Introduction
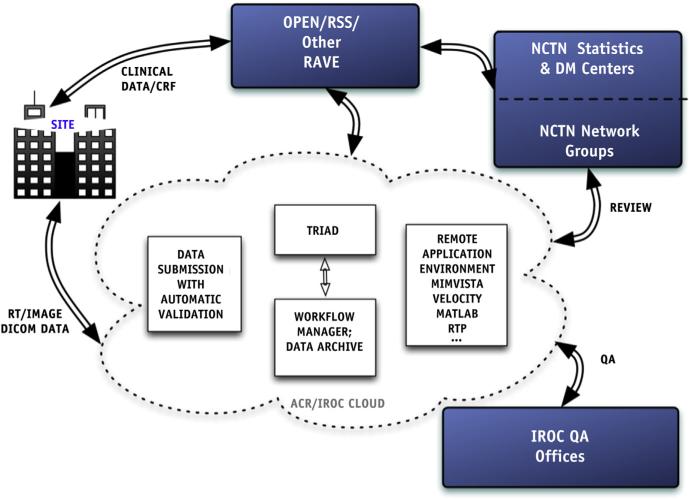
As part of the consolidation of the cooperative group clinical trial program of the National Clinical Trials Network (NCTN) of the National Cancer Institute (NCI), an Imaging and Radiation Oncology Core services organization (IROC) has been formed from current leading quality assurance (QA) centers to provide QA, along with clinical and scientific expertise, for the entire NCTN (1). An integrated information technology (IT) infrastructure, the IROC cloud, has been implemented to foster collaborative and effective interactions among participating institutions, QA centers, NCTN cooperative groups and statistics data management centers, and the IT infrastructure of the NCI (Fig. 1). An integral component of the IROC cloud is the Transfer of Images and Data (TRIAD) system designed for imaging and radiation therapy digital data transmission. The TRIAD system is now being used for digital radiation therapy and imaging data transmission for NCTN (and other) clinical trials.
Fig. 1.
IROC cloud: An IROC Information Technology (IT) infrastructure vision.
Consistency of submitted data contributes to better consistency in the treatment and review of trial data, and it facilitates scientific collaborations and also promotes safe clinical practice. The details of this data submission process are presented here.
Methods and Materials
The digital data transfer process using TRIAD includes creating a general user account, obtaining an appropriate TRIAD user role, and preparing digital data for submission following protocol specifications. Instructions can be found online (2).
The TRIAD system includes built-in functions that can be used to automate digital data QA during the transmission process. In particular, it includes an automated evaluation of the consistency between the submitted structure names and protocol requirements. To use a single set of consistent structure names, we developed a uniform radiation therapy structure name library. This library was based on common clinical radiation oncology practice, guidelines from prior publications (3) and other material (4) on this topic. In this publication, International Commission on Radiation Units and Measurements (ICRU) guidelines have been used to create standardized nomenclature for target volumes (TV), organs at risk (OAR), and planning risk volumes (PRV).
Naming conventions adopted for this library were based on the following considerations:
The technical limitations of most commercial treatment planning systems. To address these limitations, we required the following: (1) no spaces in the structure name; (2) a maximum length of 14 characters; and (3) the only special character allowed was the underscore.
Instead of using “Ipsi” and “Contra” for body side specification, we used “R” and “L” for right and left.
Structures such as “NonPTV” were created to help with dosimetric evaluation.
Accommodations were provided for implementation of modern radiation therapy technologies.
Flexibility in the expansion of names was allowed for special needs of individual trials.
Structure names were made intuitive.
Results and Discussion
The complete list of structure names is included in the supplemental material, with a brief description of each (available at www.redjournal.org). Generally, a structure name consists of the following components:
Imaging/time point, eg, CT1XXX, PT2XXX for phase 1 CT and phase 2 PET of adaptive processes
Structure description
Dose level for target volume or related nontarget volume
Structure expansion, eg, 03 for 3-mm uniform expansion and PRV for nonuniform expansion
Anatomic location, eg, L for left and Up for upper
Derivation of the structure, eg, LivermGTV for Liver minus gross tumor volume.
Conclusions
A new digital data transmission process for NCTN has been implemented using TRIAD, including a structure name library to uniformly name TV, OAR, and PRV. We plan to make it a living document to be constantly updated and available on the IROC website.
Supplementary Material
Footnotes
Conflict of interest: none.
Supplementary material for this article can be found at www.redjournal.org.
References
- 1.FitzGerald TJ. A new model for imaging and radiation therapy quality assurance in the National Clinical Trials Network of the National Cancer Institute. Int J Radiat Oncol Biol Phys. 2014;88:272–273. doi: 10.1016/j.ijrobp.2013.09.030. [DOI] [PubMed] [Google Scholar]
- 2.Radiation Therapy Oncology Group Core Lab| TRIAD. http://www.rtog.org/CoreLab/TRIAD.aspx.
- 3.Santanam L, Hurkmans C, Mutic S, et al. Standardizing naming conventions in radiation oncology. Int J Radiat Oncol Biol Phys. 2012;83:1344–1349. doi: 10.1016/j.ijrobp.2011.09.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Miller A. Standardised Nomenclature in Radiotherapy Planning. 1996 http://www.academia.edu/6221247/Standardised_Nomenclature_in_Radiotherapy_Planning.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.