Abstract
Wolbachia pipientis is a nearly ubiquitous, maternally transmitted bacterium that infects the germ line of insect hosts. Estimates are that Wolbachia infects 40 to 60% of insect species on the planet, making it one of the most prevalent infections on Earth. However, we know surprisingly little about the molecular mechanisms used by Wolbachia to infect its hosts. We passaged Wolbachia through normally restrictive Drosophila melanogaster hosts, bottlenecking Wolbachia through stochastic segregation while simultaneously selecting for mutants that could recolonize these previously restrictive hosts. Here, we show that Wolbachia alters its behavior when passaged through heterozygous mutant flies. After only three generations, Wolbachia was able to colonize the previously restrictive hosts at control titers. Additionally, the Wolbachia organisms passaged through heterozygous mutant D. melanogaster alter their pattern of tissue-specific Wsp protein production, suggesting a behavioral response to the host genotype. Using whole-genome resequencing, we identified the mutations accumulated by these lineages of Wolbachia and confirmed the existence and persistence of the mutations through clone library Sanger sequencing. Our results suggest that Wolbachia can quickly adapt to new host contexts, with genomic mutants arising after only two generations.
INTRODUCTION
Wolbachia pipientis is an obligate intracellular member of Alphaproteobacteria that forms symbioses with an extremely broad array of hosts (1). Wolbachia infections are considered a “pandemic” in insects, and estimates suggest that upwards of 40% of insect species are infected by the bacterial parasite (2, 3). Wolbachia is well known for reproductive effects induced in the host, which range from the exotic (male killing) to the most common of reproductive effects: cytoplasmic incompatibility (CI) (1). In CI, infected females can breed with either infected or uninfected males, but uninfected females cannot breed with infected males. This results in a drop in fecundity for uninfected females, allowing Wolbachia to spread effectively through new insect populations via maternal transmission (1, 4).
Because Wolbachia is obligately intracellular and not genetically tractable, the research community has turned to genomic sequencing to identify potential mechanisms behind host interaction. Even closely related Wolbachia strains (based on multilocus [MLST] profiles) exhibit significant genomic diversity (5). That is, between related hosts, Wolbachia strains can differ substantially in terms of both genomic content and divergence of orthologous genes (6). In an attempt to identify the loci involved in the induction of different reproductive phenotypes, Wolbachia strains that infect different hosts (7) or cause different reproductive phenotypes (8) have been compared, and the loci have been correlated with those phenotypes. In order to pinpoint the mechanism behind pathogen blocking, Wolbachia variants that are more or less pathogenic or protect the host against viruses to a lesser or greater extent have been sequenced (9). Additionally, even individual Wolbachia strains within individual hosts may not exist as single, clonal populations (10). The significance of this interhost diversity is still unclear, although different Wolbachia genotypes might be important during various aspects of host infection. For example, low-titer Wolbachia variants within individual hosts may become prominent in the context of colonization of new hosts (11), suggesting that Wolbachia pipientis may exist as a diverse quasispecies within a single host. All of these studies have identified interesting loci for future work and have revealed much about the basic evolutionary processes for Wolbachia. However, due to the lack of a genetic system in Wolbachia, researchers have been unable to directly confirm the hypothesis that specific loci are responsible for the phenotypes.
Although Wolbachia pipientis is not yet genetically tractable, the bacteria infect the model organism Drosophila melanogaster, which is amenable to genetic manipulation. Wolbachia relies on an ability to target the germ line and replicate within it in order to persist between generations. Wolbachia has no free-living counterpart, as far as we know, and therefore the ecology of this bacterium is the host environment. In this study, we knocked down Wolbachia's titer in Drosophila melanogaster by introducing Wolbachia into a host background that substantially reduces bacterial titer. We therefore both increased the stochastic segregation of Wolbachia and imposed a selective pressure on these Wolbachia sp. strains to adapt to a new ecological context in order to persist in the flies. Below, we characterize the passaged Wolbachia populations through molecular assays and show that after only 3 generations, Wolbachia has phenotypically adapted to a new, mutant host context.
MATERIALS AND METHODS
Drosophila stocks, crosses, and phenotypic assays.
Standard methods were used for all crosses and culturing. The following stocks were obtained from the Bloomington Drosophila Stock Center (BDSC) at Indiana University (http://flystocks.bio.indiana.edu/): stock number 145, which carries w1, was used as the Wolbachia-infected control line. Stock number 25211, an OreR-derived strain, was used as an uninfected control. One chickadee mutant stock was used in this study. The chic221 cn1/CyO; ry506 flies carry a null allele resulting from the deletion of 5′ noncoding and some chic-coding sequences (12). Wolbachia infection status for stocks acquired from the BDSC was determined via PCR and Western blotting targeting the gene wsp or its product (see below). Wolbachia bacteria were introduced into the heterozygous mutant backgrounds through crosses between w1 females (stock 145) and heterozygous males (mutant/CyO). Wolbachia was passaged in this heterozygous background for three generations, and our Wolbachia-infected lines were genotyped before and after introgression into the mutant/CyO background (see “Bioinformatic analysis of Wolbachia resequenced genomes” below). To clear flies of Wolbachia infection, they were raised for two generations on conventional fly media containing 0.25 mg/ml tetracycline (as described in reference 13). After treatment, flies were reared in bottles previously occupied by uninfected male Drosophila melanogaster flies in order to repopulate the microbiome. All flies were examined for Wolbachia infection and age matched in order to avoid confounding correlations between fly age and Wolbachia titer. We performed single-pair fly crosses in order to count the number of progeny produced. Parental flies were allowed to lay eggs in vials for 3 days and then were transferred 3 times to new vials. Progeny were counted 12 to 16 days posteclosion.
Western blots.
Western blots were used to characterize expression of the Wolbachia surface protein in flies. Flies were ground in 1.5-ml centrifuge tubes using an electric hand drill and disposable pestle in lysis buffer: 150 mM NaCl, 1% Triton X-100, 50 mM Tris-HCl (pH 8) containing Halt protease inhibitor cocktail (Thermo Scientific), and 5 mM EDTA. The lysates were centrifuged for 1 min at 8,000 × g to pellet debris. Samples were heated for 5 min at 95°C in Laemmli sample buffer containing 5% β-mercaptoethanol (Bio-Rad) prior to SDS-PAGE electrophoresis. Proteins were separated on 4 to 20% Tris-glycine NB precast gels (NuSep) in 1× Tris-glycine-SDS running buffer (Bio-Rad) and transferred to a polyvinylidene difluoride (PVDF) membrane in Tris-glycine transfer buffer with 15% methanol at 40 V on ice for 3 to 4 h. The membrane was blocked for 5 min in Starting Block T20 (TBS) blocking buffer (Thermo Scientific), followed by incubation in primary antibody (for 1 h at room temperature [RT] or overnight [O/N] at 4°C) according to standard protocols. SuperSignal West Pico chemiluminescent substrate (Thermo Scientific) was used according to the manufacturer's instructions to detect horseradish peroxidase (HRP) (after incubation with secondary antibodies) on the immunoblots. Blots were reprobed after stripping in 100 mM glycine–0.1% NP-40–1% SDS (pH 2) for 1 h at RT and then O/N at 4°C. A PageRuler prestained protein ladder (Thermo Scientific) was used as a molecular mass marker. The following antibody was obtained through BEI Resources, NIAID, NIH: monoclonal anti-Wolbachia surface protein (WSP), NR-31029; it was used at a dilution of 1:1,000. Additional antibodies were anti-actin monoclonal antibody (clone C4) at 1:10,000 (Seven Hills Bioreagents) and secondary antibodies HRP enzyme conjugates (Invitrogen) at 1:5,000.
DNA extractions and PCRs.
Quantitative PCR was used to calculate the relative titers of Wolbachia in our passaged stocks and in the ancestral line. DNA was extracted from flies by utilizing the Qiagen DNeasy blood and tissue kit (Qiagen) according to directions. DNAs were quantified using absorbance at 260 nm with an Epoch spectrophotometer (Biotek). Quantitative PCR was performed to detect the Wolbachia titer (with reference to the host) using an Applied Biosystems StepOne real-time PCR system and SybrGreen chemistry (Applied Biosystems). We used wsp primers for Wolbachia (forward, CATTGGTGTTGGTGTTGGTG; reverse, ACCGAAATAACGAGCTCCAG) and Rpl32 primers for the host (forward, CCGCTTCAAGGGACAGTATC; reverse, CAATCTCCTTGCGCTTCTTG) at 95°C for 10 min, followed by 40 cycles of 95°C for 15 s and 60°C for 1 min. Reactions were performed in a 96-well plate and calibration standards used in every run to calculate primer efficiencies. These efficiencies, along with the threshold cycle (CT) values generated by the machine, were used to calculate the relative amounts of Wolbachia using the ΔΔCT (Livak) and Pfaffl (14) methods.
Wolbachia enrichment and processing for Illumina library generation.
A simple Wolbachia enrichment protocol was used. First, 20 dissected ovaries were homogenized in GNE buffer (50 mM glycerophosphate, 10 mM NaF, 1.5 mM EGTA; pH 7.6 [with HCl]) with added dithiothreitol (DTT) (final concentration, 2 mM) using a glass dounce homogenizer on ice. These homogenates were filtered (passage through a 10-μm filter) and centrifuged at 16,000 × g for 20 min at 4°C. Samples were preserved in RNAlater solution and frozen at −80°C before DNA extraction. Genomic DNA was extracted using a Qiagen DNeasy kit, and the genetic material from each lineage was used for library generation with the Nextera DNA sample preparation kits and pooled for multiplex sequencing on a MiSeq sequencer (paired-end [PE], 300-bp reads).
Bioinformatic analysis of Wolbachia resequenced genomes.
We created a total of 10 libraries for this project: 1 for each of the passaged lines (in the mutant/CyO or the OreR background for 3 generations) and 2 for the original, ancestral line (GenBank SRA accession number SRP049613). Between 1 million and 2 million reads were achieved for each sample (see Table S1 in the supplemental material), and roughly 5% of the reads mapped to the Wolbachia wMel genome, resulting in between 20× and 60× coverage for each line. We utilized the bioinformatics pipeline Breseq (15) to identify polymorphisms and predict mutations in each resequenced genome. This pipeline is based on bowtie mapping to a reference sequence, for which we used the sequenced wMel genome (GenBank accession number NC_002978.6). Importantly, we required that indels be supported by reads in both directions, that each position be covered by 20 reads, that each single nucleotide polymorphism (SNP) investigated appear at >8% frequency in the population, and that these SNPs not appear in our ancestral sequence. Genetic mutations identified in this fashion were validated using Sanger sequencing. Primers were designed to flank two SNPs in particular (FicF, CTCGTACCAGTGCGCAA; FicR, GCACTTTATTGAAATTTGACCTAT; WD0633F, CTGTATCCTGACATACAGGAT; WD0633R, AATATTAGACTCAATATTGCG), and the resulting amplicons were cloned into pCR-TOPOII blunt cloning vectors. Twenty-four clones from each amplicon pool were sequenced, and SNPs were manually verified by examining ab1 sequence files using 4 Peaks software.
RESULTS
Heterozygous profilin flies harbor low-titer Wolbachia infections.
Wolbachia pipientis is reduced in titer when colonizing Drosophila melanogaster harboring single copies of mutant profilin (Table 1). We crossed infected, wild-type flies (stock number 145) to chic221/Cyo males and collected F1 female progeny of the chic221/+ genetic background. Compared to control flies of the same age, these heterozygous chic221/+ flies harbor one-third of the Wolbachia titer (Table 1).
TABLE 1.
Comparison of titers of Wolbachia bacteria found in F1 chic221/+ Drosophila female backgrounds and in comparable control (wild-type) fliesa
| Drosophila background | Relative quantity of gene: |
Pfaffl ratio compared to control | Relative ratio of expression | |
|---|---|---|---|---|
| wsp | rpl32 | |||
| Control (stock 145) | 13.98 | 15.57 | 4.37 | |
| F1 chic221/+ | 18.98 | 19.61 | 0.33 | 1.55 |
Relative quantity of the Wolbachia specific gene (wsp) compared to the host gene reference (rpl32) across five individual mutant flies and five individual control flies (stock 145, from which the infection within chic221/+ was derived). The relative ratio of wsp expression is statistically significantly reduced in the heterozygous mutant background (t = 15.586, df = 6, P < 0.001).
Sequential passage of Wolbachia within profilin mutant (chic221/+) and control backgrounds.
We wanted to determine if chic221/+ hosts would induce phenotypic changes in Wolbachia populations. We therefore established 20 single fly crosses between uninfected chic221/Cyo (stock 4892) male flies and infected w1 females (stock 145). As a control for the effect of bottlenecking Wolbachia in single fly lines, we also established 20 single fly crosses between uninfected wild-type males (stock 25211) and infected w1 females (stock 145). We allowed these single-pair matings to produce F1 progeny, after which females were dissected and their ovaries preserved for subsequent DNA extraction. We then selected a single, straight-winged female from each of the mutant lines (chic221/+) and mated her with the parental uninfected chic221/Cyo male flies. For our control lines, we selected a single female and mated her with the uninfected control male stock (25211). We continued the passage of Wolbachia through these host backgrounds (either chic221/+ or control) for three generations. Each of the mutant and control lines was tested for a Wolbachia infection. At that point, we had a total of 9 surviving mutant lines infected with Wolbachia, and we kept 9 control lines.
Passage of Wolbachia through mutant host backgrounds results in increased titer after only three generations.
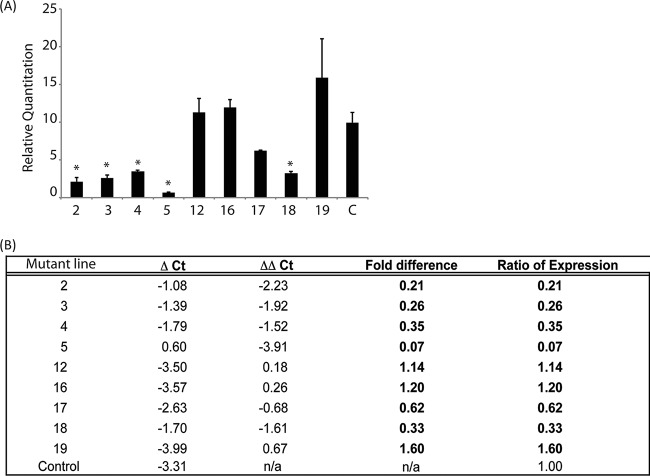
Pathogenic variants of Wolbachia tend to be of high titer compared to their less pathogenic relatives (16, 17). We therefore wondered if our passaged Wolbachia would colonize Drosophila at higher titer than expected. To characterize the titer of the Wolbachia strains in each of the passaged lines, we utilized quantitative PCR (qPCR). Flies were reared simultaneously and age matched in order to remove the confounding effects of host age on Wolbachia titer. The relative quantity of the Wolbachia-specific gene wsp was used to quantify the bacteria within each fly line (relative to the host gene rpl32). We found that many of the passaged lines within the profilin heterozygous mutant background contained significantly fewer wsp copies than did the control flies; specifically, lines 2, 3, 4, 5, and 18 displayed less than one-half of the quantity of Wolbachia organisms expected based on the control ancestral line (Fig. 1). Because these Drosophila flies are carrying the chic221 mutation and we have already shown that this mutation results in a decrease in Wolbachia titer, this finding was not surprising. However, we did identify four lines for which the quantity of wsp was not statistically significantly different from the control lines (lines 19, 17, 16, 12). In these four lines, the Wolbachia titer resembles that of the original, maternal line, although the flies carry the chic221 mutation (Fig. 1). This result suggests that Wolbachia strains within the lines have adapted to a new ecological context and are now able to colonize the mutant host at the same titer as the wild type.
FIG 1.
Wolbachia bacteria passaged within chic221/+ backgrounds are altered in titer compared to their progenitor. (A) Relative quantity of the Wolbachia specific gene (wsp) compared to the host gene reference (rpl32) across mutant lines and the control line (stock 145, labeled “C”). Statistically significant deviations from control are shown by an asterisk (*). (B) Quantitative PCR statistics based on relative quantitation of wsp compared to rpl32 calculated for each of the mutant lines and the control flies. Flies were age matched and reared simultaneously.
Passage of Wolbachia through mutant hosts induces changes in Wsp protein production.
For heterozygous mutant flies, we sought to characterize Wolbachia Wsp protein production. Wolbachia surface protein (Wsp) is an antigen found on the surface of Wolbachia and shed into the blood of vertebrate hosts infected by Wolbachia-carrying filarial nematodes (18, 19). Although its function in the bacterium is currently unknown, antibodies to the protein have been successfully used to monitor bacterial titer (20). We initially used anti-Wsp to identify titer differences in our passaged lines. However, in passaged lines we detected an alteration in tissue-specific production of Wsp (Fig. 2). In control females harboring wMel infections, production of Wsp is limited to nonovary tissues (i.e., carcasses postdissection). Although Wolbachia is known to heavily colonize the ovary in Drosophila (21–24), the bacteria do not heavily produce Wsp in that same tissue (Fig. 2A). The Wsp antibody, generated from nematode Wolbachia, may not be sensitive enough to detect very low quantities of protein expressed in this tissue. This tissue-specific protein production pattern also holds for F1 progeny resulting from crosses between males harboring single mutations in the actin binding proteins villin and profilin and control females (Fig. 2A). However, after passage of Wolbachia through heterozygous profilin mutant (chic221/+) hosts for two fly generations, Wolbachia strains that are able to colonize these hosts, regardless of titer, now alter the pattern of protein production and heavily express Wsp in the ovary (Fig. 2B).
FIG 2.
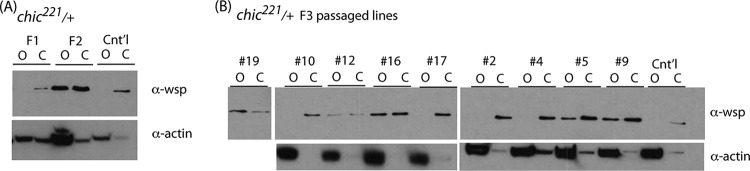
Wolbachia passaged within chic221/+ backgrounds displays altered expression of Wsp in the ovary. (A) Immunoblots of Drosophila melanogaster ovaries (O) and carcasses (C) from control and chic221/+ backgrounds, probed using an antibody against Wsp. (B) After passage for three generations through single fly crosses, some Wolbachia bacteria retain the expression of Wsp in the ovary while some revert to the control condition (expression detectable only in the carcass).
Wolbachia lineages passaged through the chic221/+ host background are more pathogenic than their progenitor.
Wolbachia strains that are overly pathogenic can have effects on host life span (25) or host health (26). As a first step to identify the pathogenic effects of passaged Wolbachia, we examined the total number of progeny produced by each of the Wolbachia-infected Drosophila lines, comparing the lines in which Wolbachia was passaged through the mutant chic221/+ hosts to those passaged through control flies. The number of progeny produced by the mutant fly lineages harboring passaged Wolbachia was bimodal in distribution: three of the mutant lines produced very few progeny (mean number of progeny, 38), while seven of the mutant lines produced many progeny (mean, 177) and were statistically similar to the control lineages (mean, 168). However, averaged overall, the number of progeny produced by the passaged lines was not significantly less than the number produced by the control lines (t = −1.407, df = 19, P = 0.176).
This result suggests that the passaged Wolbachia strains from the mutant host backgrounds might induce a more severe reproductive phenotype. However, effects observed after passage may be due to either host genetic background (the effect of genetic drift combined with deleterious homozygous alleles) or the Wolbachia infection. In order to distinguish between these two possibilities, we selected the line with the most severe phenotype (mutant 5) and cleared it of a Wolbachia infection using tetracycline (for 2 generations). We then reexamined the number of progeny that result from the previously incompatible crosses using 20 single fly matings between passaged males (from the mutant 5 line) and uninfected females (25211) and compared the number to that obtained with the analogous cross using tetracycline-cleared mutant 5 males. We saw a fecundity defect to the passaged Wolbachia infection. Mutant 5 males cleared of the Wolbachia infection produced many more progeny than mutant 5 males carrying the passaged Wolbachia infection (80% more; t = 3.064, df = 38, P = 0.004; see Table S2 in the supplemental material). This difference in thenumber of progeny was primarily due to the smaller number of female offspring produced by mutant 5 males carrying the Wolbachia infection, whereas the number of male offspring was not significantly different (for females, t = 3.104, df = 38, P = 0.004; for males, t = 1.541, df = 38, P = 0.132). This result suggests a genetic interaction between the chic null allele and Wolbachia that is currently being investigated. Therefore, the fecundity defects observed in the mutant 5 line were not a result of host loci alone but were influenced by the passaged Wolbachia infection.
Identification of the genomic loci in the passaged Wolbachia lines.
To identify Wolbachia genomic differences that might exist in our passaged lines, we selected four profilin mutant lines (low-titer line 5 and high-titer lines 12, 19, and 16), four control lines (resulting from crosses between infected control females and uninfected wild-type OreR stocks), and two parental stocks (stock 145, the source of the Wolbachia infection) for Illumina library preparation and genomic resequencing (see Table S1 in the supplemental material). We chose these particular passaged lines because they displayed production of the Wsp protein in the host ovary (lines 5, 12, 19, and 16), induced fecundity defects in the host (line 5), and/or carried low-titer or control titer Wolbachia (low titer = 5; high titer = 12, 16, 19).
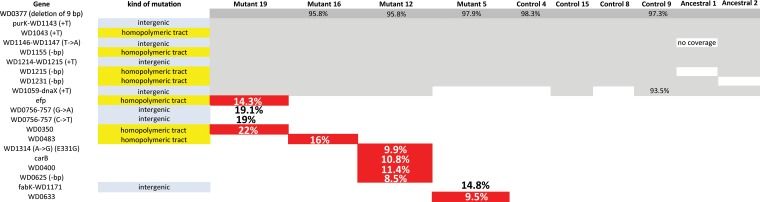
Using strict thresholds, we were able to identify our lab strain (the ancestral Wolbachia strain used in our crosses) as a wMel type, not wMel CS (using markers found in reference 15). However, our lab strain contained a number of fixed or nearly fixed differences compared to the wMel sequence deposited in GenBank (9 in total) (Fig. 3). The majority of the differences (8/9) were indels of either a single nucleotide or a stretch of 9 bp (Fig. 3). Some of the changes are expected to impact gene function (causing frameshift mutations), and some occur within homopolymeric regions in the 5′ end of the genes in question (4/9), suggesting that the changes may be important for gene regulation (e.g., phase variation).
FIG 3.
Mutations in passaged Wolbachia within chic221/+ background or control flies. Whole-genome sequencing using an Illumina MiSeq sequencer was used to identify individual mutations in each of 4 lines from chic221/+ passaged Wolbachia and compared to 4 control passaged Wolbachia as well as the ancestral stock (sequenced twice). The identified loci affected by mutation are listed, and the presence of a mutation within each lineage is depicted. If the change is predicted to alter the amino acid encoded at that position, the mutation is colored in red. If the mutation was found in <100% of the mapped reads, the percentage is shown.
We next focused on the mutations that occurred in Wolbachia within the profilin heterozygous mutant lines. The mutant lines contained the same 9 fixed differences found in the ancestral sequence, but in addition, each line was found to contain unique polymorphisms. These polymorphisms were covered by at least 20 reads and present at greater than 8% of the population of reads. Importantly, none of the new polymorphisms identified in each mutant line were shared between lines; that is, each line contained a unique set of polymorphisms. Some of the identified mutations were in intergenic regions (between fabK and WD1171 for mutant 5; between WD0756 and WD0757 for mutant 19), and although the mutations could potentially alter as-yet-unknown regulatory RNAs, they probably do not directly affect the encoded amino acids. Other identified mutations occurred within homopolymeric tracts (such as efp, WD0350 for mutant 19, WD0483 in mutant 16, and WD0625 in mutant 12) and could be important for gene regulation (e.g., phase variation). None of these mutations are in loci predicted to be essential. Four mutations occurred within coding regions and resulted in amino acid changes for mutant 12 and mutant 5. In mutant 12, we found changes in an ABC transporter (WD0400), the carbamoyl phosphate synthetase involved in production of arginine and pyrimidines (carB), and a filamentation induced by cyclic AMP (cAMP) protein (WD1314). In mutant 5, we found a single change in an ankyrin repeat containing prophage LambdaW1 protein: WD0633. We sought to confirm these nonsynonymous SNPs through PCR amplification of both WD1314 and WD0633 coupled to clone library and Sanger sequencing. We sequenced 24 clones from each of the amplified pools and recovered 1 or 2 clones with the SNP in question, supporting our observation that the Wolbachia populations do contain mutant alleles and that the observed changes were not Illumina sequencing errors.
DISCUSSION
Because Wolbachia bacteria are obligately intracellular, their ecology is their host. Here we manipulate Wolbachia's ecology by introducing the bacteria into a host background that substantially reduces titer. The rationale is that this background imposes significant selective pressure on the Wolbachia to adapt to a new ecological context in order to persist in the fly lines and also that this severe bottleneck will increase the stochastic segregation of Wolbachia each generation. We characterized the passaged Wolbachia organisms in four important ways: (i) through examination of titer using qPCR, (ii) through crosses to identify potential pathogenic effects of the passaged Wolbachia such as effects on fecundity, (iii) through quantification of Wsp protein production, and (iv) through genomic resequencing.
Wolbachia bacteria that are passaged through restrictive hosts alter their titer compared to their progenitor. In the majority of our passaged lines, we observed low-titer Wolbachia infections, as would be expected given the Wolbachia transmission defect induced by the chic221 mutation. However, in four of the passaged lines, we saw Wolbachia titers return to control values, suggesting that the bacteria are able to adapt to a new host context in only three generations. Others have selected for Wolbachia titer increases and decreases over short time scales (5 generations) and have also found the bacterial parasite to be particularly plastic with regard to this parameter (27). Also, Wolbachia lineages passaged through the chic221 heterozygous mutant hosts exhibit other phenotypic differences compared to the ancestral strain. Although the function of Wsp is not yet known, it is curious that the passaged Wolbachia alters the production of Wsp in the ovary, the reproductive tissue on which the bacteria depend for maternal transmission.
Wolbachia passaged through chic221 heterozygous mutant hosts also exhibited genomic changes. We found mutations at significant frequencies (>8%) in each of our mutant lines and confirmed the presence of two particular nonsynonymous SNPs for two of the mutant lines, showing that the Illumina resequencing correctly revealed the mutations and that they were not sequencing errors. We think it is premature to propose that these specific mutations are selected for in direct response to the host actin phenotype. However, it is curious that one of the passaged lines (mutant 12), which regained its wild-type titer, exhibited a mutation in a protein known in other systems to modify eukaryotic actin (WD1314, annotated as Fic) (Fig. 3). In sum, our results show that Wolbachia is able to adapt to a new host context on a short time scale.
What does Wolbachia heterogeneity mean for the host?
Wolbachia genetic variability within single hosts has been noted before. For example, Schneider et al. identified diverse haplotypes of Wolbachia within several insect hosts (11). Interestingly, in that study, the authors were able to shift the prevalence of different haplotypes upon transfer and maintenance in new hosts for over 100 generations (11), suggesting that Wolbachia haplotypes can shift in frequency depending on the host context. Similar to our results here, many of these mutations were found to be frameshift mutations and de novo stops (11). However, in that study, no specific phenotypic effect was attributed to the Wolbachia microheterogeneity. In our work, we were able to reveal mutations in Wolbachia in a very short number of generations (only 3). The mutations that we identified appear to be between 8.5 and 22% penetrant; the Wolbachia strains within these hosts are a pool of genetic haplotypes and do not exist as clonal isolates. We did not identify any of the SNPs in the mutant lines in the ancestral stock. However, it is possible that we lack the resolution necessary to identify rare mutants in the ancestral line. We cannot be certain if the SNPs are derived from an existing pool of genetic heterogeneity or are de novo mutations.
Here, we establish a straightforward method to generate populations of mutants in Wolbachia, an obligate intracellular parasite for which there is no genetic system. By passaging Wolbachia lineages through a host background that severely reduces titer, we enrich for or generate Wolbachia mutants. Utilizing this approach, which is analogous to forward genetics in other systems, it may be possible to learn about the biology of Wolbachia infection in a targeted and mechanistic manner.
Supplementary Material
ACKNOWLEDGMENTS
We thank two anonymous reviewers for their suggestions to improve the manuscript. We also thank Jack Werren for his invaluable feedback on this work in the early stages of development.
This research was supported by generous start-up funds from Indiana University, a Faculty Research Support Program from the Office of the Vice Provost for Research at Indiana University, and an NSF award (DBI-1219659) to I.L.G.N. Analysis of the data at Indiana University was supported by the National Center for Genome Analysis Support (NCGAS).
Stocks obtained from the Bloomington Drosophila Stock Center (NIH P40OD018537) were used in this study.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.02987-14.
REFERENCES
- 1.Werren JH, Baldo L, Clark ME. 2008. Wolbachia: master manipulators of invertebrate biology. Nat Rev Microbiol 6:741–751. doi: 10.1038/nrmicro1969. [DOI] [PubMed] [Google Scholar]
- 2.Zug R, Hammerstein P. 2012. Still a host of hosts for Wolbachia: analysis of recent data suggests that 40% of terrestrial arthropod species are infected. PLoS One 7(6):e38544. doi: 10.1371/journal.pone.0038544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.LePage D, Bordenstein SR. 2013. Wolbachia: can we save lives with a great pandemic? Trends Parasitol 29:385–393. doi: 10.1016/j.pt.2013.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Turelli M, Hoffmann AA. 1991. Rapid spread of an inherited incompatibility factor in California Drosophila. Nature 353:440–442. doi: 10.1038/353440a0. [DOI] [PubMed] [Google Scholar]
- 5.Ishmael N, Dunning Hotopp JC, Ioannidis P, Biber S, Sakamoto J, Siozios S, Nene V, Werren J, Bourtzis K, Bordenstein SR, Tettelin H. 2009. Extensive genomic diversity of closely related Wolbachia strains. Microbiology 155(Part 7):2211–2222. doi: 10.1099/mic.0.027581-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Newton RR, Newton IL. 2013. PhyBin: binning trees by topology. PeerJ 1:e187. doi: 10.7717/peerj.187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ellegaard KM, Klasson L, Naslund K, Bourtzis K, Andersson SG. 2013. Comparative genomics of Wolbachia and the bacterial species concept. PLoS Genet 9(4):e1003381. doi: 10.1371/journal.pgen.1003381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Klasson L, Westberg J, Sapountzis P, Nasiund K, Lutnaes Y, Darby AC, Veneti Z, Chen LM, Braig HR, Garrett R, Bourtzis K, Andersson SGE. 2009. The mosaic genome structure of the Wolbachia wRi strain infecting Drosophila simulans. Proc Natl Acad Sci U S A 106:5725–5730. doi: 10.1073/pnas.0810753106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chrostek E, Marialva MS, Esteves SS, Weinert LA, Martinez J, Jiggins FM, Teixeira L. 2013. Wolbachia variants induce differential protection to viruses in Drosophila melanogaster: a phenotypic and phylogenomic analysis. PLoS Genet 9(12):e1003896. doi: 10.1371/journal.pgen.1003896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Symula RE, Alam U, Brelsfoard C, Wu YN, Echodu R, Okedi LM, Aksoy S, Caccone A. 2013. Wolbachia association with the tsetse fly, Glossina fuscipes fuscipes, reveals high levels of genetic diversity and complex evolutionary dynamics. BMC Evol Biol 13:31. doi: 10.1186/1471-2148-13-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schneider DI, Riegler M, Arthofer W, Mercot H, Stauffer C, Miller WJ. 2013. Uncovering Wolbachia diversity upon artificial host transfer. PLoS One 8(12):e82402. doi: 10.1371/journal.pone.0082402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Verheyen EM, Cooley L. 1994. Profilin mutations disrupt multiple actin-dependent processes during Drosophila development. Development 120:717–728. [DOI] [PubMed] [Google Scholar]
- 13.Fry AJ, Palmer MR, Rand DM. 2004. Variable fitness effects of Wolbachia infection in Drosophila melanogaster. Heredity (Edinb) 93:379–389. doi: 10.1038/sj.hdy.6800514. [DOI] [PubMed] [Google Scholar]
- 14.Pfaffl MW. 2001. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res 29:e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Richardson MF, Weinert LA, Welch JJ, Linheiro RS, Magwire MM, Jiggins FM, Bergman CM. 2012. Population genomics of the Wolbachia endosymbiont in Drosophila melanogaster. PLoS Genet 8(12):e1003129. doi: 10.1371/journal.pgen.1003129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McGraw EA, O'Neill SL. 2004. Wolbachia pipientis: intracellular infection and pathogenesis in Drosophila. Curr Opin Microbiol 7:67–70. doi: 10.1016/j.mib.2003.12.003. [DOI] [PubMed] [Google Scholar]
- 17.Strunov A, Kiseleva E, Gottlieb Y. 2013. Spatial and temporal distribution of pathogenic Wolbachia strain wMelPop in Drosophila melanogaster central nervous system under different temperature conditions. J Invertebr Pathol 114:22–30. doi: 10.1016/j.jip.2013.05.001. [DOI] [PubMed] [Google Scholar]
- 18.Brattig NW, Bazzocchi C, Kirschning CJ, Reiling N, Buttner DW, Ceciliani F, Geisinger F, Hochrein H, Ernst M, Wagner H, Bandi C, Hoerauf A. 2004. The major surface protein of Wolbachia endosymbionts in filarial nematodes elicits immune responses through TLR2 and TLR4. J Immunol 173:437–445. doi: 10.4049/jimmunol.173.1.437. [DOI] [PubMed] [Google Scholar]
- 19.Kramer LH, Tamarozzi F, Morchon R, Lopez-Belmonte J, Marcos-Atxutegi C, Martin-Pacho R, Simon F. 2005. Immune response to and tissue localization of the Wolbachia surface protein (WSP) in dogs with natural heartworm (Dirofilaria immitis) infection. Vet Immunol Immunopathol 106:303–308. doi: 10.1016/j.vetimm.2005.03.011. [DOI] [PubMed] [Google Scholar]
- 20.Dobson SL, Bourtzis K, Braig HR, Jones BF, Zhou WG, Rousset F, O'Neill SL. 1999. Wolbachia infections are distributed throughout insect somatic and germ line tissues. Insect Biochem Mol Biol 29:153–160. doi: 10.1016/S0965-1748(98)00119-2. [DOI] [PubMed] [Google Scholar]
- 21.Clark ME, Karr TL. 2002. Distribution of Wolbachia within Drosophila reproductive tissue: implications for the expression of cytoplasmic incompatibility. Integr Comp Biol 42:332–339. doi: 10.1093/icb/42.2.332. [DOI] [PubMed] [Google Scholar]
- 22.McGraw EA, Merritt DJ, Droller JN, O'Neill SL. 2002. Wolbachia density and virulence attenuation after transfer into a novel host. Proc Natl Acad Sci U S A 99:2918–2923. doi: 10.1073/pnas.052466499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Veneti Z, Clark ME, Karr TL, Savakis C, Bourtzis K. 2004. Heads or tails: host-parasite interactions in the Drosophila-Wolbachia system. Appl Environ Microbiol 70:5366–5372. doi: 10.1128/AEM.70.9.5366-5372.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sacchi L, Genchi M, Clementi E, Negri I, Alma A, Ohler S, Sassera D, Bourtzis K, Bandi C. 2010. Bacteriocyte-like cells harbour Wolbachia in the ovary of Drosophila melanogaster (Insecta, Diptera) and Zyginidia pullula (Insecta, Hemiptera). Tissue Cell 42:328–333. doi: 10.1016/j.tice.2010.07.009. [DOI] [PubMed] [Google Scholar]
- 25.McMeniman CJ, O'Neill SL. 2010. A virulent Wolbachia infection decreases the viability of the dengue vector Aedes aegypti during periods of embryonic quiescence. PLoS Negl Trop Dis 4(7):e748. doi: 10.1371/journal.pntd.0000748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Turley AP, Moreira LA, O'Neill SL, McGraw EA. 2009. Wolbachia infection reduces blood-feeding success in the dengue fever mosquito, Aedes aegypti. PLoS Negl Trop Dis 3(9):e0000516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Perrot-Minnot MJ, Werren JH. 1999. Wolbachia infection and incompatibility dynamics in experimental selection lines. J Evol Biol 12:272–282. doi: 10.1046/j.1420-9101.1999.00025.x. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.