Abstract
Cryptococcus neoformans is a pathogen that is the most common cause of fungal meningitis. As with most fungal pathogens, the most prevalent clinical antifungal used to treat Cryptococcosis is orally administered fluconazole. Resistance to this antifungal is an increasing concern in treatment of fungal disease in general. Our knowledge of the specific determinants involved in fluconazole resistance in Cryptococcus is limited. Here we report the identification of an important genetic determinant of fluconazole resistance in Cryptococcus neoformans that encodes a basic region-leucine zipper transcription factor homologous to Saccharomyces cerevisiae Yap1. Expression of a codon-optimized form of the Cn YAP1 cDNA in S. cerevisiae complemented defects caused by loss of the endogenous S. cerevisiae YAP1 gene and activated transcription from a reporter gene construct. Mutant strains of C. neoformans lacking YAP1 were hypersensitive to a range of oxidative stress agents but importantly also to fluconazole. Loss of Yap1 homologues from other fungal pathogens like Candida albicans or Aspergillus fumigatus was previously found to cause oxidant hypersensitivity but had no detectable effect on fluconazole resistance. Our data provide evidence for a unique biological role of Yap1 in wild-type fluconazole resistance in C. neoformans.
Keywords: Cryptococcus neoformans, Yap1 transcription factor, heterologous expression, Saccharomyces cerevisiae, fluconazole resistance
1. Introduction
Fungal infections represent an increasing burden on the healthcare system (1). One example of these pathogens of emerging importance is the yeast Cryptococcus neoformans. C. neoformans is associated with 1,000,000 new cases resulting in more than 600,000 deaths yearly from meningitis caused by this organism (2). While antifungal chemotherapy is generally effective against C. neoformans, resistant isolates have been found (3). The primary and most effective drug treatment for cryptococcosis is the polyene compound amphotericin B (4). Although AmB is a potent antifungal, it is limited in its utility by a requisite intravenous route of delivery, expense and toxicity (5). In areas of the world with less developed healthcare delivery, AmB administration is not practical and the most commonly deployed antifungal is the orally deliverable azole drug fluconazole (6). FLC is also routinely used worldwide in maintenance therapy to avoid recurring bouts of cryptococcosis (4). For these reasons, FLC is the major antifungal drug used to treat cryptococcal disease.
A common issue with any antibiotic is the development of resistance. This problem is especially severe with regards to antifungal drugs as the number of different drug classes for these antibiotics is quite small (reviewed in (7)). Because of the importance of FLC as an antifungal, much work on drug resistance is focused on tolerance to this compound. Although a number of other azole compounds are approved for clinical use, all of them target the same sterol biosynthetic protein: lanosterol α-14 demethylase (8). This enzyme is encoded by the ERG11 gene in C. neoformans and mutant alleles of this gene are known to confer high level azole tolerance (9). Along with changes in the azole target enzyme, an ATP-binding cassette (ABC) transporter encoding gene called AFR1 has been reported to confer drug resistance by active efflux of these antibiotics (10). These two genes are the only known direct determinants conferring FLC resistance.
FLC resistance in C. neoformans may also be developed through an indirect mechanism in which changes in ploidy of an otherwise wild-type organism elicits drug tolerance. This phenomenon is referred to as heteroresistance and is typically associated with aneuploidies involving chromosome 1 (Chr1) (11, 12). Intriguingly, both ERG11 and AFR1 are encoded on Chr1. This suggests that the increased chromosome copy number associated with these aneuploidies may result in gene amplification and overexpression of these and other proteins, giving rise to FLC resistance (13). Heteroresistance caused by Chr1 amplification seems likely to involve participation of multiple genes for two reasons. First, even in heteroresistant strains, changes in ERG11 copy number are only two-fold (14); this alteration seems unlikely to explain the large increase in FLC tolerance. Second, loss of AFR1 from Chr1 did not eliminate the acquisition of heteroresistance (12), consistent with the belief that multiple genes are required to acquire the normal elevation in resistance seen in this genetic situation. It is important to note that detailed study of the fungal pathogen Candida albicans demonstrated a similar drug-induced, reversible aneuploidy that required the presence of both the C. albicans ERG11 gene but also a transcription factor (TAC1) controlling expression of a number of genes involved in azole resistance (15). Based on this precedent, it seems likely that heteroresistance in C. neoformans will require multiple genes on Chr1 to contribute to azole resistance.
We have recently characterized a gene encoding a transcription factor that is both carried on Chr1 and required for wild-type FLC resistance. This transcription factor is a homologue of the budding yeast Saccharomyces cerevisiae Yap1 protein (Sc Yap1) (16). Sc Yap1 was first described functionally as a high-copy-mediator of drug resistance (17) and later shown to be required for wild-type resistance to oxidative stress (18). Oxidants inhibit the nuclear export of Sc Yap1, causing this factor to accumulate in the nucleus where it can activate target gene expression (19, 20). Oxidant regulation is affected by controlling the oxidation status of cysteine residues present in the protein chain in two different clusters: the N-terminal cysteine rich domain (n-CRD) and the C-terminal cysteine rich domain (c-CRD). Mutants lacking the c-CRD are constitutively retained in the nucleus and exhibit elevated expression of some target genes (21). Surprisingly, oxidant resistance of these mutant strains is complex, with c-CRD mutants conferring hyperresistance to oxidants like diamide but hypersensitivity to H2O2 (22). Loss of the n-CRD causes hypersensitivity to H2O2 but retains diamide resistance. Data from several labs demonstrated that interdomain disulfide bonds must form in Sc Yap1 to confer normal H2O2 tolerance while simple nuclear retention caused by c-CRD mutants could explain diamide and drug hyperresistance (21, 23, 24).
The Cn Yap1 protein shares significant sequence similarity with Sc Yap1. Here we demonstrate that the cryptococcal protein can functionally replace Sc Yap1 in S. cerevisiae and that mutant C. neoformans strains lacking Cn YAP1 are hypersensitive to oxidants but also to FLC. Hyperfunctional or hypermorphic alleles of YAP1 have been seen to elicit elevated drug resistance (including FLC tolerance) but the requirement by C. neoformans for Yap1 function to maintain wild-type FLC resistance is unique. This work provides the first characterization of Cn Yap1 and evidence for its special importance in FLC resistance in this organism.
2. Materials and Methods
2.1 Strains and media
The S. cerevisiae strain used in the study was SM12 (MATα leu2-3,112::ARE-TRP5-lacZ::LEU2 ura3-52 his3-Δ200 trp1-Δ901 lys2-801 suc2-Δ9 Mel− yap1-Δ1::HIS3). This strain was used for the plasmid transformations, β-galactosidase assays, spot testing and fluorescent microscopy to characterize Cn Yap1 function in S. cerevisiae. SM12 was transformed by standard lithium acetate transformation (25). S. cerevisiae cells were grown at 30°C on rich YPD (2% yeast extract, 1% peptone, 2% glucose) medium or on synthetic complete medium lacking appropriate auxotrophic components (26). The C. neoformans wild type strain used was KN99α. Cn YAP1 in this strain background was deleted by biolistic transformation using split marker cassettes (27).
The split marker cassettes for the deletion were generated as follows: 1 kb upstream of Cn YAP1 was PCR amplified using the primer pairs (For: 5’-TTAAAAAGTTTTGTCGCTGTTGC; Rev: 5’-caatagaagctCGGGGGGGAGTGGAG) and 1 kb downstream of Cn YAP1 was amplified using the primer pairs (For: 5’-tcatctgtcccccaTATGCGGAAACGTTGGG; Rev: 5’-AGTGCAAGGCGTTTGTTGC) using KN99α genomic DNA as template. The G418R selection (28) cassette was PCR amplified from pSKB7 plasmid (provided by Stacey Klutts, University of Iowa) as split markers. The upstream split marker comprised of a 1.5kb fragment lacking the last 125 bp of the G418R gene as well as the Cn NMT terminator, and was amplified using the primer sets (For: 5’-ccactcccccccgAGCTTCTATTGTCCAGGCTG; Rev: 5’-TGTCATAGCACAGCGTTAGC). The downstream split marker comprised of a 1.7kb fragment lacking the first 80bp of the G418R gene as well as the actin promoter, and was amplified using the primer sets (For: 5’- GTGCACGGACCCTATTGTC; Rev: 5’-gtttccgcataTGGGGGACAGATGATATCC). The upstream split marker deletion construct was amplified by fusion PCR using primer pairs (For: 5’-TTAAAAAGTTTTGTCGCTGTTGC; Rev: 5’- TGTCATAGCACAGCGTTAGC) and the PCR products corresponding to 1 kb upstream of Cn YAP1 and the upstream G418R split marker were used as templates. The downstream split marker deletion construct was amplified by fusion PCR using primer pairs (For: 5’- GTGCACGGACCCTATTGTC; Rev: 5’- AGTGCAAGGCGTTTGTTGC) and the PCR products corresponding to 1 kb downstream of Cn YAP1 and the downstream G418R split marker were used as templates. The PCR amplified upstream and downstream split marker deletion constructs, which had a ~600 bp overlap, were gel purified and then used in biolistic transformation of C. neoformans. Transformants were selected on YPD supplemented with 100 µg/ml G418 disulphate and confirmed by PCR. Each yap1Δ isolate presented here derived from a separate transformation plate and thus represents an independently derived isolate.
2.2 Plasmids
DNA manipulations were done using standard procedures (29) or according to manufacturer instructions. The plasmid harboring Cn YAP1 for heterologous expression in S. cerevisiae was generated as follows. An artificial Cn YAP1 cDNA that was codon optimized for better expression in S. cerevisiae (Sc-adapted Cn YAP1) was synthesized and cloned in the plasmid vector pUC57 (Genscript, Inc.). This sequence (available on request) was designed to contain codons that were optimal for expression in S. cerevisiae but still encoded the correct 700 amino acids of the CNAG_00239 protein product. The Sc-adapted Cn YAP1 was PCR amplified using primers (For: 5’- CTCGCGAATGCATCTAGATCC; Rev: 5’- aagatcTcTCTAAAGGTGAAGAATTATTCACTGGTGTTGTCCCAATT) and cloned into pJPS10 gapped with EcoRI/ClaI by recombinational cloning in S. cerevisiae to generate pSP64. The plasmid pJPS10 contained a cDNA for the Aspergillus fumigatus ypkA gene under control of Sc CUP1 promoter and fused to a green fluorescent protein (GFP)-HA tag at its C-terminus (data not shown). The structure of pSP64 was verified by partial sequencing and multiple restriction digestions. Sc-adapted Cn YAP1 in pSP64 was under the control of the Sc CUP1 promoter and was C-terminally tagged with GFP-HA. We used pJPS10 as a control GFP-labeled protein for the fluorescent microscopy experiments. Two plasmids expressing Sc Yap1, pNT13 (pRS316 GFP-YAP1) (23) and pSEY18-R2.5 (a 2µ vector containing Sc YAP1) (30), were used as positive controls for localization and function of Sc Yap1.
2.3 Spot Assays
S. cerevisiae cultures grown overnight in SC-ura at 30°C were re-inoculated in fresh media at an A600 of 0.05 and grown to mid-log phase (A600 of around 0.5) at 30°C. 500 cells were spotted onto gradient plates and incubated at 30°C. For spot testing of C. neoformans strains, cells were grown overnight in YPD at 30°C, and were re-inoculated in fresh media at an A600 of 0.05 and grown to mid-log phase cells (A600 of around 0.8) at 30°C. 500 cells were spotted onto gradient plates at indicated oxidant/drug concentrations and incubated at 30°C. The concentration of reagent indicated on the gradient plate corresponds to the maximum reagent concentration on the plate.
2.4 β-galactosidase Assay
Yeast transformants grown overnight in SC-ura at 30°C were re-inoculated in fresh media at an A600 of 0.1 and grown to mid-log phase (A600 of around 0.8) at 30°C. An equal number of cells from each sample were harvested, washed, and assayed for β-galactosidase activity as described (31). The assay was carried out in triplicates and represented two independent trials.
2.5 Fluorescence Microscopy
SM12 strains transformed with pJPS10, pSP64 or pNT13 plasmids were grown overnight in SC-ura at 30°C, reinoculated into fresh media and grown to an A600 of 0.8–1 in SC-ura at 30°C. Cells were then incubated in 5µg/ml 4',6-diamidino-2-phenylindole (DAPI) for 30 minutes, either untreated or treated with 1mM H2O2 for 20 min, washed and resuspended in sterile water and visualized for GFP fluorescence, DAPI staining and Nomarski optics using an Olympus (Tokyo, Japan) BX-60 microscope with a 100× oil immersion objective. Images were captured using a Hamamatsu (Shizuoka, Japan) ORCA charge-coupled device camera.
2.6 Virulence Assay
C. neoformans strains were tested for virulence as described previously (32) using a mouse inhalation model. Isogenic wild-type (KN99α) and two yap1Δ::G418R derivatives were grown overnight in rich medium and washed in phosphate-buffered saline. Anesthetized female mice were inoculated with fungal strains and progress of the infection was followed daily by observation and weight monitoring. Survival data was evaluated using Kaplan-Meier estimation and P values were calculated using Prism software. All animal studies followed institutional guidelines.
3. Results
3.1 Cryptococcus neoformans encodes a Yap1 homologue
S. cerevisiae Yap1 is an important participant in stress-responsive gene transcription and required for normal oxidative stress tolerance (recently reviewed in (33)). Sc Yap1 often cooperates with a winged helix transcription factor called Skn7 to induce gene expression (34, 35). Previous studies in C. neoformans have identified a Skn7 homologue but the identity of a potential Yap1 partner protein was less clear (36). Studies on expression of the C. neoformans thioredoxin genes identified several transcription factors containing basic region-leucine zipper (bZIP) DNA-binding domains that were related to the cognate region from Sc Yap1 (37). We focused our attention on the protein that shares the most sequence similarity with Sc Yap1, encoded by the sequence designated CNAG_00239. We propose to designate this protein as the Cryptococcus neoformans Yap1 and will present the evidence in support of this assignment below.
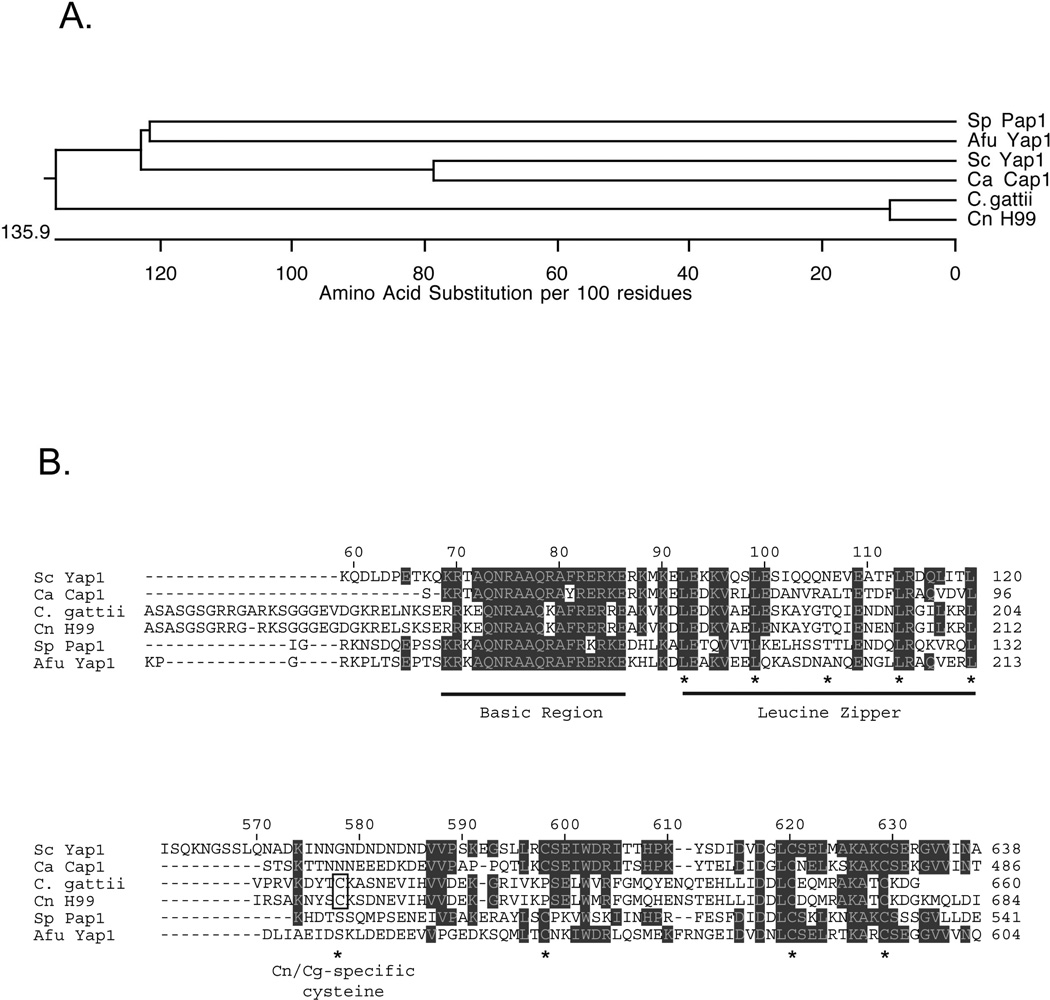
Sequence comparisons with other known fungal Yap1 homologues from Schizosaccharomyces pombe, Aspergillus fumigatus, Candida albicans, Candida glabrata and S. cerevisiae demonstrated that Cryptococcal homologues of this factor were more closely related to one another than to the other fungi (Figure 1A). Note that C. gattii also contains a similar Yap1 homologue. Comparison of the amino acid sequences between these factors identified two regions that show the most sequence conservation (Figure 1B). The highest degree of sequence conservation corresponded to the bZIP domains of these factors. This similarity suggests that all of these transcription factors recognize related DNA elements and we provide evidence in support of this suggestion for Cn Yap1 below.
Figure 1. Cryptococcal species encode a homologue of Sc Yap1.
A. An evolutionary tree diagram is shown illustrating the relatedness of different Yap1 homologues from both pathogens and nonpathogens. Sp: Schizosaccharomyces pombe; Afu: Aspergillus fumigatus; Sc: Saccharomyces cerevisiae; Ca: Candida albicans; C. gattii: Cryptococcus gattii; Cn: Cryptococcus neoformans. Note that the C. albicans Yap1 homologue is referred to as Cap1. B. Amino acid sequence alignments from the proteins described above. The numbers at the top refer to the amino acid position along the Sc Yap1 protein chain while the numbers at the right indicate position in each factor. Residues that are in shaded boxes are conserved in at least 3 different proteins. The top panel represents the basic region-leucine zipper domain of each factor with each section of this DNA-binding domain indicated and underlined. Asterisks denote the positions of the canonical leucine residues in the leucine zipper (see (64) for a review). The bottom panel represents the C-terminal cysteine-rich domain that is involved in control of nuclear localization (in all species where it has been examined). The asterisks indicate positions where cysteine residues are found. Note that the most amino-terminal cysteine is in a different position in the Cryptococcus species compared to the others. This unique cysteine residue is indicated by the open box and labeled Cn/Cg-specific cysteine.
The second conserved domain is contained at the extreme carboxy-terminus of all these proteins. This region contains several cysteine residues and is referred to in case of the Sc Yap1 as the carboxy-terminal cysteine-rich domain (c-CRD) (21). In the Yap1 homologues where it has been evaluated, the c-CRD is required for oxidant-induced nuclear localization (38, 39). Oxidation of the cysteines prevents nuclear export and leads to the factor being retained in the nucleus, triggering a rise in Yap1- dependent gene expression (21). The Cryptococcal proteins have 3 cysteine residues in their c-CRD as do the other fungal Yap1 proteins, but interestingly have shifted the position of first cysteine (cysteine 632 in the C. neoformans protein). This point will be considered further in the discussion. Importantly, the only cysteine residues present in the Cryptococcal proteins are located in the c-CRD. The other fungal Yap1 proteins possess a second cysteine rich domain (N-terminal cysteine-rich domain: n-CRD) that is lacking from both Cryptococcal proteins. Although there are some interesting differences between these fungal Yap1 proteins, the overall sequence conservation suggests that the CNAG_00239 protein might represent the Cn Yap1 homologue. We directly tested this idea by expressing this C. neoformans protein in a yap1Δ strain of S. cerevisiae
3.2 Cn Yap1 complements defects of a S. cerevisiae yap1Δ strain
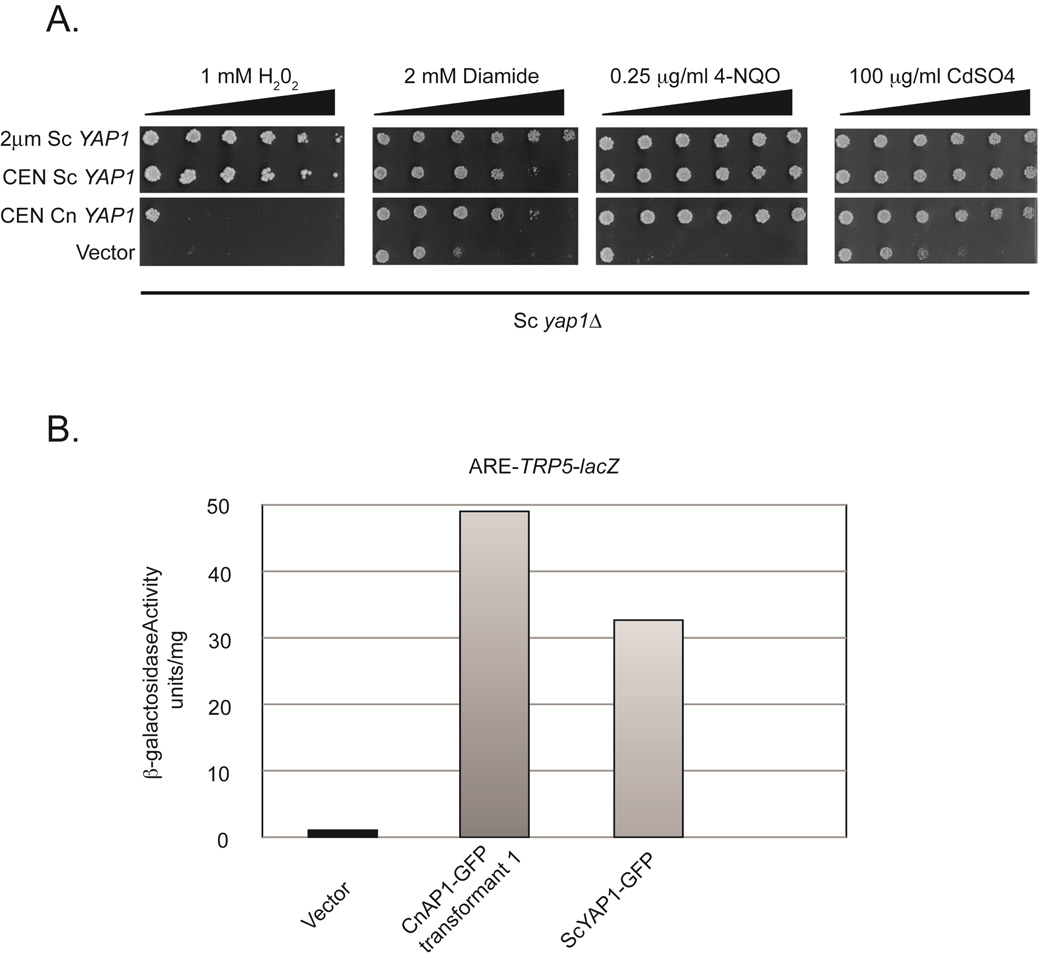
To determine if Cn Yap1 (CNAG_00239) was able to function as a Yap1 transcription factor, we expressed this protein in S. cerevisiae on a low-copy-number vector driven by the Sc CUP1 copper-inducible promoter. This plasmid was introduced into a S. cerevisiae strain constructed earlier (40) that had two key features. First, this strain lacked an endogenous copy of the Sc YAP1 gene. Second, an integrated copy of a Yap1-sensitive reporter gene was present in the genome. This reporter construct contains 3 copies of the SV40 AP-1 response element (ARE) from the early enhancer placed upstream of a translational fusion between the Sc TRP5 gene and E. coli lacZ (16). Yap1 binds to the AREs and activates expression from the TRP5-lacZ fusion because the normal TRP5 upstream activation sequences are deleted from this construct. This strain, designated SM12, was transformed with a high-copy-number plasmid expressing wild-type Sc Yap1, low-copy-number plasmids expressing either a green fluorescent protein (GFP) fusion with Sc Yap1 or the Cn Yap1-GFP fusion, or the empty vector plasmid alone. Transformants were grown to mid-log phase and then tested for oxidant resistance (Figure 2A) or TRP5-lacZ expression by β-galactosidase assay (Figure 2B).
Figure 2. Functional expression of Cn Yap1 in S. cerevisiae.
A. A S. cerevisiae strain designated SM12 (relevant genotype: yap1Δ leu2::ARE-TRP5-lacZ-LEU2) was transformed with a high-copy- (2µm Sc YAP1) or low-copy-number (CEN Sc YAP1) plasmid containing wild-type Sc YAP1, a low-copy-number plasmid carrying the CUP1-Cn YAP1-GFP fusion gene (CEN Cn YAP1) and an empty vector plasmid (Vector). Transformants were grown to mid-log phase and equal numbers of cells were then placed on media containing a gradient of the indicated oxidants (H2O2, diamide), drug (4-nitroquinoline-N-oxide: 4-NQO) or heavy metal (cadmium sulfate). Increasing concentration along the gradient is indicated by the bar of increasing width. Plates were incubated at 30°C for 48 hours and then photographed. B. Transformants from above were grown to mid-log phase and then β-galactosidase activity determined as described before (31).
Oxidant resistance was tested using a gradient plate assay in which different test compounds are present in a gradient embedded in selective media. Expression of CnYap1 in a yap1Δ strain restored growth on H2O2, diamide, 4-nitroquinoline-N-oxide and cadmium. At the concentrations tested, Cn Yap1 was able to restore growth to levels comparable to wild-type Sc Yap1 with the striking exception of H2O2. On this oxidant, the CnYap1-expressing cells grew better than cells carrying the empty vector but much less well than cells expressing the wild-type Sc Yap1. Since normal H2O2 tolerance in S. cerevisiae requires the formation of interdomain disulfide bonds between the n- and c-CRD (24), the lack of the n-CRD from Cn Yap1 may explain this selective defect in the ability to confer H2O2 resistance.
The levels of ARE-TRP5-dependent β-galactosidase activity present in the S. cerevisiae cells expressing cryptococcal Yap1 described above were also determined (Figure 2B). As anticipated from the observed complemention of the Sc yap1Δ phenotypes, expression of Cn Yap1 also induced expression of the ARE-TRP5-lacZ fusion gene to a level similar to that of native Sc Yap1.
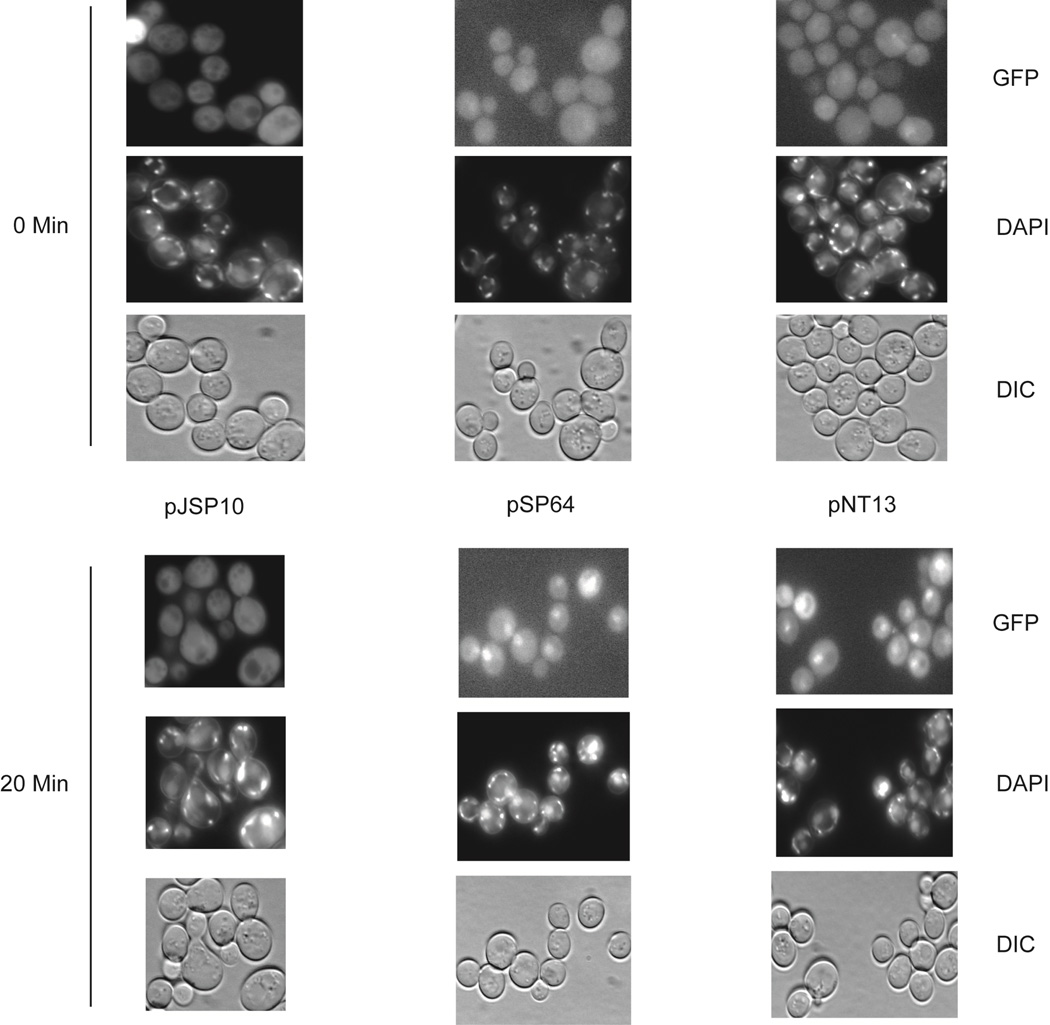
Since Cn Yap1 was expressed with a carboxy-terminal fusion to GFP, we wanted to determine if localization of this heterologous Yap1 was regulated in S. cerevisiae as is the resident Sc Yap1 protein. Transformants were grown to mid-log phase, treated with DAPI to visualize DNA and then either challenged with H2O2 or left untreated (Figure 3).
Figure 3. Regulated nuclear localization of Cn Yap1 in S. cerevisiae.
Plasmids expressing an irrelevant GFP fusion protein (pJPS10) or GFP fusion proteins forms of either Cn Yap1 (pSP64) or Sc Yap1 (pNT13) were introduced into S. cerevisiae cells containing a yap1Δ allele. Appropriate transformants were grown to mid-log phase and then incubated in the presence of DAPI for 30 minutes. Aliquots were withdrawn (0 time point) and then 1 mM H2O2 added with incubation continued for 20 minutes. Cells were visualized by fluorescence microscopy for GFP and DAPI or by Nomarski optics (DIC).
Even in the S. cerevisiae environment, Cn Yap1-GFP was enriched in the nucleus in the presence of H2O2. This closely resembles the behavior of the native Sc Yap1 and suggests the possibility that regulated nuclear export might be an important control mechanism for Cn Yap1 as it is in S. cerevisiae (19, 20). Together, these data argue that the Cn Yap1 protein functions in S. cerevisiae in a manner very much like the native protein.
3.3 Cn YAP1 is required for oxidant and antifungal drug resistance
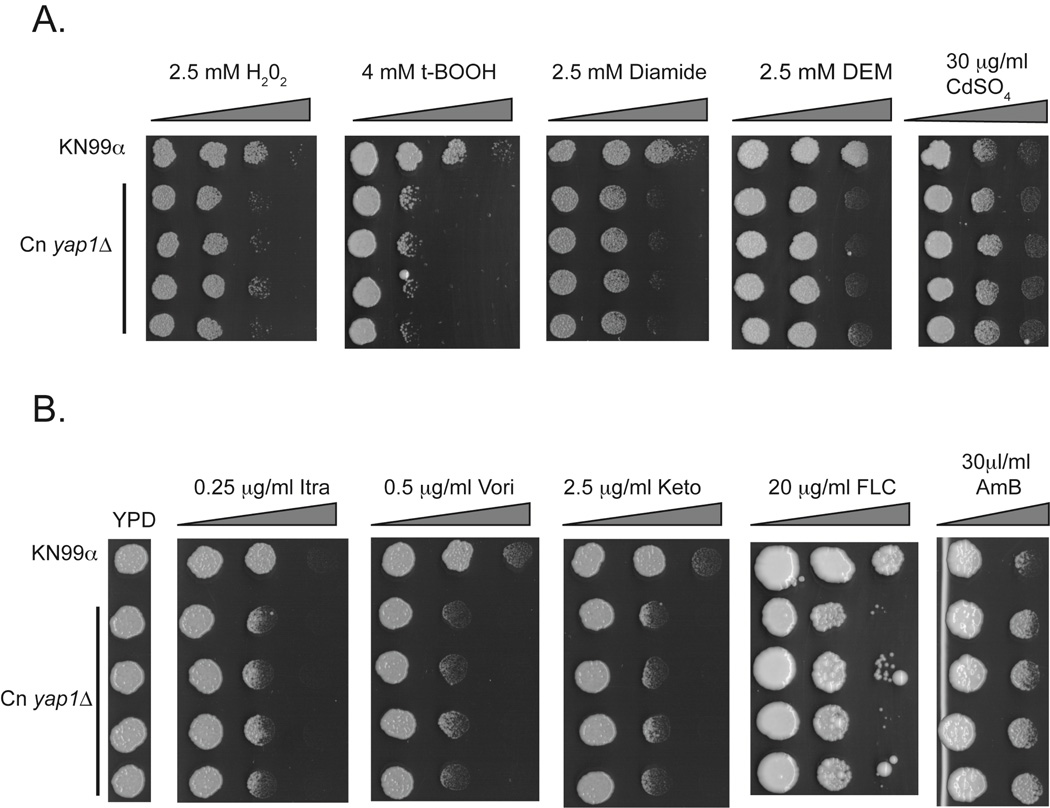
While expression of Cn Yap1 in the S. cerevisiae background provides a useful measure of the heterologous function of this transcription factor, the key issue is the role of this protein in its native environment. To examine the roles of Cn Yap1 in C. neoformans, we constructed disruption mutant strains lacking this gene. We used a split marker strategy to replace the sequence encoding Cn Yap1 with the G418R resistance gene (41). We verified gene replacement in 4 independent isolates and tested them for phenotypes of resistance to various agents that are influenced by Yap1 homologues in other fungi (Figure 4).
Figure 4. Phenotypes resulting from loss of YAP1 in C. neoformans.
A. Isogenic wild-type was compared to four independent yap1Δ::G418R strains as in Fig. 2A for growth in the presence of oxidants (A) or antifungal compounds (B). t-BOOH, t-butylhydroperoxide; DEM, diethylmaleate; AmB, amphotericin B; Itra, itraconazole; Vori, voriconazole; Keto, ketoconazole; FLC, fluconazole.
Loss of Yap1 from C. neoformans sensitized the resulting mutant strain to several different oxidative stress agents including H2O2, t-butylhydroperoxide, diamide and diethylmaleate (Figure 4A). These oxidant hypersensitivities were seen in other fungi lacking their endogenous Yap1 protein (42–44). Loss of Cn YAP1 did not detectably increase susceptibility to cadmium sulfate. Cadmium sensitivity was first seen in S. cerevisiae strains lacking YAP1 (30) and can be observed in other fungi with similar YAP1-deficient genotypes (42, 43).
We next compared the resistance of wild-type and isogenic yap1Δ::G418R strains to a variety of azole drugs and amphotericin B (AmB). For all azoles, loss of YAP1 resulted in an azole hypersensitive phenotype (Figure 4B). This phenotype is not commonly seen in other fungi lacking Yap1 and represents a unique contribution of this transcription factor to drug resistance in C. neoformans. Interestingly, loss of YAP1 increased AmB resistance.
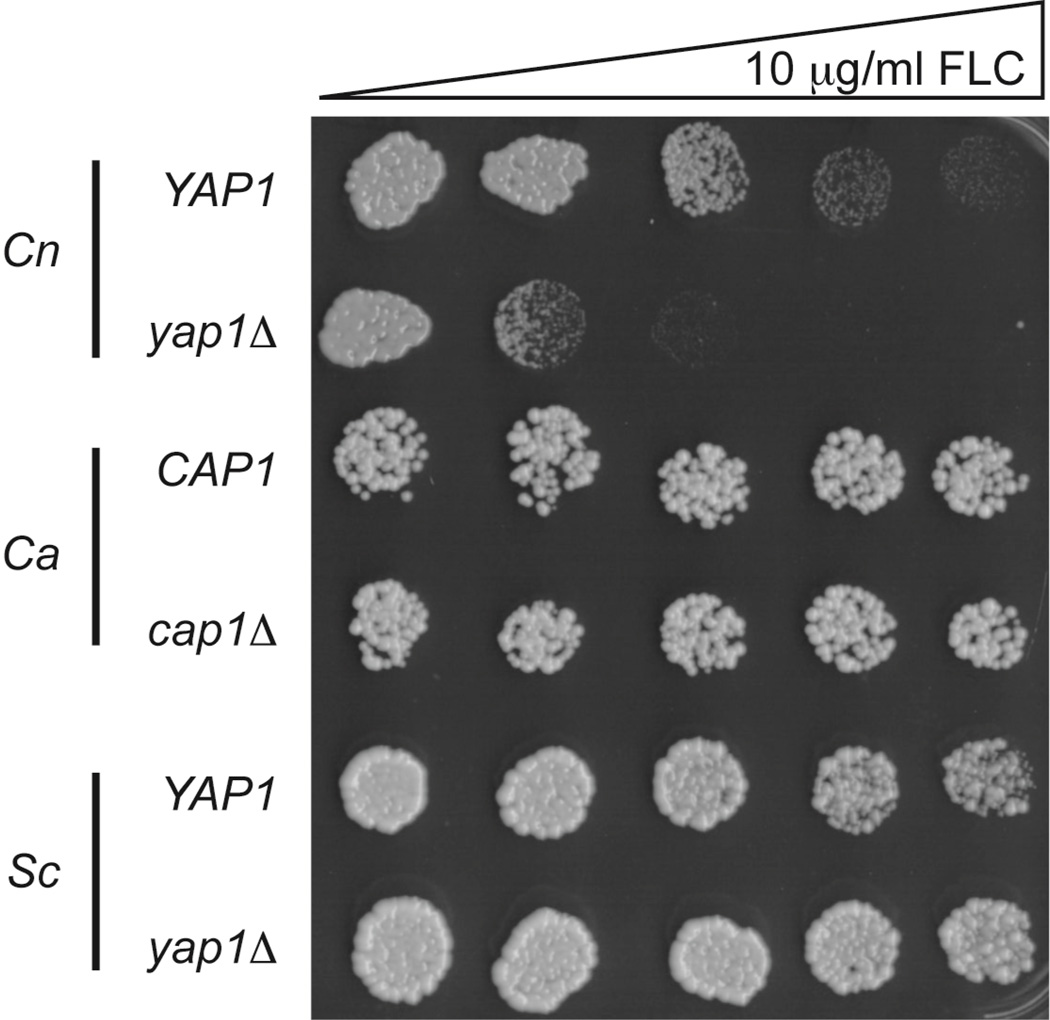
To directly compare the Yap1-dependent contribution to fluconazole resistance with that in other fungi, we used the gradient plate assay to test fluconazole resistance of isogenic wild-type and yap1D derivatives of C. neoformans, Candida albicans (YAP1 gene referred to as CAP1 in C. albicans (45)) and S. cerevisiae (Figure 5). Only in the case of yap1Δ C. neoformans was growth inhibited by FLC challenge. Removal of this gene from either C. albicans or S. cerevisiae had no detectable effect on the growth of these organisms in the presence of antifungal drug and was consistent with previous data (42) (43). These data support the view that Yap1 plays a role of special importance in normal FLC tolerance in C. neoformans.
Figure 5. A role for Yap1 in fluconazole is unique to C. neoformans.
Isogenic fungal strains containing or lacking their endogenous Yap1 genes were tested for fluconazole (FLC) resistance using a gradient plate assay. Cn: C. neoformans; Ca; Candida albicans; Sc; S. cerevisiae. The C. albicans strains used here are diploids that are either homozygous CAP1/CAP1 or cap1Δcap1Δ.
3.4 Yap1 does not play a major role in virulence of C. neoformans
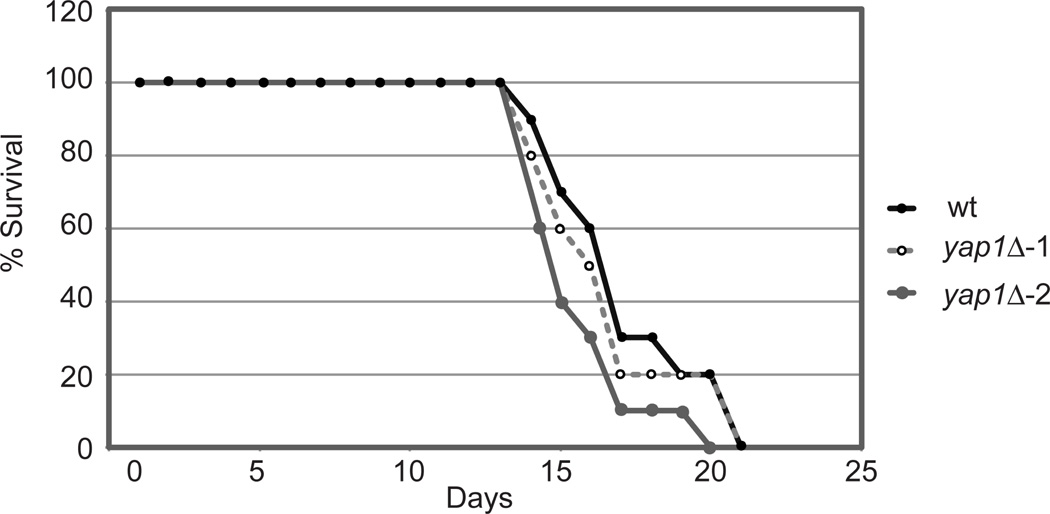
To determine if Yap1 was required for virulence of C. neoformans, we utilized a mouse inhalation model of infection (32) to compare infection isogenic wild-type C. neoformans (K99α:wt) and two independent yap1Δ derivatives. No significant difference in the virulence of the yap1Δ strains compared to wild-type could be seen as P values for the difference between wild-type and either independent yap1Δ strain were >0.01. We conclude that Yap1 is not required for virulence of C. neoformans in this experimental paradigm.
4. Discussion
Yap1 homologues are present in virtually every fungus that has been examined and are involved in oxidative stress tolerance in fungi ranging from plant to human pathogens (42–44, 46). The role of this transcription factor in controlling redox potential in fungal cells is one of its best-characterized activities (see (33, 47, 48) for reviews). Yap1 stimulates expression of antioxidant genes such as those encoding thioredoxins, glutathione peroxidases, and enzymes involved in glutathione biosynthesis (49). This type of a biological function seems almost certain to be conserved in C. neoformans as Cn yap1Δ strains are highly sensitive to a wide range of oxidants.
The unusual feature of Cn Yap1 is its critical contribution to FLC resistance at wild-type gene dosage. This phenotype is not a common attribute of other fungi lacking their respective Yap1 homologue. YAP1 hypermorphic alleles, caused by loss of the c-CRD or simply overproduction of the wild-type factor, caused increased elevated azole resistance in the resulting fungal mutant (38, 45). Organisms with a Yap1 protein that does not influence FLC resistance at its normal gene dosage/function include C. albicans (42), C. glabrata (43) and A. fumigatus (38). C. neoformans is an exception to this behavior as loss of endogenous Yap1 produces a strain that is highly sensitive to FLC compared to wild type.
It is also striking that loss of YAP1 causes C. neoformans to acquire resistance to AmB. Since AmB kills cells by binding to ergosterol in the cell membrane, it is possible that the yap1Δ cells have reduced sterol exposed to the outside of the cell. This might be triggered by reduction in sterol transport or synthesis which would also be consistent with the azole hypersensitive phenotype of these yap1Δ mutant strains. Previous exposure of C. neoformans to azole drugs prior to challenge with AmB has been found to be antagonistic in some cases (reviewed in (50)). Further analysis of this phenotype is underway.
While we do not yet understand the basis for the singular importance of Cn Yap1 in FLC resistance, a possible explanation is provided by the known cadre of target genes for Yap1-like proteins from other fungi. In S. cerevisiae, C. albicans and C. glabrata, major facilitator superfamily (MFS) transporter proteins are regulated by the respective Yap1 proteins present in these fungi (43, 45, 51, 52). MFS proteins are important determinants of drug antiporter activity and serve to reduce the intracellular drug concentration directly (reviewed in (53)). In bacteria, MFS proteins serve a more significant role in drug resistance than ABC transporters (54), a relationship apparently reversed in eukaryotic fungi (55). The importance of MFS proteins in drug resistance can be illustrated by early genetic experiments on Sc Yap1. S. cerevisiae cells lacking the PDR5 gene, a key ABC transporter involved in multidrug resistance (reviewed in (56)), are extremely sensitive to drugs. However, overproduction of Sc Yap1 was able to normally elevate drug resistance, likely through activation of expression of a MFS protein known as FLR1 (57). As mentioned above, other fungi do not show a requirement for their Yap1 homologue in terms of basal azole resistance but mutationally-activated forms of Yap1 homologues can elevate azole resistance via activation of MFS-encoding genes (45). It may be that C. neoformans places a greater importance on the role of MFS proteins in FLC tolerance. Confirmation of this suggestion requires better understanding of the comparative importance of AFR1 with the targets of Cn Yap1.
Along with its unique role in FLC resistance, the structure of Cn Yap1 is unusual among fungal Yap1 homologues. As mentioned above, Yap1 proteins typically contain two different cysteine-rich domains (reviewed in (47, 58)). Cn (and C. gattii) Yap1 lacks the amino-terminal cysteine-rich domain that is present in the other fungal Yap1 proteins shown in Figure 1. Interdomain disulfide bonds between the c-CRD and n- CRD are required for resistance to oxidative stress triggered by exposure to H2O2 (23, 24). C. neoformans also lacks a homologue of a trans-acting factor (Ybp1) that is required for oxidative folding in the presence of H2O2 (59, 60). In S. cerevisiae, this folded structure is required to recruit a transcriptional Mediator component required for H2O2 but not diamide resistance (61). The lack of the n-CRD region in the Cryptococcal proteins precludes any formation of this dually disulfide-bonded structure and argues that Cn Yap1 utilizes a different mechanism for H2O2 tolerance.
The finding that loss of Yap1 from C. neoformans had no significant effect on virulence may be explained by the selective role of this factor in FLC resistance. Growth of yap1Δ derivatives of C. neoformans was normal unless stress agents were present. Loss of Cap1 (C. albicans Yap1 homologue) did not reduce the virulence of the resulting C. albicans mutant strains in a mouse model (62, 63). It will be important to determine if C. neoformans yap1Δ mutant strains are more sensitive to azole challenge in the animal model as they are seen to be in vitro (Figure 4 and 5). The provocative location of YAP1 on Chr1 also suggests that this gene might be involved in heteroresistance, a hypothesis that can now be tested with the availability of the yap1Δ strain.
Figure 6. Yap1 is not required for virulence in a mouse inhalation model.
Mice were inoculated intranasally with 2×105 of KN99α (wild-type:wt) or two different isogenic yap1Δ isolates. Mice were weighed daily and sacrificed if their weight was found to drop to <80% of peak weight.
Highlights.
-
-
Cryptococcus neoformans expresses a functional homologue of S. cerevisiae Yap1
-
-
C. neoformans lacking YAP1 are hypersensitive to fluconazole and oxidative stress
-
-
C. neoformans Yap1 does not play a major role in fungal virulence.
Acknowledgements
This work was supported in part by NIH GM49825. We thank Jennifer Lodge for useful discussions and Matt Williams for performing animal studies.
Abbreviations
- ABC
ATP-binding cassette
- AmB
Amphotericin B
- ARE
AP-1 response element
- bZIP
basic region-leucine zipper DNA-binding domain
- Cn
Cryptococcus neoformans
- CRD
cysteine-rich domain
- FLC
fluconazole
- Sc
Saccharomyces cerevisiae
- SC-ura
synthetic complete medium lacking uracil
- YPD
yeast-peptone-dextrose growth medium
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Srinivasan A, Lopez-Ribot JL, Ramasubramanian AK. Overcoming antifungal resistance. Drug discovery today. Technologies. 2014;11:65–71. doi: 10.1016/j.ddtec.2014.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Park BJ, Wannemuehler KA, Marston BJ, Govender N, Pappas PG, Chiller TM. Estimation of the current global burden of cryptococcal meningitis among persons living with HIV/AIDS. Aids. 2009;23:525–530. doi: 10.1097/QAD.0b013e328322ffac. [DOI] [PubMed] [Google Scholar]
- 3.Sionov E, Chang YC, Garraffo HM, Dolan MA, Ghannoum MA, Kwon-Chung KJ. Identification of a Cryptococcus neoformans cytochrome P450 lanosterol 14alpha-demethylase (Erg11) residue critical for differential susceptibility between fluconazole/voriconazole and itraconazole/posaconazole. Antimicrob Agents Chemother. 2012;56:1162–1169. doi: 10.1128/AAC.05502-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gullo FP, Rossi SA, Sardi Jde C, Teodoro VL, Mendes-Giannini MJ, Fusco-Almeida AM. Cryptococcosis: epidemiology, fungal resistance, and new alternatives for treatment. European journal of clinical microbiology & infectious diseases : official publication of the European Society of Clinical Microbiology. 2013;32:1377–1391. doi: 10.1007/s10096-013-1915-8. [DOI] [PubMed] [Google Scholar]
- 5.Hamill RJ. Amphotericin B formulations: a comparative review of efficacy and toxicity. Drugs. 2013;73:919–934. doi: 10.1007/s40265-013-0069-4. [DOI] [PubMed] [Google Scholar]
- 6.Loyse A, Dromer F, Day J, Lortholary O, Harrison TS. Flucytosine and cryptococcosis: time to urgently address the worldwide accessibility of a 50-yearold antifungal. J Antimicrob Chemother. 2013;68:2435–2444. doi: 10.1093/jac/dkt221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ghannoum MA, Rice LB. Antifungal agents: mode of action, mechanisms of resistance, and correlation of these mechanisms with bacterial resistance. Clin Microbiol Rev. 1999;12:501–517. doi: 10.1128/cmr.12.4.501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Van den Bossche H, Willemsens G, Cools W, Marichal P, Lauwers W. Hypothesis on the molecular basis of the antifungal activity of N-substituted imidazoles and triazoles. Biochem Soc Trans. 1983;11:665–667. doi: 10.1042/bst0110665. [DOI] [PubMed] [Google Scholar]
- 9.Rodero L, Mellado E, Rodriguez AC, Salve A, Guelfand L, Cahn P, Cuenca-Estrella M, Davel G, Rodriguez-Tudela JL. G484S amino acid substitution in lanosterol 14-alpha demethylase (ERG11) is related to fluconazole resistance in a recurrent Cryptococcus neoformans clinical isolate. Antimicrob Agents Chemother. 2003;47:3653–3656. doi: 10.1128/AAC.47.11.3653-3656.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sanguinetti M, Posteraro B, La Sorda M, Torelli R, Fiori B, Santangelo R, Delogu G, Fadda G. Role of AFR1, an ABC transporter-encoding gene, in the in vivo response to fluconazole and virulence of Cryptococcus neoformans. Infect Immun. 2006;74:1352–1359. doi: 10.1128/IAI.74.2.1352-1359.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mondon P, Petter R, Amalfitano G, Luzzati R, Concia E, Polacheck I, Kwon-Chung KJ. Heteroresistance to fluconazole and voriconazole in Cryptococcus neoformans. Antimicrob Agents Chemother. 1999;43:1856–1861. doi: 10.1128/aac.43.8.1856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sionov E, Chang YC, Garraffo HM, Kwon-Chung KJ. Heteroresistance to fluconazole in Cryptococcus neoformans is intrinsic and associated with virulence. Antimicrob Agents Chemother. 2009;53:2804–2815. doi: 10.1128/AAC.00295-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sionov E, Lee H, Chang YC, Kwon-Chung KJ. Cryptococcus neoformans overcomes stress of azole drugs by formation of disomy in specific multiple chromosomes. PLoS Pathog. 2010;6:e1000848. doi: 10.1371/journal.ppat.1000848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sionov E, Chang YC, Kwon-Chung KJ. Azole heteroresistance in Cryptococcus neoformans: emergence of resistant clones with chromosomal disomy in the mouse brain during fluconazole treatment. Antimicrob Agents Chemother. 2013;57:5127–5130. doi: 10.1128/AAC.00694-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Selmecki A, Gerami-Nejad M, Paulson C, Forche A, Berman J. An isochromosome confers drug resistance in vivo by amplification of two genes, ERG11 and TAC1. Mol Microbiol. 2008;68:624–641. doi: 10.1111/j.1365-2958.2008.06176.x. [DOI] [PubMed] [Google Scholar]
- 16.Moye-Rowley WS, Harshman KD, Parker CS. Yeast YAP1 encodes a novel form of the jun family of transcriptional activator proteins. Genes Dev. 1989;3:283–292. doi: 10.1101/gad.3.3.283. [DOI] [PubMed] [Google Scholar]
- 17.Leppert G, McDevitt R, Falco SC, Van Dyk TK, Ficke MB, Golin J. Cloning by gene amplification of two loci conferring multiple drug resistance in Saccharomyces. Genetics. 1990;125:13–20. doi: 10.1093/genetics/125.1.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kuge S, Jones N. YAP1-dependent activation of TRX2 is essential for the response of S. cerevisiae to oxidative stress by hydroperoxides. EMBO J. 1994;13:655–664. doi: 10.1002/j.1460-2075.1994.tb06304.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yan C, Lee LH, Davis LI. Crm1p mediates regulated nuclear export of a yeast AP-1-like transcription factor. EMBO J. 1998;17:7416–7429. doi: 10.1093/emboj/17.24.7416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kuge S, Toda T, Iizuka N, Nomoto A. Crm1 (Xpo1) dependent nuclear export of the budding yeast transcription factor yAP-1 is sensitive to oxidative stress. Genes to Cells. 1998;3:521–532. doi: 10.1046/j.1365-2443.1998.00209.x. [DOI] [PubMed] [Google Scholar]
- 21.Kuge S, Jones N, Nomoto A. Regulation of yAP-1 nuclear localization in response to oxidative stress. EMBO J. 1997;16:1710–1720. doi: 10.1093/emboj/16.7.1710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wemmie JA, Steggerda SM, Moye-Rowley WS. The Saccharomyces cerevisiae AP-1 protein discriminates between oxidative stress elicited by the oxidants H2O2 and diamide. J. Biol. Chem. 1997;272:7908–7914. doi: 10.1074/jbc.272.12.7908. [DOI] [PubMed] [Google Scholar]
- 23.Coleman ST, Epping EA, Steggerda SM, Moye-Rowley WS. Yap1p activates gene transcription in an oxidant-specific fashion. Mol. Cell. Biol. 1999;19:8302–8313. doi: 10.1128/mcb.19.12.8302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Delaunay A, Isnard AD, Toledano MB. H2O2 sensing through oxidation of the Yap1 transcription factor. EMBO J. 2000;19:5157–5166. doi: 10.1093/emboj/19.19.5157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ito H, Fukuda Y, Murata K, Kimura A. Transformation of intact yeast cells treated with alkali cations. J. Bacteriol. 1983;153:163–168. doi: 10.1128/jb.153.1.163-168.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sherman F, Fink G, Hicks J. Methods in Yeast Genetics. Cold Spring Harbor, New York: Cold Spring Harbor Laboratory; 1979. [Google Scholar]
- 27.Fu J, Hettler E, Wickes BL. Split marker transformation increases homologous integration frequency in Cryptococcus neoformans. Fungal Genet Biol. 2006;43:200–212. doi: 10.1016/j.fgb.2005.09.007. [DOI] [PubMed] [Google Scholar]
- 28.Hua J, Meyer JD, Lodge JK. Development of positive selectable markers for the fungal pathogen Cryptococcus neoformans. Clinical and diagnostic laboratory immunology. 2000;7:125–128. doi: 10.1128/cdli.7.1.125-128.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sambrook J, Fritsch FE, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 30.Wu A, Wemmie JA, Edgington NP, Goebl M, Guevara JL, Moye-Rowley WS. Yeast bZIP proteins mediate pleiotropic drug and metal resistance. J. Biol. Chem. 1993;268:18850–18858. [PubMed] [Google Scholar]
- 31.Guarente L. Yeast promoter and lacZ fusions designed to study expression of cloned genes in yeast. Methods Enzymol. 1983;101:181–191. doi: 10.1016/0076-6879(83)01013-7. [DOI] [PubMed] [Google Scholar]
- 32.Wang ZA, Griffith CL, Skowyra ML, Salinas N, Williams M, Maier EJ, Gish SR, Liu H, Brent MR, Doering TL. Cryptococcus neoformans dual GDP-mannose transporters and their role in biology and virulence. Eukaryot Cell. 2014;13:832–842. doi: 10.1128/EC.00054-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Morano KA, Grant CM, Moye-Rowley WS. The response to heat shock and oxidative stress in Saccharomyces cerevisiae. Genetics. 2012;190:1157–1195. doi: 10.1534/genetics.111.128033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lee J, Godon C, Lagniel G, Spector D, Garin J, Labarre J, Toledano MB. Yap1 and Skn7 control two specialized oxidative stress response regulons in yeast. J. Biol. Chem. 1999;181:16040–16046. doi: 10.1074/jbc.274.23.16040. [DOI] [PubMed] [Google Scholar]
- 35.Morgan BA, Banks GR, Toone WM, Raitt D, Kuge S, Johnston LH. The Skn7 response regulator controls gene expression in the oxidative stress response of the budding yeast Saccharomyces cerevisiae. EMBO J. 1997;16:1035–1044. doi: 10.1093/emboj/16.5.1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wormley FL, Jr, Heinrich G, Miller JL, Perfect JR, Cox GM. Identification and characterization of an SKN7 homologue in Cryptococcus neoformans. Infect Immun. 2005;73:5022–5030. doi: 10.1128/IAI.73.8.5022-5030.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Missall TA, Lodge JK. Function of the thioredoxin proteins in Cryptococcus neoformans during stress or virulence and regulation by putative transcriptional modulators. Mol Microbiol. 2005;57:847–858. doi: 10.1111/j.1365-2958.2005.04735.x. [DOI] [PubMed] [Google Scholar]
- 38.Qiao J, Liu W, Li R. Truncated Afyap1 attenuates antifungal susceptibility of Aspergillus fumigatus to voriconazole and confers adaptation of the fungus to oxidative stress. Mycopathologia. 2010;170:155–160. doi: 10.1007/s11046-010-9309-2. [DOI] [PubMed] [Google Scholar]
- 39.Zhang X, De Micheli M, Coleman ST, Sanglard D, Moye-Rowley WS. Analysis of the oxidative stress regulation of the Candida albicans transcription factor, Cap1p. Mol. Microbiol. 2000;36:618–629. doi: 10.1046/j.1365-2958.2000.01877.x. [DOI] [PubMed] [Google Scholar]
- 40.Wemmie JA, Wu A-L, Harshman KD, Parker CS, Moye-Rowley WS. Transcriptional activation mediated by the yeast AP-1 protein is required for normal cadmium tolerance. J. Biol. Chem. 1994;269:14690–14697. [PubMed] [Google Scholar]
- 41.Kim MS, Kim SY, Jung KW, Bahn YS. Targeted gene disruption in Cryptococcus neoformans using double-joint PCR with split dominant selectable markers. Methods Mol Biol. 2012;845:67–84. doi: 10.1007/978-1-61779-539-8_5. [DOI] [PubMed] [Google Scholar]
- 42.Alarco AM, Raymond M. The bZip transcription factor Cap1p is involved in multidrug resistance and oxidative stress response in Candida albicans. J. Bacteriol. 1999;181:700–708. doi: 10.1128/jb.181.3.700-708.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chen KH, Miyazaki T, Tsai HF, Bennett JE. The bZip transcription factor Cgap1p is involved in multidrug resistance and required for activation of multidrug transporter gene CgFLR1 in Candida glabrata. Gene. 2007;386:63–72. doi: 10.1016/j.gene.2006.08.010. [DOI] [PubMed] [Google Scholar]
- 44.Lessing F, Kniemeyer O, Wozniok I, Loeffler J, Kurzai O, Haertl A, Brakhage AA. The Aspergillus fumigatus transcriptional regulator AfYap1 represents the major regulator for defense against reactive oxygen intermediates but is dispensable for pathogenicity in an intranasal mouse infection model. Eukaryot Cell. 2007;6:2290–2302. doi: 10.1128/EC.00267-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Alarco AM, Balan I, Talibi D, Mainville N, Raymond M. AP-1-mediated multidrug resistance in Saccharomyces cerevisiae requires FLR1 encoding a transporter of the major facilitator superfamily. J. Biol. Chem. 1997;272:19304–19313. doi: 10.1074/jbc.272.31.19304. [DOI] [PubMed] [Google Scholar]
- 46.Molina L, Kahmann R. An Ustilago maydis gene involved in H2O2 detoxification is required for virulence. The Plant cell. 2007;19:2293–2309. doi: 10.1105/tpc.107.052332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Herrero E, Ros J, Belli G, Cabiscol E. Redox control and oxidative stress in yeast cells. Biochim Biophys Acta. 2008;1780:1217–1235. doi: 10.1016/j.bbagen.2007.12.004. [DOI] [PubMed] [Google Scholar]
- 48.Rodrigues-Pousada C, Menezes RA, Pimentel C. The Yap family and its role in stress response. Yeast. 2010;27:245–258. doi: 10.1002/yea.1752. [DOI] [PubMed] [Google Scholar]
- 49.Gasch AP, Spellman PT, Kao CM, Carmel-Harel O, Eisen MB, Storz G, Botstein D, Brown PO. Genomic expression programs in the response of yeast cells to environmental changes. Mol. Biol. Cell. 2000;11:4241–4257. doi: 10.1091/mbc.11.12.4241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mukherjee PK, Sheehan DJ, Hitchcock CA, Ghannoum MA. Combination treatment of invasive fungal infections. Clin Microbiol Rev. 2005;18:163–194. doi: 10.1128/CMR.18.1.163-194.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Coleman ST, Tseng E, Moye-Rowley WS. Saccharomyces cerevisiae basic region-leucine zipper protein regulatory networks converge at the ATR1 structural gene. J. Biol. Chem. 1997;272:23224–23230. doi: 10.1074/jbc.272.37.23224. [DOI] [PubMed] [Google Scholar]
- 52.Schubert S, Barker KS, Znaidi S, Schneider S, Dierolf F, Dunkel N, Aid M, Boucher G, Rogers PD, Raymond M, Morschhauser J. Regulation of efflux pump expression and drug resistance by the transcription factors Mrr1, Upc2, and Cap1 in Candida albicans. Antimicrob Agents Chemother. 2011;55:2212–2223. doi: 10.1128/AAC.01343-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cannon RD, Lamping E, Holmes AR, Niimi K, Baret PV, Keniya MV, Tanabe K, Niimi M, Goffeau A, Monk BC. Efflux-mediated antifungal drug resistance. Clin Microbiol Rev. 2009;22:291–321. doi: 10.1128/CMR.00051-08. Table of Contents. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Nikaido H. Multidrug resistance in bacteria. Annu Rev Biochem. 2009;78:119–146. doi: 10.1146/annurev.biochem.78.082907.145923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Costa C, Dias PJ, Sa-Correia I, Teixeira MC. MFS multidrug transporters in pathogenic fungi: do they have real clinical impact? Frontiers in physiology. 2014;5:197. doi: 10.3389/fphys.2014.00197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Prasad R, Goffeau A. Yeast ATP-Binding Cassette Transporters Conferring Multidrug Resistance. Annu Rev Microbiol. 2012;39:63. doi: 10.1146/annurev-micro-092611-150111. [DOI] [PubMed] [Google Scholar]
- 57.Dexter D, Moye-Rowley WS, Wu A-L, Golin J. Mutations in the yeast PDR3, PDR4, PDR7 and PDR9 pleiotropic drug resistance loci affect the transcript level of an ATP binding cassette transporter encoding gene, PDR5. Genetics. 1994;136:505–515. doi: 10.1093/genetics/136.2.505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Moye-Rowley WS. Regulation of the transcriptional response to oxidative stress in fungi: similarities and differences. Eukaryot Cell. 2003;2:381–389. doi: 10.1128/EC.2.3.381-389.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gulshan K, Rovinsky SA, Moye-Rowley WS. YBP1 and its homologue YBP2/YBH1 influence oxidative-stress tolerance by nonidentical mechanisms in Saccharomyces cerevisiae. Eukaryot Cell. 2004;3:318–330. doi: 10.1128/EC.3.2.318-330.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Veal EA, Ross SJ, Malakasi P, Peacock E, Morgan BA. Ybp1 is required for the hydrogen peroxide-induced oxidation of the Yap1 transcription factor. J Biol Chem. 2003;278:30896–30904. doi: 10.1074/jbc.M303542200. [DOI] [PubMed] [Google Scholar]
- 61.Gulshan K, Rovinsky SA, Coleman ST, Moye-Rowley WS. Oxidant-specific folding of Yap1p regulates both transcriptional activation and nuclear localization. J Biol Chem. 2005;280:40524–40533. doi: 10.1074/jbc.M504716200. [DOI] [PubMed] [Google Scholar]
- 62.Patterson MJ, McKenzie CG, Smith DA, da Silva Dantas A, Sherston S, Veal EA, Morgan BA, MacCallum DM, Erwig LP, Quinn J. Ybp1 and Gpx3 signaling in Candida albicans govern hydrogen peroxide-induced oxidation of the Cap1 transcription factor and macrophage escape. Antioxid Redox Signal. 2013;19:2244–2260. doi: 10.1089/ars.2013.5199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Jain C, Pastor K, Gonzalez AY, Lorenz MC, Rao RP. The role of Candida albicans AP-1 protein against host derived ROS in in vivo models of infection. Virulence. 2013;4:67–76. doi: 10.4161/viru.22700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Miller M. The importance of being flexible: the case of basic region leucine zipper transcriptional regulators. Curr Protein Pept Sci. 2009;10:244–269. doi: 10.2174/138920309788452164. [DOI] [PMC free article] [PubMed] [Google Scholar]