011) or participation in checkpoint signaling (Somyajit et al. 2013). Another complicating matter is the variation in the RAD51 paralog group size in different branches of eukaryotes (Fig. 1), occasionally unclear phylogenetic relationships between them, and difference in severity of the mutant phenotypes. Here we focus on yeast and vertebrates, in which the paralogs are essential for homologous recombination. However in the plant, Arabidopsis thaliana, which, like vertebrates, has five RAD51 paralogs, the paralog knockouts are viable, show relatively mild sensitivity to genotoxic agents, and only the AtRad51c mutant shows a meiotic defect (Bleuyard et al. 2005; Li et al. 2005; Wang et al. 2014). This suggests that despite sequence and group composition conservation over 1.6 billion years of independent evolution, the essential function of the paralogs during replication has been lost in plants.
A number of observations indirectly suggested a role as mediator for the RAD51 paralogs. Cytological observations are consistent with a mediator function. RAD51 accumulates at a high concentration at sites of DNA damage into so called foci and this response is defective in vertebrate cells deficient for any of the five paralogs (Takata et al. 2000, 2001). The similar lack of RAD51 accumulation into DNA damage induced foci in BRCA2 deficient cells supports a mediator designation for the paralogs (Yuan et al. 1999). Furthermore, the preferential binding of the paralog complexes to ssDNA (Masson et al. 2001; Sigurdsson et al. 2001) and coimmunoprecipitation with RAD51, observed in some studies (Liu et al. 2002; Lio et al. 2003; Park et al. 2013), made them plausible candidates for a mediator role. In addition, a weak stimulatory activity of the purified RAD51B–RAD51C heterodimer in the strand-exchange assay was reported, more pronounced for the accumulation of reaction intermediates (joint molecules) than for its final product (nicked circular DNA) (Sigurdsson et al. 2001). Thus, these observations are still far from demonstrating a direct role in eliminating the inhibitory effect of RPA at DNA-strand-exchange protein filament formation stage.
With BRCA2 firmly placed in the role as mediator, how could the RAD51 paralogs influence mediation? To date, this question has not yet been answered. In vertebrate cells, BRCA2 is epistatic to the paralogs and other accessory proteins in most assays (Qing et al. 2011; Jensen et al. 2013), which supports the argument that it is the sole mediator, with the paralogs acting downstream (Jensen et al. 2013). However, BRCA2 exerts tight control over nuclear concentration of RAD51, so BRCA2-deficient cells are also essentially RAD51 deficient. Additionally, in Rad52−/− DT40 cells deletion of Xrcc3, but not Brca2 leads to lethality (Fujimori et al. 2001; Qing et al. 2011), and paralogs are partially redundant with each other (Takata et al. 2001), so inactivating one does not guarantee complete inactivation of the complex. Thus, some degree of redundancy and specialization among the five paralogs, and the role of BCRA2 in controlling localization and concentration of RAD51 prevent drawing definitive conclusions. Separation of function mutants of BRCA2 and/or a better understanding of the paralog biochemistry is required.
A case for mediator function had been made for S. cerevisiae Rad55–Rad57 as well, based on the reported stimulatory activity on joint molecule formation in reactions in which Rad51, Rad55–Rad57, and RPA were coincubated with ssDNA (Sung 1997b). Consistent with this, a screen for mutations alleviating the phenotype caused by Rad55 deficiency revealed several Rad51 mutants with increased affinity for DNA and proficiency to displace RPA from ssDNA (Fortin and Symington 2002; Malik and Symington 2008). However, accumulation of Rad55–Rad57 in DNA damage-induced foci is dependent on Rad51 and not the other way around (Lisby et al. 2004). This finding, although by no means a direct proof, can be interpreted as inconsistent with a mediator role, because the accumulation of Rad51 and all other homologous recombination proteins in foci is dependent on Rad52 (Lisby et al. 2004). A recent study provided important structural and functional insights into the role of Rad55–Rad57 heterodimer (Liu et al. 2011b). Analysis of Rad51 nucleoprotein filaments formed on ssDNA in the presence of substoichiometric amounts of Rad55–Rad57 revealed that the heterodimer generally stabilizes the filament and protects it from disassembly by the antirecombination helicase Srs2 via a specific 1:1 interaction. This is in a good agreement with the finding that srs2 mutation partially suppresses the homologous recombination defect in a rad57 mutant (Fung et al. 2009). Liu et al. (2011b) also showed that the role of Rad55–Rad57 extends beyond counteracting Srs2 activity, because neither the MMS sensitivity nor the sister chromatid recombination defect were fully restored by Srs2 deletion in the rad55 strain. Electron microphotographs showed that Rad55–Rad57 can associate with the filaments at ends, as well as internally, and provided a visual demonstration of the increased filament integrity in the presence of Srs2.
RAD51 paralogs are the only candidate mediator proteins whose evolutionary history goes beyond eukaryotes. Intriguing precedents for DNA-strand-exchange protein paralogs with accessory and possibly mediator role in HR exist in archaea (reviewed in Seitz et al. 2001; Haldenby et al. 2009; White 2011). At least one such paralog is present in all sequenced archaeal genomes (Haldenby et al. 2009; McRobbie et al. 2009). All of the archaeal paralogs contain a conserved RecA/RAD51 ATPase domain, but are smaller, lacking the amino-terminal extension present in the archaeal DNA-strand-exchange protein (RadA) and RAD51, or the carboxy-terminal domain of RecA (Rashid et al. 1996; Haldenby et al. 2009), and cannot catalyze DNA-strand exchange.
Proteins from the group aRadC (name suggested by Haldenby et al. 2009) are found broadly throughout the archaeal domain. Similar proteins (KaiC) are found in some bacteria, likely from horizontal gene transfer (Leipe et al. 2000; Haldenby et al. 2009). Limited biochemical (Sheng et al. 2008; McRobbie et al. 2009; Wang et al. 2012; Graham and Haseltine 2013; Graham et al. 2013) and genetic (Liang et al. 2013) data available suggests that aRadC proteins are involved DNA repair by HR. aRadC group members bind to ssDNA (Sheng et al. 2008; McRobbie et al. 2009; Wang et al. 2012; Graham and Haseltine 2013; Graham et al. 2013). A stimulatory or inhibitory effect of aRadC on RadA activities (strand-exchange assays, ssDNA binding, or ATPase activity) was detected in all biochemical studies in which it was analyzed (Sheng et al. 2008; McRobbie et al. 2009; Wang et al. 2012; Graham and Haseltine 2013; Graham et al. 2013), and one study reported release of the SSB inhibitory effect in a strand-exchange assay suggesting a mediator function (Sheng et al. 2008).
The other group of archaeal RadA paralogs is called RadB and is only found in euryarchaeota, which is a group more distantly related to eukarya than other archaea (Williams et al. 2013). All sequenced euryarchaeota genomes encode one RadB protein (Haldenby et al. 2009). An intriguing feature of RadB proteins was uncovered by several systematic studies of the RecA/RAD51 family evolution (Lin et al. 2006; Wu et al. 2011; Chintapalli et al. 2013). These phylogenetic reconstructions revealed that eukaryotic and archaeal RecA homologs divide into two groups: RADα, which includes archaeal RadA and eukaryotic RAD51 and DMC1 (i.e., proteins with DNA-strand-exchange activities), and RADβ, which includes archaeal RadB and eukaryotic RAD51 paralogs. RadB deletion in Haloferax volcanii sensitized cells to UV light, and no further sensitization was observed in ΔradBΔradA double mutant (Guy et al. 2006). In biochemical assay, the studied RadB proteins showed weak or undetectable ATPase activity, and ssDNA and dsDNA binding (Komori et al. 2000; Guy et al. 2006). Pyrococcus furiosus RadB inhibited RadA D-loop and strand-exchange activities (Komori et al. 2000).
Ubiquitous presence of RadA paralogs in archaea suggests that delegation of regulatory or mediator function from the DNA-strand-exchange protein to its simplified versions is one of the most conserved properties of eukaryotic HR. Reuse of the same fold may be beneficial for integration of the control functions into the nucleoprotein filament.
The Swi5–Sfr1 Complex
Among different species, subtle differences in activity of conserved mediator proteins and or RAD51 apparently demand additional proteins to effectively mediate the hand-off between RAD51 and RPA-coated ssDNA. In this respect, there is an interesting difference between the Rad52 proteins from yeasts S. cerevisiae and S. pombe. The S. cerevisiae Rad52 protein has the ability to promote Rad51-dependent DNA-strand exchange on RPA-coated DNA by itself. In contrast, S. pombe Rad52 (known as Rad22) cannot execute this reaction by itself. Although Rad22 is important to overcome the inhibitory effect of RPA, it requires the Swi5–Sfr1 complex in a downstream step to stabilize the Rad51 filament, so it can progress to strand exchange (Kurokawa et al. 2008; Kuwabara et al. 2012). Although the requirement for the Swi5–Sfr1 complex might be because of differences in Rad52 mediator activity between budding and fission yeast, it could also be related to differences in biochemical properties of the strand-exchange protein. In this respect, is it intriguing that, although S. cerevisiae Rad51 does not require a Swi5–Sfr1 activity to promote homologous recombination during mitotic growth, it does express a meiosis-specific equivalent (Sae3–Mei5). Sae3–Mei5 does stimulate the Dmc1 protein, which is the important catalytic strand-exchange protein during meiosis, whereas Rad51 only has a supportive role (Ferrari et al. 2009; Cloud et al. 2012).
Mitotic human and mouse cells also express a Swi5–Sfr1 equivalent (Akamatsu and Jasin 2010; Tsai et al. 2012). Mouse cells in which the Swi5 or Sfr1 are knocked out are viable and display no defect in homologous recombination. Thus, the impact on cell growth of the Swi5–Sfr1 complex is very different from Rad51 and Brca2, whose deletion is cell lethal. In this respect, the Swi5 and Sfr1 knockout phenotypes are reminiscent of the mouse Rad52 knockout phenotype (Rijkers et al. 1998). Interestingly, homologous recombination is further reduced in Swi5–Sfr1 mutant cells compared to wild type cells when normal Rad51 function is compromised by overexpression of a BRC domain from Brca2 (Akamatsu and Jasin 2010). This observation is consistent with, but does not prove, a function of the mammalian SWI5–SFR1 complex in the RAD52 pathway, which is uncovered when BRCA2 function is attenuated (Feng et al. 2011).
The Shu (PCSS) Complex
A set of four small (<30 kDa) S. cerevisiae proteins, Shu1, Shu2, Psy3, and Csm2, were independently identified in several screens for mutations affecting mitotic and meiotic homologous recombination (Rabitsch et al. 2001; Huang et al. 2003; Smith et al. 2004; Lee et al. 2005; Shor et al. 2005). The proteins were found to interact physically as well as genetically (Ito et al. 2001; Shor et al. 2005), forming a complex with 1:1:1:1 stoichiometry (Sasanuma et al. 2013). The complex is referred to as either Shu or PCSS. Initially, no clearly identifiable homologs were found for any of the four Shu complex proteins outside of S. cerevisiae and closely related yeasts (Shor et al. 2005). However, using loose sequence- and motif-matching criteria, the S. pombe Sws1 protein was proposed as a distant homolog of S. cerevisiae Shu2 (Martín et al. 2006). Sws1 interacts with the antirecombination helicase Srs2 and its deletion suppresses the srs2Δ and rqh1Δ (the S. pombe homolog of Sgs1 and RecQ) mutations (Martín et al. 2006). Proteomic identification of Sws1 interactors revealed Rlp1 and Rdl1, two of the four putative Rad51 paralogs encoded by S. pombe genome, sharing most sequence similarity with XRCC2 and RAD51D in humans, respectively. Importantly, S. pombe Rdl1 also shows detectable sequence similarity to S. cerevisiae Psy3, providing the second link to the S. cerevisiae Shu complex. Based on this, it was suggested that (some of) the Shu complex subunits are RAD51 paralogs. Support for this notion is provided by structure of the Psy3–Csm2 heterodimer, which is remarkably similar to the structure of the Rad51–Rad51 dimer (Tao et al. 2012; Sasanuma et al. 2013). However, amino acid similarities between the sequences are too low (∼15% identity, ∼30% similarity in pairwise alignments) to confidently determine whether Shu complex subunits and the RAD51 paralogs are indeed homologous to each other or simply share a RecA-like fold; for example, plant and vertebrate RAD51 paralogs are more similar to each other than to the Shu complex subunits.
Interactions of the Shu complex with antirecombination helicases and other genetic evidence suggested that it is involved the control of Rad51 filament stability, possibly contributing to the mediator function (Mankouri et al. 2007; Bernstein et al. 2011), and molecular details of this are beginning to emerge. Solution of the Psy3–Csm2 dimer structure by two groups revealed striking similarity to the Rad51 fold (Tao et al. 2012; Sasanuma et al. 2013), leading to the proposal that the Shu complex may integrate into the Rad51 filament and thus control its assembly (Sasanuma et al. 2013). Consistent with this, ssDNA binding by Rad51 and Psy3–Csm2 heterodimer was found to be interdependent in vitro. In the absence of Rad51, Psy3–Csm2 formed high molecular weight aggregates with ssDNA that were too big to be resolved by an electrophoretic mobility shift assay. Addition of Rad51 led to the emergence of a smaller defined cocomplex on ssDNA. On the other hand, when Psy3–Csm2 was present, Rad51 formed defined complexes with ssDNA as efficiently in the presence of ADP or without a nucleotide cofactor as it did in the presence of ATP, which can be interpreted as filament stabilization. RPA was not included in these experiments, and it is not clear how inclusion of Shu1 and Shu2 would affect the results. Involvement of the Shu complex in Rad51 filament stabilization was also supported by in vivo studies that showed its requirement for Rad51 foci assembly in meiosis and for Rad51 accumulation at a meiotic recombination hotspot (Lam and Keeney 2014; Zickler and Kleckner 2014), whereas the Shu complex itself was recruited in Rad51-independent manner (Sasanuma et al. 2013). It was also suggested that the Shu complex can stabilize Rad51 filament by counteracting Srs2 (Bernstein et al. 2011; Tao et al. 2012), in a manner similar to what has been shown for S. cerevisiae Rad55–Rad57 (Liu et al. 2011b), but such effect has not yet been shown biochemically.
The human genome encodes a likely ortholog of S. pombe Sws1. The human SWS1 protein is much smaller than Sws1 and other Shu complex members (140 vs. 210–240 amino acids) (Martín et al. 2006), and the similarity is mostly limited to the SWIM domain, an uncharacterized zinc-finger-like motif. Whether SWS1 can adopt a RecA-like fold is therefore unclear. SWS1 coimmunoprecipitated with RAD51D when both proteins were overexpressed (Martín et al. 2006), although much more extensive literature on the RAD51 paralogs and another study on human SWS1 argue against SWS1 involvement in the paralogs complexes (Liu et al. 2011c). SWS1 was found to interact with an uncharacterized protein, SWSAP1, which is predicted to share a RecA fold, contains Walker A and B motifs, and was shown to possess ssDNA-dependent ATPase activity.
CONCLUDING REMARKS
Mediator proteins have important roles in controlling homologous recombination by influencing both the efficiency of filament formation by DNA-strand-exchange proteins and the commitment to the key steps of homology recognition and strand exchange. It is intriguing to note the discrepancy between the large number of the homologous recombination accessory proteins in eukaryotes compared to the same limited number of steps in the schematic depictions used to illustrate and conceptualize the homologous recombination process for all organisms (Fig. 1). The schemes drawn for phage and bacterial homologous recombination do not differ much from those illustrating homologous recombination in human cells. The same DNA structures are present in both cases, yet the number of proteins homologous recombination critically depends on differs by an order of magnitude. Of course, these accessory proteins do not all function as mediators, as originally defined.
On the one hand, the plethora of accessory proteins may not be surprising, and plausible hypotheses explaining the parallel increase in organismal complexity and the complexity of homologous recombination can be suggested. Larger genomes may require a much more elaborate system of quality control of homologous recombination. First, more origins of replication will increase the chance of recombination malfunction, which then needs to be specifically distinguished. Second, the homology search is more challenging in a larger sequence space. Third, the increase in ploidy with the advent of sexual reproduction introduced the demand for suppression of heteroallelic recombination during mitoses and a special role for homologous recombination during meiosis. Fourth, the explosion of the noncoding part of the genome, mostly from repetitive sequences, required suppressing of nonallelic recombination.
On the other hand, the same set of biochemical assays used to define the transitions of the recombination filament in phages and bacteria is used to describe eukaryotic homologous recombination. The additional cytological techniques available in eukaryotes rely on very few endpoints, and the inferred connections between them and the molecular events can be ambiguous and unreliable. For example, oftentimes the literature equates RAD51 foci with RAD51 nucleoproteins filaments. Furthermore, extracting pre- and postsynaptic roles of accessory proteins from the study of foci is difficult. This makes defining the exact functions of the numerous homologous recombination accessory proteins a formidable challenge. Accordingly, which eukaryotic homologous recombination proteins are responsible for solving the problems hypothesized above and how exactly can they be involved is largely unclear. Understanding the advantage of additional mediators in homologous recombination may serve as an example to define fundamental aspects of increased process complexity in evolution. Developing an experimental and conceptual framework to make these connections is arguably the main challenge for the study of homologous recombination at present.
ACKNOWLEDGMENTS
This work is supported by funding from the European Community’s Seventh Framework Programme (FP7/2007–2013) (Grant No. HEALTH-F2–2010–259893) and an ECHO grant from the Netherlands Organization for Scientific Research (NWO)-Chemical Sciences.
Footnotes
Editors: Stephen Kowalczykowski, Neil Hunter, and Wolf-Dietrich Heyer
Additional Perspectives on DNA Recombination available at www.cshperspectives.org
REFERENCES
*Reference is also in this collection.
- Akamatsu Y, Jasin M 2010. Role for the mammalian Swi5-Sfr1 complex in DNA strand break repair through homologous recombination. PLoS Genet 6: e1001160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson DG, Kowalczykowski SC 1997. The translocating RecBCD enzyme stimulates recombination by directing RecA protein onto ssDNA in a χ-regulated manner. Cell 90: 77–86. [DOI] [PubMed] [Google Scholar]
- Beernink HT, Morrical SW 1999. RMPs: Recombination/replication mediator proteins. Trends Biochem Sci 24: 385–389. [DOI] [PubMed] [Google Scholar]
- Bell JC, Plank JL, Dombrowski CC, Kowalczykowski SC 2012. Direct imaging of RecA nucleation and growth on single molecules of SSB-coated ssDNA. Nature 491: 274–278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benson FE, Baumann P, West SC 1998. Synergistic actions of Rad51 and Rad52 in recombination and DNA repair. Nature 391: 401–404. [DOI] [PubMed] [Google Scholar]
- Bernstein KA, Reid RJD, Sunjevaric I, Demuth K, Burgess RC, Rothstein R 2011. The Shu complex, which contains Rad51 paralogues, promotes DNA repair through inhibition of the Srs2 anti-recombinase. Mol Biol Cell 22: 1599–1607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bianco PR, Tracy RB, Kowalczykowski SC 1998. DNA strand exchange proteins: A biochemical and physical comparison. Front Biosci 3: D570–603. [DOI] [PubMed] [Google Scholar]
- Bleuyard J-Y, Gallego ME, Savigny F, White CI 2005. Differing requirements for the Arabidopsis Rad51 paralogs in meiosis and DNA repair. Plant J 41: 533–545. [DOI] [PubMed] [Google Scholar]
- Brosey CA, Yan C, Tsutakawa SE, Heller WT, Rambo RP, Tainer JA, Ivanov I, Chazin WJ 2013. A new structural framework for integrating replication protein A into DNA processing machinery. Nucleic Acids Res 41: 2313–2327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buisson R, Dion-Côté A-M, Coulombe Y, Launay H, Cai H, Stasiak AZ, Stasiak A, Xia B, Masson J-Y 2010. Cooperation of breast cancer proteins PALB2 and piccolo BRCA2 in stimulating homologous recombination. Nat Struct Mol Biol 17: 1247–1254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cappelli E, Townsend S, Griffin C, Thacker J 2011. Homologous recombination proteins are associated with centrosomes and are required for mitotic stability. Exp Cell Res 317: 1203–1213. [DOI] [PubMed] [Google Scholar]
- Chen JJ, Silver D, Cantor S, Livingston DM, Scully R 1999. BRCA1, BRCA2, and Rad51 operate in a common DNA damage response pathway. Cancer Res 59: 1752s–1756s. [PubMed] [Google Scholar]
- Chintapalli SV, Bhardwaj G, Babu J, Hadjiyianni L, Hong Y, Todd GK, Boosalis CA, Zhang Z, Zhou X, Ma H, et al. 2013. Reevaluation of the evolutionary events within recA/RAD51 phylogeny. BMC Genomics 14: 240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cloud V, Chan YL, Grubb J, Budke B, Bishop DK 2012. Rad51 is an accessory factor for Dmc1-mediated joint molecule formation during meiosis. Science 337: 1222–1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connor F, Bertwistle D, Mee PJ, Ross GM, Swift S, Grigorieva E, Tybulewicz VL, Ashworth A 1997. Tumorigenesis and a DNA repair defect in mice with a truncating Brca2 mutation. Nat Genet 17: 423–430. [DOI] [PubMed] [Google Scholar]
- Cunningham RP, Berger H 1977. Mutations affecting genetic recombination in bacteriophage T4D: I. Pathway analysis. Virology 80: 67–82. [DOI] [PubMed] [Google Scholar]
- *.Daley JM, Gaines WA, Kwon Y, Sung P 2014. Regulation of DNA pairing in homologous recombination. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a017954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies AA, Masson J-Y, McIlwraith MJ, Stasiak AZ, Stasiak A, Venkitaraman AR, West SC 2001. Role of BRCA2 in control of the RAD51 recombination and DNA repair protein. Mol Cell 7: 273–282. [DOI] [PubMed] [Google Scholar]
- Dickey TH, Altschuler SE, Wuttke DS 2013. Single-stranded DNA-binding proteins: Multiple domains for multiple functions. Structure 21: 1074–1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dillingham MS, Kowalczykowski SC 2008. RecBCD enzyme and the repair of double-stranded DNA breaks. Microbiol Mol Biol Rev 72: 642–671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dray E, Etchin J, Wiese C, Saro D, Williams GJ, Hammel M, Yu X, Galkin VE, Liu D, Tsai M-S, et al. 2010. Enhancement of RAD51 recombinase activity by the tumor suppressor PALB2. Nat Struct Mol Biol 17: 1255–1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esser C 2004. A genome phylogeny for mitochondria among proteobacteria and a predominantly eubacterial ancestry of yeast nuclear genes. Mol Biol Evol 21: 1643–1660. [DOI] [PubMed] [Google Scholar]
- Fan J, Pavletich NP 2012. Structure and conformational change of a replication protein A heterotrimer bound to ssDNA. Genes Dev 26: 2337–2347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Z, Scott SP, Bussen W, Sharma GG, Guo G, Pandita TK, Powell SN 2011. Rad52 inactivation is synthetically lethal with BRCA2 deficiency. Proc Natl Acad Sci 108: 686–691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrari SR, Grubb J, Bishop DK 2009. The Mei5-Sae3 protein complex mediates Dmc1 activity in Saccharomyces cerevisiae. J Biol Chem 284: 11766–11770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fortin GS, Symington LS 2002. Mutations in yeast Rad51 that partially bypass the requirement for Rad55 and Rad57 in DNA repair by increasing the stability of Rad51–DNA complexes. EMBO J 21: 3160–3170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujimori A, Tachiiri S, Sonoda E, Thompson LH, Dhar PK, Hiraoka M, Takeda S, Zhang Y, Reth M, Takata M 2001. Rad52 partially substitutes for the Rad51 paralog XRCC3 in maintaining chromosomal integrity in vertebrate cells. EMBO J 20: 5513–5520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fung CW, Mozlin AM, Symington LS 2009. Suppression of the double-strand-break-repair defect of the Saccharomyces cerevisiae rad57 mutant. Genetics 181: 1195–1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Game JC, Mortimer RK 1974. A genetic study of X-ray sensitive mutants in yeast. Mutat Res 24: 281–292. [DOI] [PubMed] [Google Scholar]
- Graham WJ, Haseltine CA 2013. A recombinase paralog from the hyperthermophilic crenarchaeon Sulfolobus solfataricus enhances SsoRadA ssDNA binding and strand displacement. Gene 515: 128–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham WJ, Rolfsmeier ML, Haseltine CA 2013. An archaeal RadA paralog influences presynaptic filament formation. DNA Repair 12: 403–413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guy CP, Haldenby S, Brindley A, Walsh DA, Briggs GS, Warren MJ, Allers T, Bolt EL 2006. Interactions of RadB, a DNA repair protein in archaea, with DNA and ATP. J Mol Biol 358: 46–56. [DOI] [PubMed] [Google Scholar]
- Haldenby S, White MF, Allers T 2009. RecA family proteins in archaea: RadA and its cousins. Biochem Soc Trans 37: 102–107. [DOI] [PubMed] [Google Scholar]
- Handa N, Morimatsu K, Lovett ST, Kowalczykowski SC 2009. Reconstitution of initial steps of dsDNA break repair by the RecF pathway of E. coli. Genes Dev 23: 1234–1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris LD, Griffith JD 1989. UvsY protein of bacteriophage T4 is an accessory protein for in vitro catalysis of strand exchange. J Mol Biol 206: 19–27. [DOI] [PubMed] [Google Scholar]
- Hays SL, Firmenich AA, Massey P, Banerjee R, Berg P 1998. Studies of the interaction between Rad52 protein and the yeast single-stranded DNA-binding protein RPA. Mol Cell Biol 18: 4400–4406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holloman WK 2011. Unraveling the mechanism of BRCA2 in homologous recombination. Nat Struct Mol Biol 18: 748–754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang M-E, Rio A-G, Nicolas A, Kolodner RD 2003. A genomewide screen in Saccharomyces cerevisiae for genes that suppress the accumulation of mutations. Proc Natl Acad Sci 100: 11529–11534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito T, Chiba T, Ozawa R, Yoshida M, Hattori M, Sakaki Y 2001. A comprehensive two-hybrid analysis to explore the yeast protein interactome. Proc Natl Acad Sci 98: 4569–4574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jasin M, Rothstein R 2013. Repair of strand breaks by homologous recombination. Cold Spring Harb Perspect Biol 5: a012740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen RB 2013. BRCA2: One small step for DNA repair, one giant protein purified. Yale J Biol Med 86: 479–489. [PMC free article] [PubMed] [Google Scholar]
- Jensen RB, Carreira A, Kowalczykowski SC 2010. Purified human BRCA2 stimulates RAD51-mediated recombination. Nature 467: 678–683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen RB, Ozes A, Kim T, Estep A, Kowalczykowski SC 2013. BRCA2 is epistatic to the RAD51 paralogs in response to DNA damage. DNA Repair 12: 306–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joo C, McKinney SA, Nakamura M, Rasnik I, Myong S, Ha T 2006. Real-time observation of RecA filament dynamics with single monomer resolution. Cell 126: 515–527. [DOI] [PubMed] [Google Scholar]
- Kanaar R, Wyman C, Rothstein R 2008. Quality control of DNA break metabolism: In the “end,” it’s a good thing. EMBO J 27: 581–588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kans JA, Mortimer RK 1991. Nucleotide sequence of the RAD57 gene of Saccharomyces cerevisiae. Gene 105: 139–140. [DOI] [PubMed] [Google Scholar]
- Katagiri T, Saito H, Shinohara A, Ogawa H, Kamada N, Nakamura Y, Miki Y 1998. Multiple possible sites of BRCA2 interacting with DNA repair protein RAD51. Genes Chromosomes Cancer 21: 217–222. [PubMed] [Google Scholar]
- Kodadek T, Gan DC, Stemke-Hale K 1989. The phage T4 uvs Y recombination protein stabilizes presynaptic filaments. J Biol Chem 264: 16451–16457. [PubMed] [Google Scholar]
- Komori K, Miyata T, DiRuggiero J, Holley-Shanks R, Hayashi I, Cann IK, Mayanagi K, Shinagawa H, Ishino Y 2000. Both RadA and RadB are involved in homologous recombination in Pyrococcus furiosus. J Biol Chem 275: 33782–33790. [DOI] [PubMed] [Google Scholar]
- Krogh BO, Symington LS 2004. Recombination proteins in yeast. Annu Rev Genet 38: 233–271. [DOI] [PubMed] [Google Scholar]
- Kurokawa Y, Murayama Y, Haruta-Takahashi N, Urabe I, Iwasaki H 2008. Reconstitution of DNA strand exchange mediated by Rhp51 recombinase and two mediators. PLoS Biol 6: e88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuwabara N, Murayama Y, Hashimoto H, Kokabu Y, Ikeguchi M, Sato M, Mayanagi K, Tsutsui Y, Iwasaki H, Shimizu T 2012. Mechanistic insights into the activation of Rad51-mediated strand exchange from the structure of a recombination activator, the Swi5-Sfr1 complex. Structure 20: 440–449. [DOI] [PubMed] [Google Scholar]
- *.Lam I, Keeney S 2014. Mechanism and regulation of meiotic recombination initiation. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a016634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee W, St Onge RP, Proctor M, Flaherty P, Jordan MI, Arkin AP, Davis RW, Nislow C, Giaever G 2005. Genome-wide requirements for resistance to functionally distinct DNA-damaging agents. PLoS Genet 1: e24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leipe DD, Aravind L, Grishin NV, Koonin EV 2000. The bacterial replicative helicase DnaB evolved from a RecA duplication. Genome Res 10: 5–16. [PubMed] [Google Scholar]
- Li W, Yang X, Lin Z, Timofejeva L, Xiao R, Makaroff CA, Ma H 2005. The AtRAD51C gene is required for normal meiotic chromosome synapsis and double-stranded break repair in Arabidopsis. Plant Physiol 138: 965–976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang P-J, Han W-Y, Huang Q-H, Li Y-Z, Ni J-F, She Q-X, Shen Y-L 2013. Knockouts of RecA-like proteins RadC1 and RadC2 have distinct responses to DNA damage agents in Sulfolobus islandicus. J Genet Genomics 40: 533– 542. [DOI] [PubMed] [Google Scholar]
- Lin Z, Kong H, Nei M, Ma H 2006. Origins and evolution of the recA/RAD51 gene family: Evidence for ancient gene duplication and endosymbiotic gene transfer. Proc Natl Acad Sci 103: 10328–10333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lio Y-C, Mazin AV, Kowalczykowski SC, Chen DJ 2003. Complex formation by the human Rad51B and Rad51C DNA repair proteins and their activities in vitro. J Biol Chem 278: 2469–2478. [DOI] [PubMed] [Google Scholar]
- Lisby M, Barlow JH, Burgess RC, Rothstein R 2004. Choreography of the DNA damage response: Spatiotemporal relationships among checkpoint and repair proteins. Cell 118: 699–713. [DOI] [PubMed] [Google Scholar]
- Liu N, Lamerdin JE, Tebbs RS, Schild D, Tucker JD, Shen MR, Brookman KW, Siciliano MJ, Walter CA, Fan W, et al. 1998. XRCC2 and XRCC3, new human Rad51-family members, promote chromosome stability and protect against DNA cross-links and other damages. Mol Cell 1: 783–793. [DOI] [PubMed] [Google Scholar]
- Liu N, Schild D, Thelen MP, Thompson LH 2002. Involvement of Rad51C in two distinct protein complexes of Rad51 paralogs in human cells. Nucleic Acids Res 30: 1009–1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Doty T, Gibson B, Heyer WD 2010. Human BRCA2 protein promotes RAD51 filament formation on RPA-covered single-stranded DNA. Nat Struct Mol Biol 17: 1260–1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Ehmsen KT, Heyer WD, Morrical SW 2011a. Presynaptic filament dynamics in homologous recombination and DNA repair. Crit Rev Biochem Mol Biol 46: 240–270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Renault L, Veaute X, Fabre F, Stahlberg H, Heyer WD 2011b. Rad51 paralogues Rad55-Rad57 balance the antirecombinase Srs2 in Rad51 filament formation. Nature 479: 245–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu T, Wan L, Wu Y, Chen J, Huang J 2011c. hSWS1•SWSAP1 is an evolutionarily conserved complex required for efficient homologous recombination repair. J Biol Chem 286: 41758–41766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lok BH, Powell SN 2012. Molecular pathways: Understanding the role of Rad52 in homologous recombination for therapeutic advancement. Clin Cancer Res 18: 6400–6406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lok BH, Carley AC, Tchang B, Powell SN 2013. RAD52 inactivation is synthetically lethal with deficiencies in BRCA1 and PALB2 in addition to BRCA2 through RAD51-mediated homologous recombination. Oncogene 32: 3552–3558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovett ST 1994. Sequence of the RAD55 gene of Saccharomyces cerevisiae: Similarity of RAD55 to prokaryotic RecA and other RecA-like proteins. Gene 142: 103–106. [DOI] [PubMed] [Google Scholar]
- Lovett ST, Mortimer RK 1987. Characterization of null mutants of the RAD55 gene of Saccharomyces cerevisiae: Effects of temperature, osmotic strength and mating type. Genetics 116: 547–553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madiraju MV, Templin A, Clark AJ 1988. Properties of a mutant recA-encoded protein reveal a possible role for Escherichia coli recF-encoded protein in genetic recombination. Proc Natl Acad Sci 85: 6592–6596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madiraju MV, Lavery PE, Kowalczykowski SC, Clark AJ 1992. Enzymatic properties of the RecA803 protein, a partial suppressor of recF mutations. Biochemistry 31: 10529–10535. [DOI] [PubMed] [Google Scholar]
- Maher RL, Branagan AM, Morrical SW 2011. Coordination of DNA replication and recombination activities in the maintenance of genome stability. J Cell Biochem 112: 2672–2682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarova KS, Koonin EV 2013. Archaeology of eukaryotic DNA replication. Cold Spring Harb Perspect Biol 5: a012963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malik PS, Symington LS 2008. Rad51 gain-of-function mutants that exhibit high affinity DNA binding cause DNA damage sensitivity in the absence of Srs2. Nucleic Acids Res 36: 6504–6510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mankouri HW, Ngo HP, Hickson ID 2007. Shu proteins promote the formation of homologous recombination intermediates that are processed by Sgs1-Rmi1-Top3. Mol Biol Cell 18: 4062–4073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martín V, Chahwan C, Gao H, Blais V, Wohlschlegel J, Yates JR, McGowan CH, Russell P 2006. Sws1 is a conserved regulator of homologous recombination in eukaryotic cells. EMBO J 25: 2564–2574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masson J-Y, Tarsounas M, Stasiak AZ, Stasiak A, Shah R, McIlwraith MJ, Benson FE, West SC 2001. Identification and purification of two distinct complexes containing the five RAD51 paralogs. Genes Dev 15: 3296–3307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McRobbie A-M, Carter LG, Kerou M, Liu H, McMahon SA, Johnson KA, Oke M, Naismith JH, White MF 2009. Structural and functional characterisation of a conserved archaeal RadA paralog with antirecombinase activity. J Mol Biol 389: 661–673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Mehta A, Haber JE 2014. Sources of DNA double-strand breaks and models for recombinational DNA repair. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a016428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morimatsu K, Kowalczykowski SC 2003. RecFOR proteins load RecA protein onto gapped DNA to accelerate DNA strand exchange: A universal step of recombinational repair. Mol Cell 11: 1337–1347. [DOI] [PubMed] [Google Scholar]
- Morimatsu K, Wu Y, Kowalczykowski SC 2012. RecFOR proteins target RecA protein to a DNA gap with either DNA or RNA at the 5′ terminus: Implication for repair of stalled replication forks. J Biol Chem 287: 35621–35630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Morrical SW 2014. DNA pairing and annealing processes in homologous recombination and homology-directed repair. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a016444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortensen UH, Bendixen C, Sunjevaric I, Rothstein R 1996. DNA strand annealing is promoted by the yeast Rad52 protein. Proc Natl Acad Sci 93: 10729–10734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortensen UH, Lisby M, Rothstein R 2009. Rad52. Curr Biol 19: R676–677. [DOI] [PubMed] [Google Scholar]
- Moynahan ME, Chiu JW, Koller BH, Jasin M 1999. Brca1 controls homology-directed DNA repair. Mol Cell 4: 511–518. [DOI] [PubMed] [Google Scholar]
- Moynahan ME, Cui TY, Jasin M 2001a. Homology-directed DNA repair, mitomycin-C resistance, and chromosome stability is restored with correction of a Brca1 mutation. Cancer Res 61: 4842–4850. [PubMed] [Google Scholar]
- Moynahan ME, Pierce AJ, Jasin M 2001b. BRCA2 is required for homology-directed repair of chromosomal breaks. Mol Cell 7: 263–272. [DOI] [PubMed] [Google Scholar]
- New JH, Sugiyama T, Zaitseva E, Kowalczykowski SC 1998. Rad52 protein stimulates DNA strand exchange by Rad51 and replication protein A. Nature 391: 407–410. [DOI] [PubMed] [Google Scholar]
- Pant K, Shokri L, Karpel RL, Morrical SW, Williams MC 2008. Modulation of T4 gene 32 protein DNA-binding activity by the recombination mediator protein UvsY. J Mol Biol 380: 799–811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park MS, Ludwig DL, Stigger E, Lee SH 1996. Physical interaction between human RAD52 and RPA is required for homologous recombination in mammalian cells. J Biol Chem 271: 18996–19000. [DOI] [PubMed] [Google Scholar]
- Park J-Y, Singh TR, Nassar N, Zhang F, Freund M, Hanenberg H, Meetei AR, Andreassen PR 2013. Breast cancer-associated missense mutants of the PALB2 WD40 domain, which directly binds RAD51C, RAD51 and BRCA2, disrupt DNA repair. Oncogene 10.1038/onc.2013.421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qing Y, Yamazoe M, Hirota K, Dejsuphong D, Sakai W, Yamamoto KN, Bishop DK, Wu X, Takeda S 2011. The epistatic relationship between BRCA2 and the other RAD51 mediators in homologous recombination. PLoS Genet 7: e1002148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabitsch KP, Tóth A, Gálová M, Schleiffer A, Schaffner G, Aigner E, Rupp C, Penkner AM, Moreno-Borchart AC, Primig M, et al. 2001. A screen for genes required for meiosis and spore formation based on whole-genome expression. Curr Biol 11: 1001–1009. [DOI] [PubMed] [Google Scholar]
- Radzimanowski J, Dehez F, Round A, Bidon-Chanal A, McSweeney S, Timmins J 2013. An “open” structure of the RecOR complex supports ssDNA binding within the core of the complex. Nucleic Acids Res 41: 7972–7986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rashid N, Morikawa M, Imanaka T 1996. A RecA/RAD51 homologue from a hyperthermophilic archaeon retains the major RecA domain only. Mol Gen Genet 253: 397–400. [DOI] [PubMed] [Google Scholar]
- Rijkers T, Van Den Ouweland J, Morolli B, Rolink AG, Baarends WM, Van Sloun PP, Lohman PH, Pastink A 1998. Targeted inactivation of mouse RAD52 reduces homologous recombination but not resistance to ionizing radiation. Mol Cell Biol 18: 6423–6429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivera MC, Lake JA 2004. The ring of life provides evidence for a genome fusion origin of eukaryotes. Nature 431: 152–155. [DOI] [PubMed] [Google Scholar]
- Rivera MC, Jain R, Moore JE, Lake JA 1998. Genomic evidence for two functionally distinct gene classes. Proc Natl Acad Sci 95: 6239–6244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saeki T, Machida I, Nakai S 1980. Genetic control of diploid recovery after γ-irradiation in the yeast Saccharomyces cerevisiae. Mutat Res 73: 251–265. [DOI] [PubMed] [Google Scholar]
- Sasanuma H, Tawaramoto MS, Lao JP, Hosaka H, Sanda E, Suzuki M, Yamashita E, Hunter N, Shinohara M, Nakagawa A, et al. 2013. A new protein complex promoting the assembly of Rad51 filaments. Nat Commun 4: 1676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schild D, Lio YC, Collins DW, Tsomondo T, Chen DJ 2000. Evidence for simultaneous protein interactions between human Rad51 paralogs. J Biol Chem 275: 16443–16449. [DOI] [PubMed] [Google Scholar]
- Seitz EM, Haseltine CA, Kowalczykowski SC 2001. DNA recombination and repair in the archaea. Adv Appl Microbiol 50: 101–169. [DOI] [PubMed] [Google Scholar]
- Sharan SK, Morimatsu M, Albrecht U, Lim DS, Regel E, Dinh C, Sands A, Eichele G, Hasty P, Bradley A 1997. Embryonic lethality and radiation hypersensitivity mediated by Rad51 in mice lacking Brca2. Nature 386: 804–810. [DOI] [PubMed] [Google Scholar]
- Shen Z, Cloud KG, Chen DJ, Park MS 1996. Specific interactions between the human RAD51 and RAD52 proteins. J Biol Chem 271: 148–152. [DOI] [PubMed] [Google Scholar]
- Sheng D, Zhu S, Wei T, Ni J, Shen Y 2008. The in vitro activity of a Rad55 homologue from Sulfolobus tokodaii, a candidate mediator in RadA-catalyzed homologous recombination. Extremophiles 12: 147–157. [DOI] [PubMed] [Google Scholar]
- Shinohara A, Shinohara M, Ohta T, Matsuda S, Ogawa T 1998. Rad52 forms ring structures and co-operates with RPA in single-strand DNA annealing. Genes Cells 3: 145–156. [DOI] [PubMed] [Google Scholar]
- Shor E, Weinstein J, Rothstein R 2005. A genetic screen for top3 suppressors in Saccharomyces cerevisiae identifies SHU1, SHU2, PSY3 and CSM2: Four genes involved in error-free DNA repair. Genetics 169: 1275–1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sigurdsson S, Van Komen S, Bussen W, Schild D, Albala JS, Sung P 2001. Mediator function of the human Rad51B–Rad51C complex in Rad51/RPA-catalyzed DNA strand exchange. Genes Dev 15: 3308–3318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singleton MR, Wentzell LM, Liu Y, West SC, Wigley DB 2002. Structure of the single-strand annealing domain of human RAD52 protein. Proc Natl Acad Sci 99: 13492–13497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith S, Hwang J-Y, Banerjee S, Majeed A, Gupta A, Myung K 2004. Mutator genes for suppression of gross chromosomal rearrangements identified by a genome-wide screening in Saccharomyces cerevisiae. Proc Natl Acad Sci 101: 9039–9044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somyajit K, Basavaraju S, Scully R, Nagaraju G 2013. ATM- and ATR-mediated phosphorylation of XRCC3 regulates DNA double-strand break-induced checkpoint activation and repair. Mol Cell Biol 33: 1830–1844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song B, Sung P 2000. Functional interactions among yeast Rad51 recombinase, Rad52 mediator, and replication protein A in DNA strand exchange. J Biol Chem 275: 15895–15904. [DOI] [PubMed] [Google Scholar]
- Sugiyama T, Kowalczykowski SC 2002. Rad52 protein associates with replication protein A (RPA)-single-stranded DNA to accelerate Rad51-mediated displacement of RPA and presynaptic complex formation. J Biol Chem 277: 31663–31672. [DOI] [PubMed] [Google Scholar]
- Sung P 1997a. Function of yeast Rad52 protein as a mediator between replication protein A and the Rad51 recombinase. J Biol Chem 272: 28194–28197. [DOI] [PubMed] [Google Scholar]
- Sung P 1997b. Yeast Rad55 and Rad57 proteins form a heterodimer that functions with replication protein A to promote DNA strand exchange by Rad51 recombinase. Genes Dev 11: 1111–1121. [DOI] [PubMed] [Google Scholar]
- *.Symington LS 2014. Processing of DNA breaks: Mechanism and regulation. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a016436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takata M, Sasaki MS, Sonoda E, Fukushima T, Morrison C, Albala JS, Swagemakers SM, Kanaar R, Thompson LH, Takeda S 2000. The Rad51 paralog Rad51B promotes homologous recombinational repair. Mol Cell Biol 20: 6476–6482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takata M, Sasaki MS, Tachiiri S, Fukushima T, Sonoda E, Schild D, Thompson LH, Takeda S 2001. Chromosome instability and defective recombinational repair in knockout mutants of the five Rad51 paralogs. Mol Cell Biol 21: 2858–2866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao Y, Li X, Liu Y, Ruan J, Qi S, Niu L, Teng M 2012. Structural analysis of Shu proteins reveals a DNA-binding role essential for resisting damage. J Biol Chem 287: 20231–20239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thacker J 2005. The RAD51 gene family, genetic instability and cancer. Cancer Lett 219: 125–135. [DOI] [PubMed] [Google Scholar]
- Thompson LH 2012. Recognition, signaling, and repair of DNA double-strand breaks produced by ionizing radiation in mammalian cells: The molecular choreography. Mutat Res 751: 158–246. [DOI] [PubMed] [Google Scholar]
- Thorslund T, McIlwraith MJ, Compton SA, Lekomtsev S, Petronczki M, Griffith JD, West SC 2010. The breast cancer tumor suppressor BRCA2 promotes the specific targeting of RAD51 to single-stranded DNA. Nat Struct Mol Biol 17: 1263–1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai SP, Su GC, Lin SW, Chung CI, Xue X, Dunlop MH, Akamatsu Y, Jasin M, Sung P, Chi P 2012. Rad51 presynaptic filament stabilization function of the mouse Swi5–Sfr1 heterodimeric complex. Nucleic Acids Res 40: 6558–6569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umezu K, Chi NW, Kolodner RD 1993. Biochemical interaction of the Escherichia coli RecF, RecO, and RecR proteins with RecA protein and single-stranded DNA-binding protein. Proc Natl Acad Sci 90: 3875–3879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Giezen M, Tovar J 2005. Degenerate mitochondria. EMBO Rep 6: 525–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Heijden T, van Noort J, van Leest H, Kanaar R, Wyman C, Dekker NH, Dekker C 2005. Torque-limited RecA polymerization on dsDNA. Nucleic Acids Res 33: 2099–2105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Sheng D, Han W, Huang B, Zhu S, Ni J, Li J, Shen Y 2012. Sulfolobus tokodaii RadA paralog, stRadC2, is involved in DNA recombination via interaction with RadA and Hjc. Sci China Life Sci 55: 261–267. [DOI] [PubMed] [Google Scholar]
- Wang Y, Xiao R, Wang H, Cheng Z, Li W, Zhu G, Wang Y, Ma H 2014. The Arabidopsis RAD51 paralogs RAD51B, RAD51D and XRCC2 play partially redundant roles in somatic DNA repair and gene regulation. New Phytol 201: 292–304. [DOI] [PubMed] [Google Scholar]
- White MF 2011. Homologous recombination in the archaea: The means justify the ends. Biochem Soc Trans 39: 15–19. [DOI] [PubMed] [Google Scholar]
- Wiese C, Collins DW, Albala JS, Thompson LH, Kronenberg A, Schild D 2002. Interactions involving the Rad51 paralogs Rad51C and XRCC3 in human cells. Nucleic Acids Res 30: 1001–1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams TA, Foster PG, Cox CJ, Embley TM 2013. An archaeal origin of eukaryotes supports only two primary domains of life. Nature 504: 231–236. [DOI] [PubMed] [Google Scholar]
- Wold MS 1997. Replication protein A: A heterotrimeric, single-stranded DNA-binding protein required for eukaryotic DNA metabolism. Annu Rev Biochem 66: 61–92. [DOI] [PubMed] [Google Scholar]
- Wooster R, Bignell G, Lancaster J, Swift S, Seal S, Mangion J, Collins N, Gregory S, Gumbs C, Micklem G 1995. Identification of the breast cancer susceptibility gene BRCA2. Nature 378: 789–792. [DOI] [PubMed] [Google Scholar]
- Wu D, Wu M, Halpern A, Rusch DB, Yooseph S, Frazier M, Venter JC, Eisen JA 2011. Stalking the fourth domain in metagenomic data: Searching for, discovering, and interpreting novel, deep branches in marker gene phylogenetic trees. PloS ONE 6: e18011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyman C 2011. Mechanistic insight from chaos: How RecA mediates DNA strand exchange. Structure 19: 1031–1032. [DOI] [PubMed] [Google Scholar]
- Wyman C, Kanaar R 2006. DNA double-strand break repair: All’s well that ends well. Ann Rev Genet 40: 363–383. [DOI] [PubMed] [Google Scholar]
- Xia B, Sheng Q, Nakanishi K, Ohashi A, Wu J, Christ N, Liu X, Jasin M, Couch FJ, Livingston DM 2006. Control of BRCA2 cellular and clinical functions by a nuclear partner, PALB2. Mol Cell 22: 719–729. [DOI] [PubMed] [Google Scholar]
- Xu H, Beernink HT, Morrical SW 2010. DNA-binding properties of T4 UvsY recombination mediator protein: Polynucleotide wrapping promotes high-affinity binding to single-stranded DNA. Nucleic Acids Res 38: 4821–4833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi-Iwai Y, Sonoda E, Buerstedde JM, Bezzubova O, Morrison C, Takata M, Shinohara A, Takeda S 1998. Homologous recombination, but not DNA repair, is reduced in vertebrate cells deficient in RAD52. Mol Cell Biol 18: 6430–6435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H, Li Q, Fan J, Holloman WK, Pavletich NP 2005. The BRCA2 homologue Brh2 nucleates RAD51 filament formation at a dsDNA–ssDNA junction. Nature 433: 653–657. [DOI] [PubMed] [Google Scholar]
- Yonesaki T, Minagawa T 1985. T4 phage gene uvsX product catalyzes homologous DNA pairing. EMBO J 4: 3321–3327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yonesaki T, Minagawa T 1989. Synergistic action of three recombination gene products of bacteriophage T4, uvsX, uvsY, and gene 32 proteins. J Biol Chem 264: 7814–7820. [PubMed] [Google Scholar]
- Yuan SS, Lee SY, Chen G, Song M, Tomlinson GE, Lee EY 1999. BRCA2 is required for ionizing radiation-induced assembly of Rad51 complex in vivo. Cancer Res 59: 3547–3551. [PubMed] [Google Scholar]
- *.Zickler D, Kleckner N 2014. Recombination, pairing, and synapsis of homologs during meiosis. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a016626. [DOI] [PMC free article] [PubMed] [Google Scholar]