Abstract
IMPORTANCE
Genetic association studies of psychiatric outcomes often consider interactions with environmental exposures and, in particular, apply tests that jointly consider gene and gene-environment interaction effects for analysis. Using a genome-wide association study (GWAS) of posttraumatic stress disorder (PTSD), we report that heteroscedasticity (defined as variability in outcome that differs by the value of the environmental exposure) can invalidate traditional joint tests of gene and gene-environment interaction.
OBJECTIVES
To identify the cause of bias in traditional joint tests of gene and gene-environment interaction in a PTSD GWAS and determine whether proposed robust joint tests are insensitive to this problem.
DESIGN, SETTING, AND PARTICIPANTS
The PTSD GWAS data set consisted of 3359 individuals (978 men and 2381 women) from the Grady Trauma Project (GTP), a cohort study from Atlanta, Georgia. The GTP performed genome-wide genotyping of participants and collected environmental exposures using the Childhood Trauma Questionnaire and Trauma Experiences Inventory.
MAIN OUTCOMES AND MEASURES
We performed joint interaction testing of the Beck Depression Inventory and modified PTSD Symptom Scale in the GTP GWAS. We assessed systematic bias in our interaction analyses using quantile-quantile plots and genome-wide inflation factors.
RESULTS
Application of the traditional joint interaction test to the GTP GWAS yielded systematic inflation across different outcomes and environmental exposures (inflation-factor estimates ranging from 1.07 to 1.21), whereas application of the robust joint test to the same data set yielded no such inflation (inflation-factor estimates ranging from 1.01 to 1.02). Simulated data further revealed that the robust joint test is valid in different heteroscedasticity models, whereas the traditional joint test is invalid. The robust joint test also has power similar to the traditional joint test when heteroscedasticity is not an issue.
CONCLUSIONS AND RELEVANCE
We believe the robust joint test should be used in candidate-gene studies and GWASs of psychiatric outcomes that consider environmental interactions. To make the procedure useful for applied investigators, we created a software tool that can be called from the popular PLINK package for analysis.
Genetic studies of psychiatric disorders often conduct association analyses that consider interactions with environmental exposures of interest. Most such interaction studies have used candidate-gene designs, but psychiatric genetics is increasingly interested in using single-nucleotide polymorphism (SNP) data from genome-wide association studies (GWASs) to perform genome-wide interaction analysis.1-4 For a candidate-gene study or GWAS interaction analysis, a popular analytic tool is a joint test that considers the combined effects of SNP and SNP-environment interaction on outcome. If interactions exist, then these joint tests can yield greater power for gene mapping relative to traditional testing of marginal SNP effects alone5,6 and can resolve replication failures of marginal SNP findings.7 Many joint tests of main and interaction effects exist5,8-11 and are implemented in popular statistical software packages, such as PLINK.12 Researchers have used such procedures successfully to identify SNPs associated with various complex traits.13-15
Using joint interaction tests, we performed a genomewide interaction study of psychiatric outcomes related to post-traumatic stress disorder (PTSD) because PTSD arises from the complex interplay of genetics and environment.16 The differential risk determining those who do vs those who do not develop PTSD is multifaceted,17-25 but it is in part genetic, with approximately 30% heritability for PTSD risk after trauma.26,27 Cornelis et al28 provided an overview of genetic findings for PTSD but noted that several candidate-gene studies identified genetic variants that interact with trauma-specific environmental exposures to influence PTSD. For example, SNPs in the FKBP5 gene interact with childhood maltreatment measures to raise the risk of PTSD,18,29 whereas the effect of an insertion-deletion polymorphism in SLC6A4 on PTSD susceptibility appears to be modified by trauma type and social environment.30-32 Thus, studying genetic risk in the presence of environmental factors is important for psychiatric disorders, such as PTSD.
Motivated by the candidate-gene interaction findings for PTSD, we performed a genome-wide interaction analysis using joint interaction tests in a GWAS of PTSD and psychiatric-related outcomes collected by the Grady Trauma Project (GTP). The GTP performed genome-wide genotyping of approximately 4000 individuals and further collected psychiatric phenotype measures using outcomes such as the modified PTSD Symptom Scale (PSS)18,19,33-37 and the Beck Depression Inventory (BDI).38 The GTP also collected trauma history information from participants using measures such as the Childhood Trauma Questionnaire (CTQ)39 and the Trauma Experiences Inventory (TEI).18,35-37 Using this GTP data set, we performed genome-wide interaction analyses of quantitative psychiatric outcomes in PLINK12 with a joint SNP interaction test that considered the main effect of the SNP and its interaction with an environmental exposure (CTQ or TEI). PLINK implements this joint test using a linear regression model that regresses the psychiatric outcome on the genetic and environmental predictors as well as other covariates (such as principal components to correct for population stratification).
The results of the PLINK analyses revealed some unexpected trends. Specifically, we observed substantial genome-wide inflation in our joint tests across different outcomes and environmental exposures. At the same time, we observed no genome-wide inflation in the corresponding marginal SNP tests of each outcome that ignored the environmental exposures. Initially, it seems counterintuitive that a joint analysis of SNP and SNP-environment interaction would demonstrate genome-wide inflation, but its marginal counterpart would not. Nonetheless, subsequent research revealed a source behind our findings. For each outcome analyzed, we observed that the variance of the outcome differed by levels of the environmental exposure considered in the interaction term of the joint test. The observation that the variance of an outcome differs among levels of the trauma exposure is known as heteroscedasticity, and the phenomenon violates an important assumption of linear regression that requires the variance of trait outcome conditional on predictors to be the same across individuals (defined as homoscedasticity). Such heteroscedasticity originating from the environmental exposures leads to bias in the joint test of SNP and SNP-environment interaction but does not affect the marginal SNP test. This issue, if ignored, can lead to invalidation of joint interaction tests in candidate-gene SNP studies and larger GWAS projects.
Fortunately, we can correct for the inflation in the traditional joint tests of quantitative outcomes by modifying these tests to incorporate robust SEs40,41 that are insensitive to heteroscedasticity. These robust errors are often referred to as Huber-White, sandwich, or heteroscedasticity-consistent errors and are standard analytic tools in econometric,42 political science,43 anthropomorphic,44,45 and biomarker46 studies to deal with misspecification of mean and variance assumptions in statistical models. Such errors also were considered previously in gene-environment interaction studies to handle misspecification of the underlying mean model and violation of the homoscedasticity assumption.47,48 We found that application of this robust joint test to our PTSD interaction analyses resolved the systematic inflation that we observed in our original joint tests that used model-based SEs. We further illustrate the robust approach using simulated quantitative data under heteroscedasticity and homoscedasticity models; we observed that our robust approach (1) consistently provides valid inference in models that yield inflation for the traditional joint test of SNP and SNP-environment interaction and (2) has power similar to the traditional joint test when homoscedasticity actually holds. We therefore believe the robust joint test should be used in interaction studies to ensure valid findings in GWASs and candidate-gene studies of quantitative outcomes. To enable investigators to easily use these robust interaction tests in existing GWASs and candidate-gene studies, we implemented the approach as software that can run directly through the popular PLINK12 software package for genetic analysis.
Methods
The GTP GWAS of PTSD
The GTP completed detailed trauma interviews on a large collection of underserved adults (mean age, 40 years) with high rates of current and lifetime PTSD. Study participants were primarily female and of African American ancestry. The GTP collected a variety of self-reported measures of PTSD, depression, and trauma exposures on these individuals. In this work, we consider 2 quantitative psychiatric phenotypes: the intrusive symptoms of the PSS (PSSint) and the BDI. We also consider 2 continuous measures of trauma exposure: non–childhood abuse trauma assessed using the TEI and childhood trauma measured using the CTQ. We describe the construction of these phenotypes and trauma exposures in the eMethods in the Supplement. The institutional review board at Emory University approved the study procedures. All participants provided written informed consent before participating.
The GTP performed genome-wide SNP genotyping of 4622 individuals using the HumanOmniExpress (N = 280) and Omni1-Quad BeadChip (N = 4342) (Illumina Inc). The HumanOmniExpress interrogates 730 525 individual SNPs per sample, whereas the Omni1-Quad BeadChip interrogates 1 011 219 individual SNPs. We performed quality control on the SNP data and constructed principal components for each study participant (eMethods in the Supplement). After this process, our sample consisted of 3359 individuals (978 men and 2381 women) genotyped for 634 838 SNPs. For this article, we further filtered the SNPs considered for analysis by requiring a marker to have a minor allele frequency of 0.10 or greater in the sample. We imposed this minor allele frequency filter because an interaction test of a SNP with a minor allele frequency less than 0.10 might not follow the assumed asymptotic distribution (and therefore be invalid), because of the limited number of individuals possessing the minor allele, even if the underlying modeling assumptions hold; we wished to remove this potential confounding issue from our analysis. After this additional filtering step, the final number of SNPs used in this work decreased from 634 838 to 494 793.
Using our final sample, we first performed marginal SNP analysis of the PSSint and BDI in PLINK using a Wald test derived from a linear regression model that regressed outcome on SNP, adjusting for the effects of the top 10 principal components, sex, and genotyping chip used (HumanOmniExpress or Omni1-Quad BeadChip). We next performed joint analysis of SNP and SNP-environment interaction for various combinations of outcome (PSSint or BDI) and environmental exposure (TEI or CTQ) using the regression framework shown in equation 1 in the eMethods in the Supplement. We again adjusted for the effects of the top principal components, sex, and genotyping chip. We constructed the model-based and robust joint tests (shown in equations 3 and 5, respectively, in the eMethods in the Supplement). We performed these analyses within PLINK using an R software plugin (eMethods in the Supplement).
Results
Application of Model-Based Joint Tests in the GTP Cohort
Figure 1 shows the quantile-quantile (QQ) plots for the model-based joint analyses of SNP and SNP-environment interaction with the BDI. For comparison, Figure 1 also includes the QQ plot of the marginal SNP association results for BDI. Figure 1 clearly shows sizable inflation in P values in both joint tests that we considered. To quantify this observation, we calculated the genome-wide inflation factor (λ) for each joint analysis as the median of the observed joint tests divided by the median of a χ22 random variable (which is the asymptotic distribution of the joint Wald test under the null hypothesis). This quantity is a variation of the genomic-control inflation factor49 often applied in GWASs. We observed a genome-wide inflation factor λ = 1.21 for the model-based joint test of SNP and SNP-CTQ interaction and λ = 1.07 for the model-based joint test of SNP and SNP-TEI interaction. Applying a variation of genomic control (by dividing each test statistic by the estimated λ before calculating the P value) removed inflation at the median but did not resolve the P value inflation in these interaction tests for much of the distribution (eFigure 1 in the Supplement). In contrast, we found no such inflation in marginal SNP tests of BDI (λ = 1.01; calculated as the median of the observed marginal tests divided by the median of a χ12 random variable). The marginal result in Figure 1 did not adjust for the main effect of the environmental predictor (CTQ or TEI); however, marginal analyses that adjusted for these environmental outcomes yielded similar findings to the unadjusted analyses (eFigure 2 in the Supplement).
Figure 1.

Quantile-Quantile Plot of Observed P Values vs Expected P Values (on a −log10 Scale) for Joint and Marginal Genetic Analyses of the Beck Depression Inventory in the Posttraumatic Stress Disorder Genome-wide Association Study
Joint analyses considered environmental exposures using the Childhood Trauma Questionnaire (CTQ) and Trauma Experiences Inventory (TEI). λ denotes the inflation factor (defined as the median of the test statistic divided by the median of the asymptotic distribution of the test statistic).
Figure 2 presents analogous QQ plots for joint and marginal SNP analyses of the PSSint. As with the BDI, the QQ plots for the PSSint demonstrate genome-wide inflation of the joint SNP test (λ = 1.13 for the joint test of SNP and SNP-CTQ interaction and λ = 1.07 for the joint test of SNP and SNP-TEI interaction) but no such inflation for the marginal SNP test (λ = 1.00). As above, application of genomic control did not fully correct the P value inflation in the joint interaction tests (eFigure 1 in the Supplement). In addition, marginal SNP analyses that adjusted for the main effect of the environmental predictor yielded similar results to the unadjusted analyses that ignored the main environmental effect (eFigure 2 in the Supplement).
Figure 2.

Quantile-Quantile Plot of Observed P Values vs Expected P Values (on a −log10 Scale) for Joint and Marginal Genetic Analyses of Intrusive Symptoms of the Posttraumatic Stress Disorder (PTSD) Symptom Scale in the PTSD Genome-wide Association Study
Joint analyses considered environmental exposures using the Childhood Trauma Questionnaire (CTQ) and Trauma Experiences Inventory (TEI). λ denotes the inflation factor (defined as the median of the test statistic divided by the median of the asymptotic distribution of the test statistic).
We believe the bias we observed in the model-based joint SNP tests of main and interaction effect within the GTP was due to heteroscedasticity of the outcome variables with respect to the trauma exposure in the GTP. We estimated the variance of the PSSint and BDI across quartiles of the TEI and CTQ and reported these estimates in the Table. For the BDI, we found the variance of the outcome was statistically different across quartiles of the TEI and CTQ (P < 1 × 10−4 for each combination using a Brown-Forsythe test of variance equality50,51). For the PSSint, we also observed statistically significant differences in variance across the environmental exposures (P <1× 10−4 for each combination). Therefore, we conclude heteroscedasticity exists in the GTP data set.
Table.
Estimated Variances of Posttraumatic Stress Disorder Outcomes Across Quartiles of Trauma Exposurea
| Quartile | BDI | PSSint |
|---|---|---|
| TEI | ||
|
| ||
| First | 108.51 | 8.61 |
|
| ||
| Second | 114.26 | 12.66 |
|
| ||
| Third | 134.92 | 14.51 |
|
| ||
| Fourth | 174.50 | 18.87 |
|
| ||
| CTQ | ||
|
| ||
| First | 71.43 | 8.61 |
|
| ||
| Second | 94.00 | 11.87 |
|
| ||
| Third | 120.11 | 13.54 |
|
| ||
| Fourth | 180.72 | 18.39 |
Abbreviations: BDI, Beck Depression Index; CTQ, Childhood Trauma Questionnaire; PSSint, intrusive symptoms of the Posttraumatic Stress Disorder Symptom Scale; TEI, Trauma Experiences Inventory.
Psychiatric outcomes are the BDI and PSSint. Trauma exposures are a subscale of the TEI reflecting non–child abuse trauma exposure and the CTQ.
Application of Robust Joint Tests in the GTP Cohort
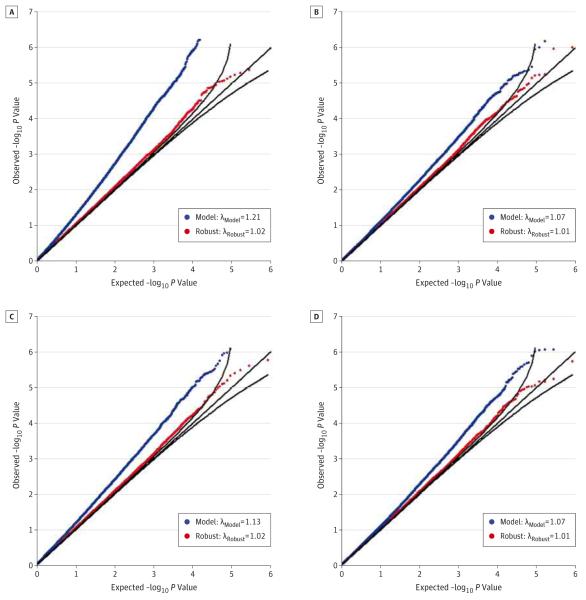
Figure 3 shows the QQ plots from the GTP GWAS comparing the expected vs observed P values (on a −log10 scale) for robust joint tests of SNP and SNP-environment interaction for the BDI and PSSint. We also provide the results for the analogous model-based joint test for comparison. For each joint analysis of SNP and SNP-environment interaction considered, the results clearly reveal that the use of the robust statistic nearly eliminates the systematic genome-wide inflation observed using the model-based statistic. For the BDI, we observed that the inflation of the joint test of the SNP and SNP-CTQ interaction decreased from λ = 1.21 when using the model-based statistic to λ = 1.02 when using the robust statistic. Similarly, genome-wide inflation of the joint test of the SNP and SNP-TEI interaction went from λ = 1.07 (model) to λ = 1.01 (robust). For the PSSint, we saw a similar reduction in the bias of the joint test of the SNP and SNP-environment interaction using the robust statistic. Once we applied the robust Huber-White variance estimators, we did not identify any joint tests that were genome-wide significant for the PSSint or BDI in the limited GTP data set.
Figure 5.
Quantile-Quantile Plot of Observed P Values vs Expected P Values (on a −log10 Scale) for Joint Analyses of the Beck Depression Inventory (BDI) and the Intrusive Symptoms of the Posttraumatic Stress Disorder (PTSD) Symptom Scale (PSSint) in the PTSD Genome-wide Association Study Using Model-Based and Robust Variance Estimators
Joint analyses considered environmental exposures using the Childhood Trauma Questionnaire (CTQ) and Trauma Experiences Inventory (TEI). λ denotes the inflation factor (defined as the median of the test statistic divided by the median of the asymptotic distribution of the test statistic). A, Joint test of the BDI (CTQ exposure); B, joint test of the BDI (TEI exposure); C, joint test of the PSSint (CTQ exposure); and D, joint test of the PSSint (TEI exposure).
Simulation Results
We further evaluated the model-based and robust versions of the joint tests using simulated data. We describe the simulation design in the eMethods in the Supplement. Figure 4 presents the QQ plots for the analysis of null data sets where neither SNP nor SNP-environment interaction effect influences phenotype. Figure 4 shows the expected vs observed P values (on a −log10 sc ale) for joint testing of SNP and SNP-environment interactions under homoscedasticity and the 2 forms of heteroscedasticity. Under homoscedasticity, the results reveal that the model-based and robust joint tests have no systematic inflation under the null hypothesis. We expect the model-based joint test to be valid in this situation because, under homoscedasticity, the modeling assumptions required for valid inference using this statistic are fulfilled. However, under heteroscedasticity, we found that the model-based joint test led to conservative or anticonservative findings depending on the underlying simulation model. When the variance of individuals with the less common environmental exposure was 3-fold greater than the trait variance for individuals with the more common exposure, we observed that the model-based joint test had systematic inflation in P values (λ = 1.33). In contrast, when the variance of individuals with the less common environmental exposure was 3-fold smaller than the trait variance for individuals with the more common exposure, we found that the model-based joint test had systematic deflation in P values (λ = 0.71). Under both heteroscedasticity models, the robust joint test of SNP and SNP-environment interaction had no such inflation (λ = 1.02 for each heteroscedasticity model). In addition to evaluating null models, we also evaluated the model-based and robust joint tests in power simulations where an SNP or SNP-environment interaction influenced phenotype. As shown in the eResults and eFigure 3 in the Supplement, we observed that the robust joint test had equivalent power to the model-based joint test when homoscedasticity holds (such that the model-based test is valid).
Figure 4.

Quantile-Quantile Plot of Observed P Values vs Expected P Values (on a −log10 Scale) for Joint Analyses of Simulated Data Using Model-Based and Robust Variance Estimators
A, Quantitative data generated assuming Var(Y|G,E = 0) = Var(Y|G,E = 1); B, quantitative data generated assuming Var(Y|G,E = 0)>Var(Y|G,E = 1); and C, quantitative data generated assuming Var(Y|G,E = 0)<Var(Y|G,E = 1). λ denotes the inflation factor (defined as the median of the test statistic divided by the median of the asymptotic distribution of the test statistic). For each simulation model, we evaluated 10 000 replicates of the data in which each data set was composed of 2000 individuals.
Discussion
Many genetic association studies of psychiatric outcomes are interested in identifying genetic variants that act in conjunction with other environmental modifiers to influence phenotype. Such interactions can resolve replication failures of marginal SNP findings7 and explain missing heritability.52 We found that a traditional joint test of SNP and SNP-environment interaction for quantitative phenotypes can result in invalid findings when the assumption of homoscedasticity (which can be assessed using the Brown-Forsythe test of variance equality) is violated. This issue affects candidate-gene association studies and GWAS projects that consider interactions of continuous phenotypes. We illustrate this point using an existing GWAS of PTSD and simulated data. To resolve this bias in the traditional joint test, we used the idea of Voorman et al48 to develop a robust joint test that uses Huber-White variance estimates to correct for heteroscedasticity and found that this approach corrects for the P value inflation (or deflation) seen in the model-based joint tests when heteroscedasticity exists while having equivalent power to the model-based test when homoscedasticity actually holds.
The observation that heteroscedasticity invalidates interaction tests has not been reported in psychiatric literature, and it is also underappreciated in nonpsychiatric genetic studies, with only a few association studies53,54 addressing and correcting for this issue using robust Huber-White variance estimators. For a binary environmental exposure, one could perform a valid joint interaction analysis in the presence of heteroscedasticity without the need of Huber-White SEs by performing a marginal genetic analysis stratified by exposure55 (because, within each exposure stratum, outcome variance is constant). However, this procedure, unlike the Huber-White SEs, is not applicable to continuous environmental predictors, such as the CTQ and TEI score that are of interest in the GTP.
Our analyses focused on joint testing of continuous phenotypes using the model of Kraft et al5; such joint tests are implemented in PLINK. Other joint interaction tests also exist,9,10,56 and because these methods use model-based SEs that assume homoscedasticity, we expect them to be invalid under heteroscedasticity. Heteroscedasticity issues likely also invalidate the more exclusive test of the gene-environment effect alone as noted by Voorman et al.48 Using the traditional 1-df Wald test for testing gene-environment interaction exclusively, we observed that heteroscedasticity led to inflated P values in the analyses of the BDI and PSSint within the GTP (eFigure 4 in the Supplement). The use of robust Huber-White SEs eliminated this inflation.
Although heteroscedasticity affects the validity of joint SNP tests of quantitative outcomes, it does not affect the validity of binary outcomes (such as disease status) because the logistic regression model for the latter outcome does not make a homoscedasticity assumption of constant variance across individuals. Nevertheless, joint SNP tests of binary outcomes using model-based variance estimates can exhibit bias if the outcome mean is misspecified; for example, one assumes a linear effect of environment on the log of the odds of disease when the relationship is truly nonlinear. Voorman et al48 and Tchetgen Tchetgen and Kraft47 noted this issue and reported that robust Huber-White SEs can be used to correct this problem and provide valid inference.
Conclusions
The robust joint test of interaction presented in this article corrects for the bias seen in the model-based joint tests when heteroscedasticity exists and remains powerful when the homoscedasticity assumption actually holds. We therefore recommend the use of the robust joint test over the model-based joint test in psychiatric candidate-gene studies and GWASs as well as studies that use nonpsychiatric outcomes. To facilitate use by the public, we implemented our procedure as a software tool (http://genetics.emory.edu/labs/epstein/software) that can be run through the PLINK software package (http://pngu.mgh.harvard.edu/~purcell/plink/; eMethods in the Supplement). We note that robust estimators for interaction analysis also are available in the ProbABEL57 package implemented in the R programming language.
Supplementary Material
Acknowledgments
Funding/Support: This study was supported by grant R01-MH071537 from the National Institute of Mental Health (Drs Almli, Binder, Bradley, Ressler, Conneely, and Epstein) and grant R01-HG007508 from the National Human Genome Research Institute (Drs Duncan and Epstein).
Role of the Funder/Sponsor: The funding sources had no role in the design and conduct of the study; collection, management, analysis, and interpretation of the data; preparation, review, or approval of the manuscript; and decision to submit the manuscript for publication.
Footnotes
Conflict of Interest Disclosures: Dr Epstein reported working as a consultant for Amnion Laboratories. No other disclosures were reported.
Disclaimer: The contents of this article do not reflect the views of the Department of Veterans Affairs or the US government.
Additional Contributions: Stephanie Sherman, PhD (Emory University), and Kristen Mercer, MPH (Emory University), provided valuable discussions on the content of the manuscript. Cheryl Strauss, BS (Emory University), provided editing assistance. No compensation was provided for such contributions. We appreciate the technical support of all of the staff and volunteers of the GTP. Most important, we are extremely indebted to and appreciative of the time and effort given from all of the participants of the GTP.
REFERENCES
- 1.Cichon S, Craddock N, Daly M, et al. Psychiatric GWAS Consortium Coordinating Committee Genomewide association studies: history, rationale, and prospects for psychiatric disorders. Am J Psychiatry. 2009;166(5):540–556. doi: 10.1176/appi.ajp.2008.08091354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Karg K, Sen S. Gene× Environment Interaction Models in Psychiatric Genetics: Behavioral Neurogenetics. Springer Publishing; New York, NY: 2012. pp. 441–462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Winham SJ, Biernacka JM. Gene-environment interactions in genome-wide association studies: current approaches and new directions. J Child Psychol Psychiatry. 2013;54(10):1120–1134. doi: 10.1111/jcpp.12114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.van Os J, Rutten BP. Gene-environment-wide interaction studies in psychiatry. Am J Psychiatry. 2009;166(9):964–966. doi: 10.1176/appi.ajp.2008.09060904. [DOI] [PubMed] [Google Scholar]
- 5.Kraft P, Yen YC, Stram DO, Morrison J, Gauderman WJ. Exploiting gene-environment interaction to detect genetic associations. Hum Hered. 2007;63(2):111–119. doi: 10.1159/000099183. [DOI] [PubMed] [Google Scholar]
- 6.Lindström S, Yen Y-C, Spiegelman D, Kraft P. The impact of gene-environment dependence and misclassification in genetic association studies incorporating gene-environment interactions. Hum Hered. 2009;68(3):171–181. doi: 10.1159/000224637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ioannidis JP. Non-replication and inconsistency in the genome-wide association setting. Hum Hered. 2007;64(4):203–213. doi: 10.1159/000103512. [DOI] [PubMed] [Google Scholar]
- 8.Wang K. Genetic association tests in the presence of epistasis or gene-environment interaction. Genet Epidemiol. 2008;32(7):606–614. doi: 10.1002/gepi.20336. [DOI] [PubMed] [Google Scholar]
- 9.Chatterjee N, Kalaylioglu Z, Moslehi R, Peters U, Wacholder S. Powerful multilocus tests of genetic association in the presence of gene-gene and gene-environment interactions. Am J Hum Genet. 2006;79(6):1002–1016. doi: 10.1086/509704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tzeng JY, Zhang D, Pongpanich M, et al. Studying gene and gene-environment effects of uncommon and common variants on continuous traits: a marker-set approach using gene-trait similarity regression. Am J Hum Genet. 2011;89(2):277–288. doi: 10.1016/j.ajhg.2011.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chatterjee N, Carroll RJ. Semiparametric maximum likelihood estimation exploiting gene-environment independence in case-control studies. Biometrika. 2005;92(2):399–418. [Google Scholar]
- 12.Purcell S, Neale B, Todd-Brown K, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81(3):559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Conti DV, Lee W, Li D, et al. Pharmacogenetics of Nicotine Addiction and Treatment Consortium. Nicotinic acetylcholine receptor β2-subunit gene implicated in a systems-based candidate gene study of smoking cessation. Hum Mol Genet. 2008;17(18):2834–2848. doi: 10.1093/hmg/ddn181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Siegert S, Hampe J, Schafmayer C, et al. Genome-wide investigation of gene-environment interactions in colorectal cancer. Hum Genet. 2013;132(2):219–231. doi: 10.1007/s00439-012-1239-2. [DOI] [PubMed] [Google Scholar]
- 15.Hamza TH, Chen H, Hill-Burns EM, et al. Genome-wide gene-environment study identifies glutamate receptor gene GRIN2A as a Parkinson’s disease modifier gene via interaction with coffee. PLoS Genet. 2011;7(8):e1002237. doi: 10.1371/journal.pgen.1002237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Koenen KC, Amstadter AB, Nugent NR. Gene-environment interaction in posttraumatic stress disorder: an update. J Trauma Stress. 2009;22(5):416–426. doi: 10.1002/jts.20435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Breslau N. The epidemiology of posttraumatic stress disorder: what is the extent of the problem? J Clin Psychiatry. 2001;62(suppl 17):16–22. [PubMed] [Google Scholar]
- 18.Binder EB, Bradley RG, Liu W, et al. Association of FKBP5 polymorphisms and childhood abuse with risk of posttraumatic stress disorder symptoms in adults. JAMA. 2008;299(11):1291–1305. doi: 10.1001/jama.299.11.1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Foa EB, Tolin DF. Comparison of the PTSD Symptom Scale-Interview Version and the Clinician-Administered PTSD scale. J Trauma Stress. 2000;13(2):181–191. doi: 10.1023/A:1007781909213. [DOI] [PubMed] [Google Scholar]
- 20.Jovanovic T, Blanding NQ, Norrholm SD, Duncan E, Bradley B, Ressler KJ. Childhood abuse is associated with increased startle reactivity in adulthood. Depress Anxiety. 2009;26(11):1018–1026. doi: 10.1002/da.20599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jovanovic T, Norrholm SD, Blanding NQ, et al. Impaired fear inhibition is a biomarker of PTSD but not depression. Depress Anxiety. 2010;27(3):244–251. doi: 10.1002/da.20663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jovanovic T, Ressler KJ. How the neurocircuitry and genetics of fear inhibition may inform our understanding of PTSD. Am J Psychiatry. 2010;167(6):648–662. doi: 10.1176/appi.ajp.2009.09071074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yehuda R, LeDoux J. Response variation following trauma: a translational neuroscience approach to understanding PTSD. Neuron. 2007;56(1):19–32. doi: 10.1016/j.neuron.2007.09.006. [DOI] [PubMed] [Google Scholar]
- 24.Morgan CA, III, Grillon C, Southwick SM, Davis M, Charney DS. Fear-potentiated startle in posttraumatic stress disorder. Biol Psychiatry. 1995;38(6):378–385. doi: 10.1016/0006-3223(94)00321-S. [DOI] [PubMed] [Google Scholar]
- 25.Rauch SL, Shin LM, Phelps EA. Neurocircuitry models of posttraumatic stress disorder and extinction: human neuroimaging research: past, present, and future. Biol Psychiatry. 2006;60(4):376–382. doi: 10.1016/j.biopsych.2006.06.004. [DOI] [PubMed] [Google Scholar]
- 26.True WR, Rice J, Eisen SA, et al. A twin study of genetic and environmental contributions to liability for posttraumatic stress symptoms. Arch Gen Psychiatry. 1993;50(4):257–264. doi: 10.1001/archpsyc.1993.01820160019002. [DOI] [PubMed] [Google Scholar]
- 27.Stein MB, Jang KL, Taylor S, Vernon PA, Livesley WJ. Genetic and environmental influences on trauma exposure and posttraumatic stress disorder symptoms: a twin study. Am J Psychiatry. 2002;159(10):1675–1681. doi: 10.1176/appi.ajp.159.10.1675. [DOI] [PubMed] [Google Scholar]
- 28.Cornelis MC, Nugent NR, Amstadter AB, Koenen KC. Genetics of post-traumatic stress disorder: review and recommendations for genome-wide association studies. Curr Psychiatry Rep. 2010;12(4):313–326. doi: 10.1007/s11920-010-0126-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xie P, Kranzler HR, Poling J, et al. Interaction of FKBP5 with childhood adversity on risk for post-traumatic stress disorder. Neuropsychopharmacology. 2010;35(8):1684–1692. doi: 10.1038/npp.2010.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kilpatrick DG, Koenen KC, Ruggiero KJ, et al. The serotonin transporter genotype and social support and moderation of posttraumatic stress disorder and depression in hurricane-exposed adults. Am J Psychiatry. 2007;164(11):1693–1699. doi: 10.1176/appi.ajp.2007.06122007. [DOI] [PubMed] [Google Scholar]
- 31.Koenen KC, Aiello AE, Bakshis E, et al. Modification of the association between serotonin transporter genotype and risk of posttraumatic stress disorder in adults by county-level social environment. Am J Epidemiol. 2009;169(6):704–711. doi: 10.1093/aje/kwn397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xie P, Kranzler HR, Poling J, et al. Interactive effect of stressful life events and the serotonin transporter 5-HTTLPR genotype on posttraumatic stress disorder diagnosis in 2 independent populations. Arch Gen Psychiatry. 2009;66(11):1201–1209. doi: 10.1001/archgenpsychiatry.2009.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Coffey SF, Dansky BS, Falsetti SA, Saladin ME, Brady KT. Screening for PTSD in a substance abuse sample: psychometric properties of a modified version of the PTSD Symptom Scale Self-Report: posttraumatic stress disorder. J Trauma Stress. 1998;11(2):393–399. doi: 10.1023/A:1024467507565. [DOI] [PubMed] [Google Scholar]
- 34.Falsetti SA, Resnick HS, Resick PA, Kilpatrick D. The Modified PTSD Symptom Scale: a brief self-report measure of postraumatic stress disorder. Behav Ther. 1993;16:161–162. [Google Scholar]
- 35.Schwartz AC, Bradley R, Penza KM, et al. Pain medication use among patients with posttraumatic stress disorder. Psychosomatics. 2006;47(2):136–142. doi: 10.1176/appi.psy.47.2.136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schwartz AC, Bradley RL, Sexton M, Sherry A, Ressler KJ. Posttraumatic stress disorder among African Americans in an inner city mental health clinic. Psychiatr Serv. 2005;56(2):212–215. doi: 10.1176/appi.ps.56.2.212. [DOI] [PubMed] [Google Scholar]
- 37.Gillespie CF, Bradley B, Mercer K, et al. Trauma exposure and stress-related disorders in inner city primary care patients. Gen Hosp Psychiatry. 2009;31(6):505–514. doi: 10.1016/j.genhosppsych.2009.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Beck AT, Ward CH, Mendelson M, Mock J, Erbaugh J. An inventory for measuring depression. Arch Gen Psychiatry. 1961;4:561–571. doi: 10.1001/archpsyc.1961.01710120031004. [DOI] [PubMed] [Google Scholar]
- 39.Bernstein DP, Fink L. Childhood Trauma Questionnaire Manual. Psychological Corporation; San Antonio, TX: 1998. [Google Scholar]
- 40.White H. A heteroskedasticity-consistent covariance matrix estimator and a direct test for heteroskedasticity. Econometrica. 1980;48(4):817–838. [Google Scholar]
- 41.Huber PJ. The behavior of maximum likelihood estimates under nonstandard conditions. In: Le Cam LM, Neyman J, editors. Proceedings of the Fifth Berkeley Symposium on Mathematical Statistics and Probability; Berkeley: University of California Press; 1967. [Google Scholar]
- 42.Green WH. Econometric Analysis. Prentice Hall; Upper Saddle River, NJ: 1999. [Google Scholar]
- 43.King G. Unifying Political Methodology: The Likelihood Theory of Statistical Inference. University of Michigan Press; Ann Arbor: 1989. [Google Scholar]
- 44.Powell LM, Auld MC, Chaloupka FJ, O’Malley PM, Johnston LD. Associations between access to food stores and adolescent body mass index. Am J Prev Med. 2007;33(4):S301–S307. doi: 10.1016/j.amepre.2007.07.007. suppl. [DOI] [PubMed] [Google Scholar]
- 45.Gao SK, Beresford SA, Frank LL, Schreiner PJ, Burke GL, Fitzpatrick AL. Modifications to the Healthy Eating Index and its ability to predict obesity: the Multi-Ethnic Study of Atherosclerosis. Am J Clin Nutr. 2008;88(1):64–69. doi: 10.1093/ajcn/88.1.64. [DOI] [PubMed] [Google Scholar]
- 46.Lakhani SR, Van De Vijver MJ, Jacquemier J, et al. The pathology of familial breast cancer: predictive value of immunohistochemical markers estrogen receptor, progesterone receptor, HER-2, and p53 in patients with mutations in BRCA1 and BRCA2. J Clin Oncol. 2002;20(9):2310–2318. doi: 10.1200/JCO.2002.09.023. [DOI] [PubMed] [Google Scholar]
- 47.Tchetgen Tchetgen EJ, Kraft P. On the robustness of tests of genetic associations incorporating gene-environment interaction when the environmental exposure is misspecified. Epidemiology. 2011;22(2):257–261. doi: 10.1097/EDE.0b013e31820877c5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Voorman A, Lumley T, McKnight B, Rice K. Behavior of QQ-plots and genomic control in studies of gene-environment interaction. PLoS One. 2011;6(5):e19416. doi: 10.1371/journal.pone.0019416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Devlin B, Roeder K, Wasserman L. Genomic control, a new approach to genetic-based association studies. Theor Popul Biol. 2001;60(3):155–166. doi: 10.1006/tpbi.2001.1542. [DOI] [PubMed] [Google Scholar]
- 50.Olkin I, Ghurye SG, Hoeffding W, Madow WG, Mann HB, editors. Contributions to Probability and Statistics: Essays in Honor of Harold Hotelling. Stanford University Press; Redwood City, CA: 1960. [Google Scholar]
- 51.Brown MB, Forsythe AB. Robust tests for the equality of variances. J Am Stat Assoc. 1974;69(346):364–367. [Google Scholar]
- 52.Eichler EE, Flint J, Gibson G, et al. Missing heritability and strategies for finding the underlying causes of complex disease. Nat Rev Genet. 2010;11(6):446–450. doi: 10.1038/nrg2809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Maxwell TJ, Ballantyne CM, Cheverud JM, Guild CS, Ndumele CE, Boerwinkle E. APOE modulates the correlation between triglycerides, cholesterol, and CHD through pleiotropy, and gene-by-gene interactions. Genetics. 2013;195(4):1397–1405. doi: 10.1534/genetics.113.157719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Manning AK, Hivert M-F, Scott RA, et al. DIAbetes Genetics Replication And Meta-analysis (DIAGRAM) Consortium; Multiple Tissue Human Expression Resource (MUTHER) Consortium. A genome-wide approach accounting for body mass index identifies genetic variants influencing fasting glycemic traits and insulin resistance. Nat Genet. 2012;44(6):659–669. doi: 10.1038/ng.2274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Aschard H, Hancock DB, London SJ, Kraft P. Genome-wide meta-analysis of joint tests for genetic and gene-environment interaction effects. Hum Hered. 2010;70(4):292–300. doi: 10.1159/000323318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wang T, Ho G, Ye K, Strickler H, Elston RC. A partial least-square approach for modeling gene-gene and gene-environment interactions when multiple markers are genotyped. Genet Epidemiol. 2009;33(1):6–15. doi: 10.1002/gepi.20351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Aulchenko YS, Struchalin MV, van Duijn CM. ProbABEL package for genome-wide association analysis of imputed data. BMC Bioinformatics. 2010;11(1):134. doi: 10.1186/1471-2105-11-134. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.