Abstract
In Alzheimer disease, the microtubule-associated protein tau dissociates from the neuronal cytoskeleton and aggregates to form cytoplasmic inclusions. Although hyper-phosphorylation of tau Ser and Thr residues is an established trigger of tau misfunction and aggregation, tau modifications extend to Lys residues as well, raising the possibility that different modification signatures depress or promote aggregation propensity depending on site occupancy. To identify Lys-residue modifications associated with normal tau function, soluble tau proteins isolated from four cognitively normal human brains were characterized by mass spectrometry methods. The major detectable Lys modification was found to be methylation, which appeared in the form of mono- and di-methyl Lys residues distributed among at least eleven sites. Unlike tau phosphorylation sites, the frequency of Lys methylation was highest in the microtubule binding repeat region that mediates both microtubule binding and homotypic interactions. When purified recombinant human tau was modified in vitro through reductive methylation, its ability to promote tubulin polymerization was retained, whereas its aggregation propensity was greatly attenuated at both nucleation and extension steps. These data establish Lys methylation as part of the normal tau post-translational modification signature in human brain, and suggest that it can function in part to protect against pathological tau aggregation.
Keywords: Tau protein, aggregation, post-translational modification, methylation, microtubule, Alzheimer’s disease, mass spectrometry
INTRODUCTION
Tau is an intrinsically disordered monomeric protein that functions in league with the neuronal cytoskeleton (reviewed in [1]). Although it interacts with diverse cytoplasmic partners through its N-terminal projection domain [2], microtubule binding and promotion of tubulin assembly remain its most widely-studied activities. These are mediated by the assembly domain located in the C-terminal half of the tau molecule, composed of up to four imperfect repeats of 31 or 32 amino acids (i.e., the microtubule-binding repeat region, MTBR4) and flanking sequences [3]. In AD, however, tau loses its microtubule binding activity, and enters a pathway that culminates in aggregation and lesion formation (reviewed in [4]). Pathological tau homotypic interactions also are mediated by MTBR residues [5], including hexapeptide motifs 275VQIINK280 (termed PHF6*) and 306VQIVYK311 (termed PHF6) located in repeats 2 and 3, respectively [6]. Completion of the aggregation pathway and lesion formation correlates with neurodegeneration [7] and cognitive decline [8], consistent with a potential role in disease pathogenesis.
The mechanisms that regulate entry into the aggregation pathway are not fully understood, but appear to involve post-translational modifications (PTMs) positioned to influence tau function, stability, and aggregation propensity [9]. The best established tau PTM is phosphorylation, which increases from ~3 mol phosphate/mol tau protein in post-mortem samples of cognitively normal brain to ~9 mol/mol in AD brain (reviewed in [10]). The location of tau phosphorylation sites has been established by a combination of antibody and LC/MS-MS analyses, revealing partial occupancy of at least 17 Ser and Thr sites in normal tau [11] and at least 40 sites in AD-derived tau aggregates (reviewed in [12]). The sites are widely distributed throughout the tau molecule, consistent with the unfolded structure of tau exposing the majority of Ser and Thr residues to phosphotransferases. Although the effects of phosphorylation on tau biology are site dependent, in general it inhibits tau function by depressing microtubule binding affinity [13–15] while increasing aggregation propensity by lowering the tau concentration required to support self-association [16].
In addition to hydroxyl amino acids, Lys residues are modified on tau protein, and these too can influence tau metabolism and aggregation so as to antagonize or synergize with the effects of phosphorylation. First, tau can be ubiquitylated, which modulates tau accumulation and aggregation [17, 18]. Five sites of ubiquitylation have been mapped by LC/MS-MS methods in filamentous tau isolated from AD brain, and these localize within the MTBR [19–21]. Second, tau can be acetylated, which can protect tau against phosphorylation of sites associated with microtubule dissociation, and thereby act as a gatekeeper for entry into the tau aggregation pathway [22]. However, occupancy of other sites can directly raise aggregation propensity [23]. Tau acetylation has been detected immunohistochemically, but the complete catalog of sites acetylated in vivo has not been established. Third, tau can be methylated, and this modification may contribute to regulation of tau metabolism by directly competing with both ubiquitylation and acetylation [21]. Seven sites of Lys monomethylation have been mapped on filamentous tau isolated from AD brain, showing that this modification overlaps with established sites of ubiquitylation [21]. However, the direct effects of methylation on tau function are unknown. For each of these three diverse Lys modifications, biological effects will depend largely on the sites on tau that they occupy in vivo. It is likely that certain combinations of PTMs preserve tau function, whereas others foster misfunction and entry into the aggregation pathway.
To test this hypothesis, we have begun mapping modifications on tau protein purified from human brain using mass spectrometry methods. Here we extend the analysis to soluble tau protein enriched from pathologically and behaviorally normal human brain. Results indicate that the major Lys modification of normal tau protein detectable by high-mass accuracy LC-MS/MS is methylation, and that this PTM appears with greatest frequency within the MTBR. The results suggest that methylation is part of the modification signature associated with preservation of normal tau function.
EXPERIMENTAL
Materials
[14C]Formaldehyde (54.8 Ci/mol) was from Perkin Elmer (Waltham, MA), purified bovine tubulin dimer from Cytoskeleton (Denver, CO), aggregation inducer Thiazine red (Chemical Abstract Service registry number 2150-33-6) from TCI America (Portland, OR), and trypsin from Promega (Madison, WI). Formvar/carbon-coated copper grids, glutaraldehyde, and uranyl acetate were from Electron Microscopy Sciences (Fort Washington, PA).
Brain-derived tau protein
This study used only archival, de-identified post mortem human brain tissue samples from autopsies performed with informed consent of each patient or relative via procedures approved by the relevant institutional committees. Tau was enriched using methods detailed previously [24]. Gray matter was homogenized in 5 v/w of homogenization buffer (20 mM MES, pH 6.8, 80 mM NaCl, 1 mM MgCl2, 2 mM EGTA, 0.1 mM EDTA, 1 mM PMSF) containing inhibitors of phosphoprotein phosphatases (10 mM sodium pyrophosphate, 20 mM NaF, 1 mM Na3VO4) and deacetylases (2 µM trichostatin A, 10 mM nicotinamde). After centrifugation (20 min at 27,000g), the supernatant was collected, adjusted to 0.5 M NaCl and 2% 2-mercaptoethanol, boiled (10 min), and recentrifuged (20 min at 27,000g). The resulting heat-stable supernatant fraction was treated with 2.5% (final concentration) of perchloric acid. After isolating the acid-soluble fraction by centrifugation (20 min at 27,000g), protein was concentrated by TCA precipitation (20% w/v) followed by two washes in cold acetone. Dry protein pellets were stored at −20°C until solubilized for LC-MS/MS.
Recombinant human tau protein
Recombinant polyhistidine-tagged 2N4R tau was prepared as described previously [25, 26], and then subjected to reductive methylation [21, 27, 28] by incubating (22°C for up to 1 h) with 0.1 M NaBH3CN and 5 mM formaldehyde in 0.1 M sodium citrate buffer, pH 6. Reactions were stopped by the addition of glycine (50 mM final concentration). Non-methylated controls were prepared under identical conditions except that formaldehyde was omitted.
For subsequent functional assays, tau reaction products were desalted (Bio-Gel P-6 resin, Bio-Rad Laboratories, Hercules, CA) in 10 mM HEPES, pH 7.4, 50 mM NaCl. Final tau protein concentration was determined by bicinchoninic acid assay (Pierce/Thermo Fisher Scientific, Rockford, IL, USA). For subsequent LC-MS/MS analysis, tau products were desalted using protein desalting spin columns (Pierce/Thermo Fisher Scientific) equilibrated in volatile buffer (50 mM NH4HCO3, pH 7.8). Tau product was then evaporated to dryness and stored at −20°C until used.
To quantify methylation stoichiometry, formaldehyde was replaced with [14C]formaldehyde at a final specific activity of 0.88 Ci/mol. Tau products were separated from radioactive reactant using protein desalting spin columns equilibrated in 10 mM HEPES, pH 7.4, 50 mM NaCl, then quantified by scintillation spectroscopy. Measurements were performed in triplicate and reported as mean ± standard deviation (SD). Methylation time course was modeled as a simple exponential growth to maximum using the function:
| (1) |
where kapp and ymax are the rate constant and maximum extent of methylation, respectively.
Tau trypsinization
Tissue-derived tau samples (400 ng) were resuspended in SDS sample buffer and fractionated by SDS-PAGE (4–20% Tris-glycine gel; Bio-Rad). Protein bands were visualized with EZ Blue Coomassie stain (Sigma). Proteins migrating with 50–75 kDa mass were excised, cut into 1 × 1mm pieces, washed with 100 mM NH4HCO3, dehydrated in acetonitrile, and dried in a vacuum centrifuge. Dried gel samples were then reduced and alkylated by rehydrating in 100 mM NH4HCO3 containing 10 mM dithiothreitol (60 min at 60°C) followed by incubation in 100 mM NH4HCO3 containing 55 mM iodoacetamide (45 min at room temperature in the dark). Samples were then washed with 100mM NH4HCO3, dehydrated in acetonitrile, and dried in a vacuum centrifuge. Reduced and alkylated samples were then rehydrated and trypsinized in digestion solution (50 mM NH4HCO3, 5 mM CaCl2, and 15 ng/µL trypsin) for 45 min at 4°C. After removal of excess digestion solution, the samples were incubated at 37°C overnight. The resulting tryptic peptides were extracted (37°C for 15 min with shaking) once with 50 mM NH4HCO3, pH 7.8, once with 50 mM NH4HCO3:acetonitrile (1:1) and twice with 5% formic acid:acetonitrile. Extracted peptides were then de-salted using C18 PepClean Spin Columns (Pierce/Thermo Fisher Scientific) according to the manufacturer instructions and dried in a vacuum centrifuge. Recombinant tau samples were handled as described above except that in some cases trypsin digestion was done in solution without preceding SDS-PAGE.
LC-MS/MS
Tryptic peptides were dissolved in 0.5% acetic acid (solvent A) and fractionated on a 10 cm PicoFrit spray tip (New Objective, Woburn, MA) packed in-house with sub-2 µm C18 resin (Prospereon Life Science, IL) using a nano-flow Xtreme simple liquid chromatography system (CVC Microtech, Fontana, CA) coupled to a hybrid linear ion trap-Orbitrap mass spectrometer (LTQ-Orbitrap, Thermo Scientific). Peptides were loaded onto the column in 98% solvent A at a flow rate of 0.6 µl/min, and eluted with a 60 min linear 2–40% solvent B (40% acetonitrile, 0.5% acetic acid) gradient at a flow rate of 0.2 µL/min. After sample elution, the column was washed with 95% solvent B and re-equilibrated with 98% solvent A.
Mass spectra were acquired in positive ion mode with a data-dependent automatic switch applied between the survey scan and MS/MS acquisition. MS1 survey scans were acquired using a mass range of m/z 400–1,600 at a resolution of 60,000 at 400 m/z, and the MS1 target value was 100,000. MS/MS scans were acquired in the linear ion trap on the five most intense ions in each survey scan with dynamic exclusion of previously selected ions using a repeat count of 1 and exclusion duration of 15 s. Fragmentation by collision-induced dissociation (normalized collision energy 35%) was performed with a target value of 30,000 counts and an ion selection threshold of 5,000 counts. Charge state screening was enabled and ions with a +1 charge state were excluded from fragmentation. Other mass spectrometric parameters were as follows: spray voltage 1.35 kV, no sheath and auxiliary gas flow, ion transfer tube temperature 180°C, activation q = 0.25 and activation time 30 ms.
Analysis of mass spectrometric data
A manually curated tau database containing all known central nervous system isoforms of Homo sapiens microtubule-associated protein tau was created. Mass spectra were searched against the database using both MASCOT and Bioworks 3.3.1 SP1 with SEQUEST algorithms. Search parameters included 20 ppm peptide mass tolerance, 1.0 amu fragment tolerance, static modification Cys + 57.02146 (carbamidomethylation) and variable modifications: Met +15.99491 (oxidation); Lys +14.01565 (monomethylation); Lys +28.03130 (dimethylation); Lys +42.04695 (trimethylation); Lys +42.01056 (acetylation); and Ser/Thr +79.96632 and −18.01054 (corresponding to phosphorylation and β-elimination, respectively). Tryptic peptides with up to three missed cleavages and charge-state dependent cross correlation (XCorr) scores ≥ 1.5, 2.5, and 3.0 for 1+, 2+, and 3+ peptides, respectively, and ΔCn > 0.1 were considered as initial positive identifications. All MS/MS and spectra of identified post-translationally modified peptides from the initial screening were subjected to location probability analysis and manual verification. For MASCOT search results, modified peptides with initial MSCORE > 20 were retained and validated by manual inspection.
CD spectroscopy
Samples were prepared for CD analysis by desalting into 100 mM NaClO4 and 20 mM H3BO3 (pH 7.4). Spectra were collected (187–245 nm) at 20°C using a Jasco J-815 CD spectrometer (0.1 cm path length). Four repetitive scans (step size 0.5 nm; bandwidth 1 nm) were recorded, averaged, and corrected for buffer-only blank without additional filtering or smoothing. Raw CD signals (in millidegrees) were converted to mean residue molar ellipticity [θ]MRW as described previously [29].
Tubulin polymerization assay
Tubulin polymerization was assayed using the light scattering method [30, 31]. Tubulin dimer (9 µM) was incubated (37°C) with either unmodified or reductively methylated tau (2 µM) in BRB80 (80 mM PIPES, pH 6.8, 1 mM EGTA, 1 mM MgCl2) containing 1 mM DTT and 1 mM GTP. Polymerization time course was followed by measuring absorbance at 340 nm every 1 min for 60 min in a Cary50 UV-Vis Spectrophotometer (Agilent, Santa Clara, CA, USA) equipped with a Peltier single-cell temperature control accessory. Assays were done in triplicate to yield parameter means ± SD. The probability (p) of differences among measured assembly parameters was assessed by one-way ANOVA and Bonferroni’s post hoc multiple comparison test.
Tau aggregation assay
2N4R tau was incubated without agitation in assembly buffer (10 mM HEPES, pH 7.4, 100 mM NaCl, and 5 mM DTT) for up to 24 h (unless otherwise specified) at 37°C. Aggregation was initiated with Thiazine red (100 µM final concentration). Aggregation products were fixed in glutaraldehyde (0.5% w/v), adsorbed to Formvar/carbon-coated copper grids, stained with 2% uranyl acetate, and viewed in a Tecnai G2 Spirit BioTWIN transmission electron microscope (FEI, Hillsboro, OR, USA) operated at 80 kV and 16,000- to 60,000-fold magnification. Individual filaments (defined as objects >10 nm in length with both ends visible in the field of view) were counted and quantified with ImageJ (National Institutes of Health). Three or more fields were captured for each condition so that at least 20 filaments were counted per condition. Total filament lengths of all resolved filaments per field are reported ± SD.
Critical concentrations (Kcrit) ± SEE were estimated from the tau concentration dependence of total filament length using linear regression as described previously [32]. To estimate the filament dissociation rate constant (ke−), tau filaments prepared as described above were diluted 10-fold into assembly buffer containing 100 µM Thiazine red and incubated at 37°C. Aliquots were removed as a function of time up to 5 h post-dilution and assayed for total filament length. Disaggregation time series were fit to an exponential decay function to obtain kapp, the pseudo-first order rate constant describing the timedependent decrease in filament length, and L0, the total filament length at time zero.
| (2) |
Rate constant ke− was estimated from kapp, L0, and the number of filaments at time zero as described previously [16, 33, 34]. The association rate constant ke+ was then estimated from the relationship [32]:
| (3) |
assuming a two state model (i.e., all tau was either monomeric or incorporated into filaments).
Aggregation lag times, defined as the time when the tangent to the point of maximum aggregation rate intersects the abscissa of the sigmoidal curve [35], were obtained ± SE from each time series by Gompertz regression as described previously [25].
The probability (p) of differences between estimated parameters (Kcrit, ke−, and ke+) was assessed by z-test:
| (4) |
where x1 ± Sx1 and x2 ± Sx2 are the pair of estimates ± SE being compared, and z is the 1-α point of the standard normal distribution using and JMP 9.0 (SAS Institute, Cary, NC). The null hypothesis was rejected at p < 0.05.
RESULTS
Post-translational modification state of tau protein isolated from pathologically normal brain tissue
To prepare material for analysis, soluble tau was isolated from the brains of four cognitively normal human males aged 55 ± 1.2 yrs (Table 1). In addition to meeting behavioral criteria, all cases were pathologically normal with respect to plaque and tangle lesion counts. Isolated tau samples were then proteolyzed with trypsin and subjected to LC-MS/MS. Resulting mass spectra were then interrogated using SEQUEST and MASCOT database search algorithms programmed to identify both unmodified peptides and peptides containing sites of phosphorylation (detected by either incorporation of a phosphoryl group or formation of β-elimination products dehydroalanine or dehydrobutyrine), mono-, di-, and tri-methylation, and acetylation. Any combination of two modifications was searched as well. Results showed that overall sequence coverage was 90% relative to longest tau isoform 2N4R, and included 90% of all Ser/Thr residues and 77.3% of all Lys residues in this isoform (Fig. 1A).
Table 1. Case demographics.
| Case (#) |
Age (yr) |
Sex | Race* | PMI† (h) |
Source§ | History‖ |
|---|---|---|---|---|---|---|
| 1 | 54 | M | C | --- | OSU | --- |
| 2 | 54 | M | AA | 10 | NICHD | HASCVD; PTSS, No history of drug, alcohol abuse |
| 3 | 55 | M | AA | 13 | NICHD | HASCVD; History of Diabetes, cocaine abuse, and hypertension |
| 4 | 57 | M | C | 5 | NICHD | Acute Bronchopneumonia/ASCVD; history of alcohol abuse |
AA, African American; C, Caucasian.
Post-mortem interval.
OSU, The Ohio State University Buckeye Brain Bank; NICHD, NICHD Brain and Tissue Bank for Developmental Disorders.
ASCVD, atherosclerotic cardiovascular disease; HASCVD, hypertensive arteriosclerotic cardiovascular disease; PTSS, Post Traumatic Stress Syndrome.
Figure 1. Summary of modification sites on human tau proteins identified by LC-MS/MS.
(A) Identified tryptic peptides in the context of the human 2N4R isoform (NCBI accession number NP_005901), where the dotted underline depicts projection domain segments n1 and n2, the solid underline identifies the MTBR (as defined by [70]), and PHF6/PHF6* mark the hexapeptide segments involved in filament nucleation. Font color depicts sequence coverage: blue, identified only in tissuederived tau, red, identified only in recombinant 2N4R tau containing near-saturating levels of methylation (21.8 mol/mol), and purple, identified in both. Phosphorylation (P) sites are marked by green circles (hollow circles, novel sites; solid circles, previously reported sites), whereas methylation sites are marked by orange (me1, monomethylation) and red (me2, dimethylation) symbols (squares for tissue tau, triangles for recombinant tau). (B) Tau methylation and phosphorylation site distribution map (2N4R tau), showing location of modification sites relative to projection domain segments n1 and n2 and MTBR repeats r1 – r4. The length of each bar corresponds to the number of cases in which the modification was found. The distribution of reported methylation sites in PHF-tau isolated from AD brain [21] is shown for comparison.
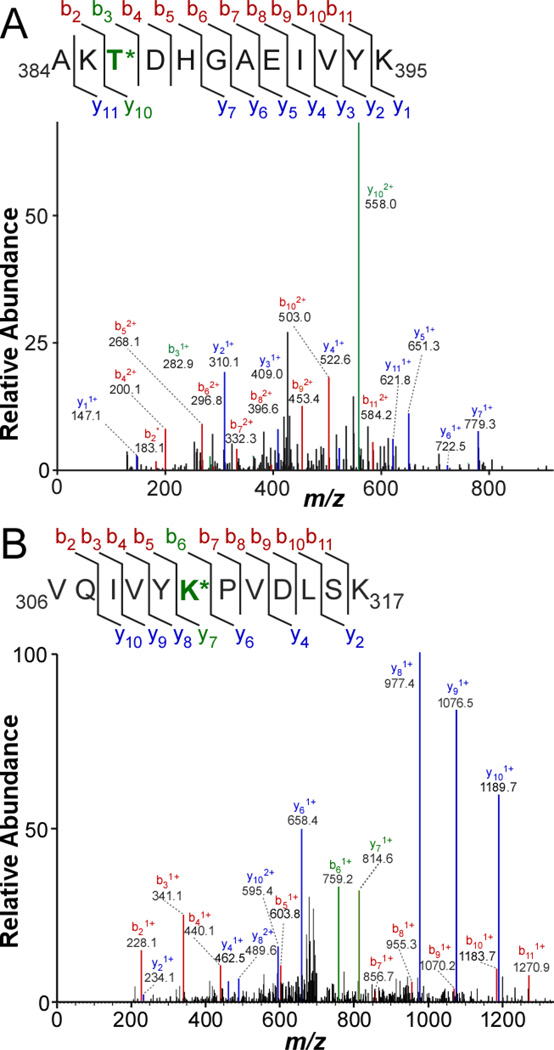
Within the coverage area, we focused first on phosphorylation site distribution, which had been reported previously at low mass accuracy for both soluble [11] and PHF-tau (reviewed in [12]). We identified 31 Ser and Thr phosphorylation sites (Table 2) distributed broadly across the molecule, but with greatest frequency in the projection domain and in the assembly domain outside the MTBR (Fig. 1B). Seven novel Thr phospho-sites were detected at T17, T39, T50, T245, T263, T361, and T386 (Table 2). Although they had been identified previously in vitro after treatment of tau with protein kinases [11], this is their first demonstration in human tissue. A representative MS/MS spectrum generated by one of them, peptide 384AKTDHGAEIVYK395 phosphorylated on T386 (0.6 ppm mass accuracy), is shown in Fig. 2A. This site lies adjacent to the S396 – S416 region previously reported to be phosphorylated in both soluble and PHF-tau (reviewed in [12]). Identification of phospho-sites varied among cases, but previously reported sites T181, S202, T231 and S404, as well as novel sites T361 and T386, were found consistently in all of them (Fig. 1B). These data confirmed that the human brainderived tau protein samples were extensively modified, and that our methods were adequate for detecting tau modifications with high mass accuracy.
Table 2. Phosphorylation sites identified on tau isolated from cognitively normal human brain.
Trypsin digests of tissue-derived tau were analyzed by LC-MS/MS, and resulting m/z data were analyzed by MASCOT and SEQUEST using ±20 ppm mass tolerance.
| aa residues* |
Peptide† | Site | Mod§ | Calculated mass |
Error ppm |
MASCOT score |
|---|---|---|---|---|---|---|
| 6–23 | QEFEVMEDHAGTYGLGDR | M11/T17 | Ox/DB | 2,051.876 | −0.6 | 21.4 |
| 25–44 | DQGGYTMHQDQEGDTDAGLK | T30/M31 | DB/Ox | 2,163.888 | −0.4 | 31.2 |
| 24–44 | KDQGGYTMHQDQEGDTDAGLK | M31/T39 | Ox/DB | 2,291.983 | −0.2 | 26.7 |
| 24–44 | KDQGGYTMHQDQEGDTDAGLK | T39 | DB | 2,275.988 | 0.3 | 21.6 |
| 25–44 | DQGGYTMHQDQEGDTDAGLK | M31/T39 | Ox/DB | 2,163.888 | −0.4 | 25.3 |
| 45–67 | ESPLQTPTEDGSEEPGSETSDAK | S46 | DA | 2,373.021 | −13.3 | 26.1 |
| 45–67 | ESPLQTPTEDGSEEPGSETSDAK | T50 | DB | 2,373.021 | −13.3 | 50.2 |
| 68–87 | STPTAEDVTAPLVDEGAPGK | T71 | P | 2,034.927 | −9.7 | 31.3 |
| 171–180 | IPAKTPPAPK | T175 | P | 1,099.591 | 1.3 | 42.7 |
| 181–190 | TPPSSGEPPK | T181 | P | 1,076.466 | 2.4 | 23.9 |
| 175–190 | TPPAPKTPPSSGEPPK | T181 | P | 1,667.804 | 0.8 | 31.9 |
| 175–190 | TPPAPKTPPSSGEPPK | S185 | P | 1,667.804 | −0.5 | 24.7 |
| 195–206 | SGYSSPGSPGTPGSR | S198 | P | 1,473.601 | −1.4 | 30.3 |
| 191–206 | SGDRSGYSSPGSPGTPGSR | S199 | P | 1,888.782 | −1.0 | 27.9 |
| 195–206 | SGYSSPGSPGTPGSR | S199 | P | 1,473.601 | −0.6 | 23.2 |
| 195–206 | SGYSSPGSPGTPGSR | S202 | P | 1,473.601 | −0.1 | 40.5 |
| 195–206 | SGYSSPGSPGTPGSRSR | S202 | P | 1,716.734 | −1.0 | 42.1 |
| 191–206 | SGDRSGYSSPGSPGTPGSR | S202 | P | 1,888.782 | 0.1 | 39.7 |
| 191–206 | SGDRSGYSSPGSPGTPGSR | T205 | P | 1,888.782 | −1.1 | 22.8 |
| 195–206 | SGYSSPGSPGTPGSR | T205 | P | 1,473.601 | −0.3 | 38.7 |
| 191–206 | SGDRSGYSSPGSPGTPGSR | S208 | P | 1,888.782 | 0.1 | 29.2 |
| 210–221 | SRTPSLPTPPTR | S214 | P | 1,389.689 | 0.4 | 45.1 |
| 212–221 | TPSLPTPPTR | S214 | P | 1,146.555 | 1.2 | 36.2 |
| 212–224 | TPSLPTPPTREPK | S214 | P | 1,500.746 | −0.9 | 27.3 |
| 212–221 | TPSLPTPPTR | T217 | P | 1,146.555 | 2.4 | 28.9 |
| 212–224 | TPSLPTPPTREPK | T217 | P | 1,500.746 | 0.9 | 20.7 |
| 226–234 | VAVVRTPPK | T231 | P | 1,046.576 | 1.4 | 27.5 |
| 231–240 | TPPKSPSSAK | S235 | P | 1,079.513 | 0.1 | 20.9 |
| 243–254 | LQTAPVPMPDLK | T245/M250 | DB/Ox | 1,307.703 | −1.9 | 20.9 |
| 243–257 | LQTAPVPMPDLKNVK | T245/M250 | DB/Ox | 1,648.909 | 1.6 | 21.4 |
| 260–267 | IGSTENLK | S262 | P | 941.434 | 0.4 | 25.0 |
| 260–267 | IGSTENLK | S262 | DA | 843.457 | 0.6 | 31.3 |
| 260–267 | IGSTENLK | T263 | DB | 843.457 | −5.5 | 25.2 |
| 282–290 | LDLSNVQSK | S285 | DA | 985.531 | 0.3 | 21.4 |
| 281–290 | KLDLSNVQSK | S289 | DA | 1,113.626 | 0.4 | 39.1 |
| 282–290 | LDLSNVQSK | S289 | DA | 985.531 | −3.8 | 22.1 |
| 282–290 | LDLSNVQSK | S289/K290 | DA/me1 | 999.547 | 0.0 | 22.8 |
| 299–317 | HVPGGGSVQIVYKPVDLSK | S305 | DA | 1,962.081 | −7.5 | 59.4 |
| 354–369 | IGSLDNITHVPGGGNK | S356 | DA | 1,560.813 | 0.7 | 29.0 |
| 354–369 | IGSLDNITHVPGGGNK | T361 | DB | 1,560.813 | 0.3 | 24.4 |
| 384–395 | AKTDHGAEIVYK | T386 | DB | 1,313.685 | 0.6 | 45.3 |
| 386–395 | TDHGAEIVYK | T386 | DB | 1,114.553 | −1.1 | 42.9 |
| 386–406 | TDHGAEIVYKSPVVSGDTSPR | S396 | P | 2,295.065 | 0.3 | 30.8 |
| 396–406 | SPVVSGDTSPR | S400 | P | 1,181.520 | −2.0 | 27.9 |
| 396–406 | SPVVSGDTSPR | S400/S404 | P/DA | 1,163.509 | 2.8 | 25.6 |
| 386–406 | TDHGAEIVYKSPVVSGDTSPR | T403 | DB | 2,197.088 | −0.6 | 34.3 |
| 396–406 | SPVVSGDTSPR | T403 | P | 1,181.520 | −0.8 | 25.7 |
| 396–406 | SPVVSGDTSPR | T403/S404 | DB/P | 1,163.509 | 0.7 | 25.5 |
| 396–406 | SPVVSGDTSPR | S404 | P | 1,181.520 | 0.2 | 37.8 |
Amino acid (aa) residue numbering conforms to human 2N4R tau.
K, methylated Lys residues.
Modified residues (Mod) are DA, dehydroalanine; DB, dehydrobutyrine; me1, monomethyl-Lys; Ox, Met-sulfoxide; P, phospho-Ser or phospho-Thr.
Figure 2. High mass accuracy determination of human tau protein post-translational modifications.
Tau proteins isolated from cognitively normal human brains (Table 1) were digested with trypsin and subjected to LC-MS/MS spectroscopy. (A) MS/MS spectrum and ion assignments for 384AKT*DHGAEIVYK395, where T* corresponds to dehydrobutyrine, a β-elimination product of phospho-Thr. The difference between expected and observed mass for this peptide was −0.00081 Da, corresponding to a mass accuracy of 0.6 ppm. (B) MS/MS spectrum and ion assignment for 306VQIVYK*PVDLSK317, where K* corresponds to dimethyl-Lys. For this methylated peptide, the difference between expected and observed mass was −0.00440 Da, corresponding to a mass accuracy of 2.2 ppm.
We next searched for Lys modifications. No evidence for K+42 modification was found, indicating that neither trimethyl-Lys nor acetyl-Lys was present in the coverage area at the level of detection available in our datasets. In contrast, search criteria identified eleven unique sites of mono- and/or dimethylation (Table 3; Fig. 1A). The methylation sites were distributed broadly across the tau molecule and varied from case to case much like phosphorylation sites. Unlike phospho-sites, however, they localized with greatest frequency inside rather than outside the MTBR (Fig. 1B). An example of a spectrum identifying K311 as a site of dimethylation at 2.2 ppm mass accuracy is shown in Fig. 2B. This residue was reported as a possible methylation site in AD-brain derived tau protein on the basis of Edman degradation years ago [36]. It resides within the “PHF6” motif of the MTBR, which has been reported to mediate the aggregation propensity of recombinant monomeric tau in vitro [6, 37]. Other methylation sites within the MTBR include K259, K290, and K353, each of which lies in a KXGS motif associated with AMP-activated protein kinase mediated regulation of microtubule binding [38]. In contrast, an assembly domain motif reported to mediate constitutive microtubule binding, 224KKVAVVR230 [39–41], was not methylated in these samples (Fig. 1). A second important binding motif, 274KVQIINKK281, was only partially resolved, but residues K280 and K281 were found to be unmethylated (Fig. 1).
Table 3. Lys methylation sites identified on tau isolated from cognitively normal human brain.
Trypsin digests of tissue-derived tau were analyzed by LC-MS/MS, and resulting m/z data were analyzed by MASCOT and SEQUEST using ±20 ppm mass tolerance as an initial search parameter.
| aa residues* |
Peptide† | Site | Mod§ | Calculated mass |
Error ppm |
MASCOT score |
|---|---|---|---|---|---|---|
| 24–44 | KDQGGYTMHQDQEGDTDAGLK | K24/M31 | me2/Ox | 2,338.025 | 2.7 | 35.5 |
| 25–44 | DQGGYTMHQDQEGDTDAGLK | M31/K44 | Ox/me2 | 2,209.930 | 0.6 | 29.2 |
| 45–67 | ESPLQTPTEDGSEEPGSETSDAK | K67 | me2 | 2,419.063 | 2.0 | 30.8 |
| 181–190 | TPPSSGEPPK | K190 | me1 | 1,010.515 | −8.9 | 20.6 |
| 258–267 | SKIGSTENLK | K259 | me1 | 1,090.610 | 1.0 | 42.8 |
| 260–267 | IGSTENLK | K267 | me1 | 875.483 | −4.6 | 29.2 |
| 260–267 | IGSTENLK | K267 | me2 | 889.499 | 0.1 | 20.1 |
| 284–290 | LDLSNVQSK | S289/K290 | DA/me1 | 999.547 | 0.0 | 22.8 |
| 284–290 | LDLSNVQSK | K290 | me2 | 1,031.573 | 17.5 | 54.9 |
| 306–317 | VQIVYKPVDLSK | K311 | me2 | 2,008.123 | −2.2 | 22.4 |
| 306–317 | VQIVYKPVDLSK | K317 | me2 | 1,832.089 | 3.4 | 3.4‖ |
| 350–369 | VQSKIGSLDNITHVPGGGNK | K353 | me2 | 2,049.109 | 3.2 | 36.0 |
| 382–395 | AKTDHGAEIVYK | K385 | me1 | 1,345.711 | −0.4 | 29.2 |
Amino acid (aa) residue numbering conforms to human 2N4R tau.
K, methylated Lys residues.
Modified residues (Mod) are DA, dehydroalanine; me1, monomethyl-Lys; me2, dimethyl-Lys; Ox, Met-sulfoxide.
SEQUEST XCorr value.
Within the N-terminal projection domain, Lys methylation was detected at K24, K44, K67, and K190 (Fig. 1). Two of these, K24 and K44, lie in close proximity to putative caspase [42, 43] and calpain-1 [44] cleavage sites, respectively. Together, these data reveal that soluble tau resides in normal brain as a methylated protein, and that its sites of methylation overlap with motifs associated with pathological aggregation and with multiple competing PTMs.
Preparation of synthetic methyl-tau proteins
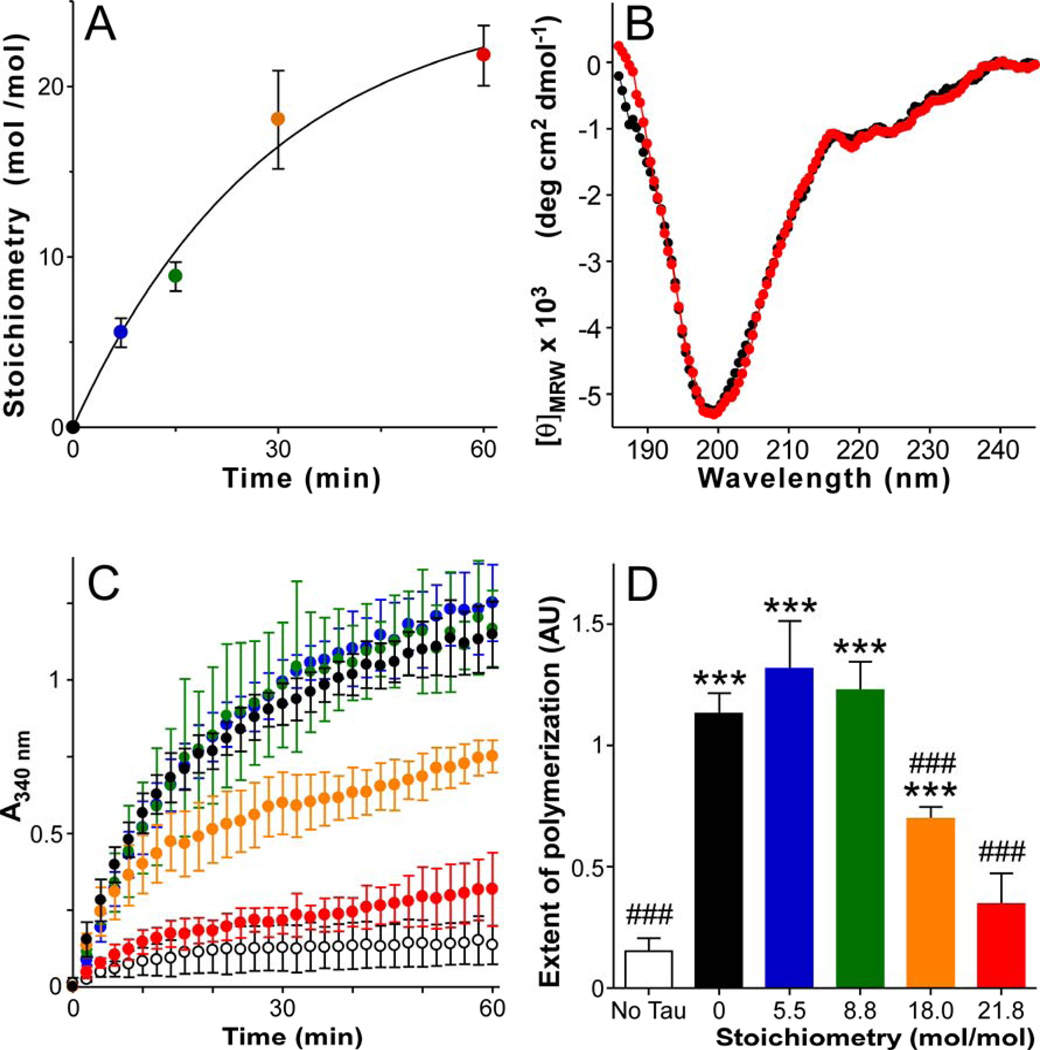
To prepare modified tau protein for functional assays, recombinant human 2N4R tau was subjected to reductive methylation (a well-characterized method for introducing Lys-specific methylation into proteins [45, 46]). The rate and extent of Lys methylation was monitored in parallel reactions containing [14C]-formaldehyde, which served to donate [14C]-labeled methyl groups to tau. After 7 min incubation, [14C]-methyl incorporation averaged 5.5 ± 0.9 mol/mol tau monomer (Fig. 3A). By 60 min incubation, incorporation averaged 21.8 ± 1.8 mol/mol and approached saturation (Fig. 3A). To identify which sites were occupied near saturation, a non-radioactive 60 min sample was subjected to trypsin digestion and LC-MS/MS analysis. Sequence coverage was 65.2% and included 68.2% of all 2N4R lysines (Fig. 1A). Methyl groups were found distributed among 23 sites in the form of mono- and di-methyl Lys residues (Table 4). Two thirds of all resolved lysines were methylated, including six of the seven sites occupied within the assembly domain of soluble brain-derived tau (Fig. 1B). These results were consistent with the exposure of most Lys residues to solvent owing to the intrinsically disordered structure of tau monomer, and with the non-specific nature of reductive methylation. It is likely, therefore, that all reductive methylation time points had similar methylation site distributions but with varying occupancies. When subjected to CD spectroscopy, all methylated tau samples yielded spectra dominated by a broad minimum of ellipticity centered at 200 nm (shown only for the 60 min sample in Fig. 3B), indicating that tau protein remained intrinsically disordered even after high-stoichiometry methylation. In all, five tau samples were prepared, containing 0 – 21.8 mol/mol methyl groups.
Figure 3. Effect of methylation on tau structure and function in vitro.
Purified recombinant human 2N4R tau protein was subjected to reductive methylation in the presence of identical concentrations of [14C]-labeled or unlabeled formaldehyde at room temperature, then assayed for methylation stoichiometry, secondary structure, and tubulin assembly promoting activity. (A) Time course of methylation determined in the presence of [14C]formaldehyde (n = 3), where the solid line represents best fit of the data with Eq. 1. [14C]methyl incorporation ranged from 0 – 21.8 mol/mol over the 60 min time course. (B) CD spectra (20°C) of methylated and non-methylated tau suspended in 100 mM sodium perchlorate, 20 mM boric acid, pH 7.4. Symbol colors correspond to stoichiometries estimated in panel A. (C) Effect of methylated tau (prepared in unlabeled formaldeyde) on tubulin assembly (37°C) as measured by absorbance (A340 nm). Each data point represents triplicate determination of A340 ± SD as a function of time. Assays were performed in the absence (white circles) or presence of tau containing 0 – 21.8 mol/mol methylation (symbol colors correspond to stoichiometries estimated in panel A). (D) Quantification of tubulin assembly at 60 min (data from Panel C). The extent of tau-mediated tubulin assembly was depressed only at high methylation stoichiometries (≥18 mol/mol). ***, p < 0.001 for comparison with no tau control (hollow bar); ###, p < 0.001 for comparison with non-methylated tau control (black bar).
Table 4. Lys methylation sites identified on recombinant tau after reductive methylation.
Trypsin digests of 2N4R tau were analyzed by LC-MS/MS, and resulting m/z data were analyzed by MASCOT and SEQUEST using ±10 ppm mass tolerance.
| aa residues* |
Peptide† | Site | Mod§ | Calculated mass |
Error ppm |
MASCOT score |
|---|---|---|---|---|---|---|
| 24–44 | KDQGGYTMHQDQEGDTDAGLK | K24 | me2 | 2,322.030 | −0.7 | 80.4 |
| 24–44 | KDQGGYTMHQDQEGDTDAGLK | K24/M31 | me1/Ox | 2,324.010 | −1.5 | 49.0 |
| 24–44 | KDQGGYTMHQDQEGDTDAGLK | K24/M31 | me2/Ox | 2,338.025 | −1.0 | 43.1 |
| 45–67 | ESPLQTPTEDGSEEPGSETSDAK | K67 | me1 | 2,405.047 | 0.0 | 4.3‖ |
| 68–87 | STPTAEDVTAPLVDEGAPGK | K87 | me1 | 1,968.976 | 0.0 | 4.6‖ |
| 144–155 | GADGKTKIATPR | K148/K150 | me1/me2 | 1,256.732 | −1.4 | 46.7 |
| 144–155 | GADGKTKIATPR | K150 | me2 | 1,242.716 | −1.6 | 50.9 |
| 156–170 | GAAPPGQKGQANATR | K163 | me2 | 1,451.771 | 0.3 | 71.5 |
| 156–170 | GAAPPGQKGQANATR | K163 | me1 | 1,437.756 | 0.8 | 62.6 |
| 170–180 | IPAKTPPAPK | K174/K180 | me2/me1 | 1,061.672 | −1.2 | 44.7 |
| 170–180 | IPAKTPPAPK | K174 | me1 | 1,033.640 | −1.4 | 43.8 |
| 175–190 | TPPAPKTPPSSGEPPK | K180 | me2 | 1,615.869 | −0.8 | 43.8 |
| 181–194 | TPPSSGEPPKSGDR | K190 | me2 | 1,439.712 | −2.1 | 41.3 |
| 232–240 | TPPKSPSSAK | K234/K240 | me2/me1 | 1,041.594 | −0.3 | 40.0 |
| 243–254 | LQTAPVPMPDLK | K254 | me1 | 1,323.734 | −0.4 | 48.7 |
| 243–254 | LQTAPVPMPDLK | M250/K254 | Ox/me1 | 1,339.729 | 0.1 | 46.8 |
| 258–274 | SKIGSTENLKHQPGGGK | K259/K267 | me2/me2 | 1,794.987 | −2.2 | 3.1‖ |
| 281–290 | KLDLSNVQSK | K281/K290 | me2/me1 | 1,173.684 | 0.1 | 55.6 |
| 281–290 | KLDLSNVQSK | K281 | me1 | 1,145.652 | −0.5 | 52.5 |
| 281–294 | KLDLSNVQSKCGSK | K281/K290 | me2/me2 | 1,619.878 | −1.1 | 49.8 |
| 281–294 | KLDLSNVQSKCGSK | K281/K290 | me1/me2 | 1,605.863 | −1.3 | 43.9 |
| 281–290 | KLDLSNVQSK | K281/K290 | me2/me2 | 1,187.699 | 0.3 | 43.1 |
| 282–290 | LDLSNVQSK | K290 | me2 | 1,031.573 | −5.1 | 57.1 |
| 282–290 | LDLSNVQSK | K290 | me1 | 1,017.557 | 0.0 | 52.5 |
| 299–317 | HVPGGGSVQIVYKPVDLSK | K311 | me1 | 1,994.107 | −1.4 | 3.7‖ |
| 350–369 | VQSKIGSLDNITHVPGGGNK | K353/K369 | me2/me1 | 2,063.124 | 1.4 | 53.4 |
| 350–370 | VQSKIGSLDNITHVPGGGNKK | K353/K370 | me2/me2 | 2,205.235 | 0.5 | 45.8 |
| 350–369 | VQSKIGSLDNITHVPGGGNK | K353 | me2 | 2,049.109 | −1.1 | 44.1 |
| 354–369 | IGSLDNITHVPGGGNK | K369 | me1 | 1,592.839 | −0.6 | 69.1 |
| 354–369 | IGSLDNITHVPGGGNKK | K370 | me2 | 1,734.950 | −2.1 | 48.3 |
| 370–379 | KIETHKLTFR | K375 | me1 | 1,286.758 | −2.2 | 48.3 |
| 371–379 | IETHKLTFR | K375 | me2 | 1,172.679 | −0.7 | 47.3 |
| 384–406 | AKTDHGAEIVYKSPVVSGDTSPR | K385/K395 | me2/me1 | 2,456.278 | 1.5 | 85.3 |
| 384–406 | AKTDHGAEIVYKSPVVSGDTSPR | K385/K395 | me1/me1 | 2,442.262 | 0.0 | 66.6 |
| 384–406 | AKTDHGAEIVYKSPVVSGDTSPR | K385/K395 | me2/me2 | 2,470.294 | 4.7 | 63.6 |
| 384–406 | AKTDHGAEIVYKSPVVSGDTSPR | K385/K395 | me1/me2 | 2,456.278 | 1.7 | 51.7 |
| 384–395 | AKTDHGAEIVYK | K385 | me1 | 1,345.711 | 0.0 | 41.5 |
| 384–406 | AKTDHGAEIVYKSPVVSGDTSPR | K385 | me1 | 2,428.247 | −0.1 | 40.9 |
| 384–395 | AKTDHGAEIVYK | K385 | me2 | 1,359.727 | −0.6 | 40.2 |
| 386–406 | TDHGAEIVYKSPVVSGDTSPR | K395 | me2 | 2,243.130 | −0.3 | 62.5 |
| 386–406 | TDHGAEIVYKSPVVSGDTSPR | K395 | me1 | 2,229.115 | 0.4 | 58.1 |
Amino acid (aa) residue numbering conforms to human 2N4R tau.
K, methylated Lys residues.
Modified residues (Mod) are me1, monomethyl-Lys; me2, dimethyl-Lys; Ox, Met-sulfoxide.
SEQUEST XCorr value.
Effects of Lys methylation on normal tau function
One of the normal functions of tau is to promote spontaneous assembly of microtubules from tubulin dimers [47–50]. Certain PTMs occurring in the assembly domain, including phosphorylation and acetylation, can modulate this interaction [13, 14, 23, 51, 52]. To determine the effect of methylation on tau-promoted microtubule assembly, purified tubulin dimers were incubated at 37°C alone or with unmodified or reductively methylated tau, and the extent of tubulin polymerization was measured spectrophotometrically. In the absence of tau, tubulin polymerized only weakly (Fig. 3CD). In contrast, the addition of unmodified tau greatly stimulated polymerization at all time points, and tau methylated at up to ~9 mol/mol promoted tubulin polymerization almost identically (Fig. 3CD). However, methylation to higher stoichiometries depressed the ability of tau to promote microtubule polymerization, with 21.8 mol/mol incorporation rendering modified tau almost completely inactive (Fig. 3CD). Together, these data indicate that Lys methylation did not modulate tau-dependent tubulin polymerization until very high-stoichiometries were incorporated.
Lys methylation modulates tau aggregation propensity
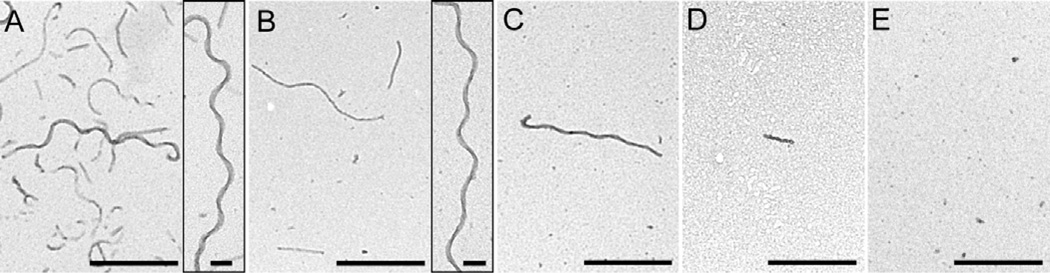
The aggregation of tau into filaments is a well-documented correlate of AD. To determine the effect of methylation on tau aggregation propensity, the methyl-tau preparations described above were incubated in the presence of Thiazine red aggregation inducer under near-physiological conditions of pH, reducing conditions, and ionic strength. Thiazine red was used as aggregation inducer because it efficiently drives aggregation of full-length 2N4R tau into filaments having mass-per-unit length similar to authentic brainderived PHFs [32, 34, 52]. The reaction approximates a homogeneous nucleation scheme characterized by initial formation of an unstable dimeric nucleus, followed by filament elongation through monomer addition [32]. Under these conditions, the inherent aggregation propensity of tau can be quantified in terms of nucleation and extension components. Tau aggregation was monitored using transmission electron microscopy, which detected both filament morphology and length. Low magnification images confirmed that unmodified recombinant tau readily aggregated (Fig. 4A), but that Lys methylation impaired total filament length in a stoichiometry-dependent manner (Fig. 4B–E). High magnification images revealed similar morphology of both unmodified and methylated tau filaments (Fig. 4AB insets), indicating that methylated tau shared the fundamental aggregation characteristics of unmodified tau, but had a lower propensity to do so.
Figure 4. Lys methylation decreases tau aggregation propensity.
Recombinant 2N4R tau (2 µM) containing zero (A), 5.5 (B), 8.8 (C), 18.0 (D), or 21.8 (E) mol/mol methylation stoichiometry was incubated (18 h at 37°C) in the presence of Thiazine red inducer, then assayed for filament formation by electron microscopy. Lys methylation did not modify tau filament morphology under these conditions, but greatly depressed aggregation propensity. Scale bars = 500 nm; Insets, scale bars = 100 nm.
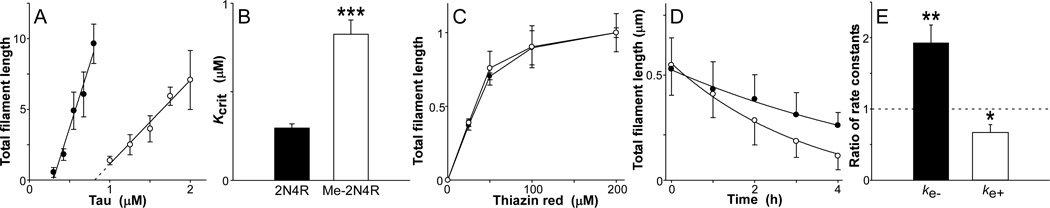
To determine the mechanism through which methylation depressed aggregation propensity, the minimal concentration of unmodified or methylated tau required to support aggregation was estimated in the presence of Thiazine red inducer. Abscissa intercepts of tau concentration dependence plots revealed that the minimal concentration for aggregation of tau methylated to 5.5 mol/mol stoichiometry increased nearly 3-fold relative to unmodified tau (Fig. 5AB). However, this methylated tau did not differ from non-methylated tau with respect to sensitivity to Thiazine red inducer (Fig. 5C). These data indicate that Lys methylation depressed the intrinsic aggregation propensity of tau, and did so in part by increasing the concentration of tau needed to support fibril formation.
Figure 5. Lys methylation modulates tau filament extension rates.
(A) Unmodified (solid circle) or 5.5 mol/mol methylated tau (hollow circle) were incubated (18 h at 37°C) at varying bulk concentrations in the presence of 100 µM Thiazine red inducer, then assayed for filament formation by electron microscopy (reported in arbitrary units). Total filament lengths were then plotted against bulk protein concentration, where each data point represents the mean ± SD of triplicate determinations and the solid lines represent best fit of the data points to linear regression. The abscissa intercept was obtained by extrapolation (dotted lines) and taken as the critical concentration (Kcrit). (B) Replot of data from panel (A), where each bar represents the Kcrit ± propagated SEE. Methylation increased Kcrit nearly 3-fold relative to unmodified tau (***, p < 0.001). (C) Unmodified (solid circle) or 5.5 mol/mol methylated tau (hollow circle) were incubated (18 h at 37°C) at constant supersaturation (i.e., 0.5 µM above Kcrit) in the presence of varying concentrations of Thiazine red, and then assayed for filament formation by electron microscopy. Each data point represents total filament lengths/field from triplicate determinations (reported in arbitrary units). (D) Filaments prepared from unmodified (solid circle) and 5.5 mol/mol methylated tau (hollow circle) as described above were diluted below Kcrit in assembly buffer, and the resultant disaggregation followed as a function of time by electron microscopy. Each data point represents total filament length per field ± SD (n = 3), whereas the solid line represents best fit of data points with Eq. 2. The first-order decay constant kapp was estimated from each regression and used in conjunction with filament length (shown in figure) and number at time t = 0 to calculate dissociate rate constant ke−. ke+ was then obtained from Eq. 3. (E) Replot of data from panel (D), where each bar represents the ratio of rate constants for filament extension (ke+) and dissociation (ke−) determined for 5.5 mol/mol methylated versus unmodified tau ± propagated SEE. A ratio of one, corresponding to no difference in rate, is marked by the dashed line. Methylation increased filament dissociation while decreasing filament extension. *, p < 0.05; **, p < 0.01 for comparison of methylated versus unmodified rate constants.
In nucleation-elongation reactions, the minimal concentration approximates Kcrit, which corresponds to the equilibrium dissociation constant for elongation, Ke [32]. It therefore reflects contributions from both the dissociation (ke−) and association (ke+) rate constants for filament elongation (Eq. 3). As a result, changes in Kcrit may stem from changes in either filament stability (ke−), efficiency of monomer association with filament ends (ke+), or both. To differentiate among these possibilities, initial filament disaggregation rates were estimated for unmodified and methylated (5.5 mol/mol stoichiometry) tau by diluting preassembled filaments below the Kcrit and measuring filament length as a function of time. Under these conditions, loss of total filament length was found to decay following first-order kinetics as predicted for endwise depolymerization from a Poisson-like length distribution [16, 33] (Fig. 5D). The dissociation constant ke− was derived from these data using the established relationship between tau mass and filament length [32]. Rate constant ke+ was then calculated from estimates of ke− and Kcrit using Eq. 3. Tau methylation was found to increase ke− nearly 2-fold and decrease ke+ ~1.5-fold (Fig. 5E), indicating that it raised minimal concentration by both weakening filament stability and slowing the rate of elongation.
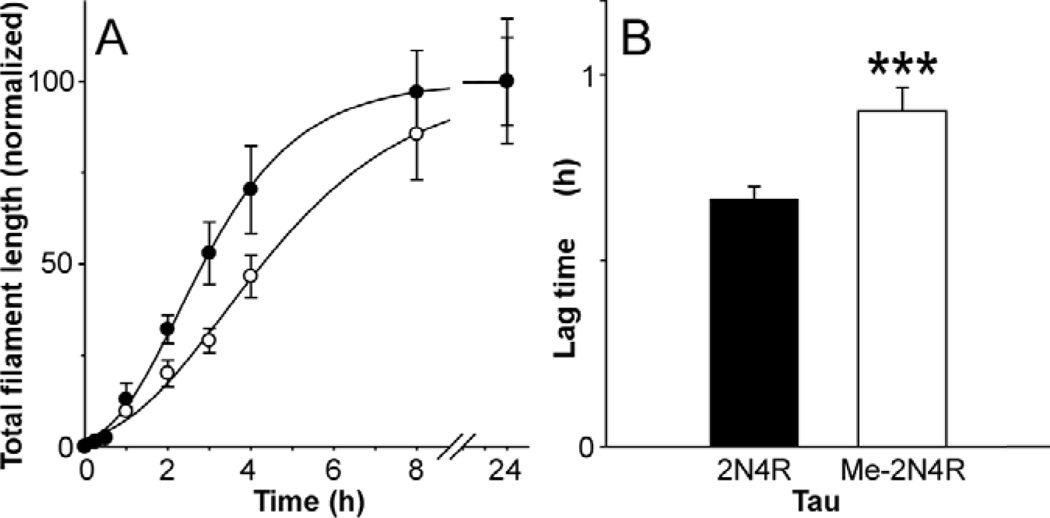
In a homogeneous nucleation dependent reaction, the rate limiting step involves formation of a thermodynamic nucleus, defined as the least stable species reversibly interconverting with monomer [53]. To determine whether methylation affected nucleation rate, tau aggregation time course was quantified for both unmodified and methylated (5.5 mol/mol stoichiometry) tau in the presence of Thiazine red. Because homogeneous nucleation rate is influenced by the degree of supersaturation (i.e., the concentration of monomer above the critical concentration; [54]), the two tau preparations were normalized for supersaturation at the start of time course measurements. When quantified over 24 h, both time series displayed lag, exponential growth, and plateau phases (Fig. 6A). Reaction lag times, which vary inversely with nucleation rate [35], were then estimated by fitting data with a 3-parameter Gompertz growth function. Under these conditions, methylated tau aggregated with a significantly longer lag time relative to unmodified tau (Fig. 6B). These data show that Lys methylation slows the nucleation phase of the tau aggregation reaction. Overall, multi-site Lys methylation at 5.5 mol/mol can depress tau aggregation at both the nucleation and elongation steps without affecting microtubule polymerizing activity.
Figure 6. Lys methylation depresses tau filament nucleation rate.
(A) Either unmodified (solid circle) or 5.5 mol/mol methylated tau (hollow circle) was incubated at constant supersaturation (i.e., 0.3 µM above Kcrit) in the presence of Thiazine red inducer, then assayed for filament formation as a function of time. Each data point represents average filament lengths/field calculated from triplicate electron microscopy images ± SD whereas normalized curve (solid lines) represents best fit of data to a three parameter Gompertz growth function [71]. Values for lag time were estimated from these plots. (B) Replot of lag times determined from data in panel (A), where each bar represents the lag time ± propagated SEE. Methylation increased lag time relative to unmodified tau (***, p < 0.001).
DISCUSSION
These findings are significant in several respects. First, they confirm that normal human tau protein is extensively phosphorylated in vivo. At least 31 Ser and Thr residues distributed broadly across projection and assembly domains were found to be modified, with local hot spots lying mostly outside the MTBR. The distribution was consistent with previous studies of normal human brain, where substantial site overlap between normal and AD-derived tau was observed [11, 55]. Including the seven new sites discovered herein, the total number of phosphorylated residues identified by all methods in normal or diseased human brain tissue now stands at 52, representing well over half of all hydroxyl amino acids in tau. The overall distribution is consistent with tau interacting with phosphotransferases in a conformation where the majority of phosphorylatable residues are accessible.
Increases in tau phosphorylation site occupancy, especially at KXGS motifs, can reduce microtubule binding function [13, 38], and therefore may represent a key step in AD pathogenesis. However, considering that hyperphosphorylation occurs in fetal development [56] and hibernation [57] without filamentous aggregation, hyperphosphorylation of tau per se does not appear to dictate entry into the tau aggregation pathway. Rather, interaction with other modifications, such as those occurring on Lys residues, may antagonize or synergize with phosphorylation to drive aggregation and lesion formation. Thus a second significant finding here is that mono- and di-methylation represent the major physiological Lys modifications in normal brain. Much like phosphorylation sites, tau methylation sites were found distributed across both projection and assembly domains, consistent with broad exposure of Lys residues to modification enzymes, and offering opportunities for cross talk with other PTMs. In the projection domain, methylation sites K24 and K44 lie adjacent to putative caspase and calpain cleavage sites, respectively. The latter cleavage event is reportedly associated with toxicity in biological models [44, 58– 60]. In the MTBR, where the majority of Lys methylation was found, sites overlapped with three of five reported ubiquitylation sites on AD-derived filamentous tau. In addition, the Lys component of three out of four KXGS motifs (K259, K290, and K353) were occupied, with the fourth, K321, lying outside the area of coverage. Thus, Lys methylation sites are positioned to potentially influence tau metabolism through cross-talk with other PTMs.
Within the tau projection domain, K163, K174, and/or K180 have been reported as acetylation sites in vivo, with occupancy detectable in normal brain and increasing with Braak stage in AD brain [61]. Within the MTBR, two sites adjacent or within the PHF6* motif (K274 and K280; [62, 63]), and AMP-dependent protein kinase recognition sites K259 and K353 [22], were reported to be acetylated as well. Although on the basis of immunoblot analysis K280 [63] and K274 [62] were unoccupied in normal brain, occupancy of K259 and K353 was reportedly highest in normal brain and should have been detected by our LC-MS/MS approach along with K163, K174, and K180. Because each of these residues was resolved in our area of coverage, failure to detect tau acetylation may reflect poor preservation of this modification during purification. Nonetheless, it was possible to detect methylation at these Lys residues, raising the potential for direct cross talk with modifications such as acetylation.
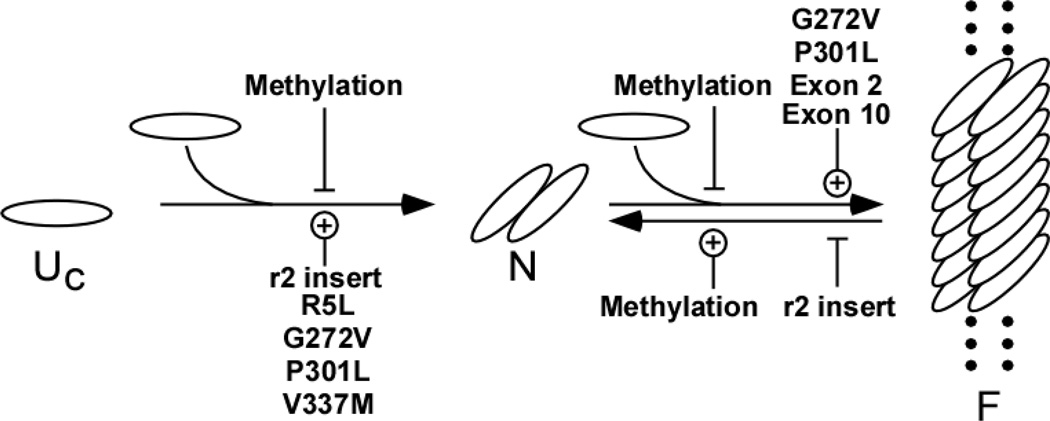
In addition to participation in cross talk, methylation of the tau assembly domain could directly affect tau biology. A third significant finding here was that mono- and di-methylation greatly suppressed tau aggregation propensity by depressing nucleation rate, inhibiting elongation rate, and destabilizing mature filaments (Fig. 7). Previously we reported that these same steps were augmented by exon 10 inclusion and certain missense tau mutations associated with frontotemporal dementia (Fig. 7). In contrast, little effect on tubulin assembly was noted until methylation stoichiometries reached very high levels. Thus, multi-site Lys methylation is a candidate modification for suppressing aggregation propensity while preserving tubulin assembly-promoting activity. This conclusion is based on the use of reductive methylation to prepare methylated tau. Because this method is non-specific, it allowed control over modification stoichiometry but not over site distribution. Although the sites modified in vivo and reductive methylation overlapped, the latter incorporated detectable levels of methylation into more sites. Therefore, the functional consequences of methylation should be considered tentative until it can be recapitulated with greater control over site specificity.
Figure 7. Effects of Lys methylation on tau aggregation pathway.
This nucleation-elongation scheme for tau aggregation was deduced on the basis of in vitro experimentation [32]. Once assembly competent tau species form (Uc), the rate-limiting step in tau fibrillization is dimer formation, which represents the thermodynamic nucleus (N). Following nucleation, extension occurs through further addition of assembly competent monomers to the filament (F) ends. Lys-methylation inhibits filament formation by slowing nucleation and extension rates, and by destabilizing mature filaments. Conversely, these same steps are reportedly augmented by missense mutants associated with frontotemporal dementia (R5L, G272V, P301L, and V337M; [72]) and by normal tau isoforms that include microtubule binding repeat r2 through alternative splicing of exon 10 [34].
The enzymes that catalyze tau methylation and dimethylation are unknown. Relative to the phosphotransferase field, the protein methylation field is young, with the first histone Lys methyltransferase and demethylase being reported only in 2000 and 2004, respectively [45, 64, 65]. Since that time, 208 putative methyltransferases have been identified in the human genome, with 57 of them containing SET domains associated with protein lysine methyltransferase activity [66]. Only a subset of these has been studied in depth, with most effort being directed toward nuclear enzymes owing to the importance of histone modification. Nonetheless, Lys methyltransferase activity has been found in the cytoplasm [67, 68]. Identification of tau methyltransferases will likely yield additional insights into the role of methylation in normal biology.
In the cell nucleus, Lys methylation of histones is a stable yet reversible mark that influences gene expression (reviewed in [69]). It remains to be seen whether the normal tau methylation pattern deduced herein is stable through development and aging. However, it does differ from the reported methylation pattern of PHF-tau isolated from AD brain [21]. With respect to methylation site distribution, PHF-tau contained fewer sites overall, with only three sites in the MTBR (Fig. 1B). Moreover, PHF-tau modification was limited to monomethylation, whereas normal tau contained dimethyl-Lys at eight of eleven identified sites. These data indicate that PHF-tau and normal soluble tau differ qualitatively with respect to Lys methylation. It will be important to determine whether the lower site frequency and bias toward monomethylation in PHF-tau is accompanied by lower site occupancy. If so, then increasing tau methylation may be a tractable approach for depressing tau aggregation and neurofibrillary lesion formation in AD.
In summary, the data identify Lys methylation as part of the normal modification signature of tau protein in human brain. In addition to indirect effects owing to PTM crosstalk, Lys methylation can directly affect tau biology by lowering aggregation propensity. It may also affect other aspects of tau biology not investigated here, such as the ability of tau to interact with its many known binding partners including signaling molecules and cytoskeletal elements (reviewed in [1]). We propose that multiple tau PTMs, including phosphorylation and methylation, act in concert to affect both normal and pathological functions of tau.
SUMMARY STATEMENT.
Diverse post-translational modifications regulate tau protein function and misfolding. Here we identified Lys methylation as a tau post-translational modification in normal human brain, and found it depressed tau aggregation propensity when modeled in vitro.
ACKNOWLEDGMENTS
We thank Dr. Douglas W. Scharre for access to the Buckeye Brain Bank and Dr. H. Ronald Zielke for access to the NICHD Brain and Tissue Bank for Developmental Disorders at the University of Maryland, Baltimore, MD (contract HHSN275200900011C, Ref. No. N01-HD-9-0011). Electron microscopy and CD resources were provided by the OSU Campus Microscopy and Imaging and Biophysical Interaction and Characterization Facilities, respectively. The mass spectrometry facility was supported by Maryland Cigarette Restitution Fund and National Cancer Institute Cancer Center Grant to Greenebaum Cancer Center, University of Maryland, Baltimore.
FUNDING
This research was supported by a grant from the National Institutes of Health (NS07741).
ABBREVIATIONS USED
- AD
Alzheimer disease
- LC-MS/MS
liquid chromatography-tandem mass spectrometry
- MTBR
microtubule binding region
- PHF
paired-helical filament
- PTM
post-translational modification
- XCorr
cross correlation value
Footnotes
AUTHOR CONTRIBUTIONS
Kristen Funk designed the experiments, purified tau from human brain, and performed microtubule assembly assays. Grace Cooper isolated recombinant tau. Kelsey Schafer performed tau aggregation experiments. Stefani Thomas, Zhongping Liao, David Clark, and Austin Yang collected and analyzed mass spectrometry data. Kristen Funk, Austin Yang, and Jeff Kuret wrote the manuscript.
REFERENCES
- 1.Morris M, Maeda S, Vossel K, Mucke L. The many faces of tau. Neuron. 2011;70:410–426. doi: 10.1016/j.neuron.2011.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brandt R, Leger J, Lee G. Interaction of tau with the neural plasma membrane mediated by tau's amino-terminal projection domain. J. Cell Biol. 1995;131:1327–1340. doi: 10.1083/jcb.131.5.1327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gustke N, Trinczek B, Biernat J, Mandelkow EM, Mandelkow E. Domains of tau protein and interactions with microtubules. Biochemistry. 1994;33:9511–9522. doi: 10.1021/bi00198a017. [DOI] [PubMed] [Google Scholar]
- 4.Spillantini MG, Goedert M. Tau pathology and neurodegeneration. Lancet Neurol. 2013;12:609–622. doi: 10.1016/S1474-4422(13)70090-5. [DOI] [PubMed] [Google Scholar]
- 5.Novak M, Kabat J, Wischik CM. Molecular characterization of the minimal protease resistant tau unit of the Alzheimer's disease paired helical filament. EMBO J. 1993;12:365–370. doi: 10.1002/j.1460-2075.1993.tb05665.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.von Bergen M, Friedhoff P, Biernat J, Heberle J, Mandelkow EM, Mandelkow E. Assembly of tau protein into Alzheimer paired helical filaments depends on a local sequence motif (306VQIVYK311) forming β structure. Proc. Natl. Acad. Sci. U. S. A. 2000;97:5129–5134. doi: 10.1073/pnas.97.10.5129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gomez-Isla T, Price JL, McKeel DW, Jr, Morris JC, Growdon JH, Hyman BT. Profound loss of layer II entorhinal cortex neurons occurs in very mild Alzheimer's disease. J. Neurosci. 1996;16:4491–4500. doi: 10.1523/JNEUROSCI.16-14-04491.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ghoshal N, Garcia-Sierra F, Wuu J, Leurgans S, Bennett DA, Berry RW, Binder LI. Tau conformational changes correspond to impairments of episodic memory in mild cognitive impairment and Alzheimer's disease. Exp. Neurol. 2002;177:475–493. doi: 10.1006/exnr.2002.8014. [DOI] [PubMed] [Google Scholar]
- 9.Martin L, Latypova X, Terro F. Post-translational modifications of tau protein: implications for Alzheimer's disease. Neurochem Int. 2011;58:458–471. doi: 10.1016/j.neuint.2010.12.023. [DOI] [PubMed] [Google Scholar]
- 10.Wang JZ, Xia YY, Grundke-Iqbal I, Iqbal K. Abnormal hyperphosphorylation of tau: sites, regulation, and molecular mechanism of neurofibrillary degeneration. J. Alzheimers Dis. 2013;33(Suppl 1):S123–S139. doi: 10.3233/JAD-2012-129031. [DOI] [PubMed] [Google Scholar]
- 11.Hanger DP, Byers HL, Wray S, Leung KY, Saxton MJ, Seereeram A, Reynolds CH, Ward MA, Anderton BH. Novel phosphorylation sites in tau from Alzheimer brain support a role for casein kinase 1 in disease pathogenesis. J. Biol. Chem. 2007;282:23645–23654. doi: 10.1074/jbc.M703269200. [DOI] [PubMed] [Google Scholar]
- 12.Hanger DP, Anderton BH, Noble W. Tau phosphorylation: the therapeutic challenge for neurodegenerative disease. Trends Mol. Med. 2009;15:112–119. doi: 10.1016/j.molmed.2009.01.003. [DOI] [PubMed] [Google Scholar]
- 13.Biernat J, Gustke N, Drewes G, Mandelkow EM, Mandelkow E. Phosphorylation of Ser262 strongly reduces binding of tau to microtubules: distinction between PHF-like immunoreactivity and microtubule binding. Neuron. 1993;11:153–163. doi: 10.1016/0896-6273(93)90279-z. [DOI] [PubMed] [Google Scholar]
- 14.Bramblett GT, Goedert M, Jakes R, Merrick SE, Trojanowski JQ, Lee VM. Abnormal tau phosphorylation at Ser396 in Alzheimer's disease recapitulates development and contributes to reduced microtubule binding. Neuron. 1993;10:1089–1099. doi: 10.1016/0896-6273(93)90057-x. [DOI] [PubMed] [Google Scholar]
- 15.Yoshida H, Ihara Y. Tau in paired helical filaments is functionally distinct from fetal tau: assembly incompetence of paired helical filament-tau. J. Neurochem. 1993;61:1183–1186. doi: 10.1111/j.1471-4159.1993.tb03642.x. [DOI] [PubMed] [Google Scholar]
- 16.Necula M, Kuret J. Site-specific pseudophosphorylation modulates the rate of tau filament dissociation. FEBS Lett. 2005;579:1453–1457. doi: 10.1016/j.febslet.2005.01.047. [DOI] [PubMed] [Google Scholar]
- 17.Petrucelli L, Dickson D, Kehoe K, Taylor J, Snyder H, Grover A, De Lucia M, McGowan E, Lewis J, Prihar G, Kim J, Dillmann WH, Browne SE, Hall A, Voellmy R, Tsuboi Y, Dawson TM, Wolozin B, Hardy J, Hutton M. CHIP and Hsp70 regulate tau ubiquitination, degradation and aggregation. Hum. Mol. Genet. 2004;13:703–714. doi: 10.1093/hmg/ddh083. [DOI] [PubMed] [Google Scholar]
- 18.Shimura H, Schwartz D, Gygi SP, Kosik KS. CHIP-Hsc70 complex ubiquitinates phosphorylated tau and enhances cell survival. J. Biol. Chem. 2004;279:4869–4876. doi: 10.1074/jbc.M305838200. [DOI] [PubMed] [Google Scholar]
- 19.Cripps D, Thomas SN, Jeng Y, Yang F, Davies P, Yang AJ. Alzheimer disease-specific conformation of hyperphosphorylated paired helical filament-Tau is polyubiquitinated through Lys-48, Lys-11, and Lys-6 ubiquitin conjugation. J. Biol. Chem. 2006;281:10825–10838. doi: 10.1074/jbc.M512786200. [DOI] [PubMed] [Google Scholar]
- 20.Morishima-Kawashima M, Hasegawa M, Takio K, Suzuki M, Titani K, Ihara Y. Ubiquitin is conjugated with amino-terminally processed tau in paired helical filaments. Neuron. 1993;10:1151–1160. doi: 10.1016/0896-6273(93)90063-w. [DOI] [PubMed] [Google Scholar]
- 21.Thomas SN, Funk KE, Wan Y, Liao Z, Davies P, Kuret J, Yang AJ. Dual modification of Alzheimer's disease PHF-tau protein by lysine methylation and ubiquitylation: a mass spectrometry approach. Acta Neuropathol. 2012;123:105–117. doi: 10.1007/s00401-011-0893-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cook C, Carlomagno Y, Gendron TF, Dunmore J, Scheffel K, Stetler C, Davis M, Dickson D, Jarpe M, Deture M, Petrucelli L. Acetylation of the KXGS motifs in tau is a critical determinant in modulation of tau aggregation and clearance. Hum. Mol. Genet. 2014;23:104–116. doi: 10.1093/hmg/ddt402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cohen TJ, Guo JL, Hurtado DE, Kwong LK, Mills IP, Trojanowski JQ, Lee VM. The acetylation of tau inhibits its function and promotes pathological tau aggregation. Nat Commun. 2011;2:252. doi: 10.1038/ncomms1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ksiezak-Reding H, Liu WK, Yen SH. Phosphate analysis and dephosphorylation of modified tau associated with paired helical filaments. Brain Res. 1992;597:209–219. doi: 10.1016/0006-8993(92)91476-u. [DOI] [PubMed] [Google Scholar]
- 25.Necula M, Kuret J. A static laser light scattering assay for surfactant-induced tau fibrillization. Anal Biochem. 2004;333:205–215. doi: 10.1016/j.ab.2004.05.044. [DOI] [PubMed] [Google Scholar]
- 26.Carmel G, Mager EM, Binder LI, Kuret J. The structural basis of monoclonal antibody Alz50's selectivity for Alzheimer's disease pathology. J Biol Chem. 1996;271:32789–32795. doi: 10.1074/jbc.271.51.32789. [DOI] [PubMed] [Google Scholar]
- 27.Geoghegan KF. Modification of amino groups. Curr Protoc Protein Sci. 2001;Chapter 15(Unit 15.2) doi: 10.1002/0471140864.ps1502s04. [DOI] [PubMed] [Google Scholar]
- 28.Jentoft N, Dearborn DG. Labeling of proteins by reductive methylation using sodium cyanoborohydride. J. Biol. Chem. 1979;254:4359–4365. [PubMed] [Google Scholar]
- 29.Yin H, Laguna KA, Li G, Kuret J. Dysbindin structural homologue CK1BP is an isoform-selective binding partner of human casein kinase-1. Biochemistry. 2006;45:5297–5308. doi: 10.1021/bi052354e. [DOI] [PubMed] [Google Scholar]
- 30.Bandyopadhyay B, Li G, Yin H, Kuret J. Tau aggregation and toxicity in a cell culture model of tauopathy. J Biol Chem. 2007;282:16454–16464. doi: 10.1074/jbc.M700192200. [DOI] [PubMed] [Google Scholar]
- 31.Mitchison T, Kirschner M. Microtubule assembly nucleated by isolated centrosomes. Nature. 1984;312:232–237. doi: 10.1038/312232a0. [DOI] [PubMed] [Google Scholar]
- 32.Congdon EE, Kim S, Bonchak J, Songrug T, Matzavinos A, Kuret J. Nucleation-dependent tau filament formation: the importance of dimerization and an estimation of elementary rate constants. J. Biol. Chem. 2008;283:13806–13816. doi: 10.1074/jbc.M800247200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kristofferson D, Karr TL, Purich DL. Dynamics of linear protein polymer disassembly. J. Biol. Chem. 1980;255:8567–8572. [PubMed] [Google Scholar]
- 34.Zhong Q, Congdon EE, Nagaraja HN, Kuret J. Tau isoform composition influences rate and extent of filament formation. J. Biol. Chem. 2012;287:20711–20719. doi: 10.1074/jbc.M112.364067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Evans KC, Berger EP, Cho CG, Weisgraber KH, Lansbury PT., Jr Apolipoprotein E is a kinetic but not a thermodynamic inhibitor of amyloid formation: implications for the pathogenesis and treatment of Alzheimer disease. Proc. Natl. Acad. Sci. U. S. A. 1995;92:763–767. doi: 10.1073/pnas.92.3.763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wischik CM, Novak M, Thogersen HC, Edwards PC, Runswick MJ, Jakes R, Walker JE, Milstein C, Roth M, Klug A. Isolation of a fragment of tau derived from the core of the paired helical filament of Alzheimer disease. Proc. Natl. Acad. Sci. U. S. A. 1988;85:4506–4510. doi: 10.1073/pnas.85.12.4506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li W, Lee VM. Characterization of two VQIXXK motifs for tau fibrillization in vitro. Biochemistry. 2006;45:15692–15701. doi: 10.1021/bi061422+. [DOI] [PubMed] [Google Scholar]
- 38.Yoshida H, Goedert M. Phosphorylation of microtubule-associated protein tau by AMPK-related kinases. J. Neurochem. 2012;120:165–176. doi: 10.1111/j.1471-4159.2011.07523.x. [DOI] [PubMed] [Google Scholar]
- 39.Goode BL, Denis PE, Panda D, Radeke MJ, Miller HP, Wilson L, Feinstein SC. Functional interactions between the proline-rich and repeat regions of tau enhance microtubule binding and assembly. Mol. Biol. Cell. 1997;8:353–365. doi: 10.1091/mbc.8.2.353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Goode BL, Feinstein SC. Identification of a novel microtubule binding and assembly domain in the developmentally regulated inter-repeat region of tau. J. Cell Biol. 1994;124:769–782. doi: 10.1083/jcb.124.5.769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Trinczek B, Biernat J, Baumann K, Mandelkow EM, Mandelkow E. Domains of tau protein, differential phosphorylation, and dynamic instability of microtubules. Mol. Biol. Cell. 1995;6:1887–1902. doi: 10.1091/mbc.6.12.1887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Guo H, Albrecht S, Bourdeau M, Petzke T, Bergeron C, LeBlanc AC. Active caspase-6 and caspase-6-cleaved tau in neuropil threads, neuritic plaques, and neurofibrillary tangles of Alzheimer's disease. Am. J. Pathol. 2004;165:523–531. doi: 10.1016/S0002-9440(10)63317-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rohn TT, Rissman RA, Davis MC, Kim YE, Cotman CW, Head E. Caspase-9 activation and caspase cleavage of tau in the Alzheimer's disease brain. Neurobiol. Dis. 2002;11:341–354. doi: 10.1006/nbdi.2002.0549. [DOI] [PubMed] [Google Scholar]
- 44.Park SY, Ferreira A. The generation of a 17 kDa neurotoxic fragment: an alternative mechanism by which tau mediates beta-amyloid-induced neurodegeneration. J. Neurosci. 2005;25:5365–5375. doi: 10.1523/JNEUROSCI.1125-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Borch RF, Bernstein MD, Durst HD. Cyanohydridoborate Anion as a Selective Reducing Agent. J. Am. Chem. Soc. 1971;93:2897–2901. [Google Scholar]
- 46.Means GE, Feeney RE. Reductive alkylation of amino groups in proteins. Biochemistry. 1968;7:2192–2201. doi: 10.1021/bi00846a023. [DOI] [PubMed] [Google Scholar]
- 47.Weingarten MD, Lockwood AH, Hwo SY, Kirschner MW. A protein factor essential for microtubule assembly. Proc Natl Acad Sci U S A. 1975;72:1858–1862. doi: 10.1073/pnas.72.5.1858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Witman GB, Cleveland DW, Weingarten MD, Kirschner MW. Tubulin requires tau for growth onto microtubule initiating sites. Proc Natl Acad Sci U S A. 1976;73:4070–4074. doi: 10.1073/pnas.73.11.4070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Esmaeli-Azad B, McCarty JH, Feinstein SC. Sense and antisense transfection analysis of tau function: tau influences net microtubule assembly, neurite outgrowth and neuritic stability. J Cell Sci. 1994;107(Pt 4):869–879. doi: 10.1242/jcs.107.4.869. [DOI] [PubMed] [Google Scholar]
- 50.Drubin DG, Kirschner MW. Tau protein function in living cells. J Cell Biol. 1986;103:2739–2746. doi: 10.1083/jcb.103.6.2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cho JH, Johnson GV. Glycogen synthase kinase 3beta phosphorylates tau at both primed and unprimed sites. Differential impact on microtubule binding. J. Biol. Chem. 2003;278:187–193. doi: 10.1074/jbc.M206236200. [DOI] [PubMed] [Google Scholar]
- 52.Chirita CN, Congdon EE, Yin H, Kuret J. Triggers of full-length tau aggregation: a role for partially folded intermediates. Biochemistry. 2005;44:5862–5872. doi: 10.1021/bi0500123. [DOI] [PubMed] [Google Scholar]
- 53.Ferrone F. Analysis of protein aggregation kinetics. Methods Enzymol. 1999;309:256–274. doi: 10.1016/s0076-6879(99)09019-9. [DOI] [PubMed] [Google Scholar]
- 54.Fesce R, Benfenati F, Greengard P, Valtorta F. Effects of the neuronal phosphoprotein synapsin I on actin polymerization. II. Analytical interpretation of kinetic curves. J. Biol. Chem. 1992;267:11289–11299. [PubMed] [Google Scholar]
- 55.Matsuo ES, Shin RW, Billingsley ML, Van deVoorde A, O'Connor M, Trojanowski JQ, Lee VM. Biopsy-derived adult human brain tau is phosphorylated at many of the same sites as Alzheimer's disease paired helical filament tau. Neuron. 1994;13:989–1002. doi: 10.1016/0896-6273(94)90264-x. [DOI] [PubMed] [Google Scholar]
- 56.Seubert P, Mawal-Dewan M, Barbour R, Jakes R, Goedert M, Johnson GV, Litersky JM, Schenk D, Lieberburg I, Trojanowski JQ, et al. Detection of phosphorylated Ser262 in fetal tau, adult tau, and paired helical filament tau. J Biol Chem. 1995;270:18917–18922. doi: 10.1074/jbc.270.32.18917. [DOI] [PubMed] [Google Scholar]
- 57.Arendt T, Stieler J, Strijkstra AM, Hut RA, Rudiger J, Van der Zee EA, Harkany T, Holzer M, Hartig W. Reversible paired helical filament-like phosphorylation of tau is an adaptive process associated with neuronal plasticity in hibernating animals. J. Neurosci. 2003;23:6972–6981. doi: 10.1523/JNEUROSCI.23-18-06972.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Amadoro G, Ciotti MT, Costanzi M, Cestari V, Calissano P, Canu N. NMDA receptor mediates tau-induced neurotoxicity by calpain and ERK/MAPK activation. Proc. Natl. Acad. Sci. U. S. A. 2006;103:2892–2897. doi: 10.1073/pnas.0511065103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Canu N, Dus L, Barbato C, Ciotti MT, Brancolini C, Rinaldi AM, Novak M, Cattaneo A, Bradbury A, Calissano P. Tau cleavage and dephosphorylation in cerebellar granule neurons undergoing apoptosis. J. Neurosci. 1998;18:7061–7074. doi: 10.1523/JNEUROSCI.18-18-07061.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Reinecke JB, DeVos SL, McGrath JP, Shepard AM, Goncharoff DK, Tait DN, Fleming SR, Vincent MP, Steinhilb ML. Implicating calpain in tau-mediated toxicity in vivo. PLoS One. 2011;6:e23865. doi: 10.1371/journal.pone.0023865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Min SW, Cho SH, Zhou Y, Schroeder S, Haroutunian V, Seeley WW, Huang EJ, Shen Y, Masliah E, Mukherjee C, Meyers D, Cole PA, Ott M, Gan L. Acetylation of tau inhibits its degradation and contributes to tauopathy. Neuron. 2010;67:953–966. doi: 10.1016/j.neuron.2010.08.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Grinberg LT, Wang X, Wang C, Sohn PD, Theofilas P, Sidhu M, Arevalo JB, Heinsen H, Huang EJ, Rosen H, Miller BL, Gan L, Seeley WW. Argyrophilic grain disease differs from other tauopathies by lacking tau acetylation. Acta Neuropathol. 2013;125:581–593. doi: 10.1007/s00401-013-1080-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Irwin DJ, Cohen TJ, Grossman M, Arnold SE, Xie SX, Lee VM, Trojanowski JQ. Acetylated tau, a novel pathological signature in Alzheimer's disease and other tauopathies. Brain. 2012;135:807–818. doi: 10.1093/brain/aws013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Rea S, Eisenhaber F, O'Carroll D, Strahl BD, Sun ZW, Schmid M, Opravil S, Mechtler K, Ponting CP, Allis CD, Jenuwein T. Regulation of chromatin structure by site-specific histone H3 methyltransferases. Nature. 2000;406:593–599. doi: 10.1038/35020506. [DOI] [PubMed] [Google Scholar]
- 65.Shi Y, Lan F, Matson C, Mulligan P, Whetstine JR, Cole PA, Casero RA. Histone demethylation mediated by the nuclear amine oxidase homolog LSD1. Cell. 2004;119:941–953. doi: 10.1016/j.cell.2004.12.012. [DOI] [PubMed] [Google Scholar]
- 66.Petrossian TC, Clarke SG. Uncovering the human methyltransferasome. Mol. Cell. Proteomics. 2011;10 doi: 10.1074/mcp.M110.000976. M110 000976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Su IH, Dobenecker MW, Dickinson E, Oser M, Basavaraj A, Marqueron R, Viale A, Reinberg D, Wulfing C, Tarakhovsky A. Polycomb group protein ezh2 controls actin polymerization and cell signaling. Cell. 2005;121:425–436. doi: 10.1016/j.cell.2005.02.029. [DOI] [PubMed] [Google Scholar]
- 68.Tee WW, Pardo M, Theunissen TW, Yu L, Choudhary JS, Hajkova P, Surani MA. Prmt5 is essential for early mouse development and acts in the cytoplasm to maintain ES cell pluripotency. Genes Dev. 2010;24:2772–2777. doi: 10.1101/gad.606110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Black JC, Van Rechem C, Whetstine JR. Histone lysine methylation dynamics: establishment, regulation, and biological impact. Mol. Cell. 2012;48:491–507. doi: 10.1016/j.molcel.2012.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Goedert M, Spillantini MG, Potier MC, Ulrich J, Crowther RA. Cloning and sequencing of the cDNA encoding an isoform of microtubule-associated protein tau containing four tandem repeats: differential expression of tau protein mRNAs in human brain. EMBO J. 1989;8:393–399. doi: 10.1002/j.1460-2075.1989.tb03390.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Necula M, Kuret J. Electron microscopy as a quantitative method for investigating tau fibrillization. Anal. Biochem. 2004;329:238–246. doi: 10.1016/j.ab.2004.02.023. [DOI] [PubMed] [Google Scholar]
- 72.Chang E, Kim S, Yin H, Nagaraja HN, Kuret J. Pathogenic missense MAPT mutations differentially modulate tau aggregation propensity at nucleation and extension steps. J. Neurochem. 2008;107:1113–1123. doi: 10.1111/j.1471-4159.2008.05692.x. [DOI] [PMC free article] [PubMed] [Google Scholar]