Abstract
Intralocus sexual conflict and intragenomic conflict both affect sex chromosome evolution and can in extreme cases even cause the complete turnover of sex chromosomes. Additionally, established sex chromosomes often become the focus of heightened conflict. This creates a tangled relationship between sex chromosomes and conflict with respect to cause and effect. To further complicate matters, sexual and intragenomic conflict may exacerbate one another and thereby further fuel sex chromosome change. Different magnitudes and foci of conflict offer potential explanations for lineage-specific variation in sex chromosome evolution and answer long-standing questions as to why some sex chromosomes are remarkably stable, whereas others show rapid rates of evolutionary change.
Sex chromosomes are hot spots for intralocus and intragenomic conflicts. These conflicts shape sex chromosome and genome evolution—and even have the power to cause the complete turnover of sex chromosomes.
Compared to the autosomes, the unique inheritance pattern of the sex chromosomes is often thought to intensify evolutionary conflict (Rice 1984; Frank 1991; Jaenike 2001). Sex chromosomes are therefore hot spots for two specific types of conflict: intralocus conflict, in which an allele confers different fitness effects depending on the sex in which it is found, and intragenomic conflict, in which selfish genetic elements (SGEs) promote their own transmission at the expense of unlinked regions of the genome. These conflicts act in distinct but complementary ways. Not only do they shape sex chromosome and genome evolution, but in some cases, they also have the power to cause complete turnover of sex chromosomes.
Unlike the autosomes, because the sex chromosomes are unevenly transmitted between males and females and are also unevenly distributed between the sexes (Fig. 1), the relative effect of male- and female-specific selection acting on them is unbalanced. The inherent differences in sex-specific selection on the sex chromosomes themselves, and between the sex chromosomes and the autosomes, form the basis of a large and often compelling body of evolutionary theory that predicts the ways that intralocus sexual conflict will arise, play out, and in some cases potentially be resolved. This theory predicts that, under some conditions, the sex chromosomes are hot spots of intralocus sexual conflict (Rice 1984; Albert and Otto 2005; Connallon and Clark 2010), and in some cases alleles that harm one sex more than they benefit the other can still reach high frequencies if they are sex-linked (Rice 1984; Dean et al. 2012). All this theory predicts that although the sex chromosomes generally represent a small proportion of the genome, they should play a disproportionately large role in sexual conflict, sexual dimorphism, and sexual selection. There is substantial empirical evidence supporting at least some of this theory (Table 1, and references therein), although there remains something of a disconnect between molecular genetic and phenotypic findings (Dean and Mank 2014).
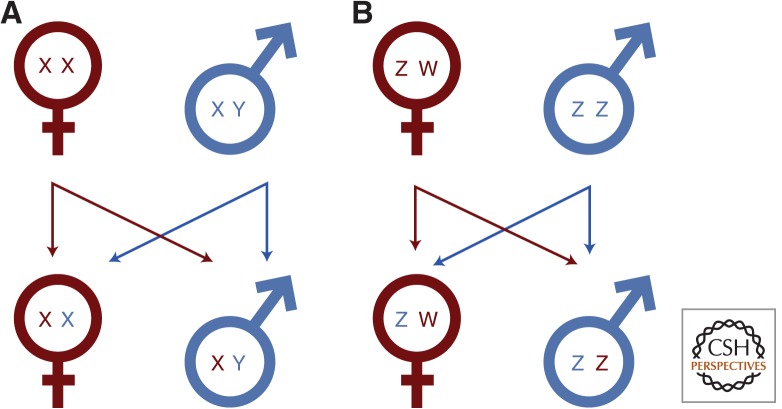
Figure 1.
Transmission of the sex chromosomes. Females are shown in red, males in blue. In male heterogamety (A), the Y chromosome is passed through the patriline and limited to males. The maternal X chromosome is passed from mother to both sons and daughters, but the paternal X can only be transmitted to daughters. Additionally, the X is present two-thirds of the time in females. In female heterogamety (B), the W chromosome is limited to females and passed solely from mother to daughter. The paternal Z chromosome can be passed from father to both daughters and sons; however, the maternal Z chromosome is only passed to sons. Converse to the X chromosome, the Z chromosome is resident in males two-thirds of the time.
Table 1.
Studies showing a disproportionate role of the sex chromosomes in sexual dimorphism, fitness, or fertility
| Male heterogamety | Female heterogamety | |||
|---|---|---|---|---|
| X chromosome | Y chromosome | Z chromosome | W chromosome | |
| Associations with sexually dimorphic phenotypic traits | Size dimorphism in red deer (Foerster et al. 2007) Sexually antagonistic fitness variation in Drosophila (Gibson et al. 2002; Innocenti and Morrow 2010) Variation in stalk-eyed fly eye span (Wolfenbarger and Wilkinson 2001) |
Male coloration in guppies (Winge 1927; Postma et al. 2011) Male mating behavior in sticklebacks (Kitano et al. 2009; although see Natri et al. 2013) |
Female mating preference in moths (Iyengar et al. 2002) Female mate choice in flycatchers (Saether et al. 2007) Male plumage traits in flycatchers (Saetre et al. 2003) |
Female benefit coloration in cichlids (Roberts et al. 2009) |
| Associations with reproduction and fertility | Male reproductive genes in mice (Mueller et al. 2008) | Male fertility genes in mammals (Lange et al. 2009) Male fitness in Drosophila (Lemos et al. 2010; Sackton et al. 2011) |
Genes related to male reproductive function in silk moth (Arunkumar et al. 2009) | Genes related to female fecundity and fertility in chickens (Moghadam et al. 2012) |
In addition to their role in sexual conflict, the sex chromosomes also experience high levels of intragenomic conflict resulting from SGEs that promote their own transmission at the expense of the rest of the genome (Burt and Trivers 2006). Of particular importance are sex-linked segregation distorters, as these generate strong selection favoring genes that suppress their action. Theory predicts that intragenomic conflict should be particularly intense when involving the sex chromosomes (Hurst and Pomiankowski 1991). This is because sex chromosomes can generate antagonistic coevolution between sex-linked segregation distorters and their suppressors (Partridge and Hurst 1998).
In addition to shaping the evolutionary properties of existing sex chromosomes, in some situations sexual and intragenomic conflict may also actually catalyze the formation of sex chromosomes (Fisher 1931; Charlesworth and Charlesworth 1980; Rice 1987; Werren and Beukeboom 1998). Conflict may also explain the rate of degradation of the sex-limited Y and W chromosomes (Bachtrog et al. 2011) and turnover of sex chromosomes (Hall 2004; van Doorn and Kirkpatrick 2007, 2010), suggesting that conflict plays a causal role in sex chromosome evolution.
There is also growing evidence of a direct relationship between SGEs and sexual conflict involving the sex chromosomes. Many SGEs increase their transmission advantage by targeting sperm, which can reduce male fertility owing to reduced overall sperm production. Lower sperm production can in turn result in reduced siring success during sperm competition (Price and Wedell 2008) and potentially favor polyandry as a female strategy to bias paternity against SGE-carrying males (Wedell 2013). As a consequence, SGEs may also influence the potential for sexual conflict as they can favor increased female mating rates, which increases the potential for conflict between the sexes.
Sexual and intragenomic conflict are therefore critical for sex chromosome evolution and, once sex chromosomes are established, can further shape their evolutionary and genomic properties. Disentangling cause and effect is difficult but crucial to understanding the role of sex chromosomes in sexual and intragenomic conflict and vice versa.
CONFLICT AS AN AGENT OF SEX CHROMOSOME TURNOVER AND CHANGE
One of the major unanswered questions in sex chromosome evolution is why some systems show long periods of relative evolutionary stasis, as exemplified by the conservation of orthologous sex chromosomes within the therian mammal (Potrzebowski et al. 2008), avian (Mank and Ellegren 2007), and Drosophila (Vicoso and Bachtrog 2013) lineages, whereas many others, described below, show rapid and repeated turnover. We also do not understand why some sex chromosome pairs diverge markedly from each other over time, whereas others exist in a state of arrested decay.
Although homologous sex chromosomes in different systems share many properties (Bachtrog et al. 2011), they also show many lineage-specific patterns, many of which have proved difficult to explain. Different conflict strengths and foci offer potential explanations for lineage-specific variation in sex chromosome evolution, as conflict can affect both rates of sex chromosome turnover and sex chromosome divergence.
Intralocus Conflict and Sex Chromosome Change
The genetic architecture and extent of intralocus sexual conflict may affect the origin of sex chromosomes in two ways. First, sexual conflict may affect the rate at which the W and Y chromosomes degenerate. The primary model for sex chromosome divergence is based on selection for suppression of recombination between the X and Y, or Z and W, attributable to linkage between the primary sex-determining gene and any nearby gene(s) with sexually antagonistic properties (Fisher 1931; Charlesworth and Charlesworth 1980; Rice 1987). Selection will act to prevent the breakup of a male-determining gene on the nascent Y chromosome from any nearby male-benefit alleles by stopping recombination between the X and Y in this region. Similarly, reduced recombination will be favored between a female-determining gene on an emergent W chromosome and nearby female-benefit alleles.
The suppression of recombination causes the Y and W chromosomes to degenerate at many loci in both gene sequence and expression, as well as allowing them to accumulate repetitive elements (Bachtrog 2013), and this is what causes the cytological and functional differences between X and Y and between Z and W chromosomes. The likelihood that a sex-determining gene will be proximate to a locus with sexually antagonistic alleles is in many ways defined by the proportion of loci in the genome with sexually antagonistic effects. The degree of conflict, and therefore the proportion of the genome with sexually antagonistic effects, may be in turn largely defined by the mating system (Charlesworth and Mank 2010). If the mating system has a major effect on sex chromosome degeneration, then we would expect monogamous species with minimal levels of sexual conflict to show slower rates of sex chromosome divergence than polyandrous or polygamous systems. However, this is complicated and may depend in part on whether the monogamy is male- or female-driven (Hosken et al. 2009).
Additionally, although sex chromosomes are relatively stable and conserved in some lineages, such as Drosophila, the therian mammals, and birds (Mank and Ellegren 2007; Potrzebowski et al. 2008; Vicoso and Bachtrog 2013), sex chromosomes in other lineages exhibit rapid turnover. For example, the sex chromosomes are not orthologous among closely related species of Oryzias rice fish (Matsuda et al. 2002; Kondo et al. 2003; Miura 2007), populations of Rana rugosa frogs (Nanda et al. 2002; Ogata et al. 2008), or in Musca domestica. In the latter case, the male-determining factor (M) has relocated in some populations from the Y to any of the five autosomes or even the X (Dübendorfer et al. 2002).
There are still many unanswered questions as to why some sex chromosome systems are highly conserved across long periods of evolutionary time, whereas others change quickly and even vary across populations of the same species. However, there are some predictions suggesting that sex chromosome turnover itself may in fact be driven by sexual conflict (van Doorn and Kirkpatrick 2007, 2010), and SGEs (see below) as the emergence of a new sex chromosome system can resolve conflict over sexually antagonistic alleles by creating a male-limited (Y) or a female-limited (W) chromosome (Roberts et al. 2009; Postma et al. 2011). Sexual conflict may also cause the formation of neosex chromosomes, via autosomal fusion to a sex chromosome. This is because sexual conflict on the autosome can select for sex-linkage, which may explain the formation of a neosex chromosome in the Japan Sea population of three-spined sticklebacks, Gasterosteus aculeatus (Kitano et al. 2009; but see Natri et al. 2013).
Population Size, Linkage, and Turnover
Population size is a major determinant in types and rates of evolution. Low effective population size (NE) reduces the efficacy of selection, and therefore a larger proportion of mutations are effectively neutral in smaller populations. Recombination affects NE, and therefore molecular evolution, as higher rates of recombination within a specific region result in a local increase in NE.
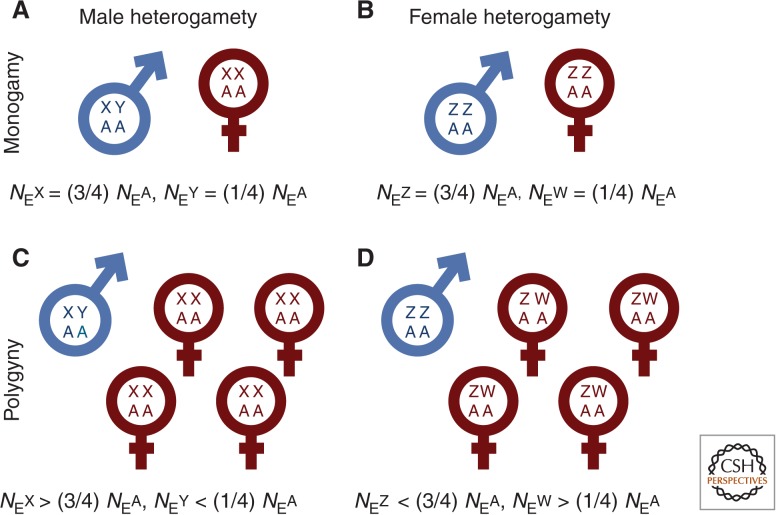
Rates of evolution of sex chromosomes also vary because of differences in relative NE. In general, and assuming an XY system and an equal population sex ratio, the census population size of the chromosomes is such that there are four copies of each autosome for every three X chromosomes and every single Y. This means that in truly monogamous populations (populations with limited sexual conflict), in which male and female reproductive skew are equal, the relative effective population size of the autosomes (NEA) is greater than that of the X chromosome (NEX), which is in turn greater than that of the Y chromosome (NEY) (Fig. 2).
Figure 2.
The effect of sexual selection on the relative effective population size. Population size of , and chromosomes are compared to the autosomes. Under strict monogamy is broadly equal to . Sexual selection (C,D) increases relative and at the expense of . These differences in effective population size may have profound effects on sex chromosome evolution and turnover.
In populations experiencing greater sexual selection and conflict—typically those with increased reproductive variance in males—relative NEY decreases even further, and NEX increases (Bachtrog et al. 2011). This is because with strong sexual selection the effective male population size is very small and autosomes are transmitted more through males than the X chromosome is. In some cases in which recombination rates are higher on the X than the autosomes because recombination is restricted to females as with Drosophila (Andolfatto 2001), NEX can even exceed NEA.
Sexual selection and its influence on the relative effective population size of the sex chromosomes is known to affect rates of evolution for X-linked genes (Charlesworth et al. 1987; Vicoso and Charlesworth 2010). However, increased sexual selection and conflict can also affect Y chromosome evolution. Sexual selection decreases NEY (Fig. 2), and this may lower the mutational input on Y chromosomes and reduce the efficacy of selection on Y-linked genes. However, the population size effects on Y-mutational input could be compensated for by male-biased mutation rates in some taxa (Sayres and Makova 2011), and the Y chromosomes’ ability to respond to selection may be offset by intrachromosome recombination and gene conversion (Lange et al. 2009; Marais et al. 2010)
Additionally, the rate of evolution of alleles on the sex chromosomes also depends on the dominance characteristics of any beneficial mutations as this affects the strength of drift the mutation is subject to (on the X), and whether or not newly beneficial alleles are already part of the standing genetic variation, which is in turn dependent on population size. For example, Y-linkage can boost the rate of increase of rare beneficial alleles and increase the purging of deleterious alleles, but it can also result in lower standing variation on which selection can act (Charlesworth et al. 1987), notwithstanding possible higher male mutation rates. Thus, it is the balance of population size, mutation rate, and dominance that determine allelic fates and sex chromosome evolution.
Similarly, the outcome of new male-determining factors depends on linkage associations between male-benefit alleles and old and neo-Y chromosomes. Depending on the detail of these associations, new sex-determining factors can be lost, retained with the loss of the old Y chromosome, or new and old can both be retained or lost (van Doorn and Kirkpatrick 2007). For example, if genes essential for male sexual fitness are closely linked to the old Y chromosome, then the spread of a new sex-determining factor can be retarded as sex chromosomes differentiate and turnover can be even more unlikely. However, with the advent of sex-limited gene expression, invasion by new sex-determining factors is easier because male/female-benefit alleles will not be inappropriately expressed. That is, female-benefit alleles (for example) will not be expressed in males with a neo-Y if the allele is already sex-limited in expression, and hence this potential avenue of selection against the neo-Y will not exist.
Sex chromosome turnover may also be facilitated by the fact that nonrecombining regions of the sex chromosomes accumulate deleterious mutations (Blaser et al. 2013). The ease of turnover depends on the magnitude of male benefits derived from alleles on the old Y chromosome, as this chromosome is typically lost in turnover events. If the Y chromosome carries numerous benefit alleles to males or, in the case of the eutherian Y, carries several genes essential to male fertility (Lange et al. 2009), loss of these genes prevents turnover as the resulting males would be sterile. With small cumulative benefits on the Y chromosome, turnover events are more likely, and this may suggest that younger Y chromosomes, which have had less time to accumulate male-benefit alleles, are more likely to undergo turnover than older Y chromosomes.
It could therefore be that small populations in general, or populations with intense sexual selection and conflict, are most likely to experience sex chromosome turnovers. However, the strong linkage effects and Hill–Robertson interference that act on Y chromosomes (Bachtrog 2013) may obscure the role of effective population size in shaping Y chromosome evolution. To date, there is little empirical data on the role of relative effective population size in sex chromosome turnover, but comparison of island versus mainland populations could be revealing.
Conflict and Sex Chromosome Dosage Compensation
Sexual conflict may also explain the curious pattern that has emerged recently regarding the difference between X and Z chromosomes in dosage compensation (Mank 2009, 2013). Once recombination is halted between the sex chromosome pair, the sex-limited Y or W degenerates in gene content and activity (Bachtrog 2013). This creates a gene imbalance between males and females, with the heterogametic sex having just one copy of X- or Z-linked genes. There is selection in the heterogametic sex to up-regulate this single copy to equilibrate dose with the two copies in the homogametic sex. Because gene expression is highly correlated between males and females (Griffin et al. 2013), dosage compensation in the heterogametic sex can cause overexpression in the homogametic sex leading to conflict between the sexes over optimal gene expression (Mank et al. 2011).
It is increasingly clear that this conflict plays out differently for X and Z chromosomes, with many male heterogametic species showing effective dosage compensation in males (Hahn and Lanzaro 2005; Ercan et al. 2007; Straub and Becker 2007; Gelbart and Kuroda 2009; Julien et al. 2012; Muyle et al. 2012; Wilkinson et al. 2013). In stark contrast, complete Z chromosome dosage compensation has yet to be seen in any female heterogametic species (Itoh et al. 2007; Vicoso and Bachtrog 2011; Harrison et al. 2012; Vicoso et al. 2013). This pattern may be attributed to the fact that sexual selection is usually stronger on males. This would mean that selection in XY males to up-regulate dosage-sensitive X-linked genes would override the cost of overexpression in XX females, but selection in ZW females to hyperexpress similar genes on the Z chromosome would be retarded by selection against overexpression in ZZ males (Mank 2013). If this is true, then sexual conflict may explain fundamental genomic properties such as sex chromosome dosage compensation.
SGEs and Sex Chromosome Turnover
SGEs can also influence sex chromosome evolution and turnover, and in many ways the effects of SGEs on sex chromosome evolution are similar to those for sexual conflict. However, the causes can differ quite substantially. For example, SGEs can also cause sex chromosome turnover. Segregation distorters such as meiotic drivers, involve two loci—a killer (drive) locus and a sensitivity (target) locus—being in strong linkage repulsion to ensure that the drive and sensitive alleles do not form a suicidal combination resulting in the driver killing itself. The reduced recombination of sex chromosomes makes them particularly prone to invasion by segregation distorters as any gene on the X/Z can drive against the Y/W (and vice versa) (Hamilton 1967; Frank 1991). Unlike distorters on autosomes, segregation distorters on sex chromosomes result in biased sex ratios, as a preponderance of X chromosome–bearing sperm results in more female offspring, or Y-bearing sperm more male offspring. This creates strong selection to rebalance sex ratios, which in turn may promote the evolution of neosex chromosomes (cf. female meiotic drive in mammals) (Yoshida and Kitano 2012) and even in the formation of entirely new sex-determination systems that supplant existing ones (Cordaux et al 2011).
Just as selection can favor reduced recombination between sex-determining genes and sexually antagonistic alleles, segregation distorters can also catalyze the silencing of sex-linked genes. However, the mechanisms by which they do this are somewhat different. One potent silencing mechanism is meiotic sex chromosome inactivation (MSCI), in which most of the genes are silenced on the sex chromosomes during meiosis in the heterogametic sex. MSCI occurs in mammals (Fernandez-Capetill et al. 2003), but remains contentious in Drosophila (Hense et al. 2007; Meiklejohn et al. 2011) and birds (Schoenmakers et al. 2009; Guioli et al. 2012; Livernois et al. 2013). One hypothesis put forward is that a key function of MSCI is to prevent SGEs from invading sex chromosomes, which may be particularly vulnerable in the heterogametic sex where they are unpaired. MSCI may therefore act to protect sex chromosomes from the invasion of sex ratio distorters (Meiklejohn and Tao 2010), although this remains to be shown. However, silencing of meiotic drivers on the sex chromosomes may also contribute to the degeneration of the Y or the W. In the fly Drosophila albomicans, for example, it is suggested that gene silencing on the Y chromosome may initiate the process of degeneration (Zhou and Bachtrog 2012). Segregation distorters may therefore have the potential to fuel the rate of sex chromosome turnover by invading and subsequently contributing to their degeneration—and hence being a potent promoter of new sex chromosomes.
SEX CHROMOSOMES AS AGENTS OF CONFLICT
Once formed, sex chromosomes may become a flashpoint within the genome for conflict. This means that conflict not only influences sex chromosome turnover and divergence but also acts more powerfully on established sex chromosomes to shape their gene content and evolution.
Sexualization of the X and Z Chromosomes
Once the X and Y (or Z and W) chromosomes have ceased recombining with each other, sexually antagonistic forces can shape both the genomic properties and even the gene content of the sex chromosomes. In theory, the genes on each of the sex chromosome types (X, Y, Z, and W) should be sexualized in unique and specific ways. Although the sex-limited Y and W chromosome often contain relatively few genes, those that remain are thought to be crucial for male (for the Y chromosome) and female (for the W chromosome) fitness, as strong sex-specific selection is needed to maintain genes on these chromosomes once recombination is suppressed (Bachtrog 2013). Evidence consistent with this is found for the D. melanogaster Y (Chippindale and Rice 2001) and the chicken W (Moghadam et al. 2012) chromosomes. Additionally, gene duplicates with sex-specific effects may be maintained on Y and W chromosomes, as relocation to the sex-limited chromosome instantly resolves sexual conflict for genes that are dispensable in the homogametic sex. This may explain the fact that the rate of gene gain on the Drosophila Y chromosome is 10 times higher than the rate of gene loss (Koerich et al. 2008), although the primate Y chromosome is more conserved for gene content (Hughes et al. 2012).
X and Z chromosomes are thought to be masculinized and feminized in different ways depending on whether alleles with sex-specific effects are dominant or recessive. In male heterogamety, the hemizygosity experienced by males may lead to the accumulation of recessive alleles with male benefits (Rice 1984; Connallon and Clark 2010; Dean et al. 2012), even if there is a substantial cost to females, as X-linked genes are always exposed to selection in males. Alternatively, the fact that the X chromosome is more often present in females may give it a larger role in dominant female-beneficial traits because the X chromosome is selected for female-benefit effects twice as often as male effects (Rice 1984; Connallon and Clark 2010). The converse predictions are expected for Z chromosomes, which should accumulate recessive female-benefit mutations or dominant male-benefit mutations.
Chromosome partitioning approaches to studies of conflict bear up these theoretical predictions, with the X chromosome in Drosophila carrying the vast majority of sexually antagonistic variation (e.g., Gibson et al. 2002). Similarly, quantitative genetic approaches show similar results, with the X chromosome contributing more to the variance than expected for sexual dimorphism in Portuguese water dogs (Chase et al. 2005) and sexually antagonistic fitness variation in red deer (Foerster et al. 2007). However, the sex chromosomes do not always seem to carry a disproportional abundance of sexually antagonistic variation (Poissant et al. 2010; Husby et al. 2013).
Predictions about the sexualization of sex chromosomes have also been used to interpret gene expression data that show nonrandom distributions of sex-biased genes. Sex-biased expression is often thought to result from selection for different transcription optima for females and males (Innocenti and Morrow 2010; Connallon and Clark 2011a; Pointer et al. 2013) and represent at least partially resolved sexual conflict (Connallon and Knowles 2005; Connallon and Clark 2011b). In this framework, genes expressed at higher levels in females (female-biased genes) are thought to confer female benefits, and genes expressed at higher levels in males (male-biased genes) are thought to confer benefits to males (Connallon and Clark 2011b; Wright et al. 2012; Pointer et al. 2013).
Using this framework, the X chromosome in both mice and Drosophila has been feminized for expression (Khil et al. 2004; Meisel et al. 2012) and the Z chromosome has been masculinized in birds (Wright et al. 2012). The Drosophila X chromosome shows weaker and less consistent evidence of demasculinization (Meisel et al. 2012), although this may be owing to the mechanism of dosage compensation in this clade (Vicoso and Charlesworth 2009). The lack of dosage compensation in birds makes it difficult to assess the degree of defeminization. Even more interesting, species with alternative reproductive cycles show deviations from the general pattern of sexualization that are entirely consistent with expectations, as observed with the masculinization of the pea aphid X chromosome, which is caused by its unique inheritance between males, sexual females, and parthenogenic females (Jaquiery et al. 2013).
Despite these broadly consistent patterns, we do not yet know the rate of sexualization of the sex chromosomes, or how the sexualization rates might be affected by feminizing or male killing endosymbionts like Wolbachia, which could be expected to act as accelerants. However, many sex chromosomes have multiple strata, or regions, that have been effective sex chromosomes for different lengths of time (Lahn and Page 1999; Bergero et al. 2007; Wright et al. 2012; Vicoso et al. 2013). Strata studies in birds suggest that masculinization of Z chromosome gene expression is much slower than the rate of mating system evolution (Wright et al. 2012).
Similarly, the sex chromosomes can become sexualized via gene duplications, as gene duplicates may find the sex chromosomes either a more favorable or a more hostile environment depending on their sex-specific effects (Gallach et al. 2010; Connallon and Clark 2011b; Gallach and Betran 2011). This may explain the nonrandom patterns of gene movement observed from and to the X chromosome in mammals (Potrzebowski et al. 2008) and Drosophila (Meisel et al. 2009; Vibranovski et al. 2009). However, no such pattern has been observed for the Z chromosome in birds or Lepidoptera (Toups et al. 2011). Furthermore, the out of X migration seen in Drosophila and mammals is a slow process (Potrzebowski et al. 2008; Vibranovski et al. 2009), suggesting that significant feminization of the X chromosome only occurs over a long period of time.
Sex Chromosomes Are Magnets for SGEs
Sex chromosomes can act as attractors of segregation distorters, and this can result in sex-ratio biases promoting rapid changes in sex-determination systems, with a concomitant selective sweep of the genomic region surrounding the new sex-determining locus (Charlesworth and Hartl 1978; Hall 2004). Evidence consistent with this scenario is that sex ratio distorters may be responsible for very rapid selective sweeps (e.g., Nolte et al 2013). Spread of segregation distorters may also result in the hitchhiking of tightly linked genes that can potentially have dramatic fitness consequences. Loci tightly linked to segregation distorters (e.g., contained within an inversion) will spread as a consequence of drive, even if they are associated with fitness costs, and can remain at high frequencies even after suppression of the distorter. Conversely, it is possible that such genetic hitchhiking also has the potential to directly affect the rate of spread of SGEs in general, providing they are tightly linked to beneficial high fitness alleles under positive selection (i.e., not opposed by the parliament of genes).
THE INTERPLAY BETWEEN SGEs AND SEXUALLY ANTAGONISTIC ALLELES
The selfish nature of SGEs generates conflict with the rest of the genome that will select for suppression and silencing. SGEs and their corresponding suppressors are therefore locked in an ongoing antagonistic arms race. For SGEs on sex chromosomes that cause sex ratio distortion, this arms race is particularly acute owing to selection to rebalance sex ratios. Counterselection to restore the sex ratio to unity in response to sex-ratio distorters can promote evolution of novel sex-determination pathways (Cordaux et al. 2011), resulting in new and different ways to determine sex with direct consequences for sexual conflict. Changes to sex determination, such as going from male heterogamety to female heterogamety (XY to ZW, or vice versa) in response to selfish distorters, will simultaneously alter the opportunity for sexual conflict (see Rice 1984). Any SGEs that are already present on a sex chromosome (or on a former autosome now involved in sex determination [e.g., Kaiser and Bachtrog 2010]) will experience a shift in the strength of sex-specific selection. In addition, they may also have different fitness effects when expressed in males and females, as many SGEs (e.g., segregation distorters, male killers, feminizers, and parthenogenesis-inducing endosymbionts) are associated with sex-ratio distortion that clearly favors one sex at the expense of the other. For example, a genome that has experienced extensive periods of feminizing selection (e.g., by feminizing, male killing, or parthenogenesis-inducing bacteria) may have accumulated female-benefit alleles that lower male fitness when expressed in “rescued” males after the evolution of suppressors of sex-ratio distortion. Sex-specific selection under periods of sex-ratio distortion may result in the accumulation of female/male-benefit alleles that are now being exposed in the opposite sex in which they were rarely expressed before suppression of sex-ratio distortion. Over time, the cost of expressing such newly exposed sexually antagonistic alleles in the “rescued” sex may be ameliorated (Bonduriansky and Chenoweth 2009). One might therefore predict the resurgence of sexually antagonistic alleles to be more prominent in populations experiencing a recent spread of a segregation-distorting suppressor allele or experiencing a shift in sex determination.
Ongoing antagonistic arms races between segregation distorters and their suppressors involving the sex chromosomes can contribute to population reproductive failure by causing male sterility and can also potentially contribute to reproductive divergence and even speciation (Meiklejohn and Tao 2010; Pregraves 2010). Reduced gene flow between populations can occur via hybrid sterility, which itself may arise for different reasons. First, it could be the consequence of a heteropopulation lacking the appropriate suppressor for segregation distorters. Second, hybrid dysfunction could arise because of coevolution between the sex ratio distorters and the MSCI silencing mechanisms. The molecular basis of MSCI may vary between populations in response to suppressing newly emerged distorters or those that have evolved to escape suppression. As a result of the conflict over transmission, incompatibilities can evolve between different components of the MSCI machinery that can result in male hybrid sterility. For example, in populations of the house mouse (Mus musculus), there is conflict between the sex chromosomes involving the Y-linked multicopy Sly gene, and the X-linked Slx and Slx1 gene complex. These genes act antagonistically during spermatogenesis with Sly promoting the transmission of the Y chromosome during MSCI, whereas Slx/Slx1 promote the transmission of the X (Cocquet et al. 2012). Interestingly, this antagonism depends on the relative expression of these genes and not on their total abundance. Nonetheless, there is evidence of a rapid expansion in copy number of these sex-linked genes during the past 3 million years, and it is suggested this is caused by the antagonistic conflict that the X and Y chromosomes are locked into (Good 2012). This is further corroborated by recurrent hybrid male sterility that is associated with overexpression of Slx/Slxl1 and Sly because of copy number variation and a concomitant biased sex ratio. There are of course other reasons for sterility such as being caused by compensatory evolution of other genes involved in gametogenesis, but it is clear that conflict involving SGEs is a major sterility source.
The rapid turnover of sex chromosomes generated by segregation-distorting SGEs and their suppressor(s) will simultaneously affect any existing sex-linked allele by altering their exposure to selection, and this will also potentially contribute to sexual conflict. There is therefore likely to be a direct link between the recurrent intragenomic conflicts generated by SGEs and their corresponding suppressors and the resurgence and exposure of sexually antagonistic alleles on the sex chromosomes. Seeing the prevalence of SGEs in nature, this source of sexual conflict is likely to be greatly overlooked.
CONCLUSIONS
It seems clear that conflict at one level can cause conflict at other levels, from genes to organelles and upward, that there can also be feedback, and that all of this interaction has implications for sex chromosome evolution. For example, SGEs such as segregation distorters, male killers, feminizers, and parthenogenesis-inducing endosymbionts can directly target sex chromosomes, thereby generating additional intragenomic and sexual conflict, whereas sexual selection can generate sexual conflict and also select against SGEs and impose strong selection on sex chromosomes. Indeed, the recognition that SGEs can be crucial for sex chromosome evolution is a recent insight that has yet to be fully explored. This is also true of population size and sex chromosome evolution. However, although there is still much to understand, the basic logic of sexual conflict and sex chromosomes has proved to be remarkably prescient and still provides a powerful lens through which sex chromosome evolution can be studied. Equally, these chromosomes provide a unique window into the genetic basis of sexual conflict.
ACKNOWLEDGMENTS
We thank Bill Rice and Sergey Gavrilets for the invitation to participate in this collection and two anonymous referees for their valuable input. J.E.M. is supported by the European Research Council (grant agreement 260233) and a short-term fellowship from the Wissenschaftskolleg zu Berlin. D.J.H. is supported by the University of Exeter, and N.W. by the University of Exeter and The Royal Society (Wolfson Award).
Footnotes
Editors: William R. Rice and Sergey Gavrilets
Additional Perspectives on The Genetics and Biology of Sexual Conflict available at www.cshperspectives.org
REFERENCES
- Albert AYK, Otto SP 2005. Sexual selection can resolve sex-linked sexual antagonism. Science 310: 119–121. [DOI] [PubMed] [Google Scholar]
- Andolfatto P 2001. Contrasting patterns of X-linked and autosomal nucleotide variation in Drosophila melanogaster and Drosophila simulans. Mol Biol Evol 18: 279–290. [DOI] [PubMed] [Google Scholar]
- Arunkumar KP, Mita K, Nagaraju J 2009. The silkworm Z chromosome is enriched in testis-specific genes. Genetics 182: 493–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachtrog D 2013. Y-chromosome evolution: Emerging insights into processes of Y-chromosome degeneration. Nat Rev Genet 14: 113–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachtrog D, Kirkpatrick M, Mank JE, McDaniel SF, Pires JC, Rice WR, Valenzuela N 2011. Are all sex chromosomes created equal? Trends Genet 27: 350–357. [DOI] [PubMed] [Google Scholar]
- Bergero R, Forrest A, Kamau E, Charlesworth D 2007. Evolutionary strata on the X chromosomes of the dioecious plant Silene latifolia: Evidence from new sex-linked genes. Genetics 175: 1945–1954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blaser O, Grossen C, Neuenschwander S, Perrin N 2013. Sex-chromosome turnovers induced by deleterious mutation load. Evolution 67: 635–645. [DOI] [PubMed] [Google Scholar]
- Bonduriansky R, Chenoweth SF 2009. Intralocus sexual conflict. Trends Ecol Evol 24: 280–288. [DOI] [PubMed] [Google Scholar]
- Burt A, Trivers R 2006. Genes in conflict: The biology of selfish genetic elements. Harvard University Press, Cambridge MA. [Google Scholar]
- Charlesworth D, Charlesworth B 1980. Sex differences in fitness and selection for centric fusions between sex chromosomes and autosomes. Genet Res 35: 205–214. [DOI] [PubMed] [Google Scholar]
- Charlesworth B, Hartl DL 1978. Population dynamics of segregation distorter polymorphism of Drosophila melanogaster. Genetics 89: 171–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlesworth D, Mank JE 2010. The birds and the bees and the flowers and the trees: Lessons from genetic mapping of sex determination in plants and animals. Genetics 186: 9–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlesworth B, Coyne JA, Barton NA 1987. The relative rates of evolution of sex chromosomes and autosomes. Am Nat 130: 113–146. [Google Scholar]
- Chase K, Carrier DR, Adler FR, Ostrander EA, Lark KG 2005. Interaction between the X chromosome and an autosome regulations size sexual dimorphism in Portugese water dogs. Genome Res 15: 1820–1824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chippindale AK, Rice WR 2001. Y chromosome polymorphism is a strong determinant of male fitness in Drosophila melanogaster. Proc Natl Acad Sci 98: 5677–5682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cocquet J, Ellis PJI, Mahadevaiah SK, Affara NA, Vaiman D, Burgoyne PS 2012. A genetic basis for a postmeiotic X versus Y chromosome intragenomic conflict in the mouse. PLoS Genet 8: e1002900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connallon T, Clark AG 2010. Sex linkage, sex-specific selection, and the role of recombination in the evolution of sexually dimorphic gene expression. Evolution 64: 3417–3442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connallon T, Clark AG 2011a. Association between sex-biased gene expression and mutations with sex-specific phenotypic consequences in Drosophila. Genome Biol Evol 3: 151–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connallon T, Clark AG 2011b. The resolution of sexual antagonism by gene duplication. Genetics 187: 919–937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connallon T, Knowles LL 2005. Intergenomic conflict revealed by patterns of sex-biased gene expression. Trends Genet 21: 495–499. [DOI] [PubMed] [Google Scholar]
- Cordaux R, Bouchon D, Grève P 2011. The impact of endosymbionts on the evolution of host sex-determination mechanisms. Trends Genet 27: 332–341. [DOI] [PubMed] [Google Scholar]
- Dean R, Mank JE 2014. The role of sex chromosomes in sexual dimorphism: Discordance between molecular and phenotypic data. J Evol Biol 10.1111/jeb.12345. [DOI] [PubMed] [Google Scholar]
- Dean R, Perry JC, Pizzari T, Mank JE, Wigby S 2012. Experimental evolution of a novel sexually antagonistic allele. PLoS Genet 8: e1002917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dübendorfer A, Hediger M, Burghardt G, Bopp D 2002. Musca domestica: A window on the evolution of sex-determining mechanisms in insects. Int J Dev Biol 46: 75–79. [PubMed] [Google Scholar]
- Ercan S, Giresi PG, Whittle CM, Zhang XM, Green RD, Lieb JD 2007. X chromosome repression by localization of the C. elegans dosage compensation machinery to sites of transcription initiation. Nat Genet 39: 403–408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez-Capetillo O, Mahadevaiah SK, Celeste A, Romanienko PJ, Camerini-Otero RD, Bonner WM, Manova K, Burgoyne P, Nussenzweig A 2003. H2AX is required for chromatin remodeling and inactivation of sex chromosomes in male mouse meiosis. Dev Cell 4: 497–508. [DOI] [PubMed] [Google Scholar]
- Fisher RA 1931. The evolution of dominance. Biol Rev 6: 345–368. [Google Scholar]
- Foerster K, Coulson T, Sheldon BC, Pemberton JM, Clutton-Brock TH, Kruuk LEB 2007. Sexually antagonistic genetic variation for fitness in red deer. Nature 447: 1107–U9. [DOI] [PubMed] [Google Scholar]
- Frank SA 1991. Divergence of meiotic drive suppression systems as an explanation for sex-biased hybrid sterility and inviability. Evolution 45: 262–267. [DOI] [PubMed] [Google Scholar]
- Gallach M, Betran E 2011. Intralocus sexual conflict resolved through gene duplication. Trends Ecol Evol 26: 222–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallach M, Chandrasekaran C, Betran E 2010. Analyses of nuclearly encoded mitochondrial genes suggest gene duplication as a mechanism for resolving intralocus sexually antagonistic conflict in Drosophila. Genome Biol Evol 2: 835–850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelbart ME, Kuroda MI 2009. Drosophila dosage compensation: A complex voyage to the X chromosome. Development 136: 1399–1410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson JR, Chippindale AK, Rice WR 2002. The X chromosome is a hot spot for sexually antagonistic fitness variation. Proc Biol Sci 269: 499–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Good JM 2012. The conflict within and the escalating war between the sex chromosomes. PLoS Genet 8: e1002955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffin RM, Dean R, Grace JL, Ryden P, Friberg U 2013. The shared genome is a pervasive constraint on the evolution of sex-biased gene expression. Mol Biol Evol 30: 2168–2176. [DOI] [PubMed] [Google Scholar]
- Guioli S, Lovell-Badge R, Turner JMA 2012. Error-prone ZW pairing and no evidence for meiotic sex chromosome inactivation in the chicken germ line. PLoS Genet 8: e1002560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn MW, Lanzaro GC 2005. Female-biased gene expression in the malaria mosquito Anopheles gambiae. Curr Biol 15: R192–R193. [DOI] [PubMed] [Google Scholar]
- Hall DW 2004. Meiotic drive and sex chromosome cycling. Evolution 58: 925–931. [DOI] [PubMed] [Google Scholar]
- Hamilton WD 1967. Extraordinary sex ratios. Science 156: 477–488. [DOI] [PubMed] [Google Scholar]
- Harrison P, Mank JE, Wedell 2012. Incomplete sex chromosome dosage compensation in the Indian meal moth, Plodia interpunctella, based on de novo transcriptome assembly. Genome Biol Evol 4: 1118–1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hense W, Baines JF, Parsch J 2007. X chromosome inactivation during Drosophila spermatogenesis. PLoS Biol 5: 2288–2295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosken DJ, Stockley P, Tregenza T, Wedell N 2009. Monogamy and the battle of the sexes. Annu Rev Entomol 54: 361–378. [DOI] [PubMed] [Google Scholar]
- Hughes JF, Skaletsky H, Brown LG, Pyntikova T, Graves T, Fulton RS, Dugan S, Ding Y, Buhay CJ, Kremitzki C, et al. 2012. Strict evolutionary conservation followed rapid gene loss on human and rhesus Y chromosomes. Nature 483: 82–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurst LD, Pomiankowski A 1991. Causes of sex-ratio bias may account for unisexual sterility in hybrids. A new explanation of Haldane's Rule and related phenomena. Genetics 128: 841–845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Husby A, Schielzeth H, Forstmeier W, Gustafsson L, Qvarnstrom A 2013. Sex chromosome linked genetic variance and the evolution of sexual dimorphism of quantitative traits. Evolution 67: 609–619. [DOI] [PubMed] [Google Scholar]
- Innocenti P, Morrow EH 2010. The sexually antagonistic genes of Drosophila melanogaster. PLoS Biol 8: e1000335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh Y, Melamed E, Yang X, Ka PK, Wang S, Yehya N, van Nas A, Replogle K, Band MR, Clayton DF, et al. 2007. Dosage compensation is less effective in birds than in mammals. J Biol 6: 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyengar VK, Reeve HK, Eisner T 2002. Paternal inheritance of a female moth’s mating preference. Nature 419: 830–832. [DOI] [PubMed] [Google Scholar]
- Jaenike J 2001. Sex chromosome meiotic drive. Annu Rev Ecol Syst 32: 25–49. [Google Scholar]
- Jaquiery J, Rispe C, Roze D, Legeai F, Le Trionnaire G, Stoeckel S, Mieuzet L, Da Silva C, Poulain J, Prunier-Leterme N, et al. 2013. Masculinization of the X chromosome in the pea aphid. PLoS Genet 9: e1003690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Julien P, Brawand D, Soumillon M, Necsulea A, Liechti A, Schutz F, Daish T, Grutzner F, Kaessmann H 2012. Mechanisms and evolutionary patterns of mammalian and avian dosage compensation. PLoS Biol 10: e100128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaiser B, Bachtrog D 2010. Evolution of sex chromosomes in insects. Annu Rev Genet 44: 91–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khil PP, Smirnova NA, Romanienko PJ, Camerini-Otero RD 2004. The mouse X chromosome is enriched for sex-biased genes not subject to selection by meiotic sex chromosome inactivation. Nat Genet 36: 642–646. [DOI] [PubMed] [Google Scholar]
- Kitano J, Ross JA, Mori S, Kume M, Jones FC, Chan YF, Absher DM, Grimwood J, Schmutz J, Myers RM, et al. 2009. A role for a neo-sex chromosome in stickleback speciation. Nature 461: 1079–1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koerich LB, Wang XY, Clark AG, Carvalho AB 2008. Low conservation of gene content in the Drosophila Y chromosome. Nature 456: 949–951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondo M, Nanda I, Hornung U, Asakawa S, Shimizu N, Mitani H, Schmid M, Shima A, Schartl M 2003. Absence of the candidate male sex-determining gene dmrt1b(Y) of medaka from other fish species. Curr Biol 13: 416–420. [DOI] [PubMed] [Google Scholar]
- Lahn BT, Page DC 1999. Four evolutionary strata on the human X chromosome. Science 286: 964–967. [DOI] [PubMed] [Google Scholar]
- Lange J, Skaletsky H, van Daalen SKM, Embry SL, Korver CM, Brown LG, Oates RD, Silber S, Repping S, Page DC 2009. Isodicentric Y chromosomes and sex disorders as byproducts of homologous recombination that maintains palindromes. Cell 138: 855–869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemos B, Branco AT, Hartl DL 2010. Epigenetic effects of polymorphic Y chromosomes modulate chromatin components, immune response, and sexual conflict. Proc Natl Acad Sci 107: 15826–15831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livernois AM, Waters SA, Deakin JE, Graves JAM, Waters PD 2013. Independent evolution of transcriptional inactivation on sex chromosomes in birds and mammals. PLoS Genet 9: e1003635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mank JE 2009. The W, X, Y and Z of sex chromosome dosage compensation. Trends Genet 25: 226–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mank JE 2013. Sex chromosome dosage compensation, definitely not for everyone. Trends Genet 29: 677–683. [DOI] [PubMed] [Google Scholar]
- Mank JE, Ellegren H 2007. Parallel divergence and degradation of the avian W sex chromosome. Trends Ecol Evol 22: 389–391. [DOI] [PubMed] [Google Scholar]
- Mank JE, Hosken DJ, Wedell N 2011. Some inconvenient trusts about sex chromosome dosage compensation and the potential role of sexual conflict. Evolution 65: 2133–2144. [DOI] [PubMed] [Google Scholar]
- Marais GAB, Campos PRA, Gordo I 2010. Can intra-Y conversion oppose the degeneration of the human Y chromosome: A simulation study. Genome Biol Evol 2: 347–357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuda M, Nagahama Y, Shinomiya A, Sato T, Matsuda C, Kobayashi T, Morrey CE, Shibata N, Asakawa S, Shimizu N, et al. 2002. DMY is a Y-specific DM-domain gene required for male development in the medaka fish. Nature 417: 559–563. [DOI] [PubMed] [Google Scholar]
- Meiklejohn CD, Tao Y 2010. Genetic conflict and sex chromosome evolution. Trends Ecol Evol 25: 215–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meiklejohn CD, Landeen EL, Cook JM, Kingan SB, Presgraves DC 2011. Sex chromosome-specific regulation in the Drosophila male germline but little evidence for chromosomal dosage compensation or meiotic inactivation. PLoS Biol 9: e1001126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meisel RP, Han MV, Hahn MW 2009. A complex suite of forces drives gene traffic from Drosophila X chromosomes. Genome Biol Evol 1: 176–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meisel RP, Malone JH, Clark AG 2012. Disentangling the relationship between sex-biased gene expression and X-linkage. Genome Res 22: 1255–1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miura I 2007. An evolutionary witness: The frog Rana rugosa underwent change of heterogametic sex from XY male to ZW female. Sex Dev 1: 323–331. [DOI] [PubMed] [Google Scholar]
- Moghadam HK, Pointer MA, Wright AE, Berlin S, Mank JE 2012. W chromosome expression responds to female-specific selection. Proc Natl Acad Sci 109: 8207–8211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller JL, Mahadevaiah SK, Park PJ, Warburton PE, Page DC, Turner JMA 2008. The mouse X chromosome is enriched for multicopy testis genes showing postmeiotic expression. Nat Genet 40: 794–799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muyle A, Zemp N, Deschamps C, Mousset S, Widmer A, Marais GAB 2012. Rapid de novo evolution of X chromosome dosage compensation in Silene latifolia, a plant with young sex chromosomes. PLoS Biol 10: e1001308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nanda I, Kondo M, Hornung U, Asakawa S, Winkler C, Shimizu A, Shan ZH, Haaf T, Shimizu N, Shima A, et al. 2002. A duplicated copy of DMRT1 in the sex-determining region of the Y chromosome of the medaka, Oryzias latipes. Proc Natl Acad Sci 99: 11778–11783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Natri HM, Shikano T, Merila J 2013. Progressive recombination suppression and differentiation in recently evolved neo-sex chromosomes. Mol Biol Evol 30: 1131–1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nolte V, Pandey RM, Kofler R, Schlotterer C 2013. Genome-wide patterns of natural variation reveal string selective sweeps and ongoing genomic conflict in Drosophila mauritiana. Genome Res 23: 99–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogata M, Hasegawa Y, Ohtani H, Mineyama M, Miura I 2008. The ZZ/ZW sex-determining mechanism originated twice and independently during evolution of the frog, Rana rugosa. Heredity 100: 92–99. [DOI] [PubMed] [Google Scholar]
- Partridge L, Hurst LD 1998. Sex and conflict. Science 281: 2003–2008. [DOI] [PubMed] [Google Scholar]
- Pointer MA, Harrison PW, Wright AE, Mank JE 2013. Masculinization of gene expression is associated with exaggeration of male sexual dimorphism. PLoS Genet 9: e1003697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poissant J, Wilson AJ, Coltman DW 2010. Sex-specific genetic variance and the evolution of sexual dimorphism: A systematic review of cross-sex genetic correlations. Evolution 64: 97–107. [DOI] [PubMed] [Google Scholar]
- Postma E, Spyrou N, Rollins LA, Brooks RC 2011. Sex-dependent selection differentially shapes genetic variation on and off the guppy Y chromosome. Evolution 65: 2145–2156. [DOI] [PubMed] [Google Scholar]
- Potrzebowski L, Vinckenbosch N, Marques AC, Chalmel F, Jegou B, Kaessmann H 2008. Chromosomal gene movements reflect the recent origin and biology of therian sex chromosomes. PLoS Biol 6: 709–716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Presgraves DC 2010. The molecular evolutionary basis of species formation. Nat Rev Genet 11: 175–180. [DOI] [PubMed] [Google Scholar]
- Price TAR, Wedell N 2004. Selfish genetic elements and sexual selection: Their impact on male fertility. Genetica 132: 295–307. [DOI] [PubMed] [Google Scholar]
- Rice WR 1984. Sex chromosomes and the evolution of sexual dimorphism. Evolution 38: 735–742. [DOI] [PubMed] [Google Scholar]
- Rice WR 1987. The accumulation of sexually antagonistic genes as a selection agent promoting the evolution of reduced recombination between primitive sex chromosomes. Evolution 41: 911–914. [DOI] [PubMed] [Google Scholar]
- Roberts RB, Ser JR, Kocher TD 2009. Sexual conflict resolved by invasion of a novel sex determiner in Lake Malawi cichlid fishes. Science 326: 998–1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sackton TB, Montenegro H, Hartl DL, Lemos B 2011. Interspecific Y chromosome introgressions disrupt testis-specific gene expression and male reproductive phenotypes in Drosophila. Proc Natl Acad Sci 108: 17046–17051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saether SA, Saetre GP, Borge T, Wiley C, Svedin N, Andersson G, Veen T, Haavie J, Servedio MR, Bures S, et al. 2007. Sex chromosome-linked species recognition and evolution of reproductive isolation in flycatchers. Science 318: 95–97. [DOI] [PubMed] [Google Scholar]
- Saetre GP, Borge T, Lindroos K, Haavie J, Sheldon BC, Primmer C, Syvanen AC 2003. Sex chromosome evolution and speciation in Ficedula flycatchers. Proc Biol Sci 270: 53–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sayres MAW, Makova KD 2011. Genome analyses substantiate male mutation bias in many species. Bioessays 33: 938–945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoenmakers S, Wassenaar E, Hoogerbrugge JW, Laven JSE, Grootegoed JA, Baarends WM 2009. Female meiotic sex chromosome inactivation in chicken. PLoS Genet 5: e100466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Straub T, Becker PB 2007. Dosage compensation: The beginning and end of generalization. Nat Rev Genet 8: 47–57. [DOI] [PubMed] [Google Scholar]
- Toups MA, Pease JB, Hahn MW 2011. No excess gene movement is detected off the avian or lepidopteran Z chromosome. Genome Biol Evol 3: 1463–1472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Doorn GS, Kirkpatrick M 2007. Turnover of sex chromosomes induced by sexual conflict. Nature 449: 909–912. [DOI] [PubMed] [Google Scholar]
- van Doorn GS, Kirkpatrick M 2010. Transitions between male and female heterogamety caused by sex-antagonistic selection. Genetics 186: 629–U294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vibranovski MD, Zhang Y, Long MY 2009. General gene movement off the X chromosome in the Drosophila genus. Genome Res 19: 897–903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vicoso B, Bachtrog D 2011. Lack of global dosage compensation in Schistosoma mansoni, a female-heterogametic parasite. Genome Biol Evol 3: 230–235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vicoso B, Bachtrog D 2013. Reversal of an ancient sex chromosome to an autosome in Drosophila. Nature 499: 332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vicoso B, Charlesworth B 2009. The deficit of male-biased genes on the D. melanogaster X chromosome is expression-dependent: A consequence of dosage compensation? J Mol Evol 68: 576–583. [DOI] [PubMed] [Google Scholar]
- Vicoso B, Charlesworth B 2010. Effective population size and the faster-X effect: An extended model. Evolution 63: 2413–2426. [DOI] [PubMed] [Google Scholar]
- Vicoso B, Emerson JJ, Zektser Y, Mahajan S, Bachtrog D 2013. Comparative sex chromosome genomics in snakes: Differentiation, evolutionary strata and lack of global dosage compensation. PLoS Biol 11: e1001643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wedell N 2013. The dynamic relationship between polyandry and selfish genetic elements. Phil Trans R Soc 368: 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werren JH, Beukeboom LW 1998. Sex determination, sex ratios, and genetic conflict. Annu Rev Ecol Syst 29: 233–261. [Google Scholar]
- Wilkinson GS, Johns PM, Metheny JD, Baker RH 2013. Sex-biased gene expression during head development in a sexually dimorphic stalk-eyed fly. PLoS ONE 8: e59826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winge Ö 1927. The location of eighteen genes in Lebistes reticulata. J Genet 18: 1–43. [Google Scholar]
- Wolfenbarger LL, Wilkinson GS 2001. Sex-linked expression of a sexually selected trait in the stalk-eyed fly, Cyrtodiopsis dalmanni. Evolution 55: 103–110. [DOI] [PubMed] [Google Scholar]
- Wright AE, Moghadam HK, Mank JE 2012. Trade-off between selection for dosage compensation and masculinization on the avian Z chromosome. Genetics 192: 1433–1445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida K, Kitano J 2012. The contribution of female meiotic drive to the evolution of neo-sex chromosomes. Evolution 66: 3198–3208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Q, Bachtrog D 2012. Chromosome-wide gene silencing initiate Y-degeneration in Drososphila. Curr Biol 22: 522–525. [DOI] [PubMed] [Google Scholar]