Abstract
Salmonella enterica is a member of the plant microbiome. Growth of S. enterica in sprouting-seed exudates is rapid; however, the active metabolic networks essential in this environment are unknown. To examine the metabolic requirements of S. enterica during growth in sprouting-seed exudates, we inoculated alfalfa seeds and identified 305 S. enterica proteins extracted 24 h postinoculation from planktonic cells. Over half the proteins had known metabolic functions, and they are involved in over one-quarter of the known metabolic reactions. Ion and metabolite transport accounted for the majority of detected reactions. Proteins involved in amino acid transport and metabolism were highly represented, suggesting that amino acid metabolic networks may be important for S. enterica growth in association with roots. Amino acid auxotroph growth phenotypes agreed with the proteomic data; auxotrophs in amino acid-biosynthetic pathways that were detected in our screen developed growth defects by 48 h. When the perceived sufficiency of each amino acid was expressed as a ratio of the calculated biomass requirement to the available concentration and compared to growth of each amino acid auxotroph, a correlation between nutrient availability and bacterial growth was found. Furthermore, glutamate transport acted as a fitness factor during S. enterica growth in association with roots. Collectively, these data suggest that S. enterica metabolism is robust in the germinating-alfalfa environment; that single-amino-acid metabolic pathways are important but not essential; and that targeting central metabolic networks, rather than dedicated pathways, may be necessary to achieve dramatic impacts on bacterial growth.
INTRODUCTION
Plants can influence both the number and species of bacteria that colonize their surfaces. The availability and types of nutrients, secretion of chemoattractants or repellents, and production of antimicrobial compounds all impact the composition of the plant-associated bacterial community. In particular, germinating seeds and roots release exudates containing complex mixtures of amino acids, sugars, organic acids, and other plant metabolites that support the growth of microbial colonists in the spermosphere and rhizosphere (1–5). Successful colonization of and persistence in these plant niches requires that bacteria acquire and metabolize these plant-derived nutrients and biosynthesize any required metabolites not sufficiently available from the plant.
The compositions of seed and root exudates are dynamic and vary based on seed and plant age, soil type, temperature, nutritional status, and crop (6, 7). The identity and quantity of nutrients in plant exudates have a profound impact on bacterial growth and metabolic activity (8). Seeds that released larger quantities of carbohydrates and amino acids supported higher Enterobacter cloacae populations, rates of bacterial growth, and metabolic activity (3, 4, 8). Nutrient identity (e.g., alanine, arginine, and aspartate), not just type (e.g., amino acids, sugars, and lipids), influences bacterial metabolism; the exudate sugars glucose and fructose stimulated higher E. cloacae metabolic activity than galactose, raffinose, and xylose, though these sugars are all present in similar quantities in cucumber seed exudates (8). Studies of plant exudates in the context of plant-bacterial interactions have typically focused on the roles of select sugars and organic acids in the rhizosphere (3, 5, 8–11). Little is known about the temporal changes in the catalog of plant-derived metabolites present in seed and early root exudates and their importance as nutrient sources for plant-colonizing bacteria. Additionally, the metabolic pathways utilized by bacteria for growth in seed/early root exudates are largely unknown. These knowledge gaps limit our understanding of spermosphere and early rhizosphere microbiology and our ability to manipulate the growth of beneficial and pathogenic bacteria in this environment.
Salmonella enterica is an enteric human pathogen that colonizes crop plants as secondary hosts (12). S. enterica has been isolated from the seeds, leaves, and fruit of a variety of fresh produce crops (13) and is the leading bacterial cause of food-borne illness linked to fresh produce consumption (14). Produce is the number one food commodity associated with food-borne-disease outbreaks (15), with an estimated 50 million cases of produce-linked food-borne illness occurring annually in the United States (16). Reports that food-borne pathogens colonize, multiply, and persist long term on plants contradict the idea that food-borne illnesses are the result of casual or accidental encounters between human pathogens and produce crops (17–19). However, it is noteworthy that S. enterica cannot liberate plant nutrients (20); instead, the bacterium must rely on freely released plant exudates (e.g., from seeds and roots) or phytopathogen or insect activity for the nutrients and energy needed for growth and reproduction (21–23).
Historical data indicate that sprouted seeds are the most common fresh produce commodity linked to S. enterica (13). Sprouted-seed-related disease outbreaks have been linked to contaminated seeds. The very low bacterial populations on contaminated seeds make detection by routine seed lot screening difficult (13), but during seed germination, the S. enterica population can grow 1 million-fold (to 109 CFU/ml) within 48 h (24). The rapid exponential growth of S. enterica on germinating seeds (24, 25) shows that they provide a very favorable environment for enteric human pathogen growth.
The plant-derived nutrients that support the rapid growth of S. enterica and the metabolic pathways the bacterium utilizes in this germinating-seed environment are both unknown. In this study, we examined how the dynamic changes in the composition of seed and early root exudates influence microbial metabolic activity and growth, using S. enterica-contaminated alfalfa seeds as a model system. We hypothesized that (i) expressed metabolic proteins indicate metabolic pathways that contribute to S. enterica growth, (ii) nutrients provided in the exudates of germinating seeds are quantitatively limiting for optimal bacterial growth, and (iii) S. enterica preferentially imports required metabolites rather than synthesizing them de novo. Our data illuminate the complex metabolic interactions that determine the success of bacteria in the spermosphere and early rhizosphere.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
The bacterial strains used in this study are listed in Table 1. Strains were cultured on Luria-Bertani (LB) agar medium at 37°C. When necessary, antibiotics were added at the following concentrations: ampicillin (Amp), 100 μg/ml; chloramphenicol (Cm), 40 μg/ml; kanamycin (Kan), 50 μg/ml; tetracycline (Tet), 15 μg/ml.
TABLE 1.
Strains and plasmids used in this study
| S. enterica strain or plasmid | Genotype | Reference or source |
|---|---|---|
| Wild type | ||
| 14028S | 14028S | |
| Amino acid auxotrophs | ||
| JDB0970 | 14028S ΔasnA::Kan ΔasnB::Cm | This work |
| DM13159 | LT2 ΔgdhA::Kan | Diana Downs |
| JDB1131 | 14028S ΔgdhA::Kan ΔgltB::Cm | This work |
| JDB0966 | 14028S ΔglnA::Cm | This work |
| DM0598 | LT2 glyA::Tn10d | Diana Downs |
| JDB1194 | 14028S ΔglyA::Tn10d | This work |
| JDB0975 | 14028S ΔhisB::Kan | This work |
| JDB0978 | 14028S ΔilvE::Cm | This work |
| JDB0979 | 14028S ΔleuB::Kan | This work |
| JDB0968 | 14028S ΔlysA::Kan | This work |
| JDB1000 | 14028S ΔmetC::Kan | This work |
| JDB0977 | 14028S ΔpheA::Kan | This work |
| TT9667 | LT2 proC693::MudA | Diana Downs |
| JDB1207 | 14028S proC693::MudA | This work |
| TT191 | LT2 thr557::Tn10 | Diana Downs |
| JDB1208 | 14028S thr557::Tn10 | This work |
| JDB0818 | 14028S ΔtrpB::Kan | 24 |
| JDB1030 | 14028S ΔtyrA::Kan | This work |
| JDB1202 | 14028S ΔpheASTM2668tyrA::Cm | This work |
| JDB1203 | 14028S ΔpheASTM2668tyrA::Cm ΔtrpB::Kan | This work |
| JDB0969 | 14028S ΔSTM2360::Cm | This work |
| JDB0972 | 14028S ΔSTM2360::Cm ΔlysA::Kan | This work |
| JDB0967 | 14028S ΔyneH::Kan | This work |
| JDB0971 | 14028S ΔyneH::Kan ΔglnA::Cm | This work |
| Transport mutants | ||
| JDB0985 | 14028S ΔbtuB::Kan | This work |
| JDB0986 | 14028S ΔfliY::Kan | This work |
| JDB0987 | 14028S ΔglnH::Kan | This work |
| JDB0988 | 14028S ΔgltI::Kan | This work |
| JDB1204 | 14028S ΔhisJ::Cm | This work |
| JDB0989 | 14028S ΔhisJ::Kan | This work |
| JDB1205 | 14028S ΔlivJ::Cm | This work |
| JDB1206 | 14028S ΔlivJ::Kan | This work |
| Plasmids | ||
| pKD3 | λ-Red recombination template with Cm cassette | 28 |
| pKD4 | λ-Red recombination template with Kan cassette | 28 |
Proteomic analysis by LC–MS-MS.
An S. enterica wild-type (WT) bacterial suspension was prepared in sterile water from an overnight LB medium streak culture and serially diluted to 103 CFU/ml. The bacterial density was verified by plating on LB medium. Three-tenths of a gram of surface-disinfested alfalfa seeds (25) were germinated in 20 ml of the 103-CFU/ml WT bacterial suspension at 24°C with gentle shaking. After 24 h, the irrigation water was removed by pipetting, and planktonic cells were harvested by centrifugation at 3,000 × g for 5 min. Total protein was extracted using the AllPrep DNA/RNA/Protein minikit (Qiagen) according to the manufacturer's instructions. The sample was submitted to the Mass Spectrometry/Proteomics Facility at the University of Wisconsin—Madison Biotechnology Center for protein identification by liquid chromatography-tandem mass spectrometry (LC–MS-MS).
Precipitated protein was solubilized overnight in 8 M urea (1.6 g/liter) at 4°C and diluted to 0.27 g/liter with 50 mM (NH4)HCO3, 4 mM Tris, pH 8. Proteins were reduced with 2 mM dithiothreitol (DTT) at 55°C for 30 min, alkylated with 5 mM iodoacetamide at 25°C in the dark for 20 min, and trypsinized (lyophilized trypsin; Promega) at an enzyme-to-substrate ratio of 1:50 at 37°C for 18 h. The products were concentrated and desalted by solid-phase extraction using Varian Omix tips according to the manufacturer's instructions. The eluates were in 2-μg/μl final concentration. LC–MS-MS was performed on a ThermoFisher Scientific Orbitrap XL mass spectrometer coupled to an Agilent 1100 series NanoLC system. The LC column was fabricated in house from a 75-μm by 360-μm fused silica capillary packed to a length of 15 cm with 3 μm C18 particles (Michrom Biosciences Magic C18AQ) and possessing an integrated electrospray emitter tip. The LC system was configured in “trap-elute” mode with a trapping column consisting of an Agilent Zorbax 300SB-C18 cartridge, 300 μm by 5 mm, packed with 5-μm particles. High-performance liquid chromatography (HPLC) solvents consisted of 0.5% acetic acid in water (A); 0.5% acetic acid, 95% acetonitrile and water (B); and 0.5% acetic acid, 1% acetonitrile in water (loading). Other LC parameters were a 5-μl injection volume and a 15-μl/min flow rate. Desalting proceeded for 20 min. At that time, the trapping-column valve was switched, bringing it in line with the binary nanopump, and the analytical column and gradient elution began. Peptides were eluted at a flow rate of 200 nl/min using a gradient of solvents A and B as follows: 0 to 40% B, 195 min; 40 to 60% B, 20 min; 60 to 100% B, 5 min; 100% B, 3 min; 100 to 0% B, 2 min; and reequilibration at 0% B, 30 min. The mass spectrometer was operated in positive ion mode with a 1.8-kV electrospray voltage and a 165°C capillary temperature. MS-MS was performed in the LTQ region of the Orbitrap XL using collision-induced dissociation (CID) fragmentation with a 2.5-m/z isolation width at a 35% normalized collision energy. Postacquisition data processing was performed using the Trans-Proteomic Pipeline (Institute for Systems Biology). Database searching utilized Mascot (Matrix Science) version 2.2. The search parameters included fully tryptic cleavage specificity (allowing 2 missed cleavages), a 30-ppm monoisotopic precursor mass error (allowing up to 2 13C atoms in the precursor ion), a 0.5-Da fragment mass error, fixed carbamidomethylation on cysteine residues, variable deamidation of asparagine and glutamine residues, and variable oxidation of methionine residues. The database consisted of Salmonella enterica serovar Typhimurium strain LT2 protein sequences to which were appended a list of common contaminant proteins, such as human keratins and trypsin, for a total of 5,507 sequence entries. Peptide and protein confidence levels were set at 95% with a requirement to observe 2 peptides for each reported protein. This yielded a peptide false-discovery rate of 1.2% and an effective protein false-discovery rate of 0%. Proteins were placed into functional metabolic categories based on their annotation in the Kyoto Encyclopedia of Genes and Genomes (KEGG) (26). Protein-associated reactions were assigned according to the method of Thiele et al. (27).
Generation of mutants.
Amino acid auxotrophs and transport mutants were generated by deletion of specific biosynthetic or transporter genes using the λ-Red recombination method (28). The primers used for targeted gene deletion and mutant verification are listed in Table 2. Mutations were P22 phage transduced into the S. enterica serovar Typhimurium 14028S background and verified using gene-flanking primers (Table 2) and the antibiotic cassette-specific primer c1 or k2 (28). Double mutants were constructed by phage transduction of single mutations into a second 14028S mutant background and verified by PCR. Alternatively, auxotrophs obtained by Tn10 or MudA transposon mutagenesis were P22 phage transduced from the S. enterica serovar Typhimurium LT2 strains listed in Table 1 into the S. enterica serovar Typhimurium 14028S background. Amino acid auxotrophy of mutants was verified by plating on minimal (M9) agar plates containing 0.4% (wt/vol) glucose, with and without addition of the appropriate amino acid(s). Amino acid crystals (<0.1 g) were sprinkled directly onto agar plates immediately before bacterial plating.
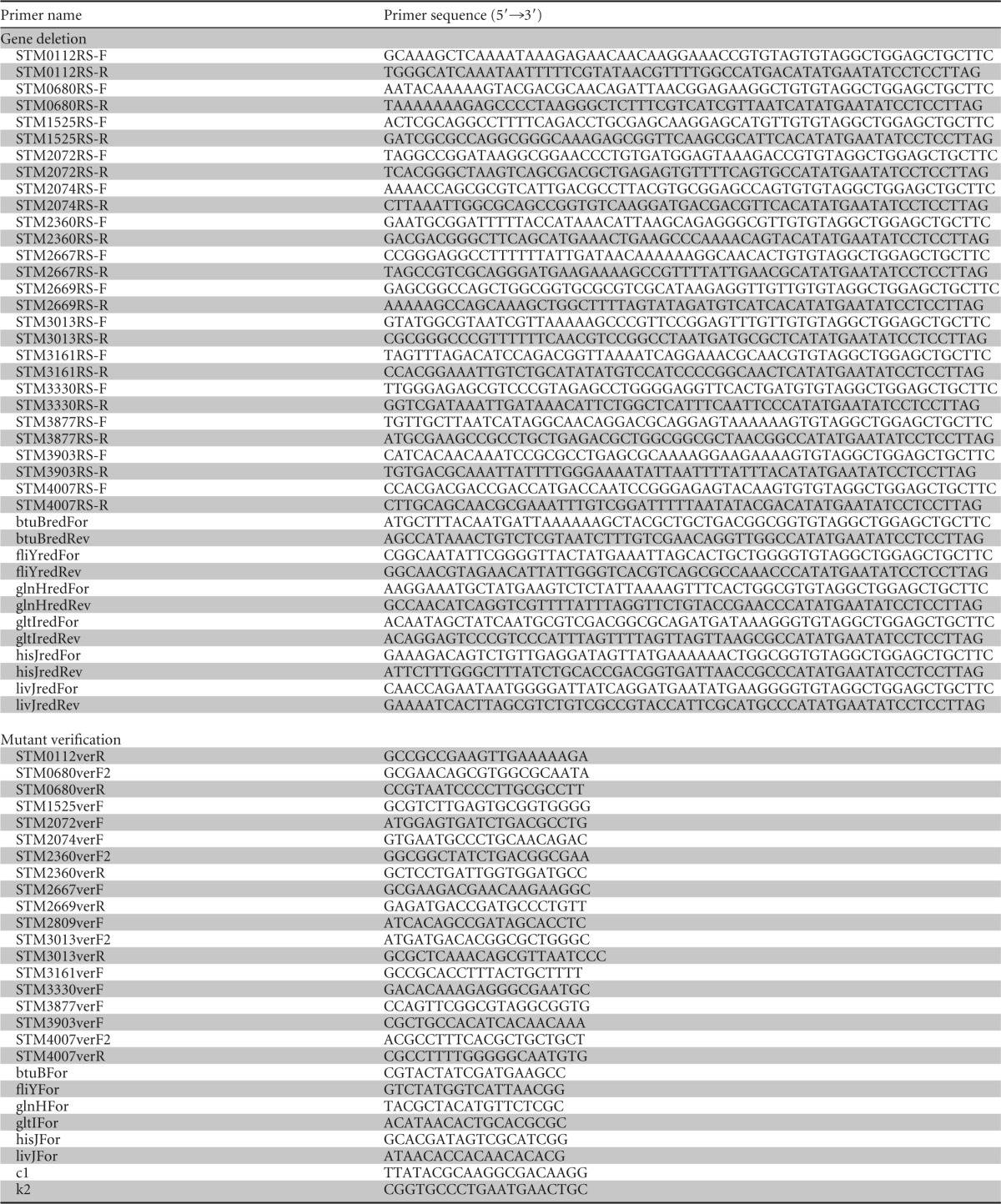
TABLE 2.
Primers used in this study
Growth assays in germinating alfalfa seedling exudate.
Bacterial growth in germinating alfalfa seedling exudates was determined as described previously (24). Overnight bacterial streak cultures were resuspended in sterile deionized water to ∼108 CFU/ml, as estimated by optical density at 600 nm and verified by dilution plating on LB medium. For single-strain assays, 0.3 g surface-disinfested alfalfa seeds were germinated in 20 ml of a 103-CFU/ml bacterial suspension for up to 72 h. For coinoculation (competition) assays, a 103-CFU/ml bacterial suspension containing a 1:1 ratio of bacterial strains was used. All germinating seedlings were incubated at 24°C with constant light and gentle shaking at ∼40 rpm. Planktonic bacterial cell populations were determined every 24 h by dilution plating on xylose-lysine-deoxycholate agar (XLD), XLD plus ampicillin (XLD+Amp), XLD+Cm, XLD+Kan, and/or XLD+Tet. The plates were incubated at 37°C overnight. There were three replicates per treatment, and each experiment was repeated at least three times.
In vivo complementation of amino acid auxotrophs.
Auxotrophs were chemically complemented by addition of 200 μl of 100 mM amino acid at 24 h to the alfalfa-bacterial culture described above. Due to lower water solubility, 400 μl of 50 mM tryptophan was used to achieve a final concentration of ∼1 mM. For each strain, replicate cultures were verified to have similar populations at 24 h prior to addition of amino acid or sterile double-distilled water (ddH2O) (control). Bacterial populations were enumerated at 48 and 72 h, as described above. There were three replicates per treatment, and each experiment was repeated at least three times.
Quantification of metabolites by liquid chromatography-mass spectrometry.
For quantification of metabolites in alfalfa exudates, 0.3 g surface-disinfested alfalfa seeds (25) were germinated in 20 ml sterile deionized water at 24°C with gentle shaking. At 0, 8, 24, 48, and 72 h, the alfalfa exudates were removed by pipetting and passed through a bottle top 0.2-μm polyethersulfone (PES) membrane filter (Corning). Exudates from three technical replicates were pooled for analysis. At least two biological replicates were analyzed for each time point.
For comparison of metabolite concentrations in the presence and absence of S. enterica, alfalfa cultures were prepared as described above, except that in the S. enterica-containing sample, 0.2 ml water was replaced by 0.2 ml of a 105-CFU/ml bacterial suspension in water. The bacterial suspension was prepared as for growth assay experiments. At 24, 48, and 72 h, 0.2 ml of irrigation water was removed from each culture by pipetting and passed through a 0.2-μm surfactant-free cellulose acetate membrane filter (Nalgene). Samples from three technical replicates were pooled for analysis, and three biological replicates were analyzed for each time point.
Samples were diluted with HPLC grade water (Chromasolv; Sigma-Aldrich) in a 1:20 ratio and analyzed using HPLC-MS consisting of a Dionex ultrahigh-performance liquid chromatograph (UHPLC) coupled to a Q Exactive Orbitrap mass spectrometer (Thermo Scientific, San Jose, CA). LC separation was achieved using a Synergi Fusion-RP 100A column (100 by 2 mm; 2.5-μm particle size; Phenomenex, Torrance, CA). The total run time was 25 min. The flow rate was 200 μl/min. Solvent A was 97:3 water/methanol with 10 mM tributylamine (TBA) and 10 mM acetic acid, pH ∼7.5; solvent B was methanol with 10 mM TBA. The gradient was 0 min, 5% B; 2.5 min, 5% B; 5 min, 20% B; 7.5 min, 20% B; 13 min, 55% B; 15.5 min, 95% B; 18.5 min, 95% B; 19 min, 0% B; 25 min, 0% B. Other LC parameters were an autosampler temperature of 4°C, an injection volume of 5 μl, and a column temperature of 25°C. Peaks were quantified based on the peak height relative to internal standards.
Statistical analyses.
Statistical analyses were performed using R software (version 2.14.1; R Development Core Team, R Foundation for Statistical Computing, Vienna, Austria [http://www.R-project.org]). The average log10-transformed bacterial population (single inoculation) or percent total population (coinoculation) was calculated for each bacterial strain at each time point for each experiment. Replicates within an experiment were treated as subsamples, and individual experiments were considered separate samples. In the single-inoculation experiments, the mean populations of wild-type and mutant were compared at each time point using paired, two-tailed t tests assuming unequal variance. For coinoculation experiments, the average population percentages of the mutant strain were compared to the expected value of μ, 50, in one-sample t tests. Since strains were inoculated at a 1:1 ratio in each coinoculation, each strain should comprise 50% of the total population if both strains are equally competitive. For complementation assays, the average populations for each treatment were calculated as described above. Differences among treatments at each time point were evaluated using Tukey's honestly significant difference (HSD) test (based on two-way analysis of variance [ANOVA], with strain and experiment as factors). Significance was set at a P value of < 0.05 for all tests. The WT bacterial growth curve was fitted to a modified Gompertz function using the nonlinear least-squares function (nls). The Gompertz function was modified as follows: y(t) = δ + αe−βe−Γt. Though the parameter β had a P value of 0.06, we still considered it significant in order to maintain the sigmoidal shape of the plotted function.
The Pearson's correlation between nutrient sufficiency and the growth phenotype was determined in R, but the plot was generated in Microsoft Excel 2010. We omitted glycine from our correlation analysis because it is readily synthesized from serine and threonine through single reactions. These chemical transformations likely occur during S. enterica growth in association with roots, as discussed above, to increase the available glycine pool. Alfalfa seedling exudates are a very poor source of glycine, and the biomass requirement for the amino acid was 2 orders of magnitude greater than that calculated for any other amino acid. The metabolic interaction between glycine, serine, and threonine is not accounted for in our calculation of nutrient sufficiency, and the true nutrient sufficiency ratio for glycine is likely lower than the value we calculated.
Differences in amino acid concentrations in the presence and absence of S. enterica were evaluated using one-sample t tests, comparing the nutrient ratio to the expected μ value of 1. If the presence of S. enterica had no effect on the nutrient concentration, then the ratio of the nutrient in the presence of S. enterica to that the absence of S. enterica should be 1. All values presented here are expressed as average ± standard deviation (SD).
RESULTS
The S. enterica active proteome indicates high metabolic activity in association with roots.
To investigate the metabolic requirements of S. enterica during root-associated growth, we used shotgun proteomics to identify expressed bacterial proteins, which indicated to us active metabolic pathways. LC–MS-MS identified 305 of the most abundant S. enterica serovar Typhimurium 14028S proteins expressed at 24 h during growth in alfalfa seedling exudates (see Table S1 in the supplemental material). Over half of these proteins (159/305; 52.1%) had known metabolic functions, based on the genome scale metabolic reconstruction of S. enterica serovar Typhimurium LT2 (27). The S. enterica serovar Typhimurium LT2 and 14028S genomes are >98% identical, and the only differences in gene content lie within prophages (29). In contrast, metabolic genes comprise roughly one-quarter of the S. enterica serovar Typhimurium LT2 chromosome (1,270/4,489 genes; 28.3%) (27). The apparent overrepresentation of metabolic genes expressed in our data, among all genes in the genome, suggests that S. enterica is in a highly metabolically active state. Furthermore, the proteins mapped to diverse metabolic networks. Proteins involved in central carbon, amino acid, nucleotide, fatty acid, cofactor, and energy metabolism were detected in our proteomic survey, in addition to those involved in transport, cell envelope biogenesis, tRNA charging, and oxidative-stress response (Fig. 1). Together, the identified metabolic proteins account for over one-quarter (561/2,023; 27.7%) of the known metabolic reactions of S. enterica serovar Typhimurium (27). While ion and metabolite transport accounted for the majority of detected reactions, a substantial number of proteins involved in central carbon and amino acid metabolism were also detected, suggesting that they may be essential metabolic networks for S. enterica growth in the spermosphere or rhizosphere. Nonmetabolic proteins, i.e., those not directly involved in nutrient utilization and biosynthesis, included those involved in genetic and environmental information processing, cell maintenance, and cell cycle control and proteins with unknown functions or network assignments (unassigned). Additionally, “unknown metabolic” proteins, e.g., those with annotated metabolic function but no known gene-protein-reaction association, were also included in the nonmetabolic set and accounted for 3.3% (10/305) of the total number of proteins identified.
FIG 1.
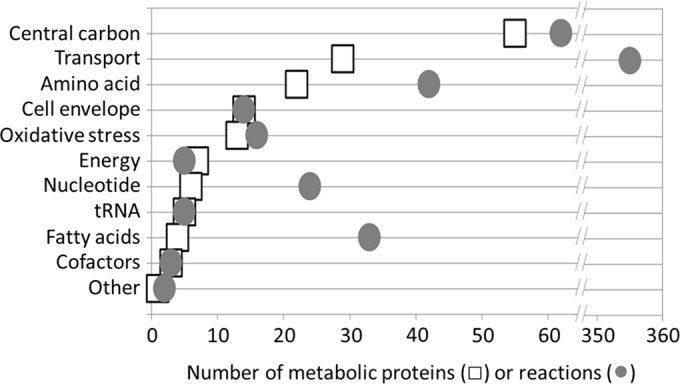
Distribution of metabolic proteins from S. enterica serovar Typhimurium growing with germinating alfalfa seeds at 24 h postinoculation. The proteins were identified using LC–MS-MS. Protein functions were categorized based on the proteins' annotation in KEGG (26). Protein-associated reactions were assigned according to the genome-scale metabolic reconstruction of S. enterica serovar Typhimurium (20).
Amino acid biosynthesis is required for optimal S. enterica growth in association with roots.
Interestingly, a majority of the proteins involved in amino acid metabolism detected in our proteomic screen were strictly biosynthetic (17/22; 77.3%). These proteins were located in the biosynthetic pathways of 15 different amino acids. These data suggested that amino acids that the bacterium cannot acquire in sufficient quantity from the plant may be important nutrients for S. enterica growth; thus, de novo biosynthesis by S. enterica is required. To test this hypothesis, amino acid auxotrophs and transport mutants were generated and tested for growth in association with germinating alfalfa sprouts.
S. enterica grows well in the spermosphere and rhizosphere of alfalfa seedlings, increasing 10,000-fold during the first 24 h and reaching populations of 109 log10 CFU/ml within 72 h (see Fig. S1 in the supplemental material). Of the 13 auxotrophs generated for amino acids, only 3 (for glycine, proline, and tryptophan) showed a growth defect relative to the WT at 24 h (Table 3). However, biosynthesis of asparagine, glutamine, lysine, methionine, phenylalanine, tryptophan, and tyrosine became necessary by 48 h and leucine by 72 h, likely because these nutrients became limiting. The auxotroph data correspond well to the proteomic data. Overall, auxotrophs corresponding to amino acids whose biosynthetic proteins were detected at 24 h exhibited growth defects at 24 h and/or 48 h. Specific amino acids became limiting at 24 h, requiring de novo biosynthesis by S. enterica. The growth defects of single-amino-acid auxotrophs were partially to fully complemented to WT levels by addition of a 1 mM concentration of the corresponding amino acid(s) to the germinating alfalfa at 24 h (see Table S2 in the supplemental material). Amino acids were applied at 24 h to reduce changes in nutrient exudation by germinating seeds (see Discussion). Chemical complementation of auxotrophs was not achieved when amino acids were provided at 0 h (data not shown), possibly because changes in seed/root exudate composition occurred in the altered chemical environment. Amino acid supplementation at either 0 or 24 h had no effect on the growth of the WT (data not shown). Collectively, the data suggest that de novo biosynthesis of certain amino acids by S. enterica is necessary because these nutrients are quantitatively limiting for bacterial growth and/or transport is metabolically costly. Though leucine, methionine, phenylalanine, and tyrosine biosynthesis was not required for WT level growth at 24 h, or leucine and glutamate biosynthesis at 48 h, auxotrophs for these amino acids were less competitive than the WT at these time points, indicating that biosynthesis of the amino acids is a fitness factor for S. enterica during different stages of plant-associated growth (see Table S3 in the supplemental material).
TABLE 3.
Protein expression data are a predictor of metabolic requirements of S. enterica during growth in germinating alfalfa exudate
| Auxotrophy | 24-h protein expression | Mutanta | Relative growth of auxotroph (log10 CFU/ml)b |
n | ||
|---|---|---|---|---|---|---|
| 24 h | 48 h | 72 h | ||||
| Arg | Biosynthesis | NA | ND | ND | ND | |
| Cys | Biosynthesis | NA | ND | ND | ND | |
| Gln | Biosynthesis | ΔglnA::Kan | +0.43 ± 0.23 | −0.20 ± 0.07 | WT | 6 |
| Ile, Leu, Val | Biosynthesis | ΔilvE::Cm | WT | WT | WT | 3 |
| Leu | Biosynthesis | ΔleuB::Kan | WT | WT | −0.46 ± 0.10 | 3 |
| Lys | Biosynthesis | ΔlysA::Kan | −0.25 ± 0.12 | −0.57 ± 0.03 | −0.56 ± 0.14 | 4 |
| Met | Biosynthesis | ΔmetC::Kan | WT | −0.90 ± 0.16 | −0.83 ± 0.19 | 4 |
| Phe | Biosynthesis | ΔpheA::Kan | WT | −0.16 ± 0.03 | −0.23 ± 0.03 | 3 |
| Ser | Biosynthesis | NA | ND | ND | ND | |
| Thr | Biosynthesis | thr557::Tn10 | +0.47 ± 0.03 | WT | WT | 3 |
| Trp | Biosynthesis | ΔtrpB::Kan | −0.40 ± 0.04 | −0.87 ± 0.15 | −0.87 ± 0.12 | 4 |
| Tyr | Biosynthesis | ΔtyrA::Kan | WT | −0.19 ± 0.07 | −0.34 ± 0.10 | 3 |
| Phe, Tyr | Biosynthesis | ΔpheASTM2668 tyrA::Cm | −0.23 ± 0.14 | WT | −0.24 ± 0.08 | 4 |
| Phe, Trp, Tyr | Biosynthesis | ΔpheASTM2668 tyrA::Cm ΔtrpB::Kan | −0.34 ± 0.23 | −0.65 ± 0.27 | −0.83 ± 0.19 | 5 |
| Asn | Catabolism | ΔasnA::Kan ΔasnB::Cm | WT | −0.25 ± 0.11 | −0.39 ± 0.12 | 4 |
| Asp | Metabolism | NA | ND | ND | ND | |
| Glu | Metabolism | ΔgdhA::Kan ΔgltB::Cm | WT | WT | WT | 3 |
| Gly | Metabolism | ΔglyA::Tn10d | −0.26 ± 0.19 | −0.24 ± 0.08 | −0.26 ± 0.16 | 5 |
| Ala | Not detected | NA | ND | ND | ND | |
| His | Not detected | ΔhisB::Kan | +0.22 ± 0.14 | WT | WT | 3 |
| Pro | Not detected | proC693::MudA | −0.59 ± 0.16 | WT | −0.16 ± 0.02 | 3 |
NA, mutant not available.
The relative growth values shown are averages ± SD and are statistically significantly different from those of the WT. Population differences between mutants and the WT that are not statistically significant are shown as WT. ND, not determined.
Specific amino acids are quantitatively limiting for S. enterica growth in association with roots.
To further test our hypothesis that amino acids are quantitatively limiting, we determined the concentrations of amino acids and their derivatives in alfalfa seedling exudate over time (see Table S4 in the supplemental material). A large proportion of amino acids were released within 8 h of seed imbibition; amino acid concentrations peaked at either 24 h, similar to results reported previously (30), or 48 h, and concentrations declined thereafter. Differences in the times of peak amino acid concentrations in plant exudates reported here and by Phillips et al. (30) are likely due to differences in seedling culture conditions. Depletion of amino acids in alfalfa exudates in the presence of S. enterica suggests that the bacteria utilize these plant-derived nutrients during growth (Fig. 2; see Table S5 in the supplemental material). Consistent with our auxotroph growth data, as the concentrations of amino acids such as tryptophan (Fig. 2A), methionine (Fig. 2B), lysine (Fig. 2C), and phenylalanine (Fig. 2D) in alfalfa seedling exudates declined over time, the growth defects of auxotrophs for these amino acids increased. The concentrations of most amino acids were depleted by 48 h, corresponding to the time at which growth defects became evident for most of the amino acid auxotrophs (Table 3). The inability to biosynthesize required nutrients as external nutrient concentrations diminished likely resulted in suboptimal growth of auxotrophs.
FIG 2.
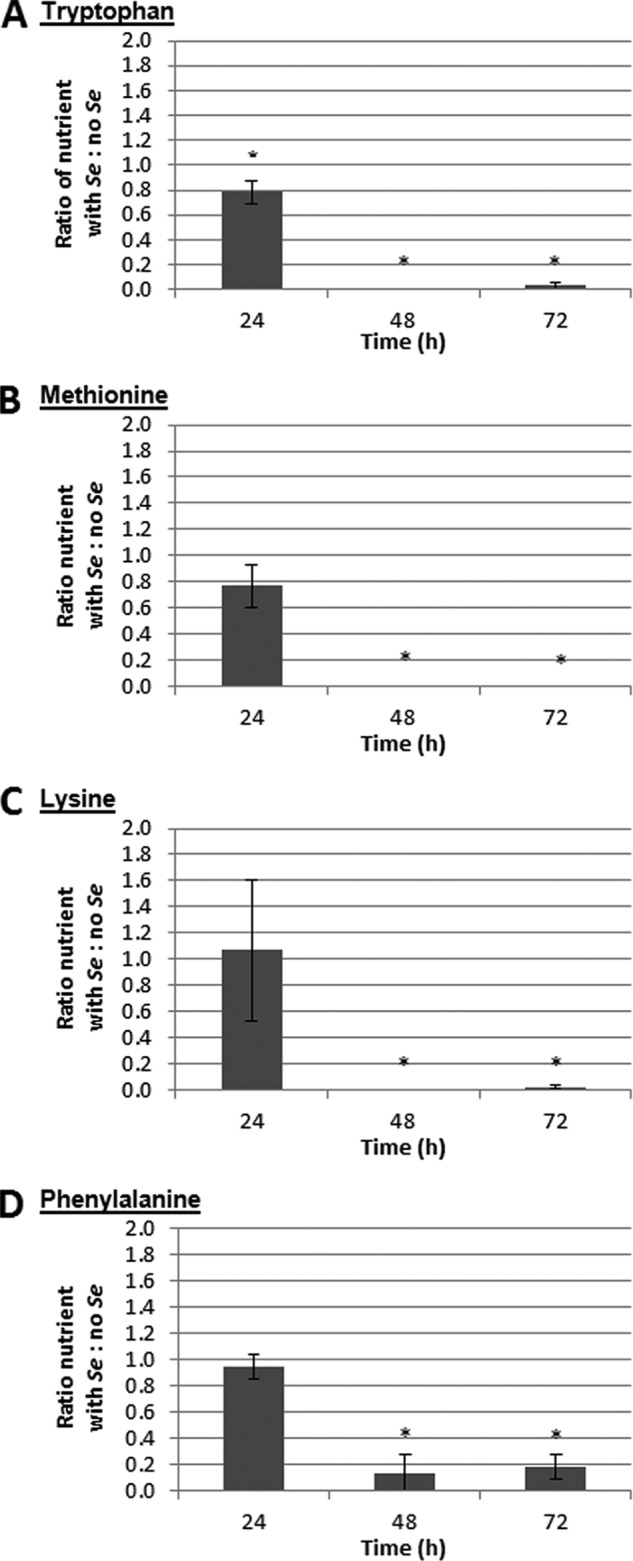
Relative concentrations of select amino acids in alfalfa seedling exudate in the presence of S. enterica (Se). The data shown are the means of three independent experiments with three replicates each. The error bars denote the standard deviations. *, the amino acid concentration in the presence of S. enterica is different from the concentration in the absence of S. enterica.
We also calculated the concentration of each of 19 proteinogenic amino acids that would be required for bacterial biomass, assuming S. enterica populations of 107, 108.5, and 109 log10 CFU/ml at 24, 48, and 72 h. These populations were the rough average populations of the WT at each time point in the singly inoculated experiments comparing WT and mutant growth (see Fig. S1 in the supplemental material). Biomass requirements were also based on the previously reported S. enterica biomass composition and biomass–viable-cell conversion factor under aerobic conditions (31, 32). The relative sufficiency of each amino acid was expressed as the ratio between the biomass requirement and the availability in alfalfa exudates (see Table S6 in the supplemental material). A higher ratio was observed for amino acids whose corresponding auxotrophs exhibited growth defects in the alfalfa seedling environment. A plot of the relative auxotroph growth at 48 h against the biomass requirement/availability ratio at 24 h revealed a moderate correlation between the relative sufficiency of each amino acid and the bacterial growth phenotype (R2 = 0.45; P = 0.01) (Fig. 3). It was expected that if an amino acid was insufficiently available in alfalfa seedling exudates, as indicated by a high biomass requirement/availability ratio, then an auxotroph incapable of biosynthesizing the required amino acid would be reduced in growth. Glycine was omitted from analysis because serine and threonine present in alfalfa exudate likely served as sources of glycine. It was notable that glycine was by far the most deficient amino acid, with biomass requirements exceeding 269 times its availability in alfalfa exudate. Linear regression indicated that nutritional status partially influences the growth phenotype of S. enterica in the alfalfa spermosphere/rhizosphere.
FIG 3.
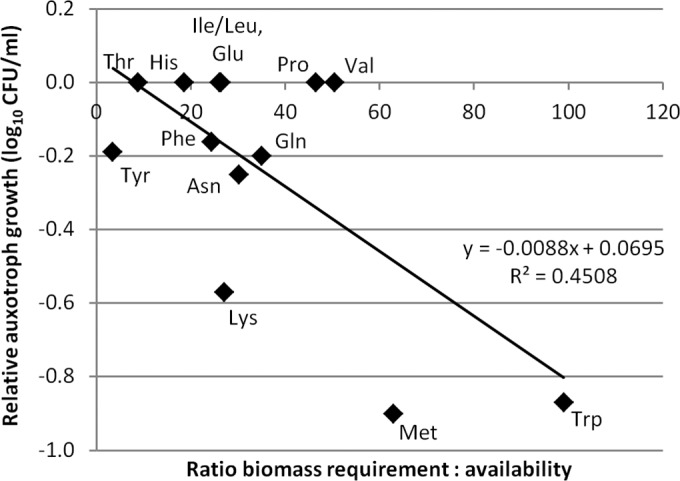
Nutrient deficiency is moderately correlated with reduced growth of S. enterica amino acid auxotrophs at 48 h postinoculation. The growth of the auxotroph is expressed relative to that of the WT and represents the mean difference of at least three experiments with three replicates each. The biomass requirement for each amino acid is the calculated amount of each nutrient present in S. enterica WT bacterial biomass at 24 h. The availability of each nutrient in alfalfa exudate at 24 h was quantified by HPLC-MS. The biomass requirement/availability ratio is a relative expression of nutrient sufficiency. The ratio at 24 h was compared to the observed auxotroph growth phenotype at 48 h, rather than growth at 24 h, because the nutrient deficiency experienced by the bacteria may not be immediately evident.
Glutamate transport acts as a fitness factor during S. enterica growth in association with roots.
Biosynthetic proteins were not detected at 24 h for amino acids (glutamate, histidine, and the branched-chain amino acids isoleucine, leucine, and valine) whose auxotrophs grew similarly to the WT over 72 h, suggesting sufficient availability of these amino acids from the environment. Instead, seven amino acid transporters were detected. To determine whether nutrient transport by these proteins was essential to S. enterica growth in association with roots, we generated deletion mutants in the corresponding genes. Mutants impaired in glutamine (ΔglnH::Kan) and glutamate (ΔgltI::Kan) transport were modestly reduced in growth in alfalfa exudates at all time points (Table 4), whereas cobalamin, cystine, histidine, and branched-chain amino acid transport mutants grew similarly to the WT. Reduced growth of the glutamine and glutamate transport mutants suggested that these amino acids were nutrients that S. enterica acquired from the plant. The reduced growth of the glutamate transport mutant, but not the glutamate auxotroph, correlated well with the metabolite quantitation data (see Table S4 in the supplemental material). These results indicated that glutamate was acquired exogenously from the plant in sufficient quantities to eliminate the need for de novo biosynthesis of the metabolite.
TABLE 4.
Impaired nutrient transport did not significantly impact S. enterica growth in germinating alfalfa exudates
| Substrate | Mutant | Relative growth of mutant (log10 CFU/ml)a |
n | ||
|---|---|---|---|---|---|
| 24 h | 48 h | 72 h | |||
| Cobalamin | ΔbtuB::Kan | WT | WT | WT | 4 |
| Cystine | ΔfliY::Kan | WT | WT | WT | 4 |
| Gln | ΔglnH::Kan | −0.28 ± 0.16 | −0.28 ± 0.11 | −0.24 ± 0.03 | 3 |
| Glu, Asp | ΔgltI::Kan | −0.26 ± 0.10 | −0.28 ± 0.19 | −0.28 ± 0.03 | 5 |
| His | ΔhisJ::Kan | +0.16 ± 0.06 | WT | WT | 4 |
| His | ΔhisJ::Cm | WT | WT | −0.21 ± 0.04 | 5 |
| Ile, Leu, Val | ΔlivJ::Kan | WT | WT | WT | 3 |
| Ile, Leu, Val | ΔlivJ::Cm | WT | WT | WT | 3 |
The relative growth values given are averages ± SD and are statistically significantly different from those of the WT. Population differences between mutants and the WT that are not statistically significant are shown as WT.
The relative importance of amino acid biosynthesis and transport is amino acid and growth stage dependent.
We posited that bacteria preferentially import required metabolites, if they are available, rather than synthesize them de novo due to the high energy cost of biosynthesis. To test this hypothesis, the auxotroph and transport mutants were coinoculated at 1:1 ratios into alfalfa seedling cultures, and populations were monitored over time. If there was no preference for biosynthesis or transport, then the auxotroph and transport mutants should each comprise 50% of the total bacterial population. However, if transport were favored over biosynthesis, then the population of the transport mutant should be less than that of the biosynthetic mutant, and vice versa. Surprisingly, our results indicate that the relative importance of amino acid biosynthesis and transport was dependent on both the specific amino acid and the growth stage of the bacterium. Data for the histidine mutants supported our hypothesis (Fig. 4A). The auxotroph, which relied on import of exogenous histidine, grew better than the transport mutant at all time points, indicating that histidine transport was more important than histidine biosynthesis for S. enterica growth in alfalfa seedling exudates. Our quantification data in Table 3 indicated that histidine was sufficiently available as a nutrient, so biosynthesis may not be required. In contrast, biosynthesis and transport of branched-chain amino acids were equally important at 24 and 48 h (Fig. 4B). Similar populations of the auxotroph and transport mutants were recovered at these time points. At 72 h, biosynthesis contributed more than transport to S. enterica growth. The transport mutant, which maintained branched-chain amino acid-biosynthetic ability, comprised a higher proportion of the total bacterial population than the auxotroph, which could not synthesize these required metabolites. The concentrations of leucine/isoleucine and valine in alfalfa exudates at 72 h were roughly half the concentrations available at 48 h (see Table S2 in the supplemental material), explaining the greater contribution of biosynthesis to bacterial fitness. Glutamine biosynthesis and transport were both important for S. enterica growth in alfalfa seedling exudates, but at different times (Fig. 4C). The transport mutant was considerably less competitive than the auxotroph at 24 h, indicating that glutamine transport was an important trait for growth in exudates released early during alfalfa seed germination. However, during later stages of seedling development, glutamine biosynthesis became more relevant. At 48 and 72 h, the glutamine transport mutant, which could synthesize glutamine, comprised a large majority of the coinoculated S. enterica population.
FIG 4.
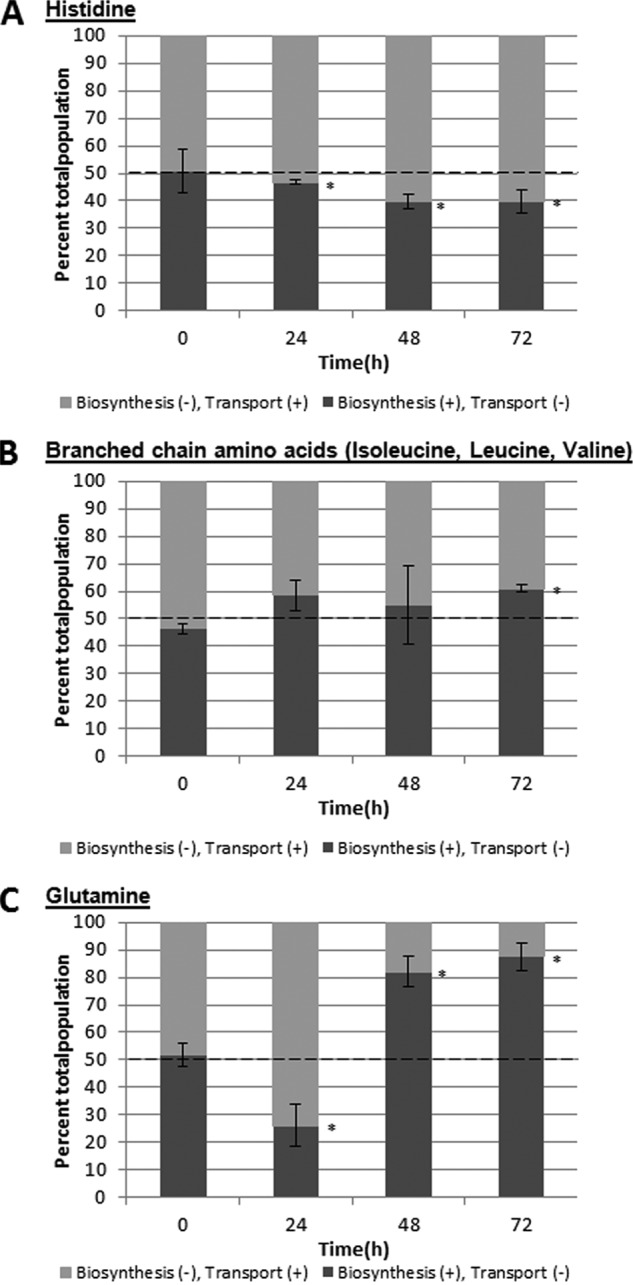
Comparison of the fitness contributions of biosynthesis and transport of specific amino acids to S. enterica growth in planta. Biosynthetic and transport mutants were coinoculated at a 1:1 ratio. The light-gray bars indicate the proportions of the total population composed of the biosynthesis (−), transport (+) ΔglnA::Cm (A), ΔhisB::Kan (B), and ΔlivE::Cm (C) mutants. The dark-gray bars indicate the proportions of the total population composed of the biosynthesis (+), transport (−) ΔglnH::Kan (A), ΔhisJ::Cm (B), and ΔlivJ::Kan (C) mutants. The data shown are the means of three independent experiments with three replicates each. The error bars indicate the standard deviations. *, the populations are statistically different from 50%.
Previously uncharacterized genes influence S. enterica growth in association with roots.
Two previously uncharacterized S. enterica genes were found to influence S. enterica growth in alfalfa exudates. We investigated these genes because their annotated functions were potentially biosynthetic for amino acids and their functional homologs were identified in the protein survey. yneH is predicted to encode glutaminase, similar to glnA. The YneH protein homolog in Escherichia coli has been biochemically confirmed to exhibit glutaminase activity, hydrolyzing l-glutamine to l-glutamate and NH4+ (33). However, the reversibility of this reaction is unknown, and we investigated the gene due to its potential to produce glutamine. While ΔglnA resulted in glutamine auxotrophy on M9 plus glucose (M9+Glc) agar, a ΔyneH mutant was prototrophic on the same medium (data not shown). This indicates that if the YneH reaction can be reversed to produce glutamine, its biosynthetic activity cannot functionally compensate for the loss of glnA. In alfalfa seedling exudates, ΔyneH::Kan had a positive effect on S. enterica WT growth. Populations of a ΔyneH::Kan mutant were higher than those of the WT at 24, 48, and 72 h (Table 5). The effects of ΔglnA and ΔyneH were additive (Tables 2 and 5).
TABLE 5.
Roles of yneH and STM2360 in S. enterica growth with germinating alfalfa seeds
| Mutant | Relative growth of mutant (log10 CFU/ml)a |
n | ||
|---|---|---|---|---|
| 24 h | 48 h | 72 h | ||
| ΔyneH::Kan | +0.32 ± 0.12 | +0.07 ± 0.05 | +0.06 ± 0.04 | 5 |
| ΔyneH::Kan ΔglnA::Cm | +0.82 ± 0.14 | WT | WT | 4 |
| ΔSTM2360::Cm | −0.29 ± 0.08 | −0.31 ± 0.05 | −0.23 ± 0.03 | 3 |
| ΔSTM2360::Cm ΔlysA::Kan | WT | −0.47 ± 0.26 | −0.49 ± 0.12 | 4 |
The relative growth values given are averages ± SD and are statistically significantly different from those of the WT. Population differences between mutants and the WT that are not statistically significant are shown as WT.
STM2360 (STM14_2907 in S. enterica 14028S) is another uncharacterized gene that may be involved in amino acid biosynthesis. It is annotated in the S. enterica genome as a gene encoding diaminopimelate decarboxylase, similar to the penultimate lysine biosynthesis gene lysA. The two corresponding proteins show 26% identity (43% similarity) over 92% of their sequences at the protein level, as determined by BLASTp (data not shown). The two genes are not homologous at the nucleotide level by BLASTn. A lysA mutant is auxotrophic for lysine, but a STM2360 mutant is not an auxotroph (data not shown). This indicates that STM2360 cannot functionally compensate for the loss of lysA despite their similar annotated functions. STM2360 contributed to S. enterica growth in alfalfa seedling exudates throughout the 72-h experimental period (Table 5). Populations of the ΔSTM2360::Cm mutant were 0.2 to 0.3 log10 CFU/ml lower than those of the WT at all sampling time points, and this growth defect could not be complemented by addition of 1 mM lysine at 24 h (data not shown). The growth defects due to the ΔlysA and ΔSTM2360 mutations were not additive (Tables 2 and 5).
DISCUSSION
Released plant exudates contain many low-molecular-weight metabolites, such as sugars and amino acids, that can be directly used by microbes for carbon and energy. These compounds are the major driver of microbial activity in the spermosphere and early rhizosphere (7), enabling members of the plant microbiome, such as S. enterica, to grow and colonize seed and root surfaces. In soil, the bulk of seed exudates are generally released within the first 12 h of imbibition (7), and enteric bacteria grow most rapidly during the first 24 h following seed imbibition (4, 24, 34). Despite the importance of this early phase of seed/root exudates in determining the behavior and composition of the plant-associated microbial community, little is known about the dynamic changes in the composition of plant exudates or the microbial metabolic requirements for growth in this environment. In this study, we determined the active proteome of S. enterica during growth in the spermosphere and early rhizosphere of germinating alfalfa seedlings to identify metabolic pathways that may be important for growth in this environment. The contributions of specific amino acid metabolic pathways to S. enterica growth were assessed by mutational analysis. We also quantified the temporal release of amino acids and their derivatives in the exudates of germinating alfalfa to correlate the S. enterica growth phenotype with its biomass requirements for these compounds.
Amino acids have previously been reported to be components of seed and root exudates (9–11, 35, 36). Intriguingly, we found that of the 22 detected proteins involved in amino acid metabolism, only one was directly involved in amino acid catabolism. In contrast, 14 detected proteins were strictly involved in the biosynthesis of specific amino acids. This finding suggested that though amino acids were present in the exudates of alfalfa seedlings, they may not be sufficiently available to meet bacterial biomass requirements. Therefore, S. enterica may require de novo biosynthesis. We investigated this hypothesis, as discussed below.
The disproportionate number of amino acid biosynthesis proteins detected in our proteomic survey, relative to the number of such proteins in the total S. enterica proteome, suggested that amino acids may be an important but limiting nutrient for S. enterica in the alfalfa spermosphere and early rhizosphere. We posited that if specific plant-derived amino acids were limiting for bacterial growth, then de novo biosynthesis by S. enterica would be required. Auxotrophs corresponding to amino acids whose biosynthetic proteins were detected at 24 h exhibited growth defects in the sprouting-alfalfa environment at 24 h and/or 48 h. These data support the notion that expressed proteins indicated metabolic pathways that contributed to S. enterica growth.
For most amino acid pathways, expressed proteins indicated that the associated pathway contributed to S. enterica growth. Disruption of these pathways reduced the S. enterica population recovered from the alfalfa spermosphere and early rhizosphere relative to the WT. However, the proteomic and growth data in association with roots did not correlate for threonine and branched-chain amino acids. Biosynthetic proteins were detected for these amino acids, but the corresponding auxotrophs were not reduced in growth throughout the 72-h experimental period. The discrepancy between the nutritional and phenotypic data, which suggested no biosynthetic requirement for threonine, and the proteomic data, which indicated threonine biosynthesis, may be explained through the metabolism of two other amino acids, glycine and methionine.
S. enterica has two pathways for glycine biosynthesis. Threonine can be converted into glycine by LtaA or TdH and Klb (26), though these proteins were not detected in our proteomic survey. Alternatively, glycine may be produced from serine by GlyA. Our data indicated that the requirement for glycine for growth far exceeds the availability of the amino acid in alfalfa exudates (see Table S6 in the supplemental material). Under such glycine starvation, the ΔglyA::Tn10d glycine auxotroph was expected to be severely reduced in growth relative to the WT. However, the minor growth defect (−0.24 to 0.26 log10 CFU/ml) observed for the mutant suggested that the bacterium was able to biosynthesize much of the glycine that it required through an alternative pathway to glyA, e.g., through conversion of threonine to glycine. Thus, the ΔglyA::Tn10d mutant was not truly auxotrophic. Auxotrophy of the ΔglyA::Tn10d mutant on M9+Glc medium suggested that the threonine-dependent pathway for glycine biosynthesis was not active; however, the pathway may be activated during growth in association with roots due to the excess availability of threonine in the exudates of germinating alfalfa. Under this hypothesis, as threonine is depleted to produce glycine, the external threonine pool would no longer be sufficient for optimal growth, and threonine biosynthesis itself would be required to meet the cellular need for both threonine and glycine. The high glycine requirement would barely be reduced in the absence of threonine biosynthesis even if all of the external threonine was converted to glycine. Taken together, glycine metabolism may explain the detection of the ThrC gene, encoding threonine synthase, when the nutritional quantification and threonine mutant data indicated that threonine was not a limiting nutrient.
Interestingly, the threonine auxotroph grew better than the WT during the first 24 h following inoculation into a germinating seed culture. The threonine-biosynthetic protein ThrB is encoded upstream of ThrC, which was detected as part of the active proteome, in an operon. ThrB and MetA, involved in methionine biosynthesis, both utilize homoserine as a substrate (37) and may compete with each other for the compound. Disruption of threonine biosynthesis might relieve this competition, allowing increased biosynthesis of methionine, a limiting nutrient in alfalfa exudates. Increased methionine may explain the improved fitness and 24-h growth of the threonine auxotroph.
The glycine and glutamate proteins (GlyA and GdhA, respectively) detected in our proteomic survey were classified as metabolic rather than biosynthetic because their roles in catabolism versus biosynthesis were unclear. The reactions that they catalyzed are reversible; glycine and glutamate can thus be used by GlyA and GdhA, respectively, as either substrates or products. The reduced growth of the ΔglyA::Tn10d mutant and the partial (48 h) to full (72 h) complementation of the growth defect by addition of 1 mM glycine suggest that GlyA produces, rather than consumes, glycine during S. enterica growth in the exudates of germinating alfalfa.
Partial to full complementation of all auxotrophs in association with roots by supplementation with a 1 mM concentration of the appropriate amino acid showed that a nutritional defect contributed to the reduced growth of these mutants. Amino acids were applied at 24 h, rather than at 0 h, to reduce changes in nutrient exudation by germinating seeds. The composition of plant exudates results from the net influx and efflux of compounds by the plant (38); these influx and efflux rates can change in response to the external chemical environment (39). However, a study by Phillips et al. (30) indicated that in germinating alfalfa, amino acid concentrations stabilize after 24 h, suggesting that by that time (i) the pool of amino acids available to microbes, i.e., S. enterica, for growth has already been released and (ii) the net concentration of plant-derived amino acids in seed/root exudates is less likely to be influenced by exogenous supplementation with the compounds. Partial, as opposed to full, chemical complementation may result from addition of an insufficient concentration of amino acid, the metabolic burden of the inserted antibiotic cassette in particular mutant backgrounds, or pleiotropic effects of specific mutations.
Quantification of amino acids in the exudates of germinating alfalfa seedlings revealed the presence of all 19 of the proteogenic amino acids detectable by our methods. The concentrations reported in this study are similar in scale to those reported for young crop seedlings (35, 36). We found no correlation between the absolute concentrations of amino acids and the growth of S. enterica amino acid auxotrophs with germinating alfalfa. However, a moderate correlation (Pearson's R2 = 0.45; P = 0.01) was discovered between the perceived sufficiency of amino acids and the amino acid auxotrophs' growth phenotypes. The perceived sufficiency of each amino acid was expressed as a ratio of the calculated biomass requirement (based on the S. enterica growth curve observed in association with roots and the biomass equation [32]) to the available concentration. This ratio is a novel approach to evaluating the nutritional status and allows the prediction of nutrients that may be limiting for bacterial growth. The statistical correlation between nutrient availability and the amino acid auxotrophs' growth phenotypes provides mathematical support for the idea that the identity of each nutrient influences bacterial metabolism and growth.
We were surprised to discover that the relative importance to S. enterica growth of biosynthesis versus transport of amino acids varied by the specific nutrient and the stage of growth. We assumed that transport would always be preferred due to the higher metabolic cost of biosynthesis. In our proteomic survey, we observed that metabolite and ion transport accounted for the majority of metabolic reactions detected. In support of our preferential-transport hypothesis, histidine auxotrophs capable of transporting histidine were more competitive than histidine transport mutants that were able to synthesize histidine. In contrast, glutamine transport was more important than biosynthesis only at 24 h, whereas the biosynthesis and transport of branched-chain amino acids (isoleucine, leucine, and valine) were equally important at 24 h and 48 h. Biosynthesis of both glutamine and the branched-chain amino acids became more important over time, presumably due to depletion of the amino acids in plant exudates. Based on our data, we reject the hypothesis that S. enterica preferentially imports required metabolites rather than synthesizing them de novo.
We discovered that two previously uncharacterized S. enterica genes, yneH and STM2360, influenced the plant-associated growth of the bacterium. In E. coli, yneH has been shown to encode a glutaminase involved in glutamine assimilation and may contribute to acid resistance (33). Glutaminase hydrolyzes glutamine to glutamate and ammonia. We initially investigated the role of this gene because the reversibility of the YneH-catalyzed reaction was unknown; acting in reverse, the reaction would be biosynthetic for glutamine. In this study, we found that even if the YneH-catalyzed reaction is reversible, leading to glutamine synthesis, it cannot functionally compensate for the loss of glutamine synthesis by glutamine synthetase (GlnA) in a ΔglnA mutant. In E. coli, deletion of yneH reduced growth by 50% in M9 medium containing glutamine as the sole carbon and nitrogen source (33). In contrast, a ΔyneH::Kan mutant of S. enterica grew roughly 2-fold better than the isogenic WT strain during the first 24 h after inoculation and similarly to the WT at 48 h and 72 h with alfalfa seedlings. While yneH promoted E. coli growth in M9+Gln, yneH negatively influenced the growth of S. enterica in association with roots. These opposing growth phenotypes may be attributed to the difference in growth conditions investigated and/or in the regulation and/or role of yneH in the two bacteria. The second uncharacterized S. enterica gene we investigated, STM2360, appears to play a positive role during growth with alfalfa seedlings. Growth of the ΔSTM2360::Cm mutant was 1.7- to 2.0-fold lower than that of the WT at all sampling time points. Though STM2360 is annotated in the S. enterica genome to have the same function as lysA, we found that the growth defect of the ΔSTM2360::Cm mutant cannot be complemented by addition of 1 mM lysine and that STM2360 could not functionally compensate for loss of lysA. The exact function of STM2360 remains elusive; biochemical and quantitative-PCR studies may reveal its activity and expression in S. enterica in the spermosphere and early rhizosphere.
During growth with germinating alfalfa, diverse S. enterica metabolic pathways are activated. Proteins involved in glycolysis, gluconeogenesis, the pentose phosphate pathway, fermentation, the tricarboxylic acid (TCA) cycle, amino acid metabolism, cofactor biosynthesis, fatty acid biosynthesis and catabolism, nutrient transport, cell envelope biogenesis, and maintenance of cellular redox potential were detected in our protein survey. A transcriptomic profile of S. enterica serovar Weltevreden during alfalfa sprout colonization compared to growth in minimal medium (40) revealed upregulation of genes in many of the same pathways. The identified S. enterica proteome indicates that S. enterica acquires and catabolizes plant-derived nutrients, as well as biosynthesizing required metabolites that are presumably absent or limiting in the spermosphere and early rhizosphere. Fructose has been identified as a major sugar in the seed and root exudates of a variety of crop plants (3, 9, 21, 35). Galactose and trehalose have also been reported in seed (7) and root (21, 35) exudates. Based on the proteomic data, we infer that S. enterica utilizes plant-derived fructose, galactose, melibiose, trehalose, and propionate. Proteins specifically involved in the catabolism of these substrates (fructose, PfkB; galactose, GalM, GalK, GalT, and GalE; melibiose, MelA; trehalose, TreA; propionate, PrpB and PrpD) and their transport (fructose, FruF; galactose, MglB) were identified in our survey. The availability of these carbon compounds in seedling and root exudates and the relative importance of each of the nutrients to S. enterica growth in the spermosphere and early rhizosphere remain to be determined.
Though other groups have previously quantified metabolites present in seed and root exudates, research has mostly focused on the roles of carbohydrates and organic acids in microbial growth and metabolic activity (3–5, 9, 11). The present study focused on the role of amino acid metabolism in bacterial growth with germinating seeds and young seedlings. We determined the dynamic changes in amino acid concentrations in the exudates of germinating seeds over the short time scale relevant for spermosphere and early rhizosphere inhabitants, including those involved in biocontrol, plant disease, and plant symbiosis. These dynamic changes and the microbial response are important aspects of spermosphere and early rhizosphere microbiology about which little is known. Early colonization facilitates microbial establishment and persistence in this niche and has human health implications. Colonization of the spermosphere and rhizosphere can lead to contamination of the entire plant (41–43), and attached S. enterica bacteria cannot be removed from crop surfaces, which are then consumed raw by humans (12).
Individual amino acids are important, but not essential, for S. enterica growth in the spermosphere and early rhizosphere. Despite the insufficient availability of many of these required nutrients in the exudates of germinating alfalfa, amino acid auxotrophs were only modestly reduced in growth. An emerging paradigm of bacterial rhizosphere competence is metabolic robustness. Pseudomonas fluorescens and E. cloacae mutants affected in single metabolic genes or functions are nonetheless able to achieve substantial growth in the rhizosphere (4, 5, 36), similar to our findings for S. enterica. To achieve dramatic reductions in bacterial growth in this environment, central metabolic networks, rather than dedicated pathways, may need to be targeted. Considering that central carbon metabolism was highly active, as indicated by our protein survey, and that central carbon metabolites serve as essential intermediates for cellular biosynthesis, perturbations in central metabolic networks may be more likely to cause pleiotropic metabolic effects that result in acute changes in growth. The metabolic robustness of S. enterica that makes metabolic manipulation difficult is the same trait that contributes to its success as a spermosphere and rhizosphere colonist.
Supplementary Material
ACKNOWLEDGMENTS
We thank Diana Downs for providing bacterial strains, Greg Barrett-Wilt and the Mass Spectrometry Facility at the University of Wisconsin—Madison Biotechnology Center for performing the LC–MS-MS proteome survey, and Kimberly Cowles and Jennifer Reed for helpful discussions.
Funding was provided by USDA-NIFA grant no. 2011-67017-30166, USDA-HATCH grant no. WIS01696, and the Food Research Institute at UW—Madison.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.02985-14.
REFERENCES
- 1.Curl EA, Truelove B. 1986. The rhizosphere. Springer-Verlag, New York, NY. [Google Scholar]
- 2.Liu S, Hu X, Lohrke SM, Baker CJ, Buyer JS, de Souze JT, Roberts DP. 2007. Role of sdhA and pfkA and catabolism of reduced carbon during colonization of cucumber roots by Enterobacter cloacae. Microbiology 153:3196–3209. doi: 10.1099/mic.0.2006/005538-0. [DOI] [PubMed] [Google Scholar]
- 3.Roberts DP, Dery PD, Yucel I, Buyer J, Holtman MA, Kobayashi DY. 1999. Role of pfkA and general carbohydrate catabolism in seed colonization by Enterobacter cloacae. Appl Environ Microbiol 65:2513–2519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Roberts DP, Dery PD, Yucel I, Buyer J, Holtman MA, Kobayashi DY. 2000. Importance of pfkA for rapid growth of Enterobacter cloacae during colonization of crop seeds. Appl Environ Microbiol 66:87–91. doi: 10.1128/AEM.66.1.87-91.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Roberts DP, McKenna LF, Lohrke SM, Rehner S, de Souza JT. 2007. Pyruvate dehydrogenase activity is important for colonization of seeds and roots by Enterobacter cloacae. Soil Biol Biochem 39:2150–2159. doi: 10.1016/j.soilbio.2007.03.027. [DOI] [Google Scholar]
- 6.Bertin C, Yang X, Weston LA. 2003. The role of root exudates and allelochemicals in the rhizosphere. Plant Soil 256:67–83. doi: 10.1023/A:1026290508166. [DOI] [Google Scholar]
- 7.Nelson EB. 2004. Microbial dynamics and interactions in the spermosphere. Annu Rev Phytopathol 42:271–309. doi: 10.1146/annurev.phyto.42.121603.131041. [DOI] [PubMed] [Google Scholar]
- 8.Roberts DP, Baker CJ, Kenna LF, Liu S, Buyer JS, Kobayashi DY. 2009. Influence of host seed on metabolic activity of Enterobacter cloacae in the spermosphere. Soil Biol Biochem 41:754–761. doi: 10.1016/j.soilbio.2009.01.010. [DOI] [Google Scholar]
- 9.Kamilova F, Kravchenko LV, Shaposhnikov AI, Azarova T, Makarova N, Lugtenberg B. 2006. Organic acids, sugars, and l-tryptophane in exudates of vegetables growing on stonewool and their effects on activities of rhizosphere bacteria. Mol Plant Microbe Interact 19:250–256. doi: 10.1094/MPMI-19-0250. [DOI] [PubMed] [Google Scholar]
- 10.Kuiper I, Kravchenko LV, Bloemberg GV, Lugtenberg BJJ. 2002. Pseudomonas putida strain PCL1444, selected for efficient root colonization and naphthalene degradation, effectively utilizes root exudate components. Mol Plant Microbe Interact 15:734–741. doi: 10.1094/MPMI.2002.15.7.734. [DOI] [PubMed] [Google Scholar]
- 11.Lugtenberg BJJ, Kravchenko LV, Simons M. 1999. Tomato seed and root exudate sugars, composition, utilization by Pseudomonas biocontrol strains and role in rhizosphere colonization. Environ Microbiol 1:439–446. doi: 10.1046/j.1462-2920.1999.00054.x. [DOI] [PubMed] [Google Scholar]
- 12.Barak JD, Schroeder B. 2012. Interrelationships of food safety and plant pathology: the life cycle of human pathogens. Annu Rev Phytopathol 50:241–266. doi: 10.1146/annurev-phyto-081211-172936. [DOI] [PubMed] [Google Scholar]
- 13.Harris LJ, Farber JN, Beuchat LR, Parish ME, Suslow TV, Garrett EH, Busta FF. 2003. Outbreaks associated with fresh produce: incidence, growth, and survival of pathogens in fresh and fresh-cut produce. Compr Rev Food Sci Food Safety 2(Suppl):78–141. doi: 10.1111/j.1541-4337.2003.tb00031.x. [DOI] [Google Scholar]
- 14.Doyle MP, Erickson MC. 2008. Summer meeting 2007: the problems with fresh produce: an overview. J Appl Microbiol 105:317–330. doi: 10.1111/j.1365-2672.2008.03746.x. [DOI] [PubMed] [Google Scholar]
- 15.Painter JA, Hoekstra RM, Ayers T, Tauze RV, Braden CR, Angulo FJ, Griffin PM. 2013. Attribution of foodborne illnesses, hospitalizations, and deaths to food commodities by using outbreak data, United States, 1998–2008. Emerg Infect Dis 19:407–415. doi: 10.3201/eid1903.111866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Centers for Disease Control and Prevention. 2013. List of selected multistate foodborne outbreaks investigations. CDC, Atlanta, GA: http://www.cdc.gov/foodsafety/outbreaks/multistate-outbreaks/outbreaks-list.html Accessed 25 November 2013. [Google Scholar]
- 17.Islam M, Doyle MP, Phatak SC, Millner P, Jiang X. 2004. Persistence of enterohemorrhagic Escherichia coli O157:H7 in soil and on leaf lettuce and parsley grown in fields treated with contaminated manure composts or irrigation water. J Food Prot 67:1365–1370. [DOI] [PubMed] [Google Scholar]
- 18.Islam M, Morgan J, Doyle MP, Phatak SC, Millner P, Jiang X. 2004. Fate of Salmonella enterica serovar Typhimurium on carrots and radishes grown in fields treated with contaminated manure composts or irrigation water. Appl Environ Microbiol 70:2497–2502. doi: 10.1128/AEM.70.4.2497-2502.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tyler HL, Triplett EW. 2008. Plants as a habitat for beneficial and/or human pathogenic bacteria. Annu Rev Phytopathol 46:53–73. doi: 10.1146/annurev.phyto.011708.103102. [DOI] [PubMed] [Google Scholar]
- 20.Teplitski M, Barak JD, Schneider KR. 2009. Human enteric pathogens in produce: un-answered ecological questions with direct implications for food safety. Curr Opin Biotechnol 20:166–171. doi: 10.1016/j.copbio.2009.03.002. [DOI] [PubMed] [Google Scholar]
- 21.Kwan G, Charkowski AO, Barak JD. 2013. Salmonella enterica suppresses Pectobacterium carotovorum subsp. carotovorum population and soft rot progression by acidifying the microaerophilic environment. mBio 4:e00557-12. doi: 10.1128/mBio.00557-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Potnis N, Soto-Arias JP, Cowles K, van Bruggen AHC, Jones JB, Barak JD. 2014. Xanthomonas perforans colonization influences Salmonella enterica in the tomato phyllosphere. Appl Environ Microbiol 80:3173–3180. doi: 10.1128/AEM.00345-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Soto-Arias JP, Groves RL, Barak JD. 2014. Transmission and retention of Salmonella enterica by phytophagous hemipteran insects. Appl Environ Microbiol 80:5447–5456. doi: 10.1128/AEM.01444-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hao LY, Willis DK, Andrews-Polymenis H, McClelland M, Barak JD. 2012. Requirement of siderophore biosynthesis for plant colonization by Salmonella enterica. Appl Environ Microbiol 78:4561–4570. doi: 10.1128/AEM.07867-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Charkowski AO, Barak JD, Sarreal CZ, Mandrell RE. 2002. Differences in growth of Salmonella enterica and Escherichia coli O157:H7 on alfalfa sprouts. Appl Environ Microbiol 68:3114–3120. doi: 10.1128/AEM.68.6.3114-3120.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kanehisa M, Goto S. 2000. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res 28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Thiele I, Hyduke DR, Steeb B, Fankam G, Allen DK, Bazzani S, Charusanti P, Chen FC, Fleming RM, Hsiung CA, De Keersmaecker SC, Liao YC, Marchal K, Mo ML, Özdemir E, Raghunathan A, Reed JL, Shin SI, Sigurbjörnsdóttir S, Steinmann J, Sudarsan S, Swainston N, Thijs IM, Zengler K, Palsson BO, Adkins JN, Bumann D. 2011. A community effort towards a knowledge-base and mathematical model of the human pathogen Salmonella Typhimurium LT2. BMC Syst Biol 5:8. doi: 10.1186/1752-0509-5-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Datsenko KA, Wanner BL. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci U S A 97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jarvik T, Smillie C, Groisman EA, Ochman H. 2010. Short-term signatures of evolutionary change in the Salmonella enterica serovar Typhimurium 14028S genome. J Bacteriol 192:560–567. doi: 10.1128/JB.01233-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Phillips DA, Fox TC, King MD, Bhuvaneswari TV, Teuber LR. 2004. Microbial products trigger amino acid exudation from plant roots. Plant Physiol 136:2887–2894. doi: 10.1104/pp.104.044222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Baumler DJ, Peplinski RG, Reed JL, Glasner JD, Perna NT. 2011. The evolution of metabolic networks of E. coli. BMC Syst Biol 5:182. doi: 10.1186/1752-0509-5-182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Raghunathan A, Reed J, Shin S, Palsson B, Daefler S. 2009. Constraint-based analysis of metabolic capacity of Salmonella typhimurium during host-pathogen interaction. BMC Syst Biol 3:38. doi: 10.1186/1752-0509-3-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Brown G, Singer A, Proudfoot M, Skarina T, Kim Y, Chang C, Dementieva I, Kuznetsova E, Gonzalez CF, Joachimiak A, Savchenko A, Yakunin AF. 2008. Functional and structural characterization of four glutaminases from Escherichia coli and Bacillus subtilis. Biochemistry 47:5724–5735. doi: 10.1021/bi800097h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stewart D, Reineke K, Ulaszek J, Fu T, Tortorello M. 2001. Growth of Escherichia coli O157:H7 during sprouting of alfalfa seeds. Lett Appl Microbiol 33:95–99. doi: 10.1046/j.1472-765x.2001.00957.x. [DOI] [PubMed] [Google Scholar]
- 35.El-Hamalawi ZA, Erwin DC. 1986. Components in alfalfa root extract and root exudate that increase oospore germination of Phytophthora megasperma f. sp. medicaginis. Phytopathology 76:508–513. doi: 10.1094/Phyto-76-508. [DOI] [Google Scholar]
- 36.Simons M, Permentier HP, de Weger LA, Wijffelman CA, Lugtenberg BJJ. 1997. Amino acid synthesis is necessary for tomato root colonization by Pseudomonas fluorescens strain WCS365. Mol Plant Microbe Interact 10:102–106. doi: 10.1094/MPMI.1997.10.1.102. [DOI] [Google Scholar]
- 37.Hacham Y, Gophna U, Amir R. 2003. In vivo analysis of various substrates utilized by cystathionine γ-synthase and O-acetylhomoserine sulfhydrylase in methionine biosynthesis. Mol Biol Evol 20:1513–1520. doi: 10.1093/molbev/msg169. [DOI] [PubMed] [Google Scholar]
- 38.Jones DL, Darrah PR. 1994. Amino-acid influx at the soil-root interface of Zea mays L. and its implications in the rhizosphere. Plant Soil 163:1–12. [Google Scholar]
- 39.Soldal T, Nissen P. 1978. Multiphasic uptake of amino acids by barley roots. Physiol Plant 43:181–188. doi: 10.1111/j.1399-3054.1978.tb02561.x. [DOI] [Google Scholar]
- 40.Brankatschk K, Kamber T, Pothier JF, Duffy B, Smits THM. 2014. Transcriptional profile of Salmonella enterica subsp. enterica serovar Weltevreden during alfalfa sprout colonization. Microb Biotechnol 7:528–544. doi: 10.1111/1751-7915.12104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cooley MB, Miller WG, Mandrell RE. 2003. Colonization of Arabidopsis thaliana with Salmonella enterica and enterohemorrhagic Escherichia coli O157:H7 and competition by Enterobacter asburiae. Appl Environ Microbiol 69:4915–4926. doi: 10.1128/AEM.69.8.4915-4926.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Guo X, van Iersel MW, Chen J, Brackett RE, Beuchat LR. 2002. Evidence of association of salmonellae with tomato plants grown hydroponically in inoculated nutrient solution. Appl Environ Microbiol 68:3639–3643. doi: 10.1128/AEM.68.7.3639-3643.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Solomon EB, Yaron S, Matthews SR. 2002. Transmission of Escherichia coli O157:H7 from contaminated manure and irrigation water to lettuce plant tissue and its subsequent internalization. Appl Environ Microbiol 68:397–400. doi: 10.1128/AEM.68.1.397-400.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


