Abstract
The human genome is replete with long non-coding RNAs (lncRNA), many of which are transcribed and likely to have a functional role. Microarray analysis of > 23 000 lncRNAs revealed downregulation of 712 (~3%) lncRNA in malignant hepatocytes, among which maternally expressed gene 3 (MEG3) was downregulated by 210-fold relative to expression in non-malignant hepatocytes. MEG3 expression was markedly reduced in four human hepatocellular cancer (HCC) cell lines compared with normal hepatocytes by real-time PCR. RNA in situ hybridization showed intense cytoplasmic expression of MEG3 in non-neoplastic liver with absent or very weak expression in HCC tissues. Enforced expression of MEG3 in HCC cells significantly decreased both anchorage-dependent and -independent cell growth, and induced apoptosis. MEG3 promoter hypermethylation was identified by methylation-specific PCR and MEG3 expression was increased with inhibition of methylation with either 5-Aza-2-Deoxycytidine, or siRNA to DNA Methyltransferase (DNMT) 1 and 3b in HCC cells. MiRNA-dependent regulation of MEG3 expression was studied by evaluating the involvement of miR-29, which can modulate DNMT 1 and 3. Overexpression of mir-29a increased expression of MEG3. GTL2, the murine homolog of MEG3, was reduced in liver tissues from hepatocyte-specific miR-29a/b1 knock-out mice compared with wild-type controls. These data show that methylation-dependent tissue-specific regulation of the lncRNA MEG3 by miR-29a may contribute to HCC growth and highlight the inter-relationship between two classes of non-coding RNA, miRNAs and lncRNAs, and epigenetic regulation of gene expression.
Keywords: MEG3, methylation, HCC, non-coding RNA
Introduction
Recent large scale complementary DNA cloning projects have identified that a large portion of the mammalian genome is transcribed, although only a small fraction of these transcripts represent protein-coding genes (Kawaji et al., 2011). The functional role for most of these transcripts remains obscure and their relevance to human physiology and disease is undefined. Although the role of protein-coding genes as oncogenes or onco-suppressors has been extensively studied, the involvement of non-protein coding genes in tumor pathogenesis and growth is less well characterized. Their role in human cancers warrants evaluation.
Amongst the non-coding RNA, microRNAs have been extensively studied and their role in carcinogenesis has been established (Croce, 2009). Several non-coding RNAs (ncRNAs) with functional involvement in the liver have been identified, although the repertoire of long ncRNA (lncRNA) is not clear. A large and growing body of literature shows the involvement of miRNAs in hepatocarcinogenesis (Meng et al., 2007; Coulouarn et al., 2009; Gramantieri et al., 2009; Ji et al., 2009; Su et al., 2009; Wang et al., 2009; Braconi et al., 2010; Pineau et al., 2010). In contrast few long non-coding RNA have been identified (reviewed in Huarte and Rinn, 2010). Long ncRNA that have been implicated as having a functional role in hepatocarcinogenesis or liver cancer growth include HULC, H19 and TUC338 (Matouk et al., 2007; Panzitt et al., 2007; Braconi et al., 2011).
The mechanisms of expression of lncRNAs are unknown, although there is evidence for regulation by similar mechanisms as for protein coding genes. Long ncRNAs frequently originate from intronic regions and are independently transcribed (Louro et al., 2009). Likewise, their functional role remains obscure. It has been postulated that ncRNAs can act transcriptionally or post-transcriptionally; however, mechanisms that underlie such regulatory behaviors still remain to be fully understood. Long ncRNAs can mediate epigenetic changes by recruiting chromatin-remodeling complexes to specific genomic loci. Other long ncRNAs have been shown to regulate transcription, whereas a few long ncRNAs are antisense transcripts, which may regulate mRNA dynamics at a post-transcriptional level (Mercer et al., 2009; Orom et al., 2010). The mechanisms by which ncRNA expression is altered in cancers are similar to those for protein coding genes, and include epigenetic mechanisms (Lujambio et al., 2007, 2008; Saito et al., 2010).
In order to begin to understand the involvement of lncRNAs in human hepatocellular cancer (HCC), we sought to identify lncRNAs that are altered in expression in HCC, and characterize their expression and functional involvement in tumor growth. We identified the maternally expressed gene-3 (MEG3), as one of the most significantly downregulated lncRNAs in malignant hepatocytes, and report a functional tissue-specific mechanism of regulation of MEG3 expression via microRNA-29a-dependent regulation of promoter methylation.
Results and discussion
Deregulated expression of the long non-coding RNA MEG3 in human HCC
To identify long non-coding RNAs that are altered in expression in malignant hepatocytes, we performed a hybridization-based microarray analysis of ncRNA expression (Arraystar, Rockville, MD, USA). The expression of several lncRNAs was noted to be altered between malignant and normal human hepatocytes (Supplementary Table 1). MEG3 was among the top most significantly downregulated lncRNAs. MEG3 is an imprinted gene maternally expressed in humans that encodes a long non-coding RNA of ~1700 nucleotides (Zhou et al., 2007). Although expressed in several diverse isoforms in fetal liver, MEG3 is expressed in adult liver as a few isoforms (Hagan et al., 2009; Zhang et al., 2010b). The potential role and involvement of MEG3 in liver cancers is unknown. Using quantitative reverse transcriptase PCR, we verified that the expression of MEG3 was reduced in four separate HCC cell lines relative to the expression in non-malignant hepatocytes (Figure 1a). Next, we assessed the expression of MEG3 in a series of human primary HCC and adjacent liver tissues. MEG3 was remarkably reduced in expression in 81% of HCC in comparison with adjacent cirrhotic tissue (Figure 1b). We also assessed expression of MEG3 in human liver tissues by in situ hybridization. MEG3 staining was intensively positive in non-neoplastic liver, whereas expression was remarkably reduced in human HCC tissues (Supplementary Figure 1). These data indicate that MEG3 expression is downregulated in malignant liver cancer cell lines and tissues.
Figure 1.
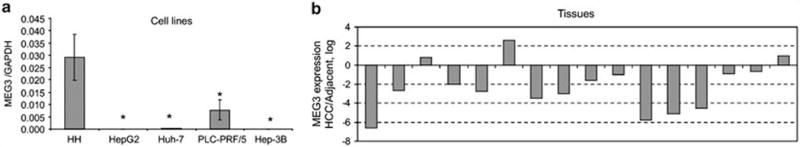
MEG3 expression is reduced in HCC. (a) MEG3 expression was assessed by real time PCR using SYBRGreen (Clontech, Mountain View, CA, USA) in human normal hepatocytes (HH) and HCC cells and expression was normalized to that of GAPDH. Bars represent mean and standard error of three samples. *P<0.05 relative to HH. (b) RNA was extracted with Trizol from human HCC and adjacent, cirrhotic tissues. MEG3 expression was assessed by real time PCR and normalized to GAPDH. Bars represent the ratio between expression in HCC and in adjacent tissue (log scale).
Functional consequences of deregulated MEG3 expression
To assess the potential functional role of MEG3 in hepatic epithelia, we overexpressed the full length of MEG3 complementary DNA in HCC cells. PLC/PRF/5 cells were transfected with MEG3-CMV6-XL5 or empty-CMV6-XL5 vectors. MEG3 expression was increased by ~eightfold after 24 h of transfection in cells transfected with vector overexpressing MEG3 in comparison with those transfected with an empty vector (Figure 2a). Cells with enforced expression of MEG3 had a decreased anchorage-dependent growth in comparison with cells transfected with empty vector (Figure 2b). The capacity of the cells to grow in absence of anchorage is a typical feature of cancer cells and was further verified by assessing the capacity of forming colonies in soft agar. Anchorage-independent growth was reduced by 40% in cells with enforced expression of MEG3 (Figure 2c). Next, we assessed the effect of MEG3 on progression through the cell cycle. Cells transfected with MEG3 showed an increased accumulation in the sub G0/1 phase without any significant modulation of the checkpoint G0/1 or G2/M (Figure 3a). These data suggested that MEG3 may modulate apoptosis. Thus we assessed the rate of late apoptosis by terminal transferase dUTP nickend labeling assay after MEG3 overexpression in HCC cells. Interestingly apoptotic cells (BrdU + /PI + ) were significantly increased after transfection with MEG3 in comparison with transfection with an empty vector (Figure 3b).
Figure 2.
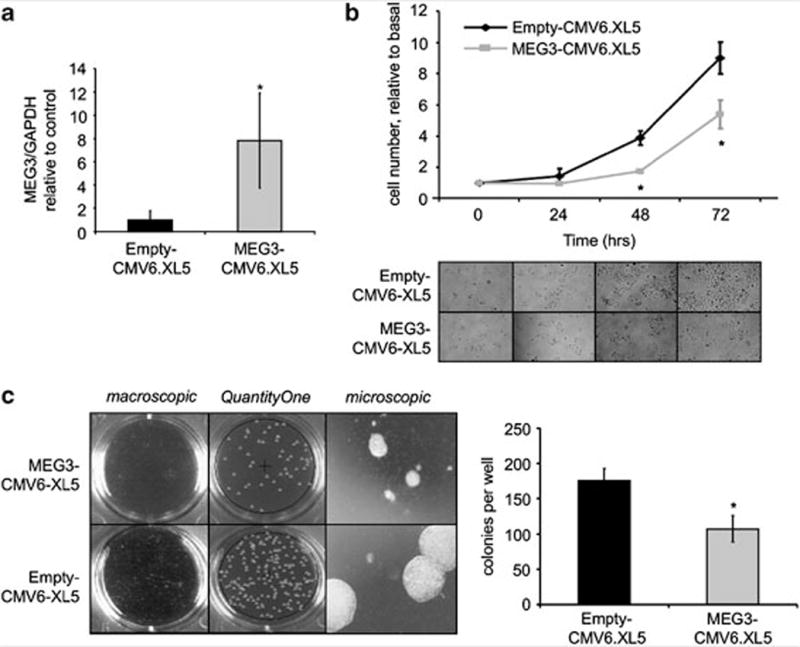
Overexpression of MEG3 decreases cell growth. PLC/PRF/5 cells were transfected with 1 μg of MEG3-CMV6.XL5 or empty vector (Origene, Rockville, MD, USA) using the Nucleofector system, solution V program T28 (Amaxa Biosystems, Koln, Germany). (a) After 24 h of transfection, cells were collected and MEG3 expression was assessed by real time PCR. Bars represent mean and standard error of three independent experiments, relative to control. *P<0.05 relative to control. (b) At 24 h after transfection, cells were plated in 12-well plates (10 000 cells/well). After 24, 48 and 72 h, cells were counted by trypan blue staining. Mean values of three independent experiments with standard errors are represented. *P<0.05 compared with control. (c) After 24 h of transfection, cells were seeded in six-well plates (5000 per well) in soft agar with 20% fetal bovine serum. The final concentration of the agar system was 1.2% for the bottom layer and 0.8% for the cell suspension layer. Colonies were imaged using the GelDoc Imaging system (Bio-Rad, Hercules, CA, USA) and quantitated using the QuantityOne software (Bio-rad). Bars represent mean and standard error of three independent experiments, relative to control. *P<0.05 relative to control.
Figure 3.
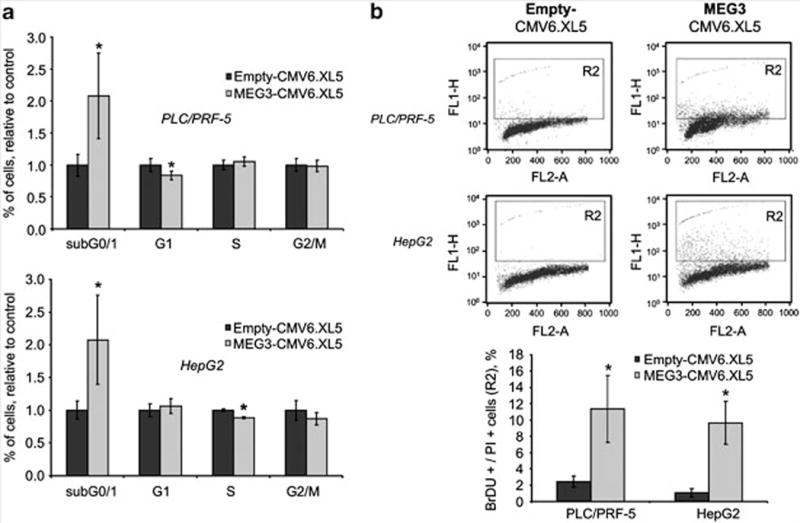
MEG3 induces apoptosis. HepG2 and PLC/PRF/5 cells were serum-starved for 48 h and then transfected with 1 μg of MEG3-CMV6.XL5 or empty vector. After 24 h cells were collected and fixed in paraformaldehyde and ethanol overnight. (a) Cells were stained by propidium iodide (PI) and analyzed by flow cytometry with a BD FACScalibur (BD Bioscience, Heidelberg, Germany). Bars represent mean and standard error of three experiments expressed as relative to empty control. *P<0.05 compared with empty controls. (b) Apoptotic cells were detected by labeling DNA breaks using the terminal deoxynucleotide transferase dUTP nick end labeling assay (Invitrogen, Carlsbad, CA, USA). Cells were processed as indicated by the manufacturer, stained by PI and bromodeoxyuridine (BrdU), and analyzed by flow cytometry with a BD FACScalibur. Bars represent mean and standard errors of three independent experiments. *P<0.05 compared with empty controls.
Deregulated expression of MEG3 in malignant hepatocytes, and in association with liver preneoplastic foci and tumors, suggests a role in tumorigenesis. There is data suggesting that MEG3 can function as a tumor-suppressor. Expression of MEG3 is lost in pituitary tumors, mengiomas and myelomas (Zhang et al., 2003, 2010a; Benetatos et al., 2008, 2010). Moreover, expression of MEG3 has been shown to suppress growth and modulate p53 expression and stimulate p53-mediated transactivation of downstream targets in colon and brain cancer cells (Zhou et al., 2007; Zhang et al., 2010a). Klibanski’s group found these effects to be only partially mediated by p53. Although we confirmed an increase of p53 in cells transfected with MEG3 (data not shown), we speculate that p53-independent mechanisms may contribute to MEG3-dependent effects in HCC. Although MEG3 expression is reduced in HCC cell lines, p53 is expressed in many of these with the exception of Hep3B. HepG2 cells retain a normal p53, PLC/PRF/5 express a functioning p53 even though at a reduced extent because of a specific mutation. Mutation in p53 gene in Huh-7 cells is responsible for a reduced half life and thus an increased expression (Bressac et al., 1990). Therefore, in view of our experimental results we postulate that the oncosuppressor effects of MEG3 are only partially mediated by p53 in HCC, and that alternative mechanisms are in place. Further studies are warranted to elucidate these alternative pathways.
Epigenetic regulation of MEG3
MEG3 is located at 14q32.3. Although a location for a tumor suppressor gene has been hypothesized in this region because it is frequently lost in cholangiocarcinoma or neuroblastoma, no protein-coding gene has been identified so far at this site (Cazals-Hatem et al., 2004). We speculate that MEG3 may be this tumor suppressor gene given our data that re-expression of MEG3 in HCC cells reduces cellular growth and induces apoptosis. As mutations are not reported at this site and a loss of heterozygosity reported in less than 10% of HCCs, mechanisms of inactivation other than genomic alterations may contribute to loss of MEG3 in HCC (Cazals-Hatem et al., 2004).
Deregulated expression of MEG3 can occur in response to diethyldithiocarbamic acid, a hepatotoxin that can induce epigenetic modifications in histones and DNA, and can be modulated by S-adenosylmethionine, a methyl donor (Oliva et al., 2009). To examine the role of aberrant methylation in deregulation of MEG3 in HCC, we evaluated the effect of the methylation-inhibitor 5-Aza-2-deoxycytidine (5-Aza-dc) on MEG3 expression. We noted that MEG3 expression was robustly increased by incubation with 5-Aza-dc 10 μM for 5 days in both Huh-7 and in HepG2 cells (Figure 4a). MEG3 expression was not modulated by 5-Aza-dc in PLC/PRF/5 cells suggesting that MEG3 expression could be modulated by mechanisms other than methylation. To better study the methylation-dependent mechanism of MEG3 alteration we further manipulated the expression of the main DNA methyltransferases (DNMT)-1 and DNMT-3B by RNA interference. In cells incubated with siRNA to DNMT-1/3B, an increase in MEG3 expression was observed (Figure 4b), even though at a lesser extent compared with 5-Aza-dc possibly due to differences in effect on DNMT activity. Methylation within the promoter region has been associated with loss of MEG3 expression in pituitary tumor and leukemia cells (Zhao et al., 2005; Benetatos et al., 2010). Thus, we next evaluated promoter methylation of MEG3 in HCC cells by performing a methylation specific PCR (Figure 4c). MEG3 is an imprinted gene expressed from the maternal allele. Thus, the normal pattern of MEG3 consists of two bands, one corresponding to the methylated paternal allele (160 bp) and a further band corresponding to the unmethylated maternal allele (120 bp). Although normal human hepatocytes have both the methylated and unmethylated forms, only the methylated form is present in HCC cells. Interestingly, re-expression of the unmethylated form occurs during exposure to 5-Aza-dc (Figure 4d). To better characterize the CpG islands that are methylated within the MEG3 promoter we performed a methylation-sequencing analysis. These experiments confirmed an increased methylation of CpG islands in HepG2 cells in comparison with non-malignant human hepatocytes (Supplementary Figure 2). In addition, methylation specific PCR analysis of DNA from HCC and adjacent cirrhotic tissues showed a marked reduction of the unmethylated form, which is responsible for the expression of MEG3, in HCC compared with adjacent tissues (Supplementary Figure 3). These data indicate that MEG3 expression can be modulated by alterations in methylation and support an epigenetic mechanism of regulation of MEG3 expression in HCC. These data are consistent with the others in the literature that provide evidence for the methylation-dependent regulation of ncRNAs (Lujambio et al., 2007, 2008; Saito et al., 2010).
Figure 4.
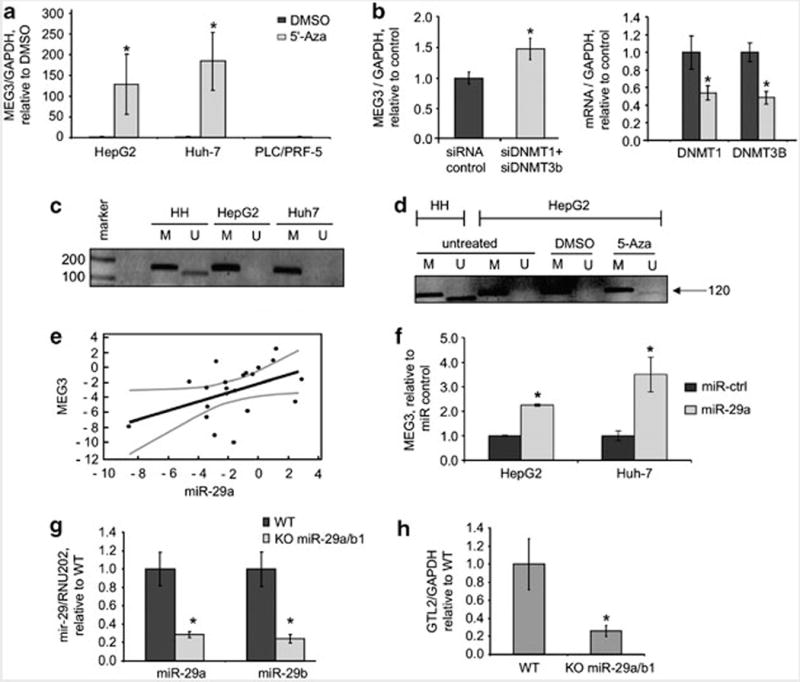
MEG3 expression is methylation dependent. (a) Cells were incubated with 5-Aza-dc (Calbiochem, Darmstadt, Germany) 10 μM for 5 days and MEG3 expression was assessed by real time PCR. Bars represent mean and standard error of three independent experiments, relative to control (dimethyl sulfoxide (DMSO)). *P<0.05 relative to controls. (b) HepG2 cells were transfected with siRNA to DNMT1 + DNMT3b or siRNA control (100nM, Dharmacon, Chicago, IL, USA) for 48 h. MEG3 and DNMT1 expression was assessed by real time PCR. DNMT1 and DNMT3b expression was reduced by ~50%. Bars represent mean and standard error of three independent experiments, relative to control. *P<0.05. (c) DNA was extracted by using phenol–chloroform and treated with bisulfite using the EzDNA Methylation kit (Zymoresearch, Orange, CA, USA). Bisulfite-treated DNA was then used for methylation specific PCR (MSP) reaction as previously described (Benetatos et al. 2010). PCR products were identified by ethidium bromide staining after 2% agarose gel electrophoresis. For each sample a primer set for the methylated (M) and unmethylated (U) copies of MEG3 gene was used. A 160 bp product represents the methylated state and a 120 bp PCR product stands for unmethylated state. (marker 100 bp: Promega, Madison, WI, USA). (d) HepG2 cells were treated with 5-Aza-dc 10 μM for 24 h and MSP was performed as described above. Treatment with 5-Aza-dc increased expression of the unmethylated form of MEG3. (e) miR-29a and MEG3 expression was assessed in non-malignant human hepatocytes (HH), HCC cells (HepG2, Huh-7, PLC/PRF/5, Hep-3B), human HCC tissues and adjacent non-tumoral tissues. Mir-29a expression was assessed by Taqman real time PCR assay and normalized to that of RNU6B (Applied Biosystem, Foster City, CA, USA). MEG3 expression was assessed by PCR using SYBRGreen and normalized to that of GAPDH. In the graph miR-29a and MEG3 are expressed relative to HH for the cell lines or to adjacent tissues for human HCC (log scale). Regression analysis was performed using the MedCalc Software (Mariakerke, Belgium). The fitted regression line (black) and 95% confidence intervals (gray) are shown. r = 0.45, 95% confidence intervals 0.03–0.79, P: 0.038. (f) HepG2 and Huh7 cells were transfected with pre-miR-29a or control (100 nM, Applied Biosystem). After 48 h, cells were collected, RNA extracted and real time PCR performed. Bars represent mean and standard error of three independent experiments, relative to controls. *P<0.05 relative to controls. (g) GTL2 is the MEG3 homologous gene in mouse genome. Liver-specific miR-29a/b1 KO mouse (C57BL/6) was developed by serial breeding of miR-29a/b1loxP/loxP and albumin-Cre (Alb-Cre) mice. All experiments were reviewed and approved by the Institutional Animal Care and Use Committee at the Ohio State University. RNA was collected from liver tissues from wild type (WT) or miR-29a/b1 KO mice. miR-29a and miR-29b were assessed by Taqman assay and normalized to that of RNU202. Bars represent mean and standard error of expression in four mice, relative to control (WT). *P<0.05 relative to controls. (h) GTL2 expression was assessed by SYBRGreen and normalized to that of mouse GAPDH. Bars represent mean and standard error of expression in four mice, relative to control (WT). *P<0.05 relative to controls.
miRNA-dependent regulation of MEG3 expression
Reduced expression of miR-29 is a frequent event in HCC and is correlated with a poor prognosis (Xiong et al., 2010). miR-29 can modulate the expression of DNMT-1 and 3B in leukemia and lung cancer cells (Fabbri et al., 2007; Garzon et al., 2009). We confirmed a similar miR-29a-dependent modulation of DNMT protein expression in HCC cells (Supplementary Figure 4). Having showed that these methyltransferases can modulate MEG3 expression, we hypothesized that deregulated miR-29 could represent a mechanism for the silencing of MEG3 in HCC through modulation of DNMTs. We focused our attention on miR-29a, the isoform mainly represented in hepatocytes (data not shown). The expression of miR-29a in several HCC cell lines and human HCC tissues correlated with expression of MEG3 with a P-value of 0.04 (Figure 4e). Over-expression of miR-29a in HCC cells increased MEG3 expression (Figure 4f). The effects of miR-29a on MEG3 expression were further evaluated in vivo using genetically engineered mice with a hepatocyte-specific knockout of miR-29 a/b1. The murine analog of MEG3, named GTL2 is located on chromosome 12. Compared with the expression in liver tissues in wild-type mice, GTL2 expression was remarkably reduced in liver tissues from miR-29 KO mice under basal conditions (Figures 4g and h). All together these studies suggest that miR-29 may indirectly modulate MEG3/GTL2 expression by acting on the methylation machinery. Xiong et al. (2010) proposed that mitochondrial apoptosis is increased in HCC cells as a result of the loss of miR-29. Our data suggest that the effects of miR-29 on apoptosis may be at least in part mediated by the modulation of MEG3.
Although the mechanisms of regulation of expression of functional lncRNAs are not known, emerging data suggest the involvement of mechanisms similar to those described for protein coding genes. Recently, Esteller’s group showed the methylation-dependent regulation of highly conserved lncRNAs (Lujambio et al., 2010), members of which have been deregulated in cancers despite a lack of mutations at their genomic site (Wojcik et al., 2010). The modulation of expression of lncRNAs by other regulatory RNAs such as miRNAs by direct mechanisms or by indirect mechanisms such as through modulation of epigenetic regulation adds considerably to the complexity of regulation of gene expression.
In summary, we have identified deregulated expression of the lncRNA MEG3 in HCC, demonstrated a functional effect in vitro, and identified a potential mechanism by which deregulated tissue-specific expression of miR-29a in HCC could epigenetically modulate MEG3 expression through promoter hypermethylation. These studies highlight the inter-relationship between two classes of non-coding RNAs, miRNAs and lncRNAs, and epigenetic regulation of gene expression. They extend the known roles of miRNA such as miR-29a in the pathobiology of HCC, and emphasize the importance and relevance of lncRNAs such as MEG3 as potential downstream targets for therapeutic intervention in HCC.
For experimental procedures, see Supplementary Materials and Methods section.
Supplementary Material
Footnotes
Conflict of interest
The authors declare no conflict of interest.
Supplementary Information accompanies the paper on the Oncogene website (http://www.nature.com/onc)
References
- Benetatos L, Dasoula A, Hatzimichael E, Georgiou I, Syrrou M, Bourantas KL. Promoter hypermethylation of the MEG3 (DLK1/MEG3) imprinted gene in multiple myeloma. Clin Lymphoma Myeloma. 2008;8:171–175. doi: 10.3816/CLM.2008.n.021. [DOI] [PubMed] [Google Scholar]
- Benetatos L, Hatzimichael E, Dasoula A, Dranitsaris G, Tsiara S, Syrrou M, et al. CpG methylation analysis of the MEG3 and SNRPN imprinted genes in acute myeloid leukemia and myelodysplastic syndromes. Leuk Res. 2010;34:148–153. doi: 10.1016/j.leukres.2009.06.019. [DOI] [PubMed] [Google Scholar]
- Braconi C, Valeri N, Gasparini P, Huang N, Taccioli C, Nuovo G, et al. Hepatitis C virus proteins modulate microRNA expression and chemosensitivity in malignant hepatocytes. Clin Cancer Res. 2010;16:957–966. doi: 10.1158/1078-0432.CCR-09-2123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braconi C, Valeri N, Kogure T, Gasparini P, Huang N, Nuovo GJ, et al. Expression and functional role of a transcribed noncoding RNA with an ultraconserved element in hepatocellular carcinoma. Proc Natl Acad Sci USA. 2011;108:786–791. doi: 10.1073/pnas.1011098108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bressac B, Galvin KM, Liang TJ, Isselbacher KJ, Wands JR, Ozturk M. Abnormal structure and expression of p53 gene in human hepatocellular carcinoma. Proc Natl Acad Sci USA. 1990;87:1973–1977. doi: 10.1073/pnas.87.5.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cazals-Hatem D, Rebouissou S, Bioulac-Sage P, Bluteau O, Blanche H, Franco D, et al. Clinical and molecular analysis of combined hepatocellular-cholangiocarcinomas. J Hepatol. 2004;41:292–298. doi: 10.1016/j.jhep.2004.04.030. [DOI] [PubMed] [Google Scholar]
- Coulouarn C, Factor VM, Andersen JB, Durkin ME, Thorgeirsson SS. Loss of miR-122 expression in liver cancer correlates with suppression of the hepatic phenotype and gain of metastatic properties. Oncogene. 2009;28:3526–3536. doi: 10.1038/onc.2009.211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croce CM. Causes and consequences of microRNA dysregulation in cancer. Nat Rev Genet. 2009;10:704–714. doi: 10.1038/nrg2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabbri M, Garzon R, Cimmino A, Liu Z, Zanesi N, Callegari E, et al. MicroRNA-29 family reverts aberrant methylation in lung cancer by targeting DNA methyltransferases 3A and 3B. Proc Natl Acad Sci USA. 2007;104:15805–15810. doi: 10.1073/pnas.0707628104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garzon R, Liu S, Fabbri M, Liu Z, Heaphy CE, Callegari E, et al. MicroRNA-29b induces global DNA hypomethylation and tumor suppressor gene reexpression in acute myeloid leukemia by targeting directly DNMT3A and 3B and indirectly DNMT1. Blood. 2009;113:6411–6418. doi: 10.1182/blood-2008-07-170589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gramantieri L, Fornari F, Ferracin M, Veronese A, Sabbioni S, Calin GA, et al. MicroRNA-221 targets Bmf in hepatocellular carcinoma and correlates with tumor multifocality. Clin Cancer Res. 2009;15:5073–5081. doi: 10.1158/1078-0432.CCR-09-0092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagan JP, O’Neill BL, Stewart CL, Kozlov SV, Croce CM. At least ten genes define the imprinted Dlk1-Dio3 cluster on mouse chromosome 12qF1. PLoS One. 2009;4:e4352. doi: 10.1371/journal.pone.0004352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huarte M, Rinn JL. Large non-coding RNAs: missing links in cancer? Hum Mol Genet. 2010;19:R152–R161. doi: 10.1093/hmg/ddq353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawaji H, Severin J, Lizio M, Forrest AR, van NE, Rehli M, et al. Update of the FANTOM web resource: from mammalian transcriptional landscape to its dynamic regulation. Nucleic Acids Res. 2011;39:D856–D860. doi: 10.1093/nar/gkq1112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji J, Shi J, Budhu A, Yu Z, Forgues M, Roessler S, et al. MicroRNA expression, survival, and response to interferon in liver cancer. N Engl J Med. 2009;361:1437–1447. doi: 10.1056/NEJMoa0901282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louro R, Smirnova AS, Verjovski-Almeida S. Long intronic noncoding RNA transcription: expression noise or expression choice? Genomics. 2009;93:291–298. doi: 10.1016/j.ygeno.2008.11.009. [DOI] [PubMed] [Google Scholar]
- Lujambio A, Ropero S, Ballestar E, Fraga MF, Cerrato C, Seti F, et al. Genetic unmasking of an epigenetically silenced microRNA in human cancer cells. Cancer Res. 2007;67:1424–1429. doi: 10.1158/0008-5472.CAN-06-4218. [DOI] [PubMed] [Google Scholar]
- Lujambio A, Calin GA, Villanueva A, Ropero S, Sanchez-Caspedes M, Blanco D, et al. A microRNA DNA methylation signature for human cancer metastasis. Proc Natl Acad Sci USA. 2008;105:13556–13561. doi: 10.1073/pnas.0803055105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lujambio A, Portela A, Liz J, Melo SA, Rossi S, Spizzo R, et al. CpG island hypermethylation-associated silencing of non-coding RNAs transcribed from ultraconserved regions in human cancer. Oncogene. 2010;29:6390–6401. doi: 10.1038/onc.2010.361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matouk IJ, Degroot N, Mezan S, Ayesh S, bu-Lail R, Hochberg A, et al. The H19 non-coding RNA is essential for human tumor growth. PLoS One. 2007;2:e845. doi: 10.1371/journal.pone.0000845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng F, Henson R, Wehbe-Janek H, Ghoshal K, Jacob ST, Patel T. MicroRNA-21 regulates expression of the PTEN tumor suppressor gene in human hepatocellular cancer. Gastroenterology. 2007;133:647–658. doi: 10.1053/j.gastro.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercer TR, Dinger ME, Mattick JS. Long non-coding RNAs: insights into functions. Nat Rev Genet. 2009;10:155–159. doi: 10.1038/nrg2521. [DOI] [PubMed] [Google Scholar]
- Oliva J, Bardag-Gorce F, French BA, Li J, French SW. The regulation of non-coding RNA expression in the liver of mice fed DDC. Exp Mol Pathol. 2009;87:12–19. doi: 10.1016/j.yexmp.2009.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orom UA, Derrien T, Beringer M, Gumireddy K, Gardini A, Bussotti G, et al. Long noncoding RNAs with enhancer-like function in human cells. Cell. 2010;143:46–58. doi: 10.1016/j.cell.2010.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panzitt K, Tschernatsch MM, Guelly C, Moustafa T, Stradner M, Strohmaier HM, et al. Characterization of HULC, a novel gene with striking up-regulation in hepatocellular carcinoma, as noncoding RNA. Gastroenterology. 2007;132:330–342. doi: 10.1053/j.gastro.2006.08.026. [DOI] [PubMed] [Google Scholar]
- Pineau P, Volinia S, McJunkin K, Marchio A, Battiston C, Terris B, et al. miR-221 overexpression contributes to liver tumorigenesis. Proc Natl Acad Sci USA. 2010;107:264–269. doi: 10.1073/pnas.0907904107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito Y, Liang G, Egger G, Friedman JM, Chuang JC, Coetzee GA, et al. Specific activation of microRNA-127 with down-regulation of the proto-oncogene BCL6 by chromatin-modifying drugs in human cancer cells. Cancer Cell. 2010;9:435–443. doi: 10.1016/j.ccr.2006.04.020. [DOI] [PubMed] [Google Scholar]
- Su H, Yang JR, Xu T, Huang J, Xu L, Yuan Y, et al. MicroRNA-101, down-regulated in hepatocellular carcinoma, promotes apoptosis and suppresses tumorigenicity. Cancer Res. 2009;69:1135–1142. doi: 10.1158/0008-5472.CAN-08-2886. [DOI] [PubMed] [Google Scholar]
- Wang B, Majumder S, Nuovo G, Kutay H, Volinia S, Patel T, et al. Role of microRNA-155 at early stages of hepatocarcinogenesis induced by choline-deficient and amino acid-defined diet in C57BL/6 mice. Hepatology. 2009;50:1152–1161. doi: 10.1002/hep.23100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wojcik SE, Rossi S, Shimizu M, Nicoloso MS, Cimmino A, Alder H, et al. Non-codingRNA sequence variations in human chronic lymphocytic leukemia and colorectal cancer. Carcinogenesis. 2010;31:208–215. doi: 10.1093/carcin/bgp209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong Y, Fang JH, Yun JP, Yang J, Zhang Y, Jia WH, et al. Effects of microRNA-29 on apoptosis, tumorigenicity, and prognosis of hepatocellular carcinoma. Hepatology. 2010;51:836–845. doi: 10.1002/hep.23380. [DOI] [PubMed] [Google Scholar]
- Zhang X, Gejman R, Mahta A, Zhong Y, Rice KA, Zhou Y, et al. Maternally expressed gene 3, an imprinted noncoding RNA gene, is associated with meningioma pathogenesis and progression. Cancer Res. 2010a;70:2350–2358. doi: 10.1158/0008-5472.CAN-09-3885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Rice K, Wang Y, Chen W, Zhong Y, Nakayama Y, et al. Maternally expressed gene 3 (MEG3) noncoding ribonucleic acid: isoform structure, expression, and functions. Endocrinology. 2010b;151:939–947. doi: 10.1210/en.2009-0657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Zhou Y, Mehta KR, Danila DC, Scolavino S, Johnson SR, et al. A pituitary-derived MEG3 isoform functions as a growth suppressor in tumor cells. J Clin Endocrinol Metab. 2003;88:5119–5126. doi: 10.1210/jc.2003-030222. [DOI] [PubMed] [Google Scholar]
- Zhao J, Dahle D, Zhou Y, Zhang X, Klibanski A. Hypermethylation of the promoter region is associated with the loss of MEG3 gene expression in human pituitary tumors. J Clin Endocrinol Metab. 2005;90:2179–2186. doi: 10.1210/jc.2004-1848. [DOI] [PubMed] [Google Scholar]
- Zhou Y, Zhong Y, Wang Y, Zhang X, Batista DL, Gejman R, et al. Activation of p53 by MEG3 non-coding RNA. J Biol Chem. 2007;282:24731–24742. doi: 10.1074/jbc.M702029200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


