Abstract
Telomeres have evolved to protect the ends of linear chromosomes from the myriad of threats posed by the cellular DNA damage signaling and repair pathways. Mammalian telomeres have to block nonhomologous end joining (NHEJ), thus preventing chromosome fusions; they need to control homologous recombination (HR), which could change telomere lengths; they have to avoid activating the ATM (ataxia telangiectasia mutated) and ATR (ATM- and RAD3-related) kinase pathways, which could induce cell cycle arrest; and they have to protect chromosome ends from hyperresection. Recent studies of telomeres have provided insights into the mechanisms of NHEJ and HR, how these double-strand break (DSB) repair pathways can be thwarted, and how telomeres have co-opted DNA repair factors to help in the protection of chromosome ends. These aspects of telomere biology are reviewed here with particular emphasis on recombination, the main focus of this collection.
Double-strand break repair pathways (e.g., nonhomologous end joining) can be deleterious at telomeres. Shelterin plays a critical role in repressing these pathways.
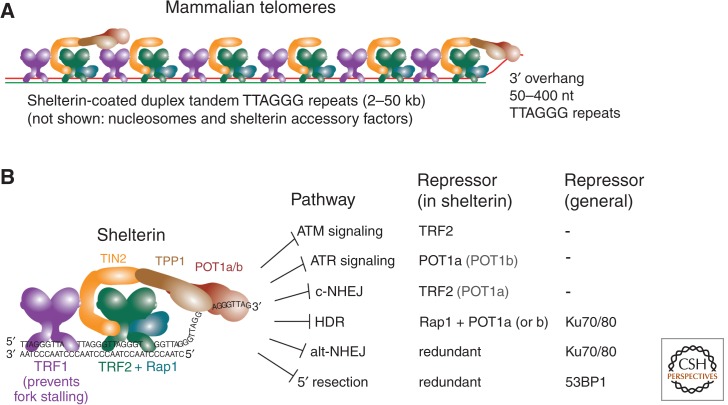
Mammalian telomeres are highly conserved in structure and function. They are built on long tandem arrays of duplex TTAGGG repeats that form the binding sites for the abundant telomere-specific protein complex, called shelterin (Fig. 1A). The telomeric repeat array ends in a 50- to 400-nt 3′ protrusion of the G-rich strand. The presence of duplex telomeric repeats, a telomere-specific protein complex, and a 3′ protrusion are general themes for all eukaryotic telomeres but the nature of the repeats and proteins vary widely. For instance, although a shelterin-like complex can be recognized in fission yeast and even trypanosomes, budding yeast telomeres function with a different set of proteins (Li et al. 2005; Lewis and Wuttke 2012). Another telomeric theme that is nearly universal is the mode of telomeric DNA maintenance, which involves the specialized reverse transcriptase, telomerase. Telomerase uses the 3′ end of the G-rich repeat strand as a primer and an internal RNA as a template to add telomeric repeats, thereby counteracting sequence loss resulting from semiconservative replication and nucleolytic processing (Blackburn and Collins 2011). Although there are organisms that maintain linear chromosomes without the help of telomerase (e.g., dipteran insects), they are rare. Mammalian cells can also maintain their telomeres in a telomerase-independent manner, using an homologous recombination (HR)-mediated process referred to as alternative lengthening of telomeres (ALT), which will be discussed in detail below.
Figure 1.
Telomeres, shelterin, and the end-protection problem. (A) The structure of mammalian telomeres, including the telomeric double-stranded DNA, the telomeric 3′ overhang, and the shelterin complex. (B) Schematic of the interactions among the six subunits that make up shelterin, their interactions with DNA, and their combined repression of the pathways that threaten telomeres (the end-protection problem).
Shelterin is the main mechanism by which telomeres solve the so-called end-protection problem. The end-protection problem refers to the collection of DNA damage response and repair pathways that can be deleterious at telomeres and therefore need to be repressed. Shelterin is composed of six structurally distinct proteins (Fig. 1B) (reviewed in Palm and de Lange 2008). TRF1 and TRF2 are diverged paralogs that have a similar domain structure, and both bind to duplex telomeric repeats as homodimers or -tetramers. Their large dimerization domains, called the TRF homology (TRFH) domains, also provide a binding surface for other shelterin components and shelterin-associated proteins. TRF1 and TRF2 are linked by TIN2, which helps to stabilize TRF1 and TRF2 on the telomeric DNA and, in addition, is crucial for the recruitment of two other shelterin proteins, TPP1 and POT1. POT1 binds to telomeric DNA in single-stranded form through an interaction of two OB-folds in the amino terminus with a 5′-(T)TAGGGTTAG-3′ recognition site. Although POT1 binds better to this site at a 3′ end, it will also associate with TTAGGG repeats when not located at a DNA end. Human cells have a single POT1 protein, whereas rodents have duplicated the POT1 gene, resulting in two functionally distinct but structurally closely related POT1 proteins at their telomeres, POT1a and POT1b. The sixth structurally distinct shelterin component is Rap1, which interacts with TRF2 but not with any of the other shelterin proteins.
Shelterin is ubiquitously expressed and sufficiently abundant to cover all telomeric repeats, even at the exceedingly long (20- to 50-kb) telomeres of Mus musculus. There is also sufficient TPP1/POT1 to engage the single-stranded telomeric DNA, although TPP1/POT1 are at least 10-fold less abundant than TRF1, TRF2, TIN2, and Rap1 (Takai et al. 2010). What regulates the abundance of shelterin is not yet known.
The repression of DNA damage signaling by shelterin is primarily the role of TRF2 and POT1 (POT1a in the mouse) (Fig. 1B). TRF2 is required to prevent the activation of the ATM kinase at chromosome ends, and the absence of POT1 leads to robust signaling by the ATR kinase. These aspects of the end-protection problem have been reviewed elsewhere (Palm and de Lange 2008; de Lange 2009). Shelterin is also involved in the regulation of telomerase-dependent maintenance of telomeres, mediating both the recruitment of telomerase and controlling the homeostasis of telomere length (Smogorzewska and de Lange 2004; Nandakumar and Cech 2013). Below, we will briefly discuss how shelterin controls classical- and alternative nonhomologous end joining (NHEJ). The major part of this article will focus on how telomeres engage the cellular HR-related pathways—including 5′ end resection—to achieve their optimal protected state, how telomeres prevent deleterious HR reactions, and what is known about the HR-dependent maintenance of telomeres in ALT cells.
PHYSIOLOGICAL AND PATHOLOGICAL RESECTION AT TELOMERES
Generating the Telomeric Overhang: Shelterin-Controlled 5′ End Processing
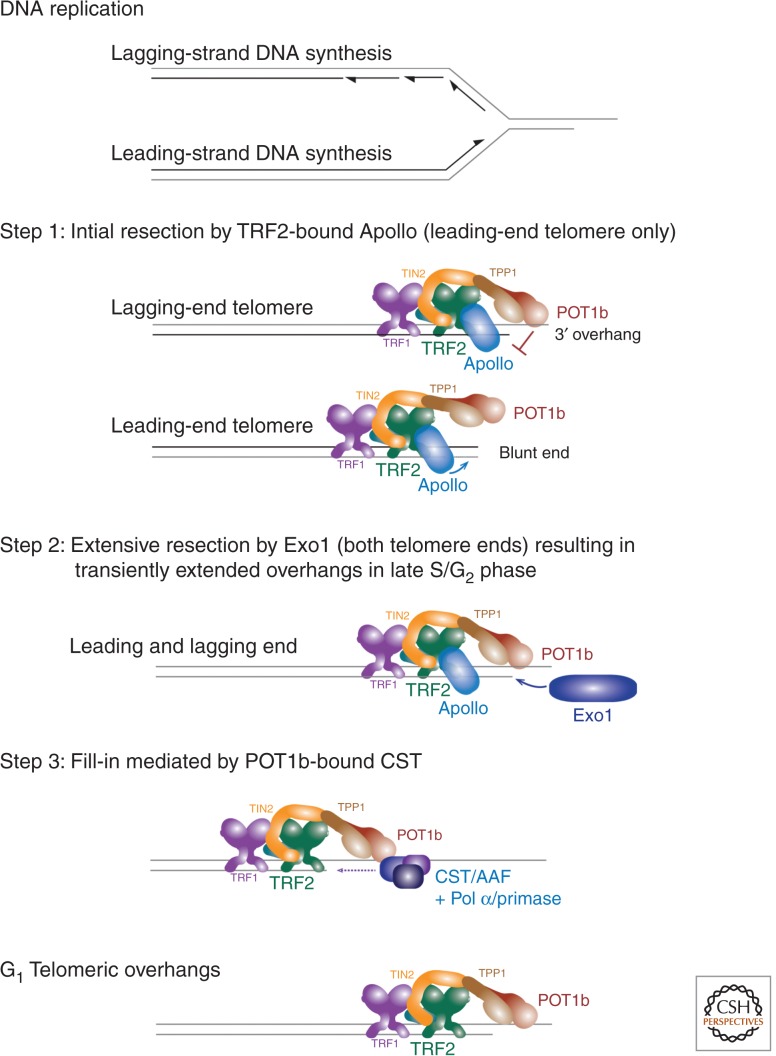
The first step in the initiation of homologous recombination and all forms of homology-directed repair is the resection of the 5′ ended strand at a DSB to generate a 3′ overhang. 5′ end resection also occurs at functional telomeres, but, interestingly, this process is very different from the processing of DSBs (for additional material on DSB end resection, see Symington [2014]). All mammalian telomeres have an overhang, implying that the telomeres formed by leading- and lagging-strand DNA synthesis (referred to as leading-end and lagging-end telomeres) acquire a 3′ protrusion after DNA replication (Fig. 2) (Makarov et al. 1997; McElligott and Wellinger 1997). This overhang is longer at lagging-end telomeres, at least in telomerase-negative human cells (Chai et al. 2006; Chow et al. 2012). Because leading-strand DNA synthesis will not generate a 3′ overhang and telomerase makes the overhangs longer but is not required for their presence (Hemann and Greider 1999; Chai et al. 2006; Wu et al. 2012), 5′ end resection is the most likely explanation for the acquisition of the 3′ overhangs at leading-end telomeres. In fact, it is now clear that 5′ end resection takes place at both leading-end telomeres and lagging-end telomeres. The generation of the 3′ overhangs has emerged as a multistep process that is closely controlled by shelterin.
Figure 2.
Generation of the telomeric 3′ overhang. Schematic of the three steps involved in the regeneration of the 3′ overhang at the telomere replicated by leading- and lagging-strand DNA synthesis. See text for details. (Figure based on data from Wu et al. 2012.)
The first indication that shelterin controls the formation of the 3′ overhang came from partial (short hairpin RNA [shRNA]) knockdown studies of human POT1, which revealed a reduction in the telomeric overhang signal and a change in the sequence at the 5′ end of telomeres. Remarkably, all human telomeres end on the sequence 3′-CCAATC-5′ (Sfeir et al. 2005) but when POT1 is depleted, this terminal specificity is lost (Hockemeyer et al. 2005). However, the partial removal of POT1 from human telomeres also activates a DNA damage response (Hockemeyer et al. 2005), confounding the interpretation of the data. In the mouse, however, the repression of ATR kinase signaling is primarily the task of POT1a, whereas POT1b is dedicated to the control of the telomeric overhang, clarifying the interpretation of phenotypes obtained with deletion of POT1b (Hockemeyer et al. 2006).
Deletion of POT1b results in a 2- to 4-fold increase in the telomeric overhang signal (Hockemeyer et al. 2006). This effect accelerates telomere shortening when there is insufficient telomerase activity in the cells, consistent with the increase in single-stranded DNA being caused by exonucleolytic attack (Hockemeyer et al. 2008; He et al. 2009). The exact nature of the defect caused by loss of POT1b became clear in recent work with compound genetic mouse cells lacking two critical nucleases, Apollo and Exonuclease 1.
Apollo is an SMN1B/PSO2-type nuclease with a role in interstrand cross-link repair that binds to the TRFH domain of TRF2 (Freibaum and Counter 2006; Lenain et al. 2006; van Overbeek and de Lange 2006; Chen et al. 2008). At telomeres, the TRF2-bound Apollo is required for the generation of the 3′ overhang at the leading ends (Lam et al. 2010; Wu et al. 2010, 2012). When Apollo is absent or cannot interact with TRF2, leading-end telomeres (separated on CsCl gradient [Wu et al. 2012]) show a diminished overhang signal, activate the ATM kinase, and fuse to other leading-end telomeres.
The resection of newly replicated telomeres by Apollo is controlled by POT1b (Wu et al. 2012). When POT1b is absent, hyperresection by Apollo occurs at both leading- and lagging-end telomeres. Presumably, POT1b will normally inhibit Apollo at lagging-end telomeres immediately after DNA replication, because they will have a short overhang to which POT1b can bind. At leading-end telomeres, POT1b is proposed to inhibit Apollo after the nuclease has generated a POT1b binding site (Fig. 2).
The second nuclease, EXO1, acts at both leading- and lagging-end telomeres, presumably after Apollo. EXO1 generates highly extended overhangs but their presence is transient (Wu et al. 2012). This transient extension of the overhangs in S phase has been observed in mouse and human cells (Dai et al. 2010; Wu et al. 2010, 2012). How EXO1 is recruited to telomeres in absence of a DNA damage response is not clear. At DSBs, EXO1 is dependent on RPA, which presumably is not available at functional/protected telomeres (Nimonkar et al. 2011; Cannavo et al. 2013; Chen et al. 2013). Perhaps TPP1/POT1a can provide this function.
Because the extended overhangs generated by EXO1 are returned to a shorter length by the time cells are in G1, it appears that a fill-in reaction is taking place. The candidate for the critical factor mediating this fill-in reaction, CST, emerged from work on budding yeast telomeres and polymerase α (Pol α)/primase accessory factor (reviewed in Price et al. 2010). Budding yeast telomeres are protected by a complex of Cdc13, Stn1, and Ten1 (CST), which prevents the activation of the ATR kinase homolog, Mec1. The RPA-like CST complex is not related to shelterin, although it uses OB-folds to bind to single-stranded DNA, as does POT1 (Gao et al. 2007; Lewis and Wuttke 2012). The mammalian ortholog of this complex was recognized by two groups as a complex of Ctc1/Stn1/Ten1 and also identified as the Pol α/primase accessory factor AAF (Casteel et al. 2009; Miyake et al. 2009; Surovtseva et al. 2009; Wan et al. 2009). In human cells, CST interacts with TPP1 (Wan et al. 2009), and knockdown of Stn1 results in an increase in the telomeric overhang (Miyake et al. 2009; Surovtseva et al. 2009), although the cell cycle effects associated with Stn1 inhibition might be a confounder. In mouse cells, CST interacts with POT1b and mutations in the POT1b residues required for this interaction also lead to extended overhangs (Wu et al. 2012). These extended overhangs arise in late S/G2 and fail to be shortened to their normal G1 length. These data suggest that the POT1b-mediated recruitment of CST is needed for a fill-in step that returns the telomeric overhangs back to their normal lengths after EXO1 processing. Essentially, the same conclusion was reached on the necessity for CST-mediated fill-in at human telomeres (Huang et al. 2012; Wang et al. 2012; Kasbek et al. 2013). Given the ability of CST/AAF to promote Pol α/primase activity on single-stranded DNA (Nakaoka et al. 2012), it is reasonable to assume that this is the way CST acts at telomeres (Fig. 2).
This highly regulated processing of the newly replicated telomere ends appears to be designed to ensure that both sister telomeres acquire an overhang. In addition, the tight regulation of telomere-end processing provides a way to control the rate of sequence loss at telomeres. Resection-dependent sequence loss ultimately determines the rate of telomere shortening in cells lacking telomerase. The gradual and progressive shortening of telomeres in the human soma is thought to represent a tumor suppressor mechanism that curbs the proliferative potential of incipient tumor cells. It may therefore be important for human somatic cells to control the rate of shortening by modulating the resection and fill-in steps.
The 3′ overhang itself is important for several aspects of telomere function. It functions as the priming site of telomerase, which is incapable of acting on a blunt end, and the 3′ overhang is thought to be crucial for the protection of chromosome ends.
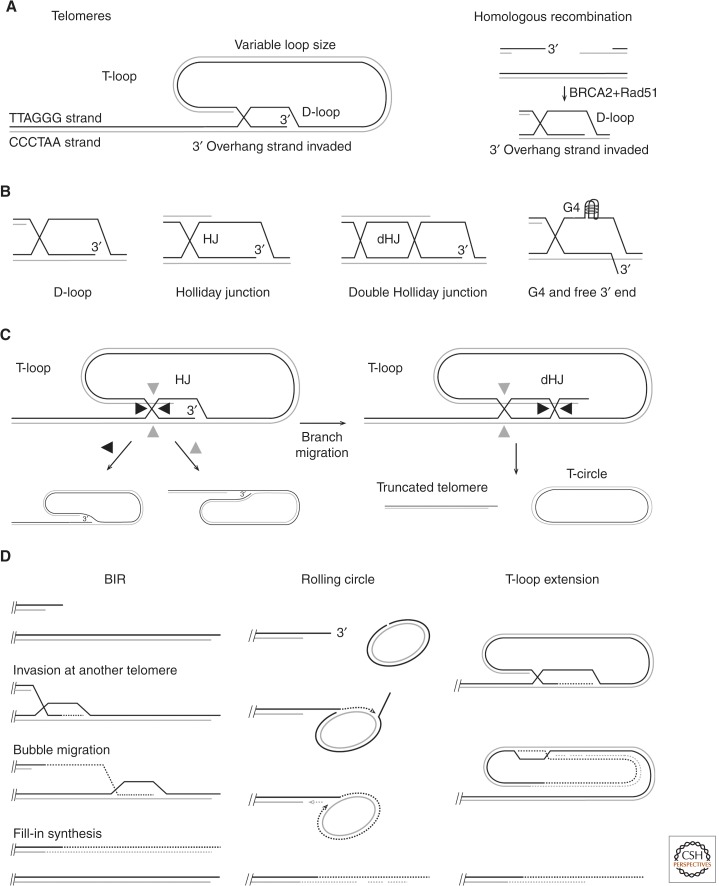
The best-understood role for the 3′ overhang is mediating the formation of the T-loop structure, by strand invasion into the double-stranded telomeric repeats (see Fig. 4) (Griffith et al. 1999). T-loops have been proposed to provide an architectural solution to the end-protection problem by sequestering the end of the chromosome from end-initiated DNA damage response pathways. T-loop formation itself could be a mechanism to block hyperresection at telomeres similarly to what happens after strand invasion in homologous recombination (Sugawara et al. 2003).
Figure 4.
Homologous recombination (HR)-related structures invoked at telomeres. (A) The structure of the T-loop compared to the strand invasion step in HR (telomeric sequences written from 5′ to 3′). (B) Possible structures at the base of the T-loop compatible with T-loop detection after psoralen cross-linking. (C) Products predicted from the indicated Holliday junction (HJ) resolution (cleavage and ligation) of the two T-loop structures shown. The products of the double Holliday junction (dHJ) schematic have been detected in cells overexpressing a mutant form of TRF2 lacking the amino-terminal basic domain (Wang et al. 2004). (D) Proposed mechanisms of telomere DNA synthesis in ALT cells. See text for discussion.
Hyperresection at Dysfunctional Telomeres
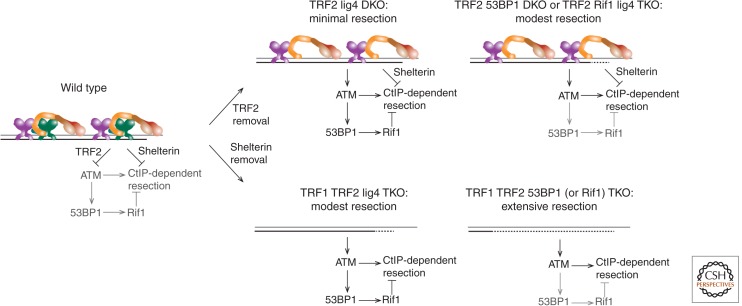
When POT1b is removed from telomeres, hyperresection of the newly replicated telomeres occurs, but this resection is distinct from resection at DSBs in that it is not directed by DNA damage signaling. ATM-dependent resection can take place at dysfunctional telomeres but only in cells that lack 53BP1, which has recently emerged as a general repressor of DSB resection in mammalian cells (Fig. 1B) (Bothmer et al. 2010; Bunting et al. 2010; Noon and Goodarzi 2011). When TRF2 is removed from telomeres in 53BP1−/− cells, telomere ends are processed in S phase by ATM- and CtIP-dependent resection (Fig. 3) (Lottersberger et al. 2013). Curiously, this resection only takes place at leading-end telomeres; the reason for this preference is not clear. The leading-end telomeres are supposedly blunt after replication and in TRF2-deficient cells they will not undergo their normal Apollo-dependent resection. Perhaps at these leading ends, which lack binding sites for POT1a and POT1b, CtIP-dependent resection can take place, whereas at the lagging-end telomeres POT1a and POT1b are protective.
Figure 3.
Repression of hyperresection at telomeres. ATM/CtIP-dependent resection at telomeres is repressed by several independent pathways. On the one hand, the 53BP1 binding partner RIF1 inhibits resection at telomeres that have become dysfunctional such that the ATM kinase pathway has been activated. On the other hand, resection is repressed by TRF2 in shelterin preventing the activation of the ATM kinase and protecting telomeres from moderate resection when 53BP1 is absent (top left and right). In addition, other components in shelterin repress resection so that extensive hyperresection only occurs in the absence of the complete shelterin complex (bottom right).
TRF2 is not the only component within shelterin required to repress hyperresection at telomeres. Modest resection also occurs after TRF1 deletion from 53BP1−/− cells (Sfeir and de Lange 2012). However, when all shelterin proteins (and all shelterin-associated factors) are removed from telomeres, the hyperresection is dramatically increased, indicating that different shelterin subunits act redundantly in blocking nucleolytic attack on chromosome ends (Fig. 3) (Sfeir and de Lange 2012). RNAi knockdown experiments have implicated CtIP, EXO1, and the BLM RECQ helicase in this resection but other factors are not excluded. Thus, the hyperresection taking place at dysfunctional telomeres that activate the ATM kinase signaling pathway bears a strong resemblance to the resection of DSBs (see Symington 2014).
Recent data has revealed how 53BP1 represses resection at telomeres and DSBs (Chapman et al. 2013; Di Virgilio et al. 2013; Escribano-Diaz et al. 2013; Feng et al. 2013; Zimmermann et al. 2013). The main factor in this process is RIF1, a protein originally identified as a part of the telomeric protein complex in budding yeast (Hardy et al. 1992). In mammals, RIF1 is not found at telomeres but localizes to DSBs in an ATM- and 53BP1-dependent manner (Silverman et al. 2004). It was found that the amino-terminal S/TQ ATM target sites of 53BP1 are required for most of 53BP1’s functions, including blocking resection in various contexts (Bothmer et al. 2011; Lottersberger et al. 2013), and for the recruitment of RIF1 to sites of DNA damage (Chapman et al. 2013; Di Virgilio et al. 2013; Escribano-Diaz et al. 2013; Feng et al. 2013; Zimmermann et al. 2013). RIF1’s role in repressing resection at telomeres was shown directly in the context of dysfunctional telomeres, thus identifying RIF1 as a factor downstream from 53BP1 in suppressing resection (Fig. 3) (Zimmermann et al. 2013). The same pathway was found to act at DSBs, further reinforcing the similarities between DSBs and dysfunctional telomeres (Chapman et al. 2013; Di Virgilio et al. 2013; Escribano-Diaz et al. 2013; Feng et al. 2013; Zimmermann et al. 2013).
HOMOLOGY-DIRECTED REPAIR REACTIONS AT TELOMERES
Formation of T-Loops and Their Function
In its simplest form, the T-loop structure resembles an early step in homologous recombination because the 3′ overhang has strand-invaded homologous sequences, resulting in the formation of a D-loop (Fig. 4A). In homologous recombination, the strand invasion is mediated by BRCA2-loaded RAD51, and the invading strand would be initially coated by RAD51. Whether BRCA2 and RAD51 are also involved in T-loop formation has not yet been established. Recent experiments have indicated that TRF2 is the main component of shelterin required for the formation/maintenance of T-loops (Doksani et al. 2013), and recombinant TRF2 has the ability to promote a loop-forming reaction on telomere model substrates in vitro (Griffith et al. 1999; Stansel et al. 2001).
How might TRF2 promote looping in vitro? One possibility is based on the inherent ability of TRF2 to change the topology of the DNA it is bound to, which results in untwisting of the surrounding sequences and therefore can stimulate strand invasion (Amiard et al. 2007; Poulet et al. 2009). In addition, TRF2 has an amino-terminal domain, referred to as the basic domain, that binds Holliday junctions (HJs) in vitro and may therefore stabilize the strand-invasion event (Fouche et al. 2006; Poulet et al. 2009). These features, together with the propensity of TRF2 to initiate binding near the end of the telomeric repeat array in vitro (Stansel et al. 2001), may explain how TRF2 can form T-loop-like structures in vitro; however, whether these features are relevant to the in vivo situation is not yet known.
It is not unlikely that TRF2 acts in conjunction with other factors to establish the T-loop structure and to protect it (see also below). It is important to note that the exact nature of the structure at the base of the T-loop is not known (Fig. 4B gives four options but others are not excluded). Given that the basic domain of TRF2 can promote the formation of HJs and stabilize them (Fouche et al. 2006; Poulet et al. 2009), it is tempting to speculate that this is the most likely configuration (Fig. 4B, second option).
T-loops have been proposed to represent an architectural solution to the end-protection problem posed by linear chromosomes (Griffith et al. 1999). It appears likely that the telomere end, when in the T-loop configuration, would be impervious to end-loading factors that initiate DNA damage response pathways. Two prominent examples are the MRN (MRE11/RAD50/NBS1)-initiated ATM kinase signaling pathway and KU70/80-dependent classical NHEJ. Neither MRN nor KU70/80 may be able to recognize the telomere for what it is, a DNA end, when the terminus is sequestered in the T-loop. Indeed, the shelterin component implicated in T-loop formation/maintenance, TRF2, also is critical for the repression of ATM signaling and classical NHEJ (de Lange 2009; Doksani et al. 2013).
It is generally assumed that T-loops are not a substrate for telomerase because the 3′ end would be base-paired. However, given that the exact structure at the base of the T-loop is not known, it is not excluded that part of the 3′ overhang is extruded and allows telomerase access (see Fig. 4B). Of course, it is also possible that telomerase gains access to the 3′ terminus when T-loops are resolved during DNA replication.
Protecting of T-Loops from Resolution and Other Steps in HR
Although the T-loop offers an architectural solution to many aspects of the end-protection problem, T-loops also create several challenges because of their structural resemblance to HR intermediates. The D-loop and possible HJ structures at the base of the loop are good substrates for nucleases and HJ resolution activities, such as MUS81/EME1, SLX4/SLX1, or GEN1 (Gaillard et al. 2003; Osman et al. 2003; Wyatt et al. 2013; Wyatt and West 2014). If a dHJ is generated by branch migration, its resolution can cleave off the loop part, generating a shortened telomere and a circular product often referred to as a T-circle (Fig. 4C, right). These products have been detected in cells induced to overexpress a TRF2 mutant lacking the amino-terminal basic domain (Wang et al. 2004). The interpretation of these findings is that the basic domain normally occupies the junction at the base of the T-loop and hence prevents HJ resolvases from gaining access and/or blocks branch migration. However, even without extensive branch migration, the D-loop itself or the single HJ intermediate will probably need protection because its resolution could give rise to unwanted products when processed by nucleases and/or resolvases (Fig. 4C, left).
A second problem inherent to the T-loop structure is that the 3′ end could be extended by polymerases, as it would be during HR (Mehta and Haber 2014). How this is prevented is not clear. It is also not clear what prevents the T-loop from forming on the sister telomere (in trans), or on chromosome-internal telomeric sequences (in trans or in cis) (e.g., Zhu et al. 2003). Finally, it has been argued that T-loops need to be resolved before the replication fork can progress through the telomeres and that the RTEL1 helicase is required for this resolution (Vannier et al. 2012). However, because the MCM replicative helicase is a 3′ to 5′ translocase (Fu et al. 2011), it should by itself be able to resolve the strand invasion that locks down the T-loop.
Repression of HR between Sister Telomeres
Sequence exchanges between sister telomeres, referred to as telomere sister chromatid exchanges or T-SCEs, are harmless as long as they are equal. If an unequal exchange occurs, however, one daughter cell will inherit a shortened telomere, which will dictate a shorter replicative life span if the cell lacks a telomere maintenance system. Because each nucleus contains a large number of telomeres, sequence exchanges between sister telomeres have to be stringently repressed to provide the population as a whole with its proper proliferative capacity. Telomeres, representing a DNA end, are inherently recombigenic. How is their recombination avoided? One obvious way is by forming a T-loop, which will sequester the telomere terminus; but genetic analysis shows that there are additional mechanisms at work. The KU70/80 heterodimer is an important repressor of HR at telomeres (Celli et al. 2006; Wang et al. 2009), analogous to its ability to repress HR at DSBs (Pierce et al. 2001). In addition, members of the shelterin complex repress the exchanges between sister telomeres. Notably, T-SCEs are only observed at substantial frequencies (>8% of telomeres) in two settings: in KU70/80-deficient mouse cells lacking either Rap1 or both POT1a and POT1b (Fig. 1B) (Palm et al. 2009; Sfeir et al. 2010). Thus, the shelterin-dependent repression of HR involves the combined action of Rap1 and one of the POT1 proteins. How these proteins block HR is not clear. In the Rap1/KU70 double-knockout cells, it was notable that there is no DNA damage signaling at telomeres, yet they undergo homologous recombination (Sfeir et al. 2010). Perhaps the lack of requirement for DNA damage signaling in this setting is because of the presence of an overhang at the telomeres, circumventing the need for ATM kinase-dependent resection.
Alternative Lengthening of the Telomeres
Telomere maintenance in the absence of telomerase was first described in budding yeast and shown to involve recombination (Lundblad and Blackburn 1993). Telomerase-independent telomere maintenance in human cells was first observed in mostly virally transformed cell populations that survived telomere crisis and became immortal (Bryan et al. 1995). Such immortal clones most often express telomerase, but a substantial number were found to maintain telomeres in the absence of detectable telomerase activity. Although first observed in cultured cells, it is now clear that the ALT pathway can sustain telomeres in a considerable subset of human cancers (reviewed in Henson and Reddel 2010).
ALT cells appear to use HR to maintain telomeric sequences. Their telomeres show an elevated rate of T-SCEs and a neo cassette embedded within one telomere was shown to spread to other telomeres (Dunham et al. 2000; Bechter et al. 2004; Londono-Vallejo et al. 2004). Furthermore, extrachromosomal telomeric DNA, including single-stranded and duplex circular telomeric DNA, is observed in ALT cells as if T-loops are more vulnerable to resolution by HJ resolvases (Tokutake et al. 1998; Cesare and Griffith 2004; Wang et al. 2004; Henson et al. 2009). ALT telomeres are also often heterogeneous in length (Bryan et al. 1995), consistent with increased recombination, and can show spreading of variant telomeric repeats through the telomeres (Varley et al. 2002; Conomos et al. 2012). Finally, the telomeric DNA in ALT cells is often associated with PML bodies and a host of recombination and DNA damage response proteins (Yeager et al. 1999).
Despite these indications that HR is unleashed at ALT telomeres, the actual mechanism by which the telomeres are maintained and how ALT is activated is unknown. The single common denominator of all ALT cells is the presence of extrachromosomal telomeric DNA that is often circular and has been proposed to function as a template for telomere elongation (Fig. 4D) (Henson et al. 2002). Telomere elongation could also occur in cis, as has been directly shown by the local amplification of a cassette inserted in a telomere (Muntoni et al. 2009). T-loop extension is a likely mechanism of telomere elongation (see Fig. 4D). The idea that the telomere maintenance in these cells is dependent on HR is consistent with the finding that diminished activity of BRCA2, MUS81, FEN1, FANCD2, FANCA, SMC5/6-dependent sumoylation, RAD50, and other recombination factors affects telomere maintenance in ALT cells (reviewed in Gocha et al. 2013).
Telomere replication problems could play an important role in inducing recombination in ALT cells. First, ALT telomeres appear to have internal nicks or gaps that would cause frequent replication fork collapse at telomeres (Nabetani and Ishikawa 2009). Second, some of the recombination proteins required for telomere maintenance in ALT cells, including MUS81, FEN1, FANCD2, and FANCA, are known to play an important role in replication-coupled repair. Accumulation of broken replication forks at telomeres would be highly recombinogenic considering the repetitive nature of telomeric DNA and thus the abundance of homology for strand invasion. A broken end could invade the sister chromatid at different positions and generate equal or unequal T-SCEs. Strand invasion could also involve a telomere of another chromosome, and, in both cases, the invading end could be involved in break-induced replication (BIR) (Fig. 4D) (Donnianni and Symington 2013; Saini et al. 2013; Wilson et al. 2013; Mehta and Haber 2014).
The only consistent genetic defect so far identified in ALT is the loss of ATRX (Heaphy et al. 2011a,b; Lovejoy et al. 2012). ATRX is a SWI/SNF-related chromatin remodeling enzyme that was shown to deposit the histone variant H3.3 into telomeric chromatin in mouse embryonic stem cells (Goldberg et al. 2010; Wong et al. 2010). How the loss of ATRX or H3.3 could lead to a higher level of telomere recombination is entirely unclear at this stage (e.g., Clynes and Gibbons 2013; Conomos et al. 2013). It is likely that in addition to ATRX deletion, a second genetic or epigenetic change is required to unleash ALT. So far, no changes in shelterin have been found in ALT cells and the mechanism of ALT activation remains elusive.
REPRESSION OF NHEJ
Classical NHEJ
Telomeres are threatened by classical KU70/80- and DNA ligase IV–dependent NHEJ (Smogorzewska et al. 2002; Celli and de Lange 2005; Celli et al. 2006). This type of DSB processing of dysfunctional telomeres takes place primarily in G1, resulting in chromosome-type fusions (the telomeres of both chromatids are fused to other telomeres) in metaphase (Konishi and de Lange 2008). However, postreplicative DNA ligase IV–dependent telomere fusions, which typically give rise to chromatid-type fusions (one chromatid fused to another) or sister telomere fusions, have also been observed (Smogorzewska et al. 2002; Dimitrova and de Lange 2009; Hsiao and Smith 2009). TRF2 is the main component of shelterin involved in blocking classical NHEJ. The mechanism by which TRF2 acts most likely involves the formation of the T-loop configuration. However, other aspects of TRF2 may be important as well because TRF2 has been reported to repress DNA repair when tethered to a nontelomeric site next to an inducible DSB, a context in which T-loop formation is impossible (Fumagalli et al. 2012). One way TRF2 may be acting in this context is through the action of its recently discovered iDDR domain (Okamoto et al. 2013). This small region in the TRF2 linker between the TRFH domain and the Myb/SANT DNA binding domain interacts with both the MRN complex and the BRCC36 deubiquitylating enzyme, and through these two modules, it inhibits the loading of 53BP1 at sites of DNA damage. In the absence of 53BP1, classical NHEJ could be thwarted both at telomeres and at other sites through induction of excessive resection.
In addition to TRF2, the engagement of the POT1 proteins on the single-stranded DNA may be acting as a deterrent to classical NHEJ. Deletion of POT1a results in a low but significant level of fusions between sister telomeres that is exacerbated when POT1b is deleted as well (Hockemeyer et al. 2006). However, it has not been established whether these are classical NHEJ events. The idea that the 3′ overhang (with POT1 proteins engaged) could block the loading of KU70/80 was also invoked to explain the paradoxical finding on TRF2 removal from telomeres in ATM-deficient cells. It was found that leading-end telomeres that had lost TRF2 were protected from NHEJ by ATM-mediated resection (Attwooll et al. 2009; Dimitrova and de Lange 2009; Lottersberger et al. 2013). Whether the 3′ overhang can really protect telomeres from NHEJ and, if so, how long the overhang needs to be and whether it has to be coated with POT1 proteins to block NHEJ remain to be determined.
Alternative NHEJ
The final DSB repair reaction that threatens telomeres is fusion through alternative NHEJ (alt-NHEJ). Hints that such alt-NHEJ might promote telomere fusions were seen in experiments in which multiple components of shelterin were targeted in KU-deficient mouse cells (Rai et al. 2010). Further insight into the type of alt-NHEJ taking place at dysfunctional telomeres recently came from experiments in which telomeres were rendered completely free of all shelterin proteins (Sfeir and de Lange 2012). When shelterin is removed from telomeres in KU-deficient cells, the telomere fusions are extremely prominent, involving up to 65% of the chromosome ends. These fusions are mediated by DNA ligase III and PARP1 and appear to rely on the microhomology provided by the TTAGGG repeats in the 3′ overhangs, which can form two base pairs per repeat. Similar to what happens at DSBs, alt-NHEJ at telomeres is repressed by KU70/80 (Wang et al. 2006). Within shelterin, multiple proteins contribute to the repression of alt-NHEJ, because the individual deletion of shelterin components in a KU-deficient background does not unleash the same dramatic alt-NHEJ phenotype as the complete removal of shelterin.
CONCLUDING REMARKS
The connection between telomere biology and DSB repair pathways is akin to the link between immunology and infectious diseases in which the study of one aspect of biology informs the other and vice versa. It is now clear that mammalian telomeres are threatened by each of the processing and repair pathways that can act on DSBs. In each case, shelterin plays a critical role in repressing these pathways, but much remains to be learned about how this complex acts. Future insights into the biochemical and structural aspects of shelterin will be helpful as will be additional information on how exactly these dangerous pathways are initiated on a substrate like an unprotected telomere. Another question that merits attention is how telomeres use DSB repair factors for protective purposes. An example is how shelterin manages to use EXO1 and Apollo to generate the 3′ overhang while avoiding hyperresection. Further insights into these types of questions will not only increase the understanding of telomeres but also further illuminate how DSB repair works.
ACKNOWLEDGMENTS
Our research is supported by grants from the National Institutes of Health (NIH), National Cancer Institute (NCI), and the Breast Cancer Research Foundation. Y.D. is an Ellison Medical Foundation/AFAR Fellow of the Life Sciences Research Foundation. T.d.L. is an American Cancer Society Research Professor.
Footnotes
Editors: Stephen Kowalczykowski, Neil Hunter, and Wolf-Dietrich Heyer
Additional Perspectives on DNA Recombination available at www.cshperspectives.org
REFERENCES
*Reference is also in this collection.
- Amiard S, Doudeau M, Pinte S, Poulet A, Lenain C, Faivre-Moskalenko C, Angelov D, Hug N, Vindigni A, Bouvet P, et al. 2007. A topological mechanism for TRF2-enhanced strand invasion. Nat Struct Mol Biol 14: 147–154. [DOI] [PubMed] [Google Scholar]
- Attwooll CL, Akpinar M, Petrini JH 2009. The mre11 complex and the response to dysfunctional telomeres. Mol Cell Biol 29: 5540–5551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bechter OE, Shay JW, Wright WE 2004. The frequency of homologous recombination in human ALT cells. Cell Cycle 3: 547–549. [PubMed] [Google Scholar]
- Blackburn EH, Collins K 2011. Telomerase: An RNP enzyme synthesizes DNA. Cold Spring Harb Perspect Biol 3: a003558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bothmer A, Robbiani DF, Feldhahn N, Gazumyan A, Nussenzweig A, Nussenzweig MC 2010. 53BP1 regulates DNA resection and the choice between classical and alternative end joining during class switch recombination. J Exp Med 207: 855–865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bothmer A, Robbiani DF, Di Virgilio M, Bunting SF, Klein IA, Feldhahn N, Barlow J, Chen HT, Bosque D, Callen E, et al. 2011. Regulation of DNA end joining, resection, and immunoglobulin class switch recombination by 53BP1. Mol Cell 42: 319–329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryan TM, Englezou A, Gupta J, Bacchetti S, Reddel RR 1995. Telomere elongation in immortal human cells without detectable telomerase activity. EMBO J 14: 4240–4248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bunting SF, Callen E, Wong N, Chen HT, Polato F, Gunn A, Bothmer A, Feldhahn N, Fernandez-Capetillo O, Cao L, et al. 2010. 53BP1 inhibits homologous recombination in Brca1-deficient cells by blocking resection of DNA breaks. Cell 141: 243–254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cannavo E, Cejka P, Kowalczykowski SC 2013. Relationship of DNA degradation by Saccharomyces cerevisiae Exonuclease 1 and its stimulation by RPA and Mre11-Rad50-Xrs2 to DNA end resection. Proc Natl Acad Sci 110: E1661–E1668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casteel DE, Zhuang S, Zeng Y, Perrino FW, Boss GR, Goulian M, Pilz RB 2009. A DNA polymerase-α·primase cofactor with homology to replication protein A-32 regulates DNA replication in mammalian cells. J Biol Chem 284: 5807–5818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celli GB, de Lange T 2005. DNA processing is not required for ATM-mediated telomere damage response after TRF2 deletion. Nat Cell Biol 7: 712–718. [DOI] [PubMed] [Google Scholar]
- Celli GB, Lazzerini Denchi E, de Lange T 2006. Ku70 stimulates fusion of dysfunctional telomeres yet protects chromosome ends from homologous recombination. Nat Cell Biol 8: 885–890. [DOI] [PubMed] [Google Scholar]
- Cesare AJ, Griffith JD 2004. Telomeric DNA in ALT cells is characterized by free telomeric circles and heterogeneous t-loops. Mol Cell Biol 24: 9948–9957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chai W, Du Q, Shay JW, Wright WE 2006. Human telomeres have different overhang sizes at leading versus lagging strands. Mol Cell 21: 427–435. [DOI] [PubMed] [Google Scholar]
- Chapman JR, Barral P, Vannier JB, Borel V, Steger M, Tomas-Loba A, Sartori AA, Adams IR, Batista FD, Boulton SJ 2013. RIF1 is essential for 53BP1-dependent nonhomologous end joining and suppression of DNA double-strand break resection. Mol Cell 49: 858–871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Yang Y, van Overbeek M, Donigian JR, Baciu P, de Lange T, Lei M 2008. A shared docking motif in TRF1 and TRF2 used for differential recruitment of telomeric proteins. Science 319: 1092–1096. [DOI] [PubMed] [Google Scholar]
- Chen H, Lisby M, Symington LS 2013. RPA coordinates DNA end resection and prevents formation of DNA hairpins. Mol Cell 50: 589–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chow TT, Zhao Y, Mak SS, Shay JW, Wright WE 2012. Early and late steps in telomere overhang processing in normal human cells: The position of the final RNA primer drives telomere shortening. Genes Dev 26: 1167–1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clynes D, Gibbons RJ 2013. ATRX and the replication of structured DNA. Curr Opin Genet Dev 23: 289–294. [DOI] [PubMed] [Google Scholar]
- Conomos D, Stutz MD, Hills M, Neumann AA, Bryan TM, Reddel RR, Pickett HA 2012. Variant repeats are interspersed throughout the telomeres and recruit nuclear receptors in ALT cells. J Cell Biol 199: 893–906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conomos D, Pickett HA, Reddel RR 2013. Alternative lengthening of telomeres: Remodeling the telomere architecture. Front Oncol 3: 27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai X, Huang C, Bhusari A, Sampathi S, Schubert K, Chai W 2010. Molecular steps of G-overhang generation at human telomeres and its function in chromosome end protection. EMBO J 29: 2788–2801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Lange T 2009. How telomeres solve the end-protection problem. Science 326: 948–952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimitrova N, de Lange T 2009. Cell cycle-dependent role of MRN at dysfunctional telomeres: ATM signaling-dependent induction of NHEJ in G1 and resection-mediated inhibition of NHEJ in G2. Mol Cell Biol 29: 5552–5563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Virgilio M, Callen E, Yamane A, Zhang W, Jankovic M, Gitlin AD, Feldhahn N, Resch W, Oliveira TY, Chait BT, et al. 2013. Rif1 prevents resection of DNA breaks and promotes immunoglobulin class switching. Science 339: 711–715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doksani Y, Wu JY, de Lange T, Zhuang X 2013. Super-resolution fluorescence imaging of telomeres reveals TRF2-dependent T-loop formation. Cell 155: 345–356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donnianni RA, Symington LS 2013. Break-induced replication occurs by conservative DNA synthesis. Proc Natl Acad Sci 110: 13475–13480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunham MA, Neumann AA, Fasching CL, Reddel RR 2000. Telomere maintenance by recombination in human cells. Nat Genet 26: 447–450. [DOI] [PubMed] [Google Scholar]
- Escribano-Diaz C, Orthwein A, Fradet-Turcotte A, Xing M, Young JT, Tkac J, Cook MA, Rosebrock AP, Munro M, Canny MD, et al. 2013. A cell cycle-dependent regulatory circuit composed of 53BP1-RIF1 and BRCA1-CtIP controls DNA repair pathway choice. Mol Cell 49: 872–883. [DOI] [PubMed] [Google Scholar]
- Feng L, Fong KW, Wang J, Wang W, Chen J 2013. RIF1 counteracts BRCA1-mediated end resection during DNA repair. J Biol Chem 288: 11135–11143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fouche N, Cesare AJ, Willcox S, Ozgur S, Compton SA, Griffith JD 2006. The basic domain of TRF2 directs binding to DNA junctions irrespective of the presence of TTAGGG repeats. J Biol Chem 281: 37486–37495. [DOI] [PubMed] [Google Scholar]
- Freibaum BD, Counter CM 2006. hSnm1B is a novel telomere-associated protein. J Biol Chem 281: 15033–15036. [DOI] [PubMed] [Google Scholar]
- Fu YV, Yardimci H, Long DT, Ho TV, Guainazzi A, Bermudez VP, Hurwitz J, van Oijen A, Scharer OD, Walter JC 2011. Selective bypass of a lagging strand roadblock by the eukaryotic replicative DNA helicase. Cell 146: 931–941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fumagalli M, Rossiello F, Clerici M, Barozzi S, Cittaro D, Kaplunov JM, Bucci G, Dobreva M, Matti V, Beausejour CM, et al. 2012. Telomeric DNA damage is irreparable and causes persistent DNA-damage-response activation. Nat Cell Biol 14: 355–365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaillard PH, Noguchi E, Shanahan P, Russell P 2003. The endogenous Mus81-Eme1 complex resolves Holliday junctions by a nick and counternick mechanism. Mol Cell 12: 747–759. [DOI] [PubMed] [Google Scholar]
- Gao H, Cervantes RB, Mandell EK, Otero JH, Lundblad V 2007. RPA-like proteins mediate yeast telomere function. Nat Struct Mol Biol 14: 208–214. [DOI] [PubMed] [Google Scholar]
- Gocha AR, Harris J, Groden J 2013. Alternative mechanisms of telomere lengthening: Permissive mutations, DNA repair proteins and tumorigenic progression. Mutat Res 743–744: 142–150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg AD, Banaszynski LA, Noh KM, Lewis PW, Elsaesser SJ, Stadler S, Dewell S, Law M, Guo X, Li X, et al. 2010. Distinct factors control histone variant H3.3 localization at specific genomic regions. Cell 140: 678–691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffith JD, Comeau L, Rosenfield S, Stansel RM, Bianchi A, Moss H, de Lange T 1999. Mammalian telomeres end in a large duplex loop. Cell 97: 503–514. [DOI] [PubMed] [Google Scholar]
- Hardy CF, Sussel L, Shore D 1992. A RAP1-interacting protein involved in transcriptional silencing and telomere length regulation. Genes Dev 6: 801–814. [DOI] [PubMed] [Google Scholar]
- He H, Wang Y, Guo X, Ramchandani S, Ma J, Shen MF, Garcia DA, Deng Y, Multani AS, You MJ, et al. 2009. Pot1b deletion and telomerase haploinsufficiency in mice initiate an ATR-dependent DNA damage response and elicit phenotypes resembling dyskeratosis congenita. Mol Cell Biol 29: 229–240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heaphy CM, de Wilde RF, Jiao Y, Klein AP, Edil BH, Shi C, Bettegowda C, Rodriguez FJ, Eberhart CG, Hebbar S, et al. 2011a. Altered telomeres in tumors with ATRX and DAXX mutations. Science 333: 425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heaphy CM, Subhawong AP, Hong SM, Goggins MG, Montgomery EA, Gabrielson E, Netto GJ, Epstein JI, Lotan TL, Westra WH, et al. 2011b. Prevalence of the alternative lengthening of telomeres telomere maintenance mechanism in human cancer subtypes. Am J Pathol 179: 1608–1615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemann MT, Greider CW 1999. G-strand overhangs on telomeres in telomerase-deficient mouse cells. Nucleic Acids Res 27: 3964–3969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henson JD, Reddel RR 2010. Assaying and investigating alternative lengthening of telomeres activity in human cells and cancers. FEBS Lett 584: 3800–3811. [DOI] [PubMed] [Google Scholar]
- Henson JD, Neumann AA, Yeager TR, Reddel RR 2002. Alternative lengthening of telomeres in mammalian cells. Oncogene 21: 598–610. [DOI] [PubMed] [Google Scholar]
- Henson JD, Cao Y, Huschtscha LI, Chang AC, Au AY, Pickett HA, Reddel RR 2009. DNA C-circles are specific and quantifiable markers of alternative-lengthening-of-telomeres activity. Nat Biotechnol 27: 1181–1185. [DOI] [PubMed] [Google Scholar]
- Hockemeyer D, Sfeir AJ, Shay JW, Wright WE, de Lange T 2005. POT1 protects telomeres from a transient DNA damage response and determines how human chromosomes end. EMBO J 24: 2667–2678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hockemeyer D, Daniels JP, Takai H, de Lange T 2006. Recent expansion of the telomeric complex in rodents: Two distinct POT1 proteins protect mouse telomeres. Cell 126: 63–77. [DOI] [PubMed] [Google Scholar]
- Hockemeyer D, Palm W, Wang RC, Couto SS, de Lange T 2008. Engineered telomere degradation models dyskeratosis congenita. Genes Dev 22: 1773–1785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsiao SJ, Smith S 2009. Sister telomeres rendered dysfunctional by persistent cohesion are fused by NHEJ. J Cell Biol 184: 515–526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C, Dai X, Chai W 2012. Human Stn1 protects telomere integrity by promoting efficient lagging-strand synthesis at telomeres and mediating C-strand fill-in. Cell Res 22: 1681–1695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasbek C, Wang F, Price CM 2013. Human TEN1 maintains telomere integrity and functions in genome-wide replication restart. J Biol Chem 288: 30139–30150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konishi A, de Lange T 2008. Cell cycle control of telomere protection and NHEJ revealed by a ts mutation in the DNA-binding domain of TRF2. Genes Dev 22: 1221–1230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam YC, Akhter S, Gu P, Ye J, Poulet A, Giraud-Panis MJ, Bailey SM, Gilson E, Legerski RJ, Chang S 2010. SNMIB/Apollo protects leading-strand telomeres against NHEJ-mediated repair. EMBO J 29: 2230–2241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenain C, Bauwens S, Amiard S, Brunori M, Giraud-Panis MJ, Gilson E 2006. The Apollo 5′ exonuclease functions together with TRF2 to protect telomeres from DNA repair. Curr Biol 16: 1303–1310. [DOI] [PubMed] [Google Scholar]
- Lewis KA, Wuttke DS 2012. Telomerase and telomere-associated proteins: Structural insights into mechanism and evolution. Structure 20: 28–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Espinal A, Cross GA 2005. Trypanosome telomeres are protected by a homologue of mammalian TRF2. Mol Cell Biol 25: 5011–5021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Londono-Vallejo JA, Der-Sarkissian H, Cazes L, Bacchetti S, Reddel RR 2004. Alternative lengthening of telomeres is characterized by high rates of telomeric exchange. Cancer Res 64: 2324–2327. [DOI] [PubMed] [Google Scholar]
- Lottersberger F, Bothmer A, Robbiani DF, Nussenzweig MC, de Lange T 2013. Role of 53BP1 oligomerization in regulating double-strand break repair. Proc Natl Acad Sci 110: 2146–2151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovejoy CA, Li W, Reisenweber S, Thongthip S, Bruno J, de Lange T, De S, Petrini JH, Sung PA, Jasin M, et al. 2012. Loss of ATRX, genome instability, and an altered DNA damage response are hallmarks of the alternative lengthening of telomeres pathway. PLoS Genet 8: 1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundblad V, Blackburn EH 1993. An alternative pathway for yeast telomere maintenance rescues est1– senescence. Cell 73: 347–360. [DOI] [PubMed] [Google Scholar]
- Makarov VL, Hirose Y, Langmore JP 1997. Long G tails at both ends of human chromosomes suggest a C strand degradation mechanism for telomere shortening. Cell 88: 657–666. [DOI] [PubMed] [Google Scholar]
- McElligott R, Wellinger RJ 1997. The terminal DNA structure of mammalian chromosomes. EMBO J 16: 3705–3714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehta A, Haber JE 2014. Sources of DNA double-strand breaks and models for recombinational DNA repair. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a016428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyake Y, Nakamura M, Nabetani A, Shimamura S, Tamura M, Yonehara S, Saito M, Ishikawa F 2009. RPA-like mammalian Ctc1-Stn1-Ten1 complex binds to single-stranded DNA and protects telomeres independently of the Pot1 pathway. Mol Cell 36: 193–206. [DOI] [PubMed] [Google Scholar]
- Muntoni A, Neumann AA, Hills M, Reddel RR 2009. Telomere elongation involves intra-molecular DNA replication in cells utilizing alternative lengthening of telomeres. Hum Mol Genet 18: 1017–1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nabetani A, Ishikawa F 2009. Unusual telomeric DNAs in human telomerase-negative immortalized cells. Mol Cell Biol 29: 703–713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakaoka H, Nishiyama A, Saito M, Ishikawa F 2012. Xenopus laevis Ctc1-Stn1-Ten1 (xCST) protein complex is involved in priming DNA synthesis on single-stranded DNA template in Xenopus egg extract. J Biol Chem 287: 619–627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nandakumar J, Cech TR 2013. Finding the end: Recruitment of telomerase to telomeres. Nat Rev Mol Cell Biol 14: 69–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nimonkar AV, Genschel J, Kinoshita E, Polaczek P, Campbell JL, Wyman C, Modrich P, Kowalczykowski SC 2011. BLM–DNA2–RPA–MRN and EXO1–BLM–RPA–MRN constitute two DNA end resection machineries for human DNA break repair. Genes Dev 25: 350–362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noon AT, Goodarzi AA 2011. 53BP1-mediated DNA double strand break repair: Insert bad pun here. DNA Repair (Amst) 10: 1071–1076. [DOI] [PubMed] [Google Scholar]
- Okamoto K, Bartocci C, Ouzounov I, Diedrich JK, Yates JR, Denchi EL 2013. A two-step mechanism for TRF2-mediated chromosome-end protection. Nature 494: 502–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osman F, Dixon J, Doe CL, Whitby MC 2003. Generating crossovers by resolution of nicked Holliday junctions: A role for Mus81-Eme1 in meiosis. Mol Cell 12: 761–774. [DOI] [PubMed] [Google Scholar]
- Palm W, de Lange T 2008. How shelterin protects mammalian telomeres. Annu Rev Genet 42: 301–334. [DOI] [PubMed] [Google Scholar]
- Palm W, Hockemeyer D, Kibe T, de Lange T 2009. Functional dissection of human and mouse POT1 proteins. Mol Cell Biol 29: 471–482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierce AJ, Hu P, Han M, Ellis N, Jasin M 2001. Ku DNA end-binding protein modulates homologous repair of double-strand breaks in mammalian cells. Genes Dev 15: 3237–3242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poulet A, Buisson R, Faivre-Moskalenko C, Koelblen M, Amiard S, Montel F, Cuesta-Lopez S, Bornet O, Guerlesquin F, Godet T, et al. 2009. TRF2 promotes, remodels and protects telomeric Holliday junctions. EMBO J 28: 641–651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price CM, Boltz KA, Chaiken MF, Stewart JA, Beilstein MA, Shippen DE 2010. Evolution of CST function in telomere maintenance. Cell Cycle 9: 3157–3165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rai R, Zheng H, He H, Luo Y, Multani A, Carpenter PB, Chang S 2010. The function of classical and alternative non-homologous end-joining pathways in the fusion of dysfunctional telomeres. EMBO J 29: 2598–2610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saini N, Ramakrishnan S, Elango R, Ayyar S, Zhang Y, Deem A, Ira G, Haber JE, Lobachev KS, Malkova A 2013. Migrating bubble during break-induced replication drives conservative DNA synthesis. Nature 502: 389–392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sfeir A, de Lange T 2012. Removal of shelterin reveals the telomere end-protection problem. Science 336: 593–597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sfeir AJ, Chai W, Shay JW, Wright WE 2005. Telomere-end processing the terminal nucleotides of human chromosomes. Mol Cell 18: 131–138. [DOI] [PubMed] [Google Scholar]
- Sfeir A, Kabir S, van Overbeek M, Celli GB, de Lange T 2010. Loss of Rap1 induces telomere recombination in the absence of NHEJ or a DNA damage signal. Science 327: 1657–1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverman J, Takai H, Buonomo SB, Eisenhaber F, de Lange T 2004. Human Rif1, ortholog of a yeast telomeric protein, is regulated by ATM and 53BP1 and functions in the S-phase checkpoint. Genes Dev 18: 2108–2119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smogorzewska A, de Lange T 2004. Regulation of telomerase by telomeric proteins. Ann Rev Biochem 73: 177–208. [DOI] [PubMed] [Google Scholar]
- Smogorzewska A, Karlseder J, Holtgreve-Grez H, Jauch A, de Lange T 2002. DNA ligase IV-dependent NHEJ of deprotected mammalian telomeres in G1 and G2. Curr Biol 12: 1635. [DOI] [PubMed] [Google Scholar]
- Stansel RM, de Lange T, Griffith JD 2001. T-loop assembly in vitro involves binding of TRF2 near the 3′ telomeric overhang. EMBO J 20: 5532–5540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugawara N, Wang X, Haber JE 2003. In vivo roles of Rad52, Rad54, and Rad55 proteins in Rad51-mediated recombination. Mol Cell 12: 209–219. [DOI] [PubMed] [Google Scholar]
- Surovtseva YV, Churikov D, Boltz KA, Song X, Lamb JC, Warrington R, Leehy K, Heacock M, Price CM, Shippen DE 2009. Conserved telomere maintenance component 1 interacts with STN1 and maintains chromosome ends in higher eukaryotes. Mol Cell 36: 207–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Symington LS 2014. Processing of DNA breaks: Mechanism and regulation. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a016436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takai KK, Hooper S, Blackwood S, Gandhi R, de Lange T 2010. In vivo stoichiometry of shelterin components. J Biol Chem 285: 1457–1467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tokutake Y, Matsumoto T, Watanabe T, Maeda S, Tahara H, Sakamoto S, Niida H, Sugimoto M, Ide T, Furuichi Y 1998. Extra-chromosomal telomere repeat DNA in telomerase-negative immortalized cell lines. Biochem Biophys Res Commun 247: 765–772. [DOI] [PubMed] [Google Scholar]
- Vannier JB, Pavicic-Kaltenbrunner V, Petalcorin MI, Ding H, Boulton SJ 2012. RTEL1 dismantles T loops and counteracts telomeric G4-DNA to maintain telomere integrity. Cell 149: 795–806. [DOI] [PubMed] [Google Scholar]
- van Overbeek M, de Lange T 2006. Apollo, an Artemis-related nuclease, interacts with TRF2 and protects human telomeres in S phase. Curr Biol 16: 1295–1302. [DOI] [PubMed] [Google Scholar]
- Varley H, Pickett HA, Foxon JL, Reddel RR, Royle NJ 2002. Molecular characterization of inter-telomere and intra-telomere mutations in human ALT cells. Nat Genet 30: 301–305. [DOI] [PubMed] [Google Scholar]
- Wan M, Qin J, Songyang Z, Liu D 2009. OB fold-containing protein 1 (OBFC1), a human homolog of yeast Stn1, associates with TPP1 and is implicated in telomere length regulation. J Biol Chem 284: 26725–26731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang RC, Smogorzewska A, de Lange T 2004. Homologous recombination generates T-loop-sized deletions at human telomeres. Cell 119: 355–368. [DOI] [PubMed] [Google Scholar]
- Wang M, Wu W, Wu W, Rosidi B, Zhang L, Wang H, Iliakis G 2006. PARP-1 and Ku compete for repair of DNA double strand breaks by distinct NHEJ pathways. Nucleic Acids Res 34: 6170–6182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Ghosh G, Hendrickson EA 2009. Ku86 represses lethal telomere deletion events in human somatic cells. Proc Natl Acad Sci 106: 12430–12435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F, Stewart JA, Kasbek C, Zhao Y, Wright WE, Price CM 2012. Human CST has independent functions during telomere duplex replication and C-strand fill-in. Cell Rep 2: 1096–1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson MA, Kwon Y, Xu Y, Chung WH, Chi P, Niu H, Mayle R, Chen X, Malkova A, Sung P, et al. 2013. Pif1 helicase and Polδ promote recombination-coupled DNA synthesis via bubble migration. Nature 502: 393–396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong LH, McGhie JD, Sim M, Anderson MA, Ahn S, Hannan RD, George AJ, Morgan KA, Mann JR, Choo KH 2010. ATRX interacts with H3.3 in maintaining telomere structural integrity in pluripotent embryonic stem cells. Genome Res 20: 351–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu P, van Overbeek M, Rooney S, de Lange T 2010. Apollo contributes to G overhang maintenance and protects leading-end telomeres. Mol Cell 39: 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu P, Takai H, de Lange T 2012. Telomeric 3′ overhangs derive from resection by Exo1 and Apollo and fill-in by POT1b-associated CST. Cell 150: 39–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyatt HDM, West SC 2014. Holliday junction resolvases. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a016485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyatt HD, Sarbajna S, Matos J, West SC 2013. Coordinated actions of SLX1-SLX4 and MUS81-EME1 for Holliday junction resolution in human cells. Mol Cell 52: 234–247. [DOI] [PubMed] [Google Scholar]
- Yeager TR, Neumann AA, Englezou A, Huschtscha LI, Noble JR, Reddel RR 1999. Telomerase-negative immortalized human cells contain a novel type of promyelocytic leukemia (PML) body. Cancer Res 59: 4175–4179. [PubMed] [Google Scholar]
- Zhu XD, Niedernhofer L, Kuster B, Mann M, Hoeijmakers JH, de Lange T 2003. ERCC1/XPF removes the 3′ overhang from uncapped telomeres and represses formation of telomeric DNA-containing double minute chromosomes. Mol Cell 12: 1489–1498. [DOI] [PubMed] [Google Scholar]
- Zimmermann M, Lottersberger F, Buonomo SB, Sfeir A, de Lange T 2013. 53BP1 regulates DSB repair using Rif1 to control 5′ end resection. Science 339: 700–704. [DOI] [PMC free article] [PubMed] [Google Scholar]