Abstract
Systems biology is an interdisciplinary effort to integrate molecular, cellular, tissue, organ, and organism levels of function into computational models that facilitate the identification of general principles. Systems medicine adds a disease focus. Systems epidemiology adds yet another level consisting of antecedents that might contribute to the disease process in populations. In etiologic and prevention research, systems-type thinking about multiple levels of causation will allow epidemiologists to identify contributors to disease at multiple levels as well as their interactions. In public health, systems epidemiology will contribute to the improvement of syndromic surveillance methods. We encourage the creation of computational simulation models that integrate information about disease etiology, pathogenetic data, and the expertise of investigators from different disciplines.
Keywords: Systems theory, systems biology, computer simulation, epidemiology
Introduction
Risk factors have always been the main focus of epidemiologists’ search for causes of disease. Nevertheless, we also have a long history of incorporating information about mechanisms of disease. Attention to lipid profiles in blood and tissue dates back to the 1950s [1]. This was soon followed by efforts to understand the role of lipids in coronary artery disease [2] and a clinical trial to test if lipids really were in the causal chain [3].
Some epidemiologists were quick to incorporate molecular information provided by microarrays [4,5]. What we offer in this paper is encouragement to continue in this tradition to improve what we do by incorporating improved sources of information.
The integration of intra-individual processes with population-based knowledge challenges the current separation of experimental and theoretical approaches. Basic laboratory research in the life sciences can be conceptualized as the search for mechanisms [6,7]. The idea is that a formal description of the mechanism underlying a biologic phenomenon will contribute to a causal explanation for its occurrence.
The notion of mechanisms is particularly dominant in systems biology, where intra- and inter-cellular processes are frequently experimentally and theoretically investigated in terms of networks and pathways. Knowledge of the structure of the network/pathway and the biophysical details of the interactions allows better understanding of the cellular mechanisms underlying cell functions [8].
The notion of mechanism appears to play a less important role in epidemiology, although this is slowly beginning to change [9]. For example, epidemiologists and those who study the epigenome are working together to gather data that are likely to allow modeling of disease occurrence based on data collected over decades [10]. Also gaining acceptance is the recognition that the contributions of social interactions to psychiatric disorders and behavioral dysfunctions are via epigenetic phenomena [11]. To contribute to advanced mechanistic knowledge in this area of study, epidemiologists need the assistance of those who are most knowledgeable about measuring epigenetic phenomena and mechanisms.
Perhaps part of their reluctance to talk about mechanisms reflects epidemiologists’ preference for the probabilistic view of causal inference [12], while causal mechanisms are often considered deterministic in biology, although computational scientists who study biological regulatory networks begin to appreciate both deterministic and stochastic approaches [13]. However, we agree with Glymour and Cheng who wrote in 1998 that “a disconnection between mechanisms, on the one hand, and probabilistic patterns, on the other, puts everything on a false footing”. [14] Still, the predominant focus on individual component causes of disease in both pathogenesis and etiology research might miss the many potential interactions between components [15]. Even gene-environment interaction studies tend not to go beyond two components. Consequently, they contribute minimally to the elucidation of network dynamics of disease causation as a whole.
Mathematical modeling can expose erroneous assumptions, define and validate working hypotheses, and guide the design of novel experiments and studies [16]. In systems biology, the structure of interaction maps encodes feedback mechanisms that can be used to explain biological phenomena in terms of the system's robustness, sensitivity, stability. To this end, the models do not have to be based on biochemical or biophysical principles but can be phenomenological [17]. The pharmacologist James Black poignantly explained in his Nobel lecture that “models [...] are not meant to be descriptions, pathetic descriptions, of nature; they are designed to be accurate descriptions of our pathetic thinking about nature.” However, it is also possible, that just as weather forecasting still has limitations despite the use of cutting edge technology and multivariable prediction algorithms, systems epidemiology will not achieve the promise that some hope for it.
In this paper, we suggest that systems epidemiology might be a promising way to supplement systems biology, with the goal of reducing disease burden at the individual [18] and population levels [19]. In essence, we propose a systems epidemiology modeled after systems biology [20-24] and conceptualized as an integration of good old-fashioned black box epidemiology [25] with mathematical modeling and computational simulation [26] (Figure 1). We define “systems epidemiology” as an epidemiologic approach to risk identification that includes (i) systems-level (e.g., omics) exposure measurements at (ii) multiple levels (sociodemographic, clinical, biological, etc), (iii) network analyses of inter-relationships among risk factors, and (iv) computational simulation of risk scenarios in parallel to data-driven biostatistical risk modeling. As such, systems epidemiology need not be seen as a replacement for conventional epidemiologic methods. Rather, systems epidemiology should be seen as a supplemental approach that has advantages that conventional epidemiologic studies lack.
Figure 1.
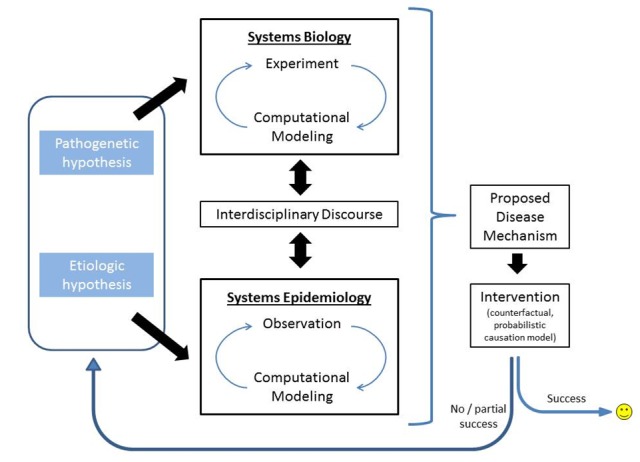
Proposed discovery process integrating pathogenetic and etiologic aspects of illness causation.
In essence, we view systems epidemiology rather broadly. Some components of studies that are needed for a full systems epidemiology study can be viewed as getting closer to that goal. For example, epidemiologic studies that collect data that can be used by molecular biologists enable the expansion and transformation of such a study from an epidemiologic study of proteins, DNA methylation, or gene activation in prospective cohorts to a systems epidemiology study that allows an evaluation of the complexity of the relationships among exposures, biomarkers, and diseases [27,28]. Even limited studies can have the capacity to evaluate relatively simple models, such as multi-hit models of disease risk [29].
In the next section, we introduce the main concepts from general systems theory, systems biology, and systems medicine, as well as define some of the words used in these fields (e.g., systems, network, complexity). In the subsequent section, we review and discuss the six publications that emphasize a conceptual definition of “systems epidemiology” by using the term in the title. In the fourth section, we offer our vision of systems epidemiology as an integration of epidemiologic methods and computational simulation.
Systems theory, biology, and medicine
Systems science is rooted in systems theory, which was developed in the first half of the 20th century (see [30] for a brief historical overview and [31] for a philosophical perspective.) A system can be defined as “a complex of interacting components together with the relationships among them that permit the identification of a boundary-maintaining entity or process”. [32] In the previous sentence, “system” is defined as a complex of components. This meaning of the word “complex” goes back to Ludwig von Bertalanffy, who coined the term “general system theory” [33] and wrote about “complexes of ‘elements’” in his chapter on mathematical underpinnings of system concepts. He considered the analysis of biological systems one of the main goals of general system theory [33]. Applied to living systems, the concepts of organization, hierarchy, and interaction of biological phenomena lead to the notions of feedback loops and mechanistic processes [30].
The method of classical science has relied on the strategy of 'reduction to components,' an approach that emphasizes the study of how one molecule, one cell, or one organ responds to a stimulus [32]. In contrast, the systems approach promotes ‘expansion to dynamics,’ which can identify how complex sets of components behave when exposed to a complex set of influences. This emphasis on relationships between system components (rather than the components themselves) gives rise to the concept of emergence, “the appearance of novel characteristics exhibited on the level of the whole ensemble, but not by the components in isolation”. [32] In essence, part of the “systems view” in science is that by studying networks of interacting phenomena, we can understand their function and their emergent properties better than by studying each component (or a few of them) in isolation.
This emphasis on relationships among components is familiar to the epidemiologist who studies context and interactions [34,35], as well as to those who study non-linear relationships [36-38]. Thus, epidemiology appears to have incorporated elements of a systems approach without calling attention to this transition. For example, the concept that obesity and other disorders can “spread through social networks” has been well recognized by those who study [39,40] and model [41] social transmission dynamics. We see the modeling of social dynamics as contributing to a better understanding of the disease occurrence in populations.
Applying a systems level approach to biological research was proposed at the turn of the millennium [20-24]. Hiroaki Kitano wrote in 2002 that “cells, tissues, organs, organisms and ecological webs are systems of components whose specific interactions have been defined by evolution; thus a system-level understanding should be the prime goal of biology. … A combination of experimental and computational approaches is expected to resolve this problem [of the intrinsic complexity of biological systems]”. [24] The goal is to achieve a systems-level understanding of both “interrelationships (organization or structure) and interactions (dynamics or behavior) of genes, proteins, and metabolites”. [20]
Thus, systems biology focuses on the interaction of components of biological systems, and their components. Moreover, it proposes that experimental and computational work be conducted in parallel. This approach has been implemented most impressively at the cellular level, for example, with biochemical receptor signaling pathways [42-44]. One recent example of a research program that goes beyond the single cell level is the integration of experimental and computational studies designed to better understand sprouting angiogenesis, a concept crucially important, for example, in developmental processes and cancer pathogenesis [45].
The application of systems biology principles in complex disease etiology research and diagnostics has been called “network medicine”. [46] Others suggest that systems biology be brought to the bedside under the name “systems medicine”. [47-50] As advances in high-throughput technologies push the many “omics”-perspectives in biomedical research from structure to function, their utilization in systems biology and systems epidemiology might help identify organizing principles [51].
Systems medicine applies the tools and concepts from systems biology and addresses complexity in two key ways. First, systems medicine uses molecular diagnostics to stratify patients and diseases to better characterize and understand disease complexity [47]. By applying a network-level view of disease to create disease networks, systems medicine will overcome current limitations in drug discovery by identifying important functional and regulatory modules within these networks [52]. Then, by analyzing and targeting hubs—the most highly interconnected nodes—within these regulatory networks, and enzymatic activity in metabolic networks, network-based approaches will be able to explore the effects of various drugs in mathematical models [53]. Such studies might improve not only our understanding of drug-response phenotypes, but will hopefully also help us tailor treatments to an individual’s drug metabolism.
“Systems epidemiology” in the literature
Often described as the “basic science” science of public health, epidemiology is concerned with the distribution, determinants and deterrents of disease occurrence in human populations [54]. With the ever growing number of newly identified risk factors, the single level approach of black-box epidemiology continues to face criticism [55,56]. By focusing on individual risk factors of disease, studies are unlikely to appreciate potentially important interactions between risk factors and the resulting alterations to network dynamics [57]. Feedback loops among risk factors, for example, might remain undetected. New analytical and systematic methods are needed to help those who want to uncover complex biochemical pathways [58]. We are confident that a systems approach to illness causation research will improve our understanding of etiologic mechanisms, and help translate complex data into knowledge relevant for medicine and public health.
While systems thinking has been embraced in biology and medicine, its adoption by public health has yet to take shape [19]. As it has surfaced in the literature, the term “systems epidemiology” has been given various meanings. Before we discuss our own vision of “systems epidemiology”, we briefly review and discuss six publications whose authors emphasize a conceptual definition of “systems epidemiology” by using the term in their title [56,57,59-62].
In what follows, we offer a few brief paragraphs trying to extract the meaning of “systems epidemiology” as used by the authors of these publications. We then attempt to integrate their commonalities into a framework for etio-pathogenetic research at the systems level.
In the field of cancer epidemiology, Lund and Dumeaux define “systems epidemiology” as “a new research discipline that seeks to integrate pathways analyses into observational study designs to improve the understanding of biological processes in the human organism”. [62] Although the authors consider systems epidemiology the “observational counterpart to systems biology”, they do not go beyond the “expansion of gene-environment epidemiologic studies with analyses of the transcriptome”. They suggest that in what they call a “globolomic design”, mRNA and miRNA analyses will “open the black box” and provide insight into biologic pathways, and even add “understanding to the concept of causality in epidemiology.” Study designs that consider the trajectory of gene expression in multistep carcinogenesis and changes in lifestyle of study participants would shed light on the interplay between risk factors and the effects that changes in risk factors have on each other.
Similar to Lund and Dumeaux in cancer research, Frank Hu proposes incorporating metabolomics data in epidemiological studies of diabetes [56]. He suggests that incorporating recent advances in high-throughput analytic techniques into human observational studies of novel metabolite biomarkers might allow a shift “from the traditional black-box strategy to a systems approach”. Nevertheless, he closes on a cautionary note by saying that “although the systems epidemiology approach can offer deeper understanding of molecular pathways underlying epidemiologic observations, whether it can improve early disease detection, clinical diagnosis, and prognosis, and contribute to personalized prevention and treatment remains to be seen.” Still, Hu and Cornelis suggest that nutritional systems epidemiology will lead to “improved personalized optimal nutrition for prevention and treatment of disease” (emphasis in original) [63].
In order to bridge the gaps between systems biology and traditional epidemiology, we must go beyond the data-rich environment of the human genome, and include data from socioeconomic and environmental levels of interaction. Infectious disease researchers have proposed a role for systems epidemiology in developing a more comprehensive understanding of tuberculosis [60,61]. While systems biology addresses biological aspects of infectious disease, systems epidemiology incorporates epidemiology, sociology, evolutionary biology, and ecology to describe physical and social environments. It also has the potential to provide a better understanding of host and pathogen interactions at levels beyond pathogenicity.
High through-put microarrays of proteins, gene expression, and epigenetic markers, as well as other biomarkers, provide the potential to extend phenotyping to include the response of a cell or tissue to stimuli. Add high-resolution imaging techniques with their capability to assess function as well as structure and you have the potential “to derive genome-wide molecular networks of genotype-phenotype associations” called ‘‘phenomics” [64] and “deep phenotyping” [57,65]. The latter term reflects a tendency in the high-tech community, to apply the adjective ‘deep’ to mean ‘comprehensive’ and ‘thorough’. One important benefit of ‘deep phenotyping’ would be its potential to reduce misclassification by refining risk classification [57].
Finally, Joffe and colleagues describe the use of diagrammatic modeling in epidemiology and propose the adoption of system-wide models to include multiple interactions in causal analyses [59]. This version of systems epidemiology addresses the philosophy of causation, a key challenge for the progress of illness causation research.
Three themes, plus one
Three themes emerge from the views of systems epidemiology provided in the previous section. Perhaps a bit surprisingly, all three are not really novel to epidemiologic theory.
First, multiple authors think that systems epidemiology will open the black box between exposure and outcome. While some have postulated that traditional epidemiology is incapable of doing this [66], others think it can [25,67,68]. In particular, we are in full agreement with Hafeman and Schwartz, who suggest that we should not discard the ‘black box’ approach altogether, but instead use existing methods to open it, e.g., mediation analysis [68]. One textbook of molecular epidemiology [69] suggests that measurement of multiple consecutive molecular markers of disease progression, together with markers of susceptibility can provide some insight into the contents of the black box [70]. Apparently, what is now proposed under the heading of systems epidemiology as “globolomics” [62] or “deep phenotyping” [57] might be not much more than molecular or genetic epidemiology, albeit at a finer resolution.
Second, at least two groups of authors stress the point that systems epidemiology should not only be thought of as the measurement of molecular markers, but should also include the study of “behavioral, sociodemographic, and group levels that may influence health and disease” [57], as well as ecologic and evolutionary factors [61]. This integration of multiple levels, layers, or scales of phenomena to be explained is familiar to epidemiologists and statisticians [71-73], as well as to those who think about general concepts of computational disease modeling [74-76].
Systems epidemiology as described in the six papers cited above is a blend of traditional genetic and molecular epidemiology that includes biomarker-environment (inter-level) interaction analyses. Although these concepts are certainly encouraging, the question remains how to design and perform analyses of intra-level interactions.
Third, the “paradox of ever-increasing measurement capabilities followed by decreasing abilities to translate basic mechanistic knowledge into clinically effective therapeutics " [57] cannot be tackled without the development of new strategies that guide us in our interpretation of data. The need to clarify how we think about complex systems studies has encouraged some to think and write about the issue of causation and causal inference in systems epidemiology [59,77]. Current work in this area [78-81] continues a longstanding tradition in epidemiologic thinking [15,82,83].
While black box elucidation, multilevel analysis, and causal inference appear to be of overall importance for systems epidemiology, one key hallmark of systems biology methods, computational modeling, is prominently absent from all six conceptions of systems epidemiology. Just as computational modeling is considered crucial for systems biology in order to compare results from top-down computational models with data from bottom-up experimental studies [20], we suggest that modeling and simulation are crucial for systems epidemiology (Figure 1).
We proceed with the expectation that computational studies will help improve our understanding of disease processes, in part, by enabling us to go back and forth between epidemiologic observation and computational model, iteratively comparing what has been simulated in silico to what we observe in vivo. The result is either a validation of the current computational model, or a modification that brings the model closer to reality. Yes, it will be difficult or even impossible to simulate multilevel study designs in detail. However, we are convinced that certain modeling techniques will be helpful. For example, the recursive Bayesian net formalism [84], although based on acyclic graph theory, allows for a causal relationship to cause the elements of a similar relationship at a different level, thereby enabling us to model quasi-feedback linkage points between levels.
Most importantly, we hold that computational studies will provide systems epidemiologists with the capacity to model the interactions between observed risk factors, thereby elucidating purportedly complex interactions among exposures and the resulting processes that lead to human disease [57]. Indeed, the idea of integrating mathematical modeling/simulation with epidemiologic studies has a longstanding tradition in infectious disease research [85-87] that continues in cancer epidemiology [88], prevention research [26], environmental epidemiology [89], and chronic disease epidemiology [90]. None of these recent citations features the term “systems epidemiology”.
Any kind of biologic simulation needs to be validated appropriately. Simple and complex ways of model verification and validation need to be considered and applied in an ongoing fashion over the model’s entire “life cycle”. [91]
How can systems biology and systems epidemiology be integrated? We expect that computational models of pathogenetic mechanisms from systems biology will soon be included in computational etiology models that simulate occurrence of disease in humans. Such an integration of methods will likely help usher in the next epoch of disease causation research, accommodating both theory and methods decades old and state of the art data analysis/modeling.
Critical to these efforts will be an ongoing interdisciplinary discourse (Figure 1). Only if systems biologists and epidemiologists intensify their conversation can all this be achieved. We acknowledge that a general level of understanding will be difficult to achieve due to the very different underlying assumptions, modes of thinking, and scientific languages spoken in wet and dry labs. Nevertheless, improved communication between experimentalists and modelers in systems biology, between epidemiologists and modelers in systems epidemiology, and among all four groups of investigators should lead to a common vocabulary and mutually-beneficial enlightenment.
Many experimentalists have a hypothesis before they do their experiments. In essence, they accept the iterative approach of modifying their hypothesis based on what they most recently found. In systems biology this process is realized in iterative cycles of data-driven modeling and model-driven experimentation. In essence, hypotheses can be formulated and validated with computational modeling. However, the goal of modeling is more than prediction. Instead, modeling provides a way of thinking, a workflow, that amongst other things guides data collection and improves understanding [92].
The term “in silico,” coined little more than two decades ago, is applied to computer simulation [93]. Two important characteristics of many of these analyses are the concepts of homeostasis and 'feedback'. Homeostasis is the regulation of a system to maintain a stable, constant condition. In order to induce, prevent or recover from a change or perturbation, information of the state of the system must be 'fed back'. Feedback therefore implies a 'before' and 'after', which is why we can only understand cellular functions and their underlying mechanisms by treating them as dynamic systems.
Homeostatic regulation allows an organism to function effectively in a broad range of environmental conditions and many diseases are considered a disturbance of homeostasis (homeostatic imbalance). In this context, allostasis is described as the process of achieving or re-establishing homeostasis through physiological change. While homeostasis emphasizes the resting condition and resistance to change, allostasis emphasizes change as a process to regain balance. In essence, allostasis is the resetting of the homeostat at a new set point.
Concepts like homeostasis, allostasis (through feedback control and regulation), and harmony (through coordination) are used to explain some form of functional stability in biological systems – the ability of a subsystem to retain its function when it is subjected to perturbations. The dynamics of (sub)systems any level of functional organization (reactions to tissue physiology) are integrated across levels of structural organization, from molecules and cells, to tissue and organ systems. A complex system, such as an organ is thus dynamic at different levels.
A more general and thus more appropriate principle to explain functional stability in organisms is homeodynamics, understood as a dynamic version of homeostasis across multiple levels of organization (e.g. from cell to tissue and organ system). Generally, a shift of focus from stasis to dynamics places the focus on adaptation, rather than stability. In epidemiology, these conceptual developments still await proper recognition [94].
Limitations of this discussion
The small number of publications on the specific topic of systems epidemiology might be seen as a limitation of this review. On the other hand, it is precisely because so little has been written that we felt the need to spread the word, and to provide our own perspective of what might contribute to advancing this field.
Perhaps the main limitation of this review is the paucity of proposed strategies that can be readily incorporated into public health research. We fully acknowledge that the proposed strategies are likely to need considerable improvement/refinement before they can be used to maximum benefit. Nevertheless, the combined “systems endeavor” discussed above holds the promise of helping to integrate pathogenetic and etiologic research elucidate illness causation mechanisms.
The epidemiologic literature in this field has been very rich and diverse [12,15,79,80,82,83,95,96], with a recent emphasis on probabilistic causation [9,78,97]. Despite progress, no agreed-upon criteria are available to help prove causation. The ongoing phenomenal successes in biomedicine despite this impossibility of causal proof suggests that going with strong correlations may be sufficient and that causal proof is not needed.
Causation can only be inferred, not observed [79]. Consequently, we have the widespread contention that observational studies cannot prove causation, but only demonstrate statistical associations. We agree with David Savitz, who suggests that ”the cliché that epidemiologic studies generate only measures of association, not causation is meaningless ... even experiments just generate measures of association as well”. [98] Perhaps part of the problem is the existence of multiple kinds of causation [99], and each might deserve its own taxonomy.
For the time being, we recommend acceptance of the currently rather successful counterfactual, probabilistic [12], and manipulationist (or interventionist) [100] causal model in clinical epidemiology, the randomized controlled trial (RCT). Such RCTs, designed to mimic a laboratory experiment, can test the hypothesis that a certain intervention does better than another one, and are clinically useful. Simulated interventions based on observational epidemiologic studies, together with observational data might offer a stronger rationale to conduct an RCT than observational data alone. In this way, a joint research program that includes both systems biology and systems epidemiology would be a rather powerful tool for causal inference.
Conclusion: Moving forward with “Systems Epidemiology”
Systems-type thinking about multiple levels of causation will allow us to better characterize the diverse range of factors influencing complex diseases. The application of complex systems methods in epidemiology is beginning to take shape. Computational models that incorporate human genomic, transcriptomic, proteomic, and metabolomic data integrated with global measurements from observational epidemiologic studies will allow epidemiologists to identify contributors to disease at multiple levels as well as their interactions. The key to success on this route will be 'integration', the integration of data from a wide range of sources, the integration of models from different levels of structural and functional organization, and the integration of expertise from different disciplines.
Acknowledgements
The authors acknowledge support from The National Institute of Neurological Disorders and Stroke (5U01NS040069-05 and 2R01NS040069 - 06A2), a grant from The National Eye Institute (1R01EY021820-01), and a center grant award from The National Institute of Child Health and Human Development (5P30HD018655-28).
References
- 1.Steiner A, Kendall FE, Mathers JA. 1952. The abnormal serum lipid pattern in patients with coronary arteriosclerosis. Circulation. 5(4), 605-08. 10.1161/01.CIR.5.4.605 [DOI] [PubMed] [Google Scholar]
- 2.Scarborough WR, Smith EW, Baker BM. 1960. Studies on subjects with and without coronary heart disease. Serum lipid, lipoprotein, and protein determinations and their relation to ballistocardiographic findings (a preliminary survey). Am Heart J. 59, 19-35. 10.1016/0002-8703(60)90381-1 [DOI] [PubMed] [Google Scholar]
- 3.Jolliffe N, Rinzler SH, Archer M. 1959. The anti-coronary club; including a discussion of the effects of a prudent diet on the serum cholesterol level of middleaged men. Am J Clin Nutr. 7, 451-62. [DOI] [PubMed] [Google Scholar]
- 4.Lieberman R, et al. 1998. Development of new cancer chemoprevention agents: role of pharmacokinetic/pharmacodynamic and intermediate endpoint biomarker monitoring. Clin Chem. 44(2), 420-27. [PubMed] [Google Scholar]
- 5.Nelson KB, et al. 1998. Neonatal cytokines and coagulation factors in children with cerebral palsy. Ann Neurol. 44(4), 665-75. 10.1002/ana.410440413 [DOI] [PubMed] [Google Scholar]
- 6.Craver CF. Explaining the brain. 2007, Oxford: Oxford University Press. [Google Scholar]
- 7.Bechtel W. 2011. Mechanism and biological explanation. Philos Sci. 78, 533-57 10.1086/661513 [DOI] [Google Scholar]
- 8.Wolkenhauer O. 2014. Why model? Front Phys. 5(21). 10.3389/fphys.2014.00021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Broadbent A. Inferring causation in epidemiology:mechanisms, black boxes, and contrasts, in Causality in the sciences, P.M. Illari, F. Russo, and J. Williamson, Editors. 2011, Oxford University Press. p. 45-69. [Google Scholar]
- 10.Bakulski KM, Fallin MD. 2014. Epigenetic epidemiology: promises for public health research. Environ Mol Mutagen. 55(3), 171-83. 10.1002/em.21850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kofink D, et al. 2013. Epigenetic dynamics in psychiatric disorders: environmental programming of neurodevelopmental processes. Neurosci Biobehav Rev. 37(5), 831-45. 10.1016/j.neubiorev.2013.03.020 [DOI] [PubMed] [Google Scholar]
- 12.Parascandola M, Weed DL. 2001. Causation in epidemiology. J Epidemiol Community Health. 55(12), 905-12. 10.1136/jech.55.12.905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ay A, Yildirim N. 2014. Dynamics matter: differences and similarities between alternatively designed mechanisms. Mol Biosyst. 10.1039/c4mb00159a [DOI] [PubMed] [Google Scholar]
- 14.Glymour C, Cheng PW. Causal Mechanism and Probability: A Normative Approach, D.o. Philosophy., Editor. 1998. [Google Scholar]
- 15.Rothman KJ. 1976. Causes. Am J Epidemiol. 104, 587-592.998606 [Google Scholar]
- 16.Wolkenhauer O. 2013. The role of theory and modeling in medical research. Front Phys. 4(377). 10.3389/fphys.2013.00377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gunawardena J. 2014. Models in biology: ‘accurate descriptions of our pathetic thinking’. BMC Biol. 12 (29). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ginsburg GS, Willard HF. 2009. Genomic and personalized medicine: foundations and applications. Transl Res. 154(6), 277-87. 10.1016/j.trsl.2009.09.005 [DOI] [PubMed] [Google Scholar]
- 19.Trochim WM, et al. 2006. Practical challenges of systems thinking and modeling in public health. Am J Public Health. 96(3), 538-46. 10.2105/AJPH.2005.066001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wolkenhauer O. 2001. Systems biology: the reincarnation of systems theory applied in biology? Brief Bioinform. 2(3), 258-70. 10.1093/bib/2.3.258 [DOI] [PubMed] [Google Scholar]
- 21.Ideker T, Galitski T, Hood L. 2001. A new approach to decoding life: systems biology. Annu Rev Genomics Hum Genet. 2, 343-72. 10.1146/annurev.genom.2.1.343 [DOI] [PubMed] [Google Scholar]
- 22.Auffray C, et al. 2003. From functional genomics to systems biology: concepts and practices. C R Biol. 326(10-11), 879-92. 10.1016/j.crvi.2003.09.033 [DOI] [PubMed] [Google Scholar]
- 23.Hood L. 2003. Systems biology: integrating technology, biology, and computation. Mech Ageing Dev. 124(1), 9-16. 10.1016/S0047-6374(02)00164-1 [DOI] [PubMed] [Google Scholar]
- 24.Kitano H. 2002. Computational systems biology. Nature. 420(6912), 206-10. 10.1038/nature01254 [DOI] [PubMed] [Google Scholar]
- 25.Savitz DA. 1994. In defense of black box epidemiology. Epidemiology. 5(5), 550-52. [PubMed] [Google Scholar]
- 26.El-Sayed AM, et al. 2013. Are Network-Based Interventions a Useful Antiobesity Strategy? An Application of Simulation Models for Causal Inference in Epidemiology. Am J Epidemiol. 10.1093/aje/kws455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pennello GA. 2013. Analytical and clinical evaluation of biomarkers assays: when are biomarkers ready for prime time? Clin Trials. 10(5), 666-76. 10.1177/1740774513497541 [DOI] [PubMed] [Google Scholar]
- 28.Dobbin KK. 2014. Statistical design and evaluation of biomarker studies. Methods Mol Biol. 1102, 667-77. 10.1007/978-1-62703-727-3_36 [DOI] [PubMed] [Google Scholar]
- 29.Leviton A, et al. 2013. Two-hit model of brain damage in the very preterm newborn: small for gestational age and postnatal systemic inflammation. Pediatr Res. 73(3), 362-70. 10.1038/pr.2012.188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Trewavas A. 2006. A brief history of systems biology. Plant Cell. 18(10), 2420-30. 10.1105/tpc.106.042267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hofkirchner, W. and M. Schafranek, General system theory. Philosophy of complex systems, 2011. 10: p. 177-194.
- 32.Laszlo A, Krippner S. 1998. Systems theories: Their origins, foundations, and development. Systems Theories and a Priori Aspects of Perception. 126, 47-74 10.1016/S0166-4115(98)80017-4 [DOI] [Google Scholar]
- 33.Bertalanffy Lv. General system theory; foundations, development, applications. 1969: p. xv, 289 p. [Google Scholar]
- 34.Albert JM, Nelson S. 2011. Generalized causal mediation analysis. Biometrics. 67(3), 1028-38. 10.1111/j.1541-0420.2010.01547.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Richiardi L, Bellocco R, Zugna D. 2013. Mediation analysis in epidemiology: methods, interpretation and bias. Int J Epidemiol. 42(5), 1511-19. 10.1093/ije/dyt127 [DOI] [PubMed] [Google Scholar]
- 36.Boeing H. 2013. Nutritional epidemiology: New perspectives for understanding the diet-disease relationship? Eur J Clin Nutr. 67(5), 424-29. 10.1038/ejcn.2013.47 [DOI] [PubMed] [Google Scholar]
- 37.Tohidi M, et al. 2013. Non-linear association between 25-hydroxyvitamin D and the incidence of type 2 diabetes: a community-based nested case-control study. Diabet Med. 30(8), 934-38. 10.1111/dme.12180 [DOI] [PubMed] [Google Scholar]
- 38.Zou L, et al. 2014. Non-linear dose-response relationship between cigarette smoking and pancreatic cancer risk: Evidence from a meta-analysis of 42 observational studies. Eur J Cancer. 10.1016/j.ejca.2013.08.014 [DOI] [PubMed] [Google Scholar]
- 39.Christakis NA, Fowler JH. 2007. The spread of obesity in a large social network over 32 years. N Engl J Med. 357(4), 370-79. 10.1056/NEJMsa066082 [DOI] [PubMed] [Google Scholar]
- 40.Huang TT, et al. 2009. A systems-oriented multilevel framework for addressing obesity in the 21st century. Prev Chronic Dis. 6(3), A82. [PMC free article] [PubMed] [Google Scholar]
- 41.Frerichs LM, Araz OM, Huang TT. 2013. Modeling social transmission dynamics of unhealthy behaviors for evaluating prevention and treatment interventions on childhood obesity. PLoS ONE. 8(12), e82887. 10.1371/journal.pone.0082887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wiley HS, Shvartsman SY, Lauffenburger DA. 2003. Computational modeling of the EGF-receptor system: a paradigm for systems biology. Trends Cell Biol. 13(1), 43-50. 10.1016/S0962-8924(02)00009-0 [DOI] [PubMed] [Google Scholar]
- 43.Oda K, et al. A comprehensive pathway map of epidermal growth factor receptor signaling. Mol Syst Biol, 2005. 1: p. 2005 0010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Oda K, Kitano H. A comprehensive map of the toll-like receptor signaling network. Mol Syst Biol, 2006. 2: p. 2006 0015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Peirce SM, Mac Gabhann F, Bautch VL. 2012. Integration of experimental and computational approaches to sprouting angiogenesis. Curr Opin Hematol. 19(3), 184-91. 10.1097/MOH.0b013e3283523ea6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Silverman EK, Loscalzo J. 2012. Network medicine approaches to the genetics of complex diseases. Discov Med. 14(75), 143-52. [PMC free article] [PubMed] [Google Scholar]
- 47.Auffray C, Chen Z, Hood L. 2009. Systems medicine: the future of medical genomics and healthcare. Genome Med. 1(1), 2. 10.1186/gm2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Federoff HJ, Gostin LO. 2009. Evolving from reductionism to holism: is there a future for systems medicine? JAMA. 302(9), 994-96. 10.1001/jama.2009.1264 [DOI] [PubMed] [Google Scholar]
- 49.Hood L, Flores M. 2012. A personal view on systems medicine and the emergence of proactive P4 medicine: predictive, preventive, personalized and participatory. New Biotechnol. 29(6), 613-24. 10.1016/j.nbt.2012.03.004 [DOI] [PubMed] [Google Scholar]
- 50.Wolkenhauer O, et al. 2013. The road from systems biology to systems medicine. Pediatr Res. 10.1038/pr.2013.4 [DOI] [PubMed] [Google Scholar]
- 51.Wolkenhauer O, Green S. 2013. The search for organizing principles as a cure against reductionism in systems medicine. FEBS J. 10.1111/febs.12311 [DOI] [PubMed] [Google Scholar]
- 52.Barabasi AL, Oltvai ZN. 2004. Network biology: understanding the cell's functional organization. Nat Rev Genet. 5(2), 101-13. 10.1038/nrg1272 [DOI] [PubMed] [Google Scholar]
- 53.Barabasi AL, Gulbahce N, Loscalzo J. 2011. Network medicine: a network-based approach to human disease. Nat Rev Genet. 12(1), 56-68. 10.1038/nrg2918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Last, J.M., A dictionary of epidemiology. 1995.
- 55.Taubes G. 1995. Epidemiology faces its limits. Science. 269(5221), 164-69. 10.1126/science.7618077 [DOI] [PubMed] [Google Scholar]
- 56.Hu FB. 2011. Metabolic profiling of diabetes: from black-box epidemiology to systems epidemiology. Clin Chem. 57(9), 1224-26. 10.1373/clinchem.2011.167056 [DOI] [PubMed] [Google Scholar]
- 57.Haring R, Wallaschofski H. 2012. Diving through the "-omics": the case for deep phenotyping and systems epidemiology. OMICS. 16(5), 231-34. 10.1089/omi.2011.0108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Thomas DC. 2005. The need for a systematic approach to complex pathways in molecular epidemiology. Cancer Epidemiol Biomarkers Prev. 14(3), 557-59. 10.1158/1055-9965.EPI-14-3-EDB [DOI] [PubMed] [Google Scholar]
- 59.Joffe M, et al. 2012. Causal diagrams in systems epidemiology. Emerg Themes Epidemiol. 9(1), 1. 10.1186/1742-7622-9-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Comas I, Gagneux S. 2011. A role for systems epidemiology in tuberculosis research. Trends Microbiol. 19(10), 492-500. 10.1016/j.tim.2011.07.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Fenner L, Egger M, Gagneux S. 2009. Annie Darwin's death, the evolution of tuberculosis and the need for systems epidemiology. Int J Epidemiol. 38(6), 1425-28. 10.1093/ije/dyp367 [DOI] [PubMed] [Google Scholar]
- 62.Lund E, Dumeaux V. 2008. Systems epidemiology in cancer. Cancer Epidemiol Biomarkers Prev. 17(11), 2954-57. 10.1158/1055-9965.EPI-08-0519 [DOI] [PubMed] [Google Scholar]
- 63.Cornelis, M.C. and F.B. Hu, Systems Epidemiology: A New Direction in Nutrition and Metabolic Disease Research. Curr Nutr Rep, 2013. 2(4). [DOI] [PMC free article] [PubMed]
- 64.Lussier YA, Liu Y. 2007. Computational approaches to phenotyping: high-throughput phenomics. Proc Am Thorac Soc. 4(1), 18-25. 10.1513/pats.200607-142JG [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Tracy RP. 2008. 'Deep phenotyping': characterizing populations in the era of genomics and systems biology. Curr Opin Lipidol. 19(2), 151-57. 10.1097/MOL.0b013e3282f73893 [DOI] [PubMed] [Google Scholar]
- 66.Skrabanek P. 1994. The emptiness of the black box. Epidemiology. 5(5), 553-55. [PubMed] [Google Scholar]
- 67.Greenland S, Gago-Dominguez M, Castelao JE. 2004. The value of risk-factor ("black-box") epidemiology. Epidemiology. 15(5), 529-35. 10.1097/01.ede.0000134867.12896.23 [DOI] [PubMed] [Google Scholar]
- 68.Hafeman DM, Schwartz S. 2009. Opening the Black Box: a motivation for the assessment of mediation. Int J Epidemiol. 38(3), 838-45. 10.1093/ije/dyn372 [DOI] [PubMed] [Google Scholar]
- 69.Schulte PA, Perera FP. Molecular Epidemiology. Principles and Practices. 1993, San Diego: Academic Press. [Google Scholar]
- 70.Wild CP. 2005. Complementing the genome with an "exposome": the outstanding challenge of environmental exposure measurement in molecular epidemiology. Cancer Epidemiol Biomarkers Prev. 14(8), 1847-50. 10.1158/1055-9965.EPI-05-0456 [DOI] [PubMed] [Google Scholar]
- 71.Merlo J, et al. 2005. A brief conceptual tutorial on multilevel analysis in social epidemiology: interpreting neighbourhood differences and the effect of neighbourhood characteristics on individual health. J Epidemiol Community Health. 59(12), 1022-28. 10.1136/jech.2004.028035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Merlo J, et al. 2005. A brief conceptual tutorial on multilevel analysis in social epidemiology: investigating contextual phenomena in different groups of people. J Epidemiol Community Health. 59(9), 729-36. 10.1136/jech.2004.023929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Merlo J, et al. 2005. A brief conceptual tutorial of multilevel analysis in social epidemiology: linking the statistical concept of clustering to the idea of contextual phenomenon. J Epidemiol Community Health. 59(6), 443-49. 10.1136/jech.2004.023473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Dammann O, Follett P. 2011. Toward multi-scale computational modeling in developmental disability research. Neuropediatrics. 42(3), 90-96. 10.1055/s-0031-1283157 [DOI] [PubMed] [Google Scholar]
- 75.Walpole J, Papin JA, Peirce SM. 2013. Multiscale Computational Models of Complex Biological Systems. Annu Rev Biomed Eng. 10.1146/annurev-bioeng-071811-150104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Tegner JN, et al. 2009. Computational disease modeling - fact or fiction? BMC Syst Biol. 3, 56. 10.1186/1752-0509-3-56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Lund E, Dumeaux V. 2010. Towards a more functional concept of causality in cancer research. Int J Mol Epidemiol Genet. 1(2), 124-33. [PMC free article] [PubMed] [Google Scholar]
- 78.Russo F, Williamson J. 2007. Interpreting Causality in the Health Sciences. Int Stud Philos Sci. 21(2), 157-70 10.1080/02698590701498084 [DOI] [Google Scholar]
- 79.Weed DL. 2008. Truth, epidemiology, and general causation. Brooklyn Law Rev. 73(3). [Google Scholar]
- 80.Galea S, Riddle M, Kaplan GA. 2010. Causal thinking and complex system approaches in epidemiology. Int J Epidemiol. 39(1), 97-106. 10.1093/ije/dyp296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kleinberg S, Hripcsak G. 2011. A review of causal inference for biomedical informatics. J Biomed Inform. 44(6), 1102-12. 10.1016/j.jbi.2011.07.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Hill AB. 1965. The environment and disease: association or causation? Proc R Soc Med. 58, 295-300. [PMC free article] [PubMed] [Google Scholar]
- 83.Susser, M.W., Causal thinking in the health sciences - Concepts and strategies of epidemiology. 1973.
- 84.Casini L, et al. 2011. Models for Prediction, Explanation and Control: Recursive Bayesian Networks. Theoria. 11(1), 5-33. [Google Scholar]
- 85.Ross R. 1905. The Logical Basis of the Sanitary Policy of Mosquito Reduction. Science. 22(570), 689-99. 10.1126/science.22.570.689 [DOI] [PubMed] [Google Scholar]
- 86.Hamer WH. 1906. The Milroy Lectures on epidemic disease in England - The evidence of variability and of persistence of type. Lancet. 167(4307), 733-39 10.1016/S0140-6736(01)80340-8 [DOI] [Google Scholar]
- 87.Kermack WO, McKendrick AG. A Contribution to the Mathematical Theory of Epidemics. Proceedings of the Royal Society of London A, 1927. 115. [Google Scholar]
- 88.Ulrich CM, Nijhout HF, Reed MC. 2006. Mathematical modeling: epidemiology meets systems biology. Cancer Epidemiol Biomarkers Prev. 15(5), 827-29. 10.1158/1055-9965.EPI-06-0252 [DOI] [PubMed] [Google Scholar]
- 89.Fabian MP, et al. 2012. The effects of indoor environmental exposures on pediatric asthma: a discrete event simulation model. Environ Health. 11, 66. 10.1186/1476-069X-11-66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Barhak J, et al. 2010. Chronic disease modeling and simulation software. J Biomed Inform. 43(5), 791-99. 10.1016/j.jbi.2010.06.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Sargent RG. Verification and validation of simulation models. Proceedings of the 2011 Winter Simulation Conference, 2011. [Google Scholar]
- 92.Epstein JM. 2008. Why Model? J Artif Soc Soc Simul. 11(4), 12. [Google Scholar]
- 93.Sieburg HB. 1990. Physiological studies in silico. Studies in the Sciences of Complexity. 12, 321-42. [Google Scholar]
- 94.Pearce N, Merletti F. 2006. Complexity, simplicity, and epidemiology. Int J Epidemiol. 35(3), 515-19. 10.1093/ije/dyi322 [DOI] [PubMed] [Google Scholar]
- 95.Maldonado G, Greenland S. 2002. Estimating causal effects. Int J Epidemiol. 31(2), 422-29. 10.1093/ije/31.2.422 [DOI] [PubMed] [Google Scholar]
- 96.Hernan MA. 2004. A definition of causal effect for epidemiological research. J Epidemiol Community Health. 58(4), 265-71. 10.1136/jech.2002.006361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Russo F. 2009. Variational causal claims in epidemiology. Perspect Biol Med. 52(4), 540-54. 10.1353/pbm.0.0118 [DOI] [PubMed] [Google Scholar]
- 98.Savitz DA. Interpreting epidemiologic evidence. 2003, Oxford: Oxford University Press. [Google Scholar]
- 99.Cartwright N. 2004. Causation: One word, many things. Philos Sci. 71, 805-19 10.1086/426771 [DOI] [Google Scholar]
- 100.Woodward, J., Making things happen - A theory of causal explanation. 2003.


