Abstract
Flavobacterium psychrophilum is an important fish pathogen in salmonid aquaculture worldwide. Due to increased antibiotic resistance, pathogen control using bacteriophages has been explored as a possible alternative treatment. However, the effective use of bacteriophages in pathogen control requires overcoming the selection for phage resistance in the bacterial populations. Here, we analyzed resistance mechanisms in F. psychrophilum after phage exposure using whole-genome sequencing of the ancestral phage-sensitive strain 950106-1/1 and six phage-resistant isolates. The phage-resistant strains had all obtained unique insertions and/or deletions and point mutations distributed among intergenic and genic regions. Mutations in genes related to cell surface properties, gliding motility, and biosynthesis of lipopolysaccharides and cell wall were found. The observed links between phage resistance and the genetic modifications were supported by direct measurements of bacteriophage adsorption rates, biofilm formation, and secretion of extracellular enzymes, which were all impaired in the resistant strains, probably due to superficial structural changes. The clustered regularly interspaced short palindromic repeat (CRISPR) region was unaffected in the resistant isolates and thus did not play a role as a resistance mechanism for F. psychrophilum under the current conditions. All together, the results suggest that resistance in F. psychrophilum was driven by spontaneous mutations, which were associated with a number of derived effects on the physiological properties of the pathogen, including reduced virulence under in vitro conditions. Consequently, phage-driven physiological changes associated with resistance may have implications for the impact of the pathogen in aquaculture, and these effects of phage resistance on host properties are therefore important for the ongoing exploration of phage-based control of F. psychrophilum.
INTRODUCTION
Flavobacterium psychrophilum is a well-known fish-pathogenic bacterium which causes the diseases rainbow trout fry syndrome (RTFS) and cold water disease (CWD). The pathogen occurs frequently in salmonid production sites and has been a problem in aquaculture worldwide since the first isolation in Washington, USA (1). Genetic characterization of F. psychrophilum strains has shown a low level of diversity with a distinct association of specific strains with particular fish species (2, 3). The complete genome sequence of the virulent F. psychrophilum strain JIPO2/86 (ATCC 49511) revealed a 2,861,988-bp circular chromosome with 2,432 predicted protein-coding genes. Among these predicted proteins, stress response mediators, gliding motility proteins, adhesins, and putative secreted proteases are probably involved in the pathogenesis of the bacterium (4).
Due to increased resistance in F. psychrophilum to applied antibiotics (5), the use of lytic bacteriophages has been proposed as an alternative method for treatment of RTFS and CWD (6). Several studies have presented detailed characterization of bacteriophages with lytic capacity against F. psychrophilum, emphasizing the potential of phages to control the pathogen and reduce its impact on fish mortality (6–8). However, as seen for treatment with antibiotics, phage control of bacteria is also challenged by the development of bacterial resistance against bacteriophages. A better understanding of the mechanisms and functional implications of phage resistance in bacteria is therefore a prerequisite for assessing the potential of a phage-based treatment of pathogens. Moreover, as phage-host interactions are believed to drive a diversification of bacterial populations through the selection for resistance (9), studies of the mechanism of phage-driven changes in bacterial populations therefore add to our understanding of the role of phages in bacterial evolution.
Bacterial populations can evolve rapidly from dominance of phage-sensitive clones to phage-resistant clones when exposed to the selection pressure from lytic phages (10, 11). Depending on the mechanism of bacterial resistance, these changes can be either spontaneous or adaptive (12). The spontaneous resistance may occur through the loss or alteration of phage receptors, and the general perception is that such changes often translate into a fitness cost, because the phage receptors are involved in nutrient uptake (9, 13). Studies of adaptive-immune mechanisms have focused on clustered regularly interspaced short palindromic repeats (CRISPRs) and the CRISPR-associated (Cas) proteins. CRISPR loci are widely distributed in Bacteria (14) and Archaea (15) and are composed of 21- to 48-bp direct repeats interspaced by nonrepetitive nucleotides called spacers, commonly flanked by a variable number of cas genes (16). Resistance is acquired by the addition of short bacteriophage DNA sequences (spacers) in genomic CRISPR loci in response to lytic proliferation of phages (17). However, the molecular antiphage mechanism of CRISPR is unclear; some studies have supported the hypothesis of a bacterial RNA interference mechanism (18), while others have indicated DNA as the target (19, 20).
The ecological and evolutionary pressures that decrease or increase the virulence of bacterial pathogens are largely unknown. In some cases, prophages contribute with lysogenic conversion genes that have profound repercussions on the properties, fitness, and virulence of the bacterial host (21). There is thus increasing evidence from bacterial pathogens that lysogeny is a driver of virulence evolution (22–24). More recently, however, studies of Flavobacterium columnare (25), Staphylococcus aureus (26), and Serratia marcescens (27) have suggested that selective pressure from lytic bacteriophages has selected for phage-resistant clones with a lower virulence. Hence, these results have shown that bacteriophages serve as a driving force in bacterial pathogenesis, affecting directly bacterial virulence at the time of infection. A recent compilation of bacteriophage susceptibility patterns in a collection of F. psychrophilum isolates which obtained resistance after exposure to specific phages revealed both cross-resistance to and loss of resistance to other bacteriophages (28). The loss of resistance was associated with the loss of a specific 6H-type prophage (29), suggesting that prophages play a role in resistance to certain phages. However, the underlying genomic mechanisms of phage resistance and their phenotypic consequences in F. psychrophilum have not been investigated in detail. In this study, we identified mutations conferring resistance in six phage-resistant F. psychrophilum strains, derived from the sensitive wild-type strain 950106-1/1, which were completely genome sequenced. In addition, we provide insights into the impact of these bacteriophage-derived genetic changes on bacterial virulence.
MATERIALS AND METHODS
Bacterial strains, media, and growth conditions.
A collection of six phage-resistant isolates (V1-20, V2-20, V3-5, V4-24, V4-28, and V4-33) were derived from the virulent F. psychrophilum strain 950106-1/1 (6) after exposure (20- to 90-h experiments) to the phage FpV4 or FpV21 or a cocktail of 11 phages (see Table S1 in the supplemental material). Phage-resistant isolates V1-20 and V3-5 were isolated in the presence of bacteriophage FpV4 in two independent cultures using multiplicities of infection (MOI) of 1 and 0.0002, respectively. In the same way, phage-resistant isolates V2-20 and V4-33 were selected in the presence of bacteriophage FpV21 with MOI of 0.3 and 0.00004, respectively. Finally, clones V4-24 and V4-28 were isolated in the presence of a cocktail of bacteriophages using MOI of 0.05 and 4, respectively (R. Christiansen, L. Madsen, I. Dalsgaard, D. Castillo, P. Kalatzis, and M. Middelboe, unpublished data). All the phage-resistant isolates were stored at −80°C in tryptone-yeast extract-salt broth (TYES-B) (0.4% tryptone, 0.04% yeast extract, 0.05% CaCl2, and 0.05% MgSO4) (30) with 15% glycerol. For growth of F. psychrophilum strains, cells were inoculated in TYES-B and incubated at 15°C with agitation for 48 to 72 h (6).
Bacteriophage preparation and host range analysis.
The 32 bacteriophages used in this study were previously isolated from fish farms in Chile and Denmark (6, 7) (see Table S2 in the supplemental material). For preparation of high-titer phage stocks, 200 μl bacteriophage solution was added to agar plates with the bacterial host embedded in a layer of top agar (TYES-B with 0.4% agar) on TYES-B with 1.1% agar (TYES-A). Following the incubation at 15°C for 4 days, bacteriophages were eluted from agar plates with confluent lyses by adding 5 ml of SM buffer (50 mM Tris-Cl [pH 7.5], 99 mM NaCl, 8 mM MgSO4) and subsequent purification by centrifugation (6,000 × g, 20 min, 4°C) and 0.2-μm-pore-size filtration.
The host range of the collection of bacteriophages on the 6 phage-resistant isolates and the original wild-type strain was determined by spotting 10 μl of bacteriophage concentrate (108 PFU ml−1) on top of a TYES-A plate freshly prepared with 4 ml top agar inoculated with 0.3 ml of the investigated strain (optical density at 525 nm [OD525] = 0.4 to 0.5) (6). The plates were examined for cell lysis (plaques) after 3 to 5 days, and the presence of a clear zone, a turbid zone, or no inhibition zone was detected. Since the reaction of the spot test can vary according to the growth condition of the host strain, these spot tests were performed three times with independent host cultures. The degree of phage resistance in the different isolates was calculated as the fraction of the total number of phages in the collection that was unable to infect the strains (i.e., no inhibition of growth).
DNA extraction and sequencing of F. psychrophilum strains.
Bacterial DNA was extracted from pelleted purified isolates using the QIAamp DNA minikit (Qiagen) according to the manufacturer's protocol. The complete genomes of the ancestral and phage-resistant clones were sequenced using Illumina HiSeq platform (BGI, China) to a depth of at least 100× per sample, with paired-end read sizes of 500 bp. Library construction, sequencing, and data pipelining were performed in accordance with the manufacturer's protocols.
Computational analyses.
The reads of the 950106-1/1 ancestral strain were aligned using the JIPO2/86 (ATCC 49511) F. psychrophilum strain as the reference genome (GenBank accession number AM398681, August 2013). The resulting sequence was then used as a reference for alignment and comparison to the phage-resistant isolates. The genome assembly process was performed using the Geneious software (31), annotation of the genomes was achieved using the RAST annotation service (32), and genome comparison was done using MAUVE v2.3.1 software (33). To determine putative subcellular localization of mutant genes, server tools were applied. Prediction of the localization of bacterial proteins was achieved using PSort v3.0b (34). Checking of transmembrane helices (TMH) was performed by TMHMM v2.0c (35). Finally, predictions of signal peptides were obtained using SignalIP v3.0 (36).
The putative CRISPR loci for the 950106-1/1 strain and the six phage-resistant isolates were identified with CRISPRfinder (http://crispr.u-psud.fr/Server/CRISPRfinder.php) (37). Repeat sequences were compared by WebLogo analysis, a Web-based application that generates graphical representations (logos) of the patterns within a multiple-sequence alignment (http://weblogo.berkeley.edu/logo.cgi) (38). Spacer sequences were aligned to the whole-genome-sequenced F. psychrophilum bacteriophages FpV4, FpV9, and FpV21 (unpublished results) and 6H (29) using the ClustalW algorithm in the Geneious 7.0.2 program (31).
Phage adsorption rates.
Bacteriophages FpV4, FpV9, and FpV21 were proliferated using the F. psychrophilum strain 950106-1/1 as a host. The adsorption constants of these bacteriophages to strain 950106-1/1 and six phage-resistant strains were determined by adding one phage to 20 ml of exponentially growing cells at a multiplicity of infection of 0.0001 and incubating the infected culture at 15°C with agitation. Samples were taken every 10 min for 1 h, centrifuged (5,000 × g, 10 min), and diluted (1:10) in SM buffer with chloroform. The phage adsorption constant was calculated from the decrease in unadsorbed phages (i.e., phages in supernatant) over time, according to the following equation: K = 2.3/(B)t × log(p0/p), where B is the concentration of bacteria (cells ml−1), p0 is the number of PFU at time zero, p is the number of PFU in supernatant (i.e., phages not adsorbed) at time t (min), and K is the velocity constant (ml min−1) (39).
Gliding assays.
The ability of strain 950106-1/1 and the six phage-resistant isolates to move by gliding was examined using the hanging drop technique (40) and glass slides, as well as agar plates as surfaces for the gliding motility. Briefly, all the isolates were grown in TYES-B for 48 to 72 h at 15°C. Glass capillary microslides (Camlab) were introduced in the bacterial cultures, and motility was observed using phase-contrast microscopy (Olympus BX61). In addition, one drop of liquid culture of each strain was deposited over a coverslip and was turned upside down and placed on tiny stands on a glass slide. Bacterial motility was observed through the coverslip (40). Finally, aliquots of 5 μl bacterial culture were spread on plates with 2×, 1×, 0.5×, and 0.1× diluted TYES-B agar (1.1%). After 72 h of incubation, the colony diameters were measured (25).
Biofilm formation assay.
Biofilm formation was quantified using the standard assay with crystal violet staining of biofilm and subsequent measurement of the optical density at 595 nm as previously described for F. psychrophilum by Álvarez et al. (41) but with some modifications. The ancestral strain 950106-1/1 and the six phage-resistant isolates were grown in half-strength TYES-B to mid-exponential phase (108 cells/ml). The cultures were diluted 1/100 in TYES-B, and then 1 ml of each dilution was inoculated in quadruplicate into polystyrene tubes (Becton Dickson, Falcon), which were incubated statically at 15°C for 5 days. Following this incubation, the supernatants were discarded, and the tubes were washed seven times with 1 ml of sterile distilled water. Then 1 ml of a 1% (wt/vol) crystal violet solution (Sigma-Aldrich) was added to each polystyrene tube containing the cells. After 45 min, the crystal violet solution was removed, the wells were washed seven times with 1 ml of sterile distilled water to remove the dye, and then 1 ml of 96% (vol/vol) ethanol was added. Biofilm formation was then quantified by measuring the optical density at 595 nm. Experiments were repeated in four independent assays.
Extracellular enzyme activity assay.
The levels of activity of the extracellular enzymes proteolysins, elastinase, and gelatinase were evaluated according to the previous tests performed with Pseudomonas aeruginosa (42) but adapted to F. psychrophilum. Aliquots (50 μl) of bacterium-free supernatant (0.45-μm-pore-size filtration) from three replicates of a 4-day liquid culture of each phage-resistant isolate and the sensitive ancestor (with similar CFU/ml) were added to holes punched into agar plates. Agar plates containing 2% skim milk were used to assess total proteolytic activity, and plates were incubated at 15°C. Gelatinase activity was determined using 2% gelatin plates and incubated at 4°C. Finally, elastinase activity was tested according to Madsen and Dalsgaard (43). Briefly, TYES-A was mixed with elastin (Sigma E-0502) to give the final concentration of 0.1%. Degradation of elastin was observed as a clearing zone surrounding the well.
Bacterial hemolytic activity was assessed using the microplate hemolysis assay described by Högfors-Rönnholm and Wiklund (44). Blood was collected in an equal volume of Alsever's solution (Sigma-Aldrich) by caudal venipuncture of rainbow trout (about 800 g). Erythrocytes were centrifuged (1,000 × g, 5 min, 4°C), washed three times with phosphate-buffered saline (PBS; pH 7.2), and used the same day. For the assay, washed and packed erythrocytes were suspended to 5% (vol/vol) in PBS. Equal amounts (30 μl) of erythrocyte and bacterial suspensions were mixed in triplicates into a U-well microtiter plate (Greiner Bio-One) and incubated for 24 h at 10°C and 400 rpm rotation. Following incubation, 150 μl 0.5% NaCl was added to the wells, and the plate was centrifuged (1,000 × g, 5 min, 4°C). The supernatants were transferred to an F-well microtiter plate (Greiner Bio-One), and the absorbance (A) was measured at 540 nm. A negative control (background [Abackground]) with only 0.5% NaCl and erythrocytes and a positive control (total hemolysis [A100%]) with distilled water and erythrocytes were included in triplicates on each plate. The hemolytic activity was calculated according to the following equation: Hemolytic activity = (A − Abackground)/(A100% − Abackground).
Statistical analysis.
A Student t test (GraphPad Prism 4 software) was used to analyze the statistical significance of the observed changes in biofilm formation and extracellular enzymatic activities between activities in the phage-resistant strains relative to the sensitive wild type. P values of <0.01 were considered statistically significant.
Nucleotide sequence accession numbers.
The genomic sequences of all strains have been deposited in the EMBL database under accession numbers CP008878 to CP008883 (see Table S1 in the supplemental material).
RESULTS
F. psychrophilum phage-resistant isolates: selection and susceptibility to phage infection.
Sixteen phage-resistant isolates derived from the wild-type F. psychrophilum strain 950106-1/1 were obtained after challenging with different bacteriophages (Christiansen et al., unpublished), and six of these strains, V1-20, V2-20, V3-5, V4-24, V4-28, and V4-33, which showed cross-resistance as well as loss of resistance to other phages in our collection of 32 bacteriophages, were selected for further characterization (Fig. 1; see also Table S1 in the supplemental material). Compared to the wild-type strain 950106-1/1, all phage-resistant isolates had increased overall resistance against phages in the collection. The phage-resistant isolates V1-20 and V2-20 showed 65% total resistance (i.e., percentage of phages for which the isolate was completely resistant), whereas isolates V4-24, V4-28, and V4-33 had become resistant to even more phages and displayed a total resistance of 100, 84, and 87%, respectively (Fig. 1). Interestingly, the isolate V3-5 had gained resistance to some phages but also lost resistance to other phages; hence, its total resistance was similar to that of the 950106-1/1 ancestral strain (31%) (Fig. 1) (28). In order to better understand the resistance mechanisms to phage infection present in F. psychrophilum, whole-genome sequencing of the 950106-1/1 susceptible strain and the six phage-resistant isolates was carried out.
FIG 1.
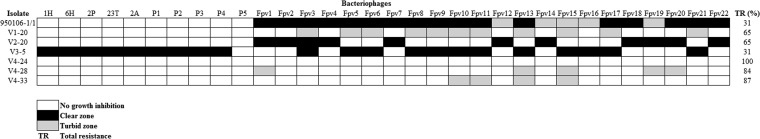
Host range of 32 bacteriophages against the 950106-1/1 ancestral sensitive strain and the six phage-resistant isolates.
Identification of CRISPR loci in F. psychrophilum strain 950106-1/1.
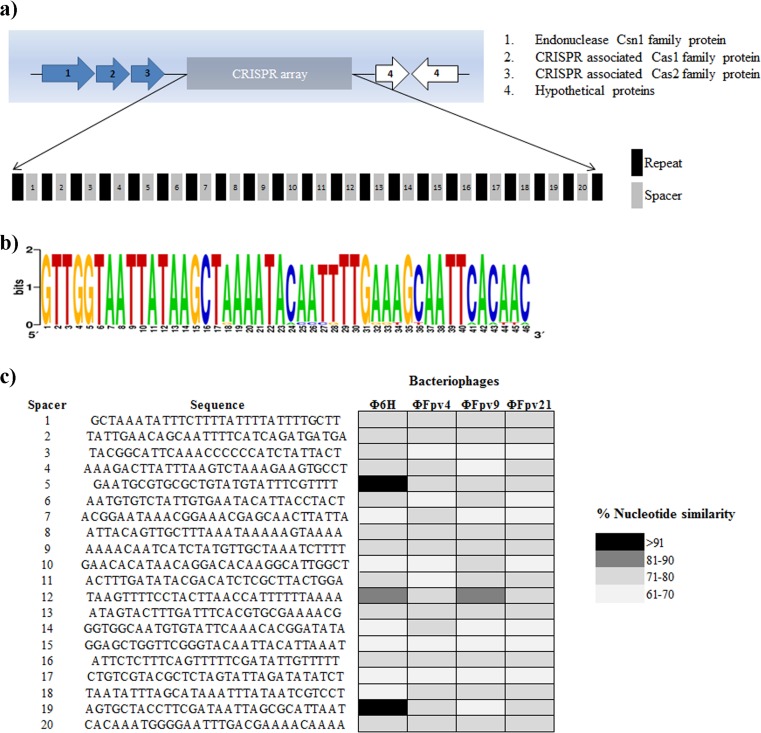
We investigated if the resistance mechanism in phage-resistant isolates was through an adaptive immune response. First, we determined the distribution of CRISPR loci in the 950106-1/1 ancestral strain. Three putative CRISPR loci were found. Two of them were, however, not associated with Cas proteins, and the repeats and spacers were located within sequences encoding proteins. One CRISPR locus (designated CRISPR1) was associated with three cas proteins, csn1, cas1, and cas2, and exhibited 20 different spacers of 29 to 30 bp and 21 repeats of 46 bp (Fig. 2a and b). Surprisingly, all the spacers derived from these putative CRISPR loci were also present in the genome of F. psychrophilum strain JIPO2/86 (ATCC 49511T) isolated in France (45). As a consequence of the association between cas genes and the size of the CRISPR1, we selected this locus to compare the spacer content between the ancestral strain 950106-1/1 and the six phage-resistant isolates.
FIG 2.
F. psychrophilum strain 950106-1/1 CRISPR1 overview. (a) Graphic representation of cas genes, spacer, and repeats of CRISPR1 in the F. psychrophilum strain 950106-1/1; (b) Web logo of nucleotide comparison repeats of CRISPR1 in F. psychrophilum strain 950106-1/1; (c) nucleotide comparison of 20 spacers of CRISPR1 in F. psychrophilum strain 950106-1/1. All the six phage-resistant isolates derived from the ancestral strain displayed the same nucleotide sequences in the spacer and repeat contents.
CRISPR1 activity in phage-resistant F. psychrophilum isolates.
Examination of the similarity of F. psychrophilum CRISPR1 spacers with sequences of known F. psychrophilum bacteriophages (unpublished data) or other sequences of extrachromosomal origin in the NCBI database revealed that 19 out of 21 spacers did not significantly match (<80% similarity) with genomic sequences from F. psychrophilum bacteriophages FpV4, FpV9, and FpV21, which were used in the selection for phage-resistant isolates, or with sequences in the NCBI database. The two exceptions were the spacers 5 and 19, which showed 100% and 97% similarity, respectively, with sequences of the temperate bacteriophage 6H (29) (Fig. 2c).
Analysis of the ability of the CRISPR1 system to incorporate novel spacers-repeats after phage exposure showed that all phage-resistant isolates, independent of the phage exposure prior to isolation, maintained a spacer composition that was identical to the phage-sensitive wild type (Fig. 2), despite their different phage susceptibility patterns.
Identification and distribution of spontaneous mutations in phage-resistant F. psychrophilum isolates.
Genome analysis revealed 130 independent mutations distributed along the genomes of all the F. psychrophilum phage-resistant isolates, of which 80% were indels (deletions and/or insertions) between 1 bp and 50 kbp and 20% were point mutations (see Table S3 in the supplemental material). The phage-resistant isolates V1-20, V2-20, and V3-5 contained 25, 18, and 30 mutations, respectively, corresponding to 19, 13, and 23% of the total mutations, while isolates V4-24, V4-28, and V4-33 contained 21, 18, and 18 mutations, corresponding to 16, 13, and 13% of the total number of mutations. We focused our attention on mutations in genes that potentially could be responsible for the observed phage resistance phenotype in F. psychrophilum.
Phage-resistant isolates showed indels in genes with functions such as cell surface protein functions, metabolism, biosynthesis, motility, genetic expression, translation, membrane transport, and hypothetical and unknown protein functions. Small indels (between 1 and 10 bp) caused amino acid changes, reading frame shifts, or anticipated stop codons (see Table S3 in the supplemental material). Of the six phage-resistant isolates, strain V3-5 (isolated in the presence of the lytic phage FpV4) accumulated the majority of mutations (23%), whereas the phage-resistant isolates V2-20 (isolated in the presence of lytic phage FpV21) and V4-28 (isolated in the presence of the cocktail with 11 different bacteriophages) showed the lowest accumulation of mutations (13% in both isolates).
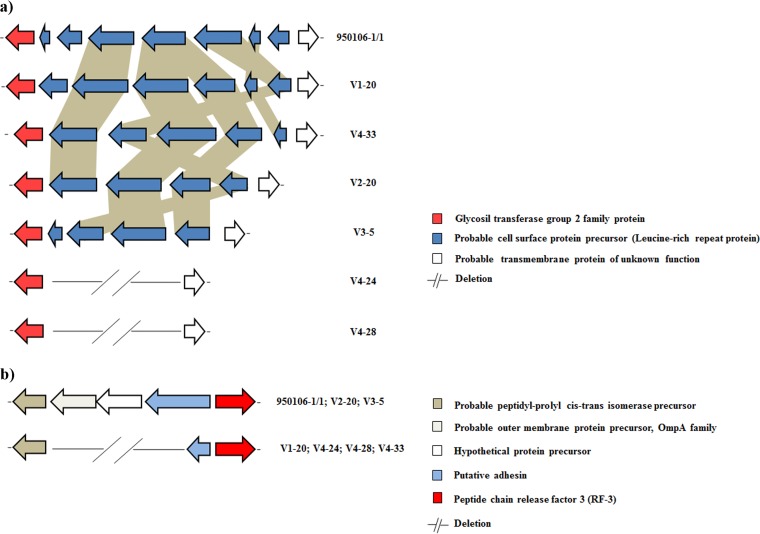
The distribution of mutations was variable among all F. psychrophilum phage-resistant isolates. However, some mutations were present in all phage-resistant clones. For example, all the isolates displayed deletions in a region encoding proteins organized in tandem with leucine-rich repeats, previously described for F. psychrophilum JIP02/86 (4). The 950106-1/1 ancestral strain displayed seven cell surface proteins tandemly organized with leucine-rich repeats that differed in amino acid sequence length according to the number of leucine units (Fig. 3a). The phage-resistant isolates showed genetic instability in this region. For example, clones V1-20, V2-20, V3-5, and V4-33 had reduced numbers of open reading frames (ORFs) and showed rearrangements in the tandem organization, whereas phage-resistant clones V4-24 and V4-28 had lost this genetic segment completely (Fig. 3a).
FIG 3.
Schematic representation of the distribution of specific mutations in F. psychrophilum phage-resistant isolates. Genes are represented as arrows, and deletion mutations are represented by short lines. Functions for genes are shown in colored boxes. (a) Leucine-rich proteins in tandem (accession numbers WP_011962354.1 to WP_011962360.1); (b) putative adhesin and outer membrane protein precursor (accession numbers WP_016361986.1 and WP_011962492.1, respectively).
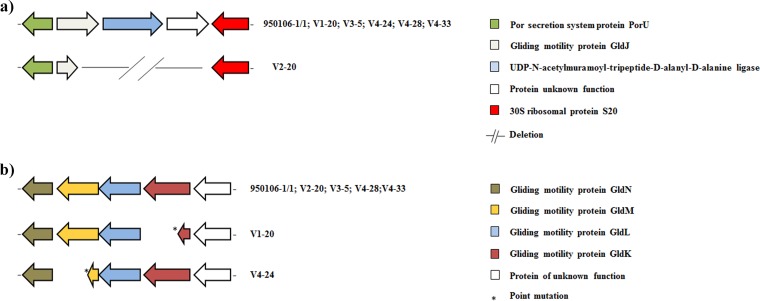
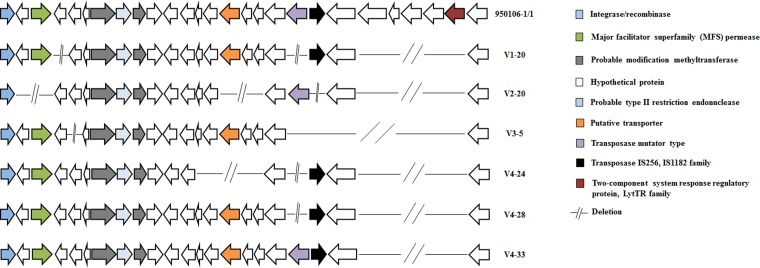
A number of mutations were specific for some phage-resistant isolates. The phage-resistant strains V1-20, V4-24, V4-28, and V4-33 were all affected by a deletion in a putative adhesion protein causing a premature stop codon, a full deletion in a probable outer membrane protein precursor OmpA family, and a deletion in a hypothetical protein (Fig. 3b). Mutant genes related to the biosynthesis of lipopolysaccharide, cell wall, and gliding motility (GldJ) were found in the phage-resistant isolate V2-20 (Fig. 4a). Moreover, the phage-resistant isolate V2-20 displayed a deletion in a permease component of a probable ABC-type transport system (Fig. 5). In this same genetic region (around 50 kbp), all phage-resistant isolates showed deletions in a putative transporter, a regulatory protein (Lyttr family), and hypothetical and unknown proteins (Fig. 5). Interestingly, many of these mutant genes, including unknown proteins, are membrane proteins associated with transmembrane helix domains or signal peptides.
FIG 4.
Schematic representation of the distribution of specific mutations in F. psychrophilum phage-resistant isolates. Genes are represented as arrows, deletion mutations are represented by short lines, and point mutations as asterisks. Functions for genes are shown in colored boxes. (a) Deletion in gliding motility protein GldJ (accession number WP_011963517.1); (b) point mutations in gliding motility proteins GldM and GldK (accession numbers WP_011964073.1 and WP_011964075.1, respectively).
FIG 5.
Schematic representation of the distribution of specific mutations in F. psychrophilum phage-resistant isolates. Genes are represented as arrows, and deletion mutations are represented by short lines. Functions for genes are shown in colored boxes. Specific region of 50 kbp presented deletions in several proteins (accession numbers according to the position: WP_011964238.1, WP_011964239.1, WP_011964240.1, WP_011964241.1, WP_011964251.1, WP_011964252.1, WP_011964253.1, WP_011964254.1, WP_011964255.1, WP_016361990.1, WP_011964257.1, WP_011964258.1, WP_011964259.1, WP_011964260.1, and WP_011964261.1).
Previous results have shown that phage-resistant isolates of F. psychrophilum derived from the 950106-1/1 strain were negative in the amplification of four genes from the specific prophage 6H using PCR, suggesting that these strains had lost this specific prophage (28). The genomic analysis of the genomes of the resistant isolates confirmed that all isolates had lost this specific genomic segment (see Table S3 in the supplemental material) and thus lost the entire prophage.
Twenty-five point mutations in specific genes were distributed among all the F. psychrophilum phage-resistant isolates, and 92% caused amino acid changes (see Table S3 in the supplemental material). Point mutations in genes, such as that encoding a probable UDP-N-acetylglucosamine acyltransferase (biosynthesis lipids), were found in the phage-resistant isolate V1-20. The isolates V1-20 and V4-24 displayed a point mutation in the gene encoding gliding motility proteins GldK and GldM, respectively, causing a premature stop codon (Fig. 4b). Likewise, several genes with point mutations (38%) were shown to be associated with membrane-associated proteins or with signal peptides (see Table S3 in the supplemental material).
Impaired bacteriophage attachment.
In order to investigate if the mutations in these proteins affected phage adsorption to phage-resistant isolates, the adsorption constants were determined (Table 1). The results showed that all phage-resistant isolates had a 50- to >500-fold reduction in their affinity for bacteriophage attachment compared with that of the sensitive wild-type strain (Table 1).
TABLE 1.
Phage adsorption constants of phages FpV4, FpV9, and FpV21 on ancestral strain and phage-resistant isolates
| Isolate | Phage adsorption constant |
||
|---|---|---|---|
| FpV4 | FpV9 | FpV21 | |
| 950106-1/1 | 3.4 × 10−8 ± 1.2 × 10−8 | 3.0 × 10−8 ± 1.0 × 10−8 | 2.7 × 10−8 ± 1.1 × 10−8 |
| V1-20 | 5.2 ×10−10 ± 1.0 × 10−10 | ||
| V2-20 | 6.1 × 10−10 ± 2.1 × 10−10 | ||
| V3-5 | 4.4 × 10−10 ± 2.0 × 10−9 | ||
| V4-24a | 3.3 × 10−10 ± 2.2 × 10−9 | 5.1 × 10−10 ± 1.0 × 10−10 | 5.9 × 10−10 ± 6.0 × 10−9 |
| V4-28a | 1.5 × 10−10 ± 3.1 × 10−9 | 6.4 × 10−10 ± 5.0 × 10−9 | 2.9 × 10−10 ± 1.0 × 10−9 |
| V4-33 | 6.7 × 10−10 ± 1.3 × 10−10 | ||
Bacteriophages Fpv4, Fpv9, and Fpv21 were selected as representatives of the cocktail of bacteriophages.
Motility, biofilm formation, and secretion of enzymes in F. psychrophilum phage-resistant isolates.
The ability of the ancestral and phage-resistant isolates to move by gliding was examined using the hanging drop technique as well as glass capillary and agar surfaces. However, motile behavior was detected neither in the wild type nor in the phage-resistant strains under any of the conditions tested (data not shown).
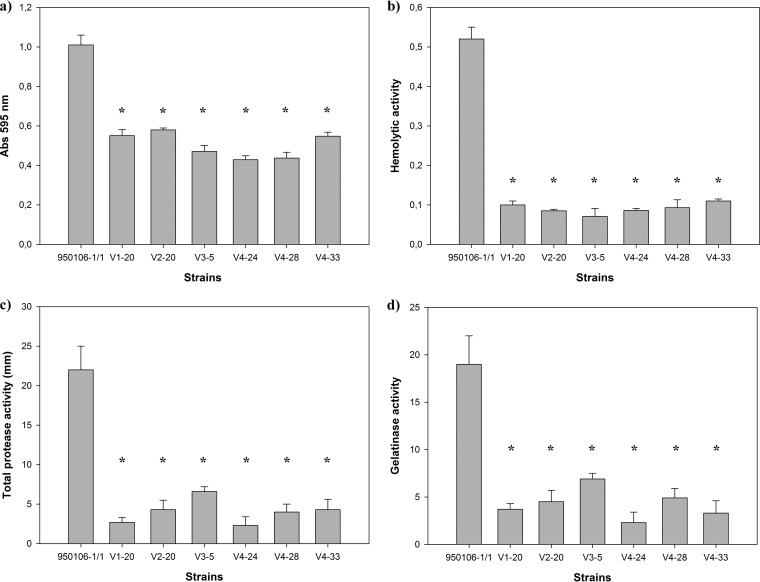
Biofilm formation arises following bacterial adhesion and is associated with the activity of a number of membrane proteins (46). Our results displayed mutations into genes encoding leucine-rich proteins, adhesin, and one regulatory system protein (Fig. 3 and 5), possibly affecting biofilm formation. Therefore, we investigated whether biofilm formation was affected in the phage-resistant isolates. This analysis revealed a significant reduction in biofilm formation in all the phage-resistant isolates compared with that of the wild-type sensitive strain (Fig. 6a). In the same way, mutations were found in gliding motility proteins, which are part of a novel protein secretion system, the Por secretion system (PorSS), identified in the phylum Bacteroidetes (47). In order to examine the effects of resistance mutations on protein secretion in general, we therefore evaluated the levels of extracellular enzymes in the supernatants of bacterial cultures. These analyses indicated that the phage-resistant isolates exhibited a lower hemolytic activity (Fig. 6b) and a reduced ability to secrete proteases (Fig. 6c), gelatinases (Fig. 6d), and elastinase (see Table S4 in the supplemental material) in comparison with those of the 950106-1/1 sensitive strain.
FIG 6.
The F. psychrophilum phage-resistant isolates displayed reduced biofilm formation and secretion of extracellular enzymes. (a) Biofilm formation of the 950106-1/1 ancestral strain and the six phage-resistant isolates; (b) hemolytic activity; (c) total protease activity on skim-milk plates; (d) gelatinase activity on gelatin plates. Results were plotted with standard deviation error bars from triplicate experiments for each isolate; the P value was calculated using a paired Student t test. Asterisks indicate values significantly lower than that of the control.
DISCUSSION
CRISPR activity in F. psychrophilum.
Studies on the effects of bacteriophages on F. psychrophilum populations have shown multiple phenotypic effects of phage exposure, including cross-resistance to some phages and loss of resistance to other phages (28), as well as the loss of a specific prophage (29) and metabolic changes (Christiansen et al., unpublished). Obviously, more knowledge about genetic properties in both phages and hosts determining susceptibility and host range properties is needed to understand the complex network of underlying genetic changes behind these phenotypic changes in F. psychrophilum following phage exposure. In order to get further insights into the resistance mechanisms in F. psychrophilum, six phage-resistant isolates were selected and complete genome sequencing was carried out.
One CRISPR loci (CRISPR1) associated with cas genes was found in F. psychrophilum strain 950106-1/1 (Fig. 2). Two of the spacers detected in the CRISPR1 loci matched with sequences of the temperate bacteriophage 6H (Fig. 2c), suggesting that the CRISPR system has previously been involved in the inactivation of this phage or a related phage. However, there are several indications that the CRISPR1 loci in F. psychrophilum do not have the ability or characteristics of an active immune system against phage infection. First, the six phage-resistant isolates and the phage-sensitive wild type had identical spacer content, independent of phage exposure (Fig. 2). Thus, even though the V3-5 resistant strain contained two spacers matching phage 6H sequences, the strain had gained sensitivity to this particular phage (28). Similarly, previous results have displayed that F. psychrophilum strain MH1, which has an identical CRISPR1 locus (unpublished results), is also sensitive to phage 6H (29). Moreover, the fact that an identical CRISPR locus was found in both the Danish F. psychrophilum strain 950106-1/1 and the previously sequenced strain JIPO2/86, isolated in France >10 years earlier (45), suggests that the CRISPR system in F. psychrophilum is not an active and dynamic system involved in phage resistance. Our observations are similar to those reported for Pseudomonas aeruginosa (48), which showed no link between resistance to phage infection and presence or absence of a specific CRISPR locus. However, the rapid dynamics of the CRISPR region in Streptococcus thermophilus (17), where bacteria responded to phage infection by integrating new spacers into the CRISPR array derived from the infecting phage genome, emphasizes that the CRISPR region does play an important role in phage resistance in other strains. Of the 20 spacers in the F. psychrophilum CRISPR locus, 18 were of unknown origin. This lack of resemblance to known sequences is probably due to the low number of sequenced F. psychrophilum bacteriophages and plasmids.
The incapacity of CRISPR1 to integrate new spacers during the selection of phage-resistant isolates is supported by previous studies which have provided evidence that CRISPR loci may be involved in functions other than phage resistance, such as regulation of the expression of endogenous genes (49) and participation in bacterial innate immune evasion and virulence (50). We do not know whether CRISPR in F. psychrophilum contributes to the regulation of endogenous bacterial genes, particularly during the interaction with fish hosts. However, the inactivity of CRISPR loci in F. psychrophilum leads us to propose that the classic way of spontaneous mutations is driving the resistance against phage infection in F. psychrophilum.
Spontaneous phage resistance mutations in F. psychrophilum and effects on virulence properties.
Our results showed that indels and point mutations affected genic regions encoding cell surface proteins, ABC-type transport components, gliding motility, and hypothetical and unknown proteins, which mostly were associated with membrane signal peptides or transmembrane helices domains (Fig. 3 to 5; see also Table S3 in the supplemental material). The observed mutations thus likely cause alterations in these genetic products, modifying the cell surface of phage-resistant isolates and possibly resulting in a cost of the resistance. Interestingly, all phage-resistant isolates showed gene loss, de novo development, and rearrangements in a specific region which encodes cell surface proteins in tandem with leucine-rich repeats, suggesting that this is a potential hot spot for spontaneous deletion formation (Fig. 3a). These cell surface proteins with leucine-rich repeats have previously been associated with bacterial adhesion in the pathogenic bacteria Bacteroides forsythus (51) and Treponema denticola (52). The observation of an impaired biofilm formation of all phage-resistant isolates in comparison to the ancestral strain (Fig. 6a) in the current study supports the hypothesis that these tandem repeat genes participate in bacterial adhesion to surfaces. However, we did not find direct relation between the number of proteins with leucine-rich repeats and the effect on biofilm formation, suggesting that other proteins were contributing to this process. Thus, the molecular mechanism of tandem repeat deletions and its effect on bacterial virulence in F. psychrophilum are unknown.
Likewise, F. psychrophilum phage-resistant isolates V1-20, V2-20, and V4-24 presented mutations in the gldJ, gldM, and gldK genes, which encode gliding motility proteins (Fig. 4). Although, our results showed that neither the wild-type strain 950106-1/1 nor the phage-resistant isolates displayed motile behavior at the tested conditions, similar results have been described in a motile Flavobacterium johnsoniae strain, where gldJ mutants were resistant to bacteriophages that infected wild-type cells (53). However, other studies in the same bacterium have identified the gliding motility proteins GldM and GldK as members of a novel protein secretion system, the Por secretion system (PorSS), involved in the secretion of virulent proteins (54–56). The phage-resistant isolate V2-20 also presented a deletion in the probable ABC-type transport system, a permease component. There is increasing evidence that these transport systems play either direct or indirect roles in the virulence of bacteria (57, 58). Members of the ABC-type transport system have been involved in the uptake of nutrients (59) and metal ions (60) and in cell attachment (61). The reduced secretion of extracellular enzymes in phage-resistant isolates in the current study thus suggested a link to the observed mutations in these genes and supports that phage resistance mutations have direct implications for the extracellular enzymatic properties and bacterial virulence of F. psychrophilum, as was previously observed in F. columnare (25).
It is likely, therefore, that these proteins participate as receptors or coreceptors in the attachment of phages to the cell surface. Further evidence supporting this suggestion was the observation of an impaired attachment of bacteriophages to phage-resistant isolates (Table 1), providing direct evidence that such mutations affect the phage adsorption. Although our observations were focused on mutations in genic regions, we cannot rule out the possibility that mutations in intergenic regions could have affected promoters and cis/trans regulatory elements, which may have limited the expression of proteins participating in the phage infection process. For example, studies using insertional mutagenesis conferred phage resistance in Listeria monocytogenes, locating a transposon insertion upstream of coding regions (62). Besides, we cannot rule out the pleiotropic effect of other mutations on phenotypic diversity (phage resistance and virulence) in F. psychrophilum, as has been observed in Pseudomonas aeruginosa (63) and Erwinia carotovora (64). Finally, it is important to note that although the mutational changes in the resistant strains point in the same direction and can be linked to potentially phage-related genes and phenotypic properties of the strains, we do not have sequenced isolates from the control cultures without phages and therefore cannot completely rule out that similar mutations may have taken place even in the absence of phages. However, based on previous work on phage-driven genetic and phenotypic diversification (65), we find this very unlikely.
Mutations that provide resistance against phage infection have previously been linked to a fitness cost in the host cells (9, 13). Observations of significantly reduced growth rates of phage-resistant F. psychrophilum isolates compared with the ancestral strain have thus been reported (Christiansen et al., unpublished). In the same way, Biolog assay of phage-resistant strains showed that some strains had lost the ability to take up specific nutrients, suggesting that putative phage receptors or coreceptors in F. psychrophilum could be involved also in the extracellular hydrolysis and subsequent uptake of nutrients (Christiansen et al., unpublished). Our results add to those earlier observations and provide evidence that mutations in cell surface proteins reduced phage attachment and bacterial virulence (Fig. 3, 4, and 6 and Table 1). Thus, cell surface properties are likely under strong selection from phage infection, as also suggested for the marine cyanobacteria Prochlorococcus (65). Consequently, bacteriophages seem to have strong effects as drivers of the phenotypic diversification in the F. psychrophilum population, with potential implications for their metabolic and virulence properties.
Implications of resistance mutations for the application of phage therapy in F. psychrophilum.
The emergence of phage-resistant clones is a common response to the selective pressure created by bacteriophages (66). For the successful application of phage therapy, it is essential to know the diverse antiphage systems present in the pathogenic bacterial population as well as the implications of these for the virulence of the pathogen. Our results show that spontaneous mutations in F. psychrophilum cells drive the resistance against lytic phage infection and suggest that proteins with leucine-rich repeats and gliding motility proteins, which were affected by indels and point mutations, probably act as phage receptors or coreceptors (Fig. 3 and 4 and Table 1). This is an important observation in the context of phage therapy, because bacterial pathogenesis has been related with these proteins (52, 55, 67), as also supported by the current study. Consequently, we suggest that F. psychrophilum phage-resistant isolates may have a reduced virulence against trout. This suggestion was supported by observations of a reduced activity of a number of traditional indicators of F. psychrophilum virulence, such as elastinase and hemolytic activity, ability to form biofilm, and ability to secrete extracellular enzymes at in vitro conditions (Fig. 6). These functional changes may thus result in inhibited adhesion, colonization, and destruction of fish tissues. Thus, although we do not have direct evidence from challenge experiments with fish, our study strongly suggests that bacteriophage resistance may be associated with reduced bacterial virulence in F. psychrophilum.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by a grant from The Danish Council for Independent Research (FNU-09-072829), The Danish Directorate for Food, Fisheries and Agri Business, and by the EU-IRSES-funded project AQUAPHAGE.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.03699-14.
REFERENCES
- 1.Nematollahi A, Decostere A, Pasmans F, Haesebrouck F. 2003. Flavobacterium psychrophilum infections in salmonid fish. J Fish Dis 26:563–574. doi: 10.1046/j.1365-2761.2003.00488.x. [DOI] [PubMed] [Google Scholar]
- 2.Valdebenito S, Avendaño-Herrera R. 2009. Phenotypic, serological and genetic characterization of Flavobacterium psychrophilum strains isolated from salmonids in Chile. J Fish Dis 32:321–333. doi: 10.1111/j.1365-2761.2008.00996.x. [DOI] [PubMed] [Google Scholar]
- 3.Nicolas P, Mondot S, Achaz G, Bouchenot C, Bernardet JF, Duchaud E. 2008. Population structure of the fish-pathogenic bacterium Flavobacterium psychrophilum. Appl Environ Microbiol 74:3702–3709. doi: 10.1128/AEM.00244-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Duchaud E, Boussaha M, Loux V, Bernardet J, Michel C, Kerouault B, Mondot S, Nicolas P, Bossy R, Caron C, Bessiéres P, Gilbrat J, Claverol S, Dumetz F, Le Hénaff M, Benmansour A. 2007. Complete genome sequence of the fish pathogen Flavobacterium psychrophilum. Nat Biotechnol 25:763–769. doi: 10.1038/nbt1313. [DOI] [PubMed] [Google Scholar]
- 5.Bruun MS, Schmidt AS, Madsen L, Dalsgaard I. 2000. Antimicrobial resistance patterns in Danish isolates of Flavobacterium psychrophilum. Aquaculture 187:201–212. doi: 10.1016/S0044-8486(00)00310-0. [DOI] [Google Scholar]
- 6.Stenholm AR, Dalsgaard I, Middelboe M. 2008. Isolation and characterization of bacteriophages infecting the fish pathogen Flavobacterium psychrophilum. Appl Environ Microbiol 74:4070–4078. doi: 10.1128/AEM.00428-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Castillo D, Higuera G, Villa M, Middelboe M, Dalsgaard I, Madsen L, Espejo RT. 2012. Diversity of Flavobacterium psychrophilum and the potential use of its phages for protection against bacterial cold water disease in salmonids. J Fish Dis 35:193–201. doi: 10.1111/j.1365-2761.2011.01336.x. [DOI] [PubMed] [Google Scholar]
- 8.Madsen L, Bertelsen SK, Dalsgaard I, Middelboe M. 2013. Dispersal and survival of Flavobacterium psychrophilum phages in vivo in rainbow trout and in vitro under laboratory conditions: implications for their use in phage therapy. Appl Environ Microbiol 79:4853–4861. doi: 10.1128/AEM.00509-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Middelboe M, Holmfeldt K, Riemann L, Nybroe O, Haaber J. 2009. Bacteriophages drive strain diversification in a marine Flavobacterium: implications for phage resistance and physiological properties. Environ Microbiol 11:1971–1982. doi: 10.1111/j.1462-2920.2009.01920.x. [DOI] [PubMed] [Google Scholar]
- 10.Middelboe M. 2000. Bacterial growth rate and marine virus-host dynamics. Microb Ecol 40:114–124. [DOI] [PubMed] [Google Scholar]
- 11.Middelboe M, Hagström A, Blackburn N, Sinn B, Fischer U, Borch NH, Pinhassi J, Simu K, Lorenz MG. 2001. Effects of bacteriophages on the population dynamics of four strains of pelagic marine bacteria. Microb Ecol 42:395–406. doi: 10.1007/s00248-001-0012-1. [DOI] [PubMed] [Google Scholar]
- 12.Lederberg J, Lederberg E. 1952. Replica plating and indirect selection of bacterial mutants. J Bacteriol 63:399–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lenski RE. 1988. Experimental studies of pleiotropy and epistasis in Escherichia coli. I Variation in competitive fitness among mutants resistant to virus T4. Evolution 42:425–432. [DOI] [PubMed] [Google Scholar]
- 14.Haft DH, Selengut J, Mongodin EF, Nelson KE. 2005. A guild of 45 CRISPR-associated (Cas) proteins families and multiple CRISPR/Cas subtypes exist in prokaryotic genomes. PLoS Comput Biol 1:474–483. doi: 10.1371/journal.pcbi.0010060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lillestøl R, Redder P, Garrett R, Brügger K. 2006. A putative viral defense mechanism in archaeal cells. Archaea 2:59–72. doi: 10.1155/2006/542818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Deveau H, Garneau JE, Moineau S. 2010. CRISPR/Cas system and its role in phage-bacteria interactions. Annu Rev Microbiol 64:475–493. doi: 10.1146/annurev.micro.112408.134123. [DOI] [PubMed] [Google Scholar]
- 17.Barrangou R, Fremaux C, Deveau H, Richards M, Boyaval P, Moineau S, Romero DA, Horvath P. 2007. CRISPR provides acquired resistance against viruses in prokaryotes. Science 315:1709–1712. doi: 10.1126/science.1138140. [DOI] [PubMed] [Google Scholar]
- 18.Makarova KS, Grishin NV, Shabalina SA, Wolf YI, Koonin EV. 2006. A putative RNA-interference-based immune system in prokaryotes: computational analysis of the predicted enzymatic machinery, functional analogies with eukaryotic RNAi, and hypothetical mechanisms of action. Biol Direct 1:7. doi: 10.1186/1745-6150-1-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Marraffini LA, Sontheimer EJ. 2008. CRISPR interference limits horizontal gene transfer in Staphylococci by targeting DNA. Science 322:1843–1845. doi: 10.1126/science.1165771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Brouns SJ, Jore MM, Lundgren M, Westra ER, Slijkhuis RJ, Snijders AP, Dickman MJ, Makarova KS, Koonin EV, van der Oost J. 2008. Small CRISPR RNAs guide antiviral defense in prokaryotes. Science 321:960–964. doi: 10.1126/science.1159689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brüssow H, Canchaya C, Hardt WD. 2004. Phages and the evolution of bacterial pathogens: from genomic rearrangements to lysogenic conversion. Microbiol Mol Biol Rev 68:560–602. doi: 10.1128/MMBR.68.3.560-602.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Davis BM, Waldor MK. 2003. Filamentous phages linked to virulence of Vibrio cholerae. Curr Opin Microbiol 6:35–42. doi: 10.1016/S1369-5274(02)00005-X. [DOI] [PubMed] [Google Scholar]
- 23.Ehrbar K, Hardt WD. 2005. Bacteriophage-encoded type III effectors in Salmonella enterica subspecies 1 serovar Typhimurium. Infect Genet Evol 5:1–9. doi: 10.1016/j.meegid.2004.07.004. [DOI] [PubMed] [Google Scholar]
- 24.Kaper JB, Nataro JP, Mobley HLT. 2004. Pathogenic Escherichia coli. Nat Rev Microbiol 2:123–140. doi: 10.1038/nrmicro818. [DOI] [PubMed] [Google Scholar]
- 25.Laanto E, Bamford JK, Laakso J, Sundberg LR. 2012. Phage-driven loss of virulence in a fish pathogenic bacterium. PLoS One 7:e53157. doi: 10.1371/journal.pone.0053157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Capparelli R, Nocerino N, Lanzetta R, Silipo A, Amoresano A, Giangrande C, Becker K, Blaiotta G, Evidente A, Cimmino A, Iannaccone M, Parlato M, Medaglia C, Roperto S, Roperto F, Ramunno L, Iannelli D. 2010. Bacteriophage-resistant Staphylococcus aureus mutant confers broad immunity against staphylococcal infection in mice. PLoS One 22:e11720. doi: 10.1371/journal.pone.0011720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Friman VP, Hiltunen T, Jalasvuori M, Lindstedt C, Laanto E, Örmälä AM, Laakso J, Mappes J, Bamford JK. 2011. High temperature and bacteriophages can indirectly select for bacterial pathogenicity in environmental reservoirs. PLoS One 6:e17651. doi: 10.1371/journal.pone.0017651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Castillo D, Christiansen RH, Espejo R, Middelboe M. 2014. Diversity and geographical distribution of Flavobacterium psychrophilum isolates and their phages: patterns of susceptibility to phage infection and phage host range. Microb Ecol 67:748–757. doi: 10.1007/s00248-014-0375-8. [DOI] [PubMed] [Google Scholar]
- 29.Castillo D, Espejo R, Middelboe M. 2014. Genomic structure of bacteriophage 6H and its distribution as prophage in Flavobacterium psychrophilum strains. FEMS Microbiol Lett 351:51–58. doi: 10.1111/1574-6968.12342. [DOI] [PubMed] [Google Scholar]
- 30.Holt RA, Rohovec JS, Fryer JL. 1993. Bacterial coldwater disease, p 3–23. In Englis V, Roberts RJ, NR Bromage (ed), Bacterial diseases of fish. Blackwell Scientific Publications, Oxford, United Kingdom. [Google Scholar]
- 31.Drummond AJ, Ashton B, Buxton S, Cheung M, Cooper A, Heled J, Kearse M, Moir R, Stones-Havas S, Sturrock S, Thierer T, Wilson A. 2010. Geneious v5.3. Bioamtters, Auckland, New Zealand. [Google Scholar]
- 32.Overbeek R, Olson R, Pusch GD, Olsen GJ, Davis JJ, Disz T, Edwards RA, Gerdes S, Parrello B, Shukla M, Vonstein V, Wattam AR, Xia F, Stevens R. 2014. The SEED and the rapid annotation of microbial genomes using subsystems technology (RAST). Nucleic Acids Res 42:D206–D214. doi: 10.1093/nar/gkt1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Darling AC, Mau B, Blattner FR, Perna NT. 2004. Mauve: multiple alignment of conserved genomic sequence with rearrangements. Genome Res 14:1394–1403. doi: 10.1101/gr.2289704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yu NY, Wagner JR, Laird MR, Melli G, Rey S, Lo R, Dao P, Sahinalp SC, Ester M, Foster LJ, Brinkman FS. 2010. PSORTb 3.0: improved protein subcellular localization prediction with refined localization subcategories and predictive capabilities for all prokaryotes. Bioinformatics 26:1608–1615. doi: 10.1093/bioinformatics/btq249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Krogh A, Larsson B, von Heijne G, Sonnhammer EL. 2001. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J Mol Biol 305:567–580. doi: 10.1006/jmbi.2000.4315. [DOI] [PubMed] [Google Scholar]
- 36.Emanuelsson O, Brunak S, von Heijne G, Nielsen H. 2007. Locating proteins in the cell using TargetP, SignalP and related tools. Nat Protoc 2:953–971. doi: 10.1038/nprot.2007.131. [DOI] [PubMed] [Google Scholar]
- 37.Grissa I, Vergnaud G, Pourcel C. 2007. CRISPRFinder: a Web tool to identify clustered regularly interspaced short palindromic repeats. Nucleic Acids Res 35:W52–W57. doi: 10.1093/nar/gkm360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Crooks GE, Hon G, Chandonia JM, Brenner SE. 2004. WebLogo: a sequence logo generator. Genome Res 14:1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hyman P, Abedon ST. 2009. Practical methods for determining phage growth paramenters, p 175-202. In Clokie MRJ, Kropinski AM (ed), Bacteriophages: methods and protocols, volume 1: isolation, characterization and interactions. Humana Press, New York, NY. [DOI] [PubMed] [Google Scholar]
- 40.Bernardet JF, Nakagawa Y, Holmes B, Subcommittee on the Taxonomy of Flavobacterium and Cytophaga-Like Bacteria of the International Committee on Systematics of Prokaryotes . 2002. Proposed minimal standards for describing new taxa of the family Flavobacteriaceae and emended description of the family. Int J Syst Evol Microbiol 52:1049–1070. doi: 10.1099/ijs.0.02136-0. [DOI] [PubMed] [Google Scholar]
- 41.Alvarez B, Secades P, Prieto M, McBride MJ, Guijarro JA. 2006. A mutation in Flavobacterium psychrophilum tlpB inhibits gliding motility and induces biofilm formation. Appl Environ Microbiol 72:4044–4453. doi: 10.1128/AEM.00128-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hosseinidoust Z, van de Ven TG, Tufenkji N. 2013. Evolution of Pseudomonas aeruginosa virulence as a result of phage predation. Appl Environ Microbiol 79:6110–6116. doi: 10.1128/AEM.01421-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Madsen L, Dalsgaard I. 1998. Characterization of Flavobacterium psychrophilum; comparison of proteolytic activity and virulence of strains from rainbow trout (Oncorhyncus mykiss), p 45–52. In Barnes AC, Davidson GA, Hiney MP, McIntosh D (ed), Methodology in fish diseases research. Fisheries Research Services, Aberdeen, Scotland. [Google Scholar]
- 44.Högfors-Rönnholm E, Wiklund T. 2010. Hemolytic activity in Flavobacterium psychrophilum is a contact-dependent, two-step mechanism and differently expressed in smooth and rough phenotypes. Microb Pathog 49:369–375. doi: 10.1016/j.micpath.2010.08.002. [DOI] [PubMed] [Google Scholar]
- 45.Bernardet JF, Kerouault B. 1989. Phenotypic and genomic studies of “Cytophaga psychrophila” isolated from diseased rainbow trout (Oncorhynchus mykiss) in France. Appl Environ Microbiol 55:1796–1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chagnot C, Zorgani MA, Astruc T, Desvaux M. 2013. Proteinaceous determinants of surface colonization in bacteria: bacterial adhesion and biofilm formation from a protein secretion perspective. Front Microbiol 14:303. doi: 10.3389/fmicb.2013.00303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.McBride MJ, Braun TF. 2004. GldI is a lipoprotein that is required for Flavobacterium johnsoniae gliding motility and chitin utilization. J Bacteriol 186:2295–2302. doi: 10.1128/JB.186.8.2295-2302.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Essoh C, Blouin Y, Loukou G, Cablanmian A, Lathro S, Kutter E, Thien HV, Vergnaud G, Pourcel C. 2013. The susceptibility of Pseudomonas aeruginosa strains from cystic fibrosis patients to bacteriophages. PLoS One 8:e60575. doi: 10.1371/journal.pone.0060575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Babu M, Beloglazova N, Flick R, Graham C, Skarina T, Nocek B, Gagarinova A, Pogoutse O, Brown G, Binkowski A, Phanse S, Joachimiak A, Koonin EV, Savchenko A, Emili A, Greenblatt J, Edwards AM, Yakunin AF. 2011. A dual function of the CRISPR-Cas system in bacterial antivirus immunity and DNA repair. Mol Microbiol 79:484–502. doi: 10.1111/j.1365-2958.2010.07465.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sampson TR, Saroj SD, Llewellyn AC, Tzeng YL, Weiss DS. 2013. A CRISPR/Cas system mediates bacterial innate immune evasion and virulence. Nature 497:254–257. doi: 10.1038/nature12048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Honma K, Kuramitsu HK, Genco RJ, Sharma A. 2001. Development of a gene inactivation system for Bacteroides forsythus: construction and characterization of a BspA mutant. Infect Immun 69:4686–4690. doi: 10.1128/IAI.69.7.4686-4690.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ikegami A, Honma K, Sharma A, Kuramitsu HK. 2004. Multiple functions of the leucine-rich repeat protein LrrA of Treponema denticola. Infect Immun 72:4619–4627. doi: 10.1128/IAI.72.8.4619-4627.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Braun TF, McBride MJ. 2005. Flavobacterium johnsoniae GldJ is a lipoprotein that is required for gliding motility. J Bacteriol 187:2628–2637. doi: 10.1128/JB.187.8.2628-2637.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.McBride MJ, Zhu Y. 2013. Gliding motility and Por secretion system genes are widespread among members of the phylum Bacteroidetes. J Bacteriol 195:270–278. doi: 10.1128/JB.01962-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sato K, Naito M, Yukitake H, Hirakawa H, Shoji M, McBride MJ, Rhodes RG, Nakayama K. 2010. A protein secretion system linked to bacteroidete gliding motility and pathogenesis. Proc Natl Acad Sci U S A 107:276–281. doi: 10.1073/pnas.0912010107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sato K, Yukitake H, Narita Y, Shoji M, Naito M, Nakayama K. 2013. Identification of Porphyromonas gingivalis proteins secreted by the Por secretion system. FEMS Microbiol Lett 338:68–76. doi: 10.1111/1574-6968.12028. [DOI] [PubMed] [Google Scholar]
- 57.Schneider E, Hunke S. 1998. ATP-binding-cassette (ABC) transport systems: functional and structural aspects of the ATP-hydrolyzing subunits/domains. FEMS Microbiol Rev 22:1–20. doi: 10.1111/j.1574-6976.1998.tb00358.x. [DOI] [PubMed] [Google Scholar]
- 58.Garmory HS, Titball RW. 2004. ATP-binding cassette transporters are targets for the development of antibacterial vaccines and therapies. Infect Immun 72:6757–6763. doi: 10.1128/IAI.72.12.6757-6763.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Coulter SN, Schwan WR, Ng EY, Langhorne MH, Ritchie HD, Westbrock-Wadman S, Hufnagle WO, Folger KR, Bayer AS, Stover CK. 1998. Staphylococcus aureus genetic loci impacting growth and survival in multiple infection environments. Mol Microbiol 30:393–404. doi: 10.1046/j.1365-2958.1998.01075.x. [DOI] [PubMed] [Google Scholar]
- 60.Fetherston JD, Bertolino VJ, Perry RD. 1999. YbtP and YbtQ: two ABC transporters required for iron uptake in Yersinia pestis. Mol Microbiol 32:289–299. doi: 10.1046/j.1365-2958.1999.01348.x. [DOI] [PubMed] [Google Scholar]
- 61.Matthysse AG, Yarnall HA, Young N. 1996. Requirement for genes with homology to ABC transport systems for attachment and virulence of Agrobacterium tumefaciens. J Bacteriol 178:5302–5308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tran HL, Fiedler F, Hodgson DA, Kathariou S. 1999. Transposon-induced mutations in two loci of Listeria monocytogenes serotype 1/2a result in phage resistance and lack of N-acetylglucosamine in the teichoic acid of the cell wall. Appl Environ Microbiol 65:4793–4798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yeom DH, Kim SK, Lee MN, Lee JH. 2013. Pleiotropic effects of acyltransferases on various virulence-related phenotypes of Pseudomonas aeruginosa. Genes Cells 18:682–693. doi: 10.1111/gtc.12076. [DOI] [PubMed] [Google Scholar]
- 64.Barnard AM, Simpson NJ, Lilley KS, Salmond GP. 2010. Mutations in rpsL that confer streptomycin resistance show pleiotropic effects on virulence and the production of a carbapenem antibiotic in Erwinia carotovora. Microbiology 156:1030–1039. doi: 10.1099/mic.0.034595-0. [DOI] [PubMed] [Google Scholar]
- 65.Avrani S, Wurtzel O, Sharon I, Sorek R, Lindell D. 2011. Genomic island variability facilitates Prochlorococcus-virus coexistence. Nature 474:604–608. doi: 10.1038/nature10172. [DOI] [PubMed] [Google Scholar]
- 66.Labrie SJ, Samson JE, Moineau S. 2010. Bacteriophage resistance mechanisms. Nat Rev Microbiol 8:317–327. doi: 10.1038/nrmicro2315. [DOI] [PubMed] [Google Scholar]
- 67.Josenhans C, Suerbaum S. 2002. The role of motility as a virulence factor in bacteria. Int J Med Microbiol 291:605–614. doi: 10.1078/1438-4221-00173. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.