Abstract
A fundamental challenge for the survival of all organisms is maintaining the integrity of the genome in all cells. Cells must therefore segregate their replicated genome equally during each cell division. Eukaryotic organisms package their genome into a number of physically distinct chromosomes, which replicate during S phase and condense during prophase of mitosis to form paired sister chromatids. During mitosis, cells form a physical connection between each sister chromatid and microtubules of the mitotic spindle, which segregate one copy of each chromatid to each new daughter cell. The centromere is the DNA locus on each chromosome that creates the site of this connection. In this review, we present a brief history of centromere research and discuss our current knowledge of centromere establishment, maintenance, composition, structure, and function in mitosis.
The centromere is the assembly site for the mitotic kinetochore. It is epigenetically defined by the presence of the histone H3 variant CENP-A.
Centromeres were first described by Walther Flemming (1882) in his pioneering characterization of cell division. Flemming noticed that the distinct threads of chromatin (later called chromosomes) each had one (and only one) site of primary constriction, where the total width of the chromosome appeared smaller than the rest of that chromosome. We now recognize that Flemming was reporting the distinct heterochromatin found specifically at centromeres. The term “chromatin” was originally proposed by Flemming as the chromosomes he was observing stain darkly when cells are treated with aniline dyes. Since then, the definition of chromatin has evolved, and now describes the histone proteins and associated proteins that package DNA.
Although more than 130 years later we lack a detailed understanding of numerous aspects of centromere biology, one universally accepted fact is that centromeres have an essential role in the segregation of chromosomes during mitosis. The centromere is a large chromatin-containing protein complex that forms the assembly site for the mitotic kinetochore, itself a megadalton protein complex that binds spindle microtubules to segregate the chromatids during anaphase (Fig. 1). In addition to microtubule attachment, kinetochores are also the site of mitotic checkpoint activation, which prevents anaphase onset in the presence of unattached kinetochores. Without the centromere, no kinetochore would form and cells could not segregate their chromosomes. Thus, the centromere is of crucial importance for chromosome segregation and mitotic control.
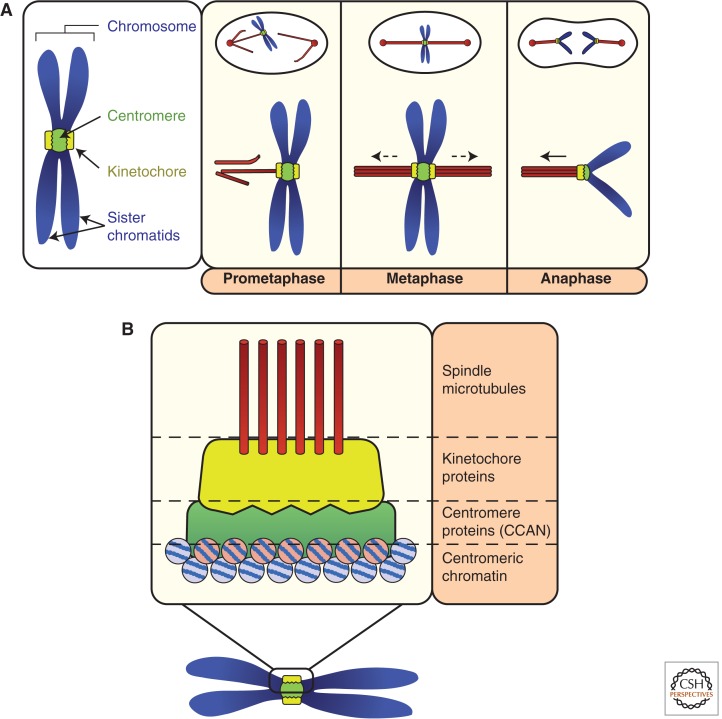
Figure 1.
Introduction to centromere function and organization during mitosis. (A) Before and during the early stages of mitosis, centromeres (green circle) recruit kinetochore proteins (yellow discs). During prometaphase, the kinetochore forms the attachment sites for spindle microtubules (red rods). Once both kinetochores of all sister chromatid pairs are stably and correctly attached to microtubules, pulling forces exerted by microtubules (dashed arrows) cause migration of linked sister chromatids to the metaphase plate. At anaphase, sister chromatid cohesion is dissolved and the centromere and kinetochore harness microtubule-dependent forces that pull each sister chromatid to opposite ends of the dividing cell. (B) Basic architecture of the centromere in mitosis. Centromeric chromatin consists of specialized nucleosomes containing the histone H3 variant centromere protein (CENP)-A. CENP-A recruits a network of centromere proteins (green) that are collectively known as the constitutive centromere associated network (CCAN). Kinetochore proteins (yellow), specifically recruited by the CCAN for mitosis, attach to spindle microtubules.
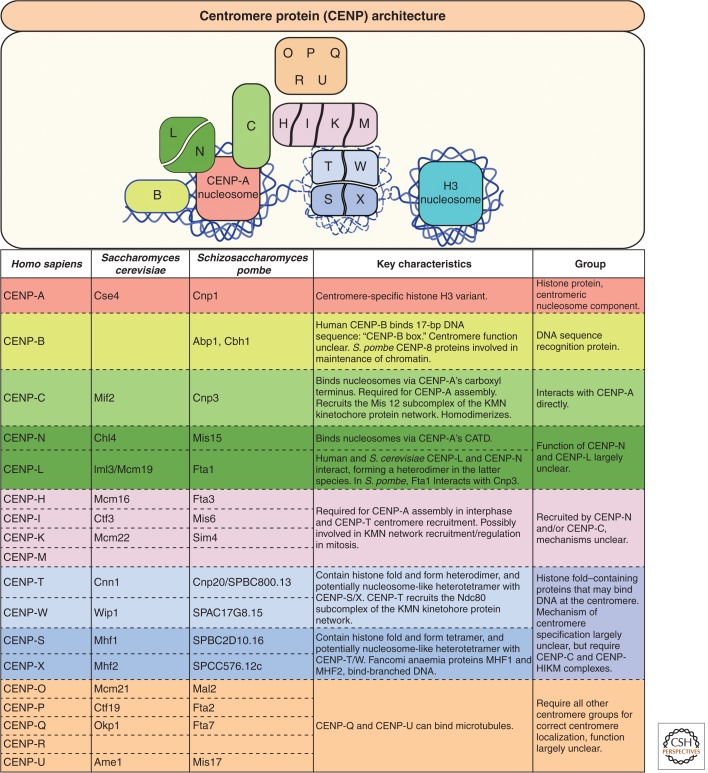
The mitotic centromere:kinetochore can be thought of as a single multiprotein complex. However, kinetochore proteins localize to centromeres before and during mitosis to enable chromosome segregation, whereas centromere proteins (CENPs) persist throughout the cell cycle. CENPs (listed in Fig. 3) are collectively called the constitutive centromere-associated network (CCAN) (Cheeseman and Desai 2008). This nomenclature system has provided some clarity in the field, but it should be noted that many CCAN proteins dynamically associate and dissociate from centromeres, and some function in microtubule regulation during mitosis. Eukaryotic centromeres vary widely in their complexity and structure, ranging from point centromeres of budding yeast that generate a single microtubule-binding site, to holocentric centromeres of nematodes that decorate the entire chromosome, to regional centromeres of vertebrates that provide a distinct attachment site for multiple microtubules. Despite this variation, the core function of the centromere, to form the kinetochore to bind microtubules so that chromosomes can be equally segregated in mitosis, is conserved. Thus, understanding the molecular underpinnings of the centromere is fundamental to establishing how cells faithfully maintain their genomes.
Figure 3.
Overview of centromere proteins and centromere architecture. Centromere proteins are grouped based on individual complexes, often based on the phenotypic consequence of protein depletion in cells and, more recently, biochemical characterization. CENP-A nucleosomes directly recruit CENP-C and possibly the CENP-N/L heterodimer. CENP-C recruits the CENP-H/I/K/M complex that in turn is required for CENP-T centromere localization. How this occurs remains unclear as specific CENP-C:CENP-H/I/K/M and CENP-H/I/K/M:CENP-T/W/S/X interactions have not been identified. The possibility that CENP-T/W/S/X proteins wrap centromeric DNA in nucleosome-like structures, and whether they associate with H3 nucleosomes at centromeres, is currently a topic of intense research. The functions and subcomplexes that comprise the remaining CENP proteins O/P/Q/R and U remain unclear. Note that both Saccharomyces cerevisiae and Saccharomyces pombe have centromere proteins not listed here (as they lack a known human homolog).
CENP-A: THE CENTROMERE-SPECIFIC HISTONE
Eukaryotic DNA is assembled around histone proteins into protein:DNA complexes known as nucleosomes. Nucleosomes throughout most of a chromosome contain two copies of each histone protein H2A, H2B, H3, and H4, together forming an octameric complex with DNA (Fig. 2). Centromeres are unique from the rest of the chromosome in that they feature nucleosomes containing a histone H3 variant, called CENP-A, in place of histone H3 (Fig. 2). CENP-A was initially discovered as a human autoantigen in CREST syndrome patients, and was subsequently shown to copurify with histones, be present in purified nucleosomes, and be essential across eukaryotic model organisms, as mutation or disruption of CENP-A causes a complete failure in centromere and kinetochore formation (Earnshaw and Rothfield 1985; Earnshaw et al. 1986; Palmer et al. 1987, 1991; Sullivan et al. 1994). In humans, functional centromeres always contain CENP-A, including “neocentromeres” that form on noncentromeric chromosomal loci (Saffery et al. 2000; Lo et al. 2001; Warburton, 2004). Experimental overexpression of Drosophila CENP-ACID causes the ectopic localization of CENP-ACID to noncentromere regions, which in turn causes ectopic kinetochore formation and numerous mitotic errors (Heun et al. 2006). When CENP-ACID is targeted to a noncentromeric region by expressing CENP-ACID-LacI fusion protein in cells containing chromosomally integrated Lac operator arrays, the localized CENP-ACID causes ectopic kinetochore formation in Drosophila (Mendiburo et al. 2011), and partial kinetochore formation in humans (Gascoigne et al. 2011). Thus, the presence of CENP-A in chromatin is the defining mark of centromeres.
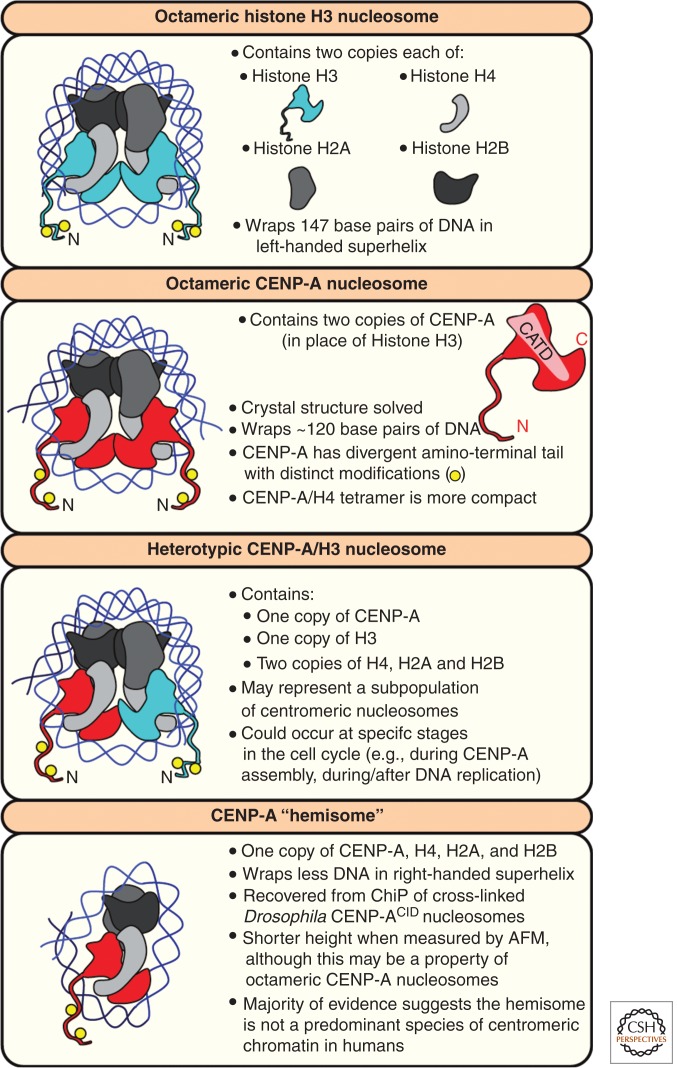
Figure 2.
CENP-A nucleosome structure and possible variants. A cartoon of a conventional octameric H3 nucleosome is shown, together with the most prominent models of CENP-A nucleosome composition, homotypic CENP-A octamers, heterotypic CENP-A/H3 octamers, and tetrameric CENP-A hemisomes. See main text for specific details.
The Epigenetic Nature of Centromeres
Understanding how CENP-A is assembled and maintained only at centromeres is central to understanding centromere and kinetochore function. A simple model for how CENP-A is targeted to the centromere is that specific centromeric DNA sequences dictate CENP-A nucleosome position. This model appears true in the “point” centromeres of some budding yeasts, including Saccharomyces cerevisiae, which has a defined 125-bp centromeric DNA sequence consisting of three centromere-DNA elements (CDEs) that are necessary and sufficient for centromere specification and kinetochore assembly (Clarke and Carbon 1980; Fitzgerald-Hayes et al. 1982; Cottarel et al. 1989). A single CENP-ACse4 nucleosome is positioned at CDEII in a centromeric DNA sequence-dependent manner (Spencer and Hieter 1992; Stoler et al. 1995; Meluh et al. 1998; Furuyama and Biggins 2007), although recent evidence suggests multiple accessory CENP-ACse4 nucleosomes, proposed to not contribute to kinetochore formation, may exist at centromeres (Coffman et al. 2011; Lawrimore et al. 2011; Haase et al. 2013). In contrast to budding yeast, fission yeast and higher eukaryotes, including humans, have “regional” centromeres that span kilo- to megabases of highly repetitive DNA. Importantly, in many eukaryotes, including humans, the underlying DNA sequence appears dispensable for centromere specification and function. Instead, CENP-A nucleosomes appear to epigenetically define the centromere.
The evidence for epigenetic specification of higher eukaryotic centromeres is compelling. Sequence-driven centromere specification would presumably impart significant selective pressure toward sequence conservation. However, higher eukaryotic centromeric DNA is the most rapidly evolving region of the genome (Csink and Henikoff 1998; Schueler et al. 2001; Padmanabhan et al. 2008), and centromeres remain the most poorly characterized loci in the human genome. The most prominent evidence for epigenetic centromere specification is the discovery of naturally occurring neocentromeres that form on a DNA locus unrelated to the endogenous chromosomal centromere. In some cases neocentromeres maintain the equal segregation of distinct chromosomal fragments where the original centromere continues to function on the original chromosome (Warburton 2004). In other cases, in which a stable neocentromere forms on a chromosome that also has its original centromere, the original centromere can be silenced and the neocentromere replaces the function of the original centromere. In either case, neocentromeres facilitate kinetochore formation, chromosome segregation, become stably inherited over multiple generations, and support development, all despite the lack of the underlying centromere-DNA sequence (Warburton et al. 1997; Saffery et al. 2000). Importantly, CENP-A is present at the site of all functional neocentromeres, but is lost from the original centromeric locus concurrent with endogenous centromere silencing (Warburton et al. 1997; Lo et al. 2001). In Schizosaccharomyces pombe, deletion of a centromere is rescued by spontaneous neocentromere formation typically close to telomeric regions, and is dependent on formation of heterochromatin (Ishii et al. 2008; Kagansky et al. 2009). Neocentromere formation has since been engineered in chicken DT40 cells, by selection of surviving clones after deletion of the endogenous Z chromosome centromere, confirming that neocentromeres maintain many features of the original centromere, without the underlying DNA (Shang et al. 2013). Taken together, these data show that the centromeres of many higher eukaryotes can be epigenetically maintained.
The Structure of CENP-A-Containing Nucleosomes
A current topic of intense study and debate is the structure and composition of CENP-A nucleosomes. Numerous CENP-A-containing nucleosome structures have been proposed, but two general models remain the most discussed (Fig. 2). One model suggests that CENP-A nucleosomes are octameric, wrap ∼120 bp of DNA, and resemble conventional H3 nucleosomes, whereas another model suggests that CENP-A wraps DNA as a hemisome, a nucleosome that contains one copy of CENP-A, H4, H2A, and H2B. Evidence supporting the presence of hemisomes comes from isolation of CENP-ACID/H4/H2A/H2B tetramers from cross-linked chromatin immunoprecipitations (ChIPs), a smaller nucleosome height than octameric H3 nucleosomes when measured by atomic force microscopy (AFM) (Dalal et al. 2007; Dimitriadis et al. 2010; Bui et al. 2012), positive supercoiling of DNA in yeast centromere plasmids (Furuyama and Henikoff 2009), and cleavage mapping of H4:DNA interactions within budding yeast nucleosomes (Henikoff et al. 2014). The reliability of AFM measurements is questionable, as reconstituted octameric CENP-A nucleosomes can appear shorter when measured by AFM (Miell et al. 2013), a result that itself has been challenged (Codomo et al. 2014; Miell et al. 2014; Walkiewicz et al. 2014). The notion that CENP-A can function as a hemisome will not be corroborated until a hemisome is successfully isolated and characterized biochemically.
In contrast to a CENP-A hemisome, the crystal structure of an octameric CENP-A nucleosome has been resolved, revealing only subtle differences to canonical octameric H3 nucleosomes (Tachiwana et al. 2011). Octameric CENP-A nucleosomes wrap slightly less DNA than H3 nucleosomes (∼120 bp vs. 147 bp), resulting in different DNA entry/exit angles from the nucleosome, and several exposed residues exist in CENP-A’s extended loop 1 region that are important for CENP-A nucleosome stability (Tachiwana et al. 2011). In support of the octameric CENP-A nucleosome model, ChIP-Seq data revealed that CENP-A mononucleosomes wrap more DNA in cells than hemisomes could wrap (Hasson et al. 2013), immunoprecipitation of Drosophila CENP-CCID mononucleosomes revealed the presence of CENP-ACID dimers (Zhang et al. 2012), and CAL1-assembled CENP-ACID nucleosomes in Drosophila are octameric (Chen et al. 2014). Finally, arrays of octameric CENP-A nucleosomes support functional centromere and kinetochore formation in vitro (Guse et al. 2011). Taken together, the most widely accepted and experimentally supported model is that CENP-A nucleosomes exist as octamers, which—other than the presence of CENP-A rather than histone H3—resemble canonical nucleosomes in their composition.
One emerging alternative model, which may reconcile the interpretation of octameric and hemisomal CENP-A nucleosomes, is that CENP-A nucleosome composition may change during progression through the cell cycle. Indeed, two recent studies in two different organisms proposed cell-cycle-dependent transitions between two CENP-A molecules per nucleosome (i.e., an octamer) to one CENP-A molecule per nucleosome (Bui et al. 2012; Shivaraju et al. 2012). These studies interpreted the changes as an octamer to hemisome transition. It is also possible a population of CENP-A nucleosomes exist as heterotypic octamers containing one molecule of CENP-A in addition to one molecule of histone H3 during specific stages of the cell cycle (Fig. 2) (Foltz et al. 2006). Overexpression of CENP-A has recently been shown to cause CENP-A/H3.3 heterotypic octameric nucleosomes assembly on chromosomal arms (Lacoste et al. 2014). However, as the predominant CENP-A nucleosome species at centromeres appears to be homotypic octamers, the functional relevance of heterotypic octamers remains unclear (Foltz et al. 2006; Kingston et al. 2011). Determining whether CENP-A nucleosomes change composition during the cell cycle and how those transitions occur is an important future goal.
DISTINGUISHING FEATURES OF CENTROMERIC CHROMATIN
How CENP-A Differs from Histone H3
The human CENP-A gene encodes a 140-residue protein (histone H3 is 136 amino acids) that is 48% identical to histone H3 overall and 62% identical across the carboxy-terminal 93 amino acids that contain the histone fold domain. Several key differences between CENP-A and H3 impart the centromere-specific functions of CENP-A. The amino-terminal 40 amino acids of CENP-A, the most divergent domain from histone H3, undergoes specific posttranslational modifications that are thought to influence CENP-A nucleosome structure and function, although their significance remains largely unclear (Zeitlin et al. 2001; Kunitoku et al. 2003; Bailey et al. 2013; Boeckmann et al. 2013). The most amino-terminal α helix in CENP-A (residues 48–56) is three residues shorter than that observed in histone H3 (residues 45–56), yet this small difference results in a loss of a DNA interaction that causes different entry/exit angles of DNA from the CENP-A nucleosome, protects less DNA in classic nucleosome micrococcal nuclease digestion assays, and most likely has a key influence of overall centromeric chromatin structure (Conde e Silva et al. 2007; Panchenko et al. 2011; Tachiwana et al. 2011). In the histone fold domain of CENP-A, a two-residue insertion (R80 and G81) in a loop domain is exposed in the CENP-A nucleosome and influences CENP-A nucleosome stability (Sekulic et al. 2010; Tachiwana et al. 2011). Importantly, a central region (residues 75–114 in humans) constitutes the CENP-A targeting domain (CATD) (Black et al. 2004) that is essential for numerous aspects of CENP-A function and structure, including binding of the CENP-A chaperone Holliday junction recognition protein (HJURP) and the centromere protein CENP-N (discussed below). Finally, CENP-A has an extended hydrophobic carboxy-terminal tail of six amino acids that binds the essential centromere protein CENP-C (Carroll et al. 2010; Guse et al. 2011; Fachinetti et al. 2013). The primary sequences of CENP-A homologs are quite divergent, suggesting the mode of CENP-A-mediated specification of centromere formation may be distinct between organisms. For example, a CENP-N homolog, which binds the CATD of CENP-A, has not been found in Drosophila or Caenorhabditis elegans.
Histone Modifications
To describe centromeric chromatin as CENP-A-containing nucleosomes is an oversimplification. Human centromeres range from 0.3 to 5 Mb in length of highly repetitive α satellite DNA. CENP-A-containing nucleosomes are thought to constitute only a subpopulation of centromeric chromatin. When centromeric chromatin is extended into long individual fibers CENP-A nucleosomes are found interspersed with H3 nucleosomes at centromeres (Zinkowski et al. 1991; Blower et al. 2002). These H3 nucleosomes are enriched in specific posttranslational modifications including histone H3 lysine 4, lysine 9, and lysine 36 dimethylation (Sullivan and Karpen 2004; Bergmann et al. 2011). In particular, H3K4me2 appears to be a centromere-specific modification that has been shown to promote CENP-A assembly into chromatin (Bergmann et al. 2011). In S. pombe, artificial tethering of the histone methyltransferase Clr4 to euchromatin promotes neocentromere formation (Ishii et al. 2008). Targeting of transcriptional activators and repressors to human artificial chromosome (HAC) centromeres influences kinetochore formation and CENP-A assembly, although the precise mechanisms remain largely unclear (Nakano et al. 2008; Cardinale et al. 2009; Ohzeki et al. 2012). In addition, tethering either the histone acetyltransferase domains of p300 and PCAF or the histone methylase Suv39H1 to HACs upsets the balance between histone H3K9 methylation and acetylation, promoting centromere formation when acetylated or acting as a barrier to CENP-A nucleosome proliferation when trimethylated (Ohzeki et al. 2012). Histone modification of non-CENP-A nucleosomes is therefore likely to have a role in centromere maintenance and function. In addition, CENP-A nucleosomes themselves are likely to be subject to modification; CENP-A ubiquitination by CUL3/RDX ubiquitin ligase has been shown to stabilize soluble CENP-A in Drosophila (Bade et al. 2014).
Other Centromere-Specific DNA-Binding Proteins
Several DNA-binding proteins, in addition to CENP-A nucleosomes, associate with centromeres and influence centromere function. The histone-fold-containing proteins CENP-T, -W, -S, and -X assemble into dimers of CENP-T/W, tetramers of CENP-S/X, and potentially a heterotetramer of CENP-T/W/S and X (Fig. 3). In vitro, a heterotetramer of CENP-T, -W, -S, and -X was proposed to protect ∼100 bp of DNA from nuclease digestion, in a similar manner to H3 and CENP-A nucleosomes (Nishino et al. 2012). More recently, a CENP-T/W/S/X octamer has been shown to bind ∼100 bp of DNA and potentially induce positive supercoils (as opposed to negative supercoils induced by nucleosomes) (Takeuchi et al. 2014). However, whether a CENP-T/W/S/X nucleosome-like species exists at centromeres remains unclear, as the path of DNA wrapping around these proteins is unknown and the nuclease protection pattern is distinct from that of a nucleosome. Histone fold proteins are not exclusively nucleosomal proteins that wrap DNA, but also include several different classes of DNA-binding factors. CENP-T/W and CENP-S/X may instead function as two independent complexes. CENP-S/X plays noncentromeric roles in DNA repair independently of CENP-T/W (Singh et al. 2010; Tao et al. 2012; Zhao et al. 2014), CENP-S/X depletion does not affect CENP-T centromere recruitment (Amano et al. 2009), and CENP-S is dispensable in engineered neocentromeres that are positive for CENP-T in chicken DT-40 cells (Shang et al. 2013). One possibility is that binding of CENP-S/X to CENP-T/W confers some centromere-specific function, but the ability of CENP-T to function independently from CENP-S suggests CENP-T/W/S/X does not form an obligate nucleosome-like particle (Hori et al. 2008a; Amano et al. 2009). Indeed, before identification of the CENP-T/W/S/X tetramer, CENP-T/W was suggested to associate primarily with histone H3 rather than CENP-A (Hori et al. 2008a; Ribeiro et al. 2010). Consistent with this, although CENP-T is lost from centromeres completely lacking CENP-A, its centromere levels are largely unaffected by a 90% CENP-A reduction (Fachinetti et al. 2013). Therefore, CENP-T/W/S/X, either as a complex or separately, may interact specifically with non-CENP-A centromeric DNA in a non-nucleosome-like manner. Recent data suggests that CENP-T and CENP-W are targeted for degradation unless they are in a heterodimeric complex with each other, suggesting that either a CENP-T/W/S/X nucleosome-like complex or separate complexes of CENP-T/W and CENP-S/X will be the functionally relevant species at centromeres (Chun et al. 2013). Ascertaining whether a CENP-T/W/S/X nucleosome exists within centromeric chromatin, and assessing its relevance to centromere function, are immediate goals for the field.
Possible Roles for Centromeric DNA Sequence?
What is the role, if any, of centromeric DNA sequence? The regional centromeres of many eukaryotes are characterized by repetitive DNA sequences. Human centromeres consist of A/T-rich 171-bp α-satellite repeats. Human centromeric DNA contains the 17-bp “CENP-B box,” which directly recruits the centromere protein CENP-B (Masumoto et al. 1989). CENP-B knockout mice are for the most part phenotypically normal (Hudson et al. 1998; Kapoor et al. 1998; Perez-Castro et al. 1998) and functional neocentromeres lack CENP-B (Saffery et al. 2000). Moreover, CENP-B is absent from the human Y chromosome (Earnshaw et al. 1987) and no CENP-B functional homologs have been identified in Xenopus, zebrafish, C. elegans, or Drosophila to date. Thus, once a centromere is formed, CENP-B appears to be dispensable for centromere function.
On the other hand, CENP-B has been suggested to promote the formation of new centromeres. Studies using HACs as a substrate for new centromere assembly showed that satellite repeats containing CENP-B box mutations were much less efficient at forming a functional centromere (Ohzeki et al. 2002). Consistent with this, fibroblasts from CENP-B knockout mice fail to form new centromeres on artificial chromosomes (Okada et al. 2007). CENP-B may also play a second role in initiating heterochromatin formation, as CENP-B box containing DNA, integrated into a chromosome arm, is specifically trimethylated on H3K9 in CENP-B containing mouse embryo fibroblasts, but not in fibroblasts from a CENP-B knockout mouse (Okada et al. 2007). The CENP-B DNA-binding domain can position CENP-A nucleosomes on DNA in vitro (Tanaka et al. 2005), and CENP-B boxes appear to phase CENP-A nucleosome position in vivo (Hasson et al. 2013).
Partial reduction in CENP-A levels has no effect on CENP-B (Fachinetti et al. 2013). However, when CENP-A is completely abolished, CENP-B centromere levels drop by around 50%, revealing a previously uncharacterized dependence of CENP-B on CENP-A nucleosomes (Fachinetti et al. 2013). Mutation of centromeric CENP-A revealed CENP-B centromere recruitment requires the CENP-A amino-terminal tail through an unknown mechanism, and CENP-B:CENP-A amino-terminal cooperation may contribute to sustaining localization of centromere proteins (Fachinetti et al. 2013). Thus, CENP-B may represent a redundant pathway for CENP-A nucleosome stabilization, positioning, and/or recruitment of other centromere-specific components. It is interesting to place this in the context of studies that find that human centromeres can remain functional even after extensive, but not complete, depletion of CENP-A (Liu et al. 2006; Fachinetti et al. 2013). CENP-B may specify the minimal sites of CENP-A nucleosomes sufficient for functional centromere formation, in this way acting as a backup to guard against CENP-A loss. Thus, in the case of severely limited CENP-A, centromere specification may no longer be epigenetically maintained, as the CENP-B box may have a role. In the same manner, an intriguing possibility is that CENP-B has a partially redundant role in maintaining CENP-A nucleosome position through DNA replication (see below). Depletion of CENP-A to a level that still maintains centromere formation, coupled with CENP-B depletion, may reveal some interesting centromere phenomena.
CENTROMERE PROTEIN RECOGNITION OF CENP-A CHROMATIN
CENP-A nucleosomes direct the assembly of functional centromeres by recruiting centromere proteins, both directly through binding and indirectly through the assembly of higher-order protein complexes. Two proteins, CENP-N and CENP-C, interact directly with CENP-A nucleosomes to promote centromere assembly (Carroll et al. 2009, 2010). Both CENP-N and CENP-C only bind nucleosomal CENP-A, and do so through recognition of different elements of the nucleosome. CENP-N binds CENP-A nucleosomes by recognizing CENP-A’s CATD, whereas CENP-C binds CENP-A’s divergent carboxy-terminal tail and the acidic patch on histones H2A and H2B (Carroll et al. 2009, 2010; Kato et al. 2013).
The functions of CENP-N are not well established. CENP-N depletion causes a loss of multiple other centromere components (Foltz et al. 2006; McClelland et al. 2007; Carroll et al. 2009), so CENP-N appears to be a key building block of the centromere. Indeed, S. cerevisiae CENP-NChl4 is required for de novo centromere formation (Mythreye and Bloom 2003). However, CENP-NChl4 is not required to sustain previously established centromeres, suggesting it may function redundantly at existing centromeres. In addition, centromeric CENP-N protein levels decrease before mitosis, suggesting that CENP-N may not be required to recruit kinetochore proteins (McClelland et al. 2007; Hellwig et al. 2011). Although CENP-N binds the CATD, this may not be necessary for CENP-N to localize to centromeres; centromeres generated by LacI-CENP-C and LacI-CENP-T fusions, which are proposed to be negative for CENP-A, are still positive for CENP-N (Gascoigne et al. 2011), and CENP-N can be recruited to H3/CENP-A chimeras that lack the CATD in vitro (Guse et al. 2011). In S. cerevisiae, CENP-NChl4 forms a heterodimer with CENP-LIml3 that interacts with CENP-CMif2, suggesting that CENP-C could mediate recruitment of CENP-N/L (Guo et al. 2013; Hinshaw and Harrison 2013).
In contrast to CENP-N, CENP-C has a clear role in both kinetochore formation and centromeric chromatin maintenance (discussed below). CENP-C is specifically recruited to centromeres through hydrophobic interactions between the CENP-A carboxyl terminus, and conserved domains in the CENP-C protein provide specificity for CENP-C recruitment to centromeres (Carroll et al. 2010; Kato et al. 2013). The nucleosome-binding domain of CENP-C contacts both the CENP-A carboxyl terminus and the acidic patch of H2A/H2B and with DNA (Kato et al. 2013). Histone H3 and CENP-A chimeras containing only the carboxy-terminal six amino acids of CENP-A are sufficient for CENP-C recruitment in Xenopus Guse et al. 2011). In human cells CENP-C centromere recruitment is reduced after replacement of CENP-A with H3/CENP-A chimeras lacking the carboxy-terminal CENP-A region (Fachinetti et al. 2013). S. cerevisiae CENP-CMif2 dimerizes through a region near its carboxyl terminus (Cohen et al. 2008), presenting the possibility of a CENP-C dimer interacting with a CENP-A octameric nucleosome, or higher-order structures involving two CENP-A nucleosomes, consistent with suggestions that CENP-C is important for higher-order centromere structure (Ribeiro et al. 2010).
Regulation of CENP-C centromere localization at endogenous centromeres is more complicated than direct recruitment by CENP-A. In cells in which CENP-A is progressively removed from centromeres, a significant population of CENP-C maintains its localization despite the absence of CENP-A (Fachinetti et al. 2013). This suggests that CENP-C may be recruited to centromeres through multiple redundant mechanisms, and also raises the possibility that CENP-C may contribute to its own stability at centromeres, which has implications for our understanding of how centromeres could be maintained through CENP-A assembly and DNA replication. As CENP-C is key for establishing the kinetochore, its resistance to decreasing CENP-A nucleosome numbers provides a potential explanation for why all but the complete depletion of CENP-A fails to result in a strong centromere defect.
KINETOCHORE AND CENTROMERE COMPOSITION
Broadly speaking, centromeric proteins have two temporally distinct roles. First, before and during mitosis, centromere proteins build the kinetochore. Second, during mitotic exit and in interphase (in humans), centromere proteins recruit the factors that replenish CENP-A nucleosomes and maintain centromeric chromatin. In this section, we briefly focus on how centromeres promote mitotic kinetochore formation, and then discuss the role of the centromere in CENP-A reassembly. For an in-depth overview of kinetochore function in mitosis, see an accompanying review of the kinetochore (Cheeseman 2014).
The hub of the mitotic kinetochore, and the machinery responsible for robust microtubule attachment, is the KMN protein network: KNL1, the Mis12 complex (Mis12, Nsl1, Nnf1, and Dsn1) and the Ndc80 complex (Ndc80/Hec1, Nuf2, Spc24, and Spc25). In all organisms, an essential function of the constitutive centromere proteins is to recruit the KMN network before and during mitosis. The amino terminus of CENP-C interacts with Nnf1 and is required for the G2 centromere targeting of the Mis12 complex in both Drosophila and human cells (Milks et al. 2009; Przewloka et al. 2011; Screpanti et al. 2011). A second essential step in KMN protein assembly at centromeres is binding of the Spc24/25 components of the Ndc80 complex by the phosphorylated amino-terminal tail of CENP-T. This conserved interaction forms stable, load-bearing attachments to microtubules via the Ndc80 complex (Bock et al. 2012; Schleiffer et al. 2012; Malvezzi et al. 2013; Nishino et al. 2013). A nonphosphorylatable CENP-T amino-terminal tail disrupts kinetochore function as it cannot fulfill these functions (Gascoigne et al. 2011). Interestingly, the Mis12 complex also binds Spc24/25 in the same manner as CENP-T, and CENP-T and Mis12 compete for Ndc80 binding, which may influence the stability of kinetochore microtubule attachments (Bock et al. 2012; Schleiffer et al. 2012). Furthermore, phosphoregulation of Mis12 and/or CENP-T could control switching between different microtubule-binding modes within kinetochores. These findings may have important implications for how kinetochores may positively select for correct microtubule attachments and destabilize erroneous attachments.
To date, 17 constitutively associated CENP proteins have been identified (Fig. 3). Broadly speaking, during mitosis the CENP proteins appear to regulate the CENP-C/T:KMN network and the KMN network:microtubule interactions. As a rule, depletion of any CENP protein impairs centromere function, causing kinetochore:microtubule attachment and chromosome congression defects (Fukagawa et al. 2001; Foltz et al. 2006; McAinsh et al. 2006; Okada et al. 2006; McClelland et al. 2007; Hori et al. 2008b; Carroll et al. 2009; Toso et al. 2009; Amaro et al. 2010). Although the function of CENP-C and CENP-T has been described in the most detail, biochemical analyses of centromere subcomplexes are also contributing to our understanding of centromere function. In addition to CENP-T/W/S/X mentioned above, CENP-LIml3 forms a heterodimer with CENP-NChl4 in S. cerevisiae, and a homologous interaction between CENP-L and the CENP-N amino terminus has been identified in human cells (Carroll et al. 2009; Hinshaw and Harrison 2013). A centromere subcomplex of CENP-H/I/K/M has also recently been reconstituted biochemically, and cell-based assays have revealed that CENP-H/I/K/M is required for CENP-C-mediated CENP-T centromere localization (A Musacchio, pers. comm.). CENP-H, CENP-I, and CENP-K were previously shown to bind KNL-1 directly (Cheeseman et al. 2008), suggesting CENP-H/I/K/M may also contribute to kinetochore formation.
The function of the remaining centromere proteins CENP-O, -P, -Q, -R, and -U remains largely unclear. In vitro, CENP-U and a CENP-Q octamer bind microtubules (Amaro et al. 2010), and CENP-U is also proposed to bind Hec1 (Hua et al. 2011). Aurora B–mediated phosphorylation of CENP-U within a CENP-O/P/Q/U complex is required for efficient recovery from spindle-damaging agents in cells (Hori et al. 2008b). Recently, fission yeast CENP-Q and CENP-O have been shown to associate with members of the Mis18 complex that regulates CENP-A assembly (see below) (Subramanian et al. 2014).
In conclusion, although centromeres are perhaps best considered as one large protein complex, the recent careful biochemical analyses of centromere subcomplexes described above have greatly aided our understanding of centromere function. Before these achievements, cell-based work had created a confusing landscape; knowing whether an observed effect is direct or indirect or affected by other consequences of the manipulation has been extremely challenging. We anticipate that further reconstitution of the centromere will continue to provide a key framework for our understanding of centromere function in cells.
CENTROMERE MAINTENANCE AND CENP-A ASSEMBLY
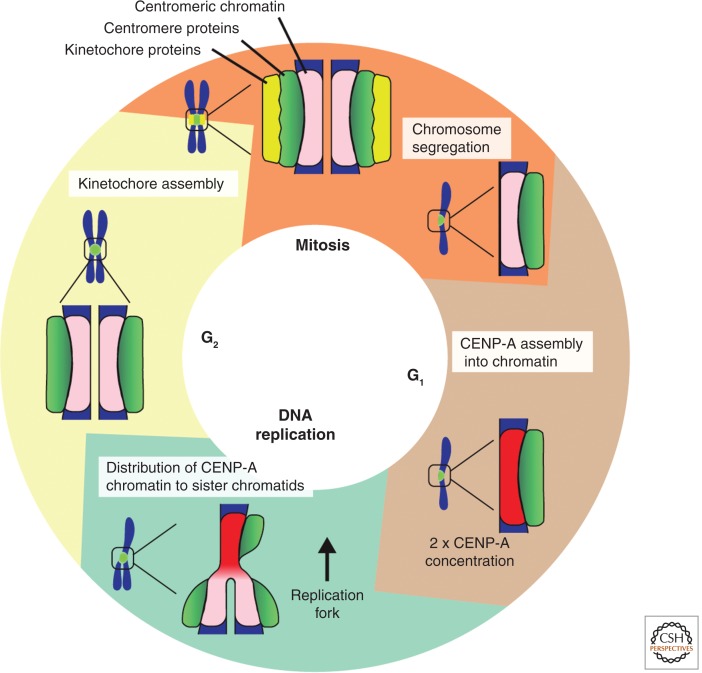
The amount of DNA within a cell doubles every cell cycle as a result of DNA replication during S phase. Conventional histones are assembled concurrently with DNA replication, with a network of histone chaperones and modifiers in place to regenerate the prereplication chromatin environment after passage of the replication fork (reviewed in Probst et al. 2009). In contrast to canonical chromatin, CENP-A nucleosome replenishment in higher eukaryotes occurs independently of DNA replication. In humans, CENP-A assembly starts after anaphase, and finishes before the end of G1 (Fig. 4) (Jansen et al. 2007). DNA replication-independent assembly is specific to CENP-A nucleosomes, and is conferred by the CATD, as pulse-labeled H3CATD chimeric proteins mimic the assembly characteristics of CENP-A, at least at endogenous centromeres that also contain CENP-A (Black et al. 2007; Bodor et al. 2013). The timing of CENP-A assembly is, in part, governed by cell-cycle kinases, including Cdk1:cyclin B. When Cdk1 is inhibited, CENP-A assembly occurs prematurely (Silva et al. 2012), and when cyclinA:Cdk1 levels or APC activity are perturbed in Drosophila, CENP-ACID assembly fails (Erhardt et al. 2008). Although CENP-A assembly remains a poorly understood process, we discuss here some of the relatively well-characterized proteins involved in CENP-A assembly in turn, before summarizing other factors that are less well understood.
Figure 4.
Functions of centromeric chromatin through the human cell cycle. Centromeric chromatin, defined by CENP-A-containing nucleosomes, is specifically assembled during mitotic exit and G1. Assembly causes the CENP-A copy number at centromeres to double (represented by transition from pink to red chromatin). During DNA replication/S phase, replication of centromere-DNA results in the distribution of CENP-A onto the two nascent DNA strands. This causes a twofold reduction in CENP-A at each centromere-DNA sequence (red to pink chromatin) (see Fig. 5 for more details). Note that it is this “diluted” centromeric chromatin (pink) that is responsible for building functional kinetochores before and during mitosis and, thus, for segregating chromosomes. Broadly speaking, centromere proteins retain localization at CENP-A chromatin through all stages of the cell cycle. How centromere proteins respond to changing CENP-A protein numbers within chromatin is not clear.
HJURP: The CENP-A Chaperone
Canonical histones require chaperones for DNA-replication-dependent assembly (Ransom et al. 2010). CENP-A, as a specialized histone, has a specialized chaperone known as HJURP in humans or Scm3 in yeast. S. cerevisae Scm3 was identified in immunopurifications of CENP-ACse4 and is required to target CENP-ACse4 to centromeres (Camahort et al. 2007; Mizuguchi et al. 2007; Stoler et al. 2007). Scm3 can assemble CENP-ACse4 into nucleosomes in vitro (Shivaraju et al. 2011). HJURP (Holliday junction recognition protein) was originally named because it was suggested to bind Holliday junctions (Kato et al. 2007) and was subsequently identified in purifications of soluble CENP-A complexes that represent a prechromatin assembly intermediate (Dunleavy et al. 2009; Foltz et al. 2009). HJURP and Scm3 bind directly to a dimer of CENP-A and histone H4 through recognition of the CATD domain of CENP-A by HJURP/Scm3’s conserved amino terminus (Foltz et al. 2009; Shuaib et al. 2010; Cho and Harrison 2011; Hu et al. 2011; Zhou et al. 2011). In addition, HJURP forms contacts with CENP-A outside of the CATD that enhance the stability of the HJURP:CENP-A:H4 complex (Bassett et al. 2012). Recognition of the CATD provides at least a partial explanation for why the CATD of CENP-A is essential for CENP-A maintenance and forms the minimal domain in CENP-A required for CENP-A assembly. A histone H3CATD chimera is recognized by HJURP and stably incorporated into centromeric chromatin (Black et al. 2004; Bodor et al. 2013). The importance of HJURP in centromere maintenance was confirmed by tethering LacI-HJURP fusion protein to Lac operators at ectopic chromosomal loci, which consequently assemble CENP-A and form largely functional kinetochores (Barnhart et al. 2011; Hori et al. 2013). In Drosophila, which lack an HJURP homolog, CAL1 assembles CENP-ACID nucleosomes in an extremely similar manner to HJURP in vertebrates (Chen et al. 2014).
The levels of soluble CENP-A:HJURP complex increase as cells enter mitosis, but this complex does not associate with centromeres until cells proceed through anaphase, concurrent with the timing of CENP-A assembly (Dunleavy et al. 2009; Foltz et al. 2009). Structural studies indicate that the HJURP:CENP-A:H4 and Scm3:Cse4:H4 trimers preclude CENP-A-H4 tetramer formation (Hu et al. 2011; Zhou et al. 2011). Thus, an extremely interesting problem is how the HJURP chaperone complex releases CENP-A to facilitate transfer of CENP-A/H4 into chromatin. Because a dimer of CENP-A:H4 is carried by each HJURP molecule, either two rounds of dimer delivery must occur to assemble an octameric nucleosome or some other intermediate containing only a CENP-A/H4 tetramer must exist. Of note, the carboxy-terminal region of HJURP contains an HJURP dimerization domain, suggesting that (HJURP:CENP-A:H4)2 exists as a soluble complex away from chromatin, and that this complex may deliver two CENP-A:H4 dimers simultaneously to sites of new CENP-A assembly (Fig. 5) (Zasadzinska et al. 2013). Similar observations have been made for Scm3 (Dechassa et al. 2014). Interestingly, HJURP recruitment to centromeres can occur without CENP-A binding or HJURP dimerization, but HJURP dimerization is required for CENP-A assembly, confirmed by elegant experiments that rescued deletion of the HJURP dimerization motif with insertion of an unrelated dimerization domain (Zasadzinska et al. 2013). Yeast Scm3 localizes to centromeres independently of CENP-ACse4/Cnp1, and also self-associates (Mizuguchi et al. 2007; Pidoux et al. 2009; Williams et al. 2009), suggesting that the mechanism of HJURP/Scm3-mediated CENP-A assembly is conserved. The major limiting factor remaining in our understanding of HJURP function is identifying how HJURP is recruited to centromeres; links between HJURP and other components of the CENP-A assembly machinery remain unclear, although Mis18β (see below) has recently been suggested to interact with HJURP (Wang et al. 2014).
Figure 5.
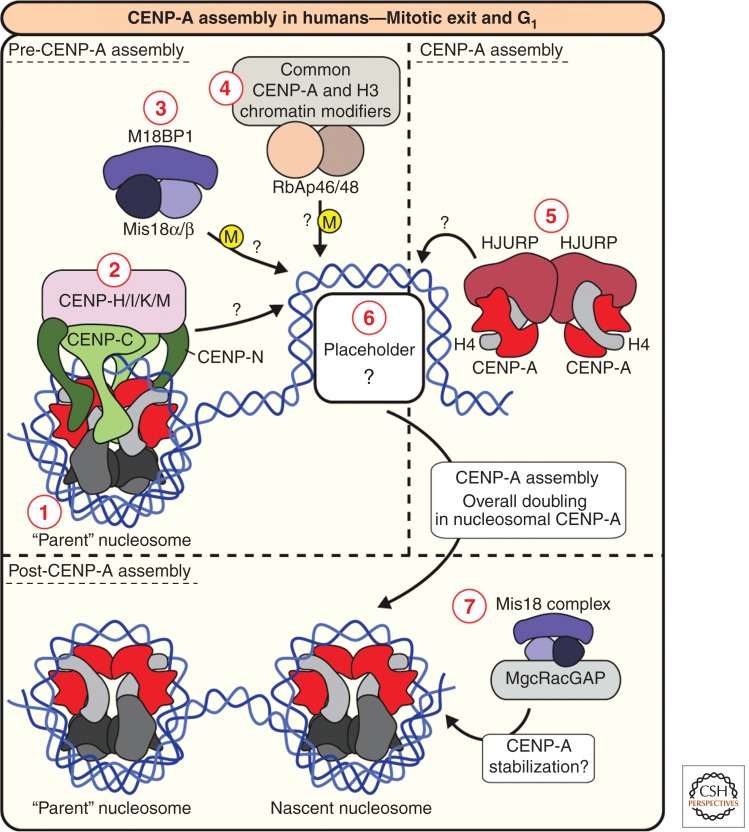
CENP-A assembly in humans. (1) “Parent” CENP-A nucleosomes (from the previous cell cycle), directly or indirectly specify the sites of new CENP-A assembly. (2) The centromere proteins shown in the figure; CENP-C, -N (which directly bind CENP-A nucleosomes), -H, -I, -K, and -M have been experimentally implicated in CENP-A assembly. (3) In addition to centromere proteins, the Mis18 complex, consisting of Mis18α, Mis18β, and M18BP1, is also required for CENP-A assembly, and most likely modifies (M) the chromatin and/or the recruitment of specialized loading factors. (4) Chromatin modifiers and chaperones, such as RbAp46 and 48, also have a role in modifying chromatin during CENP-A assembly. (5) Centromere proteins and the Mis18 complex somehow recruit HJURP, the CENP-A-specific chaperone. HJURP binding to CENP-A precludes CENP-A:H4 tetramer formation, and HJURP dimerization is required for CENP-A assembly, so one possible model is that nascent CENP-A is delivered as two dimers in a (HJURP:CENP-A:H4)2 complex, as shown. (6) The proteins or chromatin features that mark the site of new CENP-A assembly, the “placeholders,” remain unclear. (7) Finally, once CENP-A is assembled into chromatin, factors such as MgcRacGAP, which interacts with the Mis18 complex, stabilize incorporated CENP-A.
The Mis18 Complex
Mis18 was discovered in S. pombe as a protein that, when mutated, causes errors in chromosome segregation (Hayashi et al. 2004). Humans possess two homologs of Mis18, Mis18α and Mis18β, and another associating protein called Mis18-binding protein 1 (M18BP1) (Fig. 5). All three proteins interact but it is not known whether Mis18α, Mis18β, and M18BP1 function in CENP-A assembly exclusively as a complex. A homologous M18BP1 protein, Kinetochore Null 2 (KNL2), was discovered in a screen for kinetochore defects in C. elegans and M18BP1 has been identified in humans, Xenopus, and Arabidopsis (Fujita et al. 2007; Maddox et al. 2007; Moree et al. 2011; Lermontova et al. 2013). The function of the Mis18 complex remains unclear, and more proteins are likely to be involved. Indeed, two recent studies in fission yeast have independently identified Eic1/Mis19 and Eic2/Mis20 as novel Mis18 complex components (Hayashi et al. 2004; Subramanian et al. 2014). Eic1 may be the S. pombe homolog of Mis18BP1. Depletion of any one Mis18 complex component (apart from Eic2/Mis20) prevents new CENP-A assembly (Fujita et al. 2007; Maddox et al. 2007; Hayashi et al. 2014; Subramanian et al. 2014). However, in contrast to HJURP, overall cellular CENP-A levels are not diminished after Mis18 depletion, suggesting that the soluble HJURP:CENP-A:H4 remains stable without Mis18 activity and the Mis18 complex functions at chromatin rather than on soluble CENP-A.
Mis18α, Mis18β, and M18BP1 are recruited to centromeres at an early stage in CENP-A assembly, during anaphase/telophase of mitosis and in early G1 (Fujita et al. 2007; Maddox et al. 2007; Dambacher et al. 2012). The Mis18 complex is required for the recruitment of HJURP to centromeres in a number of model organisms (Pidoux et al. 2009; Williams et al. 2009; Barnhart et al. 2011; Moree et al. 2011), and in human cells, depletion of HJURP has no effect on Mis18α. Together with the early localization of the Mis18 complex to centromeres, these data have led to suggestions that the Mis18 complex is a key player in CENP-A assembly process. Artificially tethering HJURP to chromatin can bypass the requirement for M18BP1; thus, a simple model is that M18BP1 association precedes HJURP binding and is required for proper targeting of the HJURP chaperone complex to centromeres. Whether the Mis18 complex also plays an active role in the transfer of CENP-A into chromatin is unknown.
The specific activities of the Mis18 complex may involve histone and/or DNA modification. Mis18α is required for the recruitment of DNA methyltransferases to centromeres, which are in turn required for normal CENP-A centromere levels (Kim et al. 2012). Mis18 mutation in fission yeast causes an increase in centromeric H3 and H4 acetylation (Hayashi et al. 2004), and treatment with histone deacetylation inhibitors can rescue Mis18α depletion (Fujita et al. 2007). Human M18BP1 and C. elegans KNL2 have SANT/Myb DNA-binding domains, placing M18BP1/KNL2 at the necessary location to modify histones or DNA, either directly or indirectly. SANT/Myb domains are also found in histone modifiers such as Ada2 histone acetyl transferase (HAT) (Boyer et al. 2002), and the SANT/Myb domain of the telomere-binding protein TRF2 alters chromatin structure in vitro (Baker et al. 2009). Of note, the S. pombe transcriptional regulator Teb1, which also contains SANT/Myb domains, has also been shown to be required for Cnp1CENP-A assembly (Valente et al. 2013).
Constitutive Centromere Proteins and CENP-A Assembly
The fact that constitutive centromere proteins are present at centromeres during interphase (as opposed to kinetochore proteins) perhaps reflects their functions in maintaining centromeres through the cell cycle (Fig. 5). CENP-C depletion prevents CENP-A assembly onto sperm chromatin in Xenopus egg extracts, and CENP-C is required to target Xenopus M18BP1 isoform 1 to metaphase centromeres (Moree et al. 2011). An analogous system may function in Drosophila in which the CENP-A assembly factor Cal1 requires CENP-C to be recruited to centromeres in metaphase (Erhardt et al. 2008; Mellone et al. 2011). Like Mis18α, CENP-C interacts with DNA methyltransferases that may facilitate chromatin remodeling to promote CENP-A assembly (Gopalakrishnan et al. 2009). Fusing CENP-C and CENP-I proteins with LacI, when targeted to lacO sites integrated into the chromosome, support formation of centromeres that are resistant to subsequent IPTG treatment (which removes the fusion protein) (Hori et al. 2013). This indicates that functional CENP-A nucleosomes are assembled into chromatin as a consequence of CENP-C or CENP-I ectopic localization (Hori et al. 2013). Indeed, the individual CENPs that comprise the recently identified CENP-H/I/K/M complex have been implicated in CENP-A assembly in chicken and human cells (Takahashi et al. 2000; Okada et al. 2006).
Additional Factors Involved in CENP-A Assembly
Several other factors are known to function in CENP-A assembly but their activities are not exclusively devoted to centromeres. Depletion of the RbAp46/48 class of histone chaperones, Mis16 in S. pombe or RbAp46 and 48 in humans (by temperature-sensitive mutation or RNAi) causes loss of CENP-A from centromeres (Hayashi et al. 2004). In human cells, RbAp46/48 purify with the HJURP chaperone complex (Dunleavy et al. 2009) and depletion of RbAp46/48 causes a reduction in CENP-A protein levels, indicating that they play a chaperone role for CENP-A (Hayashi et al. 2004). Unlike HJURP, RbAp46/48 have important roles in regulation of conventional chromatin. RbAp46 and 48 are both subunits of the CAF-1 (chromatin assembly factor-1) complex that is required for the replication-coupled assembly of H3:H4 (Verreault et al. 1996; Tagami et al. 2004). Thus, it appears likely that CENP-A assembly shares some chromatin modifiers with conventional histone H3 assembly.
In S. cerevisiae, the E3 ubiquitin ligase Psh1 has been shown to target CENP-ACse4 for degradation (Hewawasam et al. 2010; Ranjitkar et al. 2010). Psh1 localizes to centromeres, but is thought to act on noncentromeric CENP-ACse4. Psh1 specifically acts on CENP-ACse4 as it recognizes the CATD. Because of this, Scm3 binding to the CATD protects CENP-ACse4 from Psh1-mediated degradation. Deletion of Psh1 causes assembly of CENP-ACse4 into noncentromeric chromosomal loci. These data suggest that mechanisms exist to detect and degrade Scm3-free CENP-ACse4 to prevent incorrect assembly. The role of CENP-A ubiquitination in regulating CENP-A assembly may be conserved, as ubiquitination of Drosophila CENP-ACID by the E3 ubiquitin ligases Ppa and CUL3 can destabilize or stabilize CENP-ACID, respectively (Moreno-Moreno et al. 2011; Bade et al. 2014).
At this stage, it is unclear if Psh1, or a related pathway, can act on CENP-ACse4 already within chromatin. Of note, the recent characterization of the GTP exchange factor MgcRacGAP suggests that, in addition to removal of free CENP-A, correctly assembled CENP-A nucleosomes are stabilized. Pulse labeling of SNAP-CENP-A, to selectively label “old” CENP-A nucleosomes carried over from the previous cell cycle, suggests that MgcRacGAP depletion specifically destabilizes nascent CENP-A nucleosomes (Lagana et al. 2010). However, other than the discovery that MgcRacGAP is degraded during mitotic exit (perhaps explaining its delayed centromere localization) (Nishimura et al. 2013), little is known about the role of GTP hydrolysis and MgcRacGAP in CENP-A assembly or stabilization. MgcRacGAP is also known to have important roles in cell proliferation through regulation of cytokinesis, as a member of the centralspindlin complex (White and Glotzer 2012). In addition to MgcRacGAP, subunits of the RSF complex (remodeling and spacing factor), which regulates canonical histone modification, have also been implicated in CENP-A chromatin stabilization (Obuse et al. 2004; Izuta et al. 2006; Perpelescu et al. 2009).
In summary, great progress has been made in the understanding of CENP-A assembly in the last few years, most noticeably in the identification and characterization of novel CENP-A assembly factors such as HJURP and M18BP1. We anticipate that the next few years will include consolidation of what remains a somewhat fragmented picture. As a final consideration, a comprehensive understanding of CENP-A assembly requires identification of the mechanism for targeting new CENP-A to the correct positions (Fig. 5). Several models have been proposed for a “placeholder,” a marking mechanism that identifies where to assemble new CENP-A nucleosomes, including naked DNA, histone H3.3 nucleosomes (Dunleavy et al. 2011), hybrid CENP-A/H3.3 nucleosomes, a CENP-T/W/S/X complex (Nishino et al. 2012), hemisomes (Bui et al. 2012), or an unidentified component. The chromatin-associated factors that dictate the positions of new CENP-A assembly will most likely be targets for some of the assembly factors discussed above.
CENTROMERE LONGEVITY AND CENP-A MAINTENANCE DURING DNA REPLICATION
Pulse-chase labeling of CENP-A, to specifically track incorporated CENP-A across multiple cell divisions, shows a 50% reduction in labeled CENP-A per chromosome with each replication cycle (Dunleavy et al. 2011; Bodor et al. 2013). In addition, gene deletion of CENP-A causes CENP-A centromere levels to reduce by 50% each cell cycle (Fachinetti et al. 2013). Thus, in the absence of CENP-A assembly, CENP-A is progressively depleted from centromeres after multiple cell generations (as a result of DNA replication; Fig. 6). These data highlight the remarkable persistence of existing CENP-A nucleosomes. To date, a mechanism that recycles correctly positioned, chromatinized CENP-A has not been identified. Thus, CENP-A protein deposited during early development provides the epigenetic mark of centromeres indefinitely. Moreover, histone H4 shows the same stable characteristics as CENP-A specifically at centromeres, suggesting that H4 is also stabilized as part of the CENP-A nucleosome (Bodor et al. 2013). This is not an intrinsic property of centromeric chromatin, as other nucleosomal species show no distinct centromere-specific stabilization (Bodor et al. 2013). What provides CENP-A nucleosomes with this longevity remains unclear, but histone H3CATD chimeric nucleosomes possess many of the same characteristics, suggesting that the specific nucleosome structure conferred by the CATD may have a role (Bodor et al. 2013).
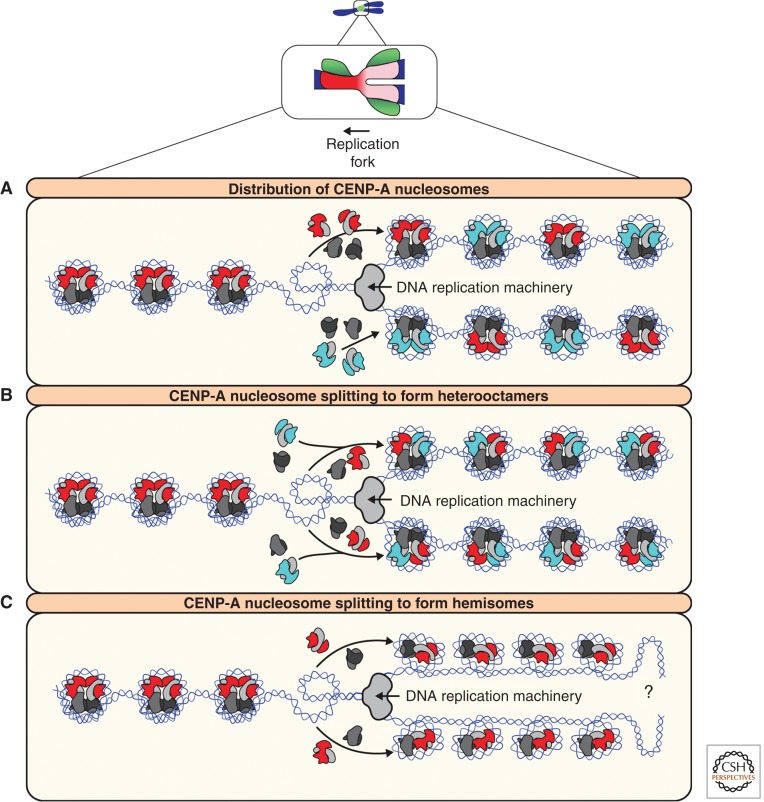
Figure 6.
Models of CENP-A nucleosome distribution during DNA replication. (A) CENP-A (red) nucleosomes remain as homotypic octamers after passage of the replication fork. In the model shown, octameric histone H3.1 (cyan) nucleosomes occupy the gaps left by the twofold reduction in CENP-A nucleosomes on each DNA strand. Other possibilities include histone H3.3, naked DNA, or other species of specialized centromeric chromatin (such as CENP-T/W/S/X). (B) Each CENP-A nucleosome is split and segregated to both DNA strands. CENP-A:H4 dimers form heterotetramers with H3/H4 dimers, resulting in heterooctamer formation. (C) Octameric CENP-A nucleosomes are split and not replenished, resulting in the formation of tetrameric hemisomes. As hemisomes are incapable of wrapping as much DNA as octameric nucleosomes, this model results in an excess of free DNA, the fate of which is unclear (?). All these models assume octameric CENP-A nucleosomes are the prereplication conformation. Note that the composition of centromeric chromatin after DNA replication becomes the template for CENP-A assembly in the next cell cycle (see Figs. 3 and 4).
Although CENP-A nucleosomes display remarkable longevity, CENP-A nucleosome disruption and reassembly on daughter DNA strands is a necessary part of centromere maintenance during DNA replication. As DNA replication proceeds, CENP-A histones are distributed equally to each new DNA strand (Figs. 4 and 6). Establishing how CENP-A nucleosomes are segregated, whether the composition of CENP-A nucleosomes changes as a result of redistribution, and how the network of centromere proteins differs between G2 and G1, are all major unanswered questions that have profound implications for understanding centromere maintenance (Fig. 6). Kinetochores are assembled on centromeres that have been reorganized as a result of DNA replication, and CENP-A assembly during mitotic exit and G1 uses a replicated centromere as a template.
The challenge of maintaining epigenetic marks on DNA as the DNA is replicated is applicable to the entire genome. Presumably, given the specialized nature of centromeric chromatin, specialized systems to maintain CENP-A likely exist. However, other than the observation that CENP-A histones are equally distributed between replicated DNA strands (Dunleavy et al. 2009), very little is known about potential changes in CENP-A nucleosome structure and/or composition. Important insights will come with a more complete understanding of the role of the centromere proteins in maintaining CENP-A nucleosomes across the DNA replication fork, as recent observations show that many centromeric proteins show distinct stability during S phase. CENP-T and CENP-W completely turn over during S phase, but increase in abundance in S/G2 relative to G1 (Prendergast et al. 2011). CENP-S and CENP-X also assemble at centromeres during S/G2 (Dornblut et al. 2014). CENP-N is stabilized specifically at the end of S phase through an unknown mechanism (McClelland et al. 2007; Hellwig et al. 2011). CENP-C and CENP-H are stabilized at centromeres during DNA replication (Hemmerich et al. 2008). As CENP-C and CENP-N, the two centromere components that directly bind CENP-A, stabilize during S phase, and CENP-C can still localize to centromeres that have completely lost CENP-A (Fachinetti et al. 2013), a speculative possibility is that the network of centromere proteins maintain centromere position during underlying passage of the replication fork and disruption of CENP-A nucleosomes. In this manner, centromere proteins may play a key role in promoting CENP-A nucleosome reformation on replicated centromeric DNA. Finally, CENP-B binding to the CENP-B box may play a role in repositioning of CENP-A nucleosomes during replication (Tanaka et al. 2005).
CONCLUDING REMARKS
Remarkable progress has been made in the understanding of centromeres since the identification of the first centromere proteins in the mid-1980s (Earnshaw and Rothfield 1985; Palmer et al. 1987). Given that the rate at which new centromere proteins are being identified appears to be slowing, we may be nearing a complete parts list of centromere proteins. How these components form a functional centromere/kinetochore is also becoming clearer. The identification of biochemically discrete complexes of centromere and kinetochore proteins, that share phenotypes when perturbed, suggests modularity in centromere assembly and functions. For example, the role of a CENP-T-containing complex in recruiting the Ndc80 complex and the function of CENP-C in recruiting of the Mis12 complex, suggests different functions of the kinetochore are brought to centromeres through different activities of core centromere proteins. However, despite this modularity, there is significant cross talk between centromere components to stably form a functional centromere and kinetochore; Mis12 and CENP-T compete for Ndc80 binding (Bock et al. 2012; Schleiffer et al. 2012; Nishino et al. 2013) and a CENP-N/L heterodimer binds CENP-C (Carroll et al. 2009; Hinshaw and Harrison 2013), presumably as part of a CENP-A nucleosome-containing complex. Pinpointing how and when different centromere modules interact remains an exciting challenge.
Significant advances in our understanding of how centromeric chromatin is specified and assembled have also been made in the past decade. Important cell and biochemical studies have led to the identification of Scm3/HJURP and the Mis18 complex as central regulators of new CENP-A assembly and have shown the distinctive cell-cycle regulation of CENP-A assembly. Establishing how centromeres direct CENP-A assembly to the correct location, to the correct level, and at the right time, are all key future goals. At this time, we lack an understanding of the changes within chromatin that occur during transfer of soluble CENP-A into nucleosomal CENP-A. This is a challenging problem, as manipulating distinct populations of CENP-A (i.e., parent nucleosomes or soluble CENP-A) in cells is not currently possible. Overcoming this obstacle will be important to establish how CENP-A nucleosomes direct CENP-A assembly.
Complementary to our understanding of centromere and kinetochore assembly, studies of the centromere have yielded new insights into epigenetic regulatory mechanisms in eukaryotes. Centromeric nucleosomes and the CENP-A histone may represent one of the best examples of true epigenetic inheritance. As we continue to make progress in our understanding of how CENP-A marks the centromere and how cells interpret that mark to maintain centromeric chromatin, we are likely to gain important insights into other epigenetic inheritance mechanisms. Furthermore, the specific recognition of centromeric chromatin to uniquely specify the functions of the centromere is an exemplar of how chromosomal loci are specialized for distinct functions.
ACKNOWLEDGMENTS
We thank members of the Straight laboratory for many helpful discussions, and Jon Pines for his helpful input during review. This work is supported by the National Institutes of Health Grant R01 GM074728 to A.F.S.
Footnotes
Editors: Mitsuhiro Yanagida, Anthony A. Hyman, and Jonathon Pines
Additional Perspectives on Mitosis available at www.cshperspectives.org
REFERENCES
*Reference is also in this collection.
- Amano M, Suzuki A, Hori T, Backer C, Okawa K, Cheeseman IM, Fukagawa T 2009. The CENP-S complex is essential for the stable assembly of outer kinetochore structure. J Cell Biol 186: 173–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amaro AC, Samora CP, Holtackers R, Wang E, Kingston IJ, Alonso M, Lampson M, McAinsh AD, Meraldi P 2010. Molecular control of kinetochore-microtubule dynamics and chromosome oscillations. Nat Cell Biol 12: 319–329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bade D, Pauleau AL, Wendler A, Erhardt S 2014. The E3 ligase CUL3/RDX controls centromere maintenance by ubiquitylating and stabilizing CENP-A in a CAL1-dependent manner. Dev Cell 28: 508–519. [DOI] [PubMed] [Google Scholar]
- Bailey AO, Panchenko T, Sathyan KM, Petkowski JJ, Pai PJ, Bai DL, Russell DH, Macara IG, Shabanowitz J, Hunt DF, et al. 2013. Posttranslational modification of CENP-A influences the conformation of centromeric chromatin. Proc Natl Acad Sci 110: 11827–11832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker AM, Fu Q, Hayward W, Lindsay SM, Fletcher TM 2009. The Myb/SANT domain of the telomere-binding protein TRF2 alters chromatin structure. Nucleic Acids Res 37: 5019–5031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnhart MC, Kuich PH, Stellfox ME, Ward JA, Bassett EA, Black BE, Foltz DR 2011. HJURP is a CENP-A chromatin assembly factor sufficient to form a functional de novo kinetochore. J Cell Biol 194: 229–243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassett EA, DeNizio J, Barnhart-Dailey MC, Panchenko T, Sekulic N, Rogers DJ, Foltz DR, Black BE 2012. HJURP uses distinct CENP-A surfaces to recognize and to stabilize CENP-A/histone H4 for centromere assembly. Dev Cell 22: 749–762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergmann JH, Rodriguez MG, Martins NM, Kimura H, Kelly DA, Masumoto H, Larionov V, Jansen LE, Earnshaw WC 2011. Epigenetic engineering shows H3K4me2 is required for HJURP targeting and CENP-A assembly on a synthetic human kinetochore. EMBO J 30: 328–340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black BE, Foltz DR, Chakravarthy S, Luger K, Woods VL Jr, Cleveland DW 2004. Structural determinants for generating centromeric chromatin. Nature 430: 578–582. [DOI] [PubMed] [Google Scholar]
- Black BE, Jansen LE, Maddox PS, Foltz DR, Desai AB, Shah JV, Cleveland DW 2007. Centromere identity maintained by nucleosomes assembled with histone H3 containing the CENP-A targeting domain. Mol Cell 25: 309–322. [DOI] [PubMed] [Google Scholar]
- Blower MD, Sullivan BA, Karpen GH 2002. Conserved organization of centromeric chromatin in flies and humans. Dev Cell 2: 319–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bock LJ, Pagliuca C, Kobayashi N, Grove RA, Oku Y, Shrestha K, Alfieri C, Golfieri C, Oldani A, Dal Maschio M, et al. 2012. Cnn1 inhibits the interactions between the KMN complexes of the yeast kinetochore. Nat Cell Biol 14: 614–624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodor DL, Valente LP, Mata JF, Black BE, Jansen LE 2013. Assembly in G1 phase and long-term stability are unique intrinsic features of CENP-A nucleosomes. Mol Biol Cell 24: 923–932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boeckmann L, Takahashi Y, Au WC, Mishra PK, Choy JS, Dawson AR, Szeto MY, Waybright TJ, Heger C, McAndrew C, et al. 2013. Phosphorylation of centromeric histone H3 variant regulates chromosome segregation in Saccharomyces cerevisiae. Mol Biol Cell 24: 2034–2044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyer LA, Langer MR, Crowley KA, Tan S, Denu JM, Peterson CL 2002. Essential role for the SANT domain in the functioning of multiple chromatin remodeling enzymes. Mol Cell 10: 935–942. [DOI] [PubMed] [Google Scholar]
- Bui M, Dimitriadis EK, Hoischen C, An E, Quenet D, Giebe S, Nita-Lazar A, Diekmann S, Dalal Y 2012. Cell-cycle-dependent structural transitions in the human CENP-A nucleosome in vivo. Cell 150: 317–326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camahort R, Li B, Florens L, Swanson SK, Washburn MP, Gerton JL 2007. Scm3 is essential to recruit the histone h3 variant cse4 to centromeres and to maintain a functional kinetochore. Mol Cell 26: 853–865. [DOI] [PubMed] [Google Scholar]
- Cardinale S, Bergmann JH, Kelly D, Nakano M, Valdivia MM, Kimura H, Masumoto H, Larionov V, Earnshaw WC 2009. Hierarchical inactivation of a synthetic human kinetochore by a chromatin modifier. Mol Biol Cell 20: 4194–4204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll CW, Silva MC, Godek KM, Jansen LE, Straight AF 2009. Centromere assembly requires the direct recognition of CENP-A nucleosomes by CENP-N. Nat Cell Biol 11: 896–902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll CW, Milks KJ, Straight AF 2010. Dual recognition of CENP-A nucleosomes is required for centromere assembly. J Cell Biol 189: 1143–1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Cheeseman IM 2014. The kinetochore. Cold Spring Harb Perspect Biol 6: a015826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheeseman IM, Desai A 2008. Molecular architecture of the kinetochore-microtubule interface. Nat Rev Mol Cell Biol 9: 33–46. [DOI] [PubMed] [Google Scholar]
- Cheeseman IM, Hori T, Fukagawa T, Desai A 2008. KNL1 and the CENP-H/I/K complex coordinately direct kinetochore assembly in vertebrates. Mol Biol Cell 19: 587–594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CC, Dechassa ML, Bettini E, Ledoux MB, Belisario C, Heun P, Luger K, Mellone BG 2014. CAL1 is the Drosophila CENP-A assembly factor. J Cell Biol 204: 313–329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho US, Harrison SC 2011. Recognition of the centromere-specific histone Cse4 by the chaperone Scm3. Proc Natl Acad Sci 108: 9367–9371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chun Y, Lee M, Park B, Lee S 2013. CSN5/Jab1 interacts with the centromeric components CENP-T and CENP-W and regulates their proteasome-mediated degradation. J Biol Chem 288: 27208–27219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke L, Carbon J 1980. Isolation of a yeast centromere and construction of functional small circular chromosomes. Nature 287: 504–509. [DOI] [PubMed] [Google Scholar]
- Codomo CA, Furuyama T, Henikoff S 2014. CENP-A octamers do not confer a reduction in nucleosome height by AFM. Nat Struct Mol Biol 21: 4–5. [DOI] [PubMed] [Google Scholar]
- Coffman VC, Wu P, Parthun MR, Wu JQ 2011. CENP-A exceeds microtubule attachment sites in centromere clusters of both budding and fission yeast. J Cell Biol 195: 563–572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen RL, Espelin CW, De Wulf P, Sorger PK, Harrison SC, Simons KT 2008. Structural and functional dissection of Mif2p, a conserved DNA-binding kinetochore protein. Mol Biol Cell 19: 4480–4491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conde e Silva N, Black BE, Sivolob A, Filipski J, Cleveland DW, Prunell A 2007. CENP-A-containing nucleosomes: Easier disassembly versus exclusive centromeric localization. J Mol Biol 370: 555–573. [DOI] [PubMed] [Google Scholar]
- Cottarel G, Shero JH, Hieter P, Hegemann JH 1989. A 125-base-pair CEN6 DNA fragment is sufficient for complete meiotic and mitotic centromere functions in Saccharomyces cerevisiae. Mol Cell Biol 9: 3342–3349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Csink AK, Henikoff S 1998. Something from nothing: The evolution and utility of satellite repeats. Trends Genet 14: 200–204. [DOI] [PubMed] [Google Scholar]
- Dalal Y, Wang H, Lindsay S, Henikoff S 2007. Tetrameric structure of centromeric nucleosomes in interphase Drosophila cells. PLoS Biol 5: e218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dambacher S, Deng W, Hahn M, Sadic D, Frohlich J, Nuber A, Hoischen C, Diekmann S, Leonhardt H, Schotta G 2012. CENP-C facilitates the recruitment of M18BP1 to centromeric chromatin. Nucleus 3: 101–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dechassa ML, Wyns K, Luger K 2014. Scm3 deposits a (Cse4-H4)2 tetramer onto DNA through a Cse4-H4 dimer intermediate. Nucleic Acids Res 42: 5534–5542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimitriadis EK, Weber C, Gill RK, Diekmann S, Dalal Y 2010. Tetrameric organization of vertebrate centromeric nucleosomes. Proc Natl Acad Sci 107: 20317–20322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dornblut C, Quinn N, Monajambashi S, Prendergast L, van Vuuren C, Munch S, Deng W, Leonhardt H, Cardoso MC, Hoischen C, et al. 2014. A CENP-S/X complex assembles at the centromere in S and G2 phases of the human cell cycle. Open Biol 4: 130229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunleavy EM, Roche D, Tagami H, Lacoste N, Ray-Gallet D, Nakamura Y, Daigo Y, Nakatani Y, Almouzni-Pettinotti G 2009. HJURP is a cell-cycle-dependent maintenance and deposition factor of CENP-A at centromeres. Cell 137: 485–497. [DOI] [PubMed] [Google Scholar]
- Dunleavy EM, Almouzni G, Karpen GH 2011. H3.3 is deposited at centromeres in S phase as a placeholder for newly assembled CENP-A in G1 phase. Nucleus 2: 146–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Earnshaw WC, Rothfield N 1985. Identification of a family of human centromere proteins using autoimmune sera from patients with scleroderma. Chromosoma 91: 313–321. [DOI] [PubMed] [Google Scholar]
- Earnshaw W, Bordwell B, Marino C, Rothfield N 1986. Three human chromosomal autoantigens are recognized by sera from patients with anti-centromere antibodies. J Clin Invest 77: 426–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Earnshaw WC, Machlin PS, Bordwell BJ, Rothfield NF, Cleveland DW 1987. Analysis of anticentromere autoantibodies using cloned autoantigen CENP-B. Proc Natl Acad Sci 84: 4979–4983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erhardt S, Mellone BG, Betts CM, Zhang W, Karpen GH, Straight AF 2008. Genome-wide analysis reveals a cell cycle-dependent mechanism controlling centromere propagation. J Cell Biol 183: 805–818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fachinetti D, Diego Folco H, Nechemia-Arbely Y, Valente LP, Nguyen K, Wong AJ, Zhu Q, Holland AJ, Desai A, Jansen LE, et al. 2013. A two-step mechanism for epigenetic specification of centromere identity and function. Nat Cell Biol 15: 1056–1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitzgerald-Hayes M, Clarke L, Carbon J 1982. Nucleotide sequence comparisons and functional analysis of yeast centromere DNAs. Cell 29: 235–244. [DOI] [PubMed] [Google Scholar]
- Flemming W 1882. Zellsubstanz, kern und zelltheilung. F.C.W. Vogel, Leipzig. [Google Scholar]
- Foltz DR, Jansen LE, Black BE, Bailey AO, Yates JR III, Cleveland DW 2006. The human CENP-A centromeric nucleosome-associated complex. Nat Cell Biol 8: 458–469. [DOI] [PubMed] [Google Scholar]
- Foltz DR, Jansen LE, Bailey AO, Yates JR III, Bassett EA, Wood S, Black BE, Cleveland DW 2009. Centromere-specific assembly of CENP-A nucleosomes is mediated by HJURP. Cell 137: 472–484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita Y, Hayashi T, Kiyomitsu T, Toyoda Y, Kokubu A, Obuse C, Yanagida M 2007. Priming of centromere for CENP-A recruitment by human hMis18α, hMis18β, and M18BP1. Dev Cell 12: 17–30. [DOI] [PubMed] [Google Scholar]
- Fukagawa T, Mikami Y, Nishihashi A, Regnier V, Haraguchi T, Hiraoka Y, Sugata N, Todokoro K, Brown W, Ikemura T 2001. CENP-H, a constitutive centromere component, is required for centromere targeting of CENP-C in vertebrate cells. EMBO J 20: 4603–4617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furuyama S, Biggins S 2007. Centromere identity is specified by a single centromeric nucleosome in budding yeast. Proc Natl Acad Sci 104: 14706–14711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furuyama T, Henikoff S 2009. Centromeric nucleosomes induce positive DNA supercoils. Cell 138: 104–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gascoigne KE, Takeuchi K, Suzuki A, Hori T, Fukagawa T, Cheeseman IM 2011. Induced ectopic kinetochore assembly bypasses the requirement for CENP-A nucleosomes. Cell 145: 410–422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gopalakrishnan S, Sullivan BA, Trazzi S, Della Valle G, Robertson KD 2009. DNMT3B interacts with constitutive centromere protein CENP-C to modulate DNA methylation and the histone code at centromeric regions. Hum Mol Genet 18: 3178–3193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Q, Tao Y, Liu H, Teng M, Li X 2013. Structural insights into the role of the Chl4-Iml3 complex in kinetochore assembly. Acta Crystallogr D Biol Crystallogr 69: 2412–2419. [DOI] [PubMed] [Google Scholar]
- Guse A, Carroll CW, Moree B, Fuller CJ, Straight AF 2011. In vitro centromere and kinetochore assembly on defined chromatin templates. Nature 477: 354–358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haase J, Mishra PK, Stephens A, Haggerty R, Quammen C, Taylor RM II, Yeh E, Basrai MA, Bloom K 2013. A 3D map of the yeast kinetochore reveals the presence of core and accessory centromere-specific histone. Curr Biol 23: 1939–1944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasson D, Panchenko T, Salimian KJ, Salman MU, Sekulic N, Alonso A, Warburton PE, Black BE 2013. The octamer is the major form of CENP-A nucleosomes at human centromeres. Nat Struct Mol Biol 20: 687–695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi T, Fujita Y, Iwasaki O, Adachi Y, Takahashi K, Yanagida M 2004. Mis16 and Mis18 are required for CENP-A loading and histone deacetylation at centromeres. Cell 118: 715–729. [DOI] [PubMed] [Google Scholar]
- Hayashi T, Ebe M, Nagao K, Kokubu A, Sajiki K, Yanagida M 2014. Schizosaccharomyces pombe centromere protein Mis19 links Mis16 and Mis18 to recruit CENP-A through interacting with NMD factors and the SWI/SNF complex. Genes Cells 19: 541–554. [DOI] [PubMed] [Google Scholar]
- Hellwig D, Emmerth S, Ulbricht T, Doring V, Hoischen C, Martin R, Samora CP, McAinsh AD, Carroll CW, Straight AF, et al. 2011. Dynamics of CENP-N kinetochore binding during the cell cycle. J Cell Sci 124: 3871–3883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemmerich P, Weidtkamp-Peters S, Hoischen C, Schmiedeberg L, Erliandri I, Diekmann S 2008. Dynamics of inner kinetochore assembly and maintenance in living cells. J Cell Biol 180: 1101–1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henikoff S, Ramachandran S, Krassovsky K, Bryson TD, Codomo CA, Brogaard K, Widom J, Wang JP, Henikoff JG 2014. The budding yeast Centromere DNA Element II wraps a stable Cse4 hemisome in either orientation in vivo. eLife 3: e01861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heun P, Erhardt S, Blower MD, Weiss S, Skora AD, Karpen GH 2006. Mislocalization of the Drosophila centromere-specific histone CID promotes formation of functional ectopic kinetochores. Dev Cell 10: 303–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hewawasam G, Shivaraju M, Mattingly M, Venkatesh S, Martin-Brown S, Florens L, Workman JL, Gerton JL 2010. Psh1 is an E3 ubiquitin ligase that targets the centromeric histone variant Cse4. Mol Cell 40: 444–454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinshaw SM, Harrison SC 2013. An Iml3-Chl4 heterodimer links the core centromere to factors required for accurate chromosome segregation. Cell Rep 5: 29–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hori T, Amano M, Suzuki A, Backer CB, Welburn JP, Dong Y, McEwen BF, Shang WH, Suzuki E, Okawa K, et al. 2008a. CCAN makes multiple contacts with centromeric DNA to provide distinct pathways to the outer kinetochore. Cell 135: 1039–1052. [DOI] [PubMed] [Google Scholar]
- Hori T, Okada M, Maenaka K, Fukagawa T 2008b. CENP-O class proteins form a stable complex and are required for proper kinetochore function. Mol Biol Cell 19: 843–854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hori T, Shang WH, Takeuchi K, Fukagawa T 2013. The CCAN recruits CENP-A to the centromere and forms the structural core for kinetochore assembly. J Cell Biol 200: 45–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu H, Liu Y, Wang M, Fang J, Huang H, Yang N, Li Y, Wang J, Yao X, Shi Y, et al. 2011. Structure of a CENP-A-histone H4 heterodimer in complex with chaperone HJURP. Genes Dev 25: 901–906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua S, Wang Z, Jiang K, Huang Y, Ward T, Zhao L, Dou Z, Yao X 2011. CENP-U cooperates with Hec1 to orchestrate kinetochore-microtubule attachment. J Biol Chem 286: 1627–1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson DF, Fowler KJ, Earle E, Saffery R, Kalitsis P, Trowell H, Hill J, Wreford NG, de Kretser DM, Cancilla MR, et al. 1998. Centromere protein B null mice are mitotically and meiotically normal but have lower body and testis weights. J Cell Biol 141: 309–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishii K, Ogiyama Y, Chikashige Y, Soejima S, Masuda F, Kakuma T, Hiraoka Y, Takahashi K 2008. Heterochromatin integrity affects chromosome reorganization after centromere dysfunction. Science 321: 1088–1091. [DOI] [PubMed] [Google Scholar]
- Izuta H, Ikeno M, Suzuki N, Tomonaga T, Nozaki N, Obuse C, Kisu Y, Goshima N, Nomura F, Nomura N, et al. 2006. Comprehensive analysis of the ICEN (interphase centromere complex) components enriched in the CENP-A chromatin of human cells. Genes Cells 11: 673–684. [DOI] [PubMed] [Google Scholar]
- Jansen LE, Black BE, Foltz DR, Cleveland DW 2007. Propagation of centromeric chromatin requires exit from mitosis. J Cell Biol 176: 795–805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagansky A, Folco HD, Almeida R, Pidoux AL, Boukaba A, Simmer F, Urano T, Hamilton GL, Allshire RC 2009. Synthetic heterochromatin bypasses RNAi and centromeric repeats to establish functional centromeres. Science 324: 1716–1719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapoor M, Montes de Oca Luna R, Liu G, Lozano G, Cummings C, Mancini M, Ouspenski I, Brinkley BR, May GS 1998. The cenpB gene is not essential in mice. Chromosoma 107: 570–576. [DOI] [PubMed] [Google Scholar]
- Kato T, Sato N, Hayama S, Yamabuki T, Ito T, Miyamoto M, Kondo S, Nakamura Y, Daigo Y 2007. Activation of Holliday junction recognizing protein involved in the chromosomal stability and immortality of cancer cells. Cancer Res 67: 8544–8553. [DOI] [PubMed] [Google Scholar]
- Kato H, Jiang J, Zhou BR, Rozendaal M, Feng H, Ghirlando R, Xiao TS, Straight AF, Bai Y 2013. A conserved mechanism for centromeric nucleosome recognition by centromere protein CENP-C. Science 340: 1110–1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim IS, Lee M, Park KC, Jeon Y, Park JH, Hwang EJ, Jeon TI, Ko S, Lee H, Baek SH, et al. 2012. Roles of Mis18α in epigenetic regulation of centromeric chromatin and CENP-A loading. Mol Cell 46: 260–273. [DOI] [PubMed] [Google Scholar]
- Kingston IJ, Yung JS, Singleton MR 2011. Biophysical characterization of the centromere-specific nucleosome from budding yeast. J Biol Chem 286: 4021–4026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunitoku N, Sasayama T, Marumoto T, Zhang D, Honda S, Kobayashi O, Hatakeyama K, Ushio Y, Saya H, Hirota T 2003. CENP-A phosphorylation by Aurora-A in prophase is required for enrichment of Aurora-B at inner centromeres and for kinetochore function. Dev Cell 5: 853–864. [DOI] [PubMed] [Google Scholar]
- Lacoste N, Woolfe A, Tachiwana H, Garea AV, Barth T, Cantaloube S, Kurumizaka H, Imhof A, Almouzni G 2014. Mislocalization of the centromeric histone variant CenH3/CENP-A in human cells depends on the chaperone DAXX. Mol Cell 53: 631–644. [DOI] [PubMed] [Google Scholar]
- Lagana A, Dorn JF, De Rop V, Ladouceur AM, Maddox AS, Maddox PS 2010. A small GTPase molecular switch regulates epigenetic centromere maintenance by stabilizing newly incorporated CENP-A. Nat Cell Biol 12: 1186–1193. [DOI] [PubMed] [Google Scholar]
- Lawrimore J, Bloom KS, Salmon ED 2011. Point centromeres contain more than a single centromere-specific Cse4 (CENP-A) nucleosome. J Cell Biol 195: 573–582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lermontova I, Kuhlmann M, Friedel S, Rutten T, Heckmann S, Sandmann M, Demidov D, Schubert V, Schubert I 2013. Arabidopsis kinetochore null2 is an upstream component for centromeric histone H3 variant cenH3 deposition at centromeres. Plant Cell 25: 3389–3404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu ST, Rattner JB, Jablonski SA, Yen TJ 2006. Mapping the assembly pathways that specify formation of the trilaminar kinetochore plates in human cells. J Cell Biol 175: 41–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo AW, Craig JM, Saffery R, Kalitsis P, Irvine DV, Earle E, Magliano DJ, Choo KH 2001. A 330 kb CENP-A binding domain and altered replication timing at a human neocentromere. EMBO J 20: 2087–2096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maddox PS, Hyndman F, Monen J, Oegema K, Desai A 2007. Functional genomics identifies a Myb domain-containing protein family required for assembly of CENP-A chromatin. J Cell Biol 176: 757–763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malvezzi F, Litos G, Schleiffer A, Heuck A, Mechtler K, Clausen T, Westermann S 2013. A structural basis for kinetochore recruitment of the Ndc80 complex via two distinct centromere receptors. EMBO J 32: 409–423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masumoto H, Masukata H, Muro Y, Nozaki N, Okazaki T 1989. A human centromere antigen (CENP-B) interacts with a short specific sequence in alphoid DNA, a human centromeric satellite. J Cell Biol 109: 1963–1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McAinsh AD, Meraldi P, Draviam VM, Toso A, Sorger PK 2006. The human kinetochore proteins Nnf1R and Mcm21R are required for accurate chromosome segregation. EMBO J 25: 4033–4049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClelland SE, Borusu S, Amaro AC, Winter JR, Belwal M, McAinsh AD, Meraldi P 2007. The CENP-A NAC/CAD kinetochore complex controls chromosome congression and spindle bipolarity. EMBO J 26: 5033–5047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellone BG, Grive KJ, Shteyn V, Bowers SR, Oderberg I, Karpen GH 2011. Assembly of Drosophila centromeric chromatin proteins during mitosis. PLoS Genet 7: e1002068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meluh PB, Yang P, Glowczewski L, Koshland D, Smith MM 1998. Cse4p is a component of the core centromere of Saccharomyces cerevisiae. Cell 94: 607–613. [DOI] [PubMed] [Google Scholar]
- Mendiburo MJ, Padeken J, Fulop S, Schepers A, Heun P 2011. Drosophila CENH3 is sufficient for centromere formation. Science 334: 686–690. [DOI] [PubMed] [Google Scholar]
- Miell MD, Fuller CJ, Guse A, Barysz HM, Downes A, Owen-Hughes T, Rappsilber J, Straight AF, Allshire RC 2013. CENP-A confers a reduction in height on octameric nucleosomes. Nat Struct Mol Biol 20: 763–765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miell MD, Straight AF, Allshire RC 2014. Reply to “CENP-A octamers do not confer a reduction in nucleosome height by AFM.” Nat Struct Mol Biol 21: 5–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milks KJ, Moree B, Straight AF 2009. Dissection of CENP-C-directed centromere and kinetochore assembly. Mol Biol Cell 20: 4246–4255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuguchi G, Xiao H, Wisniewski J, Smith MM, Wu C 2007. Nonhistone Scm3 and histones CenH3-H4 assemble the core of centromere-specific nucleosomes. Cell 129: 1153–1164. [DOI] [PubMed] [Google Scholar]
- Moree B, Meyer CB, Fuller CJ, Straight AF 2011. CENP-C recruits M18BP1 to centromeres to promote CENP-A chromatin assembly. J Cell Biol 194: 855–871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno-Moreno O, Medina-Giro S, Torras-Llort M, Azorin F 2011. The F box protein partner of paired regulates stability of Drosophila centromeric histone H3, CenH3(CID). Curr Biol 21: 1488–1493. [DOI] [PubMed] [Google Scholar]
- Mythreye K, Bloom KS 2003. Differential kinetochore protein requirements for establishment versus propagation of centromere activity in Saccharomyces cerevisiae. J Cell Biol 160: 833–843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakano M, Cardinale S, Noskov VN, Gassmann R, Vagnarelli P, Kandels-Lewis S, Larionov V, Earnshaw WC, Masumoto H 2008. Inactivation of a human kinetochore by specific targeting of chromatin modifiers. Dev Cell 14: 507–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura K, Oki T, Kitaura J, Kuninaka S, Saya H, Sakaue-Sawano A, Miyawaki A, Kitamura T 2013. APC(CDH1) targets MgcRacGAP for destruction in the late M phase. PLoS ONE 8: e63001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishino T, Takeuchi K, Gascoigne KE, Suzuki A, Hori T, Oyama T, Morikawa K, Cheeseman IM, Fukagawa T 2012. CENP-T-W-S-X forms a unique centromeric chromatin structure with a histone-like fold. Cell 148: 487–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishino T, Rago F, Hori T, Tomii K, Cheeseman IM, Fukagawa T 2013. CENP-T provides a structural platform for outer kinetochore assembly. EMBO J 32: 424–436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obuse C, Yang H, Nozaki N, Goto S, Okazaki T, Yoda K 2004. Proteomics analysis of the centromere complex from HeLa interphase cells: UV-damaged DNA binding protein 1 (DDB-1) is a component of the CEN-complex, while BMI-1 is transiently co-localized with the centromeric region in interphase. Genes Cells 9: 105–120. [DOI] [PubMed] [Google Scholar]
- Ohzeki J, Nakano M, Okada T, Masumoto H 2002. CENP-B box is required for de novo centromere chromatin assembly on human alphoid DNA. J Cell Biol 159: 765–775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohzeki J, Bergmann JH, Kouprina N, Noskov VN, Nakano M, Kimura H, Earnshaw WC, Larionov V, Masumoto H 2012. Breaking the HAC barrier: Histone H3K9 acetyl/methyl balance regulates CENP-A assembly. EMBO J 31: 2391–2402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada M, Cheeseman IM, Hori T, Okawa K, McLeod IX, Yates JR III, Desai A, Fukagawa T 2006. The CENP-H-I complex is required for the efficient incorporation of newly synthesized CENP-A into centromeres. Nat Cell Biol 8: 446–457. [DOI] [PubMed] [Google Scholar]
- Okada T, Ohzeki J, Nakano M, Yoda K, Brinkley WR, Larionov V, Masumoto H 2007. CENP-B controls centromere formation depending on the chromatin context. Cell 131: 1287–1300. [DOI] [PubMed] [Google Scholar]
- Padmanabhan S, Thakur J, Siddharthan R, Sanyal K 2008. Rapid evolution of Cse4p-rich centromeric DNA sequences in closely related pathogenic yeasts, Candida albicans and Candida dubliniensis. Proc Natl Acad Sci 105: 19797–19802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer DK, O’Day K, Wener MH, Andrews BS, Margolis RL 1987. A 17-kD centromere protein (CENP-A) copurifies with nucleosome core particles and with histones. J Cell Biol 104: 805–815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer DK, O’Day K, Trong HL, Charbonneau H, Margolis RL 1991. Purification of the centromere-specific protein CENP-A and demonstration that it is a distinctive histone. Proc Natl Acad Sci 88: 3734–3738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panchenko T, Sorensen TC, Woodcock CL, Kan ZY, Wood S, Resch MG, Luger K, Englander SW, Hansen JC, Black BE 2011. Replacement of histone H3 with CENP-A directs global nucleosome array condensation and loosening of nucleosome superhelical termini. Proc Natl Acad Sci 108: 16588–16593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Castro AV, Shamanski FL, Meneses JJ, Lovato TL, Vogel KG, Moyzis RK, Pedersen R 1998. Centromeric protein B null mice are viable with no apparent abnormalities. Dev Biol 201: 135–143. [DOI] [PubMed] [Google Scholar]
- Perpelescu M, Nozaki N, Obuse C, Yang H, Yoda K 2009. Active establishment of centromeric CENP-A chromatin by RSF complex. J Cell Biol 185: 397–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pidoux AL, Choi ES, Abbott JK, Liu X, Kagansky A, Castillo AG, Hamilton GL, Richardson W, Rappsilber J, He X, et al. 2009. Fission yeast Scm3: A CENP-A receptor required for integrity of subkinetochore chromatin. Mol Cell 33: 299–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prendergast L, van Vuuren C, Kaczmarczyk A, Doering V, Hellwig D, Quinn N, Hoischen C, Diekmann S, Sullivan KF 2011. Premitotic assembly of human CENPs-T and -W switches centromeric chromatin to a mitotic state. PLoS Biol 9: e1001082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Probst AV, Dunleavy E, Almouzni G 2009. Epigenetic inheritance during the cell cycle. Nat Rev Mol Cell Biol 10: 192–206. [DOI] [PubMed] [Google Scholar]
- Przewloka MR, Venkei Z, Bolanos-Garcia VM, Debski J, Dadlez M, Glover DM 2011. CENP-C is a structural platform for kinetochore assembly. Curr Biol 21: 399–405. [DOI] [PubMed] [Google Scholar]
- Ranjitkar P, Press MO, Yi X, Baker R, MacCoss MJ, Biggins S 2010. An E3 ubiquitin ligase prevents ectopic localization of the centromeric histone H3 variant via the centromere targeting domain. Mol Cell 40: 455–464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ransom M, Dennehey BK, Tyler JK 2010. Chaperoning histones during DNA replication and repair. Cell 140: 183–195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribeiro SA, Vagnarelli P, Dong Y, Hori T, McEwen BF, Fukagawa T, Flors C, Earnshaw WC 2010. A super-resolution map of the vertebrate kinetochore. Proc Natl Acad Sci 107: 10484–10489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saffery R, Irvine DV, Griffiths B, Kalitsis P, Wordeman L, Choo KH 2000. Human centromeres and neocentromeres show identical distribution patterns of >20 functionally important kinetochore-associated proteins. Hum Mol Genet 9: 175–185. [DOI] [PubMed] [Google Scholar]
- Schleiffer A, Maier M, Litos G, Lampert F, Hornung P, Mechtler K, Westermann S 2012. CENP-T proteins are conserved centromere receptors of the Ndc80 complex. Nat Cell Biol 14: 604–613. [DOI] [PubMed] [Google Scholar]
- Schueler MG, Higgins AW, Rudd MK, Gustashaw K, Willard HF 2001. Genomic and genetic definition of a functional human centromere. Science 294: 109–115. [DOI] [PubMed] [Google Scholar]
- Screpanti E, De Antoni A, Alushin GM, Petrovic A, Melis T, Nogales E, Musacchio A 2011. Direct binding of Cenp-C to the Mis12 complex joins the inner and outer kinetochore. Curr Biol 21: 391–398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekulic N, Bassett EA, Rogers DJ, Black BE 2010. The structure of (CENP-A-H4)2 reveals physical features that mark centromeres. Nature 467: 347–351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang WH, Hori T, Martins NM, Toyoda A, Misu S, Monma N, Hiratani I, Maeshima K, Ikeo K, Fujiyama A, et al. 2013. Chromosome engineering allows the efficient isolation of vertebrate neocentromeres. Dev Cell 24: 635–648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shivaraju M, Camahort R, Mattingly M, Gerton JL 2011. Scm3 is a centromeric nucleosome assembly factor. J Biol Chem 286: 12016–12023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shivaraju M, Unruh JR, Slaughter BD, Mattingly M, Berman J, Gerton JL 2012. Cell-cycle-coupled structural oscillation of centromeric nucleosomes in yeast. Cell 150: 304–316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shuaib M, Ouararhni K, Dimitrov S, Hamiche A 2010. HJURP binds CENP-A via a highly conserved N-terminal domain and mediates its deposition at centromeres. Proc Natl Acad Sci 107: 1349–1354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva MC, Bodor DL, Stellfox ME, Martins NM, Hochegger H, Foltz DR, Jansen LE 2012. Cdk activity couples epigenetic centromere inheritance to cell cycle progression. Dev Cell 22: 52–63. [DOI] [PubMed] [Google Scholar]
- Singh TR, Saro D, Ali AM, Zheng XF, Du CH, Killen MW, Sachpatzidis A, Wahengbam K, Pierce AJ, Xiong Y, et al. 2010. MHF1-MHF2, a histone-fold-containing protein complex, participates in the Fanconi anemia pathway via FANCM. Mol Cell 37: 879–886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spencer F, Hieter P 1992. Centromere DNA mutations induce a mitotic delay in Saccharomyces cerevisiae. Proc Natl Acad Sci 89: 8908–8912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoler S, Keith KC, Curnick KE, Fitzgerald-Hayes M 1995. A mutation in CSE4, an essential gene encoding a novel chromatin-associated protein in yeast, causes chromosome nondisjunction and cell cycle arrest at mitosis. Genes Dev 9: 573–586. [DOI] [PubMed] [Google Scholar]
- Stoler S, Rogers K, Weitze S, Morey L, Fitzgerald-Hayes M, Baker RE 2007. Scm3, an essential Saccharomyces cerevisiae centromere protein required for G2/M progression and Cse4 localization. Proc Natl Acad Sci 104: 10571–10576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian L, Toda NR, Rappsilber J, Allshire RC 2014. Eic1 links Mis18 with the CCAN/Mis6/Ctf19 complex to promote CENP-A assembly. Open Biol 4: 140043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan BA, Karpen GH 2004. Centromeric chromatin exhibits a histone modification pattern that is distinct from both euchromatin and heterochromatin. Nat Struct Mol Biol 11: 1076–1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan KF, Hechenberger M, Masri K 1994. Human CENP-A contains a histone H3 related histone fold domain that is required for targeting to the centromere. J Cell Biol 127: 581–592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tachiwana H, Kagawa W, Shiga T, Osakabe A, Miya Y, Saito K, Hayashi-Takanaka Y, Oda T, Sato M, Park SY, et al. 2011. Crystal structure of the human centromeric nucleosome containing CENP-A. Nature 476: 232–235. [DOI] [PubMed] [Google Scholar]
- Tagami H, Ray-Gallet D, Almouzni G, Nakatani Y 2004. Histone H3.1 and H3.3 complexes mediate nucleosome assembly pathways dependent or independent of DNA synthesis. Cell 116: 51–61. [DOI] [PubMed] [Google Scholar]
- Takahashi K, Chen ES, Yanagida M 2000. Requirement of Mis6 centromere connector for localizing a CENP-A-like protein in fission yeast. Science 288: 2215–2219. [DOI] [PubMed] [Google Scholar]
- Takeuchi K, Nishino T, Mayanagi K, Horikoshi N, Osakabe A, Tachiwana H, Hori T, Kurumizaka H, Fukagawa T 2014. The centromeric nucleosome-like CENP-T-W-S-X complex induces positive supercoils into DNA. Nucleic Acids Res 42: 1644–1655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka Y, Tachiwana H, Yoda K, Masumoto H, Okazaki T, Kurumizaka H, Yokoyama S 2005. Human centromere protein B induces translational positioning of nucleosomes on α-satellite sequences. J Biol Chem 280: 41609–41618. [DOI] [PubMed] [Google Scholar]
- Tao Y, Jin C, Li X, Qi S, Chu L, Niu L, Yao X, Teng M 2012. The structure of the FANCM-MHF complex reveals physical features for functional assembly. Nat Commun 3: 782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toso A, Winter JR, Garrod AJ, Amaro AC, Meraldi P, McAinsh AD 2009. Kinetochore-generated pushing forces separate centrosomes during bipolar spindle assembly. J Cell Biol 184: 365–372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valente LP, Dehe PM, Klutstein M, Aligianni S, Watt S, Bahler J, Promisel Cooper J 2013. Myb-domain protein Teb1 controls histone levels and centromere assembly in fission yeast. EMBO J 32: 450–460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verreault A, Kaufman PD, Kobayashi R, Stillman B 1996. Nucleosome assembly by a complex of CAF-1 and acetylated histones H3/H4. Cell 87: 95–104. [DOI] [PubMed] [Google Scholar]
- Walkiewicz MP, Dimitriadis EK, Dalal Y 2014. CENP-A octamers do not confer a reduction in nucleosome height by AFM. Nat Struct Mol Biol 21: 2–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Liu X, Dou Z, Chen L, Jiang H, Fu C, Fu G, Liu D, Zhang J, Zhu T, et al. 2014. Mitotic regulator Mis18β interacts with and specifies the centromeric assembly of molecular chaperone Holliday junction recognition protein (HJURP). J Biol Chem 289: 8326–8336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warburton PE 2004. Chromosomal dynamics of human neocentromere formation. Chromosome Res 12: 617–626. [DOI] [PubMed] [Google Scholar]
- Warburton PE, Cooke CA, Bourassa S, Vafa O, Sullivan BA, Stetten G, Gimelli G, Warburton D, Tyler-Smith C, Sullivan KF, et al. 1997. Immunolocalization of CENP-A suggests a distinct nucleosome structure at the inner kinetochore plate of active centromeres. Curr Biol 7: 901–904. [DOI] [PubMed] [Google Scholar]
- White EA, Glotzer M 2012. Centralspindlin: At the heart of cytokinesis. Cytoskeleton 69: 882–892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams JS, Hayashi T, Yanagida M, Russell P 2009. Fission yeast Scm3 mediates stable assembly of Cnp1/CENP-A into centromeric chromatin. Mol Cell 33: 287–298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zasadzinska E, Barnhart-Dailey MC, Kuich PH, Foltz DR 2013. Dimerization of the CENP-A assembly factor HJURP is required for centromeric nucleosome deposition. EMBO J 32: 2113–2124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeitlin SG, Shelby RD, Sullivan KF 2001. CENP-A is phosphorylated by Aurora B kinase and plays an unexpected role in completion of cytokinesis. J Cell Biol 155: 1147–1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Colmenares SU, Karpen GH 2012. Assembly of Drosophila centromeric nucleosomes requires CID dimerization. Mol Cell 45: 263–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Q, Saro D, Sachpatzidis A, Singh TR, Schlingman D, Zheng XF, Mack A, Tsai MS, Mochrie S, Regan L, et al. 2014. The MHF complex senses branched DNA by binding a pair of crossover DNA duplexes. Nat Commun 5: 2987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Z, Feng H, Zhou BR, Ghirlando R, Hu K, Zwolak A, Miller Jenkins LM, Xiao H, Tjandra N, Wu C, et al. 2011. Structural basis for recognition of centromere histone variant CenH3 by the chaperone Scm3. Nature 472: 234–237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zinkowski RP, Meyne J, Brinkley BR 1991. The centromere-kinetochore complex: A repeat subunit model. J Cell Biol 113: 1091–1110. [DOI] [PMC free article] [PubMed] [Google Scholar]