Abstract
Streptococcus suis is an emerging zoonotic pathogen causing severe infections in pigs and humans. In previous studies, 33 serotypes of S. suis have been identified using serum agglutination. Here, we describe a novel S. suis strain, CZ130302, isolated from an outbreak of acute piglet meningitis in eastern China. Strong pathogenicity of meningitis caused by strain CZ130302 was reproduced in the BALB/c mouse model. The strain showed a high fatality rate (8/10), higher than those for known virulent serotype 2 strains P1/7 (1/10) and 9801 (2/10). Cell adhesion assay results with bEnd.3 and HEp2 cells showed that CZ130302 was significantly close to P1/7 and 9801. Both the agglutination test and its complementary test showed that strain CZ130302 had no strong cross-reaction with the other 33 S. suis serotypes. The multiplex PCR assays revealed no specified bands for all four sets used to detect the other 33 serotypes. In addition, genetic analysis of the whole cps gene clusters of all serotypes was performed in this study. The results of comparative genomics showed that the cps gene cluster of CZ130302, which was not previously reported, showed no homology to the gene sequences of the other strains. Especially, the wzy, wzx, and acetyltransferase genes of strain CZ130302 are phylogenetically distinct from strains of the other 33 serotypes. Therefore, this study suggested that strain CZ130302 represents a novel variant serotype of S. suis (designated serotype Chz) which has a high potential to be virulent and associated with meningitis in animals.
INTRODUCTION
Streptococcus suis causes meningitis and septicemia in pigs and is also known as a zoonotic agent (1). Human infections of S. suis were first reported in Denmark in 1968 (2). Since then, this pathogen has spread all over the world. The human Streptococcus suis was epidemic in most Europe countries (3, 4), as well as in Asian countries, such as Vietnam and Thailand (5–7). In China, two outbreaks of human streptococcosis have occurred, affecting more than 100 people and causing 39 deaths (8). More and more S. suis infections from China, Thailand, Hong Kong, Taiwan, and Singapore have been reported, which indicates that S. suis has been an important cause of adult meningitis, endocarditis, septicemia, and arthritis in Asia (9).
The serotyping of S. suis isolates rests on the basis of the antigenicity of their capsular polysaccharides (CPs); 35 serotypes have been identified by agglutination tests (10). With the development of sequence analysis of 16S rRNA and cpn60 genes in S. suis, the original S. suis serotypes 32 and 34 were reclassified as Streptococcus orisratti (11). Phylogenetic analyses of the cps gene cluster, conserved Wzy polymerase, Wzx flippase, and glycosyltransferase are all taken as important means of classifying a novel serotype (12). Multiplex PCR assays against the specific genes of the cps clusters have also been developed to identify serotypes in S. suis (13–15).
From March to May 2013, strain CZ130302 caused an outbreak of streptococcosis in piglets at multiple large-scale pig farms in Jiangsu Province, China. This pathogenic bacterium induced meningitis in 30-day-old piglets, with a total morbidity rate of 25% to 35%. The fatality rate of diseased piglets could reach 65%. We identified that the agent responsible for meningitis and septicemia in piglets as S. suis (CZ130302), and the strong pathogenicity of meningitis was reproduced successfully in a BALB/c mouse model. Follow-up identification and characteristic analysis of the serotype of the CZ130302 strain were performed. Interestingly, this strain did not belong to any known S. suis serotype. All the results suggested that the strain was a novel serotype which was probably responsible for the new round of emerging zoonosis in the swine industry.
MATERIALS AND METHODS
Ethics statement.
Five-week-old male germfree BALB/c mice and New Zealand White rabbits were purchased from the Comparative Medicine Center of Yangzhou University. All animal experiments were approved by Department of Science and Technology of Jiangsu Province [license number SYXK (SU) 2010-0005].
Bacterial strains and growth conditions.
The important strains used in this study are listed in Table 1. The new variant strain CZ130302 was isolated from an outbreak of acute piglet meningitis in 2013. The S. suis reference serotypes 1 to 5, 7 to 12, 16, 17, 20 to 24, 26, 32, 33, and 34 are stored in our laboratory; serotypes 6, 13, 15, 18, 19, 23, 25, 27, 30, and 31 are preserved in China Animal Health and Epidemiology Center. In addition, a total of 254 S. suis strains isolated from different sources and regions, at different times, and of different serotypes were included in this study. The bacteria were grown in Todd-Hewitt broth (THB; BD) and plated on Todd-Hewitt agar (THA) containing 7.5% (vol/vol) sheep blood at 37°C.
TABLE 1.
Bacterial strains and cell lines used in this study
| Strain or cell designation | Characteristic or function | Reference |
|---|---|---|
| Strains | ||
| P1/7 | European classical highly virulent strain, isolated from a pig dying from meningitis | 21 |
| 9801 | Virulent strain of serotype 2 isolated from a pig that died with acute septicemia, China, 1998 | 22 |
| CZ130302 | A novel variant serotype Chz of S. suis which caused acute meningitis in piglets, China, 2013 | This study |
| CZ110902 | Serotype Chz, clinical isolate JiangSu province, China, 2011 | This study |
| HN136 | Serotype Chz, clinical isolate HeNan province, China, 2006 | This study |
| AH681 | Serotype Chz, clinical isolate AnHui province, China, 2006 | This study |
| Cells | ||
| HEp-2 | Human laryngeal cancer epithelial cell line, widely used to evaluate the pathogenicity of S. suis isolates | 19 |
| bEnd.3 | Mouse brain microvascular endothelial cell line | This study |
Identification of bacteria.
Regular bacterial isolation was performed. The isolate was identified as S. suis using the Vitek 2 system (bioMérieux Vitek, Inc., Hazelwood, MO). The isolates were also confirmed as S. suis by 16S rRNA gene sequencing and gdh-specific PCR (Table 2) (11, 16). The capsule of S. suis CZ130302 was observed by transmission electron microscopy (see Fig. S1B in the supplemental material).
TABLE 2.
Primers used for PCR amplification
| Primer | Sequence (5′–3′) | Product size (bp) | Tma (°C) | Comment |
|---|---|---|---|---|
| 16S-rRNA-F | AGAGTTTGATCGTGGCTCA | 1,500 | 55 | Domain-specific 16S primers |
| 16S-rRNA-R | TACGGTTACCTTGTTACGACTT | |||
| gdh-F | CCATGGACAGATAAAGATGG | 688 | 54 | Primers for S. suis identification |
| gdh-R | GCAGCGTATTCTGTCAAACG | |||
| Chz-M-F | AATGAATAAGGAACTTGAACTA | 424 | 59.8 | Constructed in this study |
| Chz-M-R | CGTATCATCTGTATTAGCTAAA |
Tm, melting temperature.
Phylogenetic relationships of Streptococcus spp. based on a 552-bp segment of the cpn60 gene were determined by following the procedures outlined by Hill et al. (11). A ClustalW alignment with default parameters was used with 552-bp nucleic acid sequences. Similarly, sequencing analyses of sodA and recN were performed. The phylogenetic tree was constructed with the MEGA (v.5.0.3) software package using the neighbor-joining method, with P-distance, complete gap deletion, and bootstrapping (n = 500) parameters.
Agglutination tests.
Serological typing was carried out by agglutination performed as described earlier (17). The serotyping antiserum produced by rabbits was prepared by reported methods. All reference serotypes were tested for reactivity with CZ130302 antiserum. Correspondingly, the reference antisera of serotypes 1, 1/2, and 2 to 34 were used to agglutinate the variant CZ130302. Positive results were recorded when a strong reaction was obtained within 1 min. The judgment standards are shown in Fig. 1.
FIG 1.
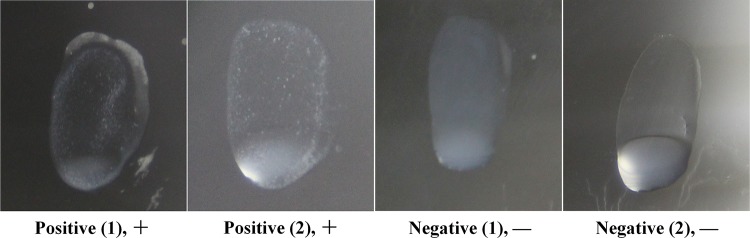
Judgment standards of the agglutination assay.
Multiplex PCR.
The serotyping primers were designed based on the sequences of capsule loci (cps), which were described in earlier reports (13, 14). Using the multiplex PCR system, four reactions were developed to detect all serotypes of S. suis. Furthermore, a cross-hybridization experiment was performed to screen the new specific gene in CZ130302. Primers for cps chzM were designed for monitoring known serotypes and clinical isolates that were nontypeable (Table 2). All primers were produced by Life Technologies and dissolved in Tris-EDTA (TE) buffer.
Genetic typing analyses of the cps gene cluster.
The complete genome sequence of S. suis strain CZ130302 was determined by Solexa pyrosequencing at BGI (Shenzhen, China), and the cps gene cluster was obtained. Maps of the new strain gene cluster were constructed manually in the VECTOR NTI program. Visual representation of the alignments using nucleotide similarities (tblastx) of all cps gene clusters was carried out with the Artemis comparison tool (ACT) (18). Conserved Wzy polymerase, Wzx flippase, and glycosyltransferase genes as the serotype-specific genes were analyzed by MEGA (v.5.0.3).
MLST.
All the isolates in this study were typed using multilocus sequence typing (MLST). The seven housekeeping genes (dpr, mutS, cpn60, thrA, recA, aroA, and gki) were amplified by PCR, and internal fragments sequences were obtained as described previously (5). For each isolate, the allele numbers and sequence types (STs) were defined by analysis of the allele sequences in the MLST database (http://ssuis.mlst.net/). The results were analyzed by eBURST (version 3).
Assessment of pathogenicity of S. suis CZ130302 in mouse infection model.
The BALB/c mouse infection model (19, 20) was used to compare the virulence of the new serotype isolate with that of two known virulent serotype 2 S. suis strains, P1/7 and 9801 (21, 22). A total of 5 × 107 CFU/mouse of each strain was injected intraperitoneally (10 mice per strain) to obtain the survival curve. The groups were observed throughout a 7-day period, and survival condition was recorded every day. The blank-control group was injected with sterile phosphate-buffered saline (PBS). The mice were observed for 7 days until survival rates were steady. A total of 2 × 107 CFU/mouse (∼10× the 50% lethal dose [LD50]) of S. suis CZ130302 was injected intraperitoneally into 50 mice. Five symptomatic mice were selected for euthanization and dissection every 24 h. Bacteria were isolated from the hearts, kidneys, lungs, brains, and urine homogenate by plating 10-fold serial dilutions on THA. The number of bacteria colonizing the organs of the mice during systemic infection was obtained.
Adhesion assays with HEp-2 and bEnd.3 cells.
In accordance with the bacterial colonization capacity in vivo, we used the human laryngeal carcinoma cell line HEp-2 and mouse brain microvascular endothelial cells (bEnd.3) as in the models of bacterial colonization (19, 20, 23) and meningitis (24, 25) in vitro, respectively. The adherence assays were performed as previously described (26). To release all bacteria, the monolayers were disrupted by adding sterile water for HEp-2 cells or 0.01% Triton X-100 for bEnd.3 cells after digestion with trypsin.
Cytotoxicity assays.
In order to further confirm the ability to cause meningitis, the cytotoxic effect of bacteria was evaluated along with bEnd.3 cell adhesion by lactate dehydrogenase (LDH) measurement using the CytoTox 96 nonradioactive cytotoxicity assay (Promega Corporation, USA) (27, 28). The percent cytotoxicity was calculated as [(sample OD490 − bacterial spontaneous OD490 − cell spontaneous OD490)/(cell maximum OD490 − cell spontaneous OD490)] × 100, where OD490 is optical density at 490 nm. LDH release was measured after different durations of incubation (1 h to 4 h) at a bacterium-cell ratio of 1:1 at 37°C.
Statistical analysis.
Statistical analysis for in vitro and in vivo experiments was carried out using Prism 5 (GraphPad Software, La Jolla, CA). One-way analysis of variance (ANOVA) was used in the analysis of the cell adherence assay results. Student's t tests were applied for comparison of serum IgG levels, and mouse survival data were analyzed by the Kaplan-Meier estimation method (29). A difference with a P value of <0.05 was considered significant, and a P value of <0.01 was considered greatly significant.
Nucleotide sequence accession number.
For meningitis-associated S. suis isolate CZ130302, a cps cluster sequence of 28,481 bp was obtained. The DNA sequence was deposited in GenBank under accession number KJ669337.
RESULTS
Isolation and identification of bacteria.
The significant beta-hemolytic zones were created by piglet meningitis-associated strain CZ130302 (see Fig. S1A in the supplemental material). The capsule of S. suis CZ130302 was observed by transmission electron microscopy (see Fig. S1B). Further identification of the organism as S. suis was confirmed at the OIE Reference Laboratory for Swine Streptococcosis in Nanjing Agricultural University by the Vitek 2 system (bioMérieux Vitek), the result of which was completely consistent with S. suis (see Fig. S2). 16S rRNA gene sequencing was also performed; the results showed >99% homology with the S. suis European classical strain P1/7 (1,524/1,528 bases) and Chinese epidemic strain SC84 (1,523/1,528 bases).
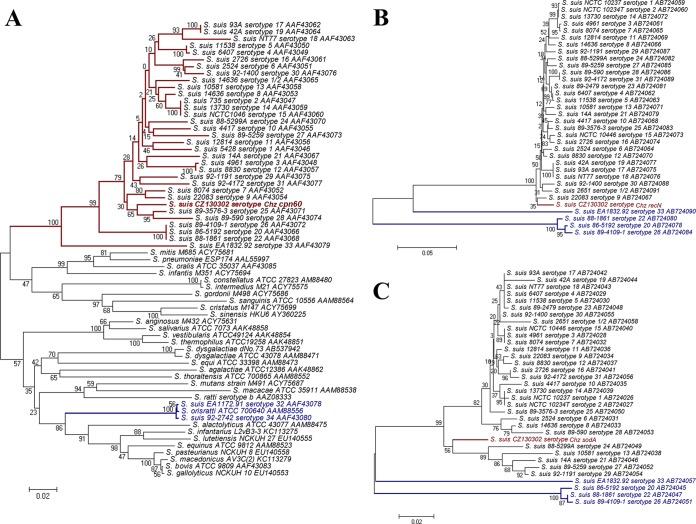
Phylogenetic relationships of the cpn60, recN, and sodA genes in all serotypes were demonstrated in cladograms (Fig. 2). In a phylogenetic tree of partial cpn60, S. orisratti and S. suis serotypes 32 and 34 are located in a group including S. equinus, S. alactolyticus, and so on, while strain CZ130302 and all other serotypes of S. suis are found together in a separate and distinct cluster (Fig. 2A). Likewise, the phylogenetic trees of sodA and recN showed that serotypes 20, 22, 26, and 33 located outside a clade formed by 29 other serotypes and strain CZ130302 (Fig. 2B and C). These result indicated that serotype Chz was a veritable emerging serotype of S. suis.
FIG 2.
Phylogenetic analysis of three key genes in Streptococcus. (A) Phylogenetic relationships of most Streptococcus spp. based on a 552-bp segment of the cpn60 gene. The tree was constructed with the MEGA (v.5.0.3) software package using the neighbor-joining method, with P-distance, complete gap deletion, and bootstrapping (n = 500) parameters. Accession numbers for sequences used in this analysis are shown after the strain names. The cpn60 segments of S. suis strains are highlighted with red branches, and the red type indicates the cpn60 segment from S. suis serotype Chz. The cpn60 segments of previous S. suis serotypes 32 and 34 and S. orisratti are highlighted with blue branches and red type. (B) Phylogenetic relationships of 35 serotypes of S. suis based on a 1,056-bp segment of the recN gene. The red type indicates the recN segment from S. suis serotype Chz. The recN segments of previous S. suis serotypes 20, 22, 26, and 33 are highlighted with blue branches and type. (C) Phylogenetic relationships of 35 serotypes of S. suis based on a 409-bp segment of the sodA gene. The red type indicates the sodA segment from S. suis serotype Chz. The sodA segments of S. suis serotypes 20, 22, 26, and 33 are highlighted with blue branches and type.
Agglutination tests.
Agglutination tests of isolate CZ130302 showed negative reactions with all 33 serotypes. Correspondingly, the reversed agglutination tests between the existing S. suis serotypes and the new serotyping antiserum produced by rabbits showed no strong positive result for any of the 33 serotype reference strains (see Table S1 in the supplemental material).
Identification of serotypes by multiplex PCR.
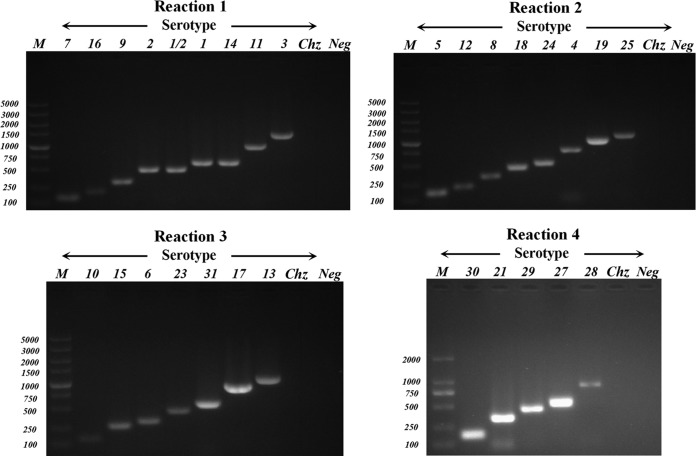
Specific PCRs for the 33 known serotypes were performed, and they confirmed a negative result for the novel variant CZ130302 (Fig. 3). Every serotype reference strain was used as a positive control. The results indicated that no serotype-specific genes of known serotypes were found in strain CZ130603, whose cps gene cluster was highly differential.
FIG 3.
Multiplex PCR (4 reaction sets) products of S. suis new serotype CZ130302 and 29 reference strains. PCR products were electrophoresed on a 1.5% (wt/vol) agarose gel, stained with GoldView, and photographed under UV light. Serotypes are indicated above the lanes. Lanes M, 5,000-bp DNA ladder markers (Biomed, Beijing, China); sizes (bp) are indicated on the left.
The cps cluster of strain CZ130302.
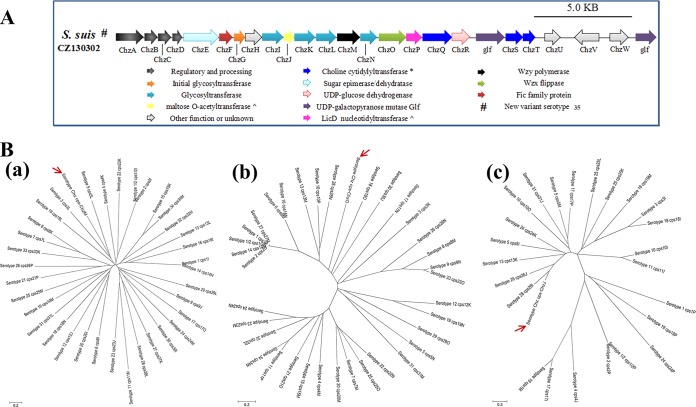
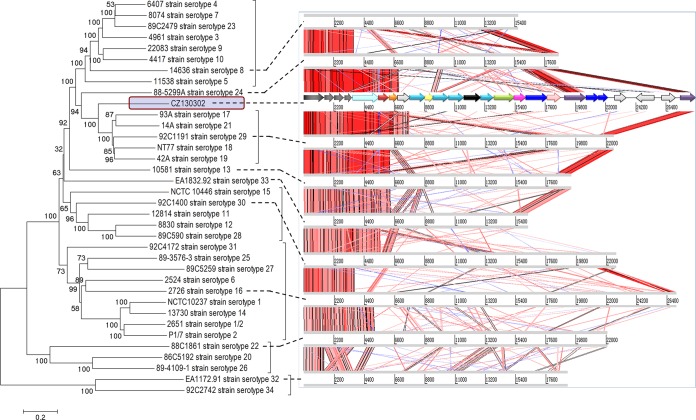
The CZ130302 CP genes are named chzA-chzW, corresponding to the regulation portions (cpsA-cpsW) of the chromosome (cps gene cluster portions) in S. suis (Fig. 4A) (12). In order to identify whether this strain represented an emerging serotype of S. suis, genetic analysis of whole cps gene clusters of all serotypes was performed in this study. The results of comparative genomics showed that the cps gene cluster of CZ130302 lacked homology with the sequences of other known strains; no cps cluster has been seen to lack homology to other such sequences previously (Fig. 5). The novel serotype shares homologous wzg, wzd, wze, and wzh sequences with all known serotypes of S. suis in their cps gene clusters, whereas it contains other unique key genes from chzI (7,735 bp) to chzW (28,481 bp) (Fig. 5; see also Table S2 in the supplemental material), such as the polymerase (wzy), flippase (wzx), glycosyltransferase, and acetyltransferase genes (Fig. 4B).
FIG 4.
The cps cluster of strain CZ130302 and its key genes. (A) Schematic diagram of the genetic organization of the S. suis serotype Chz strain CZ130302 cps gene cluster. Genes encoding conserved domain proteins are represented by the same colors. White arrows refer to other genes in the cps gene clusters that were not identified as part of the conserved core described by Okura et al. (12). The direction of the arrows indicates the direction of transcription. The color key for the functional classes of genes in the cps cluster is shown at the bottom. (B) Sequence relationship of Wzy, Wzx, and acetyltransferase of all S. suis serotypes. Three neighbor-joining trees (bootstrap n = 1,000; Poisson correction) were constructed based on the ClustalW alignments of the Wzy, Wzx, and acetyltransferase amino acid sequences from all of S. suis serotypes. Wzy, Wzx, and acetyltransferase from S. suis strains CZ130302 are indicated by red arrows.
FIG 5.
Comparative genome alignments of 35 cps gene clusters. The color key for the functional classes of genes and relevant information is the same as shown in Fig. 6A. Phylogenetic relationships of the cps gene clusters were obtained using a neighbor-joining tree (bootstrap n = 1,000) based on a ClustalW alignment of the complete cluster sequences. Visual representation of the alignments using nucleotide similarities (tblastx) of the cps gene clusters as determined with the Artemis comparison tool (ACT) (18).
Some of the CZ130302 cps genes were predicted to encode modifying enzymes (such as acetyltransferase [chzJ], nucleotidyltransferase [chzP], choline phosphate cytidylyltransferase [chzQ], UDP-glucose dehydrogenase [chzR], and phosphocholine cytidylyltransferase [chzT]), which are involved in the biosynthesis and addition to other components on CPs (such as glycerol and choline) (Fig. 4A; see also Table S2 in the supplemental material). The novel S. suis cps gene cluster also has a disrupted gene encoding a protein in the transposase family (chzW) in the 3′ region, similar to most of the cps gene clusters. The cps gene cluster of the new serotype has a >65% specific sequence, which guides the synthesis of the characteristic capsule of isolate CZ130302 (see Table S2).
Development of novel serotype-specific PCR.
We selected oligonucleotide primers within the cps chzM gene to generate specific amplicons of 424 bp according to the cross-hybridization results. A total of 45 nontypeable strains of S. suis isolated from China were used to check the cps chzM gene; 3 (HN136, AH681, and CZ110902) of them showed 424-bp bands. Agglutination tests of two isolates (HN136 and CZ110902) showed classical positivity with the CZ130302 antiserum (see Table S3 in the supplemental material). The sequencing results for their cps chzM genes showed >99% homology with the cps gene of reference strain CZ130302. The results demonstrate that they all belong to this novel serotype.
MLST typing.
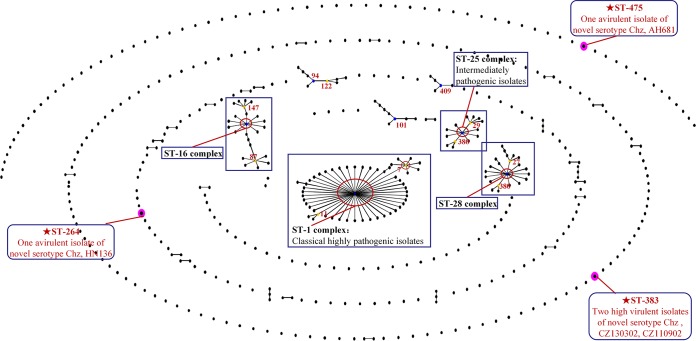
All 4 isolates of the novel serotype were characterized using MLST. Two isolates were classified as ST 383 and showed strong pathogenicity in the piglet and BALB/c mouse models (see Table S4 in the supplemental material). Avirulent strain HN136 was classified as ST 264, and AH681 was ST 475. None of these three STs could be grouped in any clonal complexes (CC) according to eBURST analysis (Fig. 6).
FIG 6.
eBURST diagram of the S. suis population. Population snapshots of S. suis of related STs within the entire S. suis MLST database were constructed. Each ST is represented as a dot. Two dots separated by one node represent a single-locus variation between two STs (a single-locus variant). The STs positioned centrally in the clonal complex (CC) are primary founders (blue) or subgroup founders (yellow). STs in purple circles are those identified in this study. For clarity, labels of STs have been removed, except related STs and founders in CCs. Some STs are labeled with blue boxes to emphasize their importance. The eBURST diagram does not show the genetic distance between unlinked STs and CCs.
Evaluation of pathogenicity of the novel serotype strains in the BALB/c mouse model.
The mouse model has been demonstrated to be a useful tool for evaluating the virulence of S. suis. The mortality of BALB/c mice was observed for 7 days after the challenge. The survival curve for strain CZ130302 was significantly lower than for strains P1/7, 9801, and HN136 (P < 0.01) (Fig. 7A). These results confirmed that strain CZ130302 showed high virulence and pathogenicity in the BALB/c mouse model. However, strain HN136 was avirulent in this study (Fig. 7A; see also Table S4 in the supplemental material).
FIG 7.
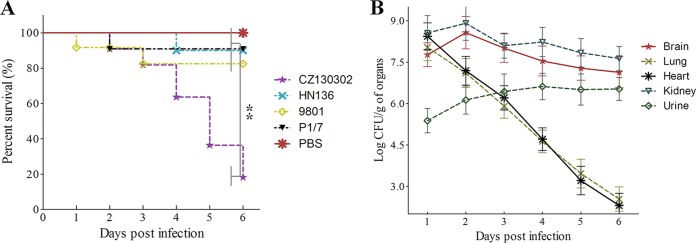
Challenge studies in the BALB/c mouse model and cell adhesion assay. (A) Mortality curve of lethal challenge with S. suis strains. A total of 5 × 107CFU/mouse of each strain was injected intraperitoneally (10 mice per strain) to obtain the survival curve. The groups were observed throughout a 7-day period, and survival condition was recorded every day. (B) A total of 2 × 107 CFU/mice (∼10× LD50) of S. suis CZ130302 was injected intraperitoneally into 50 mice. Five symptomatic mice were euthanized to perform reisolation of S. suis every 24 h by plating 10-fold serial dilutions on THA (**, P < 0.01; *, P < 0.05).
Strong virulence of the isolate CZ130302 associated with acute meningitis.
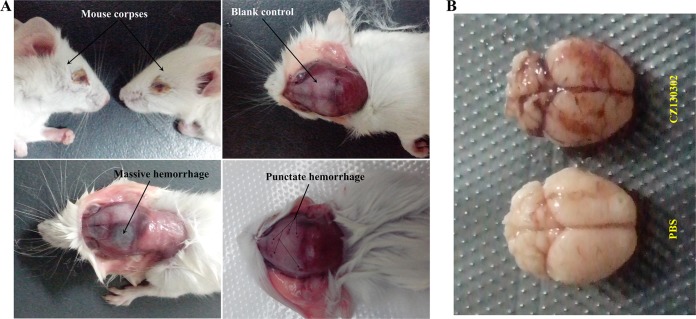
The novel serotype isolate CZ130302 caused a large outbreak of piglet meningitis in eastern China. This strong pathogenicity of meningitis was reproduced successfully in the BALB/c mouse model. More than 60% (19/30) of mice infected with CZ130302 (5 × 105 CFU/mouse) showed neurological symptoms (see Video S1 in the supplemental material), and many survivors had sequelae, including tetraplegia, paraplegia, neck-crooking, circling, etc. The density of CZ130302 was able to reach 1 × 108 CFU/g in the brains and kidneys of dying mice 3 days after challenge, with 1 × 105 CFU/g or less in other organs (Fig. 7B). The pathological observation of brain tissue showed obvious abscess and bleeding (Fig. 8). These results demonstrated that isolate CZ130302 had a strong capacity to cause meningitis in BALB/c mice.
FIG 8.
Autopsy images of mouse brain.
High virulence of cerebral infection verified by host cell adhesion assay.
The capacities of adhesion to host cells were compared among the CZ130302, 9801, and P1/7 strains under the same conditions. As shown in Fig. 9A, the bEnd.3 cell adhesion for strains P1/7, 9801, and CZ130302 was significantly higher than for HN136 (P < 0.01) (Fig. 9A). The HEp2 cell adhesion for strain CZ130302 was significantly lower than for strain P1/7 (P < 0.01) and not significantly different from that of strain 9801 (Fig. 9A). These results suggested that strain CZ130302 had stronger capacity than strains HN136 in the adhesion of HEp2 and bEnd.3 cells, with a bit weaker capacity for strain P1/7. These findings confirmed the notion that the pathogenesis of new isolate CZ130302 might be associated with bacterial colonization in respiratory tract and brain tissue.
FIG 9.
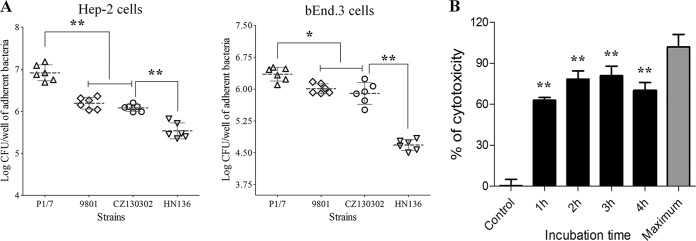
Cell adhesion and cytotoxicity assay. All assays were run in triplicate. Statistical significance was determined by Student's t test (**, P < 0.01; *, P < 0.05). (A) The assessment of cell adhesion ability of S. suis serotype Chz. Virulent strain CZ130302 shows a strong capacity of adhesion to bEnd.3 and HEp2 cells (MOI, 100). (B) Assessment of the cytotoxicity of strain CZ130302. An MOI of 1 bacterium/cell (2 × 105 CFU bacteria/well) was chosen to study the kinetics of cytotoxicity by S. suis. Strain CZ130302 was able to significantly damage mouse brain microvascular endothelial cells (bEnd.3).
Strain CZ130302 can damage bEnd.3 cells.
A multiplicity of infection (MOI) of 1 bacterium/cell (2 × 105 CFU/well) was chosen to study the kinetics of cytotoxicity by S. suis. Maximal cytotoxic levels were observed at the third hour of bacterium-cell contact (Fig. 9B). The kinetics of cell damage fell between 60% and 80% (Fig. 9B). These results suggested that strain CZ130302 was able to kill mouse brain microvascular endothelial cells (bEnd.3) due to bacterial colonization in the brain tissue.
DISCUSSION
S. suis is increasingly recognized as a significant zoonotic agent. Increasing awareness of S. suis infection is expected to help counter animal or human streptococcosis. In this study, obvious neurological symptoms were observed in piglets infected by strain CZ130302, such as walking in circles and single-side neck crooking. The mouse model also replicated the classical symptom. Previous studies have shown that meningitis was mainly caused by serotypes 2, 9, and 14 (30–32), of which the presenting features were generally similar to those of pyogenic meningitis caused by other bacteria. Acute meningitis caused by a novel variant serotype has never been reported previously. This potentially underrecognized hazard of the swine industry is demonstrated by this study.
Serological typing is the foundation of S. suis serotyping (17, 33). The antiserum of the novel serotype prepared can provide reliable and original results for identification of this novel serotype. PCR typing assays provide a fast and cost-effective way to determine the serotypes of isolates. The multiplex PCR method has been developed in recent years (13, 14, 34); the specific cps genes are planned for use in typing. This gene cluster strongly supports the results of agglutination assays and avoids the false positivity of naked-eye observation in the agglutination test. Inevitably, cross-antigenicity happened between the novel antiserum and some serotypes (serotypes 15, 13, 26, 6, 19, 24, and 25). For increased assurance, we designed the cps chzM gene primers to differentiate this novel serotype from all exciting serotypes. Three positive strains were searched from the 45 nontypeable S. suis strains stored in our laboratory. These positive strains were from different areas and periods, but they were all recently isolated strains from eastern China.
MLST has been widely used to study genetic diversity, population structure, and molecular epidemiology in S. suis (5). None of the three STs of the novel serotype was linked with any highly virulent STs by virologists, whereas ST 383 isolates were strongly associated with high pathogenicity in an animal model (35, 36). Additionally, the complete sequence of the cps locus of CZ130302 was obtained in subsequent research. Capsular polysaccharides are an extremely diverse range of molecules that may differ not only by monosaccharide units but also in how these units are joined together (37). CPs of all S. suis serotypes are synthesized by the Wzx/Wzy pathway, which recognizes common oligosaccharide structures conserved in the different repeat units (12).
The results of this study demonstrate that strain CZ130302 belongs to a novel serotype (Chz) of S. suis, based on sequencing of the cps gene cluster, PCR, and agglutination typing. MLST analysis and Wzx/Wzy phylogenetic tree profiling also prove to be useful in establishing the serotype.
Supplementary Material
ACKNOWLEDGMENTS
This research was supported by the Special Fund for Public Welfare Industry of the Chinese Ministry of Agriculture (2013003041), the 948 Major Project and Industry Project from the Chinese Ministry of Agriculture (2014-S11), and a project funded by the Priority Academic Program Development of Jiangsu Higher Education Institutions (PAPD).
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.02962-14.
REFERENCES
- 1.Fittipaldi N, Segura M, Grenier D, Gottschalk M. 2012. Virulence factors involved in the pathogenesis of the infection caused by the swine pathogen and zoonotic agent Streptococcus suis. Future Microbiol 7:259–279. doi: 10.2217/fmb.11.149. [DOI] [PubMed] [Google Scholar]
- 2.Perch B, Kristjansen P, Skadhauge K. 1968. Group R streptococci pathogenic for man. Two cases of meningitis and one fatal case of sepsis. Acta Pathol Microbiol Scand 74:69–76. [PubMed] [Google Scholar]
- 3.Demar M, Belzunce C, Simonnet C, Renaux A, Abboud P, Okandze A, Marois-Crehan C, Djossou F. 2013. Streptococcus suis meningitis and bacteremia in man, French Guiana. Emerg Infect Dis 19:1545–1546. doi: 10.3201/eid1909.121872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zalas-Wiecek P, Michalska A, Grabczewska E, Olczak A, Pawlowska M, Gospodarek E. 2013. Human meningitis caused by Streptococcus suis. J Med Microbiol 62:483–485. doi: 10.1099/jmm.0.046599-0. [DOI] [PubMed] [Google Scholar]
- 5.King SJ, Leigh JA, Heath PJ, Luque I, Tarradas C, Dowson CG, Whatmore AM. 2002. Development of a multilocus sequence typing scheme for the pig pathogen Streptococcus suis: identification of virulent clones and potential capsular serotype exchange. J Clin Microbiol 40:3671–3680. doi: 10.1128/JCM.40.10.3671-3680.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Suankratay C, Intalapaporn P, Nunthapisud P, Arunyingmongkol K, Wilde H. 2004. Streptococcus suis meningitis in Thailand. Southeast Asian J Trop Med Public Health 35:868–876. [PubMed] [Google Scholar]
- 7.Takamatsu D, Wongsawan K, Osaki M, Nishino H, Ishiji T, Tharavichitkul P, Khantawa B, Fongcom A, Takai S, Sekizaki T. 2008. Streptococcus suis in humans, Thailand. Emerg Infect Dis 14:181–183. doi: 10.3201/eid1401.070568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yu H, Jing H, Chen Z, Zheng H, Zhu X, Wang H, Wang S, Liu L, Zu R, Luo L, Xiang N, Liu H, Liu X, Shu Y, Lee SS, Chuang SK, Wang Y, Xu J, Yang W. 2006. Human Streptococcus suis outbreak, Sichuan, China. Emerg Infect Dis 12:914–920. doi: 10.3201/eid1206.051194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mai NT, Hoa NT, Nga TV, Linh le D, Chau TT, Sinh DX, Phu NH, Chuong LV, Diep TS, Campbell J, Nghia HD, Minh TN, Chau NV, de Jong MD, Chinh NT, Hien TT, Farrar J, Schultsz C. 2008. Streptococcus suis meningitis in adults in Vietnam. Clin Infect Dis 46:659–667. doi: 10.1086/527385. [DOI] [PubMed] [Google Scholar]
- 10.Palmieri C, Varaldo PE, Facinelli B. 2011. Streptococcus suis, an emerging drug-resistant animal and human pathogen. Front Microbiol 2:235. doi: 10.3389/fmicb.2011.00235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hill JE, Gottschalk M, Brousseau R, Harel J, Hemmingsen SM, Goh SH. 2005. Biochemical analysis, cpn60 and 16S rDNA sequence data indicate that Streptococcus suis serotypes 32 and 34, isolated from pigs, are Streptococcus orisratti. Vet Microbiol 107:63–69. doi: 10.1016/j.vetmic.2005.01.003. [DOI] [PubMed] [Google Scholar]
- 12.Okura M, Takamatsu D, Maruyama F, Nozawa T, Nakagawa I, Osaki M, Sekizaki T, Gottschalk M, Kumagai Y, Hamada S. 2013. Genetic analysis of capsular polysaccharide synthesis gene clusters from all serotypes of Streptococcus suis: potential mechanisms for generation of capsular variation. Appl Environ Microbiol 79:2796–2806. doi: 10.1128/AEM.03742-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kerdsin A, Akeda Y, Hatrongjit R, Detchawna U, Sekizaki T, Hamada S, Gottschalk M, Oishi K. 2014. Streptococcus suis serotyping by a new multiplex PCR. J Med Microbiol 63:824–830. doi: 10.1099/jmm.0.069757-0. [DOI] [PubMed] [Google Scholar]
- 14.Liu Z, Zheng H, Gottschalk M, Bai X, Lan R, Ji S, Liu H, Xu J. 2013. Development of multiplex PCR assays for the identification of the 33 serotypes of Streptococcus suis. PLoS One 8:e72070. doi: 10.1371/journal.pone.0072070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang K, Sun X, Lu C. 2012. Development of rapid serotype-specific PCR assays for eight serotypes of Streptococcus suis. J Clin Microbiol 50:3329–3334. doi: 10.1128/JCM.01584-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Okwumabua O, O'Connor M, Shull E. 2003. A polymerase chain reaction (PCR) assay specific for Streptococcus suis based on the gene encoding the glutamate dehydrogenase. FEMS Microbiol Lett 218:79–84. doi: 10.1111/j.1574-6968.2003.tb11501.x. [DOI] [PubMed] [Google Scholar]
- 17.Higgins R, Gottschalk M, Boudreau M, Lebrun A, Henrichsen J. 1995. Description of six new capsular types (29–34) of Streptococcus suis. J Vet Diagn Invest 7:405–406. doi: 10.1177/104063879500700322. [DOI] [PubMed] [Google Scholar]
- 18.Carver TJ, Rutherford KM, Berriman M, Rajandream MA, Barrell BG, Parkhill J. 2005. ACT: the Artemis Comparison Tool. Bioinformatics 21:3422–3423. doi: 10.1093/bioinformatics/bti553. [DOI] [PubMed] [Google Scholar]
- 19.Si Y, Yuan F, Chang H, Liu X, Li H, Cai K, Xu Z, Huang Q, Bei W, Chen H. 2009. Contribution of glutamine synthetase to the virulence of Streptococcus suis serotype 2. Vet Microbiol 139:80–88. doi: 10.1016/j.vetmic.2009.04.024. [DOI] [PubMed] [Google Scholar]
- 20.Wichgers Schreur PJ, Rebel JM, Smits MA, van Putten JP, Smith HE. 2011. TroA of Streptococcus suis is required for manganese acquisition and full virulence. J Bacteriol 193:5073–5080. doi: 10.1128/JB.05305-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Allen AG, Bolitho S, Lindsay H, Khan S, Bryant C, Norton P, Ward P, Leigh J, Morgan J, Riches H, Eastty S, Maskell D. 2001. Generation and characterization of a defined mutant of Streptococcus suis lacking suilysin. Infect Immun 69:2732–2735. doi: 10.1128/IAI.69.4.2732-2735.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ju CX, Gu HW, Lu CP. 2012. Characterization and functional analysis of atl, a novel gene encoding autolysin in Streptococcus suis. J Bacteriol 194:1464–1473. doi: 10.1128/JB.06231-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vanier G, Segura M, Friedl P, Lacouture S, Gottschalk M. 2004. Invasion of porcine brain microvascular endothelial cells by Streptococcus suis serotype 2. Infect Immun 72:1441–1449. doi: 10.1128/IAI.72.3.1441-1449.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.El-Assaad F, Wheway J, Mitchell AJ, Lou J, Hunt NH, Combes V, Grau GE. 2013. Cytoadherence of Plasmodium berghei-infected red blood cells to murine brain and lung microvascular endothelial cells in vitro. Infect Immun 81:3984–3991. doi: 10.1128/IAI.00428-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huang SH, Wang L, Chi F, Wu CH, Cao H, Zhang A, Jong A. 2013. Circulating brain microvascular endothelial cells (cBMECs) as potential biomarkers of the blood-brain barrier disorders caused by microbial and non-microbial factors. PLoS One 8:e62164. doi: 10.1371/journal.pone.0062164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wertheim HF, Nghia HD, Taylor W, Schultsz C. 2009. Streptococcus suis: an emerging human pathogen. Clin Infect Dis 48:617–625. doi: 10.1086/596763. [DOI] [PubMed] [Google Scholar]
- 27.Behl C, Davis JB, Lesley R, Schubert D. 1994. Hydrogen peroxide mediates amyloid beta protein toxicity. Cell 77:817–827. doi: 10.1016/0092-8674(94)90131-7. [DOI] [PubMed] [Google Scholar]
- 28.Segura M, Gottschalk M. 2002. Streptococcus suis interactions with the murine macrophage cell line J774: adhesion and cytotoxicity. Infect Immun 70:4312–4322. doi: 10.1128/IAI.70.8.4312-4322.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Langová K, Gallo J. 2010. Is Kaplan-Meier statistics the most appropriate tool for survivorship measurement of outcomes in orthopaedics? Acta Chir Orthop Traumatol Cech 77:118–123. (In Czech.) [PubMed] [Google Scholar]
- 30.Fowler HN, Brown P, Rovira A, Shade B, Klammer K, Smith K, Scheftel J. 2013. Streptococcus suis meningitis in swine worker, Minnesota, USA. Emerg Infect Dis 19:330–331. doi: 10.3201/eid1902.120918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wu Z, Zhang W, Lu C. 2008. Comparative proteome analysis of secreted proteins of Streptococcus suis serotype 9 isolates from diseased and healthy pigs. Microb Pathog 45:159–166. doi: 10.1016/j.micpath.2008.04.009. [DOI] [PubMed] [Google Scholar]
- 32.Haleis A, Alfa M, Gottschalk M, Bernard K, Ronald A, Manickam K. 2009. Meningitis caused by Streptococcus suis serotype 14, North America. Emerg Infect Dis 15:350–352. doi: 10.3201/eid1502.080842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gottschalk M, Higgins R, Jacques M, Beaudoin M, Henrichsen J. 1991. Characterization of six new capsular types (23 through 28) of Streptococcus suis. J Clin Microbiol 29:2590–2594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kerdsin A, Dejsirilert S, Akeda Y, Sekizaki T, Hamada S, Gottschalk M, Oishi K. 2012. Fifteen Streptococcus suis serotypes identified by multiplex PCR. J Med Microbiol 61:1669–1672. doi: 10.1099/jmm.0.048587-0. [DOI] [PubMed] [Google Scholar]
- 35.Ye C, Bai X, Zhang J, Jing H, Zheng H, Du H, Cui Z, Zhang S, Jin D, Xu Y, Xiong Y, Zhao A, Luo X, Sun Q, Gottschalk M, Xu J. 2008. Spread of Streptococcus suis sequence type 7, China. Emerg Infect Dis 14:787–791. doi: 10.3201/eid1405.070437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhu W, Wu C, Sun X, Zhang A, Zhu J, Hua Y, Chen H, Jin M. 2013. Characterization of Streptococcus suis serotype 2 isolates from China. Vet Microbiol 166:527–534. doi: 10.1016/j.vetmic.2013.06.009. [DOI] [PubMed] [Google Scholar]
- 37.Roberts IS. 1996. The biochemistry and genetics of capsular polysaccharide production in bacteria. Annu Rev Microbiol 50:285–315. doi: 10.1146/annurev.micro.50.1.285. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.