Abstract
Recognition of an invading pathogen is critical to elicit protective responses. Certain microbial structures and molecules, which are crucial for their survival and virulence, are recognized by different families of evolutionarily conserved pattern recognition receptors (PRRs). This recognition initiates a signaling cascade that leads to the transcription of inflammatory cytokines and chemokines to eliminate pathogens and attract immune cells, thereby perpetuating further adaptive immune responses. Considerable research on the molecular mechanisms underlying host–pathogen interactions has resulted in the discovery of multifarious PRRs. In this review, we discuss the recent developments in microbial recognition by Toll-like receptors (TLRs) and intracellular nucleic acid sensors and the signaling pathways initiated by them.
Various pattern recognition receptors, including Toll-like receptors and intracellular DNA sensors, recognize structures and molecules of microbial pathogens and initiate signaling pathways that culminate in inflammation.
Microbes are disease-causing entities that have exerted powerful selective pressure throughout the evolution of eukaryotes (Beutler 2009). As a consequence, eukaryotes have evolved a complex immune system to counteract microbial attacks. This immune system has two arms, termed innate immunity and adaptive immunity. Initially, innate immunity was considered to be nonspecific and less complex. However, this notion was changed by the revolutionary discovery of the first Toll-like receptor (TLR) in the mid-1990s. Since then, there has been tremendous growth in our understanding of the previously understated roles of the innate immune system in the recognition of microbial pathogens, their elimination, and inflammation. It has been established that innate immune recognition engages germline-encoded pattern recognition receptors (PRRs) that recognize conserved microbial structures of pathogens known as pathogen-associated molecular patterns (PAMPs) (Akira et al. 2001; Janeway and Medzhitov 2002) and activate the expression of major histocompatibility (MHC) proteins, costimulatory molecules, and inflammatory mediators in the form of cytokines and chemokines by macrophages, dendritic cells (DCs), neutrophils, and other nonprofessional immune cells (Table 1). These processes not only trigger immediate and early mechanisms of host defense, but also prime and orchestrate antigen-specific adaptive immune responses.
Table 1.
Pattern recognition receptors (PRRs) that recognize conserved microbial structures of pathogens
| PRRs | Localization | PAMP recognized | Key adaptors | Effector response |
|---|---|---|---|---|
| TLRs | ||||
| TLR1 | Cell surface | Triacylated lipopeptides | MyD88 | IL-6, TNF-α |
| TLR2 | Cell surface | Di/triacylated lipopeptides | MyD88, TIRAP | IL-6, TNF-α, IL-8, MCP-1, RANTES |
| TLR3 | Endosomes | dsRNA | TRIF | IFN-β |
| TLR4 | Cell surface | LPS | MyD88, TRIF, TIRAP, TRAM | IL-6, TNF-α, IFNβ, IP-10 |
| TLR5 | Cell surface | Flagellin | MyD88 | TNF-α |
| TLR6 | Cell surface | Diacylated lipopeptides | MyD88, TIRAP | TNF-α, IL-6, IL-8, MCP-1, RANTES |
| TLR7 | Endosomes | ssRNA | MyD88 | IFN-α |
| TLR8 | Endosomes | ssRNA | MyD88 | IFN-α |
| TLR9 | Endosomes | CpG DNA | MyD88 | IFN-α |
| TLR11 | Endosomes | Profilin, flagellin | MyD88 | IL-12, TNF-α |
| TLR12 | Endosomes | Profilin | MyD88 | IL-12p40, IFN-α |
| TLR13 | Endosomes | 23s rRNA | MyD88 | IL-6, IL-12p40 |
| RLRs | ||||
| RIG-I | Cytoplasm | Short dsRNA, ssRNA | IPS-1, STING | IFN-β, IL-6 |
| MDA5 | Cytoplasm | Long dsRNA | IPS-1 | IFN-β |
| LGP2 | Cytoplasm | dsRNA | IPS-1 | IFN-β |
| DDX3 | Cytoplasm | Viral RNA | IPS-1 | IFN-β |
| Cytosolic DNA sensors | ||||
| DAI | Cytoplasm | dsDNA | STING | IFN-β |
| RNA Pol III | AT rich dsDNA | IPS1 | IFN-β | |
| IFI16 | Nucleus and cytoplasm | dsDNA | STING | IFN-β, IP-10, IL-6, IL-1β |
| AIM2 | Cytoplasm | dsDNA | ASC | IL-1β, IL-18 |
| Ku70 | Cytoplasm | dsDNA | ? | IFN-γ |
| MRE11 | dsDNA, ISD | STING | IFN-β, IL-6, IP-10 | |
| cGAS | Cytoplasm | dsDNA | STING | IFN-β |
| LRRFIP1 | Cytoplasm | dsDNA, dsRNA | β-catenin | IFN-β |
| DHX36 | Cytoplasm | dsDNA | MyD88 | TNF-α |
| DHX9 | Cytoplasm | dsDNA | MyD88 | TNF-α |
| DDX41 | Cytoplasm | c-di-GMP, c-di-AMP | STING | IFN-α, IFN-β |
| STING | Cytoplasm | c-di-GMP | IFN-β | |
| HMGB | Cytoplasm | dsDNA, ssDNA | ? | IFN-β, IL-6, RANTES |
| Histone H2B | Nucleus and cytoplasm | Poly (dA:dT), genomic DNA | IPS1 | IFN-β |
Innate immune recognition engages germline-encoded pattern recognition receptors (PRRs) that recognize conserved microbial structures of pathogens known as pathogen-associated molecular patterns (PAMPs) and activate the expression of major histocompatibility (MHC) proteins, costimulatory molecules, and inflammatory mediators in the form of cytokines and chemokines by macrophages, dendritic cells (DCs), neutrophils, and other nonprofessional immune cells.
IL, interleukin; IFN, interferon; TNF, tumor necrosis factor.
Microbial species, such as bacteria, viruses, fungi, and parasites, are uniquely accessorized with diverse kinds of PAMPs, which are fundamentally important for their survival. PAMPs can vary in their molecular nature from lipids, lipoproteins, and proteins to nucleic acids (Akira et al. 2006) and are redundantly or nonredundantly detected by specific PRRs. PRRs belong to several classes depending on their structural similarities, including TLRs, RIG-I-like receptors (RLRs), NOD-like receptors (NLRs), C-type lectin receptors (CLRs), and cytosolic DNA sensors. Recently, PRRs have also been reported to recognize host-derived danger signals, which are known as damage-associated molecular patterns (DAMPs) (Tang et al. 2012).
This article reviews the current information on the detection of microbial pathogens by PRRs, especially TLRs, RLRs, and intracellular DNA sensors and their signaling mechanisms that culminate in inflammation.
MICROBIAL SENSING BY TLRs
To date, 10 and 12 members of the TLR family have been identified in humans and mice, respectively. TLRs are type I transmembrane proteins composed of extracellular leucine-rich repeats (LRRs) that mediate recognition of PAMPs, transmembrane domains, and cytoplasmic Toll interleukin (IL)-1 receptor (TIR) domains that interact with downstream adaptor proteins required for signaling. TLR1 to TLR9 are conserved in both mice and humans, whereas mouse TLR10 is nonfunctional and TLR11, TLR12, and TLR13 have been deleted from the human genome (Kawai and Akira 2010). TLRs exist as either heterodimers or homodimers, and ligand binding to TLRs induces conformational changes for their activation. TLRs are broadly classified into two categories depending on their cellular localizations and the PAMPs they recognize. TLR1, TLR2, TLR4, TLR5, TLR6, and TLR10 are located on the plasma membrane and recognize lipids, lipoproteins, and proteins, whereas TLR3, TLR7, TLR8, TLR9, TLR11, TLR12, and TLR13 are localized in endosomal compartments where they recognize microbial nucleic acids and, under some special conditions (autoimmunity), self-nucleic acids (Kawai and Akira 2010; Celhar et al. 2012; Oldenburg et al. 2012; Koblansky et al. 2013). On specific ligand recognition, TLRs activate multiple signaling pathways by recruiting adaptor proteins, which initiate signal transduction pathways that culminate in the activation of transcription factors, such as nuclear factor-kappa B (NF-κB), mitogen-activated protein kinases (MAPKs), and members of the interferon (IFN) regulatory factor family, to regulate the expressions of cytokines, chemokines, and IFNs that eventuate in the host defense to microbial infection. TLRs are localized in different subcellular locations to provide optimum access to their ligands, and this specific localization is very important for the precise signaling of TLRs.
Cell Surface TLRs—Expression, Structure, and Ligands
TLR4 was identified as a receptor for bacterial lipopolysaccharide (LPS), a cell wall component of Gram-negative bacteria known to cause septic shock (Kawai and Akira 2010). TLR4 associates with myeloid differentiation factor 2 (MD2) on the cell surface to recognize LPS. Insights from a crystal structure study of the TLR4–MD2–LPS complex revealed that two copies of TLR4–MD2–LPS interact symmetrically to form a TLR4 homodimer (Park et al. 2009). After ligand binding, TLR4 translocates to the endosome through a dynein-dependent mechanism to induce a TRIF-dependent pathway (Kagan et al. 2008). TLR4 also recognizes the F protein of RSV (Kurt-Jones et al. 2000), mouse mammary tumor virus envelope proteins, Streptococcus pneumoniae pneumolysin, paclitaxel, and glycoinositol phosphate from Trypanosoma spp. (Kawai and Akira 2010; Broz and Monack 2013).
TLR2 has specificity for multiple microbial components derived from bacteria, fungi, viruses, and mycoplasma. It senses lipoproteins, peptidoglycans, lipotechoic acids from Gram-positive bacteria, zymosan, mannan, tGPI-mucin from Trypanosoma cruzi, and hemagglutinin of measles virus (Schwandner et al. 1999; Underhill et al. 1999; Kawai and Akira 2010). TLR2-mediated recognition of PAMPs and subsequent signaling occur via heterodimerization of TLR2 with either TLR1 or TLR6 on the plasma membrane. TLR2/TLR1 and TLR2/TLR6 recognize triacylated lipoproteins and diacylated lipoproteins, respectively, and induce the production of various proinflammatory cytokines, but not type I IFNs. The crystal structures of the two heterodimers revealed that each heterodimer forms an “m”-shaped complex with its ligand, thereby stabilizing the two receptors (Jin et al. 2007; Oliveira-Nascimento et al. 2012). A recent study showed the existence of TLR2/TLR10 preformed dimers, although their function is unknown (Guan et al. 2010). Many accessory molecules and coreceptors concentrate microbial products on the cell surface or inside phagosomes to facilitate TLR2 responses. One such coreceptor is CD36, which binds to ligands and transfers them to an accessory molecule, CD14, which finally loads the ligands onto TLR2/TLR6 heterodimers (FSL-1, MALP-2, and LTA) or TLR2/TLR1 heterodimers (lipomannan) (Hoebe et al. 2005; Jimenez-Dalmaroni et al. 2009). In a cell type–specific manner, TLR2 can trigger type I IFN production in response to Vaccinia viruses specifically by inflammatory monocytes (Barbalat et al. 2009).
TLR5 is a receptor for flagellin, a protein component of bacterial flagella (Akira et al. 2006). TLR5 is highly expressed in CD11c+ CD11b+ lamina propria DCs (LPDCs) in the small intestine. Following flagellin recognition, LPDCs induce the differentiation of naïve B cells into IgA-producing plasma cells and promote the differentiation of naïve T cells into IL-17-producing helper T cells (TH17) and T helper type 1 (TH1) cells (Uematsu et al. 2008). TLR11 shares a close homology with TLR5 and recognizes flagellin independently of TLR5 (Mathur et al. 2012). It also recognizes an unknown proteinaceous component of uropathogenic Escherichia coli (UPEC) (Zhang et al. 2004) and a profilin-like molecule derived from Toxoplasma gondii (Yarovinsky et al. 2005).
Endosomal TLRs—Expression, Structure, and Ligands
The nucleic acid–recognizing TLRs are TLR3, TLR7, TLR8, TLR9, and TLR13, which recognize DNA or RNA derived from bacteria and viruses as well as self-nucleic acids in autoimmune conditions. TLR3 is highly expressed in innate immune cells, except for neutrophils and plasmacytoid DCs (pDCs), and recognizes viral double-stranded RNA (dsRNA) and a synthetic analogue of dsRNA polyinosinic–polycytidylic acid (poly I:C) in endolysosomes (Takeuchi and Akira 2010; Thompson et al. 2011). Cocrystallization studies of a TLR3–dsRNA complex revealed that TLR3 has a horseshoe-shaped solenoid structure and that dsRNA binds to the amino-terminal and carboxy-terminal portions on the lateral convex surface of the TLR3 ectodomain (Choe et al. 2005; Liu et al. 2008). Recently, Bruton’s tyrosine kinase (BTK) was shown to phosphorylate the cytoplasmic domain of TLR3, particularly the Tyr759 residue, following ligand binding to initiate downstream signaling (Lee et al. 2012). TLR3 is one of the major RNA sensors, and recognizes a number of microbial RNAs including genomic RNA of reoviruses, dsRNA produced during replication of single-stranded RNA (ssRNA) of RSV, encephalomyocarditis virus (EMCV), and West Nile virus (WNV), some small-interfering RNAs, murine cytomegalovirus, and herpes simplex virus type I (HSV-1) (Zhang et al. 2007; Kawai and Akira 2010).
TLR7 and TLR8 are closely related and confined to endosomal compartments where they recognize ssRNA, mainly viral RNAs. TLR7 is predominantly expressed in pDCs and can recognize small purine analog imidazoquinoline derivatives imiquimod and resiquimod (R848), guanine analogues, and uridine or uridine/guanosine-rich ssRNA. The viral ssRNAs recognized by TLR7 are from vesicular stomatitis virus (VSV), influenza A virus, human immunodeficiency virus, and coxsackievirus B (Wang et al. 2007). RSV, Sendai virus (SV) partially, and human metapneumovirus (HMPV) are also recognized by TLR7 in a cell-specific manner (Melchjorsen et al. 2005; Lee et al. 2007; Phipps et al. 2007; Goutagny et al. 2010). Additionally, RNA from streptococcus B bacteria is sensed by TLR7 in conventional DCs (cDCs) (Mancuso et al. 2009). Although little is known about the ligands recognized by TLR8, human TLR8 was shown to recognize R848 and viral ssRNA, whereas TLR8-deficient mice responded normally to these ligands. Other ligands, like Vaccinia viral RNA, were reported to be TLR8 agonists, but remain controversial (Cervantes et al. 2012). Moreover, several studies have shown that human TLR8 responds to total bacterial RNA (Broz and Monack 2013). Crystal structure studies of unliganded and ligand-induced activated human TLR8 revealed that, the unliganded TLR8 showed a preformed dimeric form and the Z-loop between LRR14 and LRR15 was cleaved, whereas amino- and carboxy-terminal halves remained associated and confer ligand recognition and dimerization. On ligand binding, the preformed TLR8 dimer undergoes reorganization to bring the two carboxyl termini to close proximity to enable subsequent dimerization with TIR domain and downstream signaling (Tanji et al. 2013).
Notably, likewise TLR5, TLR11 recognizes flagellin, but is localized in different subcellular compartment than TLR5 (i.e., TLR5 is on the cell membrane whereas TLR11 in endolysosomes) (Mathur et al. 2012). TLR12 is predominantly expressed in myeloid cells and is highly homologous to TLR11. It can recognize profilin from T. gondii (Koblansky et al. 2013). TLR12 can function either as a heterodimer with TLR11 or alone (Broz and Monack 2013).
Recent studies have shed some light on TLR13 ligands. It was reported that TLR13 recognizes a conserved CGGAAAGACC motif in Staphylococcus aureus 23S rRNA and that E. coli 23S rRNA can induce a TLR13-dependent transcriptional response resulting in pro-IL-1β induction (Li and Chen 2012; Oldenburg et al. 2012). Similarly, both heat-killed and live Streptococcus pyogenes are recognized by a TLR13-dependent pathway (Hidmark et al. 2012). Because both TLR8 and TLR13 can recognize bacterial RNAs, the possibility of redundancy between TLR8 and TLR13 is a question for future studies.
Another member of the TLR family recognizing nucleic acids is TLR9, which, unlike other TLRs, recognizes bacterial and viral DNA that is rich in unmethylated CpG DNA motifs. TLR9 is highly expressed in pDCs, macrophages, and B cells, and can be activated by synthetic CpG oligonucleotides. Compared with the sequence-independent recognition of the 2′-deoxyribose sugar backbone of natural phosphodiester oligodeoxynucleotides by TLR9, the CpG DNA motif is required for recognition of synthetic phosphorothioate oligodeoxynucleotides (Haas et al. 2008; Wagner 2008). Viral DNAs recognized by TLR9 arise from murine cytomegalovirus (MCMV), HSV-1, HSV-2, and adenoviruses (Gurtler and Bowie 2013). Apart from its specificity for DNA, TLR9 has been shown to directly recognize hemozoin, an insoluble crystalline byproduct generated by Plasmodium falciparum during the process of detoxification after host hemoglobin is digested (Coban et al. 2010). Additionally, TLR3, TLR7, TLR9, and UNC93B1 render host resistance to Leishmania major infection (Schamber-Reis et al. 2013).
Proper cellular localization is critical for efficient signaling by nucleic acid-recognizing TLRs. It is also important for avoiding recognition of host self-DNA, which, if recognized by TLRs, can lead to autoimmunity. TLR9 and TLR7 are localized in the endoplasmic reticulum (ER) in unstimulated cells, but are recruited to endolysosomes after ligand stimulation (Kim et al. 2008). UNC93B1 is an important protein that controls the trafficking of TLR3, TLR7, and TLR9 from the ER to endolysosomes (Tabeta et al. 2006). Further, UNC93B1 actively and continuously regulates excessive TLR7 activation of immune cells by using TLR9 to counteract TLR7 as shown by mice harboring an amino acid substitution (D34 to A) in UNC93B1, that shows TLR7-hyperreactive and TLR9-hyporeactive phenotype and subsequent TLR7-dependent systemic lethal inflammation, thereby supporting the understanding that an opposing relationship between TLR7 and TLR9 is a potential mechanism regulating autoimmunity (Fukui et al. 2011).
In endolysosomes, TLR9 undergoes proteolytic cleavage by cathepsins B, S, L, H, and K and asparginyl endopeptidase to acquire a functional form that mediates ligand recognition and initiates subsequent signaling cascades (Asagiri et al. 2008; Ewald et al. 2008; Matsumoto et al. 2008; Park et al. 2008; Sepulveda et al. 2009). However, this functional cleavage of TLR9 remains controversial based on the importance of the amino-terminal region of TLR9 for CpG DNA recognition and binding (Peter et al. 2009). Interestingly, a study reported TLR9 to be expressed in the cell surface of murine splenic DCs using TLR9N- and TLR9C-specific antibodies and further explained that, succeeding the cleavage of TLR9 in the endolysosome, the amino-terminal cleaved fragment (TLR9N) remains associated with truncated TLR9 forming the complex TLR9+C, which acts as a bona fide DNA sensor; however, TLR9C alone could not recognize DNA (Onji et al. 2013).
TLR SIGNALING
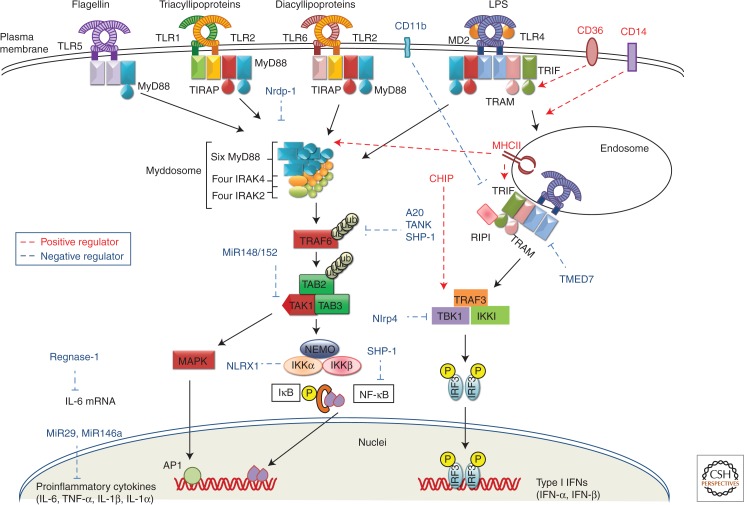
TLR signaling starts with the organization and recruitment of four TIR domain–containing adaptor proteins differentially. They are myeloid differentiation factor 88 (MyD88), MyD88 adaptor-like protein (MAL/TIRAP), TIR domain–containing adaptor–inducing IFN-β (TRIF; also known as TICAM-1), and TRIF-related adaptor molecule (TRAM) (Takeuchi and Akira 2010). TLR signaling is majorly divided into two pathways: MyD88 dependent and TRIF dependent, depending on the primary adaptor usage. With the exception of TLR3, all TLRs use MyD88 for activation of NF-κB and MAPKs to induce proinflammatory genes. TLR2 and TLR4 signaling depends on TIRAP, which mediate interaction between MyD88 and activated TLRs. The promiscuity of lipid binding by phosphoinositide-binding domain of TIRAP allows it to sample multiple compartments for the presence of activated TLRs and, hence, diversifies the subcellular sites of TLR signal transduction (Bonham et al. 2014). MyD88 recruitment activates a series of IL-1R1-associated protein kinases (IRAKs), IRAK4, IRAK2, and IRAK1, to form “Myddsome,” which further interacts with tumor necrosis factor (TNF)-α receptor–associated factor 6 (TRAF6) (Lin et al. 2010). TRAF6 activates TAK1 complex, which further phosphorylates IκB kinase (IKK)-β and MAP kinase (MAPKs). IKK complex (IKK-α/IKK-β/NEMO) catalyzes the phosphorylation of NF-κB inhibitory protein IκBα, which undergoes proteasome degradation to render NF-κB free to translocate into nucleus and induce proinflammatory gene expression (Fig. 1) (Kawai and Akira 2010).
Figure 1.
Signaling pathway of cell surface TLRs. TLR4 recognizes LPS in complex with MD2 to start a signaling cascade by recruiting adaptors MyD88 and TIRAP and forming a complex of IRAK4, 1, and 2, and TRAF6. TRAF6 catalyzes formation of K-63 linked polyubiquitin chains on TRAF6 itself and generates an unconjugated polyubiquitin chain. TRAF6 activates TAK1 complex, which further activates IKK complex and MAP kinases. IKK complex phosphorylates IκB, which undergoes proteosomal degradation to release NF-κB for further translocation to nucleus and subsequent induction of proinflammatory cytokines. MAP kinases phosphorylate Jun kinases (JNK), p38 kinase, extracellular signal regulated kinase 1(ERK1), and ERK2. TLR4 also signals through TRIF-dependent pathways with the help of adaptor TRAM after translocation to endosome and activates IRF3 for type I IFN production. TLR1/2 and TLR6/2 heterodimers recognize triacylated and diacylated lipoproteins, respectively, whereas TLR5 recognizes flagellin and all of them initiate a MyD88-dependent signaling pathway that culminates in the induction of proinflammatory cytokines. TLR signaling is modulated by CD14, CD36, Nrdp1, CHIP, MHCII, Regnase-1, A20, SHP-1, TMED7, CD11b, NLRP4, NLRX1, and miRNAs (miR-146a, miR-29, miR-148/152).
TLR3 as well as TLR4 use a TRIF-dependent pathway that results in activation of IRF3 and NF-κB for subsequent induction of type I interferons and inflammatory cytokines, respectively (Akira et al. 2006). TLR4 requires TRAM to interact with TRIF; however, to directly interact with TRIF, the two tyrosine residues in the cytoplasmic domain of TLR3, Tyr858, and Tyr759 are phosphorylated by the epidermal growth factor ErbB1 and Btk, respectively (Lee et al. 2012; Yamashita et al. 2012). TRIF interacts with TRAF6 and activates TAK1 to activate NF-κB in a manner similar to MyD88. TRIF also recruits TRAF3, which activates TANK-binding kinase 1 (TBK1) and IKKi to phosphorylate IRF3 and IRF7 for subsequent induction of type I IFN gene expression (Hacker et al. 2006; Kawai and Akira 2010)
Continuing studies have uncovered plenty of regulators critical for TLR signaling that differ in their cellular location and mode of action (Qian and Cao 2013). Membrane-resident molecules, such as CD14 and CD36, modulate TLR signaling. CD14, a glycophosphotidylinositol-anchored protein, which acts as a coreceptor for LPS along with TLR4 and MD2, induces ITAM-mediated Syk and PLC-γ2-dependent endocytosis, thus promoting TLR4 internalization to endosomes to facilitate TRIF-dependent signaling (Zanoni et al. 2011). CD14 is also necessary for TLR7- and TLR9-dependent induction of proinflammatory cytokines (Baumann et al. 2010). CD36, a protein of class B scavenger receptor family, acts as a coreceptor for oxidized low-density lipoprotein (LDL) and amyloid β peptide and enables formation of TLR4/6 heterodimer through Src kinases ensuing to sterile proinflammatory events (Stewart et al. 2010). Ubiquitin-modifying enzymes are also emerging as important regulators of TLR signaling. Nrdp-1, an E3 ligase, directly binds and polyubiquitinates MyD88 and TBK1, inducing degradation of former and augmenting activation of later to attenuate production of inflammatory cytokines and promote preferential type I IFN production, respectively (Wang et al. 2009). Furthermore, heat shock cognate 70 (HSC70)-interacting protein (CHIP) can enhance TLR signaling by recruiting, polyubiquinating and activating the tyrosine kinase Src and atypical protein kinase C ζ (PKCζ) to the TLR complex, henceforth, leading to activation of IRAK-1, TBK1, IRF3, and IRF7 (Fig. 1) (Yang et al. 2011). Mitochondrial ubiquitin ligase MARCH5 positively regulates TLR7 signaling and catalyzes the K63-linked ubiquitination of TANK, making it incapable of inhibiting TRAF3 (Fig. 2) (Shi et al. 2011).
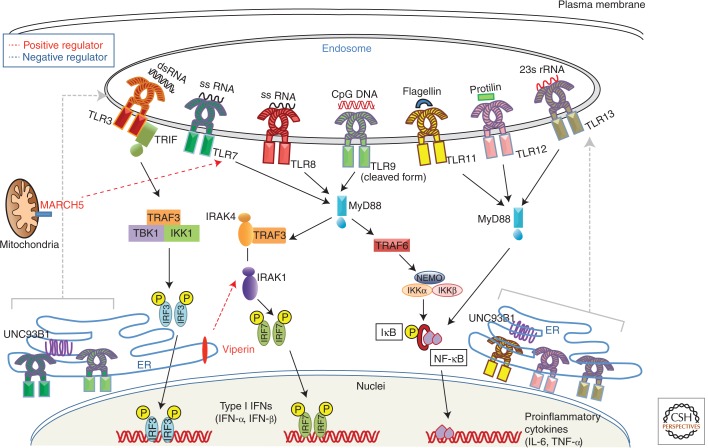
Figure 2.
Signaling pathway of endosomal TLRs. Endosomal residents of TLR family are TLR3, TLR7, TLR8, TLR9, TLR11, TLR12, and TLR13. dsRNA recognition by TLR3 recruits TRIF to initiate signaling cascade that further activates TBK1 to induce IRF3-mediated type I IFN production. Concurrently, TRIF can also interact with TRAF6 to activate NF-κB for transcription of proinflammatory cytokines. TLR7 and TLR8 recognize viral ssRNA, whereas TLR9 recognizes CpG DNA from both bacteria and viruses. Ligand stimulation facilitates UNC93B1-dependent trafficking of TLR7 and TLR9 from ER to endosomes, where TLR9 undergoes proteolytic cleavage. Following this TLR7 and TLR9 recruit MyD88 to activate NF-κB and IRF7 to induce transcription of proinflammatory cytokines and type I IFN genes, respectively. TLR11, TLR12, and TLR13 recognize flagellin, profilin, and bacterial 23sRNA, respectively, and induce NF-κB signaling through MyD88 and TIRAP. Endosomal TLR signaling can be modulated by MARCH5 and viperin.
A novel function of MHC class II molecules is reported, where intracellular MHC class II molecules interacted with Btk via the costimulatory molecule CD40 and maintained Btk activation. Activated Btk interacts with MyD88 and TRIF to promote enhanced production of inflammatory cytokines and type I IFNs (Liu et al. 2011). Antiviral protein viperin mediates antiviral function through TLR7/9- IRAK1 signaling axis, wherein, viperin that is induced after TLR7 or TLR9 stimulation interacts with IRAK1 and TRAF6 to recruit them to lipid bodies and to facilitate K63-linked ubiquitination of IRAK1, which induces nuclear translocation of IRF7 and subsequent type I IFN production by pDCs (Saitoh et al. 2011). Utilizing a combinatorial approach including transcriptomics, genetic/chemical perturbations, and phosphoproteomics, signaling components involved in the TLR response in dendritic cells were discovered systematically, particularly, Polo-like kinases (Plks) 2 and 4 were found to be important for antiviral responses both in vitro and in vivo (Chevrier et al. 2011).
TLR signaling is negatively regulated by various molecules at numerous levels to keep check on the excessive immune responses that can lead to detrimental consequences. TANK, IRAK-M, Atg16L, Regnase-1, tristeraprolin, A20, SHP-1, SENP6, GOLD domain–containing TMED7, Trim 30α, CD11b, NLRP4, NLRX1, and miRNAs (miR-146a, miR-199a, miR-155, miR-126, miR-21, miR-29, miR-148/152, miR-466l) negatively regulate TLR signaling by different mechanisms (Fig. 1) (Kawai and Akira 2010; Doyle et al. 2012; Kondo et al. 2012; Liu et al. 2013; Olivieri et al. 2013; Qian and Cao 2013).
INTRACELLULAR VIRAL RECOGNITION AND SIGNALING BY RLRs
Viral RNAs are also recognized by another family of cytoplasmic receptors known as RLRs. The three members that constitute the RLR family are retinoic acid-inducible gene I (RIG-I), melanoma differentiated gene 5 (MDA5), and laboratory of genetics and physiology 2 (LGP2). RLRs are expressed in a variety of cell types, including myeloid cells, epithelial cells, fibroblasts, and cells of the central nervous system, although RLR function is not necessary for IFN production by pDCs despite their expression in this cell type (Loo and Gale 2011). RLRs are structurally similar and harbor three distinct domains: an amino-terminal region composed of tandem caspase activation and recruitment domains (CARDs), a central DEAD box helicase/ATPase domain, and a carboxy-terminal regulatory domain. LGP2 is homologous to RIG-I and MDA5, but lacks the amino-terminal CARD domain, and functions as a regulator of RIG-I and MDA5 signaling (Fig. 3) (Saito et al. 2007; Yoneyama and Fujita 2008).
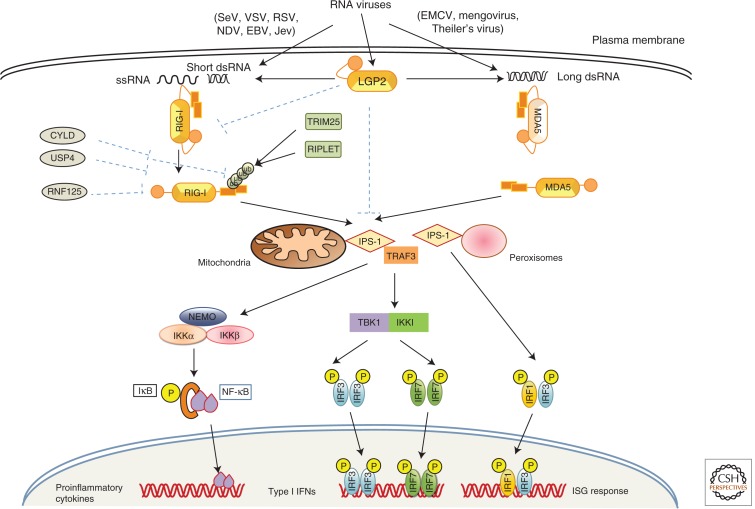
Figure 3.
RLR Signaling pathway. RLR family contains of RIG-I, MDA5, and LGP2. RIG-I and MDA5 recognize viruses differentially. RIG-I recognizes SeV, VSV, RSV, NDV, EBV, and JEV, whereas MDA5 recognizes viruses of picornavirus family members. After virus infection, long viral dsRNA or short dsRNA activates MDA5 or RIG-I, respectively, and they undergo conformational change to obtain an open confirmation and their CARD domain interacts with CARD domain of IPS-1 localized in mitochondria and peroxisomes to start signaling. Mitochondrial IPS-1 signaling leads to activation of NF-κB and RF3/IRF7 resulting in the production of type I interferons and proinflammatory cytokines, whereas peroxisomal IPS-1 activates IRF1- and IRF3-dependent signaling leading to interferon stimulatory genes (ISGs) expression. LGP2 is a positive regulator of RIG-I and MDA5. RIG-I signaling is also positively regulated by Trim25 and RIPLET, whereas it is negatively regulated by CYLD, USP4, and RNF125.
A number of studies have shown that RIG-I recognizes members of the Paramyxoviridae, Rhabdoviridae, Orthomyxoviridae, and flavivirus genera, and, consequently, RIG-I-deficient cells have impaired type I and inflammatory cytokine production in response to NDV, SV, VSV, and JEV (Kato et al. 2006). RIG-I also recognizes influenza A virus, measles virus, and Ebola virus (Thompson et al. 2011). Listeria monocytogenes actively secretes small RNAs via a SecA2 secretion system, thereby triggering strong RIG-I activation that leads to type I IFN production (Abdullah et al. 2012; Hagmann et al. 2013). Compared with RIG-I, MDA5 recognizes a different class of viruses, such as Picornaviridae, and MDA5-deficient mice show abrogated type I IFN production in response to EMCV, Theiler’s virus, hepatitis C virus, mengovirus, and murine norovirus (Kato et al. 2006; McCartney et al. 2008). Moreover, dengue virus, WNV, and reovirus are recognized by both RIG-I and MDA5 in concert (Fredericksen et al. 2008; Loo et al. 2008). The preferences for target RNA recognition differ between RIG-I and MDA5. RIG-I preferentially recognizes short dsRNA (from 19- or 21-mers to 1 kb) with 5′-triphosphorylated ssRNA, a common characteristic of viral RNA (Hornung et al. 2006; Pichlmair et al. 2006; Takeuchi and Akira 2010; Thompson et al. 2011). However, MDA5 recognizes high molecular weight dsRNA- (long dsRNA of more than 2 kb) like poly I:C (Kato et al. 2006). LGP2 was initially suggested to function as a negative regulator of RIG-I and MDA5 signaling (Yoneyama et al. 2005; Saito et al. 2007). In contrast, LGP2-deficient mice and cells expressing an LGP2 mutant with a point mutation, D30A, that abrogates the ATPase activity, had impaired type I IFN production in response to RIG-I and MDA5 ligands, implying positive regulation of RIG-I and MDA5 by LGP2 (Satoh et al. 2010).
Structural studies have shown that RIG-I in unstimulated cells is sequestered in the cytoplasm in an inactive (“closed” conformation) form that is autoinhibited by its regulatory domain (Saito et al. 2007). However, after virus infection, RIG-I undergoes conformational changes to an “open” conformation to become active and multimerizes in an ATP-dependent manner (Scott 2010). The CARD domains of activated RIG-I and MDA5 then homotypically interact with the CARD of interferon promoter stimulator 1 (IPS-1; also known as MAVS, VISA, or Cardif) (Kawai and Akira 2006), which is localized in the outer membranes of mitochondria and the peroxisomal membranes (Dixit et al. 2010; Scott 2010). This interaction relocates RLRs to IPS-1-associated membranes to form an IPS-1 signalosome with other downstream molecules (Loo and Gale 2011). To elucidate the activation mechanism of MAVS, it was shown that viral infection induces a prion-like conformational switch forming very large functional aggregates of MAVS-like prions on the mitochondrial membrane catalyzed by RIG-I in the presence of unanchored K63 ubiquitin chains, thereby activating and propagating antiviral signaling cascade (Hou et al. 2011).
RLR signaling is tightly regulated both positively and negatively by differential ubiquitination. TRIM25 and RNF135 (Riplet or REUL) are E3 ubiquitin ligases that enhance RIG-I activation by K63 polyubiquitination (Gack et al. 2007; Pichlmair et al. 2009). In contrast, RNF125 inhibits RIG-I signaling by K48 polyubiquitination and proteasome-mediated degradation of RIG-I (Arimoto et al. 2007). CYLD (cylindromatosis) and ubiquitin-specific protease 4 (USP4) are deubiquitinases that remove K63-linked or K48-linked polyubiquitin chains, respectively, from RIG-I to inhibit RIG-I-dependent IFN production (Friedman et al. 2008; Wang et al. 2013). Additionally, several microRNAs, such as miR-146a, miR-4661, miR-122, and miR-24, have been shown to regulate the RIG-I pathway. Intriguingly, recently reported that MAVS mRNA is bicistronic and codes for both full-length MAVS and a truncated variant, miniMAVS. miniMAVS inhibits MAVS-induced antiviral responses, however, both proteins positively regulate cell death (Brubaker et al. 2014).
Recently, many studies have shown that cytosolic synthetic dsRNA, poly I:C, virus-derived RNA, and bacteria-derived RNA can activate the NLRP3 inflammasome independently of RLRs (Kanneganti et al. 2006; Rajan et al. 2010; Eigenbrod et al. 2012). DExD/H box helicase family member DHX33 was identified as an RNA sensor that activates the NLRP3 inflammasome in response to cytosolic poly I:C, reoviral RNA, and bacterial RNA (Mitoma et al. 2013).
INTRACELLULAR DNA SENSING
DNA was initially shown to be recognized by TLR9. However, several studies using TLR9 antagonists, TLR9, and DNase II–deficient mice soon suggested the presence of additional DNA sensors and pathways that function independently of TLR9 and are present in the cytoplasm. New advances are revealing a whole repertoire of much sought-after cytosolic DNA sensors, and the list is continuing to expand. These DNA sensors recognize microbial or self-DNA present in the cytoplasm as a sign of infection or cell damage and induce the production of type I IFNs, type III IFNs, or IL-1β. Most of the DNA sensors recognize foreign DNA in the cytoplasm and generally utilize stimulator of interferon genes (STING; also known as TMEM173, MPYS, MITA, and ERIS) and TBK1 to induce type I IFN production (Fig. 4) (Ishii et al. 2006; Ishikawa and Barber 2008).
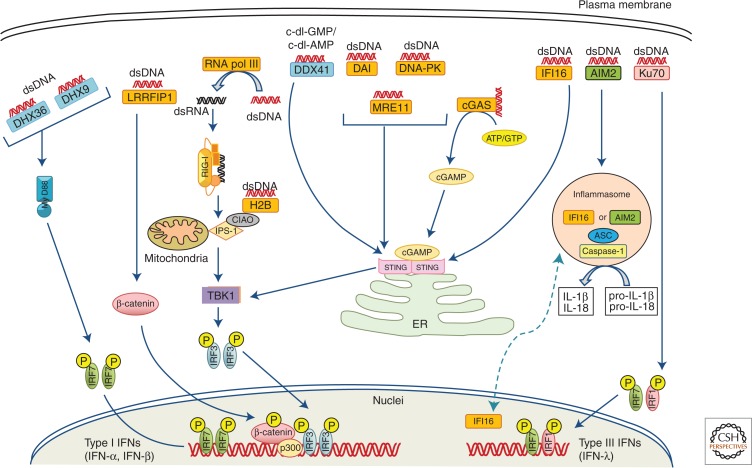
Figure 4.
Cytosolic DNA sensors and their signaling pathways. DNA present in the cytoplasm derived from either hosts or microbes are recognized by various kinds of DNA sensors. DDX41, DAI, DNA-PK, IFI16, MRE11, and cGAS recognize dsDNA and induce type I IFN production through STING via TBK1- and IRF3-dependent pathways. AT-rich dsDNA recognized by RNA polymerase III is transcribed to 5′-triphosphate RNA to be further sensed by RIG-I to induce IPS-1-dependent type I production; similarly, H2B also recognizes dsDNA and signals through IPS1. LRRFIP1 binds both dsDNA and dsRNA and activates β-catenin, which enhances a TBK1-independent and IRF3-dependent IFN-β transcription. DHX9 and DHX36 are MyD88-dependent DNA sensors. AIM2 and nuclear IFI16 can form inflammasomes together with ASC and caspase-1 on recognition of dsDNA to mediate a caspase-1-dependent cleavage of IL-1β and IL-18 from pro-IL-1β and pro-1L-18. Finally, Ku-70 recognizes dsDNA to induce IRF1/IRF7-dependent type III IFN.
DNA-dependent activator of IRFs (DAI) (Takaoka et al. 2007; Ishii et al. 2008), RNA polymerase III (Pol III) (Ablasser et al. 2009; Chiu et al. 2009), IFN-γ-inducible protein 16 (IFI16) (Unterholzner et al. 2010), leucine-rich repeat flightless-interacting protein 1 (LRRFP1) (Yang et al. 2010), extrachromosomal histone H2B (Kobiyama et al. 2010), DNA-PK (Ferguson et al. 2012), and MRE11 (Kondo et al. 2013) recognize dsDNA to induce type I IFN production. Members of the DExD/H-box helicase family, DHX9, DHX36, and DDX41, were also identified as candidate DNA sensors, although they have different specificities for microbial DNA recognition (Kim et al. 2010). Unlike other cytosolic DNA sensors, AIM2 and nuclear IFI16 recognize cytosolic DNA and form an active inflammasome by recruiting ASC and pro-caspase-1, which cleaves pro-IL-1β and pro-IL-18 to their mature forms for secretion (Fernandes-Alnemri et al. 2010; Kerur et al. 2011). Ku70 senses cytosolic DNA and induces type III IFN production (Zhang et al. 2011).
Recently, by using biochemical strategies, a crucial study by Chen and colleagues showed that cyclic GMP-AMP (cGAMP) synthase (cGAS), a member of the nucleotidyltransferase family acts as a novel sensor for cytosolic DNA and provides the endogenous second messenger cGAMP for STING activation. On encountering DNA in the cytoplasm, cGAS synthesizes cyclic-di-GMP-AMP (c-di-GAMP) from ATP and GTP, which then binds to STING to induce IRF3-mediated IFN-β production. cGAS have been shown to directly interact with dsDNA independently of its sequence through the amino-terminal domain and activates the synthesis of cGAMP. cGAS knockdown leads to impaired IFN-β induction and IRF3 activation in response to DNA transfection or DNA virus infection (Sun et al. 2013; Wu et al. 2013). Furthermore, by generating cGAS knockout mice, the Chen group showed that multiple cell types including fibroblasts, macrophages, and DCs from cGAS-deficient mice had impaired type I IFN and other proinflammatory cytokine production in response to transfected immunostimulatory DNA or DNA virus (HSV-1, VACV) infection, whereas the responses to poly I:C, poly dAdT, and RNA virus infection remained intact. Moreover, cGAS-deficient mice were more susceptible to lethal HSV-1 infection than wild-type mice. cGAMP also acts as an immune adjuvant to stimulate antigen-specific T cells and antibody responses in a STING-dependent manner (Li et al. 2013). Previous studies have shown that a mutant allele of murine STING (R231A) was unresponsive to cyclic dinucleotides (CDNs) but normally responsive to dsDNA, thus presenting a paradox (Burdette et al. 2011). Later, following the discovery of endogenous cGAMP, the R231A mutant was found to respond normally to endogenous cGAMP, but did not respond to the microbial counterpart cGAMP synthesized by cGAS from Vibrio cholera or chemically synthesized cGAMP (Davies et al. 2012; Ablasser et al. 2013; Diner et al. 2013). Taken together, these observations suggested that a clear mechanism exists for classification of endogenous cGAMP and other microbial CDNs. In a series of elegant biochemical, biophysical, and structural studies, four independent groups showed that endogenous cGAMP synthesized by cGAS is a noncanonical CDN with unique phosphodiester linkages between 2′OH of GMP and 5′-phosphate of AMP and between 3′OH of AMP and 5′-phosphate of GMP, thus referred to as 2′3′-cGAMP, and that 2′3′-cGAMP is a far more potent stimulator of STING than canonical CDNs from microbial origin that have 3′–5′ linkages (Ablasser et al. 2013; Diner et al. 2013; Gao et al. 2013; Zhang et al. 2013). These structural studies and in vitro enzymatic reactions further showed that cGAS first catalyzes the synthesis of a linear 2′–5′-linked dinucleotide, followed by its cGAS-dependent cyclization in a second step through a 3′–5′ phosphodiester linkage to form 2′3′-cGAMP (Ablasser et al. 2013; Gao et al. 2013; Xiao and Fitzgerald 2013). Further investigation of the structural insights revealed that cGAS contains a less conserved amino-terminal stretch, followed by a highly conserved Mab21 domain of the nucleotidyl transferase (NTase) superfamily having a bilobal scaffold, and that a unique zinc-binding motif called a “zinc thumb” is present across both lobes of cGAS, providing an essential B-DNA-binding platform (Civril et al. 2013).
STING Is Cardinal for Intracellular DNA Sensing
With continuing research, STING has emerged as the key adaptor in the cytosolic DNA-sensing pathway, as several studies have shown its indispensable roles in innate immune responses to DNA derived from viral pathogens, bacterial pathogens, and eukaryotic pathogens, thus ingraining STING as a prime signaling adaptor in the intracellular DNA-sensing pathway (Burdette and Vance 2013). STING is also implicated in certain autoimmune diseases and adjuvanticity of DNA vaccines (Ishikawa et al. 2009; Gall et al. 2012). Structurally, STING is composed of an amino-terminal domain consisting of four transmembrane domains that anchor STING in the ER and a carboxy-terminal domain (CTD) that is speculated to be cytosolic (Burdette and Vance 2013; Paludan and Bowie 2013). In resting cells, STING is localized in the mitochondrial-associated membrane (MAM)/ER, but after HSV-1 infection, it rapidly traffics via unclear mechanisms to an uncharacterized perinuclear region, although this endosomal compartment containing STING and TBK1 does involve autophagy-related gene 9a (Atg9a) (Ishikawa et al. 2009; Saitoh et al. 2010). Furthermore, on recognition of DNA by upstream DNA sensors, STING binds to both IRF3 and TBK1 via its carboxy-terminal region and thus provides a scaffold to promote TBK1-mediated phosphorylation of IRF3 (Tanaka and Chen 2012). Ubiquitination is also suggested to be important for STING signaling, as the ubiquitin ligases TRIM56 and RNF5 have been shown to regulate STING signaling positively and negatively, respectively (Zhong et al. 2009; Tsuchida et al. 2010). Furthermore, STING was reported to directly recognize CDNs, such as c-di-GMP from bacteria (Burdette et al. 2011). Structural studies of human STING by different research groups have shown that the STING CTD exists in a symmetrical dimeric state when unbound to CDNs and that CDN binding does not bring about any conformational change of the CTD, meaning that a definite mechanism for how STING is activated to move from the ER to engage with TBK1 remains elusive (Paludan and Bowie 2013). Besides providing a scaffold for TBK1 and IRF3 on virus infection or nucleic acid stimulation, STING regulates a unique antiviral pathway by recruiting STAT6 to the ER for subsequent phosphorylation by TBK1, independently of Janus kinases (JAKs), thereby inducing STAT6-specific target genes for immune cell homing (Chen et al. 2011). Recently, negative feedback control of STING activity by CDNs was reported. After initial activation of the STING–TBK1–IRF3 axis, STING is phosphorylated by UNC-51-like kinase (ULK1/ATG1), which suppresses IRF3 activity to avoid sustained production of inflammatory cytokines. ULK1 activation is triggered by CDNs generated by cGAS (Konno et al. 2013).
CONCLUDING REMARKS
Studies on pathogen recognition by the host innate immune system have come a long way since the first description of PRRs. Diverse kinds of PRR families, such as TLRs, RLRs, NLRs, CLRs, AIM2-like receptors (ALRs), peptidoglycan recognition proteins (PGRPs), and an expanding number of cytosolic DNA sensors have been identified, each having specificity for distinct PAMPs. This review has refurbished the present knowledge on pathogen sensing by PRRs, as well as their ligands, structures, expression patterns, and signaling mechanisms that are crucial for host defense. Nevertheless, a thrust area for research on pathogen recognition requires investigation of multiple PRRs activated in response to pathogens and the crosstalk among these PRRs that coordinates host immune responses. Furthermore, in the case of cytosolic DNA sensors, with the identification of a number of DNA sensor candidates, the scope of the questions is increasing with regard to the extent of redundancy between these receptors, their expression patterns in different cell types, and the mechanisms by which they recognize different kinds of dsDNA (i.e., their ligand specificity). Hence, detailed research encompassing the molecular and cellular mechanisms of PRR signaling will provide a better understanding, thereby helping to combat microbial infections effectively.
Footnotes
Editor: Ruslan M. Medzhitov
Additional Perspectives on Innate Immunity and Inflammation available at www.cshperspectives.org
REFERENCES
- Abdullah Z, Schlee M, Roth S, Mraheil MA, Barchet W, Bottcher J, Hain T, Geiger S, Hayakawa Y, Fritz JH, et al. 2012. RIG-I detects infection with live Listeria by sensing secreted bacterial nucleic acids. EMBO J 31: 4153–4164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ablasser A, Bauernfeind F, Hartmann G, Latz E, Fitzgerald KA, Hornung V 2009. RIG-I-dependent sensing of poly(dA:dT) through the induction of an RNA polymerase III–transcribed RNA intermediate. Nat Immunol 10: 1065–1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ablasser A, Goldeck M, Cavlar T, Deimling T, Witte G, Rohl I, Hopfner KP, Ludwig J, Hornung V 2013. cGAS produces a 2′-5′-linked cyclic dinucleotide second messenger that activates STING. Nature 498: 380–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akira S, Takeda K, Kaisho T 2001. Toll-like receptors: Critical proteins linking innate and acquired immunity. Nat Immunol 2: 675–680. [DOI] [PubMed] [Google Scholar]
- Akira S, Uematsu S, Takeuchi O 2006. Pathogen recognition and innate immunity. Cell 124: 783–801. [DOI] [PubMed] [Google Scholar]
- Arimoto K, Takahashi H, Hishiki T, Konishi H, Fujita T, Shimotohno K 2007. Negative regulation of the RIG-I signaling by the ubiquitin ligase RNF125. Proc Natl Acad Sci 104: 7500–7505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asagiri M, Hirai T, Kunigami T, Kamano S, Gober HJ, Okamoto K, Nishikawa K, Latz E, Golenbock DT, Aoki K, et al. 2008. Cathepsin K-dependent toll-like receptor 9 signaling revealed in experimental arthritis. Science 319: 624–627. [DOI] [PubMed] [Google Scholar]
- Barbalat R, Lau L, Locksley RM, Barton GM 2009. Toll-like receptor 2 on inflammatory monocytes induces type I interferon in response to viral but not bacterial ligands. Nat Immunol 10: 1200–1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumann CL, Aspalter IM, Sharif O, Pichlmair A, Bluml S, Grebien F, Bruckner M, Pasierbek P, Aumayr K, Planyavsky M, et al. 2010. CD14 is a coreceptor of Toll-like receptors 7 and 9. J Exp Med 207: 2689–2701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beutler B 2009. Microbe sensing, positive feedback loops, and the pathogenesis of inflammatory diseases. Immunol Rev 227: 248–263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonham KS, Orzalli MH, Hayashi K, Wolf AI, Glanemann C, Weninger W, Iwasaki A, Knipe DM, Kagan JC 2014. A promiscuous lipid-binding protein diversifies the subcellular sites of Toll-like receptor signal transduction. Cell 156: 705–716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broz P, Monack DM 2013. Newly described pattern recognition receptors team up against intracellular pathogens. Nat Rev Immunol 13: 551–565. [DOI] [PubMed] [Google Scholar]
- Brubaker SW, Gauthier AE, Mills EW, Ingolia NT, Kagan JC 2014. A bicistronic MAVS transcript highlights a class of truncated variants in antiviral immunity. Cell 156: 800–811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burdette DL, Vance RE 2013. STING and the innate immune response to nucleic acids in the cytosol. Nat Immunol 14: 19–26. [DOI] [PubMed] [Google Scholar]
- Burdette DL, Monroe KM, Sotelo-Troha K, Iwig JS, Eckert B, Hyodo M, Hayakawa Y, Vance RE 2011. STING is a direct innate immune sensor of cyclic di-GMP. Nature 478: 515–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celhar T, Magalhaes R, Fairhurst AM 2012. TLR7 and TLR9 in SLE: When sensing self goes wrong. Immunol Res 53: 58–77. [DOI] [PubMed] [Google Scholar]
- Cervantes JL, Weinerman B, Basole C, Salazar JC 2012. TLR8: The forgotten relative revindicated. Cell Mol Immunol 9: 434–438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Sun H, You F, Sun W, Zhou X, Chen L, Yang J, Wang Y, Tang H, Guan Y, et al. 2011. Activation of STAT6 by STING is critical for antiviral innate immunity. Cell 147: 436–446. [DOI] [PubMed] [Google Scholar]
- Chevrier N, Mertins P, Artyomov MN, Shalek AK, Iannacone M, Ciaccio MF, Gat-Viks I, Tonti E, DeGrace MM, Clauser KR, et al. 2011. Systematic discovery of TLR signaling components delineates viral-sensing circuits. Cell 147: 853–867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu YH, Macmillan JB, Chen ZJ 2009. RNA polymerase III detects cytosolic DNA and induces type I interferons through the RIG-I pathway. Cell 138: 576–591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choe J, Kelker MS, Wilson IA 2005. Crystal structure of human toll-like receptor 3 (TLR3) ectodomain. Science 309: 581–585. [DOI] [PubMed] [Google Scholar]
- Civril F, Deimling T, de Oliveira Mann CC, Ablasser A, Moldt M, Witte G, Hornung V, Hopfner KP 2013. Structural mechanism of cytosolic DNA sensing by cGAS. Nature 498: 332–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coban C, Igari Y, Yagi M, Reimer T, Koyama S, Aoshi T, Ohata K, Tsukui T, Takeshita F, Sakurai K, et al. 2010. Immunogenicity of whole-parasite vaccines against Plasmodium falciparum involves malarial hemozoin and host TLR9. Cell Hos Microbe 7: 50–61. [DOI] [PubMed] [Google Scholar]
- Davies BW, Bogard RW, Young TS, Mekalanos JJ 2012. Coordinated regulation of accessory genetic elements produces cyclic di-nucleotides for V. cholerae virulence. Cell 149: 358–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diner EJ, Burdette DL, Wilson SC, Monroe KM, Kellenberger CA, Hyodo M, Hayakawa Y, Hammond MC, Vance RE 2013. The innate immune DNA sensor cGAS produces a noncanonical cyclic dinucleotide that activates human STING. Cell Rep 3: 1355–1361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixit E, Boulant S, Zhang Y, Lee AS, Odendall C, Shum B, Hacohen N, Chen ZJ, Whelan SP, Fransen M, et al. 2010. Peroxisomes are signaling platforms for antiviral innate immunity. Cell 141: 668–681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle SL, Husebye H, Connolly DJ, Espevik T, O’Neill LA, McGettrick AF 2012. The GOLD domain-containing protein TMED7 inhibits TLR4 signalling from the endosome upon LPS stimulation. Nat Commun 3: 707. [DOI] [PubMed] [Google Scholar]
- Eigenbrod T, Franchi L, Munoz-Planillo R, Kirschning CJ, Freudenberg MA, Nunez G, Dalpke A 2012. Bacterial RNA mediates activation of caspase-1 and IL-1β release independently of TLRs 3, 7, 9 and TRIF but is dependent on UNC93B. J Immunol 189: 328–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewald SE, Lee BL, Lau L, Wickliffe KE, Shi GP, Chapman HA, Barton GM 2008. The ectodomain of Toll-like receptor 9 is cleaved to generate a functional receptor. Nature 456: 658–662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferguson BJ, Mansur DS, Peters NE, Ren H, Smith GL 2012. DNA-PK is a DNA sensor for IRF-3-dependent innate immunity. eLife 1: e00047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandes-Alnemri T, Yu JW, Juliana C, Solorzano L, Kang S, Wu J, Datta P, McCormick M, Huang L, McDermott E, et al. 2010. The AIM2 inflammasome is critical for innate immunity to Francisella tularensis. Nat Immunol 11: 385–393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fredericksen BL, Keller BC, Fornek J, Katze MG, Gale M Jr 2008. Establishment and maintenance of the innate antiviral response to West Nile Virus involves both RIG-I and MDA5 signaling through IPS-1. J Virol 82: 609–616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman CS, O’Donnell MA, Legarda-Addison D, Ng A, Cardenas WB, Yount JS, Moran TM, Basler CF, Komuro A, Horvath CM, et al. 2008. The tumour suppressor CYLD is a negative regulator of RIG-I-mediated antiviral response. EMBO Rep 9: 930–936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukui R, Saitoh S, Kanno A, Onji M, Shibata T, Ito A, Matsumoto M, Akira S, Yoshida N, Miyake K 2011. Unc93B1 restricts systemic lethal inflammation by orchestrating Toll-like receptor 7 and 9 trafficking. Immunity 35: 69–81. [DOI] [PubMed] [Google Scholar]
- Gack MU, Shin YC, Joo CH, Urano T, Liang C, Sun L, Takeuchi O, Akira S, Chen Z, Inoue S, et al. 2007. TRIM25 RING-finger E3 ubiquitin ligase is essential for RIG-I-mediated antiviral activity. Nature 446: 916–920. [DOI] [PubMed] [Google Scholar]
- Gall A, Treuting P, Elkon KB, Loo YM, Gale M Jr, Barber GN, Stetson DB 2012. Autoimmunity initiates in nonhematopoietic cells and progresses via lymphocytes in an interferon-dependent autoimmune disease. Immunity 36: 120–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao P, Ascano M, Wu Y, Barchet W, Gaffney BL, Zillinger T, Serganov AA, Liu Y, Jones RA, Hartmann G, et al. 2013. Cyclic [G(2′,5′)pA(3′,5′)p] is the metazoan second messenger produced by DNA-activated cyclic GMP-AMP synthase. Cell 153: 1094–1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goutagny N, Jiang Z, Tian J, Parroche P, Schickli J, Monks BG, Ulbrandt N, Ji H, Kiener PA, Coyle AJ, et al. 2010. Cell type-specific recognition of human metapneumoviruses (HMPVs) by retinoic acid-inducible gene I (RIG-I) and TLR7 and viral interference of RIG-I ligand recognition by HMPV-B1 phosphoprotein. J Immunol 184: 1168–1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan Y, Ranoa DR, Jiang S, Mutha SK, Li X, Baudry J, Tapping RI 2010. Human TLRs 10 and 1 share common mechanisms of innate immune sensing but not signaling. J Immunol 184: 5094–5103. [DOI] [PubMed] [Google Scholar]
- Gurtler C, Bowie AG 2013. Innate immune detection of microbial nucleic acids. Trends Microbiol 21: 413–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas T, Metzger J, Schmitz F, Heit A, Muller T, Latz E, Wagner H 2008. The DNA sugar backbone 2′ deoxyribose determines toll-like receptor 9 activation. Immunity 28: 315–323. [DOI] [PubMed] [Google Scholar]
- Hacker H, Redecke V, Blagoev B, Kratchmarova I, Hsu LC, Wang GG, Kamps MP, Raz E, Wagner H, Hacker G, et al. 2006. Specificity in Toll-like receptor signalling through distinct effector functions of TRAF3 and TRAF6. Nature 439: 204–207. [DOI] [PubMed] [Google Scholar]
- Hagmann CA, Herzner AM, Abdullah Z, Zillinger T, Jakobs C, Schuberth C, Coch C, Higgins PG, Wisplinghoff H, Barchet W, et al. 2013. RIG-I detects triphosphorylated RNA of Listeria monocytogenes during infection in non-immune cells. PLoS ONE 8: e62872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hidmark A, von Saint Paul A, Dalpke AH 2012. Cutting edge: TLR13 is a receptor for bacterial RNA. J Immunol 189: 2717–2721. [DOI] [PubMed] [Google Scholar]
- Hoebe K, Georgel P, Rutschmann S, Du X, Mudd S, Crozat K, Sovath S, Shamel L, Hartung T, Zahringer U, et al. 2005. CD36 is a sensor of diacylglycerides. Nature 433: 523–527. [DOI] [PubMed] [Google Scholar]
- Hornung V, Ellegast J, Kim S, Brzozka K, Jung A, Kato H, Poeck H, Akira S, Conzelmann KK, Schlee M, et al. 2006. 5′-Triphosphate RNA is the ligand for RIG-I. Science 314: 994–997. [DOI] [PubMed] [Google Scholar]
- Hou F, Sun L, Zheng H, Skaug B, Jiang QX, Chen ZJ 2011. MAVS forms functional prion-like aggregates to activate and propagate antiviral innate immune response. Cell 146: 448–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishii KJ, Coban C, Kato H, Takahashi K, Torii Y, Takeshita F, Ludwig H, Sutter G, Suzuki K, Hemmi H, et al. 2006. A Toll-like receptor-independent antiviral response induced by double-stranded B-form DNA. Nat Immunol 7: 40–48. [DOI] [PubMed] [Google Scholar]
- Ishii KJ, Kawagoe T, Koyama S, Matsui K, Kumar H, Kawai T, Uematsu S, Takeuchi O, Takeshita F, Coban C, et al. 2008. TANK-binding kinase-1 delineates innate and adaptive immune responses to DNA vaccines. Nature 451: 725–729. [DOI] [PubMed] [Google Scholar]
- Ishikawa H, Barber GN 2008. STING is an endoplasmic reticulum adaptor that facilitates innate immune signalling. Nature 455: 674–678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa H, Ma Z, Barber GN 2009. STING regulates intracellular DNA-mediated, type I interferon-dependent innate immunity. Nature 461: 788–792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janeway CA Jr, Medzhitov R 2002. Innate immune recognition. Ann Rev Immunol 20: 197–216. [DOI] [PubMed] [Google Scholar]
- Jimenez-Dalmaroni MJ, Xiao N, Corper AL, Verdino P, Ainge GD, Larsen DS, Painter GF, Rudd PM, Dwek RA, Hoebe K, et al. 2009. Soluble CD36 ectodomain binds negatively charged diacylglycerol ligands and acts as a co-receptor for TLR2. PLoS ONE 4: e7411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin MS, Kim SE, Heo JY, Lee ME, Kim HM, Paik SG, Lee H, Lee JO 2007. Crystal structure of the TLR1-TLR2 heterodimer induced by binding of a tri-acylated lipopeptide. Cell 130: 1071–1082. [DOI] [PubMed] [Google Scholar]
- Kagan JC, Su T, Horng T, Chow A, Akira S, Medzhitov R 2008. TRAM couples endocytosis of Toll-like receptor 4 to the induction of interferon-β. Nat Immunol 9: 361–368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanneganti TD, Body-Malapel M, Amer A, Park JH, Whitfield J, Franchi L, Taraporewala ZF, Miller D, Patton JT, Inohara N, et al. 2006. Critical role for Cryopyrin/Nalp3 in activation of caspase-1 in response to viral infection and double-stranded RNA. J Biol Chem 281: 36560–36568. [DOI] [PubMed] [Google Scholar]
- Kato H, Takeuchi O, Sato S, Yoneyama M, Yamamoto M, Matsui K, Uematsu S, Jung A, Kawai T, Ishii KJ, et al. 2006. Differential roles of MDA5 and RIG-I helicases in the recognition of RNA viruses. Nature 441: 101–105. [DOI] [PubMed] [Google Scholar]
- Kawai T, Akira S 2006. Innate immune recognition of viral infection. Nat Immunol 7: 131–137. [DOI] [PubMed] [Google Scholar]
- Kawai T, Akira S 2010. The role of pattern-recognition receptors in innate immunity: Update on Toll-like receptors. Nat Immunol 11: 373–384. [DOI] [PubMed] [Google Scholar]
- Kerur N, Veettil MV, Sharma-Walia N, Bottero V, Sadagopan S, Otageri P, Chandran B 2011. IFI16 acts as a nuclear pathogen sensor to induce the inflammasome in response to Kaposi Sarcoma-associated herpesvirus infection. Cell Host Microbe 9: 363–375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim YM, Brinkmann MM, Paquet ME, Ploegh HL 2008. UNC93B1 delivers nucleotide-sensing Toll-like receptors to endolysosomes. Nature 452: 234–238. [DOI] [PubMed] [Google Scholar]
- Kim T, Pazhoor S, Bao M, Zhang Z, Hanabuchi S, Facchinetti V, Bover L, Plumas J, Chaperot L, Qin J, et al. 2010. Aspartate-glutamate-alanine-histidine box motif (DEAH)/ RNA helicase A helicases sense microbial DNA in human plasmacytoid dendritic cells. Proc Natl Acad Sci 107: 15181–15186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobiyama K, Takeshita F, Jounai N, Sakaue-Sawano A, Miyawaki A, Ishii KJ, Kawai T, Sasaki S, Hirano H, Ishii N, et al. 2010. Extrachromosomal histone H2B mediates innate antiviral immune responses induced by intracellular double-stranded DNA. J Virol 84: 822–832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koblansky AA, Jankovic D, Oh H, Hieny S, Sungnak W, Mathur R, Hayden MS, Akira S, Sher A, Ghosh S 2013. Recognition of profilin by Toll-like receptor 12 is critical for host resistance to Toxoplasma gondii. Immunity 38: 119–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondo T, Kawai T, Akira S 2012. Dissecting negative regulation of Toll-like receptor signaling. Trends Immunol 33: 449–458. [DOI] [PubMed] [Google Scholar]
- Kondo T, Kobayashi J, Saitoh T, Maruyama K, Ishii KJ, Barber GN, Komatsu K, Akira S, Kawai T 2013. DNA damage sensor MRE11 recognizes cytosolic double-stranded DNA and induces type I interferon by regulating STING trafficking. Proc Natl Acad Sci 110: 2969–2974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konno H, Konno K, Barber GN 2013. Cyclic dinucleotides trigger ULK1 (ATG1) phosphorylation of STING to prevent sustained innate immune signaling. Cell 155: 688–698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurt-Jones EA, Popova L, Kwinn L, Haynes LM, Jones LP, Tripp RA, Walsh EE, Freeman MW, Golenbock DT, Anderson LJ, et al. 2000. Pattern recognition receptors TLR4 and CD14 mediate response to respiratory syncytial virus. Nat Immunol 1: 398–401. [DOI] [PubMed] [Google Scholar]
- Lee HK, Lund JM, Ramanathan B, Mizushima N, Iwasaki A 2007. Autophagy-dependent viral recognition by plasmacytoid dendritic cells. Science 315: 1398–1401. [DOI] [PubMed] [Google Scholar]
- Lee KG, Xu S, Kang ZH, Huo J, Huang M, Liu D, Takeuchi O, Akira S, Lam KP 2012. Bruton’s tyrosine kinase phosphorylates Toll-like receptor 3 to initiate antiviral response. Proc Natl Acad Sci 109: 5791–5796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li XD, Chen ZJ 2012. Sequence specific detection of bacterial 23S ribosomal RNA by TLR13. eLife 1: e00102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li XD, Wu J, Gao D, Wang H, Sun L, Chen ZJ 2013. Pivotal roles of cGAS-cGAMP signaling in antiviral defense and immune adjuvant effects. Science 341: 1390–1394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin SC, Lo YC, Wu H 2010. Helical assembly in the MyD88-IRAK4-IRAK2 complex in TLR/IL-1R signalling. Nature 465: 885–890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L, Botos I, Wang Y, Leonard JN, Shiloach J, Segal DM, Davies DR 2008. Structural basis of toll-like receptor 3 signaling with double-stranded RNA. Science 320: 379–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Zhan Z, Li D, Xu L, Ma F, Zhang P, Yao H, Cao X 2011. Intracellular MHC class II molecules promote TLR-triggered innate immune responses by maintaining activation of the kinase Btk. Nat Immunol 12: 416–424. [DOI] [PubMed] [Google Scholar]
- Liu X, Chen W, Wang Q, Li L, Wang C 2013. Negative regulation of TLR inflammatory signaling by the SUMO-deconjugating enzyme SENP6. PLoS Pathog 9: e1003480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loo YM, Gale M Jr 2011. Immune signaling by RIG-I-like receptors. Immunity 34: 680–692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loo YM, Fornek J, Crochet N, Bajwa G, Perwitasari O, Martinez-Sobrido L, Akira S, Gill MA, Garcia-Sastre A, Katze MG, et al. 2008. Distinct RIG-I and MDA5 signaling by RNA viruses in innate immunity. J Virol 82: 335–345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mancuso G, Gambuzza M, Midiri A, Biondo C, Papasergi S, Akira S, Teti G, Beninati C 2009. Bacterial recognition by TLR7 in the lysosomes of conventional dendritic cells. Nat Immunol 10: 587–594. [DOI] [PubMed] [Google Scholar]
- Mathur R, Oh H, Zhang D, Park SG, Seo J, Koblansky A, Hayden MS, Ghosh S 2012. A mouse model of Salmonella typhi infection. Cell 151: 590–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto F, Saitoh S, Fukui R, Kobayashi T, Tanimura N, Konno K, Kusumoto Y, Akashi-Takamura S, Miyake K 2008. Cathepsins are required for Toll-like receptor 9 responses. Biochem Biophys Res Commun 367: 693–699. [DOI] [PubMed] [Google Scholar]
- McCartney SA, Thackray LB, Gitlin L, Gilfillan S, Virgin HW, Colonna M 2008. MDA-5 recognition of a murine norovirus. PLoS Pathog 4: e1000108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melchjorsen J, Jensen SB, Malmgaard L, Rasmussen SB, Weber F, Bowie AG, Matikainen S, Paludan SR 2005. Activation of innate defense against a paramyxovirus is mediated by RIG-I and TLR7 and TLR8 in a cell-type-specific manner. J Virol 79: 12944–12951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitoma H, Hanabuchi S, Kim T, Bao M, Zhang Z, Sugimoto N, Liu YJ 2013. The DHX33 RNA helicase senses cytosolic RNA and activates the NLRP3 inflammasome. Immunity 39: 123–135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oldenburg M, Kruger A, Ferstl R, Kaufmann A, Nees G, Sigmund A, Bathke B, Lauterbach H, Suter M, Dreher S, et al. 2012. TLR13 recognizes bacterial 23S rRNA devoid of erythromycin resistance-forming modification. Science 337: 1111–1115. [DOI] [PubMed] [Google Scholar]
- Oliveira-Nascimento L, Massari P, Wetzler LM 2012. The role of TLR2 in infection and immunity. Frontiers Immunol 3: 79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olivieri F, Rippo MR, Prattichizzo F, Babini L, Graciotti L, Recchioni R, Procopio AD 2013. Toll like receptor signaling in “inflammaging”: MicroRNA as new players. Immun Ageing 10: 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onji M, Kanno A, Saitoh S, Fukui R, Motoi Y, Shibata T, Matsumoto F, Lamichhane A, Sato S, Kiyono H, et al. 2013. An essential role for the N-terminal fragment of Toll-like receptor 9 in DNA sensing. Nat Commun 4: 1949. [DOI] [PubMed] [Google Scholar]
- Paludan SR, Bowie AG 2013. Immune sensing of DNA. Immunity 38: 870–880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park B, Brinkmann MM, Spooner E, Lee CC, Kim YM, Ploegh HL 2008. Proteolytic cleavage in an endolysosomal compartment is required for activation of Toll-like receptor 9. Nat Immunol 9: 1407–1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park BS, Song DH, Kim HM, Choi BS, Lee H, Lee JO 2009. The structural basis of lipopolysaccharide recognition by the TLR4-MD-2 complex. Nature 458: 1191–1195. [DOI] [PubMed] [Google Scholar]
- Peter ME, Kubarenko AV, Weber AN, Dalpke AH 2009. Identification of an N-terminal recognition site in TLR9 that contributes to CpG-DNA-mediated receptor activation. J Immunol 182: 7690–7697. [DOI] [PubMed] [Google Scholar]
- Phipps S, Lam CE, Mahalingam S, Newhouse M, Ramirez R, Rosenberg HF, Foster PS, Matthaei KI 2007. Eosinophils contribute to innate antiviral immunity and promote clearance of respiratory syncytial virus. Blood 110: 1578–1586. [DOI] [PubMed] [Google Scholar]
- Pichlmair A, Schulz O, Tan CP, Naslund TI, Liljestrom P, Weber F, Reis e Sousa C 2006. RIG-I-mediated antiviral responses to single-stranded RNA bearing 5′-phosphates. Science 314: 997–1001. [DOI] [PubMed] [Google Scholar]
- Pichlmair A, Schulz O, Tan CP, Rehwinkel J, Kato H, Takeuchi O, Akira S, Way M, Schiavo G, Reis e Sousa C 2009. Activation of MDA5 requires higher-order RNA structures generated during virus infection. J Virol 83: 10761–10769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian C, Cao X 2013. Regulation of Toll-like receptor signaling pathways in innate immune responses. Ann NY Acad Sci 1283: 67–74. [DOI] [PubMed] [Google Scholar]
- Rajan JV, Warren SE, Miao EA, Aderem A 2010. Activation of the NLRP3 inflammasome by intracellular poly I:C. FEBS Lett 584: 4627–4632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito T, Hirai R, Loo YM, Owen D, Johnson CL, Sinha SC, Akira S, Fujita T, Gale M Jr 2007. Regulation of innate antiviral defenses through a shared repressor domain in RIG-I and LGP2. Proc Natl Acad Sci 104: 582–587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitoh T, Fujita N, Yoshimori T, Akira S 2010. Regulation of dsDNA-induced innate immune responses by membrane trafficking. Autophagy 6: 430–432. [DOI] [PubMed] [Google Scholar]
- Satoh T, Kato H, Kumagai Y, Yoneyama M, Sato S, Matsushita K, Tsujimura T, Fujita T, Akira S, Takeuchi O 2010. LGP2 is a positive regulator of RIG-I- and MDA5-mediated antiviral responses. Proc Natl Acad Sci 107: 1512–1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitoh T, Satoh T, Yamamoto N, Uematsu S, Takeuchi O, Kawai T, Akira S 2011. Antiviral protein viperin promotes Toll-like receptor 7- and Toll-like receptor 9-mediated type I interferon production in plasmacytoid dendritic cells. Immunity 34: 352–363. [DOI] [PubMed] [Google Scholar]
- Schamber-Reis BL, Petritus PM, Caetano BC, Martinez ER, Okuda K, Golenbock D, Scott P, Gazzinelli RT 2013. UNC93B1 and nucleic acid-sensing Toll-like receptors mediate host resistance to infection with Leishmania major. J Biol Chem 288: 7127–7136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwandner R, Dziarski R, Wesche H, Rothe M, Kirschning CJ 1999. Peptidoglycan- and lipoteichoic acid-induced cell activation is mediated by toll-like receptor 2. J Biol Chem 274: 17406–17409. [DOI] [PubMed] [Google Scholar]
- Scott I 2010. The role of mitochondria in the mammalian antiviral defense system. Mitochondrion 10: 316–320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sepulveda FE, Maschalidi S, Colisson R, Heslop L, Ghirelli C, Sakka E, Lennon-Dumenil AM, Amigorena S, Cabanie L, Manoury B 2009. Critical role for asparagine endopeptidase in endocytic Toll-like receptor signaling in dendritic cells. Immunity 31: 737–748. [DOI] [PubMed] [Google Scholar]
- Shi HX, Liu X, Wang Q, Tang PP, Liu XY, Shan YF, Wang C 2011. Mitochondrial ubiquitin ligase MARCH5 promotes TLR7 signaling by attenuating TANK action. PLoS Pathog 7: e1002057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart CR, Stuart LM, Wilkinson K, van Gils JM, Deng J, Halle A, Rayner KJ, Boyer L, Zhong R, Frazier WA, et al. 2010. CD36 ligands promote sterile inflammation through assembly of a Toll-like receptor 4 and 6 heterodimer. Nat Immunol 11: 155–161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun L, Wu J, Du F, Chen X, Chen ZJ 2013. Cyclic GMP-AMP synthase is a cytosolic DNA sensor that activates the type I interferon pathway. Science 339: 786–791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabeta K, Hoebe K, Janssen EM, Du X, Georgel P, Crozat K, Mudd S, Mann N, Sovath S, Goode J, et al. 2006. The Unc93b1 mutation 3d disrupts exogenous antigen presentation and signaling via Toll-like receptors 3, 7 and 9. Nat Immunol 7: 156–164. [DOI] [PubMed] [Google Scholar]
- Takaoka A, Wang Z, Choi MK, Yanai H, Negishi H, Ban T, Lu Y, Miyagishi M, Kodama T, Honda K, et al. 2007. DAI (DLM-1/ZBP1) is a cytosolic DNA sensor and an activator of innate immune response. Nature 448: 501–505. [DOI] [PubMed] [Google Scholar]
- Takeuchi O, Akira S 2010. Pattern recognition receptors and inflammation. Cell 140: 805–820. [DOI] [PubMed] [Google Scholar]
- Tanaka Y, Chen ZJ 2012. STING specifies IRF3 phosphorylation by TBK1 in the cytosolic DNA signaling pathway. Sci Signal 5: ra20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang D, Kang R, Coyne CB, Zeh HJ, Lotze MT 2012. PAMPs and DAMPs: Signal 0s that spur autophagy and immunity. Immunol Rev 249: 158–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanji H, Ohto U, Shibata T, Miyake K, Shimizu T 2013. Structural reorganization of the Toll-like receptor 8 dimer induced by agonistic ligands. Science 339: 1426–1429. [DOI] [PubMed] [Google Scholar]
- Thompson MR, Kaminski JJ, Kurt-Jones EA, Fitzgerald KA 2011. Pattern recognition receptors and the innate immune response to viral infection. Viruses 3: 920–940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuchida T, Zou J, Saitoh T, Kumar H, Abe T, Matsuura Y, Kawai T, Akira S 2010. The ubiquitin ligase TRIM56 regulates innate immune responses to intracellular double-stranded DNA. Immunity 33: 765–776. [DOI] [PubMed] [Google Scholar]
- Uematsu S, Fujimoto K, Jang MH, Yang BG, Jung YJ, Nishiyama M, Sato S, Tsujimura T, Yamamoto M, Yokota Y, et al. 2008. Regulation of humoral and cellular gut immunity by lamina propria dendritic cells expressing Toll-like receptor 5. Nat Immunol 9: 769–776. [DOI] [PubMed] [Google Scholar]
- Underhill DM, Ozinsky A, Smith KD, Aderem A 1999. Toll-like receptor-2 mediates mycobacteria-induced proinflammatory signaling in macrophages. Proc Natl Acad Sci 96: 14459–14463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unterholzner L, Keating SE, Baran M, Horan KA, Jensen SB, Sharma S, Sirois CM, Jin T, Latz E, Xiao TS, et al. 2010. IFI16 is an innate immune sensor for intracellular DNA. Nat Immunol 11: 997–1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner H 2008. The sweetness of the DNA backbone drives Toll-like receptor 9. Curr Opin Immunol 20: 396–400. [DOI] [PubMed] [Google Scholar]
- Wang JP, Asher DR, Chan M, Kurt-Jones EA, Finberg RW 2007. Cutting Edge: Antibody-mediated TLR7-dependent recognition of viral RNA. J Immunol 178: 3363–3367. [DOI] [PubMed] [Google Scholar]
- Wang C, Chen T, Zhang J, Yang M, Li N, Xu X, Cao X 2009. The E3 ubiquitin ligase Nrdp1 “preferentially” promotes TLR-mediated production of type I interferon. Nat Immunol 10: 744–752. [DOI] [PubMed] [Google Scholar]
- Wang L, Zhao W, Zhang M, Wang P, Zhao K, Zhao X, Yang S, Gao C 2013. USP4 positively regulates RIG-I-mediated antiviral response through deubiquitination and stabilization of RIG-I. J Virol 87: 4507–4515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J, Sun L, Chen X, Du F, Shi H, Chen C, Chen ZJ 2013. Cyclic GMP-AMP is an endogenous second messenger in innate immune signaling by cytosolic DNA. Science 339: 826–830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao TS, Fitzgerald KA 2013. The cGAS-STING pathway for DNA sensing. Mol Cell 51: 135–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashita M, Chattopadhyay S, Fensterl V, Saikia P, Wetzel JL, Sen GC 2012. Epidermal growth factor receptor is essential for Toll-like receptor 3 signaling. Sci Signal 5: ra50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang P, An H, Liu X, Wen M, Zheng Y, Rui Y, Cao X 2010. The cytosolic nucleic acid sensor LRRFIP1 mediates the production of type I interferon via a β-catenin-dependent pathway. Nat Immunol 11: 487–494. [DOI] [PubMed] [Google Scholar]
- Yang M, Wang C, Zhu X, Tang S, Shi L, Cao X, Chen T 2011. E3 ubiquitin ligase CHIP facilitates Toll-like receptor signaling by recruiting and polyubiquitinating Src and atypical PKCζ. J Exp Med 208: 2099–2112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yarovinsky F, Zhang D, Andersen JF, Bannenberg GL, Serhan CN, Hayden MS, Hieny S, Sutterwala FS, Flavell RA, Ghosh S, et al. 2005. TLR11 activation of dendritic cells by a protozoan profilin-like protein. Science 308: 1626–1629. [DOI] [PubMed] [Google Scholar]
- Yoneyama M, Fujita T 2008. Structural mechanism of RNA recognition by the RIG-I-like receptors. Immunity 29: 178–181. [DOI] [PubMed] [Google Scholar]
- Yoneyama M, Kikuchi M, Matsumoto K, Imaizumi T, Miyagishi M, Taira K, Foy E, Loo YM, Gale M Jr, Akira S, et al. 2005. Shared and unique functions of the DExD/H-box helicases RIG-I, MDA5, and LGP2 in antiviral innate immunity. J Immunol 175: 2851–2858. [DOI] [PubMed] [Google Scholar]
- Zanoni I, Ostuni R, Marek LR, Barresi S, Barbalat R, Barton GM, Granucci F, Kagan JC 2011. CD14 controls the LPS-induced endocytosis of Toll-like receptor 4. Cell 147: 868–880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D, Zhang G, Hayden MS, Greenblatt MB, Bussey C, Flavell RA, Ghosh S 2004. A toll-like receptor that prevents infection by uropathogenic bacteria. Science 303: 1522–1526. [DOI] [PubMed] [Google Scholar]
- Zhang SY, Jouanguy E, Ugolini S, Smahi A, Elain G, Romero P, Segal D, Sancho-Shimizu V, Lorenzo L, Puel A, et al. 2007. TLR3 deficiency in patients with herpes simplex encephalitis. Science 317: 1522–1527. [DOI] [PubMed] [Google Scholar]
- Zhang X, Brann TW, Zhou M, Yang J, Oguariri RM, Lidie KB, Imamichi H, Huang DW, Lempicki RA, Baseler MW, et al. 2011. Cutting edge: Ku70 is a novel cytosolic DNA sensor that induces type III rather than type I IFN. J Immunol 186: 4541–4545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Shi H, Wu J, Sun L, Chen C, Chen ZJ 2013. Cyclic GMP-AMP containing mixed phosphodiester linkages is an endogenous high-affinity ligand for STING. Mol Cell 51: 226–235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong B, Zhang L, Lei C, Li Y, Mao AP, Yang Y, Wang YY, Zhang XL, Shu HB 2009. The ubiquitin ligase RNF5 regulates antiviral responses by mediating degradation of the adaptor protein MITA. Immunity 30: 397–407. [DOI] [PubMed] [Google Scholar]