Abstract
The plant response to sulfur deficiency includes extensive metabolic changes which can be monitored at various levels (transcriptome, proteome, metabolome) even before the first visible symptoms of sulfur starvation appear. Four members of the plant-specific LSU (response to Low SUlfur) gene family occur in Arabidopsis thaliana (LSU1-4). Variable numbers of LSU genes occur in other plant species but they were studied only in Arabidopsis and tobacco. Three out of four of the Arabidopsis LSU genes are induced by sulfur deficiency. The LSU-like genes in tobacco were characterized as UP9 (UPregulated by sulfur deficit 9). LSU-like proteins do not have characteristic domains that provide clues to their function. Despite having only moderate primary sequence conservation they share several common features including small size, a coiled–coil secondary structure and short conserved motifs in specific positions. Although the precise function of LSU-like proteins is still unknown there is some evidence that members of the LSU family are involved in plant responses to environmental challenges, such as sulfur deficiency, and possibly in plant immune responses. Various bioinformatic approaches have identified LSU-like proteins as important hubs for integration of signals from environmental stimuli. In this paper we review a variety of published data on LSU gene expression, the properties of lsu mutants and features of LSU-like proteins in the hope of shedding some light on their possible role in plant metabolism.
Keywords: Arabidopsis, tobacco, coiled coil, SALK mutants, gene expression, OAS, ethylene, UP9
INTRODUCTION
The first global analyzes of gene expression profiles under sulfur deficiency stress in Arabidopsis appeared in Hirai et al. (2003); Maruyama-Nakashita et al. (2003) and Nikiforova et al. (2003), however, these studies focused on genes encoding proteins with known functions. Two years later LSU1 (At3g49580) and LSU2 (At5g24660) were identified as two out of 15 sulfur-responsive genes which were significantly up-regulated in roots as early as 2 h (LSU1) or 4 h (LSU2) after plants were transferred to sulfur-free medium; a sulfur-responsive element (SURE) was identified in their promoter regions (Maruyama-Nakashita et al., 2005). In the same year the At3g49580 gene appeared on the list of important network elements identified in a pioneering study involving reconstruction of the gene-metabolite network involved in the plant response to sulfur deficiency stress (Nikiforova et al., 2005). At the same time the tobacco UP9 gene was independently shown to be strongly and specifically up-regulated by sulfur deficiency (-S) using an unbiased suppression subtractive hybridization approach (Wawrzyńska et al., 2005). Since then rather few studies focusing on LSU-like genes and proteins have been published; however, several reports presented results of high throughput experiments which included also data related to the regulation of expression and phenotypes of the Arabidopsis lsu mutants. The systematic review of available data presented below provides clear evidence of the importance of this family of proteins and, hopefully contributes to uncovering their function.
LSU/UP9 GENES AND THEIR EXPRESSION
LSU GENES IN Arabidopsis
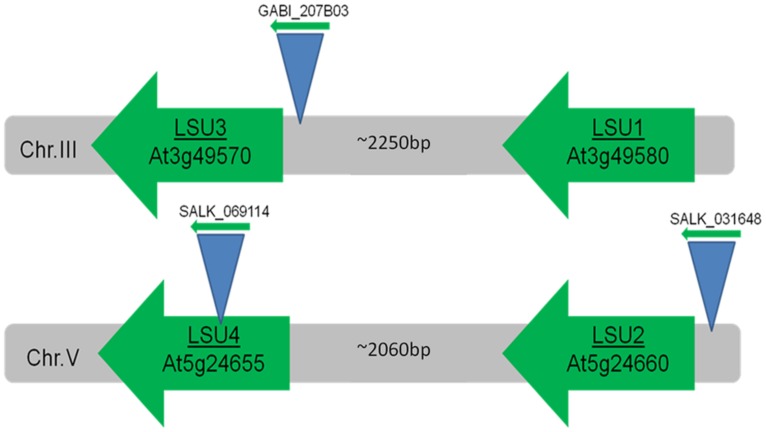
Arabidopsis thaliana contains four LSU genes (LSU1–LSU4) which are localized in pairs of direct repeats on two chromosomes (Figure 1). The nucleotide sequences of chromosome III corresponding to LSU1 and LSU3 transcripts are separated by about 2250 bp; the distance between LSU2 and LSU4 is slightly shorter (about 2060 bp). The open reading frames (ORFs) are relatively small and consist of about 280 bp. Most LSU genes have no introns; however, a spliced variant of LSU1 (At3g49580.2) encoding a protein with internal deletion of 19 amino acids was reported [http://www.arabidopsis.org]. Searches of publicly available microarrays using the Genvestigator platform (Zimmermann et al., 2004, 2008) showed that LSU1 and LSU2 are strongly expressed under -S but LSU4 appears not to be induced by -S. LSU3 was not included in these microarrays. Expression of LSU1 and LSU2 is induced not only by -S but also by other stressful environmental conditions such as salt stress and AgNO3 treatment.
FIGURE 1.
Localization of ‘response to Low Sulfur’ (LSU) genes in the Arabidopsis genome. Number of base pairs (bp) between LSU open reading frames is indicated. Positions of T-DNA inserts are shown schematically.
Several global analyzes of Arabidopsis gene expression in various growth conditions and developmental stages provide valuable information about expression of LSUs. Most of these data relate to LSU2, suggesting that this member of the family is preferentially involved in the plant response to certain stresses or certain processes. Expression of LSU2 is induced during oxidative stress (Davletova et al., 2005) and at the beginning of an extended night, which may indicate that it is induced by carbon starvation and in response to sugar (Usadel et al., 2008). Recently it has been shown that expression of LSU2 is induced by a combination of light and plastid signaling (Ruckle et al., 2012); these authors identified LSU2 as one of seven so-called END (enhanced de-etiolation) genes. They went on to characterize some of the end mutants, including lsu2 (SALK_031648), and showed that expression of some photosynthesis-related genes (Lhcb1.4, PsbS, RbcS1, and CHS) was attenuated in them. The mechanisms responsible for the regulation of transcription in end mutants remain unclear; the expression of END genes is regulated by a variety of signals besides light and plastid signals, so it may be different for different mutants. Ruckle et al. (2012) concluded that the products of END genes contribute to a complex network responsible for optimization of chloroplast function during chloroplast biogenesis, and probably during periods of chloroplast dysfunction. The link between LSU2 and chloroplasts was also emphasized in a recent report, where LSU2 was identified as one of 39 genes that were differentially expressed in six independent microarray experiments using plants with the provoked retrograde signaling in response to disturbances of chloroplast performance by chemical treatment or mutation of some metabolic pathways (Glasser et al., 2014).
In addition LSU2 was tentatively identified as one of the genes involved in the crosstalk between several signals (nitrate, sulfur, iron, and hormones) from analysis of transcriptome data for Arabidopsis plants grown under sulfur and iron restriction, and various nitrate and stress hormone treatments (Omranian et al., 2012).
Somewhat surprisingly expression of LSU1 was found to be repressed during infection with cabbage leaf curl virus (CaLCuV), whereas LSU2 expression was apparently unaffected (Ascencio-Ibanez et al., 2008). The LSU1 gene was also shown to be constitutively (phase-independently) expressed during pollen germination and tube growth (Wang et al., 2008).
Analysis of publicly available data from two sets of high-throughput experiments led to the identification of LSU1 as a member of a six-gene cluster responding to O-acetylserine (OAS) levels in shoots (Hubberten et al., 2012b). One set of data was from experiments on diurnal oscillations of genes and metabolites (Espinoza et al., 2010); the second set was from studies of plants during the light-dark transition (Caldana et al., 2011). OAS was one of the compounds most affected by changes in conditions in both studies. Hubberten et al. (2012b) confirmed that regardless of temperature (20 or 4°C), the level of OAS (and the expression of the above-mentioned genes) increased during the night and decreased during the day. Increased expression of LSU1 (and the other five genes) was also observed following induction of the chemically inducible ectopic copy of SERAT (encodes serine acetyltransferase, which is involved in OAS synthesis) in sulfur-sufficient transgenic plants (Hubberten et al., 2012b). The same group used a split-root approach to explore further the role of OAS in the regulation of plant S-status in Arabidopsis. One half of the root was exposed to -S, whilst the other half of the root of the same plant was grown in sulfur-sufficient conditions. OAS levels were low in both halves of the split root, and expression of previously mentioned OAS-responsive genes, including LSU1 was also low (Hubberten et al., 2012a).
It has recently been reported that expression of LSU1 (and BGLU28 [At2g44460], SDI1 [At5g48850] and SULTR4;2 [At3g12520]) is much less affected by S availability in the sultr1;2 mutants than in the wild type (Zhang et al., 2014). This observation is not strictly related to the function of LSU/UP9 proteins, nevertheless it is worth noting because it makes an important contribution to understanding the plant mechanisms responsible for sensing S availability and thus also S status-dependent regulation of gene expression.
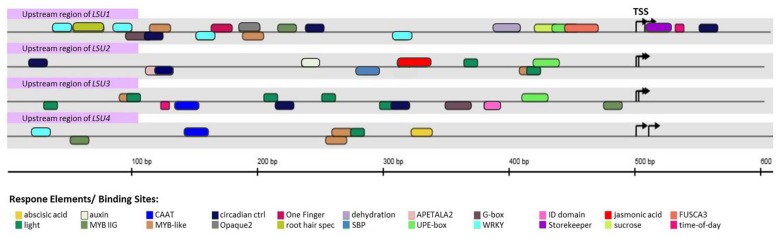
The results of in silico analysis of the promoter regions of the LSUs are shown in Figure 2. Analyzes of the 500 bp upstream transcription start site (TSS) demonstrate the potential for differential expression of each LSU gene. In all but the LSU4 promoter, there is an element specific for induction in -S, UPE-box (Wawrzyńska et al., 2010). Additional sulfur-responsive elements (SURE boxes) which are not included in Figure 2 have previously been identified in the promoter regions of LSU1 and LSU2 (Maruyama-Nakashita et al., 2005). The LSU1 promoter contains the largest number of potential regulatory elements. Only LSU1 has consensuses for FUSCA3 and OPAQUE2-like factors, both of which are essential for seed-specific expression (Moreno-Risueno et al., 2008) and the cis-elements related to response to dehydration and sucrose. The binding site for the bZIP transcription factors (TFs; G-box) is present in the LSU1 and LSU3 promoters, whilst the consensus for binding the WRKY TFs is present in LSU1 and LSU4. The promoter regions of LSU2, LSU3, and LSU4 (but not LSU1) have sequences for binding the light-responsive factors. The LSU2 promoter contains sequences responsive to auxins and jasmonic acid as well as sequences for APETALA2 and SQUAMOSA promoter-binding protein (SBP), indicating that LSU2 may play an important role in ontogenesis. The LSU3 promoter contains a specific sequence which binds the INDETERMINATE1 (ID domain) responsible for the transition to flowering; the LSU4 promoter has a cis-acting sequence responsive to abscisic acid (ABA). The putative roles of these elements in the regulation of LSU gene expression should be verified experimentally.
FIGURE 2.
Depiction of transcription factor (TF) binding sites found in the upstream regions of LSU1-4. The sequences were scanned for matches to TFs binding sites using MatInspector, part of the Genomatix Software Suite (www.genomatix.de). A match is represented by a round-ended rectangle. Matches with the positive and negative strands are depicted above or below the sequence line, respectively. The arrow symbol on the sequence stands for a transcription start site (TSS); note that there are several putative TSSs for each LSU.
UP9 GENES IN TOBACCO
The tobacco LSU-like proteins were grouped into six clusters (UP9A to UP9F); however, the exact number of such genes in tobacco remains unclear (Lewandowska et al., 2010). Only one of these genes, UP9C, has been investigated further. An increase in the level of the UP9C transcript was observed just 2 days after transferring plants from sulfur-sufficient to sulfur-deficient medium in all parts tested (roots, young leaves, mature leaves, stems) and a further increase in transcript level was observed after additional days under -S. Analysis of the promoter region of UP9C indicated that it has only one TSS located 109 bp upstream of the translational start site (Wawrzyńska et al., 2010). The same study also reported the presence of an interesting motif, UPE-box, in the UP9C promoter. The authors used the DNA fragment containing UPE-box (from the promoter region of UP9C) in a yeast-one-hybrid experiment and identified NtEIL2, a tobacco member of the EIL family, as a TF which bound to the UPE-box (Wawrzyńska et al., 2010). Transient expression assays in Nicotiana benthamiana plants indicated that NtEIL2 was responsible for the UPE-box-dependent up-regulation of the reporter gene in -S conditions. Interestingly, an Arabidopsis homolog of NtEIL2, SLIM1, which has been identified earlier as a critical transcriptional regulator of plant sulfur response and sulfur metabolism (Maruyama-Nakashita et al., 2006), was also able to bind to UP9C promoter containing UPE-box. Mutations in UPE-box affect the binding of both factors, NtEIL2 and AtSLIM1; however, in the presence of SLIM1 the promoter was constitutively active, regardless of the plants’ sulfur status (Wawrzyńska et al., 2010). In conclusion, UP9C seems to be regulated directly by NtEIL2, in a sulfur-dependent manner. Some as yet unidentified species-specific factors guarantee the specificity of the NtEIL2-dependent up-regulation of the UP9C gene (and possibly other genes containing UPE-box) in -S conditions. Further in silico analysis of the promoter region showed that the UP9C promoter has elements which are potentially responsive to light, salt stress and phytohormones such as ABA, ethylene and cytokines as well as the above-mentioned SURE located 350 bp upstream of the start codon. The biological significance of these cis-factors is unknown.
UPE-box is also present in the promoters of several Arabidopsis genes (Wawrzyńska et al., 2010). A search of the genome sequence revealed that it was present in the promoter regions of LSU1 (At3g49580), LSU2 (At5g24660), and LSU3 (At3g49570; but not LSU4) and also in several other genes which are up-regulated in -S. Interestingly the set of genes containing UPE-box in promoter appears to be very similar to the OAS cluster genes (Hubberten et al., 2012b).
PHENOTYPES OF THE MUTANTS
ANALYSIS OF Arabidopsis SALK MUTANTS
One of the difficulties in determining the function of proteins from the LSU family is that information about the phenotypes of knock-out (KO) and knock-down (KD) mutants is scarce. There are T-DNA insertional mutants for LSU2 (e.g., SALK_31648, SALK_070105), LSU3 (e.g., GABI_207B03), and LSU4 (e.g., SALK_069114) but not for LSU1 (Figure 1). The high probability of functional overlap makes it desirable to test multiple lsu KO or KD mutants, but so far no data have been published. Most available data relate to lsu2 mutants, for example an interesting report on the functional characterization of abiotic stress response proteins with unknown function was published recently (Luhua et al., 2013). These authors tested the response to treatments such as salinity, oxidative, osmotic, heat, cold, and hypoxia stress of 1007 T-DNA insertional mutants in genes with unknown function. The lsu2 mutant (SALK_31648C) was one of 69 genes with an unknown function that seemed to be more tolerant of osmotic stress than the wild type; responses to other stresses did not appear to be altered. Another study reported that lsu2 mutants (SALK_031648, SALK_070105) exhibited enhanced susceptibility to two evolutionarily distinct pathogens, Pseudomonas syringae and Hyaloperonospora arabidopsidis (Mukhtar et al., 2011). According to the authors, LSU2 (and other proteins, for example JAZ3) has some effect on the functioning of the NB-LRR (nucleotide binding leucine-rich repeat) intracellular immune receptors with particular emphasis on the RPS2 (Resistance to Pseudomonas syringae 2) protein. Activation of NB-LRR proteins is responsible for robust disease-resistance responses such as host cell death and systemic defense signaling.
Defects in flower and inflorescence development were observed in the insertion mutant lsu4 (SALK_069114) when grown under short-day conditions (Myakushina et al., 2009). Mutation of the LSU4 gene caused delayed flowering and disturbances in the formation of flower organs. There were also significant changes in the expression of many regulatory genes, including down-regulation of LEAFY (LFY), APETALA1 (AP1), APETALA3 (AP3), PISTILLATA (PI) and SEPALLATA3 (SEP3) and up-regulation of APETALA2 (AP2), AGAMOUS (AG) and SEPALLATA (SEP2). It is worth mentioning that the authors noted that LSU4 expression increased two to threefold under phosphorus, nitrogen, potassium, or iron deficiency.
SILENCING OF UP9 IN TOBACCO
Analysis of the tobacco antisense UP9C transformants (KD) revealed no evidence of phenotypic differences from the wild type, although the KD transformants did have a different metabolite profile from wild type plants (Lewandowska et al., 2010). The metabolite profiles of KDs grown in -S were more similar to the profiles of parental line plants grown in sulfur-sufficient conditions, suggesting that the KD lines failed to adjust their metabolism to the -S conditions. In addition the level of non-protein thiols (consisting mostly of glutathione) in mature leaves and roots, but not in young leaves, was different in KD plants. Wild type plants showed the expected reduction in glutathione levels in mature leaves 2 days after transfer to -S, but there was no change in the KDs, which had a high level of glutathione in the mature leaves regardless of the conditions. The mutants did, however, have low levels of glutathione in the roots, particularly under -S; mutants also had lower levels of sulfur in the roots under -S than the wild type. Another interesting observation was that under -S several genes were misregulated in the mutants; usually the level of transcription was lower in the KDs than the wild type. It must be remembered, however, that only a limited selection of genes was tested and no high-throughput analysis was performed in this study.
Ethylene plays a very important role in plant response to several stresses and regulates many processes (Adie et al., 2007; Lin et al., 2009). In -S conditions ethylene levels increase in wild type tobacco. KD plants have lower levels of ethylene than wild type plants in -S conditions (Moniuszko et al., 2013), but in sulfur-sufficient conditions the UP9C-silenced line produced slightly more ethylene than the wild type. Transcriptome analysis revealed significant changes in the gene expression pattern of the KD line relative to the wild type; only 130 of the 360 genes up-regulated in the wild type in -S were also up-regulated in the mutants and only 14 of 91 genes down-regulated in the wild type were also down-regulated in the mutant. Some genes were regulated in the mutant but not in the wild type. Differences in the expression profiles of the mutants and wild type may provide clues to function. Gene ontology (GO) analysis indicated clearly that UP9C does not participate in sulfur deficiency-dependent regulation of genes encoding isoforms of APS reductase (APR) or genes encoding S-adenosylmethionine synthase (SAMS) as these genes were induced in -S in both the mutant and the wild type. Genes from several categories including ‘response to hormone stimulus,’ ‘signal transduction,’ ‘defense response’ and ‘regulation of transcription’ genes were, however, misregulated in the mutant. Although many genes had different expression profiles in the KD several genes related to ethylene signaling (homologs of Arabidopsis EIN3-BINDING F BOX PROTEIN 1 (EBF1), ETHYLENE INSENSITIVE 4 (EIN4) and ETHYLENE RESPONSE SENSOR 1 (ERS1)) and ABA- and cytokine-mediated signaling (homologs of ARABIDOPSIS THALIANA HOMEOBOX 7 (ATHB-7) and HISTIDINE-CONTAINING PHOSPHOTRANSMITTER 1 (AHP1)) attracted particular attention (Moniuszko et al., 2013). The expression of these genes was slightly higher in the KD than in the wild type in sulfur-sufficient medium, but the most interesting effect was the very low expression of these genes in the KD line when plants were transferred to -S conditions. In Arabidopsis EBF1 is important for proteosomal degradation of ETHYLENE-INSENSITIVE3 (EIN3), the positive regulator of ethylene-responsive genes, whilst EIN4 and ERS1 are genes for ethylene receptors (Wang et al., 2006). These observations, along with the reduced ethylene level in the mutant grown in -S, prompted the authors to hypothesize that UP9C is involved in modulation of the ethylene signaling pathway, which is important in plant response to -S conditions. The main conclusion to be drawn from this work is that one of the functions of UP9C - and possibly also other LSU-like proteins - in plant response to -S may be related to the involvement of LSU-like proteins in tuning up ‘hormone stimulus’ signals induced by -S conditions. Although the authors focused on ethylene it is likely that other hormone signaling systems, possibly those involved in -S response, are also affected in the mutant.
LSU-LIKE PROTEINS AND THEIR POTENTIAL INTERACTING PARTNERS
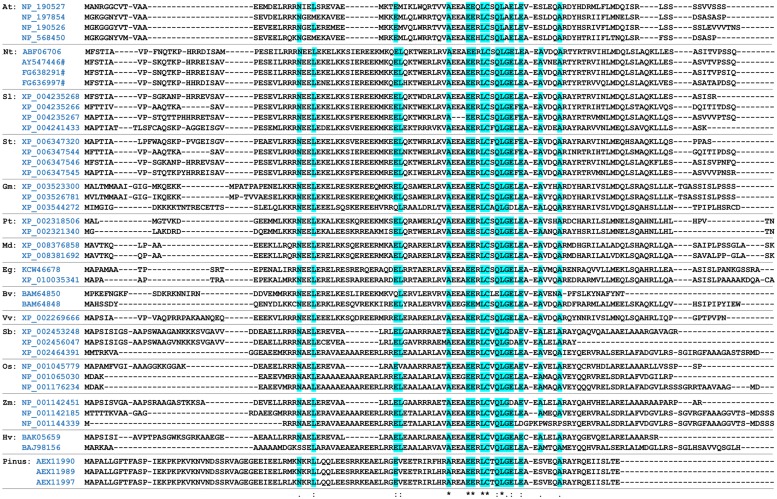
LSU/UP9 family proteins are small (10–13 kDa) and consist of about 100 amino acids (Figure 3). A BLAST (blastp) search of non-redundant protein sequences revealed multiple homologs of LSU in various plant species, both monocotyledons and dicotyledons, including Solanum lycopersicum (4 homologs), Solanum tuberosum (4), Glycine max (3), Populus trichocarpa (3), Zea mays (3), Hordeum vulgare (2), Oryza sativa (3), Beta vulgaris (2) and many others. The LSU-like proteins are also present in gymnosperms, like Pinus sp. We believe that so far only Arabidopsis LSUs and tobacco UP9s have been analyzed. Computer analysis and the circular dichroism spectra indicated that UP9C has an alpha-helical structure (Lewandowska et al., 2010). The presence of two stranded coiled–coil regions in UP9C (Lewandowska et al., 2010) is strongly suggestive of multimer formation; UP9C-UP9C interactions were observed in yeast two-hybrid (Y2H) experiments. Interestingly, despite relatively weak conservation of the primary sequence, both homologous UP9C-UP9C and heterologous LSU-UP9C (cross-species) interactions were observed. A potential nuclear localization signal was found in UP9C using the MOTIFSCAN program; according to PSORT UP9C has a cytosol-nuclear localization. No nuclear localization motifs have been identified in Arabidopsis LSU proteins. Nuclear localization of UP9C was reported (Lewandowska et al., 2010), but more recent experimental data suggest that it is present in both cytoplasm and nucleus (Moniuszko et al., 2013). Because they are small proteins it is likely that LSU-like proteins can cross the nuclear pores without a specific transport mechanism. There are no specific motifs or domains in LSU/UP9 proteins that suggest their function. The significance of the short, strongly evolutionarily conserved region in the members of this family (Figure 3) is unknown.
FIGURE 3.
Alignment of the selected LSU-like proteins. The evolutionary conserved amino acids identified by the MAFT alignment software [http://mafft.cbrc.jp] are highlighted. The accession numbers of the protein sequences are provided. # denotes the accession number to the corresponding nucleotide sequence; At, Arabidopsis thaliana; Nt, Nicotiana tabacum; Sl, Solanum lycopersicum; St, Solanum tuberosum; Gm, Glycine max; Pt, Populus trichocarpa; Md, Malus domesticus; Eg, Eucalyptus grandis; Bv, Beta vulgaris; Vv, Vitis vinifera; Sb, Sorghum bicolor; Os, Oryza sativa; Zm, Zea mays; Hv, Hordeum vulgare; Pinus, Pinus taeda.
The LSU/UP9 proteins seem to be involved in multiple protein–protein contacts (Table 1; Figure 4). Data from tobacco are limited; however, some of the interacting partners identified by the Y2H approach have been confirmed using other methods. For example, UP9C interacts with ACO2A, an enzyme which converts 1-aminocyclopropane-1-carboxylate (ACC) to ethylene; it was therefore proposed that ethylene production might be controlled by UP9C through its interactions with ACO2A (Moniuszko et al., 2013). Joka2/NBR1 functions as a cargo receptor in selective autophagy (Zientara-Rytter et al., 2011) and is another protein which is unquestionably involved in interactions with UP9/LSU; however, the biological significance of these interactions is as yet unexplained.
Table 1.
Tobacco proteins found as interacting with tobacco UP9C.
| Accession number | Clone name | Number of amino acids in the clone | Identification/Function | Corresponding A. thaliana gene |
|---|---|---|---|---|
| Library from Nicotiana plumbaginifolia seedlings grown in S-sufficient conditions | ||||
| ABF06703 | NpJoka2 | 467 | NBR1-like, cargo receptor of selective autophagy | At4g24690 |
| ABF06705 | NpJoka8 | 360 | HLH superfamily; bHLH66 | At2g24260 |
| ABF06704 | NpJoka20 | 161 | Ribosomal L7/L12 | At3g27850 At4g36420 At4g37660 |
| Library from 3-month-old Nicotiana tabacum plants transfered for 2 days into S-deficient conditions | ||||
| GU066878 | Joka 31A | 117 | ACC oxidase | At1g05010 |
| GU066879 | Joka 31B | 56 | ACC oxidase | At1g05010 |
| GU066880 | Joka 32 | 253 | PRP11; ZnF-U1 – splicing factor | At2g32600 |
| GU066881 | Joka 33 | 245 | TIM50 (mt-inner membrane) | At1g55900 |
| GU066882 | Joka 34 | 376 | RING-finger-containing E3 ubiquitin ligase | At3g58030 |
| GU066883 | Joka 35 | 147 | RING-finger-containing E3 ubiquitin ligase | At3g16720 |
| GU066884 | Joka 36 | 314 | Apetala 2-like (tran At4g36920 At5g67180 At5g60120 |
|
| GU066885 | Joka 37 | 110 | Function unknown; Involucin repeat; phosphoenolopyruvate carboxylase; E2-enzyme | At2g28540 At3g55720 |
| GU066886 | Joka 38 | 144 | DUF248/methyltransferase | At4g18030 At1g26850 |
| GU066887 | Joka 39 | 119 | DUF632/Function unknown, leucine zipper | At2g27090 |
| GU066888 | Joka 40 | 515 | Function unknown, nucleoporin-like | At4g37130 |
| GU066889 | Joka 41 | 99 | Poly A binding | At1g49760 At4g34110 At2g23350 At1g22760 At1g71770 |
| GU066890 | Joka 42 | 77 | FtsH protease | At2g26140 |
| GU066891 | Joka 43 | 128 | Unknown | At3g24506 At2g17240 |
| GU066892 | Joka 44 | 75 | Microtubule-associated MAP65-1a | At5g55230 At4g26760 |
| GU066893 | Joka 46 | 184 | CHORD, PBS2, RAR1, interacts with SGT1; Rar1/TMV resistance | At5g51700 |
| GU066894 | Joka 47 | 200 | JAZ1 (transcription factor) | At1g19180 |
FIGURE 4.

Venn diagram of potential LSU1 and LSU2 interacting partners (Arabidopsis Interactome Mapping Consortium, 2011).
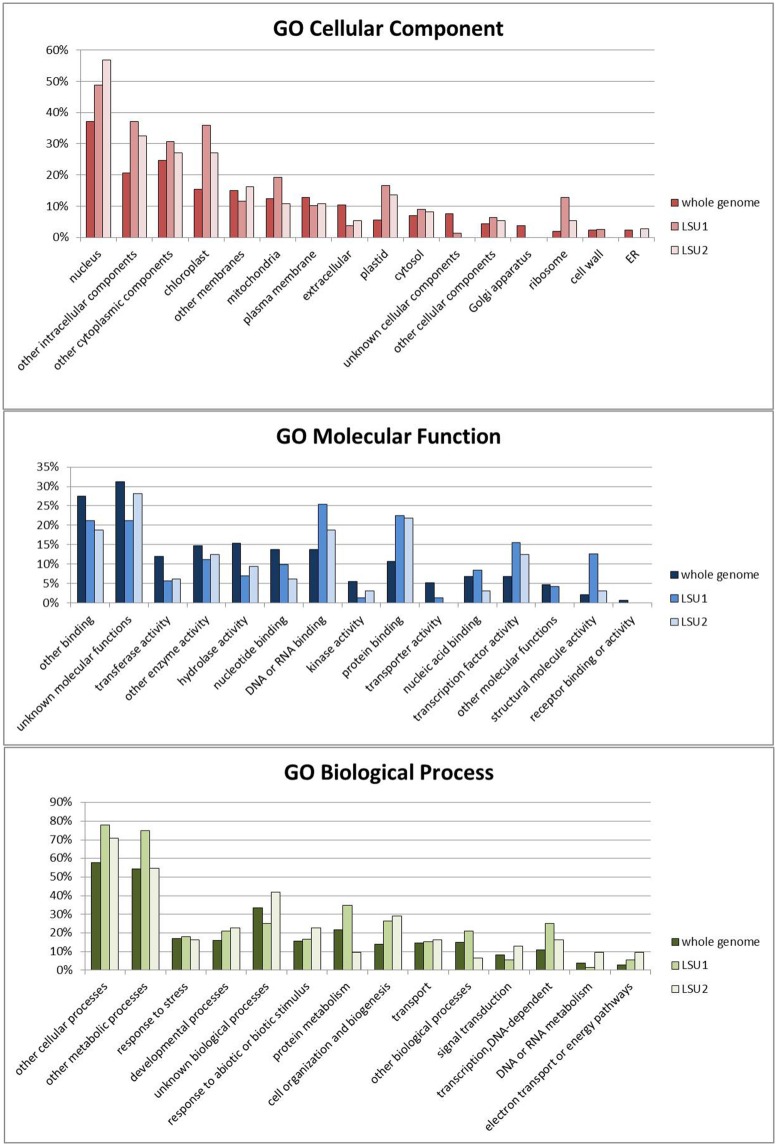
Mapping of the Arabidopsis interactome based on the Y2H system (Arabidopsis Interactome Mapping Consortium, 2011) has revealed numerous partners of LSU1 and LSU2; unfortunately LSU3 and LSU4 were not included in the experiments. The lists of proteins which potentially interact with LSU1 or LSU2 are quite long (80 and 37 proteins, respectively) and include 17 elements common to both proteins (Figure 4). Functional categorization of potential interacting partners using GO analysis indicated some changes in the distribution of gene product locations, molecular functions and biological processes relative to those for the genome as a whole (Figure 5). Both groups (LSU1 and LSU2 interacting partners) were more likely than average to be located in the nucleus, chloroplasts (plastids) or ribosomes. Nuclear proteins which are LSU1 or LSU2 interacting partners include members of the JAZ family of repressors. It is worth noting that it has been demonstrated that the tobacco homolog of JAZ interacts with UP9C (Table 1). Molecular Functions GO categories such as ‘DNA or RNA binding,’ ‘protein binding’ and ‘TF activity’ are over-represented among LSU1 and LSU2 interacting partners, whereas categories related to some enzymatic activities are under-represented.
FIGURE 5.
Functional categorization of the potential LSU1 and LSU2 interacting partners for GO Cellular Component, GO Molecular Function and GO Biological Process. The analysis was done using the Gene ontology tools available at TAIR [http://www.arabidopsis.org].
We have also noticed that the group of LSU1 interacting partners includes a relatively high proportion of proteins from the Molecular Function GO category ‘structural molecule activity’ (all are ribosomal proteins). Overrepresentation of any Biological Process GO category was less apparent; ‘cell organization and biogenesis’ and ‘DNA-dependent transcription’ and perhaps the ‘protein metabolism’ and ‘transcription, DNA-dependent’ categories were only slightly overrepresented among LSU1 partners. The category of ‘cell organization and biogenesis’ proteins which interact with LSU1 includes some ribosomal proteins, chaperones and members of RING superfamily (potential E3 ubiquitin ligases). LSU1 partners include, amongst others, members of the ERF/AP2, bHLH and myb-like HTH families of transcriptional factors.
It has also been demonstrated that LSU2 protein interacts with the pathogenic effectors of two different plant pathogens, the bacterium P. syringae and the oocyte Hyaloperonospora arabidopsidis (Mukhtar et al., 2011). The involvement of LSU2 in the immune response to these pathogens was verified by the same authors through the demonstration of enhanced susceptibility in lsu2 mutants (see also above).
CONCLUDING REMARKS
It is unclear why plants have several isoforms of LSU. The proteins have probably partially overlapping functions; however, the data reported above suggest some functional specificity. LSU1, LSU2, and LSU3 genes from Arabidopsis are induced by sulfur deficiency; however, only LSU2 has been shown to be involved in retrograde signaling associated with chloroplast malfunction. The molecular role of LSU-like family members remains unclear although an increasing amount of evidence links the family with complex intracellular regulatory functions and coordination of organellar and cytosolic metabolism. It is possible that LSU/UP9 proteins modulate degradation of some specific “strategic” targets (such as TFs) in response to environmental stresses or are (directly or indirectly) involved in regulation of cellular degradation machinery. Although there is no clear evidence that LSU-like family members play such roles their interactions with presumed E3 ubiquitin ligases, chaperons (DnaJ-domain, Hsp60) and particularly with NBR1 (a selective autophagy cargo receptor) make the hypothesis plausible.
AUTHOR CONTRIBUTIONS
Agnieszka Sirko and Anna Wawrzynska drafted the manuscript. Milagros Collados Rodríguez and Pawel, Skektas contributed to the writing process and preparation of figures. All authors were involved in preparing the final version.
Conflict of Interest Statement
The Associate Editor Stanislav Kopriva declares that, despite having edited a Research Topic with the author Agnieszka Sirko, the review process was handled objectively and no conflict of interest exists. The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
Research in our laboratory is supported by grants from the Polish Ministry of Science and Higher Education (W16/7.PR/2011), the Polish National Science Centre (2011/03/B/NZ1/01243) and EC FP7-PEOPLE (BIONUT-ITN; project ref.: 264296).
REFERENCES
- Adie B., Chico J. M., Rubio-Somoza I., Solano R. (2007). Modulation of plant defenses by ethylene. J. Plant Growth Regul. 26 160–177 10.1007/s00344-007-0012-6 [DOI] [Google Scholar]
- Arabidopsis Interactome Mapping Consortium. (2011). Evidence for network evolution in an Arabidopsis interactome map. Science 333 601–607 10.1126/science.1203877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ascencio-Ibanez J. T., Sozzani R., Lee T. J., Chu T. M., Wolfinger R. D., Cella R., et al. (2008). Global analysis of Arabidopsis gene expression uncovers a complex array of changes impacting pathogen response and cell cycle during geminivirus infection. Plant Physiol. 148 436–454 10.1104/pp.108.121038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caldana C., Degenkolbe T., Cuadros-Inostroza A., Klie S., Sulpice R., Leisse A., et al. (2011). High-density kinetic analysis of the metabolomic and transcriptomic response of Arabidopsis to eight environmental conditions. Plant J. 67 869–884 10.1111/j.1365-313X.2011.04640.x [DOI] [PubMed] [Google Scholar]
- Davletova S., Schlauch K., Coutu J., Mittler R. (2005). The zinc-finger protein Zat12 plays a central role in reactive oxygen and abiotic stress signaling in Arabidopsis. Plant Physiol. 139 847–856 10.1104/pp.105.068254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espinoza C., Degenkolbe T., Caldana C., Zuther E., Leisse A., Willmitzer L., et al. (2010). Interaction with diurnal and circadian regulation results in dynamic metabolic and transcriptional changes during cold acclimation in Arabidopsis. PLoS ONE 5:e14101 10.1371/journal.pone.0014101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glasser C., Haberer G., Finkemeier I., Pfannschmidt T., Kleine T., Leister D., et al. (2014). Meta-analysis of retrograde signaling in Arabidopsis thaliana reveals a core module of genes embedded in complex cellular signaling networks. Mol. Plant 7 1167–1190 10.1093/mp/ssu042 [DOI] [PubMed] [Google Scholar]
- Hirai M. Y., Fujiwara T., Awazuhara M., Kimura T., Noji M., Saito K. (2003). Global expression profiling of sulfur-starved Arabidopsis by DNA macroarray reveals the role of O-acetyl-l-serine as a general regulator of gene expression in response to sulfur nutrition. Plant J. 33 651–663 10.1046/j.1365-313X.2003.01658.x [DOI] [PubMed] [Google Scholar]
- Hubberten H. M., Drozd A., Tran B. V., Hesse H., Hoefgen R. (2012a). Local and systemic regulation of sulfur homeostasis in roots of Arabidopsis thaliana. Plant J. 72 625–635 10.1111/j.1365-313X.2012.05105.x [DOI] [PubMed] [Google Scholar]
- Hubberten H. M., Klie S., Caldana C., Degenkolbe T., Willmitzer L., Hoefgen R. (2012b). Additional role of O-acetylserine as a sulfur status-independent regulator during plant growth. Plant J. 70 666–677 10.1111/j.1365-313X.2012.04905.x [DOI] [PubMed] [Google Scholar]
- Lewandowska M., Wawrzynska A., Moniuszko G., Lukomska J., Zientara K., Piecho M., et al. (2010). A contribution to identification of novel regulators of plant response to sulfur deficiency: characteristics of a tobacco gene UP9C, its protein product and the effects of UP9C silencing. Mol. Plant 3 347–360 10.1093/mp/ssq007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Z., Zhong S., Grierson D. (2009). Recent advances in ethylene research. J. Exp. Bot. 60 3311–3336 10.1093/jxb/erp204 [DOI] [PubMed] [Google Scholar]
- Luhua S., Hegie A., Suzuki N., Shulaev E., Luo X., Cenariu D., et al. (2013). Linking genes of unknown function with abiotic stress responses by high-throughput phenotype screening. Physiol. Plant. 148 322–333 10.1111/ppl.12013 [DOI] [PubMed] [Google Scholar]
- Maruyama-Nakashita A., Inoue E., Watanabe-Takahashi A., Yamaya T., Takahashi H. (2003). Transcriptome profiling of sulfur-responsive genes in Arabidopsis reveals global effects of sulfur nutrition on multiple metabolic pathways. Plant Physiol. 132 597–605 10.1104/pp.102.019802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maruyama-Nakashita A., Nakamura Y., Tohge T., Saito K., Takahashi H. (2006). Arabidopsis SLIM1 is a central transcriptional regulator of plant sulfur response and metabolism. Plant Cell 18 3235–3251 10.1105/tpc.106.046458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maruyama-Nakashita A., Nakamura Y., Watanabe-Takahashi A., Inoue E., Yamaya T., Takahashi H. (2005). Identification of a novel cis-acting element conferring sulfur deficiency response in Arabidopsis roots. Plant J. 42 305–314 10.1111/j.1365-313X.2005.02363.x [DOI] [PubMed] [Google Scholar]
- Moniuszko G., Skoneczny M., Zientara-Rytter K., Wawrzynska A., Glow D., Cristescu S. M., et al. (2013). Tobacco LSU-like protein couples sulphur-deficiency response with ethylene signalling pathway. J. Exp. Bot. 64 5173–5182 10.1093/jxb/ert309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno-Risueno M. A., Gonzalez N., Diaz I., Parcy F., Carbonero P., Vicente-Carbajosa J. (2008). FUSCA3 from barley unveils a common transcriptional regulation of seed-specific genes between cereals and Arabidopsis. Plant J. 53 882–894 10.1111/j.1365-313X.2007.03382.x [DOI] [PubMed] [Google Scholar]
- Mukhtar M. S., Carvunis A. R., Dreze M., Epple P., Steinbrenner J., Moore J., et al. (2011). Independently evolved virulence effectors converge onto hubs in a plant immune system network. Science 333 596–601 10.1126/science.1203659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myakushina Y. A., Milyaeva E. L., Romanov G. A., Nikiforova V. Y. (2009). Mutation in LSU4 gene affects flower development in Arabidopsis thaliana. Doklady Biochem. Biophys. 428 257–260 10.1134/S1607672909050093 [DOI] [PubMed] [Google Scholar]
- Nikiforova V. J., Daub C. O., Hesse H., Willmitzer L., Hoefgen R. (2005). Integrative gene-metabolite network with implemented causality deciphers informational fluxes of sulphur stress response. J. Exp. Bot. 56 1887–1896 10.1093/jxb/eri179 [DOI] [PubMed] [Google Scholar]
- Nikiforova V., Freitag J., Kempa S., Adamik M., Hesse H., Hoefgen R. (2003). Transcriptome analysis of sulfur depletion in Arabidopsis thaliana: interlacing of biosynthetic pathways provides response specificity. Plant J. 33 633–650 10.1046/j.1365-313X.2003.01657.x [DOI] [PubMed] [Google Scholar]
- Omranian N., Mueller-Roeber B., Nikoloski Z. (2012). PageRank-based identification of signaling crosstalk from transcriptomics data: the case of Arabidopsis thaliana. Mol. Biosyst. 8 1121–1127 10.1039/c2mb05365a [DOI] [PubMed] [Google Scholar]
- Ruckle M. E., Burgoon L. D., Lawrence L. A., Sinkler C. A., Larkin R. M. (2012). Plastids are major regulators of light signaling in Arabidopsis. Plant Physiol. 159 366–390 10.1104/pp.112.193599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Usadel B., Blasing O. E., Gibon Y., Retzlaff K., Hohne M., Gunther M., et al. (2008). Global transcript levels respond to small changes of the carbon status during progressive exhaustion of carbohydrates in Arabidopsis rosettes. Plant Physiol. 146 1834–1861 10.1104/pp.107.115592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W., Esch J. J., Shiu S. H., Agula H., Binder B. M., Chang C., et al. (2006). Identification of important regions for ethylene binding and signaling in the transmembrane domain of the ETR1 ethylene receptor of Arabidopsis. Plant Cell 18 3429–3442 10.1105/tpc.106.044537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Zhang W. Z., Song L. F., Zou J. J., Su Z., Wu W. H. (2008). Transcriptome analyses show changes in gene expression to accompany pollen germination and tube growth in Arabidopsis. Plant Physiol. 148 1201–1211 10.1104/pp.108.126375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wawrzyńska A., Lewandowska M., Hawkesford M. J., Sirko A. (2005). Using a suppression subtractive library-based approach to identify tobacco genes regulated in response to short-term sulphur deficit. J. Exp. Bot. 56 1575–1590 10.1093/jxb/eri152 [DOI] [PubMed] [Google Scholar]
- Wawrzyńska A., Lewandowska M., Sirko A. (2010). Nicotiana tabacum EIL2 directly regulates expression of at least one tobacco gene induced by sulphur starvation. J. Exp. Bot. 61 889–900 10.1093/jxb/erp356 [DOI] [PubMed] [Google Scholar]
- Zhang B., Pasini R., Dan H., Joshi N., Zhao Y., Leustek T., et al. (2014). Aberrant gene expression in the Arabidopsis SULTR1;2 mutants suggests a possible regulatory role for this sulfate transporter in response to sulfur nutrient status. Plant J. 77 185–197 10.1111/tpj.12376 [DOI] [PubMed] [Google Scholar]
- Zientara-Rytter K., Łukomska J., Moniuszko G., Gwozdecki R., Surowiecki P., Lewandowska M., et al. (2011). Identification and functional analysis of Joka2, a tobacco member of the family of selective autophagy cargo receptors. Autophagy 7 1145–1158 10.4161/auto.7.10.16617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann P., Hirsch-Hoffmann M., Hennig L., Gruissem W. (2004). GENEVESTIGATOR. Arabidopsis microarray database and analysis toolbox. Plant Physiol. 136 2621–2632 10.1104/pp.104.046367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann P., Laule O., Schmitz J., Hruz T., Bleuler S., Gruissem W. (2008). Genevestigator transcriptome meta-analysis and biomarker search using rice and barley gene expression databases. Mol. Plant 1 851–857 10.1093/mp/ssn048 [DOI] [PubMed] [Google Scholar]