Abstract
AMPA receptors (AMPARs) play a critical role in excitatory glutamatergic neurotransmission. The number and subunit composition of AMPARs at synapses determines the dynamics of fast glutamatergic signalling. Functional AMPARs on the cell surface are tetramers. Thus tetrameric assembly of AMPARs represents a promising target for modulating AMPAR-mediated signalling in health and disease. Multiple structural domains within the receptor influence AMPAR assembly. In a proposed model for AMPAR assembly, the amino-terminal domain underlies the formation of a dimer pool. The transmembrane domain facilitates the formation and enhances the stability of the tetramer. The ligand-binding domain influences assembly through a process referred to as ‘domain swapping’. We propose that this core AMPAR assembly process could be regulated by neuronal signals and speculate on possible mechanisms for such regulation.
Introduction
In the central nervous system, fast excitatory glutamatergic neurotransmission is mediated by ligand-gated or ionotropic glutamate receptors (iGluRs), predominantly the α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid (AMPA) receptor subtype. There are four AMPAR subunits (GluA1–4) encoded by separate genes, which are further diversified and differentiated by alternative splicing and mRNA editing (Traynelis et al. 2010). AMPARs are tetrameric complexes assembled from four identical (homomeric) or similar (heteromeric) subunits. The number and subunit composition of AMPARs determine the efficiency and dynamics of AMPAR-mediated synaptic signalling. Different subunit compositions confer distinct biophysical and molecular properties to AMPARs. For example, GluA2-containing receptors, which predominate in principal neurons, display a linear current–voltage relationship and are impermeable to Ca2+ due to constitutive Q/R (glutamine/arginine) editing of the GluA2 subunit (Isaac et al. 2007). In contrast, GluA2-lacking receptors, which are often found in non-principal cells, display a double-rectification, have higher single-channel conductances and are Ca2+ permeable (Geiger et al. 1995; Cull-Candy et al. 2006). Both the number and the subunit compositions of AMPARs vary amongst different brain regions/neuronal cell types (Martin et al. 1993; Cull-Candy et al. 2006) and change during development (Kumar et al. 2002), synaptic plasticity (Collingridge et al. 2004; Wang et al. 2012), and pathological states such as epilepsy (Loddenkemper et al. 2014).
The composition of AMPARs on the plasma membrane is not solely dictated by the expression pattern of the four AMPAR genes. Rather, there is preferential assembly between particular subunits and/or splice variants in the endoplasmic reticulum (ER) (Greger et al. 2002, 2003; Brorson et al. 2004; Coleman et al. 2006; Penn et al. 2012). For example, GluA3 subunits can form homo-tetramers when expressed alone (Suzuki et al. 2008), but in the presence of other subunits such as GluA1 and GluA2 (and possibly GluA4 as well), GluA3 forms obligate heteromers (Rossmann et al. 2011). To gain insight into the mechanisms underlying the preferential assembly of AMPAR subunits and its regulation, it is important to understand how various structural elements influence AMPAR assembly.
In this review, we propose a hypothetical model of AMPAR tetrameric assembly (Fig. 1A) encompassing the intrinsic interactions amongst subunits. We provide an overview of the experimental evidence supporting this model. In addition, we will speculate on how assembly may be regulated and briefly explore the possibility of modulating AMPAR assembly for basic research and clinical applications.
Figure 1. Model of AMPAR assembly.
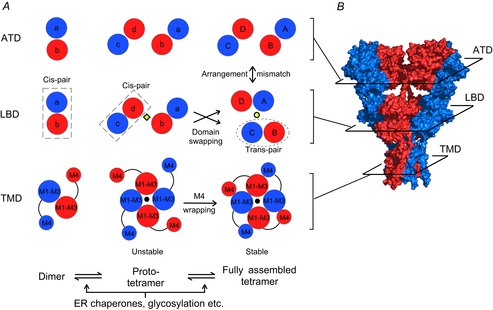
A, dimer formation is driven by associations between the ATDs (left panels). At this stage the LBDs within the dimer form a cis-LBD pair (dashed rectangles). A pair of dimers forms an unstable intermediate (referred to as a ‘proto-tetramer’). In the proto-tetramer, the M1 and M3 helices, as well as the M2 re-entrant loops, form the ion channel core (middle panels). The black dot denotes the ion channel pore. At this stage the cis-LBD pair adopts the same arrangement as the ATD dimers. The interface between two LBD pairs (yellow diamond) is between the B and D subunits. We propose two processes that may drive the proto-tetramer to a relatively stable tetramer (right panels): (i) at the level of the TMD the M4 helix from each subunit associates with the ion channel core (M1–M3) of an adjacent subunit, a process referred to as ‘M4 wrapping’; (ii) at the level of the LBD, the cis-pairs dissociate, allowing changes in subunit arrangement and the formation of trans-pairs (dotted ellipse). After this exchange (referred to as ‘domain swapping’), the dimer–dimer interface (yellow dot) is between the A and C subunits. Both processes may introduce new inter-molecular interactions and/or relieve structural tension present in the proto-tetramer, leading to a decrease in free energy and stabilization of the tetrameric complex. The assembly process could be subject to controls by ER chaperones, glycosylation etc. B, crystal structure of homomeric GluA2 receptor (3KG2) (Sobolevsky et al. 2009). Subunits are coloured as in A. Subunit arrangement at the level of each domain (planar sections) is shown in the right panels of A.
Mechanistic model of AMPAR assembly
iGluR subunits have a modular structure containing four discrete domains: an amino-terminal domain (ATD) and a ligand-binding domain (LBD), both located extracellularly, a transmembrane domain (TMD) consisting of hydrophobic segments M1–M4, and an intracellular carboxy-terminal domain (CTD) (Traynelis et al. 2010). The ATD, LBD and TMD all impact tetrameric assembly of AMPARs (Nakagawa, 2010; Sukumaran et al. 2012). At present, there is no evidence for a role of the CTD in AMPAR assembly.
The near-full-length crystal structure of GluA2 (Fig. 1B) revealed a number of interesting features about AMPARs. For example, the ATD and LBD each possess a 2-fold rotational symmetry, where subunits of like conformation are positioned opposite to each other (Fig. 1A, right panels) (Sobolevsky et al. 2009). In contrast, the TMD displays 4-fold symmetry. Further, there is a mismatch in subunit arrangement between the ATD and LBD. In the fully assembled receptor (Fig. 1A, right panels), subunits that are proximal to each other (B and D) at the ATD level are located distally at the LBD level, and vice versa.
Figure 1A illustrates a hypothetical model of AMPAR assembly highlighting the intrinsic interactions among subunits at levels of the ATD (upper panels), the LBD (middle panels), and the TMD (lower panels). In this model, the ATD facilitates the formation of dimers (left panels). At the dimer stage, the LBDs of the two participating subunits might associate to form an LBD pair (dubbed ‘cis-LBD pair’). Dimers then associate with each other via interactions within the ion channel core to form an unstable intermediate termed a ‘proto-tetramer’ (central panels). Cis-LBD pairs persist into the proto-tetramer stage leading to an arrangement at the level of the LBD matching that of the ATD. The M4 transmembrane segment of each subunit then wraps around the ion channel core (M1–M3) of an adjacent subunit – a process we term ‘M4 wrapping’ – and stabilizes the tetramer. At some point during this process, the LBDs separate and exchange their interacting partners to form ‘trans-LBD pairs’ leading to the subunit arrangement mismatch between the ATD and the LBD observed in the fully assembled tetrameric complex (Sobolevsky et al. 2009). We refer to this process as ‘domain swapping’. M4 wrapping in the TMD, along with domain swapping, presumably contributes to the overall intrinsic energetics of tetramerization. Additionally extrinsic factors such as ER chaperones and glycosylation could also have important, albeit undefined, modulatory effects on these subunit interactions facilitating tetramerization.
Dimer formation driven by ATD
AMPAR assembly presumably involves a dimer intermediate based on results from single-particle electron microscopy (Tichelaar et al. 2004; Shanks et al. 2010), size-exclusive chromatography (Shanks et al. 2010), as well as blue-native polyacrylamide gel electrophoresis (BN-PAGE) (Greger et al. 2003; Salussolia et al. 2013). The composition of the dimer pool impacts the eventual subunit composition of the fully assembled receptors (Ayalon & Stern-Bach, 2001; Rossmann et al. 2011). Analytical ultra-centrifugation studies have measured the selectivity and affinity of the dimerization of isolated ATDs from AMPARs as well as kainate receptors. These measured parameters are at least partially predictive of the readily observed subunit compositions of full-length receptors (Kumar et al. 2011; Rossmann et al. 2011).
The GluA1/GluA2 heteromeric receptor is one of the major subunit compositions at synapses (Lu et al. 2009), which is consistent with the high affinity between the isolated ATDs of GluA1 and GluA2 (Rossmann et al. 2011). GluA1/GluA2 hetero-tetramers are apparently centrally symmetrical, with like subunits positioned oppositely (1-2-1-2) (Mansour et al. 2001), similar to the subunit arrangement of N-methyl-d-aspartate receptors (NMDARs) (Salussolia et al. 2011b; Riou et al. 2012). How this subunit arrangement arises is unclear. It is possible that the tetramer is assembled from a pair of GluA1/GluA2 hetero-dimers. However, since the ATD is translated first, its folding and dimerization might occur before the entire polypeptide is synthesized and released from the translation machinery, as occurs in the case of potassium channels (Lu et al. 2001; Phartiyal et al. 2007). As polypeptides translated from the same gene are locally concentrated at a polyribosome, this initial dimerization process might yield a significant amount of homo-dimers instead of hetero-dimers. The relative significance of the population of subunits assembled co-translationally into homo-dimers, if they do exist, is unknown. Also undefined is whether this population can still participate in the formation of hetero-tetramers. If they can, these homo-dimers must dissociate at some point during the assembly process so that they can rearrange to adopt the 1-2-1-2 arrangement (Rossmann et al. 2011; Herguedas et al. 2013).
Transmembrane interactions impacting tetramer formation
When isolated, neither ATDs nor LBDs form tetramers in solution (Weston et al. 2006; Zhao et al. 2012). Further, AMPARs lacking the ATD form functional receptors (Pasternack et al. 2002; Moykkynen et al. 2014). Hence the ATD and the LBD are not sufficient for the formation of a tetrameric complex. In a mature receptor the most extensive inter-subunit molecular contacts occur at the TMD level (Sobolevsky et al. 2009). Ayalon & Stern-Bach (2001) showed that the TMD affects the subunit composition of surface-expressed iGluRs. Recent studies have directly demonstrated that molecular interactions in the TMD are critical to AMPAR tetramerization (Salussolia et al. 2013).
One factor stabilizing the tetrameric complex involves the M4 transmembrane segment, which is peripheral to the ion channel core (M1–M3) (Fig. 1, lower panels). The M4 segment is a critical component of AMPAR tetramerization (Salussolia et al. 2011a, 2013). Without the M4 segment, GluA1 as well as GluA2 subunits fail to form homo-tetramers (Fig. 2A) or hetero-tetramers (Salussolia et al. 2013). In the GluA2 structure (Sobolevsky et al. 2009), the M4 from each subunit fits tightly into a groove formed by the M1 and M3 transmembrane helices of an adjacent subunit (Fig. 2E). Tryptophan substitutions in this ‘M4 interaction face’ (e.g. V795W and E813W) but not on the opposite side of the helix (e.g. L811W) disrupt tetramerization as assayed using blue-native PAGE (Fig. 2B) and fluorescent size-exclusive chromatography (Fig. 2C). These experimental results, combined with structural information, indicate that the interactions between one face of the M4 helix and the outer surface of the ion channel core (M1–M3) stabilize the tetrameric complex (Fig. 2D and E).
Figure 2. M4 helix is necessary for AMPAR tetramerization.
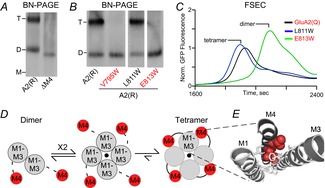
A, blue-native PAGE (BN-PAGE) illustrating that the deletion of M4 (ΔM4) yields only dimers in contrast to wild-type GluA2(R) (A2(R)). Tetramer (T) and dimer (D) bands are based on apoferritin and NativeMark standards. B, example BN-PAGE for single Trp (W) substitutions of the M4 helix in GluA2(R). Trp substitutions at certain positions (V795W and E813W) had the same effect on tetramerization as the deletion of the entire M4, while others (L811W) had no effect. C, fluorescent size-exclusion chromatography (FSEC) of GluA2(Q)–enhanced green fluorescent protein (EGFP) with single M4 substitutions correlated well with results from BN-PAGE. Peaks corresponding to different oligomeric species are indicated. Data from Salussolia et al. (2013). D, putative model for iGluR assembly at the level of the transmembrane domain where a pair of dimers forms a tetrameric complex, presumably stabilized by interactions between the M4 of a subunit and M1–M3 of an adjacent subunit. E, positions in the M4 segment where single Trp substitutions block tetramerization are located along a specific face of the helix (red), referred to as ‘VLGAVE’ (V, valine; L, leucine; G, glycine; A, alanine; V, valine; E, glutamate). The VLGAVE face of the M4 segment from one subunit (dark grey) fits into a groove formed by M1 and M3 from an adjacent subunit (light grey).
Such a mechanism would require the packing of M1 and M3 helices from all four subunits into an ion channel before the M4 even comes into play (Fig. 1, central panels). Interestingly, the Q/R editing site in the pore region regulates AMPAR assembly and trafficking, with the editing of glutamine to arginine resulting in reduced tetramerization and ER export of homomeric GluA2 receptors (Greger et al. 2002, 2003). The constitutive Q/R editing of GluA2 presumably causes GluA2 subunits to assemble less efficiently, therefore contributing to GluA2's longer ER dwell time (Greger et al. 2003).
The involvement of the ion channel core in AMPAR assembly is not surprising since the homologous region is also involved in the assembly of potassium channels (Splitt et al. 2000; Wang et al. 2009). Moreover, the ‘lurcher’ mutation, which is located within the highly conserved ‘SYTANLAAF’ motif near the M3 bundle crossing, also makes tetramerization less efficient (Kim et al. 2010). We therefore hypothesize that interactions within the ion channel core facilitate the formation of the proto-tetramer (Fig. 1, central panel), while the wrapping of the M4 segment stabilizes the proto-tetramer and drives its transition into a mature tetramer (Fig. 1, right panel).
Domain swapping
The LBD, which drives receptor gating, also influences AMPAR and kainate receptor trafficking (Mah et al. 2005; Penn et al. 2008; Coleman et al. 2009, 2010), possibly by modulating tetrameric assembly. For example, the R/G (arginine/glycine) editing site in the LBD and the flip-flop alternative splicing site, which includes parts of the LBD and the LBD–M4 linker, both regulate AMPAR assembly (Greger et al. 2006; Penn et al. 2012). In our proposed model, domain swapping in the LBD is a factor facilitating the transition from the proto-tetramer to a fully assembled tetramer (Fig. 1A).
The proposed cis-LBD pair (Fig. 1A, middle panel) has not been experimentally observed, suggesting that the association must be transient if it exists at all (Shanks et al. 2010; Sukumaran et al. 2012). Nevertheless, there is circumstantial evidence for the existence of such a configuration. An L-to-Y (leucine to tyrosine) mutation that stabilizes the LBD dimer interface (Sun et al. 2002) reduces tetramer formation (Greger et al. 2006; Shanks et al. 2010). One explanation for the mutation's negative effect on assembly may be that it traps the LBDs in the ‘cis-LBD pair’ configuration, reducing the rate of ‘domain swapping’ (Nakagawa, 2010). Since flip-flop alternative splicing targets part of the LBD, it may also influence AMPAR assembly and forward trafficking (Brorson et al. 2004; Coleman et al. 2006; Penn et al. 2012) by modulating ‘domain swapping’. Further supporting this idea, in the GluK2 kainate receptor, which displays an ATD–LBD arrangement mismatch similar to AMPARs (Das et al. 2010), stabilizing the LBD dimer interface with disulfide cross-links leads to a drastic reduction in the receptor's ER export (Priel et al. 2006).
How ‘domain swapping’ might influence tetramer formation remains unclear. One possibility is that domain swapping influences how AMPARs interact with ER-associated proteins (e.g. chaperone proteins, cornichons, etc.). Alternatively domain swapping might affect the intrinsic energetics of the receptor. During ‘domain swapping’, structural tensions in the LBD–M4 linker and/or steric clashes at the LBD dimer–dimer interface (Fig. 1A) that might exist at the proto-tetramer stage are relaxed, allowing tighter packing in the TMD and resulting in a negative ΔG. In this scenario, it is unclear whether domain swapping is influenced by M4 wrapping or vice versa.
Regulation of AMPAR assembly: evidence and possibilities
Changes in the number and subunit composition of surface-expressed AMPARs underlie many forms of synaptic plasticity (Collingridge et al. 2004; Wang et al. 2012). Changes in synaptic strength during synaptic plasticity require a reserve pool of iGluRs that are fully assembled and ready for rapid plasma membrane insertion (Granger et al. 2013). Endocytic recycling of AMPARs contributes significantly to this reserve pool (Park et al. 2004; Petrini et al. 2009) and could largely account for the change in the number of iGluRs. However, in addition to the number of iGluRs, the subunit compositions also change during synaptic plasticity and disease states. For example, the proportion of GluA2-lacking AMPARs in dopaminergic neurons in the ventral tegmental area increases in response to cocaine exposure (Bowers et al. 2010). Also, in patients with refractory epilepsy, there is a significant loss of AMPARs containing the GluA1 subunit in multiple brain regions (Grigorenko et al. 1997). How exactly these changes in AMPAR subunit composition are achieved is still unclear. Subunit compositions of AMPARs in this intracellular pool may in part be established by a constitutive supply of readily assembled receptors – GluA2-containing as well as GluA2-lacking – flowing in from the ER regardless of synaptic activities. Alterations in the subunit compositions of surface-expressed receptors would then result from activity-dependent regulations on exocytosis, endocytosis and synaptic targeting, as well as lysosomal degradation (Derkach et al. 2007; Makino & Malinow, 2009). Complementing these mechanisms, the subunit composition of the reserve pool could vary in an activity-dependent manner due to changes in the receptor availability from the ER, where translation of individual AMPAR subunits and their tetrameric assembly occurs. Such a scenario requires regulations at stages prior to the receptors’ exit from the ER. In neurons, the ER network extends into the dendritic tree (Cui-Wang et al. 2012). In addition, mRNAs encoding for AMPARs accumulate in the dendritic ER and are translated locally (Grooms et al. 2006; Mameli et al. 2007). Here we describe a number of possible mechanisms through which AMPAR biogenesis may be regulated (Fig. 3).
Figure 3. Possible mechanisms underlying the regulation of AMPAR assembly.
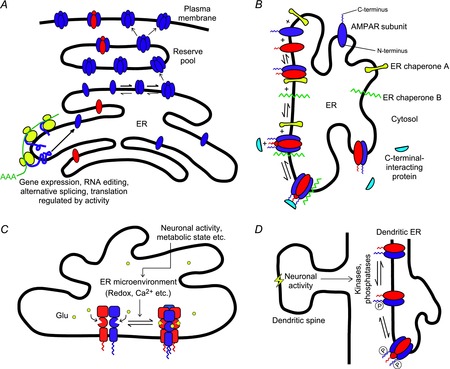
A, the level of gene expression, alternative splicing and/or translation of a particular type of AMPAR subunit (blue) is modulated by an activity-dependent signal, thereby shifting the subunit composition of the reserve receptor pool. B, two hypothetical ER chaperones ‘A’ and ‘B’ bind to and stabilize dimers and tetramers of a particular subunit composition (red and blue), respectively, thereby modulating the equilibrium of tetramerization. C-terminal-interacting proteins might also take part in this process. C, the equilibrium between receptor dimers and tetramers may be influenced by the microenvironment in the ER lumen (e.g. redox, Ca2+, glutamate concentration, etc.), which could in turn be altered by changes in neuronal activity and metabolic state. D, since the C-termini of AMPAR subunits in the ER are exposed to the cytoplasm, their phosphorylation states could be modified by cytoplasmic kinases and phosphatases, whose activities could be influenced by neuronal signals from nearby dendritic spines. The phosphorylation level of the CTD could in turn modulate preferential assembly of different subunits, leading to changes in subunit composition.
(1) The overall and relative abundance of AMPAR subunits and splice variants may be altered by neuronal activity – through gene expression, RNA splicing/alternative splicing, translation etc. – leading to a change in receptor number and composition (Fig. 3A). For example, chronic activity block in cultured CA1 hippocampal neurons leads to an increase in local dendritic synthesis of GluA1 subunits, but not GluA2, causing the subunit composition of synaptic receptors to shift towards those lacking GluA2 (Ju et al. 2004). Alternatively, this could be a result of alternative mRNA splicing or editing. For instance, activity block in cultured CA1 neurons results in an increase in the percentage of the flop isoform of both GluA1 (A1o) and GluA2 (A2o) (Penn et al. 2012). Since GluA2 subunits have a longer ER residence time than GluA1 (Greger et al. 2003), the newly emerged GluA1o subunits are more likely to recruit the lingering GluA2i subunits into forming A1o/A2i heteromers.
(2) Native AMPARs in neurons form complexes with transmembrane auxiliary subunits such as transmembrane auxillary regulatory proteins (TARPs), cornichons, CKAMP44 and SynDIG1 (Jackson & Nicoll, 2011). Some of these complexes might form in the ER during or following tetrameric assembly (Standley et al. 2000; Greger et al. 2002). Cornichons, in addition to being auxiliary subunits that modulate AMPAR function, may also act as ER chaperones that enhance the forward trafficking of receptors out of the ER (Shi et al. 2010). Bona fide ER chaperones such as calnexin and BiP, which are involved in protein folding and trafficking (Ellgaard & Helenius, 2003), associate with AMPARs (Rubio & Wenthold, 1999) possibly in a subunit-specific manner (Fukata et al. 2005). Additionally, AMPARs also associate, directly or indirectly, with a number of PDZ domain proteins such as SAP97, GRIP and PICK1 (Anggono & Huganir, 2012). For example, in hippocampal neurons, SAP97 associates with GluA1 early in the secretory pathway while the receptors are in the ER or the cis-Golgi apparatus (Sans et al. 2001). PICK1, on the other hand, associates with GluA2 subunits residing in the ER and together with Ca2+–calmodulin-dependent protein kinase II (CaMKII) regulates the ER export of GluA2-containing receptors (Greger et al. 2002; Lu et al. 2014). Such associations may stabilize or destabilize specific oligomeric species (Fig. 3B) by altering the intrinsic energetics of certain structural changes involved in the assembly process. This could occur in an activity-dependent manner, shifting the dimer/tetramer equilibrium and/or the predominance of subunit combinations in the ER during synaptic plasticity.
(3) The extracellular domains of the receptor, which are located in the ER lumen during assembly (Fig. 3C), influence receptor assembly. As such, the ER microenvironment as well as molecular processes that occur within the ER lumen could impact assembly through these domains. The binding of intraluminal glutamate is required for efficient ER exit of iGluRs (Mah et al. 2005; Fleck, 2006; Penn et al. 2008). One possibility is that the conformational changes in the LBD induced by glutamate influence the rate of formation and/or stability of the tetrameric complex, possibly by altering the efficiency of ‘domain swapping’. Fluctuations in luminal glutamate concentration might therefore regulate AMPAR assembly. Additionally, Ca2+ concentration and redox states within the ER lumen, which are subject to changes depending on the cell's metabolic state (Montero et al. 1995; Avezov et al. 2013), also represent possible routes of regulation on receptor assembly under disease states. According to the crystal structure, there are two intra-subunit disulfide bonds in each GluA2 subunit (one in each of the ATD and LBD) (Sobolevsky et al. 2009), which could be important to the proper folding of these domains. The redox state in the ER lumen might therefore affect the structural integrity of the extracellular domains via the process of oxidative protein folding (Tu & Weissman, 2004), and in turn influence assembly.
An additional ER-related process that might influence AMPAR assembly is N-glycosylation of the ATD. N-Glycosylation of certain ion channels such as acetylcholine receptors and voltage-gated sodium channels (Merlie et al. 1982; Schmidt & Catterall, 1986) are required for their proper assembly. Furthermore, N-glycosylation of many neuronal proteins, such as the sodium channel β4 subunit and GABAA receptor subunits, is developmentally regulated or might change in pathological states (Zhou et al. 2012; Mueller et al. 2014). While it is imaginable that N-glycosylation might represent a pathway for regulating AMPAR assembly as well, its actual involvement is controversial (Kawamoto et al. 1995; Everts et al. 1997). Nevertheless, the role of N-glycosylation in AMPAR assembly remains an intriguing issue that warrants further investigation.
(4) Activity-dependent modifications of C-terminal sites such as phosphorylation, palmitoylation, O-glycosylation and S-nitrosylation (Kanno et al. 2010; Traynelis et al. 2010; Selvakumar et al. 2013) could influence the subunit composition of AMPARs. Although phosphorylation of the C-terminus is known to affect AMPAR trafficking in a subunit composition-dependent manner (Henley et al. 2011), whether it could occur early in the secretory pathway is unknown. Still, the C-terminal domains of AMPARs in the ER are exposed to the cytoplasm, where cellular kinases and phosphatases are located. Protein kinase C-mediated phosphorylations in the C-terminal domains of NMDAR do indeed occur in the ER and regulate the receptor's export (Scott et al. 2001). In cardiomyocytes, protein kinase A phosphorylation of HERG channels in the ER augments its synthesis and export (Sroubek & McDonald, 2011). Thus, a similar mechanism involving C-terminal phosphorylation (or other types of modifications) could influence the assembly of AMPARs in the ER as well, either by directly altering the interactions between subunits or by modulating the functions of C-terminus-associating proteins (Fig. 3D).
Understanding the regulation of AMPAR assembly could have a widespread impact on both basic and clinical research. One could utilize this knowledge to design artificial AMPAR constructs that tetramerize on-demand in response to physiological manipulations (e.g. application of drugs, electric stimulations, etc.). One possible strategy to achieve this is to substitute pairs of amino acid residues involved in receptor assembly with unnatural amino acids that cross-link in response to UV light (e.g. BpA; Klippenstein et al. 2014). The resultant receptor would only tetramerize upon UV exposure. Such a construct, if achievable, would be a powerful tool for studying the dynamics of AMPAR assembly and trafficking. On the other hand, small molecules that modulate the assembly of AMPAR and other iGluRs would have great potential as therapeutic agents. For instance, AMPAR antagonists NBQX and GYKI-54266, in addition to their functional effects on the receptor, can also rescue the forward trafficking deficit of the L-to-Y mutant of GluA4 (Coleman et al. 2009). This finding opens up the possibility for designing ‘pharmacological chaperones’ that penetrate into the ER to target the assembly process in a subunit-specific manner.
Acknowledgments
We thank Drs Mark Bowen and Rashek Kazi, and Camilo Ferrer for helpful discussions and/or comments on the manuscript.
Glossary
- AMPA
α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid
- AMPAR
AMPA receptor
- ATD
amino-terminal domain
- BN-PAGE
blue-native polyacrylamide gel electrophoresis
- CTD
carboxy-terminal domain
- ER
endoplasmic reticulum
- iGluR
ionotropic glutamate receptor
- LBD
ligand-binding domain
- NMDAR
N-methyl-d-aspartate receptor
- TMD
transmembrane domain
Biographies
Quan Gan acquired his BSc degree in 2010 at the Chinese University of Hong Kong, majoring in Biochemistry. He joined the Graduate Program in Neuroscience at Stony Brook University the same year. His thesis research focuses on mechanisms governing AMPAR assembly.
Catherine Salussolia received her BA in Psychology from Swarthmore College (2004), her MS in Neuroscience and Neuropharmacology at Albany Medical College (2006), and her MD (2014)/PhD in Neuroscience (2012) at Stony Brook University. Catherine is currently completing a Pediatric Neurology residency.
Lonnie Wollmuth received his PhD in the Department of Physiology and Biophysics at the University of Washington in 1992. After postdoctoral work at the Max Planck Institute for Medical Research in Heidelberg, Germany, he moved to the Department of Neurobiology and Behaviour at Stony Brook University in 1998, where he holds the rank of Professor.
Additional information
Competing interests
None declared.
Funding
This work was supported by a grant from the Thomas Hartman Center for Parkinson's Research (L.P.W.), a SBU-CSHL Collaborative Grant (L.P.W.), an NIH National Research Service Award (NRSA) from NINDS (NS073382) (C.L.S.) and an American Heart Association pre-doctoral fellowship (Q.G.).
Author's present address
C. L. Salussolia: Department of Pediatrics, Stony Brook Children's Hospital, Stony Brook, NY, USA.
References
- Anggono V. Huganir RL. Regulation of AMPA receptor trafficking and synaptic plasticity. Curr Opin Neurobiol. 2012;22:461–469. doi: 10.1016/j.conb.2011.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avezov E, Cross BC, Kaminski Schierle GS, Winters M, Harding HP, Melo EP, Kaminski CF. Ron D. Lifetime imaging of a fluorescent protein sensor reveals surprising stability of ER thiol redox. J Cell Biol. 2013;201:337–349. doi: 10.1083/jcb.201211155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayalon G. Stern-Bach Y. Functional assembly of AMPA and kainate receptors is mediated by several discrete protein-protein interactions. Neuron. 2001;31:103–113. doi: 10.1016/s0896-6273(01)00333-6. [DOI] [PubMed] [Google Scholar]
- Bowers MS, Chen BT. Bonci A. AMPA receptor synaptic plasticity induced by psychostimulants: the past, present, and therapeutic future. Neuron. 2010;67:11–24. doi: 10.1016/j.neuron.2010.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brorson JR, Li D. Suzuki T. Selective expression of heteromeric AMPA receptors driven by flip-flop differences. J Neurosci. 2004;24:3461–3470. doi: 10.1523/JNEUROSCI.5023-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coleman SK, Moykkynen T, Cai C, von Ossowski L, Kuismanen E, Korpi ER. Keinanen K. Isoform-specific early trafficking of AMPA receptor flip and flop variants. J Neurosci. 2006;26:11220–11229. doi: 10.1523/JNEUROSCI.2301-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coleman SK, Moykkynen T, Hinkkuri S, Vaahtera L, Korpi ER, Pentikainen OT. Keinanen K. Ligand-binding domain determines endoplasmic reticulum exit of AMPA receptors. J Biol Chem. 2010;285:36032–36039. doi: 10.1074/jbc.M110.156943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coleman SK, Moykkynen T, Jouppila A, Koskelainen S, Rivera C, Korpi ER. Keinanen K. Agonist occupancy is essential for forward trafficking of AMPA receptors. J Neurosci. 2009;29:303–312. doi: 10.1523/JNEUROSCI.3953-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collingridge GL, Isaac JT. Wang YT. Receptor trafficking and synaptic plasticity. Nat Rev Neurosci. 2004;5:952–962. doi: 10.1038/nrn1556. [DOI] [PubMed] [Google Scholar]
- Cui-Wang T, Hanus C, Cui T, Helton T, Bourne J, Watson D, Harris KM. Ehlers MD. Local zones of endoplasmic reticulum complexity confine cargo in neuronal dendrites. Cell. 2012;148:309–321. doi: 10.1016/j.cell.2011.11.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cull-Candy S, Kelly L. Farrant M. Regulation of Ca2+-permeable AMPA receptors: synaptic plasticity and beyond. Curr Opin Neurobiol. 2006;16:288–297. doi: 10.1016/j.conb.2006.05.012. [DOI] [PubMed] [Google Scholar]
- Das U, Kumar J, Mayer ML. Plested AJ. Domain organization and function in GluK2 subtype kainate receptors. Proc Natl Acad Sci U S A. 2010;107:8463–8468. doi: 10.1073/pnas.1000838107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derkach VA, Oh MC, Guire ES. Soderling TR. Regulatory mechanisms of AMPA receptors in synaptic plasticity. Nat Rev Neurosci. 2007;8:101–113. doi: 10.1038/nrn2055. [DOI] [PubMed] [Google Scholar]
- Ellgaard L. Helenius A. Quality control in the endoplasmic reticulum. Nat Rev Mol Cell Biol. 2003;4:181–191. doi: 10.1038/nrm1052. [DOI] [PubMed] [Google Scholar]
- Everts I, Villmann C. Hollmann M. N-Glycosylation is not a prerequisite for glutamate receptor function but is essential for lectin modulation. Mol Pharmacol. 1997;52:861–873. doi: 10.1124/mol.52.5.861. [DOI] [PubMed] [Google Scholar]
- Fleck MW. Glutamate receptors and endoplasmic reticulum quality control: looking beneath the surface. Neuroscientist. 2006;12:232–244. doi: 10.1177/1073858405283828. [DOI] [PubMed] [Google Scholar]
- Fukata Y, Tzingounis AV, Trinidad JC, Fukata M, Burlingame AL, Nicoll RA. Bredt DS. Molecular constituents of neuronal AMPA receptors. J Cell Biol. 2005;169:399–404. doi: 10.1083/jcb.200501121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geiger JR, Melcher T, Koh DS, Sakmann B, Seeburg PH, Jonas P. Monyer H. Relative abundance of subunit mRNAs determines gating and Ca2+ permeability of AMPA receptors in principal neurons and interneurons in rat CNS. Neuron. 1995;15:193–204. doi: 10.1016/0896-6273(95)90076-4. [DOI] [PubMed] [Google Scholar]
- Granger AJ, Shi Y, Lu W, Cerpas M. Nicoll RA. LTP requires a reserve pool of glutamate receptors independent of subunit type. Nature. 2013;493:495–500. doi: 10.1038/nature11775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greger IH, Akamine P, Khatri L. Ziff EB. Developmentally regulated, combinatorial RNA processing modulates AMPA receptor biogenesis. Neuron. 2006;51:85–97. doi: 10.1016/j.neuron.2006.05.020. [DOI] [PubMed] [Google Scholar]
- Greger IH, Khatri L, Kong X. Ziff EB. AMPA receptor tetramerization is mediated by Q/R editing. Neuron. 2003;40:763–774. doi: 10.1016/s0896-6273(03)00668-8. [DOI] [PubMed] [Google Scholar]
- Greger IH, Khatri L. Ziff EB. RNA editing at Arg607 controls AMPA receptor exit from the endoplasmic reticulum. Neuron. 2002;34:759–772. doi: 10.1016/s0896-6273(02)00693-1. [DOI] [PubMed] [Google Scholar]
- Grigorenko E, Glazier S, Bell W, Tytell M, Nosel E, Pons T. Deadwyler SA. Changes in glutamate receptor subunit composition in hippocampus and cortex in patients with refractory epilepsy. J Neurol Sci. 1997;153:35–45. doi: 10.1016/s0022-510x(97)00180-9. [DOI] [PubMed] [Google Scholar]
- Grooms SY, Noh KM, Regis R, Bassell GJ, Bryan MK, Carroll RC. Zukin RS. Activity bidirectionally regulates AMPA receptor mRNA abundance in dendrites of hippocampal neurons. J Neurosci. 2006;26:8339–8351. doi: 10.1523/JNEUROSCI.0472-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henley JM, Barker EA. Glebov OO. Routes, destinations and delays: recent advances in AMPA receptor trafficking. Trends Neurosci. 2011;34:258–268. doi: 10.1016/j.tins.2011.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herguedas B, Krieger J. Greger IH. Receptor heteromeric assembly–how it works and why it matters: the case of ionotropic glutamate receptors. Prog Mol Biol Transl Sci. 2013;117:361–386. doi: 10.1016/B978-0-12-386931-9.00013-1. [DOI] [PubMed] [Google Scholar]
- Isaac JT, Ashby MC. McBain CJ. The role of the GluR2 subunit in AMPA receptor function and synaptic plasticity. Neuron. 2007;54:859–871. doi: 10.1016/j.neuron.2007.06.001. [DOI] [PubMed] [Google Scholar]
- Jackson AC. Nicoll RA. The expanding social network of ionotropic glutamate receptors: TARPs and other transmembrane auxiliary subunits. Neuron. 2011;70:178–199. doi: 10.1016/j.neuron.2011.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ju W, Morishita W, Tsui J, Gaietta G, Deerinck TJ, Adams SR, Garner CC, Tsien RY, Ellisman MH. Malenka RC. Activity-dependent regulation of dendritic synthesis and trafficking of AMPA receptors. Nat Neurosci. 2004;7:244–253. doi: 10.1038/nn1189. [DOI] [PubMed] [Google Scholar]
- Kanno T, Yaguchi T, Nagata T, Mukasa T. Nishizaki T. Regulation of AMPA receptor trafficking by O-glycosylation. Neurochem Res. 2010;35:782–788. doi: 10.1007/s11064-010-0135-1. [DOI] [PubMed] [Google Scholar]
- Kawamoto S, Hattori S, Sakimura K, Mishina M. Okuda K. N-linked glycosylation of the α-amino-3-hydroxy-5-methylisoxazole-4-propionate (AMPA)-selective glutamate receptor channel α2 subunit is essential for the acquisition of ligand-binding activity. J Neurochem. 1995;64:1258–1266. doi: 10.1046/j.1471-4159.1995.64031258.x. [DOI] [PubMed] [Google Scholar]
- Kim KS, Yan D. Tomita S. Assembly and stoichiometry of the AMPA receptor and transmembrane AMPA receptor regulatory protein complex. J Neurosci. 2010;30:1064–1072. doi: 10.1523/JNEUROSCI.3909-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klippenstein V, Ghisi V, Wietstruk M. Plested AJ. Photoinactivation of glutamate receptors by genetically encoded unnatural amino acids. J Neurosci. 2014;34:980–991. doi: 10.1523/JNEUROSCI.3725-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar J, Schuck P. Mayer ML. Structure and assembly mechanism for heteromeric kainate receptors. Neuron. 2011;71:319–331. doi: 10.1016/j.neuron.2011.05.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar SS, Bacci A, Kharazia V. Huguenard JR. A developmental switch of AMPA receptor subunits in neocortical pyramidal neurons. J Neurosci. 2002;22:3005–3015. doi: 10.1523/JNEUROSCI.22-08-03005.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loddenkemper T, Talos DM, Cleary RT, Joseph A, Sanchez Fernández I, Alexopoulos A, Kotagal P, Najm I. Jensen FE. Subunit composition of glutamate and gamma-aminobutyric acid receptors in status epilepticus. Epilepsy Res. 2014;108:605–615. doi: 10.1016/j.eplepsyres.2014.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu J, Robinson JM, Edwards D. Deutsch C. T1–T1 interactions occur in ER membranes while nascent Kv peptides are still attached to ribosomes. Biochemistry. 2001;40:10934–10946. doi: 10.1021/bi010763e. [DOI] [PubMed] [Google Scholar]
- Lu W, Khatri L. Ziff EB. Trafficking of α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid receptor (AMPA) receptor subunit GluA2 from the endoplasmic reticulum is stimulated by a complex containing Ca2+/calmodulin-activated kinase II (CaMKII) and PICK1 protein and by release of Ca2+ from internal stores. J Biol Chem. 2014;289:19218–19230. doi: 10.1074/jbc.M113.511246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu W, Shi Y, Jackson AC, Bjorgan K, During MJ, Sprengel R, Seeburg PH. Nicoll RA. Subunit composition of synaptic AMPA receptors revealed by a single-cell genetic approach. Neuron. 2009;62:254–268. doi: 10.1016/j.neuron.2009.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mah SJ, Cornell E, Mitchell NA. Fleck MW. Glutamate receptor trafficking: endoplasmic reticulum quality control involves ligand binding and receptor function. J Neurosci. 2005;25:2215–2225. doi: 10.1523/JNEUROSCI.4573-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makino H. Malinow R. AMPA receptor incorporation into synapses during LTP: the role of lateral movement and exocytosis. Neuron. 2009;64:381–390. doi: 10.1016/j.neuron.2009.08.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mameli M, Balland B, Luján R. Lüscher C. Rapid synthesis and synaptic insertion of GluR2 for mGluR-LTD in the ventral tegmental area. Science. 2007;317:530–533. doi: 10.1126/science.1142365. [DOI] [PubMed] [Google Scholar]
- Mansour M, Nagarajan N, Nehring RB, Clements JD. Rosenmund C. Heteromeric AMPA receptors assemble with a preferred subunit stoichiometry and spatial arrangement. Neuron. 2001;32:841–853. doi: 10.1016/s0896-6273(01)00520-7. [DOI] [PubMed] [Google Scholar]
- Martin LJ, Blackstone CD, Levey AI, Huganir RL. Price DL. AMPA glutamate receptor subunits are differentially distributed in rat brain. Neuroscience. 1993;53:327–358. doi: 10.1016/0306-4522(93)90199-p. [DOI] [PubMed] [Google Scholar]
- Merlie JP, Sebbane R, Tzartos S. Lindstrom J. Inhibition of glycosylation with tunicamycin blocks assembly of newly synthesized acetylcholine receptor subunits in muscle cells. J Biol Chem. 1982;257:2694–2701. [PubMed] [Google Scholar]
- Montero M, Brini M, Marsault R, Alvarez J, Sitia R, Pozzan T. Rizzuto R. Monitoring dynamic changes in free Ca2+ concentration in the endoplasmic reticulum of intact cells. EMBO J. 1995;14:5467–5475. doi: 10.1002/j.1460-2075.1995.tb00233.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moykkynen T, Coleman SK, Semenov A. Keinanen K. The N-terminal domain modulates α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid (AMPA) receptor desensitization. J Biol Chem. 2014;289:13197–13205. doi: 10.1074/jbc.M113.526301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller TM, Haroutunian V. Meador-Woodruff JH. N-Glycosylation of GABAA receptor subunits is altered in schizophrenia. Neuropsychopharmacology. 2014;39:528–537. doi: 10.1038/npp.2013.190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa T. The biochemistry, ultrastructure, and subunit assembly mechanism of AMPA receptors. Mol Neurobiol. 2010;42:161–184. doi: 10.1007/s12035-010-8149-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park M, Penick EC, Edwards JG, Kauer JA. Ehlers MD. Recycling endosomes supply AMPA receptors for LTP. Science. 2004;305:1972–1975. doi: 10.1126/science.1102026. [DOI] [PubMed] [Google Scholar]
- Pasternack A, Coleman SK, Jouppila A, Mottershead DG, Lindfors M, Pasternack M. Keinanen K. α-Amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid (AMPA) receptor channels lacking the N-terminal domain. J Biol Chem. 2002;277:49662–49667. doi: 10.1074/jbc.M208349200. [DOI] [PubMed] [Google Scholar]
- Penn AC, Balik A, Wozny C, Cais O. Greger IH. Activity-mediated AMPA receptor remodeling, driven by alternative splicing in the ligand-binding domain. Neuron. 2012;76:503–510. doi: 10.1016/j.neuron.2012.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penn AC, Williams SR. Greger IH. Gating motions underlie AMPA receptor secretion from the endoplasmic reticulum. EMBO J. 2008;27:3056–3068. doi: 10.1038/emboj.2008.222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrini EM, Lu J, Cognet L, Lounis B, Ehlers MD. Choquet D. Endocytic trafficking and recycling maintain a pool of mobile surface AMPA receptors required for synaptic potentiation. Neuron. 2009;63:92–105. doi: 10.1016/j.neuron.2009.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phartiyal P, Jones EM. Robertson GA. Heteromeric assembly of human ether-à-go-go-related gene (hERG) 1a/1b channels occurs cotranslationally via N-terminal interactions. J Biol Chem. 2007;282:9874–9882. doi: 10.1074/jbc.M610875200. [DOI] [PubMed] [Google Scholar]
- Priel A, Selak S, Lerma J. Stern-Bach Y. Block of kainate receptor desensitization uncovers a key trafficking checkpoint. Neuron. 2006;52:1037–1046. doi: 10.1016/j.neuron.2006.12.006. [DOI] [PubMed] [Google Scholar]
- Riou M, Stroebel D, Edwardson JM. Paoletti P. An alternating GluN1-2-1-2 subunit arrangement in mature NMDA receptors. PLoS One. 2012;7:e35134. doi: 10.1371/journal.pone.0035134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossmann M, Sukumaran M, Penn AC, Veprintsev DB, Babu MM. Greger IH. Subunit-selective N-terminal domain associations organize the formation of AMPA receptor heteromers. EMBO J. 2011;30:959–971. doi: 10.1038/emboj.2011.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubio ME. Wenthold RJ. Calnexin and the immunoglobulin binding protein (BiP) coimmunoprecipitate with AMPA receptors. J Neurochem. 1999;73:942–948. doi: 10.1046/j.1471-4159.1999.0730942.x. [DOI] [PubMed] [Google Scholar]
- Salussolia CL, Corrales A, Talukder I, Kazi R, Akgul G, Bowen M. Wollmuth LP. Interaction of the M4 segment with other transmembrane segments is required for surface expression of mammalian α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid (AMPA) receptors. J Biol Chem. 2011a;286:40205–40218. doi: 10.1074/jbc.M111.268839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salussolia CL, Gan Q, Kazi R, Singh P, Allopenna J, Furukawa H. Wollmuth LP. A eukaryotic specific transmembrane segment is required for tetramerization in AMPA receptors. J Neurosci. 2013;33:9840–9845. doi: 10.1523/JNEUROSCI.2626-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salussolia CL, Prodromou ML, Borker P. Wollmuth LP. Arrangement of subunits in functional NMDA receptors. J Neurosci. 2011b;31:11295–11304. doi: 10.1523/JNEUROSCI.5612-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sans N, Racca C, Petralia RS, Wang YX, McCallum J. Wenthold RJ. Synapse-associated protein 97 selectively associates with a subset of AMPA receptors early in their biosynthetic pathway. J Neurosci. 2001;21:7506–7516. doi: 10.1523/JNEUROSCI.21-19-07506.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt JW. Catterall WA. Biosynthesis and processing of the α subunit of the voltage-sensitive sodium channel in rat brain neurons. Cell. 1986;46:437–444. doi: 10.1016/0092-8674(86)90664-1. [DOI] [PubMed] [Google Scholar]
- Scott DB, Blanpied TA, Swanson GT, Zhang C. Ehlers MD. An NMDA receptor ER retention signal regulated by phosphorylation and alternative splicing. J Neurosci. 2001;21:3063–3072. doi: 10.1523/JNEUROSCI.21-09-03063.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selvakumar B, Jenkins MA, Hussain NK, Huganir RL, Traynelis SF. Snyder SH. S-Nitrosylation of AMPA receptor GluA1 regulates phosphorylation, single-channel conductance, and endocytosis. Proc Natl Acad Sci U S A. 2013;110:1077–1082. doi: 10.1073/pnas.1221295110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shanks NF, Maruo T, Farina AN, Ellisman MH. Nakagawa T. Contribution of the global subunit structure and stargazin on the maturation of AMPA receptors. J Neurosci. 2010;30:2728–2740. doi: 10.1523/JNEUROSCI.5146-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y, Suh YH, Milstein AD, Isozaki K, Schmid SM, Roche KW. Nicoll RA. Functional comparison of the effects of TARPs and cornichons on AMPA receptor trafficking and gating. Proc Natl Acad Sci U S A. 2010;107:16315–16319. doi: 10.1073/pnas.1011706107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sobolevsky AI, Rosconi MP. Gouaux E. X-ray structure, symmetry and mechanism of an AMPA-subtype glutamate receptor. Nature. 2009;462:745–756. doi: 10.1038/nature08624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Splitt H, Meuser D, Borovok I, Betzler M. Schrempf H. Pore mutations affecting tetrameric assembly and functioning of the potassium channel KcsA from Streptomyces lividans. FEBS Lett. 2000;472:83–87. doi: 10.1016/s0014-5793(00)01429-0. [DOI] [PubMed] [Google Scholar]
- Sroubek J. McDonald TV. Protein kinase A activity at the endoplasmic reticulum surface is responsible for augmentation of human ether-a-go-go-related gene product (HERG) J Biol Chem. 2011;286:21927–21936. doi: 10.1074/jbc.M110.201699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Standley S, Roche KW, McCallum J, Sans N. Wenthold RJ. PDZ domain suppression of an ER retention signal in NMDA receptor NR1 splice variants. Neuron. 2000;28:887–898. doi: 10.1016/s0896-6273(00)00161-6. [DOI] [PubMed] [Google Scholar]
- Sukumaran M, Penn AC. Greger IH. AMPA receptor assembly: atomic determinants and built-in modulators. Adv Exp Med Biol. 2012;970:241–264. doi: 10.1007/978-3-7091-0932-8_11. [DOI] [PubMed] [Google Scholar]
- Sun Y, Olson R, Horning M, Armstrong N, Mayer M. Gouaux E. Mechanism of glutamate receptor desensitization. Nature. 2002;417:245–253. doi: 10.1038/417245a. [DOI] [PubMed] [Google Scholar]
- Suzuki E, Kessler M. Arai AC. The fast kinetics of AMPA GluR3 receptors is selectively modulated by the TARPs γ4 and γ8. Mol Cell Neurosci. 2008;38:117–123. doi: 10.1016/j.mcn.2008.01.018. [DOI] [PubMed] [Google Scholar]
- Tichelaar W, Safferling M, Keinanen K, Stark H. Madden DR. The three-dimensional structure of an ionotropic glutamate receptor reveals a dimer-of-dimers assembly. J Mol Biol. 2004;344:435–442. doi: 10.1016/j.jmb.2004.09.048. [DOI] [PubMed] [Google Scholar]
- Traynelis SF, Wollmuth LP, McBain CJ, Menniti FS, Vance KM, Ogden KK, Hansen KB, Yuan H, Myers SJ. Dingledine R. Glutamate receptor ion channels: structure, regulation, and function. Pharmacol Rev. 2010;62:405–496. doi: 10.1124/pr.109.002451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu BP. Weissman JS. Oxidative protein folding in eukaryotes: mechanisms and consequences. J Cell Biol. 2004;164:341–346. doi: 10.1083/jcb.200311055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G, Gilbert J. Man HY. AMPA receptor trafficking in homeostatic synaptic plasticity: functional molecules and signaling cascades. Neural Plast. 2012;2012:825364. doi: 10.1155/2012/825364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Alimi Y, Tong A, Nichols CG. Enkvetchakul D. Differential roles of blocking ions in KirBac1.1 tetramer stability. J Biol Chem. 2009;284:2854–2860. doi: 10.1074/jbc.M807474200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weston MC, Schuck P, Ghosal A, Rosenmund C. Mayer ML. Conformational restriction blocks glutamate receptor desensitization. Nat Struct Mol Biol. 2006;13:1120–1127. doi: 10.1038/nsmb1178. [DOI] [PubMed] [Google Scholar]
- Zhao H, Berger AJ, Brown PH, Kumar J, Balbo A, May CA, Casillas E, Jr, Laue TM, Patterson GH, Mayer ML. Schuck P. Analysis of high-affinity assembly for AMPA receptor amino-terminal domains. J Gen Physiol. 2012;139:371–388. doi: 10.1085/jgp.201210770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou TT, Zhang ZW, Liu J, Zhang JP. Jiao BH. Glycosylation of the sodium channel β4 subunit is developmentally regulated and involves in neuritic degeneration. Int J Biol Sci. 2012;8:630–639. doi: 10.7150/ijbs.3684. [DOI] [PMC free article] [PubMed] [Google Scholar]


