Abstract
Sequencing and analyses of 16S rRNA gene amplicons were performed to estimate the composition of the rumen methanogen community in 252 samples from eight cohorts of sheep and cattle, separated into 16 different sample groups by diet, and to determine which methanogens are most prominent in the rumens of farmed New Zealand ruminants. Methanobacteriales (relative abundance ± standard deviation, 89.6% ± 9.8%) and Methanomassiliicoccales (10.4% ± 9.8%) were the two major orders and contributed 99.98% (±0.1%) to the rumen methanogen communities in the samples. Sequences from Methanobacteriales were almost entirely from only four different species (or clades of very closely related species). Each was detectable in at least 89% of the samples. These four species or clades were the Methanobrevibacter gottschalkii clade and Methanobrevibacter ruminantium clade with a mean abundance of 42.4% (±19.5% standard deviation) and 32.9% (±18.8%), respectively, and Methanosphaera sp. ISO3-F5 (8.2% ± 6.7%) and Methanosphaera sp. group5 (5.6% ± 5.7%). These four species or clades appeared to be primarily represented by only one or, in one case, two dominant sequence types per species or clade when the sequences were grouped into operational taxonomic units (OTUs) at 99% sequence identity. The mean relative abundance of Methanomassiliicoccales in the samples was relatively low but exceeded 40% in some of the treatment groups. Animal feed affected the apparent methanogen community structure of both orders, as evident from differences in relative abundances of the major OTUs in animals under different feeding regimens.
INTRODUCTION
Methane is the second most significant anthropogenic greenhouse gas, after carbon dioxide, and its concentration in the atmosphere is increasing (1). One major contributor to this is an intensification and expansion of global agricultural activities, in particular increased ruminant livestock numbers. Ruminants, such as cattle, sheep, and domesticated deer, produce large amounts of methane via enteric fermentation and are the most numerous farm animals in New Zealand. It is estimated that methane from these animals accounts for almost 30% of New Zealand's total anthropogenic greenhouse gas emissions (2).
Methane is produced in the rumen by methanogenic archaea, here referred to as methanogens. Methanogens comprise a phylogenetically diverse group of microorganisms that are present in the rumen at up to ∼1010 cells/g rumen contents (3). Previous analyses have shown that members of the orders Methanobacteriales and Methanomassiliicoccales are dominant in the rumens of domesticated ruminants in New Zealand and many other geographic locations around the world (4). The order name Methanomassiliicoccales has only recently been proposed (5). These methanogens are closely related to the Thermoplasmatales (6) and were previously known as rice cluster III (RC-III) archaea (7), rumen cluster C (RCC) (4), or Methanoplasmatales (8). Within the order Methanobacteriales, hydrogenotrophic methanogens of the genus Methanobrevibacter appear to be most abundant in the rumen (4). Species of the second most abundant genus in the order Methanobacteriales, Methanosphaera, have been characterized as methylotrophs (9, 10). Little is known about substrate requirements of members of the order Methanomassiliicoccales, but evidence is accumulating that some members of this order may grow methylotrophically as well (5, 11–13).
Altogether, at least 40 different strains of methanogens have been isolated from the rumen or other intestinal environments, representing a range of species. Cultivation-independent studies suggest that other potentially important species are yet to be cultured and characterized. Assessing the diversity by analyzing the rumen microbiota from a large number of animals has become feasible with the development of next-generation sequencing techniques, bioinformatics tools for the analysis of such data, and the recent development of a highly resolving taxonomic framework for rumen and intestinal methanogens (14). The identification of dominant rumen methanogens and an understanding of rumen methanogen population structure and its variations are the first steps toward the generation of targeted collections of genome sequences (15). These, in turn, are an important basis for the development of methane mitigation tools, such as vaccines and small-molecule inhibitors, as they enable targeted development of antigens in vaccines and the design of inhibitory compounds that target rumen-inhabiting methanogens.
In this study, the rumen methanogen microbiota from eight cohorts of sheep and cattle was analyzed using high-throughput 454 Titanium sequencing. The animals were maintained under different farming conditions and stemmed from different geographic locations around the country. These analyses provide insight into the composition and diversity of the methanogen component of the New Zealand rumen microbiota at unprecedented depth, define the key members of rumen methanogen communities, and identify their closest cultivated relatives.
MATERIALS AND METHODS
Collection of samples from ruminant animals and DNA extraction.
The use of animals, including welfare, husbandry, experimental procedures, and collection of the rumen samples used for this study, was approved by the AgResearch Grasslands and Ruakura Animal Ethics Committees and complied with the institutional Codes of Ethical Conduct for the Use of Animals in Research, Testing and Teaching, as prescribed in the Animal Welfare Act of 1999 and its amendments. Unless indicated otherwise, animals were fed with pasture consisting predominantly of perennial rye grass (Lolium perenne) and white clover (Trifolium repens). Details of the sample groups are given in Table 1, and accession numbers of the sequencing data sets are given in Table S1 in the supplemental material. All animals were sampled via oral stomach tubing to minimize bias in the community structure due to different sampling methods (16). Data sets analyzed in this study have been used previously as a test data set for taxonomic assignments of V6-to-V8 amplicon sequencing reads with RIM-DB, a taxonomic framework for methanogens from the rumen and other intestinal environments (14).
TABLE 1.
Detailed information on host species, number of animals, sampling points, and feed for the animals sampleda
| Animal cohort | Sample set | Ruminant species | No. of animals sampled | Sampling date | Feed | Location (New Zealand) |
|---|---|---|---|---|---|---|
| ARC | ARC | Cow | 38 | October 2010 | Pasture | Hawera, Taranaki |
| ARS | P1.ARS | Sheep | 8 | December 2010 | Pasture | Aorangi, Manawatu |
| P2.ARS | 8 | February 2011 | Pelleted lucerne | Palmerston North, Manawatu | ||
| P3.ARS | 8 | April 2011 | Pasture | Aorangi, Manawatu | ||
| BRA | BRA | Sheep | 10 | June 2011 | Pasture | Palmerston North, Manawatu |
| CGP | G1.CGP | Sheep | 19 | June 2010 | Pasture | Palmerston North, Manawatu |
| G2.CGP | 18 | July 2010 | Wheat grain-lucerne hay pellets | Palmerston North, Manawatu | ||
| DAG | P1.DAG | Cow | 15 | April 2010 | Pasture | Hamilton, Waikato |
| P2.DAG | 16 | September 2010 | Pasture | Hamilton, Waikato | ||
| P3.DAG | 16 | October 2010 | Pasture | Hamilton, Waikato | ||
| EAR | P1.EAR | Cow | 25 | November 2010 | Pasture | Palmerston North, Manawatu |
| P2.EAR | 23 | March 2011 | Pasture | Takapau, Hawkes Bay | ||
| RAP | RAP | Sheep | 18 | August 2011 | Pasture | Palmerston North, Manawatu |
| VAC | P1.VAC | Sheep | 11 | May 2012 | Lucerne chaffage | Palmerston North, Manawatu |
| P2.VAC | 8 | July 2012 | Pasture | Aorangi, Manawatu | ||
| P3.VAC | 11 | August 2012 | Lucerne chaffage | Palmerston North, Manawatu |
A total of 252 samples from the eight different cohorts were analyzed. Depending on age, diet, or sampling time point, the cohorts were further divided into sample sets (between one and three sample sets per cohort).
DNA extraction and amplification of target genes and pyrosequencing of amplicons.
DNA was extracted from 30 mg of homogenized, freeze-dried, and ground rumen sample as described by Rius et al. (17). Amplicons of ∼492 bp in length, spanning variable regions V6 to V8 of the 16S rRNA gene, were generated as described previously (18) using archaeon-specific primers Ar915aF (5′-CCATCTCATCCCTGCGTGTCTCCGACTCAGTCAGGAATTGGCGGGGGAGCAC-3′), which is preceded by 454 Titanium adapter A (underlined) and a two-base linker (bold italic), and Ar1386R (5′-CCTATCCCCTGTGTGCCTTGGCAGTCTCAGNNNNNNNNNNNNCAGCGGTGTGTGCAAGGAGC-3′), which is preceded by 454 Titanium adapter B (underlined); a unique, error-correcting 12-mer barcode (indicated as NNNNNNNNNNNN); and a two-base linker (bold italic).
PCR products were then processed for sequencing as described previously (18).
Amplicon sequence data analyses.
Data were processed using the QIIME package, version 1.8 (19). Reads were quality filtered and assigned to the corresponding samples by barcodes using the split_library.py script with the options “-r -z truncate_remove -l 350 -L 1000.” For error correction, the split_library.py script outputs of all experiments were concatenated and processed together with concatenated quality (“qual”) files, using Acacia with default settings (20). Error-corrected sequence reads were chimera checked with the QIIME script parallel_identify_chimeric_seqs.py, using the parameters –d 4 and –n 2 and RIM-DB as reference set (14). Sequences identified as chimeras were removed from the sequence data using the QIIME filter_fasta.py script. Clustering of operational taxonomic units (OTUs) was done by pick_otus.py using the default clustering method UCLUST (21). The similarity threshold used for clustering of amplicon reads was 0.99 unless noted otherwise. Taxonomic assignment of representative sequences for each identified OTU was done using the parallel_assign_taxonomy_blast.py script with RIM-DB as reference database for taxonomic assignment. OTU tables were generated using the make_otu_table.py script. For the analysis of non-error-corrected sequence data, the error correction step was omitted.
Shannon diversity indices were calculated according to the work of Shannon (22), and Pielou's evenness index was calculated according to the work of Pielou (23). Mean relative abundances of OTUs per sample group were used for hierarchical clustering of samples. Clustering was performed using ‘complete linkage’ and the hclust function of the stats package in R (24). Statistical significance of clusters was determined by using pvclust (25).
Alignment, tree construction, and taxonomic assignment.
16S rRNA gene sequences were aligned to the SILVA database version 111 using the SINA aligner with default settings (26). Sequences were then imported into ARB (27), and the alignment was checked manually and curated when necessary. Aligned sequences were exported in PHYLIP format to construct phylogenetic trees using the maximum likelihood method in RAxML version 7.0.3 (28). The parameters “-m GTRGAMMA -# 500 -f a -x 2 -p 2” were used.
RESULTS
Diversity of methanogenic archaea in rumen samples from New Zealand.
A range of rumen samples was analyzed to identify the major archaeal representatives of the rumen microbiota in New Zealand ruminants. A total of 703 OTUs were identified, which were present with at least two reads in the 252 samples analyzed (using error-corrected sequences clustered at a similarity threshold of 99%, rarefied to 190 reads per sample) in the 16 different sample groups from 8 cohorts of animals (Table 1 has details). The analysis revealed that, for every sample group, the microbiota was dominated by only two orders, the Methanobacteriales (mean relative abundance ± standard deviation, 89.6% ± 9.8%) and the Methanomassiliicoccales (10.4% ± 9.8%). Differences in diversity and evenness between orders were determined by a species-level-based calculation of the Shannon index and Pielou's evenness index. Sequences assigned to the order Methanobacteriales were slightly more diverse (Shannon index, 0.96 ± 0.23) and more even (Pielou's evenness index, 0.65 ± 0.14) than sequences assigned to the order Methanomassiliicoccales (Shannon index, 0.77 ± 0.46; Pielou's evenness index, 0.61 ± 0.32). Considerable differences in the indices existed between sample groups (see Fig. S1 in the supplemental material).
Identification and phylogenetic analysis of dominant species in Methanobacteriales.
Nearly all Methanobacteriales sequences (89.1% ± 9.8% of all reads) were assignable to only four different species (or clades of very closely related species that cannot be distinguished even with almost-full-length 16S rRNA gene sequences [14]). The two most abundant were the Methanobrevibacter gottschalkii clade and the Methanobrevibacter ruminantium clade, with mean relative abundances of 42.4% (±19.5%) and 32.9% (±18.8%), respectively. Both clades were highly prevalent and were detected in 99.1% and 100% of all 252 samples, respectively. Less abundant, but also highly prevalent in the samples, were two Methanosphaera species, Methanosphaera sp. ISO3-F5 (mean relative abundance, 8.2% ± 6.7%; prevalence, 94.8%) and Methanosphaera sp. group5 (5.5% ± 5.7%; prevalence, 91.3%). The latter species has recently been defined (14) and does not yet have an identified cultured representative.
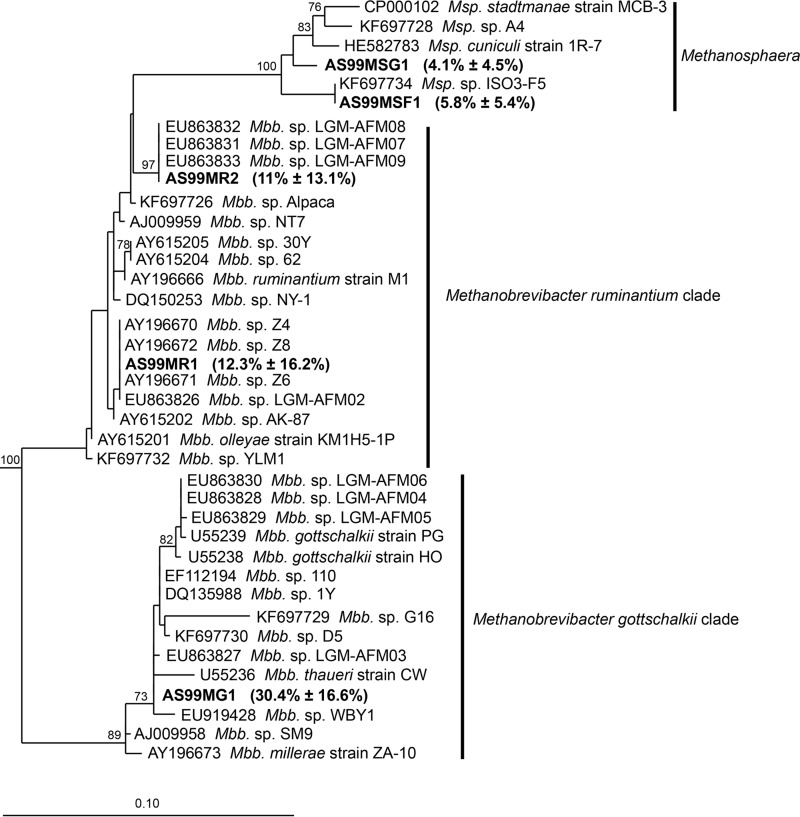
The presence of only four major species or clades of Methanobacteriales prompted us to analyze the diversity of sequence types within each of these species. Rank abundance curves for operational taxonomic units (OTUs) were constructed from error-corrected sequences and a sequence similarity threshold of 99%. These revealed a high relative abundance of only one or two of the OTUs within each of the four major Methanobacteriales species or clades (see Fig. S2 in the supplemental material). In total, five different OTUs were identified that together comprised 63.6% (±11.3%) of the total relative abundance. Among these OTUs were one Methanobrevibacter gottschalkii clade OTU (30.4% ± 16.6%; prevalence, 100%), two Methanobrevibacter ruminantium clade OTUs (12.3% ± 16.2%; prevalence, 81.8%; and 11% ± 13.1%; prevalence, 87.3%), one Methanosphaera sp. ISO3-F5 OTU (5.8% ± 5.4%; prevalence, 91.7%) and one Methanosphaera sp. group5 OTU (4.1% ± 4.5%; prevalence, 84.1%). Representative sequences of these five most abundant Methanobacteriales OTUs and the corresponding region (V6 to V8) of 16S rRNA gene sequences of 34 Methanobacteriales isolates and enrichment cultures were used to construct a phylogenetic tree and to identify the closest isolates (Fig. 1; see Table S2 in the supplemental material for similarities between sequences from the major OTUs and Methanobacteriales isolates). This analysis revealed several isolate sequences that were highly similar or identical to the most abundant Methanobrevibacter gottschalkii and Methanobrevibacter ruminantium clade OTU sequences.
FIG 1.
Phylogeny of Methanobacteriales isolates and highly abundant Methanobacteriales OTUs. Representative sequences of highly abundant OTUs (AS99MG1, AS99MR1, AS99MR2, AS99MSF1, and AS99MSG1) were aligned to the ARB database, version 111, using SINA-aligner (26). Phylogenetic analysis was performed on the V6-to-V8 region of the 16S rRNA gene of the aligned OTUs and 34 Methanobacteriales strains and enrichment cultures from intestinal environments. The tree was resampled 500 times, and only bootstrap values of ≥70% are shown. The dendrogram was rooted with five Methanopyrus sequences. The scale bar indicates 0.10 inferred nucleotide substitutions per position. Mbb., Methanobrevibacter; Msp., Methanosphaera.
The most abundant Methanobrevibacter gottschalkii OTU, AS99MG1, shared high sequence similarity (>99.5%) with three strains, Methanobrevibacter sp. strain 1Y (DQ135988; 99.78%), isolated from the rumen of an Australian sheep; Methanobrevibacter sp. strain 110 (EF112194; 99.78%), isolated from the rumen of a New Zealand calf on pasture; and Methanobrevibacter sp. strain LGM-AFM03 (EU863827; 99.76%), isolated from feces of a Chinese buffalo. Other cultured close relatives from the rumen were Methanobrevibacter sp. strain D5 (KF697730) and Methanobrevibacter sp. strain G16 (KF697729). OTU AS99MG1 was also identical to partial sequences of Methanobrevibacter strains YE300 (GQ906572), YE301 (GQ906575), YE302 (GQ906573), YE303 (GQ906574), and YE304 (GQ906576), which were isolated from bovine rumen contents in Australia (29). However, it should be noted that these sequences (GQ906572 to GQ906576) have only an ∼130-bp-long overlap (primarily in the V6 region) and may not be identical throughout the full V6-to-V8 region.
Cultured representatives were found that had 100% 16S rRNA gene sequence identity (over the V6-to-V8 region) to both of the highly abundant Methanobrevibacter ruminantium OTUs, referred to as AS99MR1 and AS99MR2. For AS99MR1, these were Methanobrevibacter sp. strains Z4 (AY196670), Z6 (AY196671), and Z8 (AY196672) from bovine rumen contents, and for AS99MR2, these were Methanobrevibacter sp. strains LMG-AFM07 (EU863831), LMG-AFM08 (EU863832), and LMG-AFM09 (EU863833), which were cultured from camel feces (5). Two further partial sequences of isolates had 100% sequence identity with AS99MR2—Methanobrevibacter sp. strains YE286 (GQ906569) and YE287 (GQ906570), from Australian bovine rumen contents—although these sequences were short and the overlap of available sequence information was only over ∼130 bp (primarily in the V6 region).
Of the two dominant Methanosphaera OTUs identified, AS99MSF1 and AS99MSG1, only AS99MSF1 had a 100% match to a sequence from an isolate, Methanosphaera sp. strain ISO3-F5 (KF697734), which was isolated from a New Zealand sheep rumen. OTU AS99MSG1 was not identical to any isolate sequences but was most similar to gene sequences from Methanosphaera sp. strain ISO3-F5 (97.3%) and Methanosphaera sp. strain A4 (97.1%; KF697728). This group, containing AS99MSG1, is potentially a species without cultured isolates.
The OTU-level analyses were extended by reanalyzing the data using different cutoffs for sequence clustering (99% and 100%, respectively) prior to and after error correction of sequence reads. This was performed to determine if these high-abundance OTUs could be observed regardless of the type of data processing or whether they were technical artifacts. Each of the four different methods resulted in construction of the same five highly abundant OTUs, and the representative sequences from these shared 100% sequence similarity regardless of the clustering method used. The sum of the abundance of the five dominant OTUs varied between 17.9% ± 10.3% (reads not error corrected, 100% similarity for sequence clustering) and 63.6% ± 11.3% (reads error corrected, 99% similarity for sequence clustering) of the total relative abundance. More detailed results from the different processing methods are given in Fig. S3 and Table S3 in the supplemental material.
Identification and phylogenetic analysis of dominant OTUs in Methanomassiliicoccales.
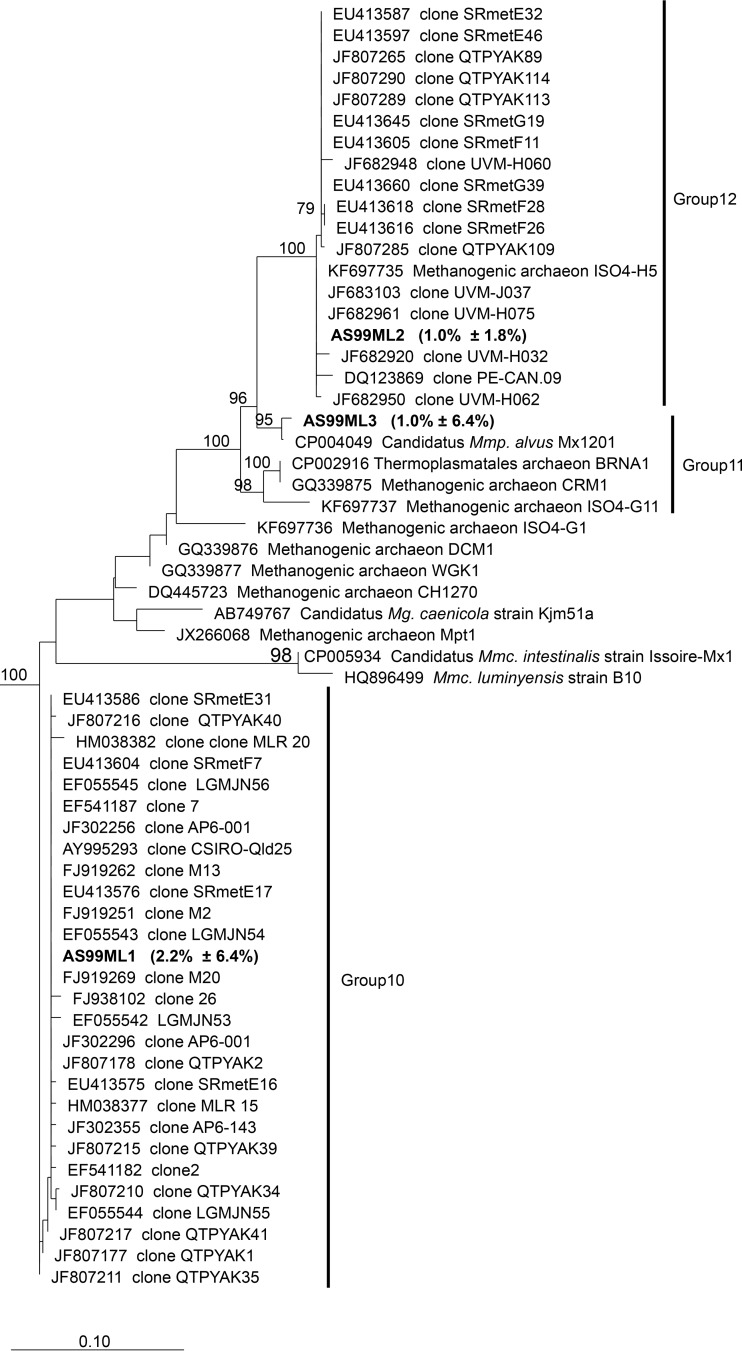
Members of the order Methanomassiliicoccales made up a smaller proportion of the methanogen community (mean relative abundance, 10.2% ± 9.8%; prevalence, 96.0%). The three most abundant Methanomassiliicoccales OTUs contributed 4.2% (±6.8%) to the relative abundance of the methanogen community. A phylogenetic analysis on representative sequences from the most dominant Methanomassiliicoccales OTUs was performed and identified several closely related sequences, mostly of uncultivated representatives (Fig. 2; see also Table S4 in the supplemental material). OTU AS99ML1 had the highest mean abundance (2.2% ± 3.0%; prevalence, 71.0%), and fell into Methanomassiliicoccales group10 (14), which currently contains no cultured representatives. However, identical OTUs have been detected in cultivation-independent studies analyzing a variety of different host species from different geographic locations, e.g., Chinese sheep (FJ919251, FJ919262, and FJ919269), Chinese cattle (EF541187, EF055545, and EF055543), alpaca in the United States (JF302296 and JF302256), Chinese yak (JF807178), Norwegian reindeer (EU413604, EU413586, and EU413576), and Australian sheep (AY995293). The two next most abundant OTUs, which contributed each about 1% to the total methanogen relative abundance, were OTUs AS99ML2 (1.0 ± 1.8%; prevalence, 53.2%) and AS99ML3 (1.0% ± 6.4%; prevalence, 5.5%). The sequence from OTU AS99ML2 fell into Methanomassiliicoccales group12 (14). It was identical to the cultured rumen Methanomassiliicoccales sp. strain ISO4-H5 (KF697735) and to two sequences from uncultured archaea in the bovine rumen (JF682961 and JF683103). The sequence from OTU AS99ML3 in Methanomassiliicoccales group11 (14) was most similar to the 16S rRNA gene sequence of “Candidatus Methanomethylophilus alvus” Mx1201, an isolate from the human gut (98.9% sequence identity; see Table S4) (30). It is noteworthy that OTU AS99ML3 was detected in only one cohort (CGP), where it reached a particularly high abundance (15.6% ± 20.7%; maximal relative abundance, 68.4% in one sample) in sheep fed a 2:3 concentrate/forage ratio (wheat grain/lucerne hay; fresh basis) pelleted diet. When the same animals were fed on pasture, they harbored only low counts of this OTU (0.03% ± 0.12%).
FIG 2.
Phylogeny of Methanomassiliicoccales sequences and highly abundant Methanomassiliicoccales OTUs. Representative sequences of highly abundant OTUs (AS99ML1, AS99ML2, and AS99ML3) were aligned to the ARB database, version 111, using SINA-aligner (26). Fifty-seven sequences of group10, group11, and group12 and of isolates and enrichment cultures were selected in RIM-DB (14). Phylogenetic analysis was performed on the V6-to-V8 region of the 16S rRNA gene of the aligned OTUs and selected database sequences. The tree was resampled 500 times, and only bootstrap values of ≥70% are shown. The dendrogram was rooted with five Methanopyrus sequences. The scale bar indicates 0.10 inferred nucleotide substitutions per position. Mmc., Methanomassiliicoccus; Mmp., Methanomethylophilus; Mg., Methanogranum.
Other orders of archaea.
Methanobacteriales and Methanomassiliicoccales constituted the largest proportion of the New Zealand rumen methanogen community, and sequences from other orders were detected at only very low relative abundances (Methanosarcinales, 0.02% ± 0.1%; prevalence, 2.8%; Methanopyrales, 0.004% ± 0.05%; prevalence, 0.4%; and Methanomicrobiales, 0.002% ± 0.03%; prevalence, 0.4%). The sequences of reads classified in the order Methanosarcinales were assigned to Methanimicrococcus blatticola, reads in the order Methanopyrales were assigned to Methanopyrus kandleri, and reads in the order Methanomicrobiales were assigned to Methanocorpusculum sp. The amplification primers used in this study were searched for mismatches to the primer binding sites, to analyze if a primer bias could cause over- or underrepresentation of some methanogen orders. The analysis revealed that the numbers of mismatches were comparable across all different orders (see Table S5 in the supplemental material), and no conserved mismatches were identified.
Factors influencing methanogen community structure.
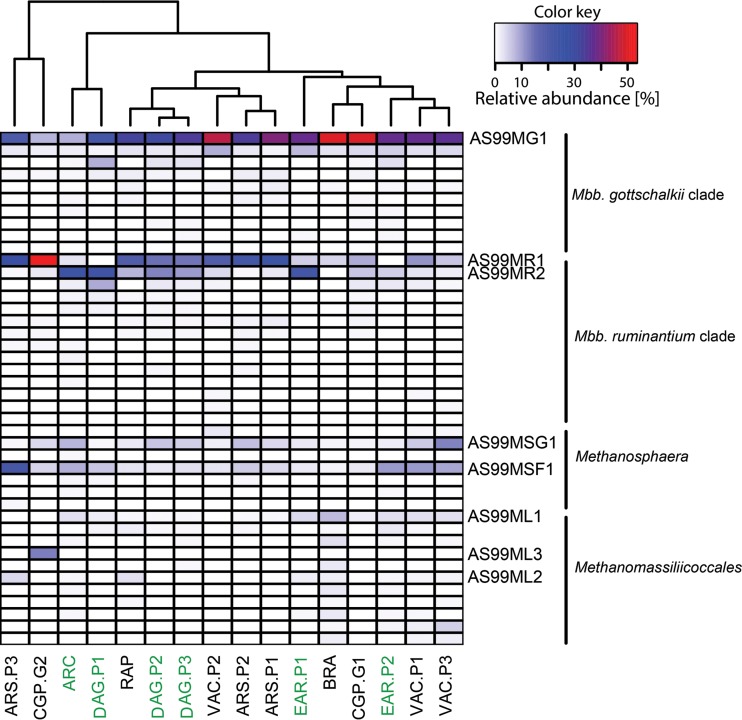
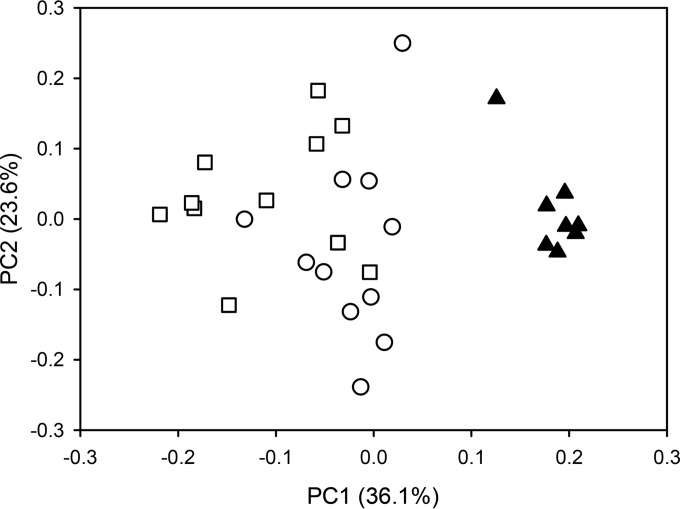
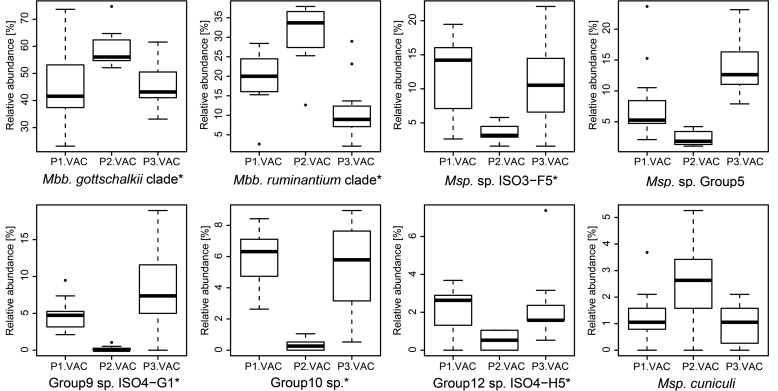
The relative abundance of the methanogen OTUs varied considerably across all samples. Hierarchical clustering of the different sample groups was performed to determine if common factors, such as host species or diet, shaped the composition of the methanogen community. Hierarchical clustering resulted in three major clusters that were supported by high approximate unbiased P values (P > 0.95, calculated with pvclust). However, none of these clusters contained sample groups of animals that were exclusively fed the same diet or that came from the same host species, indicating that neither of these factors strongly affected the community structure across all sample groups (Fig. 3). Effects of diet were mainly visible within cohorts of animals that were split up into different sample groups and fed different diets at different times. For example, animals of the VAC cohort that were maintained either on pasture or on lucerne chaffage were clearly separated by the clustering. Animals of this cohort were maintained on a lucerne chaffage feed in a first period, switched to pasture in a second period, and returned to lucerne chaffage in a final, third period (Table 1). The methanogen community changed when the animals were changed to a pasture diet but changed back again when they were fed lucerne chaffage again (Fig. 4). A one-way analysis of variance (ANOVA) followed by a Tukey honestly significant difference (HSD) post hoc test was used to determine which species-level OTUs are responsible for these diet-dependent changes. It revealed statistically significant differences (P < 0.05) for six of the eight most abundant species-level OTUs (defined as >1.5% relative abundance in any sample of this trial) in the sample groups (Fig. 5). Species-level OTUs of the most highly abundant hydrogenotrophic Methanobrevibacter spp. appeared to be predominant in pasture-fed animals, while methylotrophic Methanosphaera and Methanomassiliicoccales OTUs were highly abundant in lucerne-fed animals.
FIG 3.
Hierarchical clustering of sample groups based on the mean of the relative abundance of each OTU in each sample group. Samples were rarefied to 190 reads, and only OTUs with a mean relative abundance of at least 0.02 were included. These OTUs were 10 Methanobrevibacter gottschalkii clade OTUs, 14 Methanobrevibacter ruminantium clade OTUs, seven Methanosphaera OTUs, and 11 Methanomassiliicoccales OTUs. Names of animal cohorts are shown below the heat map, with the color of the font indicating the host species (green = cow, black = sheep). Heat map colors represent relative abundance of OTUs (in percent).
FIG 4.
Effects of diet on methanogen community structure. A principal coordinate analysis plot of Bray-Curtis dissimilarities is shown. The open circles represent samples taken in period 1 (sample group P1.VAC), when sheep were fed lucerne chaffage; the closed triangles represent samples from the same sheep taken in period 2 (P2.VAC), when they were fed pasture; and the open squares represent samples from these animals in period 3 (P3.VAC), when they had been fed lucerne chaffage again. Samples were rarefied to 190 reads, and calculations of Bray-Curtis-dissimilarities were performed on OTU tables where OTUs had had been summarized to the species level.
FIG 5.
Effect of diet on the relative abundance of methanogens. Shown are changes in the relative abundance of methanogens during serial treatments with different feeds (P1.VAC, lucerne chaffage; P2.VAC, pasture; P3.VAC, lucerne chaffage). Asterisks after the species names indicate species that are statistically significantly different between P2.VAC and the two other periods, determined by one-way ANOVA followed by a Tukey HSD post hoc test (P < 0.05).
A less pronounced separation of communities was observed for animals of the DAG cohort, which indicated a seasonal effect (see Fig. S4 in the supplemental material). These animals were sampled in April, September, and October. Hierarchical clustering and principal coordinate analysis based on Bray-Curtis dissimilarities indicated that communities of samples taken in September and October were more similar to each other than to the samples taken in April (Fig. 3; see also Fig. S4A). One-way ANOVA followed by a Tukey HSD post hoc test showed that the relative abundance of two Methanosphaera species differed significantly (P < 0.05) between the April and the September and October sampling points, while no other significant differences could be determined (Fig. S4B). The abundance of Methanosphaera sp. ISO3-F5 was highest in April and low in September and October, while Methanosphaera sp. group5 was lowest in April and highest in September and October (Fig. S4B). There is currently no cultured representative for Methanosphaera group5, but this study suggests that Methanosphaera group5 occupies a niche that is distinct from that of other known Methanosphaera isolates.
Overall, the results of this study suggest that the microbiota composition of animals from different cohorts grazing on pasture can be as dissimilar to each other as to animals fed a completely different diet.
DISCUSSION
The aim of this study was to obtain an insight into the diversity of New Zealand rumen methanogens, determine if there were common or abundant taxa, and see how variable the composition of the methanogen community structure is.
Core methanogen microbiota in New Zealand ruminants.
The rumen archaeal component of the New Zealand ruminant microbiota is dominated by members of the order Methanobacteriales (predominately two Methanobrevibacter spp. and two Methanosphaera spp.) and Methanomassiliicoccales. These results are consistent with results from previous, smaller-scale studies on methanogens in New Zealand ruminants (18, 31). In this more comprehensive analysis of New Zealand rumen methanogens, members of the order Methanomicrobiales were detected at only very low abundances. This finding is in accordance with a number of studies that analyzed rumen methanogens in different locations around the world (4) but contrasts with findings from studies carried out on some Korean and Japanese cows (6, 32) and Indian water buffaloes (33), where these organisms were found to be major constituents of rumen methanogen communities. Whether these different observations are due to differences in community structure or the consequence of technical artifacts remains to be shown, but it is difficult to compare studies that have used different methodologies for data generation. For example, the choice of specific amplification primers may introduce a bias toward some organism groups and may discriminate against others (both by primer sequence and by target region of the 16S rRNA gene for amplification) (34, 35). Thus far, studies investigating primer bias have not been performed on rumen methanogen mock communities of known composition. These types of analyses would allow quantitative estimates of the biases and would identify suitable methods to correct these. Mismatches of amplification primers do, however, indicate that bias against specific groups of methanogens can be expected, as has been recently shown by Tymensen and McAllister (35). Our own analysis of the primers that we used against a comprehensive curated database of methanogen 16S rRNA gene sequences revealed very few mismatches. The mismatches that were identified did not indicate any systematic exclusion of specific taxa, and the few mismatches that were found appeared to be randomly distributed throughout the primer binding site.
OTU-level analysis of methanogens in New Zealand ruminants.
One of the major findings of this study was that a large proportion of the rumen methanogen community was represented by just five OTUs out of a total of 703. Community structure analysis at the OTU level has to be interpreted carefully, as sequencing errors and sequence similarity thresholds may affect the construction and abundance of OTUs. Clustering of sequence reads at maximal similarity thresholds and the clustering of erroneous sequence reads will artificially inflate the number of OTUs, while error correction or denoising of sequencing reads and the use of low sequence similarity thresholds for clustering of sequence reads may have the opposite effect. Therefore, a comparison of the abundance of OTUs before and after error correction of sequencing reads and by using similarity thresholds of 99% and 100% for sequence clustering was performed. Even when using uncorrected sequence reads and a similarity threshold of 100%, the five abundant OTUs still accounted for 17.9% ± 10.3% of the total relative abundance. However, the abundance of the same five OTUs accounted for 63.6% ± 11.3% after error correction of sequence reads and with a similarity threshold of 99%. Both results indicate that the five analyzed OTUs contributed a considerable proportion to the community and that it is not an artifact of data processing, but the current technical limitations do not allow an exact determination of their abundance.
The limited species and OTU-level diversity has environmental and economic implications, as it strongly encourages the ongoing efforts to develop mitigation technologies targeting ruminant methane. It will still be necessary to determine if these highly abundant strains are clonal or if significant genomic diversity is misleadingly masked by identical regions of the 16S rRNA gene. However, the low diversity means that initial efforts to develop methane mitigation technologies based on small-molecule inhibitors and vaccines that target a small amount of diversity could inhibit a large part of the methanogens in the community. The dominance of a few methanogens in all sample groups suggests that they are the best adapted at filling their niches, even when feeds are changed. A concern is that eliminating these dominant taxa will result in the elaboration of minor populations to fill the vacated niche space. It can be assumed, however, that those species that might take over are less well adapted and may be even easier to eliminate with further refinement of the technologies, because smaller impacts might make them uncompetitive in the rumen. If the minor components of the community occupy specialized niches, then their proliferation is even less likely.
The most abundant Methanobrevibacter gottschalkii and Methanobrevibacter ruminantium OTUs shared high sequence identity with strains isolated from camel feces (36), which indicates that these OTUs may be widely spread among different host species. Similarly, the most abundant Methanomassiliicoccales OTU identified in this study has also been identified in different host species and in a variety of geographic locations around the world. Currently, few genomes of ruminal methanogen species are publicly available (37–39), which prevents extensive comparative genomic analyses. Future studies may elucidate whether genetic factors influence host specificity of rumen methanogens or if these strains are equally well adapted to the rumen environment regardless of the host species. Large-scale isolation efforts will be required to obtain more cultured representatives of the target species. These novel strains would need to be characterized phenotypically and genomically to gather information about their uniformity. A similar approach has been undertaken for Methanobrevibacter smithii, the most-abundant methanogen of the human gut (40). Hansen et al. isolated 20 different Methanobrevibacter smithii strains from fecal samples of mothers and their mono- and dizygotic twins (41). The full-length 16S rRNA genes of the 20 Methanobrevibacter smithii isolates have a minimum sequence similarity of >99.5% to each other and represent in this respect a situation similar to the high-abundance OTUs in the rumen. Hansen et al. sequenced and analyzed the genomes of the isolates and found a core set of 987 genes and 1,860 variably represented genes (41). The results also revealed a higher genomic similarity of strains that were isolated from members of the same family. The latter observation is of particular interest, if these findings were translatable to herds of livestock that share a highly similar genetic background and that are kept in close proximity to each other. A highly similar or even clonal population of rumen methanogen strains would enable the identification of widely conserved potential targets and facilitate the development of effective vaccines and inhibitors. One drawback of the large-scale isolation efforts may be a biased isolation of strains that have similar phenotypic and genotypic characteristics. The recent advancements in single-cell genomics (42) hold promise to overcome this bias and may provide more significant insights into the genomic diversity of uncultured strains at the inter- and intra-animal level.
Factors influencing methanogen communities in the rumen.
The results of this study have also enhanced our understanding of how host species and diet may affect the diversity of rumen methanogens in New Zealand. The methanogen communities from different host species (sheep or cattle) were not clearly separated by hierarchical clustering, and the most abundant OTUs appeared to be present across all different cohorts, indicating that, in this study, host-specific community types of rumen methanogen strains either may not exist or may not be resolvable at the level of the (partial) 16S rRNA gene across different diets. It is possible that subtle differences may exist and be detectable when animals with different phenotypes or genotypes are fed the same diet under controlled conditions (43, 44).
Our analysis revealed that diet affected the methanogen community structure within a single group of animals. These differences in methanogen community composition are likely to be mediated by changes in the community structure of the other prominent microbial organism groups (bacteria, protozoa, and fungi) in the rumen, as methanogens use the fermentation by-products (e.g., H2, formate, and methanol) of fermenting microorganisms (45). Changes in the profiles of feed fermentation caused by a different diet would therefore induce changes in methanogen community structure, as has been shown previously (13, 46). However, the results did not reveal a strong clustering that would indicate that the methanogen community structure of pasture-fed animals is very different from that of animals fed other feeds. This could have several causes: (i) the rumen environment in animals provided different feed may not differ sufficiently to cause changes in the community structure that would be visible by clustering, (ii) animals grazing on pasture consume a relatively undefined diet that undergoes seasonal changes and may be different for individual animals because of preferential selection of different plants in the pasture, and (iii) the limited range of substrates and niches constrains the potential diversity of methanogens and only the relative proportions of the different substrates control the ultimate abundance within a very small diversity.
In conclusion, the results of this study have shown that the New Zealand ruminants display a surprisingly limited diversity at the 16S rRNA gene level. These findings have significance for the development of effective vaccines or inhibitors to decrease methane emissions from ruminants. Further analyses of the rumen microbial diversity at a global scale are under way (www.globalrumencensus.org.nz) and will allow further comparisons of rumen community types and dominant OTUs.
Supplementary Material
ACKNOWLEDGMENTS
We thank David Pacheco, Cesar Pinares-Patiño, Xuezhao Sun, Garry Waghorn, and Neil Wedlock for facilitating access to rumen samples. We are also grateful to Gemma Henderson, Michelle Kirk, and Faith Cox for generating some of the sequence data sets. Wayne Young and Bill Kelly provided valuable criticisms of the manuscript.
This work was funded under contract by the New Zealand Pastoral Greenhouse Gas Research Consortium (PGgRc) and the New Zealand Agricultural Greenhouse Gas Research Centre (NZAGRC) and by AgResearch Core Funding.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.03018-14.
REFERENCES
- 1.Yusuf RO, Noor ZZ, Abba AH, Hassan MAA, Din MFM. 2012. Methane emission by sectors: a comprehensive review of emission sources and mitigation methods. Renew Sustain Energy Rev 16:5059–5070. doi: 10.1016/j.rser.2012.04.008. [DOI] [Google Scholar]
- 2.Clark H, Kelliher F, Pinares-Patino C. 2011. Reducing CH4 emissions from grazing ruminants in New Zealand: challenges and opportunities. Asian Australas J Anim Sci 24:295–302. doi: 10.5713/ajas.2011.r.04. [DOI] [Google Scholar]
- 3.Joblin KN. 2005. Methanogenic archaea, p 47–53. In Makkar HPS, McSweeney CS (ed), Methods in gut microbial ecology for ruminants. Springer, Dordrecht, The Netherlands. [Google Scholar]
- 4.Janssen PH, Kirs M. 2008. Structure of the archaeal community of the rumen. Appl Environ Microbiol 74:3619–3625. doi: 10.1128/AEM.02812-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Iino T, Tamaki H, Tamazawa S, Ueno Y, Ohkuma M, Suzuki K, Igarashi Y, Haruta S. 2013. Candidatus Methanogranum caenicola: a novel methanogen from the anaerobic digested sludge, and proposal of Methanomassiliicoccaceae fam. nov. and Methanomassiliicoccales ord. nov., for a methanogenic lineage of the class Thermoplasmata. Microbes Environ 28:244–250. doi: 10.1264/jsme2.ME12189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tajima K, Nagamine T, Matsui H, Nakamura M, Aminov RI. 2001. Phylogenetic analysis of archaeal 16S rRNA libraries from the rumen suggests the existence of a novel group of archaea not associated with known methanogens. FEMS Microbiol Lett 200:67–72. doi: 10.1111/j.1574-6968.2001.tb10694.x. [DOI] [PubMed] [Google Scholar]
- 7.Kemnitz D, Kolb S, Conrad R. 2005. Phenotypic characterization of rice cluster III archaea without prior isolation by applying quantitative polymerase chain reaction to an enrichment culture. Environ Microbiol 7:553–565. doi: 10.1111/j.1462-2920.2005.00723.x. [DOI] [PubMed] [Google Scholar]
- 8.Paul K, Nonoh JO, Mikulski L, Brune A. 2012. “Methanoplasmatales,” Thermoplasmatales-related archaea in termite guts and other environments, are the seventh order of methanogens. Appl Environ Microbiol 78:8245–8253. doi: 10.1128/AEM.02193-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fricke WF, Seedorf H, Henne A, Krüer M, Liesegang H, Hedderich R, Gottschalk G, Thauer RK. 2006. The genome sequence of Methanosphaera stadtmanae reveals why this human intestinal archaeon is restricted to methanol and H2 for methane formation and ATP synthesis. J Bacteriol 188:642–658. doi: 10.1128/JB.188.2.642-658.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Miller TL, Wolin MJ. 1985. Methanosphaera stadtmaniae gen. nov., sp. nov.: a species that forms methane by reducing methanol with hydrogen. Arch Microbiol 141:116–122. doi: 10.1007/BF00423270. [DOI] [PubMed] [Google Scholar]
- 11.Borrel G, Harris HM, Parisot N, Gaci N, Tottey W, Mihajlovski A, Deane J, Gribaldo S, Bardot O, Peyretaillade E, Peyret P, O'Toole PW, Brugere JF. 2013. Genome sequence of “Candidatus Methanomassiliicoccus intestinalis” Issoire-Mx1, a third Thermoplasmatales-related methanogenic archaeon from human feces. Genome Announc 1:e00453-13. doi: 10.1128/genomeA.00453-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dridi B, Fardeau ML, Ollivier B, Raoult D, Drancourt M. 2012. Methanomassiliicoccus luminyensis gen. nov., sp. nov., a methanogenic archaeon isolated from human faeces. Int J Syst Evol Microbiol 62:1902–1907. doi: 10.1099/ijs.0.033712-0. [DOI] [PubMed] [Google Scholar]
- 13.Poulsen M, Schwab C, Jensen BB, Engberg RM, Spang A, Canibe N, Højberg O, Milinovich G, Fragner L, Schleper C. 2013. Methylotrophic methanogenic Thermoplasmata implicated in reduced methane emissions from bovine rumen. Nat Commun 4:1428. doi: 10.1038/ncomms2432. [DOI] [PubMed] [Google Scholar]
- 14.Seedorf H, Kittelmann S, Henderson G, Janssen PH. 2014. RIM-DB: a taxonomic framework for community structure analysis of methanogenic archaea from the rumen and other intestinal environments. PeerJ 2:e494. doi: 10.7717/peerj.494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Leahy S, Kelly W, Ronimus R, Wedlock N, Altermann E, Attwood G. 2013. Genome sequencing of rumen bacteria and archaea and its application to methane mitigation strategies. Animal 7:235–243. doi: 10.1017/S1751731113000700. [DOI] [PubMed] [Google Scholar]
- 16.Henderson G, Cox F, Kittelmann S, Miri VH, Zethof M, Noel SJ, Waghorn GC, Janssen PH. 2013. Effect of DNA extraction methods and sampling techniques on the apparent structure of cow and sheep rumen microbial communities. PLoS One 8:e74787. doi: 10.1371/journal.pone.0074787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rius A, Kittelmann S, Macdonald K, Waghorn G, Janssen P, Sikkema E. 2012. Nitrogen metabolism and rumen microbial enumeration in lactating cows with divergent residual feed intake fed high-digestibility pasture. J Dairy Sci 95:5024–5034. doi: 10.3168/jds.2012-5392. [DOI] [PubMed] [Google Scholar]
- 18.Kittelmann S, Seedorf H, Walters WA, Clemente JC, Knight R, Gordon JI, Janssen PH. 2013. Simultaneous amplicon sequencing to explore co-occurrence patterns of bacterial, archaeal and eukaryotic microorganisms in rumen microbial communities. PLoS One 8:e47879. doi: 10.1371/journal.pone.0047879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Pena AG, Goodrich JK, Gordon JI, Huttley GA, Kelley ST, Knights D, Koenig JE, Ley RE, Lozupone CA, McDonald D, Muegge BD, Pirrung M, Reeder J, Sevinsky JR, Turnbaugh PJ, Walters WA, Widmann J, Yatsunenko T, Zaneveld J, Knight R. 2010. QIIME allows analysis of high-throughput community sequencing data. Nat Methods 7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bragg L, Stone G, Imelfort M, Hugenholtz P, Tyson GW. 2012. Fast, accurate error-correction of amplicon pyrosequences using Acacia. Nat Methods 9:425–426. doi: 10.1038/nmeth.1990. [DOI] [PubMed] [Google Scholar]
- 21.Edgar RC. 2010. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26:2460–2461. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- 22.Shannon CE. 1948. A mathematical theory of communication. Bell Syst Tech J 27:379–423. doi: 10.1002/j.1538-7305.1948.tb01338.x. [DOI] [Google Scholar]
- 23.Pielou E. 1966. The measurement of diversity in different types of biological collections. J Theor Biol 13:131–144. doi: 10.1016/0022-5193(66)90013-0. [DOI] [Google Scholar]
- 24.R Development Core Team. 2011. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. [Google Scholar]
- 25.Suzuki R, Shimodaira H. 2006. Pvclust: an R package for assessing the uncertainty in hierarchical clustering. Bioinformatics 22:1540–1542. doi: 10.1093/bioinformatics/btl117. [DOI] [PubMed] [Google Scholar]
- 26.Pruesse E, Peplies J, Glöckner FO. 2012. SINA: accurate high-throughput multiple sequence alignment of ribosomal RNA genes. Bioinformatics 28:1823–1829. doi: 10.1093/bioinformatics/bts252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ludwig W, Strunk O, Westram R, Richter L, Meier H, Buchner A, Lai T, Steppi S, Jobb G, Förster W. 2004. ARB: a software environment for sequence data. Nucleic Acids Res 32:1363–1371. doi: 10.1093/nar/gkh293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stamatakis A. 2006. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22:2688–2690. doi: 10.1093/bioinformatics/btl446. [DOI] [PubMed] [Google Scholar]
- 29.Gilbert RA, Ouwerkerk D, Zhang LH, Klieve AV. 2010. In vitro detection and primary cultivation of bacteria producing materials inhibitory to ruminal methanogens. J Microbiol Methods 80:217–218. doi: 10.1016/j.mimet.2009.12.004. [DOI] [PubMed] [Google Scholar]
- 30.Borrel G, Harris HM, Tottey W, Mihajlovski A, Parisot N, Peyretaillade E, Peyret P, Gribaldo S, O'Toole PW, Brugère J-F. 2012. Genome sequence of “Candidatus Methanomethylophilus alvus” Mx1201, a methanogenic archaeon from the human gut belonging to a seventh order of methanogens. J Bacteriol 194:6944–6945. doi: 10.1128/JB.01867-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jeyanathan J, Kirs M, Ronimus RS, Hoskin SO, Janssen PH. 2011. Methanogen community structure in the rumens of farmed sheep, cattle and red deer fed different diets. FEMS Microbiol Ecol 76:311–326. doi: 10.1111/j.1574-6941.2011.01056.x. [DOI] [PubMed] [Google Scholar]
- 32.Shin EC, Choi BR, Lim WJ, Hong SY, An CL, Cho KM, Kim YK, An JM, Kang JM, Lee SS. 2004. Phylogenetic analysis of archaea in three fractions of cow rumen based on the 16S rDNA sequence. Anaerobe 10:313–319. doi: 10.1016/j.anaerobe.2004.08.002. [DOI] [PubMed] [Google Scholar]
- 33.Chaudhary P, Sirohi S. 2009. Dominance of Methanomicrobium phylotype in methanogen population present in Murrah buffaloes (Bubalus bubalis). Lett Appl Microbiol 49:274–277. doi: 10.1111/j.1472-765X.2009.02654.x. [DOI] [PubMed] [Google Scholar]
- 34.Kim M, Morrison M, Yu Z. 2011. Evaluation of different partial 16S rRNA gene sequence regions for phylogenetic analysis of microbiomes. J Microbiol Methods 84:81–87. doi: 10.1016/j.mimet.2010.10.020. [DOI] [PubMed] [Google Scholar]
- 35.Tymensen LD, McAllister TA. 2012. Community structure analysis of methanogens associated with rumen protozoa reveals bias in universal archaeal primers. Appl Environ Microbiol 78:4051–4056. doi: 10.1128/AEM.07994-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jin W, Cheng Y-F, Mao S-Y, Zhu W-Y. 2011. Isolation of natural cultures of anaerobic fungi and indigenously associated methanogens from herbivores and their bioconversion of lignocellulosic materials to methane. Bioresour Technol 102:7925–7931. doi: 10.1016/j.biortech.2011.06.026. [DOI] [PubMed] [Google Scholar]
- 37.Leahy SC, Kelly WJ, Altermann E, Ronimus RS, Yeoman CJ, Pacheco DM, Li D, Kong Z, McTavish S, Sang C, Lambie SC, Janssen PH, Dey D, Attwood GT. 2010. The genome sequence of the rumen methanogen Methanobrevibacter ruminantium reveals new possibilities for controlling ruminant methane emissions. PLoS One 5:e8926. doi: 10.1371/journal.pone.0008926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Leahy SC, Kelly WJ, Li D, Li Y, Altermann E, Cox F, Attwood GT. 2013. The complete genome sequence of Methanobrevibacter sp. AbM4. Stand Genomic Sci 8:215–227. doi: 10.4056/sigs.3977691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lee J-H, Rhee M-S, Kumar S, Lee G-H, Chang D-H, Kim D-S, Choi S-H, Lee D-W, Yoon M-H, Kim B-C. 2013. Genome sequence of Methanobrevibacter sp. strain JH1, isolated from rumen of Korean native cattle. Genome Announc 1:e00002-13. doi: 10.1128/genomeA.00002-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dridi B, Henry M, El Khéchine A, Raoult D, Drancourt M. 2009. High prevalence of Methanobrevibacter smithii and Methanosphaera stadtmanae detected in the human gut using an improved DNA detection protocol. PLoS One 4:e7063. doi: 10.1371/journal.pone.0007063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hansen EE, Lozupone CA, Rey FE, Wu M, Guruge JL, Narra A, Goodfellow J, Zaneveld JR, McDonald DT, Goodrich JA, Heath AC, Knight R, Gordon JI. 2011. Pan-genome of the dominant human gut-associated archaeon, Methanobrevibacter smithii, studied in twins. Proc Natl Acad Sci U S A 108:4599–4606. doi: 10.1073/pnas.1000071108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rinke C, Schwientek P, Sczyrba A, Ivanova NN, Anderson IJ, Cheng JF, Darling A, Malfatti S, Swan BK, Gies EA, Dodsworth JA, Hedlund BP, Tsiamis G, Sievert SM, Liu WT, Eisen JA, Hallam SJ, Kyrpides NC, Stepanauskas R, Rubin EM, Hugenholtz P, Woyke T. 2013. Insights into the phylogeny and coding potential of microbial dark matter. Nature 499:431–437. doi: 10.1038/nature12352. [DOI] [PubMed] [Google Scholar]
- 43.King EE, Smith RP, St-Pierre B, Wright A-DG. 2011. Differences in the rumen methanogen populations of lactating Jersey and Holstein dairy cows under the same diet regimen. Appl Environ Microbiol 77:5682–5687. doi: 10.1128/AEM.05130-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kittelmann S, Pinares-Patiño CS, Seedorf H, Kirk MR, Ganesh S, McEwan JC, Janssen PH. 2014. Two different bacterial community types are linked with the low-methane emission trait in sheep. PLoS One 9:e103171. doi: 10.1371/journal.pone.0103171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wolin MJ. 1980. The rumen fermentation: a model for microbial interactions in anaerobic ecosystems. Adv Microb Ecol 3:49–77. [Google Scholar]
- 46.Zhou M, Hernandez-Sanabria E, Guan LL. 2010. Characterization of variation in rumen methanogenic communities under different dietary and host feed efficiency conditions, as determined by PCR-denaturing gradient gel electrophoresis analysis. Appl Environ Microbiol 76:3776–3786. doi: 10.1128/AEM.00010-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.