Abstract
We demonstrate that recent observed increases in cell permeation activity of cyclic peptides via transporter sequences is due to the underlying phase behavior of peptide-lipid complexes and its relation to the topological requirement of membrane permeation. We also show how these effects can be amplified by incorporating hydrophobicity in these sequences.
Cyclic peptides exhibit strong enhancement in receptor-binding affinity, specificity, and stability against enzymatic degradation relative to their linear counterparts, which is due in part to their reduced conformational freedom.1–5 In general, however, the translocation efficiency across cell membranes for such cyclic peptides is low, thereby limiting their applications. Recent work has shown that a significant increase in cyclic peptide translocation efficiency is possible when cationic and hydrophobic residues are incorporated into cyclic peptides, notably arginine and tryptophan.6–9 However, at present there is no detailed molecular understanding of these effects. The relative importance of peptide cyclicity and hydrophobicity in these transporter sequences is not known, nor the precise relation between these cyclic transporter sequences and linear cell penetrating peptides (CPP) such as HIV TAT, ANTP penetratin, or oligoarginine.10–18
In this work, we show that observed increases in cyclic peptide cell permeation activity correlates strongly with the phase behavior of the underlying peptide-lipid system, and is related to the induction of negative Gaussian membrane (or ‘saddle-splay’) curvature, which is topologically necessary for pore formation and other membrane destabilization mechanisms.11 Furthermore, we apply recently identified sequence rules for cell penetrating peptides and show using high resolution synchrotron small angle x-ray scattering (SAXS) how the presence of cationic charge, hydrophobicity, and peptide cyclicity work together to amplify the membrane curvature generation required for membrane permeation, and quantitatively delineate the relative importance of each using mutant sequences. We demonstrate that cyclic peptides, even short peptides that exhibit no activity in their linearized versions, can have increased tendency for generating negative Gaussian curvature. We show that this design criterion can improve a peptide’s ability to penetrate cells as measured by cell uptake experiments. The detailed agreement between biophysical measurements and cell uptake experiments suggests that deterministic molecular design of optimized transporter sequences are possible.
To isolate the synergistic effects of peptide cyclicity, hydrophobicity, and arginine content, four peptides are synthesized: two cyclic peptides, cyc-R4 (cyclo(Arg-Arg-Arg-Arg-Gln)) and cyc-R4F2 (cyclo(Phe-Phe-Arg-Arg-Arg-Arg-Gln)), and their linear counterparts, lin-R4 (Arg-Arg-Arg-Arg) and lin-R4F2 (Phe-Phe-Arg-Arg-Arg-Arg-Gln). (In the synthesis of cyclic peptides, Gln serves as a convenient anchor for attachment to the solid phase as well as the point of N-to-C peptide cyclization.7) Arg is chosen as the cationic component since it is a principal component in prototypical cell penetrating peptides such as HIV TAT, ANTP, and polyArg. In addition, since Arg is expected to generate membrane curvature via multidentate H-bonding near the membrane surface, and since aromatic residues can hydrogen bond at polar-apolar interfaces,19–21 Phe is chosen as the hydrophobic amino acid for this study. Details on the materials and methods, including SAXS, confocal microscopy, cell culture, peptide uptake, and cytotoxicity measurements, are included in supplementary materials.
Eukaryotic membranes are complex, with protein as well as lipid components, with different headgroups, chain lengths, and architectures. We have chosen to work with ternary mixtures of DOPS/DOPE/DOPC lipids (DOPS: 1,2-dioleoyl-sn-glycero-3-phospho-L-serine; DOPE:1,2-dioleoyl-sn-glycero-3-phospho-ethanolamine; DOPC:1,2-dioleoyl-sn-glycero-3-phosphocholine). Depending on cell type, these comprise ~70% of lipids in most eukaryotic cells.22 A ternary mixture of DOPS/DOPE/DOPC allows us to independently vary the charge density and the intrinsic curvature of the membrane, both of which are important parameters for the present problem.23 It should be noted that even studies with ternary membranes can only give a baseline picture, due to the existence of specialized amphiphiles such as sterols in eukaryotic membranes, which play important roles in raft formation, protein sorting and cell signaling.24
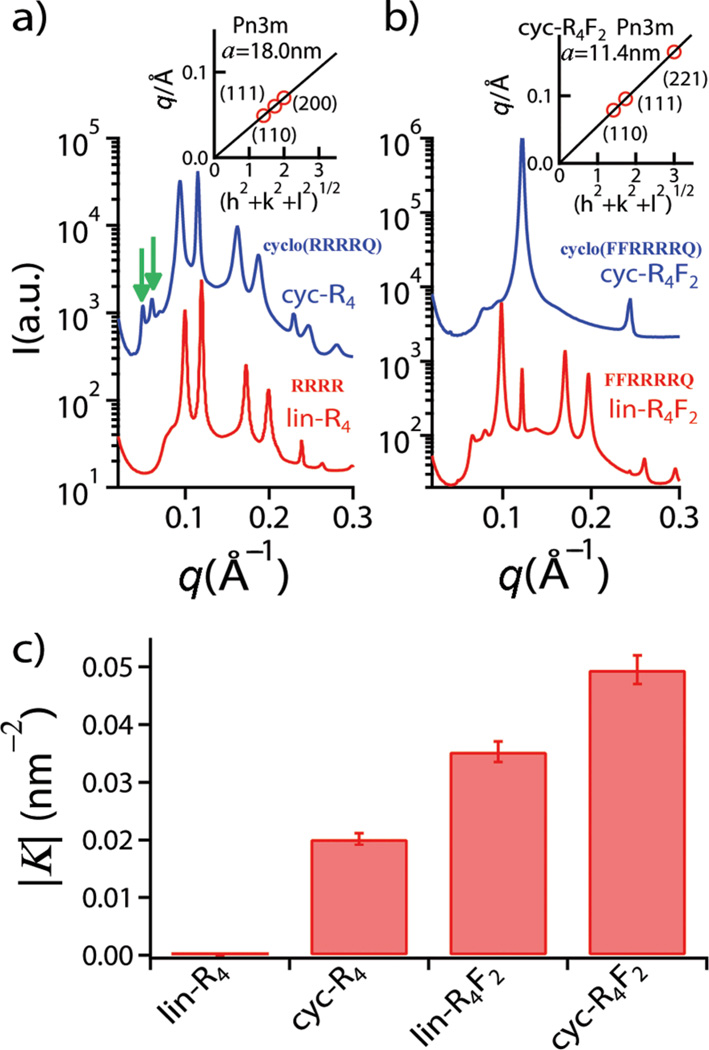
Rather than directly solving the structure of a CPP induced pore, we map out the conditions under which CPPs induce topologically active membrane curvature at specific global peptide/lipid (P/L) molar ratios using SAXS‡. It is well established10–12 that lin-R4 has negligible cell penetrating activity, so we perform a baseline comparison of lin-R4 with cyc-R4. When exposed to lin-R4, small unilamellar vesicles (SUVs) were observed to undergo a structural phase transition to inverted hexagonal (HII) and lamellar (Lα) phases (Fig. 1a) due to strong electrostatic interactions. In contrast, when exposed to cyc-R4, an additional phase is observed. By indexing the peaks of the diffracted intensity curve, we find that the new phase is a cubic Pn3m ‘double-diamond’ structure, as indicated by the characteristic correlation peaks with ratios √2: √3: √4: √6. The first two peaks of Pn3m are indicated by the green arrows in Fig. 1a for the cyc-R4 curve, whereas for the lin-R4 curve, there are no such peaks. By fitting the slopes of the measured q values at every peak, we calculate the lattice parameter to be a =18.0 nm (Fig. 1a inset). This is consistent with the behavior of the HIV TAT peptide.15 The Pn3m is a bicontinuous cubic phase where two non-intersecting water channels are separated by a lipid bilayer.25 The bilayer traces out a minimal surface, and indicates that negative Gaussian curvature exists at every point of each of the two constituent monolayer leaflets, the type of curvature necessary for ‘hole’ formation in a membrane. This indicates that the peptide clearly induces negative Gaussian curvature strain on a membrane surface. In addition to being a necessary condition for pore formation, negative Gaussian curvature is also required for processes such as budding or blebbing, which is observed in macropinocytosis.26 Interestingly, although negative Gaussian curvature on a monolayer leaflet (as in a transmembrane pore) is distinct from negative Gaussian curvature in a bilayer (as in a bud or bleb), there is strong correlation between peptide induced membrane permeation and peptide induced formation of cubic phases,15, 16, 23 which suggests that the root phenomenon of negative Gaussian membrane curvature generation by peptides can be expressed as different structural outcomes in isolated membranes under different conditions.
Fig. 1.
Comparison between cyclic peptides (cyc-R4, cyc-R4F2) and their linear counterparts (lin-R4, lin-R4F2). a) SAXS measurement for cyc-R4 and lin-R4; b) SAXS measurement for cyc-R4F2 and lin-R4F2 (DOPS/DOPE/DOPC = 20/80/00, peptide/lipid molar ratio P/L = 1/10). The SAXS intensity curves in a) and b) are shifted for clarity. c) Calculated average magnitude of Gaussian curvature |K| from the lattice constants of Pn3m cubic phase. Insets in a) and b) show indexation of the Pn3m cubic phase by agreement between the measured peak q positions and the Miller indices h, k, l, with relationship q = 2π(h2+k2+l2)1/2/a, for a cubic phase with lattice constant, a. The arrows (green) in a) indicate the first two peaks of Pn3m cubic phase.
The role of hydrophobicity in cell penetrating peptides is complex.16 The inclusion of a single hydrophobic amino acid can change the translocation mechanism. Moreover, the amino acid content of natural cell penetrating peptides and antimicrobial peptides follow basic design rules that are rooted in the need for generating negative Gaussian membrane curvature, so that the addition of hydrophobicity can either enhance or suppress activity, depending on the lysine and arginine content.16, 23 Following those design rules, we add two phenylalanines to the baseline R4 peptide to make a new peptide R4F2. Whereas lin-R4 does not induce a cubic phase, and does not exhibit cell penetrating activity, both lin-R4F2 and cyc-R4F2 induce Pn3m cubic phases rich in negative Gaussian curvature (alin-R4F2 = 13.6 nm and acyc-R4F2 = 11.4 nm, Fig. 1b), illustrating the important role of both hydrophobicity and cyclicity. The average Gaussian curvature induced by those peptides per unit cell can be quantified via <K>=2πχ/a2A0, where a is the cubic lattice constant, χ is the Euler characteristic, and A0 is the surface area per unit cell (χ = −2 and A0 = 1.919 for Pn3m). This provides a way to directly compare the relative effects of hydrophobicity and peptide cyclicity under the same conditions (target lipid composition PS/PE/PC=20/80/00, P/L=1/10). The average magnitudes of the Gaussian curvature generated by cyc-R4 and lin-R4F2 are |K| ~ 0.020 nm−2 and |K| ~ 0.035 nm−2, respectively (Fig. 1c), indicating that the contribution to negative Gaussian curvature generation from hydrophobicity in these peptides is greater than that from peptide cyclicity. Interesting, when both of these enhancement mechanisms are combined in cyc-R4F2, the induced negative Gaussian curvature is |K| ~ 0.050 nm−2, which is significantly higher than either mechanism alone, suggesting that the mechanisms can be additive and that it is possible to amplify cell penetrating activity by combining both mechanisms: By adding appropriate hydrophobic amino acids, we increase the negative Gaussian curvature generation ability of cyc-R4F2 by almost 3 fold relative to cyc-R4, which is itself already much more active than lin-R4. This will be confirmed by cell uptake experiments described below.
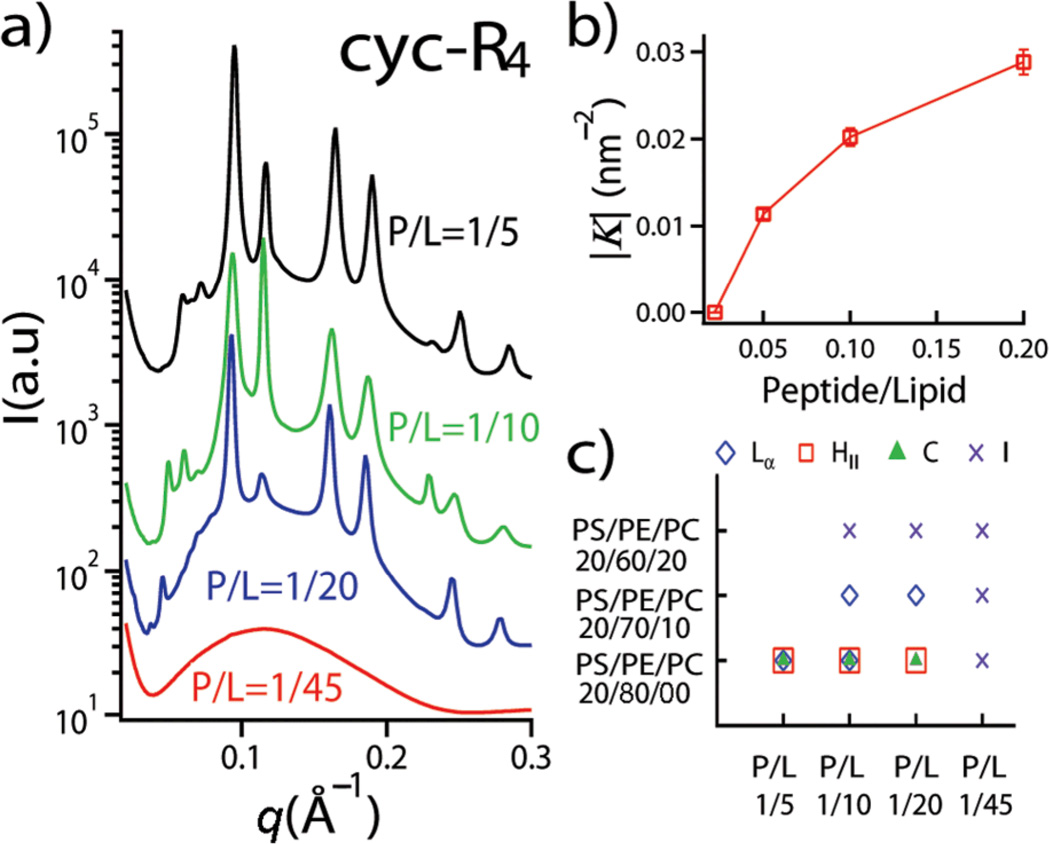
It is in principle possible to tune the activity of cell penetrating peptides with different membrane compositions, since the generation of negative Gaussian curvature is affected by the lipid compositions of the target membrane and peptide/lipid molar ratios (Fig. 2). For a fixed lipid composition PS/PE/PC=20/80/00 at P/L=1/45, SAXS measurements reveal that the system consists of only a unilamellar membrane form factor, indicating that no significant restructuring of the SUVs has taken place. As the P/L ratio is increased, new phases appear. For example, above P/L=1/10, there are three coexisting phases, a lamellar phase (Lα), an inverted hexagonal phase (HII), and a Pn3m cubic phase (C) (Fig. 2a). Interestingly, the magnitude of negative Gaussian curvature generated in the membrane can be quantitatively controlled by the P/L ratio (Fig. 2b). High P/L ratios induce small lattice constants and consequently more negative Gaussian curvature per unit cell |K|. By varying peptide/lipid ratios and lipid compositions, we map out a phase diagram for the mixture of cyc-R4 and lipid membranes (Fig. 2c). In order to isolate the effects of membrane intrinsic curvature from electrostatic effects, we systematically vary the fraction of DOPE and DOPC while keeping the fraction of DOPS constant in a tertiary PS/PE/PC membrane, so that the intrinsic curvature of the membrane is varied without significantly changing the charge density. As in the case for cell penetrating peptides, such as HIV TAT, polyR, and ANTP penetratin,11, 15, 16 we find that the cubic phase is favored for membrane compositions rich in negative intrinsic curvature lipids (such as PS/PE/PC = 20/80/00), suggesting that they share common mechanisms, and indicating that a high local concentration of DOPE is necessary for peptide induced membrane restructuring. This suggests that lipid microphase separation is important for forming the high curvatures needed in pore formation, a behavior that is consistent with recent mass spectrometry imaging of mating pores in tetrahymena.27
Fig. 2.
Effect of lipid composition and peptide/lipid molar ratio on the membrane activity of cyc-R4. a) SAXS data show different phase behaviors of the cyc-R4 peptide-lipid system at different peptide/lipid molar ratios for PS/PE/PC = 20/80/00. The SAXS intensity curves are shifted for clarity. b) Gaussian curvature obtained at different P/L molar ratios for PS/PE/PC = 20/80/00; c) Phase diagram for the cyc-R4 peptide-lipid system as a function of lipid composition and peptide/lipid molar ratios. The symbols denote the following: Lα (diamond): lamellar phase; HII (square): inverted hexagonal phase; C (triangle): Pn3m cubic phase; I (cross): form factor.
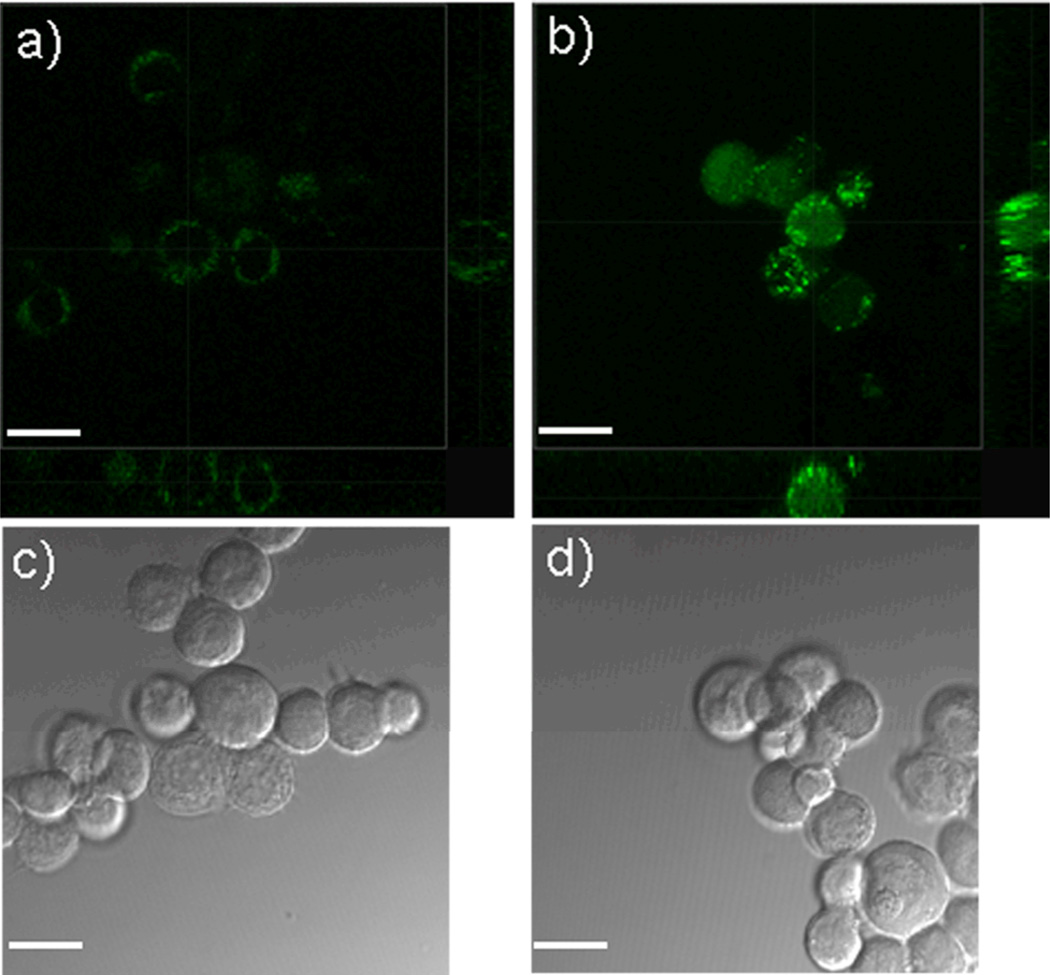
We perform cell uptake experiments in order to test translocation efficiencies for cyclic peptides across membranes. After incubating MCF7 cells with lin-R4F2-FITC (Ac-Phe-Phe-Arg-Arg-Arg-Arg-Glu[Lys(FITC)-NH2]-OAll) and cyc-R4F2-FITC (Cyclo{Phe-Phe-Arg-Arg-Arg-Arg-Glu[Lys(FITC)-NH2]}), which are the FITC-labeled versions of lin-R4F2 and cyc-R4F2 peptides, respectively, we examined the cells using laser scanning confocal microscopy (LSCM), as shown in Fig. 3. The fluorescence intensity from lin-R4F2-FITC treated cells (Fig. 3a) is significantly weaker than that from cyc-R4F2-FITC treated cells (Fig. 3b). This suggests that cyc-R4F2-FITC is able to enter cells more efficiently than lin-R4F2-FITC. In addition, the spatial distribution of the fluorescence signal inside the MCF7 cells is also different between the two peptides, as lin-R4F2-FITC is mostly located near the membrane periphery, while cyc-R4F2-FITC is distributed throughout the intracellular environment. The three-dimensional reconstructions of the LSCM images in Fig. 3b confirm that the cyclic peptides have translocated to the cell interior, rather than remaining on the cell surface.
Fig. 3.
Internalization of peptides into MCF7 cells in vitro. MCF cells are incubated with lin-R4F2-FITC (a, c) or cyc-R4F2-FITC (b, d) at 10 µM concentration for 5 hrs without serum. a), b): three-dimensional reconstructions of the LSCM images of cells after incubation with lin-R4F2-FITC (green) and cyc-R4F2-FITC (green), respectively, showing greater internalization for the latter case; c), d): the corresponding differential interference contrast images of a), b) (Scale bar = 40 µm).
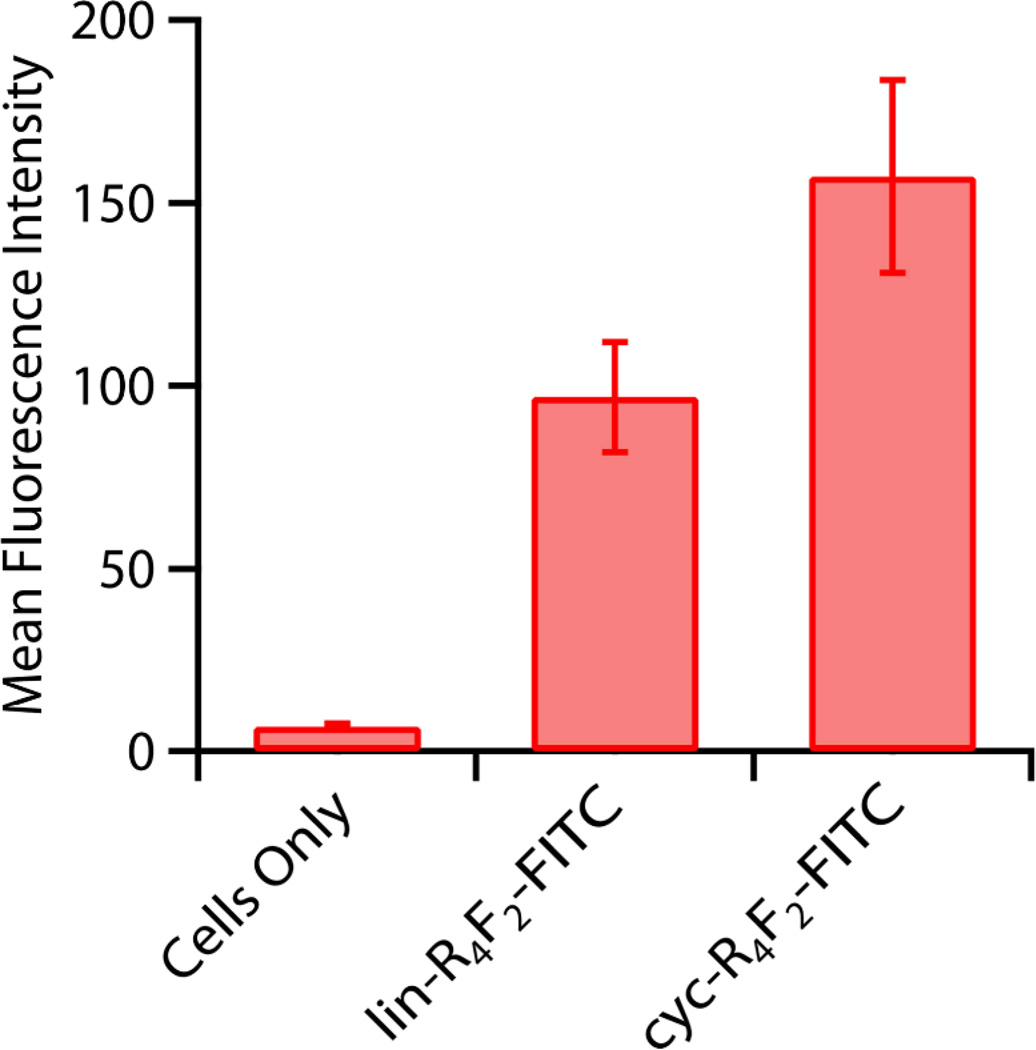
In order to obtain a quantitative and statistical measure of comparative peptide translocation efficiency, Fluorescence-Activated Cell Sorting (FACS) was performed with the MCF7 cells incubated with lin-R4F2-FITC and cyc-R4F2-FITC. Fig. 4 shows the mean fluorescence intensity of cell populations internalized with the different peptides. Here, the mean fluorescence intensity represents the quantitative extent of peptide uptake by each cell population. The result demonstrates a strong increase in the mean fluorescence intensity for cyc-R4F2-FITC treated cells compared with lin-R4F2-FITC treated cells, and confirms the trends observed from the LSCM images and peptide-lipid phase behavior from SAXS measurements. MTS cell proliferation assays28 confirm that these peptides are essentially non-cytotoxic under the experimental conditions (Figure S1, Supplementary materials).
Fig. 4.
FACS analysis showing quantitative uptake of peptides into MCF7 cells, showing the progressive increase from incorporation of peptide hydrophobicity and cyclicity. Cells are incubated with 5 µM peptides for 5 hrs without serum. Error bars represent standard errors of the mean.
Negative Gaussian membrane curvature is a necessary ingredient for membrane permeation. It has been recently shown that generation of this type of membrane curvature necessitates specific amino acid compositions in pore forming peptides.11 Arginine can create negative membrane curvature via induced electrostatic wrapping of the cationic peptide by anionic cell membranes. However, it can also create some positive curvature in the perpendicular direction via steric effects from multidentate hydrogen bonding to bulking lipid headgroups along the chain, thus resulting in negative Gaussian curvature.16 We have shown that cyc-R4 has an enhanced activity for generating curvature relative to lin-R4. This is consistent with the higher density presentation of guanidinium groups and to the reduced conformational freedom in the cyclic arginine peptide. Both of these effects enhance the crowding of peptide-associated lipid headgroups. We have also shown that by adding two hydrophobic phenylalanines to the cyclic peptide, the resultant cyc-R4F2 can generate more negative Gaussian curvature, which empirically translates into a higher cell penetrating efficiency from direct measurement. We hypothesize that the additional hydrophobic residues reinforce the weak positive curvature generation from short tracts of arginine, since short polyR do not induce the same degree of excluded-volume interactions in associated lipid heads as long polyR.16 This hypothesis is consistent with the well known observation that insertion of hydrophobic domains into membranes can result in positive membrane curvature.11, 16, 29–32 Taken together, these general principles can potentially guide the design of transporter sequences for circular peptides.
Supplementary Material
Acknowledgements
We thank Dr. Tao Liu and Prof. Dehua Pei at the Ohio State University for providing us the peptides used in this study. We also thank Nathan W. Schmidt, Abhijit Mishra and Ghee Hwee Lai for useful discussions and suggestions. X-ray work was performed at the Stanford Synchrotron Radiation Lab (SSRL) and the Advanced Photon Source (APS). SSRL is supported by DOE and NIH. Use of the APS is supported by DOE DE-AC02-06CH11357. This work is supported by NIH grant 1UO1AI082192-01, NSF grant DMR1106106, and the Department of Defense Prostate Cancer Research Program under award number W81XWH-09-1-0584.
Footnotes
Electronic Supplementary Information (ESI) available. See DOI: 10.1039/b000000x/
The ratio of peptide to lipid in the complex will be determined by thermodynamics, and thereby be affected by the binding energy of the peptide to the membrane. We estimate the free energy change upon binding by estimating the entropy gain of counterions.33, 34 For R9, we estimate ~< 13.5kT per peptide, compared to the measured value of 11.4kT.18 For R4, we estimate ~6kT per peptide.
Notes and references
- 1.Trabi M, Craik DJ. Trends Biochem. Sci. 2002;27:132–138. doi: 10.1016/s0968-0004(02)02057-1. [DOI] [PubMed] [Google Scholar]
- 2.Craik DJ. Science. 2006;311:1563–1564. doi: 10.1126/science.1125248. [DOI] [PubMed] [Google Scholar]
- 3.Saether O, Craik DJ, Campbell ID, Sletten K, Juul J, Norman DG. Biochemistry. 1995;34:4147–4158. doi: 10.1021/bi00013a002. [DOI] [PubMed] [Google Scholar]
- 4.Gudmundsson OS, Pauletti GM, Wang W, Shan DX, Zhang HJ, Wang BH, Borchardt RT. Pharm. Res. 1999;16:7–15. doi: 10.1023/a:1018828207920. [DOI] [PubMed] [Google Scholar]
- 5.Pomilio AB, Battista ME, Vitale AA. Curr. Org. Chem. 2006;10:2075–2121. [Google Scholar]
- 6.Mandal D, Shirazi AN, Parang K. Angew Chem Int Edit. 2011;50:9633–9637. doi: 10.1002/anie.201102572. [DOI] [PubMed] [Google Scholar]
- 7.Liu T, Liu Y, Kao HY, Pei D. J. Med. Chem. 2010;53:2494–2501. doi: 10.1021/jm901778v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lattig-Tunnemann G, Prinz M, Hoffmann D, Behlke J, Palm-Apergi C, Morano I, Herce HD, Cardoso MC. Nat Commun. 2011;2:453. doi: 10.1038/ncomms1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu T, Qian Z, Liu Y-y, Jhiang SM, Pei D. P Natl Acad Sci USA. 2012 submitted. [Google Scholar]
- 10.Futaki S, Suzuki T, Ohashi W, Yagami T, Tanaka S, Ueda K, Sugiura Y. J Biol Chem. 2001;276:5836–5840. doi: 10.1074/jbc.M007540200. [DOI] [PubMed] [Google Scholar]
- 11.Schmidt N, Mishra A, Lai GH, Wong GCL. FEBS Lett. 2010;584:1806–1813. doi: 10.1016/j.febslet.2009.11.046. [DOI] [PubMed] [Google Scholar]
- 12.El-Sayed A, Futaki S, Harashima H. AAPS J. 2009;11:13–22. doi: 10.1208/s12248-008-9071-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Derossi D, Joliot AH, Chassaing G, Prochiantz A. J Biol Chem. 1994;269:10444–10450. [PubMed] [Google Scholar]
- 14.Torchilin VP, Rammohan R, Weissig V, Levchenko TS. P Natl Acad Sci USA. 2001;98:8786–8791. doi: 10.1073/pnas.151247498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mishra A, Gordon VD, Yang L, Coridan R, Wong GCL. Angew Chem Int Edit. 2008;47:2986–2989. doi: 10.1002/anie.200704444. [DOI] [PubMed] [Google Scholar]
- 16.Mishra A, Lai GH, Schmidt NW, Sun VZ, Rodriguez AR, Tong R, Tang L, Cheng J, Deming TJ, Kamei DT, Wong GCL. P Natl Acad Sci USA. 2011;108:16883–16888. doi: 10.1073/pnas.1108795108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Su Y, Waring AJ, Ruchala P, Hong M. Biochemistry. 2010;49:6009–6020. doi: 10.1021/bi100642n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ziegler A. Adv Drug Deliver Rev. 2008;60:580–597. doi: 10.1016/j.addr.2007.10.005. [DOI] [PubMed] [Google Scholar]
- 19.White SH, Wimley WC. Annu. Rev. Biophys. Biomol. Struct. 1999;28:319–365. doi: 10.1146/annurev.biophys.28.1.319. [DOI] [PubMed] [Google Scholar]
- 20.Hessa T, Meindl-Beinker NM, Bernsel A, Kim H, Sato Y, Lerch-Bader M, Nilsson I, White SH, von Heijne G. Nature. 2007;450:1026–1030. doi: 10.1038/nature06387. [DOI] [PubMed] [Google Scholar]
- 21.Wimley WC, White SH. Nat. Struct. Biol. 1996;3:842–848. doi: 10.1038/nsb1096-842. [DOI] [PubMed] [Google Scholar]
- 22.Lipowsky R, Sackmann E. Structure and dynamics of membranes. Amsterdam; New York: Elsevier Science; 1995. [Google Scholar]
- 23.Schmidt NW, Mishra A, Lai GH, Davis M, Sanders LK, Tran D, Garcia A, Tai KP, McCray PB, Ouellette AJ, Selsted ME, Wong GCL. J Am Chem Soc. 2011;133:6720–6727. doi: 10.1021/ja200079a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mouritsena OG, Zuckermannb MJ. Lipids. 2004;39:1101–1113. doi: 10.1007/s11745-004-1336-x. [DOI] [PubMed] [Google Scholar]
- 25.Schwarz US, Gompper G. Phys Rev Lett. 2000;85:1472–1475. doi: 10.1103/PhysRevLett.85.1472. [DOI] [PubMed] [Google Scholar]
- 26.Mercer J, Helenius A. Science. 2008;320:531–535. doi: 10.1126/science.1155164. [DOI] [PubMed] [Google Scholar]
- 27.Kurczy ME, Piehowski PD, Van Bell CT, Heien ML, Winograd N, Ewing AG. Proc Natl Acad Sci U S A. 2010;107:2751–2756. doi: 10.1073/pnas.0908101107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Berg K, Zhai L, Chen M, Kharazmi A, Owen TC. Parasitol. Res. 1994;80:235–239. doi: 10.1007/BF00932680. [DOI] [PubMed] [Google Scholar]
- 29.Matsuzaki K, Sugishita K, Ishibe N, Ueha M, Nakata S, Miyajima K, Epand RM. Biochemistry. 1998;37:11856–11863. doi: 10.1021/bi980539y. [DOI] [PubMed] [Google Scholar]
- 30.Ford MG, Mills IG, Peter BJ, Vallis Y, Praefcke GJ, Evans PR, McMahon HT. Nature. 2002;419:361–366. doi: 10.1038/nature01020. [DOI] [PubMed] [Google Scholar]
- 31.McMahon HT, Gallop JL. Nature. 2005;438:590–596. doi: 10.1038/nature04396. [DOI] [PubMed] [Google Scholar]
- 32.Matsuzaki K. Biochimica et Biophysica Acta. 1999;1462:1–10. doi: 10.1016/s0005-2736(99)00197-2. [DOI] [PubMed] [Google Scholar]
- 33.Yang L, Liang H, Angelini TE, Butler J, Coridan R, Tang JX, Wong GCL. Nat Mater. 2004;3:615–619. doi: 10.1038/nmat1195. [DOI] [PubMed] [Google Scholar]
- 34.Wong GCL, Pollack L. Annu Rev Phys Chem. 2010;61:171–189. doi: 10.1146/annurev.physchem.58.032806.104436. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.