Abstract
Repair of DNA double-strand breaks via homologous recombination can produce double Holliday junctions (dHJs) that require enzymatic separation. Topoisomerase IIIα (TopIIIα) together with RMI1 disentangles the final hemicatene intermediate obtained once dHJs have converged. How binding of RMI1 to TopIIIα influences it to behave as a hemicatenane dissolvase, rather than as an enzyme that relaxes DNA topology, is unknown. Here, we present the crystal structure of human TopIIIα complexed to the first oligonucleotide binding domain (OB-fold) of RMI1. TopIIIα assumes a toroidal type 1A topoisomerase fold. RMI1 attaches to the edge of the gate in TopIIIα through which DNA passes. RMI1 projects a 23-residue loop into the TopIIIα gate, thereby influencing the dynamics of its opening and closing. Our results provide a mechanistic rationale for how RMI1 stabilizes TopIIIα gate opening to enable dissolution and illustrate how binding partners modulate topoisomerase function.
Introduction
Homologous recombination (HR) is a central pathway for the repair of DNA double-strand breaks and single-stranded gaps, and is required for the maintenance and restart of stalled replication forks1,2. HR uses a homologous DNA sequence as a template for the repair of the damaged DNA strand. In canonical HR, following DNA synthesis and ligation, a double Holliday junction (dHJ) intermediate is produced, which typically interconnects two sister chromatids3,4. These dHJ intermediates are processed, in either a crossover or a non-crossover fashion5–7 to produce recombinant products. Several nucleases are able to cleave the symmetrical Holliday junction, yielding an equal number of crossover or non-crossover products. Mitotic crossovers, however, carry the risk of loss of heterozygosity (LOH), a known driver of oncogenic transformation8,9. In mitotic cells, LOH appears to be rare, contributing to only 2.5% of HR mediated double-strand breakage repair events10. Alternative pathways must therefore exist to bias HR events towards a non-crossover outcome.
The DNA molecules within the dHJ intermediate are topologically linked. The disjunction of the entangled chromosomes can be accomplished through the duplex unwinding activity of a specific helicase, coupled to DNA strand unlinking by a type 1A topoisomerase. The interplay between RecQ helicases and topoisomerases, which is conserved in all kingdoms of life, exclusively generates non-crossover products11 via a process termed dHJ dissolution. In humans, TopIIIα acts with BLM11, a RecQ helicase that is mutated in the cancer predisposition disorder Bloom’s syndrome. The yeast counterparts of these proteins are Top3 topoisomerase and Sgs1 helicase12–14. While Sgs1(BLM) and Top3(TopIIIα) are required to process various topologically linked substrates15,16, they act in concert with an additional essential component; Rmi1, in yeast17,18, and the RMI1-RMI2 complex in human cells19–21. Together, the RecQ helicase, Top3(TopIIIα) and Rmi1(RMI1) proteins form the minimal dHJ dissolvasome. Loss or mutation of any component of this complex results in genomic instability18,22,23.
dHJ dissolution requires a branch migration step that collapses the two HJs into a hemi-catenane, followed by a decatenation step that unhooks the hemi-catenane intermediate into two separate DNA duplexes24. In yeast, where the reaction has been biochemically dissected in detail25–27 convergence of the two HJs is driven by Sgs1 in an ATP dependent process. Top3 provides the enzymatic unlinking activity for helicase mediated dHJ migration, and, together with Rmi1, unhooks the final hemi-catene25,26.
TopIIIα is a member of the type 1A family of topoisomerases, which form a toroidal structure and introduce a transient single stranded nick in one DNA strand (cut strand: C-strand), allowing a second ssDNA (single strand DNA) strand to be transferred reversibly through the nick (transfer strand: T-strand). When T- and C-strands belong to separate duplexes the outcome is decatenation or catenation (hemicatenation if only one strand is moved), or in case of a HJ substrate dHJ dissolution or formation. Should the T- and C-strands belong to the same, plectonemically-linked duplex, relaxation then occurs. The -in and out-movement of a DNA single-strand (T-strand) is made possible by a gate of intertwined β-sheets allowing reversible opening and closing of the torus28. In this context, the C-strand is cut in a reversible transesterification process giving rise to a 5’-phosphotyrosine ssDNA intermediate, with the T-strand being passed through the nick29. Top3 and TopIIIα, in isolation, are moderately effective in relaxing negatively supercoiled plasmids, but do possess modest ssDNA decatenases activity30. In yeast as in human21, efficient decatenation requires the presence of Rmi1, which also serves to inhibit the relaxation activity25. Biochemical analysis of this regulatory behavior led to the discovery that Rmi1 increases the steady-state level of the so-called open complex of Top3 and suggested a model where Rmi1 stabilizes the open conformation of the topoisomerase gate25. The structural basis by which RMI1(Rmi1) modulate gating of strand passage by TopIIIα(Top3) and bestow efficient decatenation functionality that allows dHJ dissolution has remained elusive.
We set out to define how RMI1(Rmi1) modulates TopIIIα(Top3) function using structural and biochemical approaches. We show that RMI1 modulates the dynamics of the topoisomerase action through a loop lining the topoisomerase gate.
Results
Construct design of the TopIIIα–RMI1complex
Human TopIIIα and yeast Top3 carry distinct sequence signatures that distinguish them from their prokaryotic Top3 counterparts (Supplementary fig. 1,2). In the absence of structural information, it is unclear to what extent the eukaryotic TopIIIα(Top3) decatenases have diverged from their prokaryotic counterparts, and how RMI1 modulates topoisomerase function by promoting decatenase activity. In an effort to address these questions, we determined the structure of the TopIIIα-RMI1 complex. We obtained crystals of the human TopIIIα-RMI1 complex were obtained after 4–5 days for human TopIIIα (residues 1 to 753 following deletion of residues 754 to 1001) in complex with human RMI1 (DUF1767+OBN: residues 1 to 219) (fig. 1a, b) in the presence of Mg2+ (200 mM MgCl2) or Ca2+ (200 mM calcium citrate) and belonged to space group P41212 with one heterodimer in the crystallographic asymmetric unit (Supplementary fig. 3). The TopIIIα (1–753) and RMI1 (1–219) constructs used in crystallization, when tested together with BLM, remained active in dHJ dissolution (Supplementary fig. 4a) arguing that these boundaries are sufficient for dissolution. The structure of Mg2+-bound TopIIIα in complex with RMI1 (TopIIIα-Mg2+-RMI1) was solved by molecular replacement, and refined to a maximal resolution of 2.85 Å. The model has good agreement with the diffraction data (Rwork = 19.7%, Rfree = 23.1%) and excellent validation statistics. The structure of a crystal grown in the presence of Ca2+, TopIIIα-Ca2+-RMI1, was also determined at 3.25 Å resolution (Rwork = 19.7%, Rfree = 22.5%) and has similar validation statistics. The domain boundaries for the human TopIIIα-RMI1 (OBN) complex used for crystallization are equivalent to those of full-length S. cerevisiae Top3-Rmi1. Human RMI1 encompasses a DUF1767 three α-helix bundle followed by an N- (OBN) and C- terminal (OBC) oligo-nucleotide binding domain17,20 (fig. 1a, b). OBC, which is absent in yeast, is responsible for RMI2 binding in the human complex31,32. Deletion of human RMI1-OBC has no adverse effect on dHJ dissolution in vitro making TopIIIα-BLM-RMI1 (DUF1767+OBN) the minimal functional dissolvasome complex32.
Figure 1. Overall architecture and TopIIIα-RMI1 binding interface.
a) Domain organization of the human TopIIIα-RMI1 complex. b) Overall architecture of the crystallized complex: (I) Toprim domain (cyan); (II) gate domain with the two topo-fold domains (green); (III) catalytic 5Y-CAP (blue), (IV) non-catalytic CAP domain (grey) with RMI1 shown in red. The catalytic metal is represented by a red sphere. c) Hydrophobic zipper between RMI1 residues originating from OBN and residues originating from the α12 and β10–α12 gate of TopIIIα. d) As in c) but depicting the corresponding hydrogen bonding network. e) Interaction between the RMI1 loop segment and the hydrophobic patch of the TopIIIα gate (helix α19) (near proposed pivot point see Fig 5). RMI1 P98 is engaged in hydrophobic stacking with TopIIIα residues F94, neighboring RMI1 Y100 stacks with TopIIIα F262. f) similar to e) highlighting the corresponding hydrogen-bonding and salt bridge network. RMI1 Q113 is at the center of a hydrogen-bonding network binding TopIIIα E254, H530 and K529, while ionic interaction between RMI1 E119 and TopIIIα R338 lock down the RMI1 insertion loop in position.
Overview of the TopIIIα-RMI1 complex
The overall shape of the TopIIIα–RMI1 complex resembles a brass doorknocker (fig. 1b), with TopIIIα forming the ring and RMI1 acting as its base. In the crystal, RMI1 makes distinct interactions with two TopIIIα molecules, which let us to define the biological TopIIIα-Rmi1 unit by single particle electron microscopy (EM) as depicted in fig. 1b (Supplementary fig. 3).
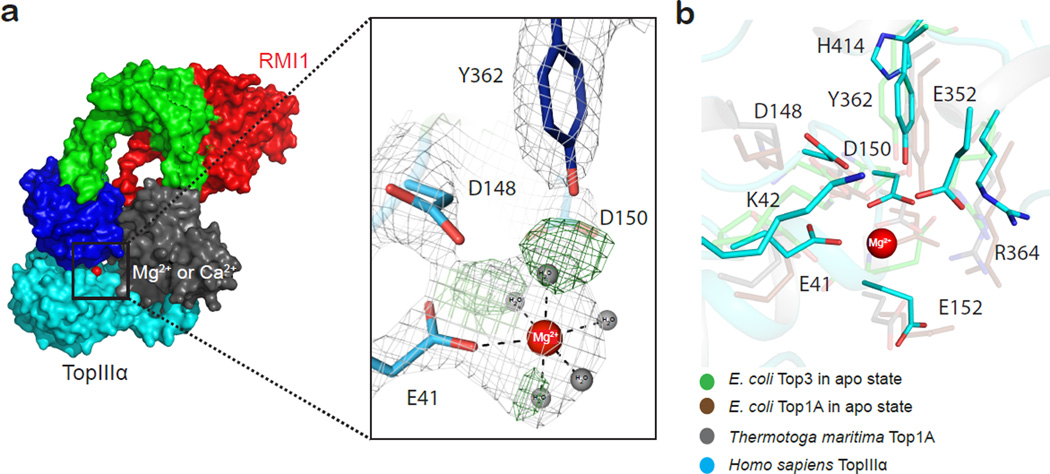
Human TopIIIα displays the canonical toroidal topoisomerase shape found in all type 1A family members solved to date (fig. 1b)33,34,35. Members of this family comprise five characteristic domains: domain I, or toprim (topoisomerase-primase), harbors an acidic cluster (residues D148, D150 and E152) that binds the metal ion cofactor required for C-strand cleavage28,36; domain II creates the ~26 Å in diameter topoisomerase gate formed with two antiparallel topo-fold domains37; domains III forms winged-helix folds38,39 and carries the catalytic tyrosine (Y362) needed for ssDNA cleavage and formation of the 5’-phosphotyrosine ssDNA intermediate29; domain IV shares the same winged-helix fold with domain III; domain V in the C-terminal region (residues 655 to 1001) includes a predicted zinc finger motif (residues 655 to 694). While domains I to IV are well defined in our structure, density for the N-terminal part of domain V (residues 638 to 753) could not be unambiguously assigned and has been omitted from the model. No detectable Zn2+ anomalous signal was observed (Unpublished data; N.B and R.D.B).
RMI1 binds the toroidal TopIIIα structure via its OBNβ-barrel domain at a site diametrically opposite to the toprim ssDNA binding site, with the RMI1 N-terminus projecting away from the topoisomerase. RMI1 attaches to the top of the topoisomerase (domain II) gate, utilizing an interface frequently used by other OB-fold containing proteins when binding protein ligands, sugars or oligonucleotides40 (fig. 1c,d). RMI1 projects a large insertion loop, located between β1–β2, into the central gate of the topoisomerase (fig. 1b,e,f, Supplementary fig. 4b,c). In the structure of isolated RMI1, this loop was found partially disordered, and shown to be involved in TopIIIα binding32. The RMI1-TopIIIα complex arrangement places the RMI1 insertion loop which is not stabilized by crystal contacts at the center of the topoisomerase gate (Supplementary fig. 4c).
Mechanism of single-strand DNA cleavage
TopIIIα showed a strong positive mFo-DFc difference electron density at the toprim acidic residues cluster (fig. 2a) and an anomalous signal peak at the same location corresponding to one calcium ion in TopIIIα-Ca2+-RMI1. We note that Mg2+ and Ca2+ permitted crystallization and support catalysis41, but neither Zn2+or Cs+ allowed crystallization. Due to the Mg2+ dependence of DNA relaxation, the coordination chemistry observed and the proximity of the density to the acidic cluster, we assigned the mFo-DFc electron density as Mg2+ in TopIIIα-Mg2+-RMI1 (Ca2+ in TopIIIα-Ca2+-RMI1). The metal-ion is octahedrally coordinated by five water molecules and one carboxylic oxygen of E41. The hydroxyl group of Y362 and carboxylic oxygen atoms of E352 point towards the water molecules, well located to potentially form hydrogen bonds. D150 provide additional hydrogen bonds to bridging water molecules within the hydration shell (fig. 2a). E352, conserved in eukaryotic orthologues (fig. 2b and Supplementary fig. 1 and 2a) is not part of the previously identified acidic cluster in Escherichia coli topoisomerases, where it is substituted by a glutamine. Key residues serving comparable roles in eukaryotic and prokaryotic type IA enzymes include R364, which is involved in pentavalent phosphate transition-state stabilization, and H414, which is implicated in anchoring cut ssDNA to the (−1) position42,43 (fig. 2b).
Figure 2. Catalytic site of human TopIIIα.
a) TopIIIα catalytic site residues and the Mg2+ binding site. Mg2+ is shown as a red sphere, with corresponding water molecules depicted as grey spheres. Simulated annealing composite 2mFo-DFc omit electron density map contoured at 1.5σ (gray). The omit difference map shows peaks (green) for water molecules prior to their inclusion into the model. E41 serves as the sole residue directly contacting Mg2+ while D150 bridges a water molecule from the hydration shell. b) Structural alignment of different catalytic centers taken from the available type IA topoisomerases structures: E. coli Top3 in apo state (PDB code 1D6M35) in cyan, E. coli Top1A in the apo state (PDB code 1ECL33) in magenta, Top1A from Thermotoga maritima (PDB code 2GAI34) in grey, with human TopIIIα (pdb 4CGY) shown in green.
As previous structures of prokaryotic type 1A topoisomerases did not show signs of bound catalytic metal ion, the TopIIIα-RMI1 structure now provides insight into type 1A metal mediated ssDNA cleavage. We find one Mg2+ or Ca2+ ion in our structures, with no indication of further binding sites. Instead, a lysine residue (K42), conserved within the topoisomerase type 1A family (fig. 2b) is situated at the site equivalent to the second metal-ion in TopII44. K42 is ideally positioned to stabilize the negatively charged pentavalent phosphate transition state arising during transesterification, thus serving a functionally equivalent role to the second metal ion. Our findings, therefore, favor a one-metal catalytic mechanism within the type 1A topoisomerase family, as proposed previously44.
TopIIIα resembles a prokaryotic type1A relaxase
TopIIIα domains I, III and IV share close structural similarity with the Top1A relaxases from E. coli and Thermatoga maritima (rmsd: 1.6 Å over 480 Cα positions, Supplementary fig. 1 and 2a), while the similarity with the E. coli Top3 decatenase is less pronounced (rmsd: 2.7 Å) (fig. 3a). The most apparent differences between TopIIIα and the prokaryotic Top3 decatenases (encoded by topB), however, are the absence of two loops equivalent to E. coli 241–255 (E. coli Top3 numbering: domain II) and 502–519 (domain IV), both lining the central cavity of the topoisomerase (fig. 3a). The latter has been implicated in decatenation in the prokaryotic Top3 family (and is henceforth referred to as the decatenation loop)35,45. Thus TopIIIα, at the structural level, has close resemblance to a relaxase, and does not display obvious structural features characteristic of prokaryotic type 1A decatenases.
Figure 3. The RMI1 decatenation loop.
a) Comparison between E. coli Top3 (shaded grey) and human TopIIIα (green). RMI1 (shaded red) inserts motifs at sites where E. coli Top3 diverges from human TopIIIα. The two unique E. coli Top3 loop segments shown are the loop between residues 241 and 255, and the decatenation loop (502–519). b) The final TOPO-Ca2+-RMI1 model overlaid with an unbiased 2mFo-DFc electron density envelope calculated prior to modeling the RMI1 decatenation loop, contoured at 1σ. c) Sequence alignments between different yeast strains and the human RMI1. Region in the black box within the S. cerevisiae sequence have been randomized in the rlRmi1 construct (see Methods).
The RMI1-TopIII interface
RMI1 OBN attaches to the TopIIIα gate and acts as the major binding interface, occupying a total surface area of 1565 Å2. This interface is mainly hydrophobic in nature with alkyl chains from RMI1 L177, L181 (β4–β5A loop), V179 (β5A strand), the aromatic ring of Y151 (β3–β4 loop) and methionines M134, M136 and M149 (αI, β2 strand and β3 strands respectively) forming a hydrophobic zipper with TopIIIα L302, L297, V298, L299 and A295 on α12, and L290 on the β10–α12 loop (fig. 1c). These residues form a hydrophobic groove that accommodates TopIIIα helix α12. RMI1 OBN-domain residues K78 and R176 (β4–β5A) form a bifurcated salt bridge with TopIIIα E305 and further stabilize binding (fig. 1d).
The RMI1 insertion loop inserts into the TopIIIα gate
The RMI1 loop, residues 94 to 134, which is inserted between RMI1 strands β1 and β2, lines the gate of the topoisomerase (fig. 1b, 3b). In the two TopIIIα-RMI1 complex structures, the loop differs slightly: while being less ordered in TopIIIα-Mg2+-RMI1 (Supplementary fig. 4c, d and e) with an apparent discontinuity of the electron density around residues 120 and 131, the RMI1 insertion loop can be fully traced in the electron density maps obtained with the TopIIIα-Ca2+-RMI1 data (fig. 3b). Hence, the TopIIIα-Ca2+-RMI1 complex is subsequently be used as the basis for discussion. The RMI1 insertion loop emanates from β1 (D94), and protrudes into the TopIIIα gate creating two layers of aligned and parallel rings (fig. 1b). Residues from the insertion loop of RMI1 form tight intercalated hydrophobic stacking interactions with the TopIIIα gate. Most prominently RMI1 P98 and Y100, stack against TopIIIα F94, F262 and V263, respectively (fig. 1e, f). This region in TopIIIα, by structural analogy to the E. coli topoisomerase 1, is expected to serve as pivot-point for gate opening46.
In the conformational state trapped in the crystal, the RMI1 insertion loop restricts the size of the central opening in the TopIIIα gate from 25 Å to 13 Å. As the ssDNA has to be moved into the central cavity through an opening between domains I and III, the topoisomerase gate is effectively closed when occupied by the RMI1 insertion loop (fig. 1b).
When examining the structure of the TopIIIα–RMI1 complex, we note that in regions where the TopIIIα structure diverges from that of the E. coli Top3 decatenase, RMI1 donates structural motifs in trans (fig. 3a, b). We therefore focused on the RMI1 insertion loop, examining whether decatenation ability could principally be provided by RMI1 structural motifs.
Decatenation and dHJ dissolution in vitro
To functionally dissect the contribution of the RMI1 insertion loop within the TopIIIα-RMI1 complex we switched to yeast as a model system, as the yeast Top3-Rmi1 mediated dissolution reaction has been biochemically dissected in detail25,26. The S. cerevisiae Rmi1 retains the overall domain organization of the metazoan orthologs (DUF1767 + OBN + insertion loop) (fig. 3c and Supplementary fig. 2), with the corresponding insertion loop segment extended by ca. 39 residues relative to the 23 residues present in the human RMI1 (fig. 3c).
We first tested whether the S. cerevisiae Rmi1 decatenation loop could be deleted, or excised using flanking tobacco etch virus protease (TEV) recognition sites. All constructs tested resulted in Rmi1 derivatives with limited solubility and defective Top3 binding. We then replaced the 58 amino acid yeast Rmi1 insertion loop (residues D87 to V145) with 23 randomly scrambled residues taken from the human RMI1 loop (fig. 3c). We found that this Rmi1 construct, designated randomized loop (rlRmi1) is proficient in yeast Top3 binding (Supplementary fig. 5a) and forms the basis for subsequent biochemical experiments.
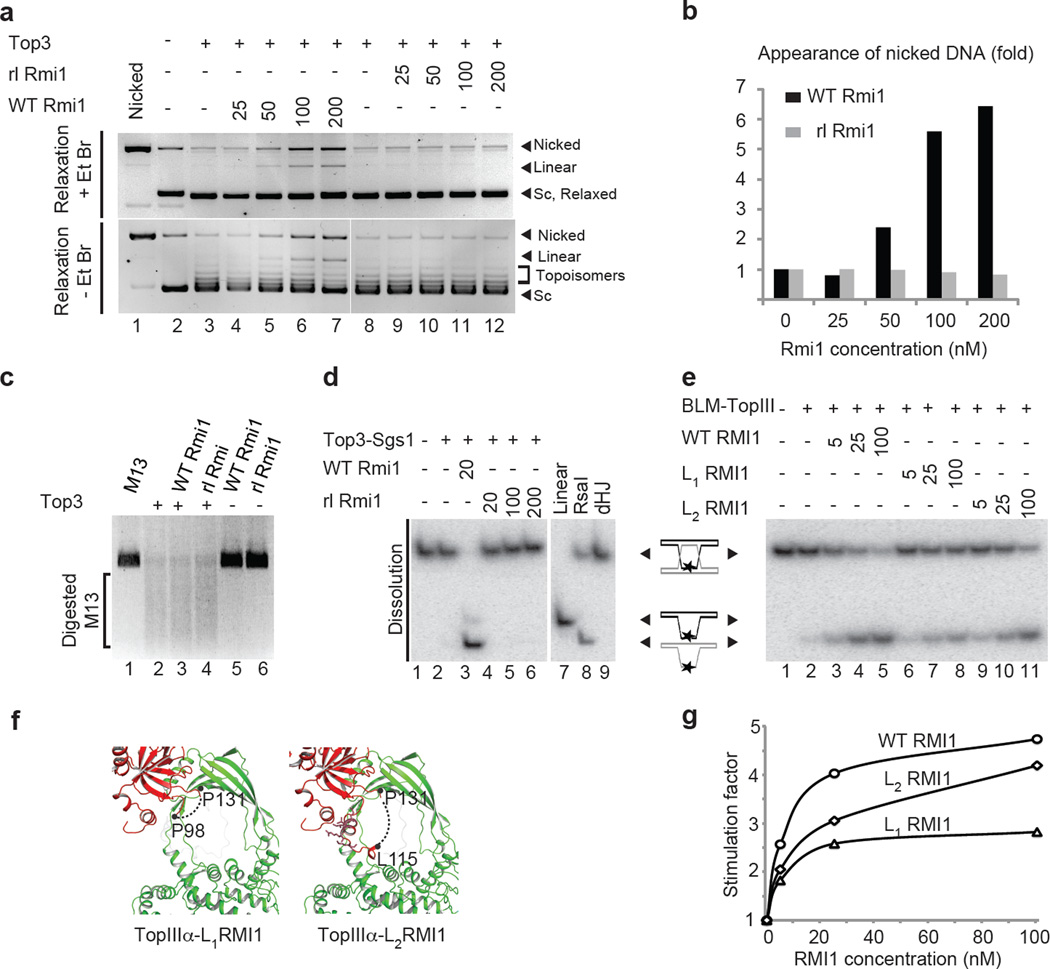
Isolated wild-type Top3 is able to relax negatively supercoiled DNA (scDNA) in presence of Mg2+ producing multiple topoisomers (partially relaxed DNA) detectable on ethidium bromide (EtBr) free gels (fig. 4a, −EtBr: compare lanes 2 and 3). Running gels in the presence of EtBr, on the other hand, allows nicked circular DNA to be electrophoretically separated from fully relaxed DNA. This analysis (fig. 4a, + EtBr: lanes 4 to 7) and quantification (fig. 4b) showed that there was a 6-fold increase in the accumulation of nicked DNA, as a function of the concentration of wild-type Rmi1 added. Previous work demonstrated that this accumulation of nicked species corresponds to Top3 being stabilized in an ssDNA-bound open form, with the scissile phosphate covalently attached to the catalytic tyrosine of domain III25,26. When using rlRmi1, however, we did not observe any increase in nicked DNA (fig. 4a, + EtBr: lanes 9 to 12; fig. 4b for quantification).
Figure 4. The RMI1 decatenation loop stabilizes the open Top3 and stimulates dHJ dissolution.
a) Yeast Top3 relaxation assay using pUC19 DNA to assess the effect of yeast Rmi1 wild-type (WT Rmi1) versus rlRmi1. Increasing concentrations of WT Rmi1 (lanes 4–7) and rlRmi1 (lanes 9–12) (Supplementary fig. 6a) b) Quantitation of the nicked DNA band intensity for WT Rmi1 (grey) and rlRmi1 (black) on EtBr stained gels as shown in a). c) Digest of single stranded M13 substrate with the different complexes following overnight incubation (Supplementary fig. 6b). d) dHJs dissolution assay of a short synthetic dHJ junction, using the yeast Top3, Sgs1 and Rmi1. dHJ substrate was incubated in the absence (lane 1) or presence of Top3 and Sgs1 alone (lane 2). Lanes 3–6 as lane 2, but with addition of GST-WT Rmi1 (lane 3), or increasing concentrations of GST-rlRmi1. A dHJ substrate digested with RsaI (Lane 8) was used as a migration marker indicating the product of dHJ dissolution (Supplementary fig. 6c). e) as in d) with the dHJ substrate incubated in the absence (lane 1) or presence of human TopIIIα and BLM alone (lane 2). Lanes 6–9 were as lane 2, but following addition of WT RMI1 (lanes 3, 4 and 5), L1RMI1(lanes 6, 7 and 8) or L2RMI1 (lanes 9, 10 and 11) at increasing concentrations (Supplementary fig. 6d) f) Scheme representation of the human L1RMI1 and L2RMI1 constructs. Residues conserved residues between yeast and human are represented as sticks. g) Stimulation factor quantitation of e).
To rule out nonspecific inactivation of Top3 by rlRmi1; for example through aggregation, we next examined whether rlRmi1 also interferes with Top3 mediated single strand DNA cleavage. For this, we incubated Top3-rlRmi1 complexes with M13 viral ssDNA at 400 nM, the highest Rmi1 concentration used in relaxation assays (fig. 4c). Top3 alone was able to effectively cleave ssDNA into smaller fragments. In the presence of wild-type or rlRmi1, we observed comparable levels of M13 ssDNA cleavage after 1 h (Supplementary fig. 5b) or following incubation overnight (fig. 4c). As judged by ssDNA cleavage, rlRmi1 itself is thus not deleterious for Top3 stability. We therefore conclude that the effects on Top3 catalysis observed in the presence of rlRmi1 (fig. 4a) are likely a direct consequence of loop sequence scrambling and shortening. These experiments demonstrate that the Rmi1 insertion loop is involved in stabilizing Top3 in a nicked, open conformation, while not interfering with the principal ability of Rmi1 to inhibit Top3 mediated relaxation and single strand cleavage. We conclude that the insertion loop in Rmi1 hence likely directly influences the open-closing dynamics of the Top3 gate.
The insertion loop is key for dHJ dissolution in vitro
We next examined the concerted action of Top3 and Sgs1 in dissolution of a synthetic dHJ (fig. 4d). This substrate allowed us to examine the contribution of Rmi1 to the decatenation of the final hemicatene intermediate. Wild-type Rmi1 strongly stimulated the dissolution of dHJ by the Top3-Rmi1 complex, as previously observed26,27. However, when wild-type Rmi1 was substituted by rlRmi1, the dissolution efficiency was greatly diminished. Similarly, we found the rlRmi1 construct to be defective in a plasmid catenation and decatenation26 assay, a related process dependent on Sgs1, E. coli single strand binding protein (SSB) that can replace yeast RPA protein (the cognate ssDNA-binding protein) and Top3 (Supplementary fig. 5c). These data suggest strongly that the Rmi1 loop and its ability to stabilize the open-state of Top3 is also required for decatenation and dHJ dissolution in vitro. We subsequently refer to the Rmi1 insertion loop (residues 94 to 134) as the RMI1 decatenation loop.
In an effort to further define the region within the Rmi1 decatenation loop required for dHJ dissolution, we reverted to the human TopIIIα-RMI1 system, and engineered two deletion constructs: L1RMI1, with a Ser-Gly-Ser-Gly insertion between residues P98 and P131, and L2RMI1 with a Ala-Gly-Gly-Ser-Gly-Ser-Gly linker inserted between L115 and P131 (fig. 4e, f): L1RMI1 links the loop at its most narrow point retaining only the interaction between the RMI1-OBN and the TopIIIα gate hence removing the decatenation loop, while L2RMI1 retains TopIIIα interaction with RMI1-OBN and the N-terminal half of the RMI1 loop (fig. 1e, f), the latter being also the most conserved portion of the decatenation loop between yeast and human proteins (fig. 4f). While L2RMI1 had near wild-type ability to stimulate dHJ dissolution, L1RMI1 showed an impairment of approximately 50% compared to wild-type, which could not be rescued by addition of higher concentrations of RMI1 (fig. 4e and fig. 4g for quantification). We conclude therefore that human RMI1 loop residues 99 to 116 contribute to the decatenation process, and that the RMI1 decatenation loop is required for TopIIIα-Rmi1 mediated dissolution in the human and yeast complexes.
Discussion
We have determined the structure of the human TopIIIα-RMI1(OBN) complex, and find strong resemblance between TopIIIα and prokaryotic type 1A relaxases. Dissolution of a synthetic Holliday junction is dependent on the RMI1 decatenation loop segment, which is donated in trans into the central gate of the topoisomerase. To illustrate what we propose happens in the TopIIIα-RMI1 reaction cycle, we built a TopIIIα model representing different stages of decatenation, based on known structures and simulations available for E. coli Top1A46,47. This model depicts domain III and the gate in a presumed open conformation (fig. 5), and suggests that the gate may create an opening of up to 30 Å in width analogous to what has been suggested for E. coli TopIA46. For the topoisomerase cycle to be completed, the T-strand has to be moved through the C-strand nick, followed by C-strand religation and subsequent DNA release (fig. 5). A certain degree of plasticity within the decatenation loop is thus required to accommodate the ssDNA upon gate closure, as the cavity between the decatenation loop and the topoisomerase gate only measures around 11 Å at its widest point (with the diameter of ssDNA being approximately 7 Å). This is in line with the plasticity of the decatenation loop observed when comparing the Mg2+- and Ca2+-bound crystal forms (fig. 3b, Supplementary fig. 4d and e). In the Mg2+-bound structure, the loop segment is disordered around this presumed ssDNA T-strand binding site (residues 116 to 131). This site is also the locus where the additional 39 residues within the S. cerevisiae Rmi1 loop are inserted further supporting the notion that conformation variability exists at this site (fig. 3c). As truncation or substitution of the tip of the loop in the human L2Rmi1 mutant only gave rise to very minor dHJ dissolution defects in vitro (fig. 4e), we conclude that this part of the loop, which is likely to be in direct contact with DNA, exerts neither crucial attractive (through charged or hydrophobic residues) nor repulsive (through steric crowding) interactions with DNA. Despite the necessary plasticity of the loop in this region, the presence of the RMI1 OBN-fold and its attachment points within the gate (fig. 2b) impart steric constraints that are expected to disfavor dsDNA accommodation within the TopIIIα gate. This is in agreement with the low efficiency (~10-fold less efficient than ssDNA) by which dsDNA is accommodated by the Top3-Rmi1 complex in decatenation experiments25, and contrasts with the behavior seen with prokaryotic Top3 enzymes48.
Figure 5. Model for the final step of dHJ dissolution catalyzed by TopIIIα-RMI1.
A model depicting the TopIIIα-RMI1 catalytic cycle with the decatenation of a hemicatene, the final product of the dHJ dissolution process, taken as an example: TopIIIα (in green) loads onto the DNA substrate (step 1) and introduces a nick in the C-strand (step 2), generating a gap through which the T-strand can be passaged following opening of the gate (step 3). RMI1 (in red) stabilizes step 3 allowing sufficient time to move a DNA strand (T-strand) originating from a second, separate DNA duplex into the gate. Following gate closure (step 4), the substrate is released (step 5) and the decatenation process is completed.
The RMI decatenation loop modulates the gate dynamic
In the model of the open form (fig. 5, #3), we observed that the pivot region of the gate is in close proximity to the beginning of the RMI1 decatenation loop involving residues around P98-K109 (fig. 1e). Experimentally we find that human RMI1 residues 96 to 116 are required for effective decatenation (fig. 4 e, g). In addition, RMI1 residues ranging from P96 to K109 are also reasonably conserved among orthologues (fig. 3c). We therefore hypothesize that this contact region between the RMI1 loop and TopIIIα contributes to the modulation of TopIIIα opening-closing equilibrium.
Towards a unified mechanism for decatenation
Analysis of the TopIIIα-RMI1 system has the great advantage that decatenation is stimulated by RMI1 addition in trans, allowing one to directly probe the effect of RMI1 on decatenation without manipulating the topoisomerase itself. Prokaryotic type IA topoisomerases implicated in relaxation and those involved in decatenation (e.g.: E. coli Top3) share almost identical enzymatic cores with only minimal structural differences. The most striking difference is the presence or absence of the decatenation loop lining the gate, which is found exclusively in the prokaryotic decatenases35. Deletion of this loop in E. coli Top3 results in almost complete loss of decatenation activity, albeit with relaxation activity also being impaired45. The structural equivalent of this loop in the eukaryotic Top3(TopIIIα) decatenases is absent, which has previously been used as an argument against loop-mediated decatenation. We now find that RMI1 inserts a loop segment in trans, positioned in proximity to the two loci of the prokaryotic decatenation loop. Moreover, this loop in RMI1 is required for effective decatenation and dHJ resolution.
Based on our new data, we propose a model (fig. 5) whereby, following the binding and scission of the single stranded C-strand, the topoisomerase gate opens. While C-strand cleavage is not dependent on RMI1, the gate once open is stabilized through structural elements originating from the RMI1 loop (or possibly the functionally equivalent loops found in prokaryotic type IA enzymes). The structural and biochemical data supports a model where the gate in its stabilized open state will provide the topoisomerase with sufficient time to accommodate distant ssDNA T-strands originating from different duplexes (e.g. in dHJ dissolution). This permits the movement of these strands in and out of the gate, thereby driving decatenation (as an ‘inter-molecular process’)25. In its relaxation mode, on the other hand, where the T-strand belongs to the same duplex as the C-strand (working in an intra-molecular process) movement of DNA strands likely occurs over shorter distances and hence does not require extended gate opening times.
TopIIIα-RMI1 function in the dissolvasome
dHJ dissolution is dependent on the presence of a complete dissolvasome consisting, minimally, of TopIIIα(Top3), Rmi1(RMI1) and a RecQ helicase. In yeast, the Sgs1 RecQ helicase serves both a catalytic and non-catalytic scaffolding role. We now find that RMI1 does not fundamentally change the mode of topoisomerase action, but rather modulates its dynamics and specificity, allowing the decatenation of a dHJ hemicatene rather than relaxation. Further studies are required to understand how TopIIIα-RMI1 functionally and structurally integrates within the RecQ-containing dissolvasome.
Conclusion
In the cell, the majority of proteins function in the context of macromolecular complexes. This work serves as an example of how binding proteins, such as RMI1, modulate topoisomerase function within their large assembles (also referred as toposomes49). Epitopes such as the RMI1 loop, which are unstructured in isolation, can nonetheless assume crucial catalytic and regulatory roles when put in the context of these macromolecular machines50.
Online methods
Protein expression prior crystallization
Human Strep-TopIIIα (residues 1–753)-RMI1 (residues 1–219) complex was co-expressed in High Five insect cells using a pFastBac dual transfer vector (Bac-to-Bac system, Invitrogen). Cells are infected at a concentration of 4 × 106 cells/ml and harvested after 48 h after infection. Lysis in 50mM Tris-HCl pH 8.0, 200 mM NaCl, 1 mM β-mercaptoethanol, 1 mM PMSF and protease inhibitor cocktail (Sigma) was performed by sonication followed by clarification of the lysate by ultracentrifugation. Streptactin affinity purification was performed as a first strep affinity step, followed by POROSQ anion exchange chromatography (buffered by 50 mM Tris pH8.0, 0.2 mM TCEP) to remove traces of endogenous DNA. As a final step, the purified protein was subjected to size exclusion chromatography on a Superdex 200 column (GE Healthcare) (50 mM HEPES pH7.4, 300 mM NaCl, 0.2 mM TCEP). Fractions containing the heterodimeric complex were pooled and subjected to crystallization experiments.
Complex crystallization
The complex (8 mg/ml) was crystallized in 8–12% (w/v) PEG 2000, 100 mM Tris-HCl pH 7.0, 200 mM MgCl2. Crystals grew only in the presence of Mg2+ or Ca2+. The conditions of crystallization for the crystals grown in presence of Ca2+ were: Tris pH 7.0, 8% (w/v) PEG 5000 MME, 200 mM calcium citrate. Crystals in either Mg2+ or Ca2+ appeared as square based rods growing to maximal dimension within 4–5 days. Cryopreservation of crystals was performed by supplementation of the growth condition with 35% ethylene glycol prior to flash cooling in liquid nitrogen.
Data collection and processing
X-ray diffraction data were recorded using a Pilatus 6M detector (Dectris) at beamline PXII of the Swiss Light Source (Villigen, Switzerland) using a wavelength of 1.0 Å. Data sets from crystals grown in the presence of Mg2+ (TOPIIIα-Mg2+-RMI1) and Ca2+ (TOPIIIα-Ca2+-RMI1) were collected. Data were processed with XDS51 and POINTLESS52 was used for space group prediction and format conversion prior to additional scaling and merging with AIMLESS52. Averaged intensities were converted to structure factor amplitudes by TRUNCATE52.
Both data sets are strongly anisotropic. Analyzed in the true space group, P41212, the resolution at which 〈I/σ〉 = 2 for the TopIIIα-Mg2+–RMI1 data is 3.16 Å along the ab plane and 2.63 Å along the c axis, the two principle directions for anisotropy in tetragonal crystals, giving a difference between them when represented as a thermal ellipsoid (DeltaB) is 26.6 Å2. The TOPIIIα-Ca2+-RMI1 data have a DeltaB = 24.0 Å2 and resolution limits of 3.64 Å along the ab plane and 2.88 Å along the c axis. The anisotropy resulted in high merging R factors for both data sets, but over the resolution range defined the anisotropic 〈I/σ〉 = 2 cutoff the CC1/2 (correlation coefficient between random half data sets53) remained greater than 0.4 for suggesting that there might be significant signal at high resolution the potential of which could be validated in refinement. Final data processing statistics are given in (Table 1). 96.7% and 96.0% of residues were in the favored regions of the Ramachandran plot for TOPIIIα–Mg2+–RMI1 and TOPIIIα-Ca2+–RMI1, respectively, with 0% and 0.1% outliers, respectively.
Table 1.
Data collection and structure refinement statistics.
| TOPIIIα-Mg2+–RMI1 PDB: 4CGY |
TOPIIIα-Ca2+–RMI1 PDB: 4CHT |
|
|---|---|---|
| Data collection | ||
| Space group | P41212 | P41212 |
| Wavelength (Å) | 1.0 | 1.0 |
| Cell dimensions | ||
| a, b, c (Å) | 94.00, 94.00, 331.72 | 93.38, 93.38, 331.62 |
| α, β, γ (°) | 90, 90, 90 | 90, 90, 90 |
| Resolution range (Å)1 | 62.78–2.85 (2.97–2.85) | 65.05–3.25 (3.45–3.25) |
| Unique reflections | 41328 (4527) | 27765 (4404) |
| Rmerge (%) | 14.9 (163) | 16.1 (78.1) |
| Rmeas(%) | 16.0 (175) | 19.7 (96.2) |
| Rpim (%) | 4.2 (44.8) | 8.4(41.8) |
| CC1/2 outer shell;number of pairs2 | 0.814; n =4527 | 0.727; n = 4328 |
| I / σI | 16.7 (2.2) | 8.2 (2.0) |
| Completeness (%) | 100 (100) | 99.8 (99.9) |
| Redundancy | 14.1 (14.7) | 5.3(5.3) |
| Refinement | ||
| Resolution (Å) | 2.85 Å | 3.25 Å |
| Rwork / Rfree | 19.7 / 23.1 (24.0 / 27.6) | 19.7 / 22.5 (25.1 / 27.8) |
| No. atoms | ||
| Protein | 6557 | 6632 |
| Ion | 1 | 1 |
| Water | 45 | 5 |
| Average B-factors (Å2) | ||
| Protein | 80.3 | 83.5 |
| Ligand/ion | 62.3 | 81.8 |
| Water | 59.0 | 78.9 |
| R.m.s. deviations | ||
| Bond lengths (Å) | 0.009 | 0.010 |
| Bond angles (°) | 1.04 | 1.06 |
| Ramachandram plot (Molprobity) | ||
| Favored (%) | 96.7 | 96.0 |
| Outliers (%) | 0 | 0.1 |
Values in parentheses are for highest-resolution shell.
Random half-set (Pearson) correlation coefficient as defined in (Karplus & Diederichs, 2012)
Structure determination and refinement
The TopIIIα-Mg2+–RMI1 structure was solved by molecular replacement (MR) in space group P41212 using PHASER54 with E. coli Top1A33 search model proposed by BALBES and prepared with CHAINSAW and a search model based on hRMI1 (PDB code 3NBI).
The initial TopIIIα-Mg2+–RMI1 model was iteratively rebuilt with COOT55 and refined with PHENIX.REFINE56 or REFMAC57 at 3.3 Å resolution, at which the outermost resolution shell Rmerge was less than 50%. After effort to improve the initial 3.3-Å model stalled, the data were reassessed using ‘paired-refinement’ strategy of Karplus and Diederichs53 using autoBUSTER and an optimal high resolution limit of 2.85 Å was found. At this resolution, the outermost resolution shell (2.97–2.85 Å) in the direction of the ab plane CC1/2 is 0.30, along the c axis the CC1/2 is 0.97 giving an overall CC1/2 of 0.81 and a DeltaB of 25.9 Å2. Inclusion of the weak high-resolution data in refinement significantly improved the quality of the refined models and the interpretability of the election density maps; revealing mFo-DFc difference map peaks for the water molecules coordinated by the Mg2+ ion, an additional 39 water molecules and enabled the correction of several side chain rotamers. The final TopIIIα-Mg2-RMI1 model was produced after iterative model building with COOT and refinement with autoBUSTER58 (http://www.globalphasing.com/buster/) using data to 2.85 Å resolution, one TLS group per chain. The TopIIIα-Ca2+-RMI1 crystal structure was determined by rigid body fitting of the TopIIIα-Mg2+-RMI1 to data at 3.25 Å resolution selected using similar CC1/2 cutoffs derived for TopIIIα-Mg2+–RMI1 (in the outer resolution shell (3.45–3.25 Å) a CC1/2 of 0.24 in the direction of the ab plane, CC1/2 0.97 in the direction of the c axis giving an overall CC1/2 of 0.73 and a DeltaB of 35.5 Å2). The TopIIIα-Ca2+-RMI1 model was rebuilt and refined in the same way as TopIIIα-Mg2+-RMI1 with the addition of local structure similarity restraints imposed to the final TopIIIα-Mg2+-RMI1 model. The metal sites in both structures were restrained to maintain octahedral geometry and appropriate bond lengths (Mg2+⋯OH2 = 2.09 Å and Ca2+⋯OH2 = 2.42 Å both with σ = 0.02 Å), and the same randomly assigned set of 5% of all reflections was omitted from refinement for cross validation. Molprobity59 was used to analyze model quality and places it in the 100th percentile, among the best for this resolution.
The RMI1 decatenation loop (residues 116–130) was ordered in the TopIIIα-Ca2+-RMI1 crystal as opposed to TopIIIα-Mg2+-RMI1 allowing the loop to be traced. The final refined contain residues RMI1 2–216 and TopIIIα 20–637 with one hydrated Mg2+/Ca2+ ion bound to Glu41 in monodentate mode. Several side chains were truncated at Cβ due to a lack of electron density and additional residues of the TopIIIα N- and C-terminus that could not be unequivocally built were omitted from the model. Refinement statistics for the final models are given in Table 1.
Electron Microscopy
Sample used for crystal growth was diluted to a concentration of 200 nM in 25 mM HEPES pH 7.4, 100 mM NaCl and 0.2 mM TCEP immediately before applying it to glow-discharged holey carbon grids (Cu 400 mesh grids, Quantifoil Micro Tool GmbH). The protein was adsorbed to the EM grids for 45 seconds and negatively stained with 2% (m/w) uranyl acetate solution. The grid was examined using a Philips CM10 electron microscope equipped with a LaB6 filament and operated at 80 kV. Images were recorded as CCD micrographs at 130000× nominal magnification resulting in a pixel size of 3.7 Å. A total of 4000 particles were picked and subsequently analyzed using EMAN2. To avoid model bias, the initial model used for angular refinements was a featureless Gaussian blob with dimension estimated from the crystal structure of TopIIIα-RMI1, generated with the command makeinitialmodel.py as implemented in EMAN. After several cycles of refinement convergence to the final model shown in Supplementary fig. 3 was attained. The angular spacing used for orientation generation was set to 10 deg. Smaller increment did not improve the model further.
Relaxation, M13 digest assays, and catenation-decatenation assays with yeast proteins
Yeast Top3, Sgs1 and bacterial SSB are were expressed and purified as outlined in23. Yeast Rmi1 WT and rl constructs were expressed in insect cells (High Five) as GST N-terminal fusion proteins and C-terminal His6 tag following a previously described protocol23 to avoid nuclease contamination. Proteins were stored in 50 mM Tris pH 7.0, 150 mM NaCl, 1 mM β-mercaptoethanol, 10% glycerol (final buffer). For the relaxation assay, proteins (Top3 and different Rmi1 constructs) were incubated in presence of substrate PUC19 (200 ng per reaction) and incubated 1 h at 42°C in reaction buffer (25 mM HEPES pH 7, 100 µg/ml, 100 µg/ml BSA, 32% glycerol, 5 mM NaOc, 0.5 mM MgOAc). The reactions were stopped by the addition of EDTA and Proteinase K and incubation for 1 h at 37°C. The samples are loaded on 1% agarose gel poured in presence or absence of EtBr.
M13 (200 ng per reaction, M13 mp18 phage ssDNA from Clontech) digestions were performed for 1 h, 3 h and overnight duration in a reaction buffer 25 mM HEPES pH 7.4, 100 µg/ml BSA, 32% glycerol, 5 mM NaOc, 2 mM MgOAc. The reactions were stopped by incubation with EDTA and Proteinase K for 1 h at 37°C. The gels were stained with SYBR green II. For the catenation assays, 100nM Top3, 60nM Sgs1 and 1µM E. coli SSB (Sigma) were incubated alone or with 200nM GST-wtRmi1 or GST-rlRmi1 in the presence of 100ng pUC19 as described previoulsy25. The decatenation assays were performed as the catenation assay but in the presence of 100ng of kDNA (Topogen). Products were loaded on a 1% agarose gel in the presence of ethidium bromide.
Double Holliday junction dissolution
The human L1 and L2 RMI1 loop variants were generated by a site-directed mutagenesis strategy, cloned in pGEX plasmids and produced in E. coli ROSETTA strain (overnight expression at 20°C following induction with 0.2 mM IPTG). The cells were harvested and lysed by sonification. After ultracentrifugation clarification, a GST sepharose affinity step was performed followed by overnight TEV cleavage (4°C, 2% of TEV protease). RMI1 protein is further purified by anion exchange chromatography (POROS Q resin, GE healthcare) allowing the separation of GST tag (elution around 140 mM NaCl) from RMI1 (elution around 200 mM NaCl). The proteins were dialyzed overnight against final buffer.
The preparation of the dHJ substrate and dissolution reactions were carried out as described in reference60 with the exception that the reaction products were separated using 8% native acrylamide gel electrophoresis run at 4°C. After drying, the gels were exposed overnight to a phosphorImager screen, and the image was recorded using a Typhoon scanner. The stimulation factor represented on figure 4g is calculated as the percentage of product formed in the presence of RMI1 divided by the percentage of product observed in the absence of RMI1.
Supplementary Material
Acknowledgments
This work was supported by the Novartis Research Foundation (to N.H.T) and grants from Krebsforschung Schweiz (KFS-2986-08-2012) (to N.H.T), Marie Curie Fellowship (FP7-PEOPLE-2009-IEF 253555-TOPO) (to N.B), European Molecular Biology Organization long term Fellowship (EMBO ALTF 693–2009) (to N.B) and European Research Council (to N.H.T), the Nordea Foundation, The Villum Kann Rasmussen Fund (to I.D.H), and the U.S. National Institutes of Health (GM-41347 and GM-62653) (to S.C.K). We are grateful to H. Stahlberg and team for use of the EM facilities at Center for Cellular Imaging and NanoAnalytics (c-CINA). We thank S. Gasser, U. Rass, K. Shimada, J. Keusch, E. Fischer, A. Scrima, D. Hess, K. Boehm, D. Klein, H. Gut and M. Renatus for help and fruitful discussion. We would also like to thank A. Costa and colleagues at the London Research Institute for sharing results prior publication. Part of this work was performed at beamline PXII of the Swiss Light Source at the Paul Scherrer Institute, Villigen, Switzerland.
Footnotes
Accession codes
Coordinates of the models were deposited in the PDB under the following accession codes: 4CGY for TOPIIIα–RMI1 structure bound to a Mg2+ ion and 4CHT for TOPIIIα–RMI1 structure bound to a Ca2+ ion.
Author contributions
N.B carried out molecular biology, protein expression and purification, crystallization, structure determination and model refinement. R.D.B carried out final stages of model refinement and maps improvements. A.H.B. performed the relaxation, catenation and decatenation assays, with contributions by W.A. and N.B., and dissolution assays. W.A. did the M13-digest assay. N.B. performed the construct design, expression and purification of human RMI1-mutant proteins for dissolution assays. P.C provided yeast Top3 and Sgs1 proteins as well as SSB protein. S.C carried out single particle electron microscopy and 3D model reconstruction. M.F and N.B.L performed yeast strain constructions and drop assay tests. N.B and N.H.T designed the project with the help of P.C, S.C.K and I.D.H. N.B and N.H.T wrote the paper with the help of the other co-authors. N.H.T supervised the project.
References of the main text
- 1.Petermann E, Helleday T. Pathways of mammalian replication fork restart. Nature reviews. Molecular cell biology. 2010;11:683–687. doi: 10.1038/nrm2974. [DOI] [PubMed] [Google Scholar]
- 2.Moynahan ME, Jasin M. Mitotic homologous recombination maintains genomic stability and suppresses tumorigenesis. Nature reviews. Molecular cell biology. 2010;11:196–207. doi: 10.1038/nrm2851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Holliday R. A mechanism for gene conversion in fungi. Genetical research. 2007;89:285–307. doi: 10.1017/S0016672308009476. [DOI] [PubMed] [Google Scholar]
- 4.Szostak JW, Orr-Weaver TL, Rothstein RJ, Stahl FW. The double-strand-break repair model for recombination. Cell. 1983;33:25–35. doi: 10.1016/0092-8674(83)90331-8. [DOI] [PubMed] [Google Scholar]
- 5.Heyer WD, Ehmsen KT, Solinger JA. Holliday junctions in the eukaryotic nucleus: resolution in sight? Trends in biochemical sciences. 2003;28:548–557. doi: 10.1016/j.tibs.2003.08.011. [DOI] [PubMed] [Google Scholar]
- 6.Matos J, Blanco MG, Maslen S, Skehel JM, West SC. Regulatory control of the resolution of DNA recombination intermediates during meiosis and mitosis. Cell. 2011;147:158–172. doi: 10.1016/j.cell.2011.08.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Heyer WD. Recombination: Holliday junction resolution and crossover formation. Current biology : CB. 2004;14:R56–R58. doi: 10.1016/j.cub.2003.12.043. [DOI] [PubMed] [Google Scholar]
- 8.Cavenee WK, et al. Expression of recessive alleles by chromosomal mechanisms in retinoblastoma. Nature. 1983;305:779–784. doi: 10.1038/305779a0. [DOI] [PubMed] [Google Scholar]
- 9.Thiagalingam S, et al. Mechanisms underlying losses of heterozygosity in human colorectal cancers. Proceedings of the National Academy of Sciences of the United States of America. 2001;98:2698–2702. doi: 10.1073/pnas.051625398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.LaRocque JR, et al. Interhomolog recombination and loss of heterozygosity in wild-type and Bloom syndrome helicase (BLM)-deficient mammalian cells. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:11971–11976. doi: 10.1073/pnas.1104421108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wu L, Hickson ID. The Bloom's syndrome helicase suppresses crossing over during homologous recombination. Nature. 2003;426:870–874. doi: 10.1038/nature02253. [DOI] [PubMed] [Google Scholar]
- 12.Gangloff S, McDonald JP, Bendixen C, Arthur L, Rothstein R. The yeast type I topoisomerase Top3 interacts with Sgs1, a DNA helicase homolog: a potential eukaryotic reverse gyrase. Molecular and cellular biology. 1994;14:8391–8398. doi: 10.1128/mcb.14.12.8391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bennett RJ, Noirot-Gros MF, Wang JC. Interaction between yeast sgs1 helicase and DNA topoisomerase III. The Journal of biological chemistry. 2000;275:26898–26905. doi: 10.1074/jbc.M003137200. [DOI] [PubMed] [Google Scholar]
- 14.Fricke WM, Kaliraman V, Brill SJ. Mapping the DNA topoisomerase III binding domain of the Sgs1 DNA helicase. The Journal of biological chemistry. 2001;276:8848–8855. doi: 10.1074/jbc.M009719200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Harmon FG, DiGate RJ, Kowalczykowski SC. RecQ helicase and topoisomerase III comprise a novel DNA strand passage function: a conserved mechanism for control of DNA recombination. Molecular cell. 1999;3:611–620. doi: 10.1016/s1097-2765(00)80354-8. [DOI] [PubMed] [Google Scholar]
- 16.Cejka P, Kowalczykowski SC. The full-length Saccharomyces cerevisiae Sgs1 protein is a vigorous DNA helicase that preferentially unwinds holliday junctions. The Journal of biological chemistry. 2010;285:8290–8301. doi: 10.1074/jbc.M109.083196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xu D, et al. RMI, a new OB-fold complex essential for Bloom syndrome protein to maintain genome stability. Genes & development. 2008;22:2843–2855. doi: 10.1101/gad.1708608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mullen JR, Nallaseth FS, Lan YQ, Slagle CE, Brill SJ. Yeast Rmi1/Nce4 controls genome stability as a subunit of the Sgs1-Top3 complex. Molecular and cellular biology. 2005;25:4476–4487. doi: 10.1128/MCB.25.11.4476-4487.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Singh TR, et al. BLAP18/RMI2, a novel OB-fold-containing protein, is an essential component of the Bloom helicase-double Holliday junction dissolvasome. Genes & development. 2008;22:2856–2868. doi: 10.1101/gad.1725108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Raynard S, Bussen W, Sung P. A double Holliday junction dissolvasome comprising BLM, topoisomerase IIIalpha, and BLAP75. The Journal of biological chemistry. 2006;281:13861–13864. doi: 10.1074/jbc.C600051200. [DOI] [PubMed] [Google Scholar]
- 21.Wu L, et al. BLAP75/RMI1 promotes the BLM-dependent dissolution of homologous recombination intermediates. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:4068–4073. doi: 10.1073/pnas.0508295103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lai MS, Seki M, Ui A, Enomoto T. Rmi1, a member of the Sgs1-Top3 complex in budding yeast, contributes to sister chromatid cohesion. EMBO reports. 2007;8:685–690. doi: 10.1038/sj.embor.7401000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chang M, et al. RMI1/NCE4, a suppressor of genome instability, encodes a member of the RecQ helicase/Topo III complex. The EMBO journal. 2005;24:2024–2033. doi: 10.1038/sj.emboj.7600684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mankouri HW, Hickson ID. The RecQ helicase-topoisomerase III-Rmi1 complex: a DNA structure-specific 'dissolvasome'? Trends in biochemical sciences. 2007;32:538–546. doi: 10.1016/j.tibs.2007.09.009. [DOI] [PubMed] [Google Scholar]
- 25.Cejka P, Plank JL, Dombrowski CC, Kowalczykowski SC. Decatenation of DNA by the S. cerevisiae Sgs1-Top3-Rmi1 and RPA complex: a mechanism for disentangling chromosomes. Molecular cell. 2012;47:886–896. doi: 10.1016/j.molcel.2012.06.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cejka P, Plank JL, Bachrati CZ, Hickson ID, Kowalczykowski SC. Rmi1 stimulates decatenation of double Holliday junctions during dissolution by Sgs1-Top3. Nature structural & molecular biology. 2010;17:1377–1382. doi: 10.1038/nsmb.1919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen CF, Brill SJ. Binding and activation of DNA topoisomerase III by the Rmi1 subunit. The Journal of biological chemistry. 2007;282:28971–28979. doi: 10.1074/jbc.M705427200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Corbett KD, Berger JM. Structure, molecular mechanisms, and evolutionary relationships in DNA topoisomerases. Annual review of biophysics and biomolecular structure. 2004;33:95–118. doi: 10.1146/annurev.biophys.33.110502.140357. [DOI] [PubMed] [Google Scholar]
- 29.Tse YC, Kirkegaard K, Wang JC. Covalent bonds between protein and DNA. Formation of phosphotyrosine linkage between certain DNA topoisomerases and DNA. The Journal of biological chemistry. 1980;255:5560–5565. [PubMed] [Google Scholar]
- 30.Yang J, Bachrati CZ, Ou J, Hickson ID, Brown GW. Human topoisomerase IIIalpha is a single-stranded DNA decatenase that is stimulated by BLM and RMI1. The Journal of biological chemistry. 2010;285:21426–21436. doi: 10.1074/jbc.M110.123216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hoadley KA, et al. Structure and cellular roles of the RMI core complex from the bloom syndrome dissolvasome. Structure. 2010;18:1149–1158. doi: 10.1016/j.str.2010.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang F, et al. Crystal structures of RMI1 and RMI2, two OB-fold regulatory subunits of the BLM complex. Structure. 2010;18:1159–1170. doi: 10.1016/j.str.2010.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lima CD, Wang JC, Mondragon A. Three-dimensional structure of the 67K N-terminal fragment of E. coli DNA topoisomerase I. Nature. 1994;367:138–146. doi: 10.1038/367138a0. [DOI] [PubMed] [Google Scholar]
- 34.Hansen G, Harrenga A, Wieland B, Schomburg D, Reinemer P. Crystal structure of full length topoisomerase I from Thermotoga maritima. Journal of molecular biology. 2006;358:1328–1340. doi: 10.1016/j.jmb.2006.03.012. [DOI] [PubMed] [Google Scholar]
- 35.Mondragon A, DiGate R. The structure of Escherichia coli DNA topoisomerase III. Structure. 1999;7:1373–1383. doi: 10.1016/s0969-2126(00)80027-1. [DOI] [PubMed] [Google Scholar]
- 36.Aravind L, Leipe DD, Koonin EV. Toprim--a conserved catalytic domain in type IA and II topoisomerases, DnaG-type primases, OLD family nucleases and RecR proteins. Nucleic acids research. 1998;26:4205–4213. doi: 10.1093/nar/26.18.4205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Duguet M, Serre MC, Bouthier de La Tour C. A universal type IA topoisomerase fold. Journal of molecular biology. 2006;359:805–812. doi: 10.1016/j.jmb.2006.04.021. [DOI] [PubMed] [Google Scholar]
- 38.Schultz SC, Shields GC, Steitz TA. Crystal structure of a CAP-DNA complex: the DNA is bent by 90 degrees. Science. 1991;253:1001–1007. doi: 10.1126/science.1653449. [DOI] [PubMed] [Google Scholar]
- 39.Brennan RG, Matthews BW. The helix-turn-helix DNA binding motif. The Journal of biological chemistry. 1989;264:1903–1906. [PubMed] [Google Scholar]
- 40.Flynn RL, Zou L. Oligonucleotide/oligosaccharide-binding fold proteins: a growing family of genome guardians. Critical reviews in biochemistry and molecular biology. 2010;45:266–275. doi: 10.3109/10409238.2010.488216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Goulaouic H, et al. Purification and characterization of human DNA topoisomerase IIIalpha. Nucleic acids research. 1999;27:2443–2450. doi: 10.1093/nar/27.12.2443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Changela A, DiGate RJ, Mondragon A. Crystal structure of a complex of a type IA DNA topoisomerase with a single-stranded DNA molecule. Nature. 2001;411:1077–1081. doi: 10.1038/35082615. [DOI] [PubMed] [Google Scholar]
- 43.Changela A, DiGate RJ, Mondragon A. Structural studies of E. coli topoisomerase III-DNA complexes reveal a novel type IA topoisomerase-DNA conformational intermediate. Journal of molecular biology. 2007;368:105–118. doi: 10.1016/j.jmb.2007.01.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schmidt BH, Burgin AB, Deweese JE, Osheroff N, Berger JM. A novel and unified two-metal mechanism for DNA cleavage by type II and IA topoisomerases. Nature. 2010;465:641–644. doi: 10.1038/nature08974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li Z, Mondragon A, Hiasa H, Marians KJ, DiGate RJ. Identification of a unique domain essential for Escherichia coli DNA topoisomerase III-catalysed decatenation of replication intermediates. Molecular microbiology. 2000;35:888–895. doi: 10.1046/j.1365-2958.2000.01763.x. [DOI] [PubMed] [Google Scholar]
- 46.Feinberg H, Lima CD, Mondragon A. Conformational changes in E. coli DNA topoisomerase I. Nature structural biology. 1999;6:918–922. doi: 10.1038/13283. [DOI] [PubMed] [Google Scholar]
- 47.Xiong B, et al. The type IA topoisomerase catalytic cycle: A normal mode analysis and molecular dynamics simulation. Proteins. 2008;71:1984–1994. doi: 10.1002/prot.21876. [DOI] [PubMed] [Google Scholar]
- 48.Li Z, Mondragon A, DiGate RJ. The mechanism of type IA topoisomerase-mediated DNA topological transformations. Molecular cell. 2001;7:301–307. doi: 10.1016/s1097-2765(01)00178-2. [DOI] [PubMed] [Google Scholar]
- 49.Viard T, de la Tour CB. Type IA topoisomerases: a simple puzzle? Biochimie. 2007;89:456–467. doi: 10.1016/j.biochi.2006.10.013. [DOI] [PubMed] [Google Scholar]
- 50.Bolanos-Garcia VM, et al. Spatial and temporal organization of multi-protein assemblies: achieving sensitive control in information-rich cell-regulatory systems. Philosophical transactions. Series A, Mathematical, physical, and engineering sciences. 2012;370:3023–3039. doi: 10.1098/rsta.2011.0268. [DOI] [PubMed] [Google Scholar]
References for online methods
- 51.Kabsch W. Automatic Processing of Rotation Diffraction Data from Crystals of Initially Unknown Symmetry and Cell Constants. Journal of Applied Crystallography. 1993;26:795–800. [Google Scholar]
- 52.Winn MD, et al. Overview of the CCP4 suite and current developments. Acta crystallographica. Section D, Biological crystallography. 2011;67:235–242. doi: 10.1107/S0907444910045749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Karplus PA, Diederichs K. Linking crystallographic model and data quality. Science. 2012;336:1030–1033. doi: 10.1126/science.1218231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mccoy AJ, et al. Phaser crystallographic software. Journal of Applied Crystallography. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta crystallographica. Section D, Biological crystallography. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Adams PD, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallographica Section D-Biological Crystallography. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallographica Section D-Biological Crystallography. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 58.Bricogne G, et al. BUSTER version X.Y.Z. Cambridge, United Kingdom: Global Phasing Ltd.; 2011. [Google Scholar]
- 59.Chen VB, et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta crystallographica. Section D, Biological crystallography. 2010;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bachrati CZ, Hickson ID. Dissolution of double Holliday junctions by the concerted action of BLM and topoisomerase IIIalpha. Methods in molecular biology. 2009;582:91–102. doi: 10.1007/978-1-60761-340-4_8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.