Abstract
We examined the immune microenvironment of primary colorectal cancer (CRC) using immunohistochemistry, laser capture microdissection/qRT-PCR, flow cytometry and functional analysis of tumor infiltrating lymphocytes. A subset of CRC displayed high infiltration with activated CD8+ CTL as well as activated Th1 cells characterized by IFN-γ production and the Th1 transcription factor Tbet. Parallel analysis of tumor genotypes revealed that virtually all of the tumors with this active Th1/CTL microenvironment had defects in mismatch repair, as evidenced by microsatellite instability (MSI). Counterbalancing this active Th1/CTL microenvironment, MSI tumors selectively demonstrated highly up-regulated expression of multiple immune checkpoints, including five – PD-1, PD-L1, CTLA-4, LAG-3 and IDO – currently being targeted clinically with inhibitors. These findings link tumor genotype with the immune microenvironment, and explain why MSI tumors are not naturally eliminated despite a hostile Th1/CTL microenvironment. They further suggest that blockade of specific checkpoints may be selectively efficacious in the MSI subset of CRC.
Keywords: PD-1, LAG-3, IFN-γ, Tumor microenvironment, Immunotherapy
Introduction
An increasing body of research has revealed that the immune microenvironment of cancer is unique and complex; indeed, tumors can be viewed as distinct immunologic organs. Pathologists have long recognized the diversity of immune infiltration into tumors, invoking terms like “lymphocyte poor” and “inflammatory”. For some cancer types, it is becoming clear that simple quantitation of lymphocyte infiltration has prognostic significance, suggesting that lymphocyte infiltration is not passive but may actively promote or inhibit tumor growth (1). In the case of colorectal cancer (CRC), Galon and colleagues demonstrated that levels of lymphocyte infiltration into primary tumors represented a strong independent predictor of relapse and overall survival, with high lymphocyte infiltration being a positive prognostic factor (2). Using expression profiling of CRCs, they further defined the relevance of specific immune signatures, demonstrating that T helper1 cells (Th1) type, IFN-γ dominant immune profiles signified an improved prognosis while Th17 type, IL-17 dominant immune profiles signified a poor prognosis (3). These findings are in concordance with multiple studies in murine models demonstrating that Th1 / cytotoxic T lymphocyte (CTL) dominant immune responses are anti-tumorigenic (4) while Th17 dominant responses are typically pro-carcinogenic (5).
The relevance of molecular regulation of lymphocyte function in the tumor microenvironment (TME) has been further highlighted by the emerging clinical experience in treatment of cancer with antibody blockade of the PD-1 pathway. Anti-PD-1 and anti-PD-L1 antibodies have been demonstrated to induce significant durable tumor regressions in patients with melanoma, renal cancer and non-small cell lung cancer (6-9). The finding that expression of PD-L1 in tumor biopsies is a predictor of response to PD-1 pathway blocking antibodies (6) supports conclusions from murine studies that this pathway plays a major role in blocking anti-tumor immune responses directly within the TME (10). With increasing efforts to develop antibody blockers of multiple additional immune inhibitory ligands and receptors (termed checkpoints) for cancer immunotherapy, understanding the expression patterns and functions of these molecules in the context of the TME will be critical in selecting patient populations most likely to benefit from their application (11). In contrast to melanoma, renal and lung cancer, CRC cohorts demonstrated a very low response rate to PD-1 or PD-L1 blockade and have been considered a non-responding histology to the PD-1 pathway blockade (6-8). However, there were insufficient patients to potentially define subsets of CRC that might be more amenable to checkpoint blockade.
With these considerations in mind, we have undertaken a comprehensive analysis of the immune microenvironment of CRC, using a combination of immunohistochemistry (IHC), laser capture microdissection combined with quantitative reverse transcription PCR (LCM/qRT-PCR), multiparameter flow cytometry (MFC) and functional analysis of purified tumor infiltrating lymphocytes (TIL) from a large set of surgically resected primary CRC. These analyses revealed, in an unbiased fashion, that a subset of CRC possesses a highly activated Th1- and CTL-rich microenvironment. Through concomitant genetic analysis, we found that this subset of tumors harbored microsatellite instability (MSI), indicating that they were mismatch repair deficient (12). This finding was in accordance with numerous prior studies indicating that MSI tumors are highly infiltrated with T-cells including CTLs (13-16). How could tumors persist in this hostile microenvironment? To find out, we carried out an extensive investigation of the regulatory factors operative in this microenvironment. We found that there was a dramatic overexpression of immune checkpoint-related proteins in the microenvironment of MSI tumors, thus explaining a long-standing enigma and suggesting that immunotherapeutic interventions involving checkpoint blockade might be selectively effective in this important subset of cancers. Based on these findings, clinical trials have been initiated to test PD-1 blockade in CRC patients selected to be MSI.
Results
We began our analysis of primary sporadic CRC by performing quantitative IHC for standard T cell subsets using antibodies to CD3, CD4, CD8 and Foxp3. We distinguished three compartments – true TIL representing lymphocytes intercalated within the glandular or medullary epithelial component of the tumor, T cells in the tumor stroma surrounding the epithelial component of the tumor and the invasive front where the tumor invades into the lamina propria (Fig. S1). Infiltration by these T cell types in each of the compartments was quantitated numerically by blinded analysis of 5 high power fields (hpf; 0.0028mm2/hpf) and displayed as average number of stained cells per hpf. We found a subset of tumors (roughly 20%) that had large infiltrates of total (CD3+ T cells) as well as CD4+ and CD8+ T cells in all three compartments with CD8+ T cells most dramatically increased. We wondered whether this subset of tumors was MSI, given prior studies indicating lymphocyte infiltrations in such tumors (13-16). Indeed, analysis of microsatellites showed that nearly all of the tumors with the infiltrates were MSI. We therefore focused on differences between MSI versus MSS tumors (Table S1) for the in-depth analysis of the immune TME of CRC.
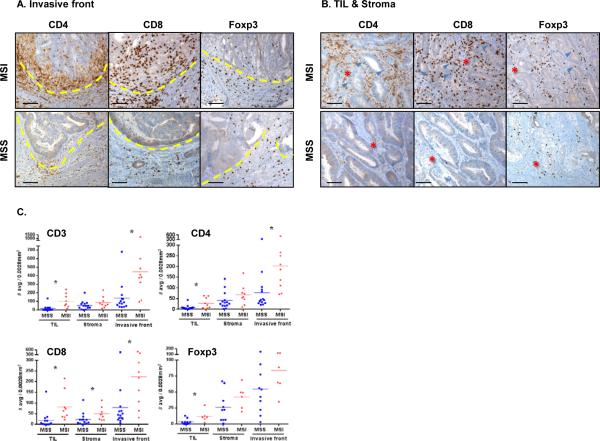
Figures 1A & B show typical examples of IHC staining and Fig.1C shows the quantitative analyses demonstrating statistical differences between MSI and MSS tumors for the total CD3+ T cells and CD4+ T cells among TIL and invasive front as well as CD8+ T cells among all three compartments. Although it did not reach statistical significance in tumor stroma and the invasive front, we observed higher Foxp3+ cell infiltrates, representative of regulatory T cells (Treg), in MSI compared to MSS tumors. Of note, a single outlier included in our MSS cohort displayed a high infiltration of each T cell subset, similar in range to the one of MSI tumors (Fig. 1C, S2 and S3A).
Figure 1. Geographic distribution in situ of MSI and MSS CRC-infiltrating lymphocytes.
FFPE tissue sections were characterized by IHC for CD4+, CD8+ and Foxp3+ cell infiltration. Three distinct histological areas designated as tumor infiltrating lymphocytes (TIL), tumor stroma and invasive front (where tumor invaded normal lamina propria) were histologically identified and separately analyzed. Invasive front (A) and TIL/Stroma (B) areas of a representative MSS (bottom panel) and MSI (top panel) are shown (20X magnification). Dashed lines in (A) delineate the invasive front with the tumor tissue on the top side and the normal tissue on the bottom side. Red stars and blue arrows in (B) indicate the tumor stroma and tumor epithelium-infiltrating immune cells, respectively. Scale bars, 100μm. C, cell density was scored in 14 MSS (blue) and 9 MSI (red) specimens by determining the average number of stained cells in 5 distinct hpf (0.0028mm2/hpf). The graphs display the mean for each group and * indicates statistically significant differences between MSS and MSI (p<0.05 using Mann-Whitney U test).
We further explored the nature of the T cell infiltrates by performing LCM on MSI and MSS tumors, separately dissecting the TIL, stroma and invasive front compartments (Fig. S1) and then performed qRT-PCR (Fig. 2A-C) for selected genes encoding signature T cell cytokines as well as core transcription factors for each of the three major T helper subsets, Th1/Tc1 (type 1 CTL; TBX21 and IFNG are common to Th1 and Tc1), Th2 and Th17. We additionally analyzed genes associated with CTL and Treg and also general inflammatory cytokines. Finally, we analyzed expression of genes encoding both co-inhibitory membrane ligands and receptors (commonly termed checkpoints) and metabolic enzymes that have been shown to regulate lymphocyte activity; these serve feedback inhibitory functions in normal physiology but can represent important mechanisms of adaptive immune resistance by tumors in the face of an endogenous anti-tumor T cell repertoire (11).
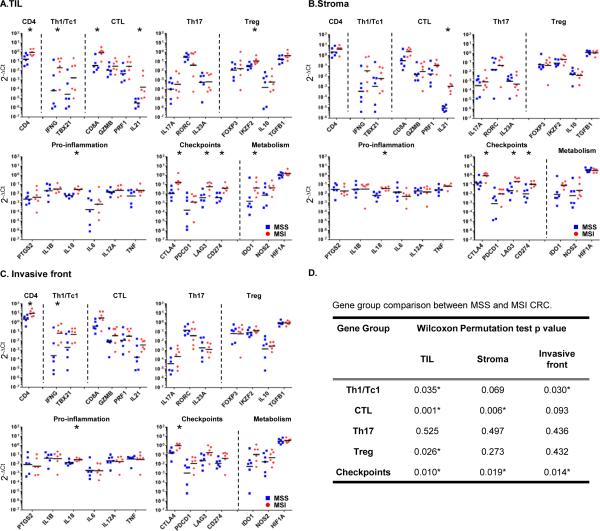
Figure 2. Th1 and CTL based immune signature and elevated checkpoint expression in MSI CRC.
RNA was extracted from tissue samples laser-microdissected representing TIL in tumor nests (A), stroma surrounding tumor (B) and invasive front (C) areas of MSS (blue squares) and MSI (red circles) CRC specimens. Immune-related gene expression profiles were assessed using Taqman-based qRT-PCR for selected genes. Sets of genes were defined by functional relevance (Th1/Tc1, CTL, Th17, Treg, pro-inflammation, immune checkpoints and metabolism). The Y axis represents an arbitrary unit of expression 2−ΔCt, ΔCt representing cycle threshold (Ct) of the gene of interest normalized by Ct of ubiquitous genes (GUSB, GAPDH). The graphs display the geometric means. Their differential representation between MSS and MSI specimens was analyzed using adjusted Wilcoxon Mann Whitney test as described in Methods section. *, Wilcoxon p-value < 0.05. D, gene group comparison in TIL, tumor stroma and invasive front areas between MSS and MSI specimens.Permutation Test Results based on the maximum Wilcoxon Mann Whitney test statistic within the gene groups Th1/Tc1, CTL, Th17 and immune checkpoints. * indicates statistically significant differences between MSS and MSI (p<0.05).
We found that the expression of the gene encoding IFN-γ (IFNG) the canonical Th1/Tc1 cytokine is higher in all three compartments of MSI compared to MSS tumors (Fig. 2A-C; the differences reach statistical significance in the TIL and invasive front areas with Wilcoxon test p values = 0.041, for both of them). The expression of TBX21 encoding Tbet, the Th1/Tc1 canonical transcription factor is similarly increased in MSI tumors, though differences did not reach statistical significance among the cohort analyzed. The CD8A gene, mainly expressed by CTL is highly differentially expressed in the TIL regions of MSI relative to MSS tumors (Fig. 2A; p=0.004), in concordance with significantly higher CD8 infiltration observed by IHC (Fig. 1C). In marked contrast to Th1 and CTL genes, expression of Th17 genes is virtually identical between MSI and MSS tumors for all compartments. IL-13 and IL-4 (the canonical Th2-type cytokines) were undetectable in most of the MSI and MSS samples for each of the TME regions analyzed and GATA3 (Th2 transcription factor) expression was not different between MSS and MSI (data not shown). Treg-associated genes, including FOXP3, were similar between MSI and MSS tumors. Gene group comparison analysis using Wilcoxon-Mann-Whitney Permutation test confirms that Th1/Tc1 (TBX21 and IFNG) and CTL (CD8A, GZMB, PRF1, IL21) groups but not the Th17 (RORC, IL17A and IL23) group show statistical differences in their representation between TIL compartments of MSI and MSS CRC (Fig. 2D). In summary, MSI tumors have a selective Th1 and CTL infiltration and activation relative to MSS tumors. The highest value for IFNG and TBX21 expression in TIL from a single MSS sample (detailed in Fig. S3B) represents the same outlier observed in the quantitative analysis of CD4 and CD8 TIL infiltration from Fig. 1C. Among genes encoding inflammatory cytokines, IL18, which is generally associated with Th1 responses and selectively promotes IFN-γ production by T cells, is more highly expressed in MSI tumors in all three compartments (p<0.05) whereas, genes encoding IL-1 and IL-6, which selectively promote Th17 responses, are not (Fig. 2A-C). The expression of PTGS2 (encoding COX2), IL12A (encoding IL12p35) and TNF (encoding TNF-α) did not differ between MSS and MSI specimens.
We next analyzed the expression of genes encoding checkpoint receptors. We found that all three of the clinically targeted checkpoint receptors - CTLA-4 (CTLA4), PD-1 (PDCD1) and LAG-3 (LAG3) - were expressed at considerably higher levels in MSI compared to MSS tumors (p<0.05 in all compartments for CTLA4, in TIL and invasive front for LAG3; p>0.05 in all compartments for PDCD1). The gene encoding PDL1 (CD274), a major IFN-γ-inducible PD-1 ligand expressed in the epithelial cells of many solid tumors (17) was unexpectedly expressed in TIL and stroma of MSI tumors (Wilcoxon test p values are 0.009 and 0.015, respectively; Fig. 2A&B). Among inhibitory metabolic enzymes, the IFN-γ-induced gene encoding indolamine 2’3’ dioxygenase (IDO), which metabolizes tryptophan to kynurenine, demonstrates significantly higher expression in TIL of MSI tumors (p=0.041; Fig. 2A). In contrast, arginase (ARG1), a myeloid enzyme induced by the Th2 cytokines IL-4 and IL-13 and up-regulated in M2 macrophages, was generally not detectable in either MSI or MSS tumors (data not shown). Thus, along with the Th1 and CTL genes, multiple immune-inhibitory genes, including a number that are IFN-γ-induced, are preferentially expressed in the TILs and stroma of MSI tumors. To note, these results reflect a combination of higher immune infiltration and cellular upregulation in MSI compared to MSS tumors.
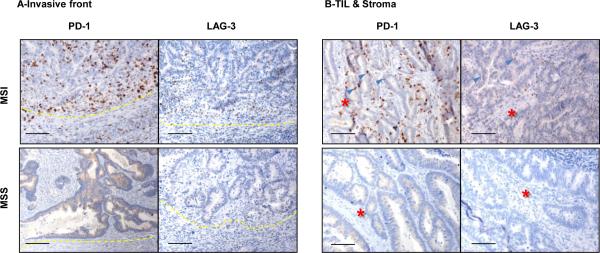
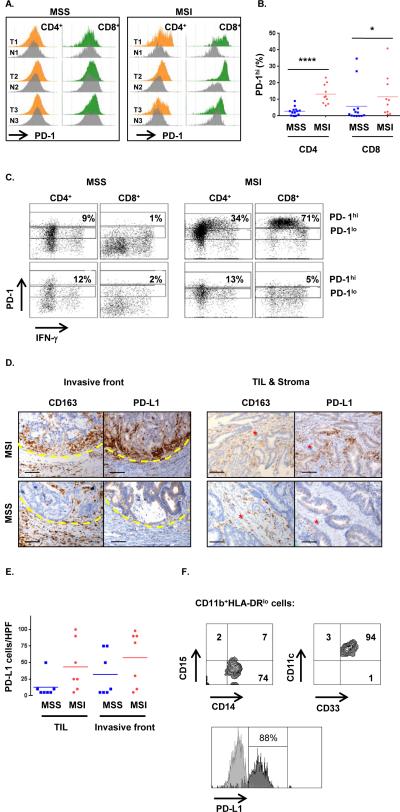
Because differences in checkpoints expression could have significant implications in defining patient subgroups potentially responsive to checkpoint blockade, we sought to determine whether differences at the RNA level were mirrored at the protein level. Indeed, IHC for both PD-1 and LAG-3 demonstrated a robust expression of each of them in lymphocytes of MSI tumors, whereas very little was observed in MSS tumors (Fig. 3A-B). The qRT-PCR/LCM analyses shown in Fig. 2 therefore underestimated the differences between MSI and MSS tumors with respect to PD-1 protein surface expression by TIL. Multi-parameter flow cytometry of freshly isolated lymphocytes from tumors demonstrated that a large proportion of both CD4+ and CD8+ T cells infiltrating MSI tumors express high levels of PD-1 (Fig. 4A-B). This PD-1hi population was largely absent in MSS tumors (except two MSS specimens, one of which was the highly infiltrated MSS outlier described above) and the normal mucosa adjacent to MSI tumors. The T-bet downregulation and PD-1 upregulation by T cells is typically found in chronic viral infection and termed “exhaustion” (18). Since the presence of PD-1hi T cell infiltrate in MSI tumors is concomitant with the detection of high IFN-γ (MFC in Fig.4C and qRT-PCR in Fig.2) and high T-bet (qRT-PCR in Fig.2), further investigation should be conducted to determine the co-expression of these molecules at the single cell level and formally rule out the classic “exhaustion” phenotype of these TILs. Of note, the two MSS tumors exhibiting an unusually strong proportion of PD1hi CD8+ T cells (Fig.4B), were also characterized by a high proportion of IFN-γ-producing CD8+ T cells (Fig.S3 and data not shown). Our findings suggest that a small proportion of MSS tumors are characterized by the concordant detection of Th1/CTL infiltration and immune checkpoint expression that is found in all MSI tumors.
Figure 3. PD-1 and LAG-3 expression in MSI and MSS CRC specimens.
IHC analysis of PD-1 and LAG-3 expression in invasive front (A) and TIL/Stroma (B) areas was performed on FFPE tissue sections of a representative set of MSI (top panel) and MSS (bottom panel) CRC specimens. Magnification, X20; Scale bars, 100μM; Red stars and blue arrows in (B) indicate the tumor stroma and tumor epithelium infiltrating immune cells, respectively.
Fig. 4. MSI CRC are characterized by IFN-γ-producing PD1hi TIL and PDL-1+ tumor-infiltrating myeloid cells.
A, Freshly dissociated MSS and MSI colon tumors (T) as well as patient-matched normal tissue (N) were assessed by MFC for the expression of PD-1 on infiltrating CD4+ and CD8+ T cells. PD-1 expression in tumor was normalized to the normal tissue ran simultaneously and both histograms were aligned to delineate in tumor samples the PD1hi cells when compared to normal tissue. B, proportion of PD-1hi CD4+ and CD8+ cells among CD3+ lymphocytes infiltrating MSS (blue squares) and MSI (red circles) specimens. In each group the mean is indicated and * represents statistically significant differences between MSS and MSI (*p<0.05, ****p<0.0001; non-parametric Mann-Whitney U test). C, representative intracellular staining for IFN-γ production by in vitro PMA/ionomycin activated T cells (3hrs). The dot plots show the co-expression of PD-1 and IFN-γ in CD4+ T cells and CD8+ T cells in a representative set of MSS (Left) and MSI (Right) CRC (Top) and patient-matched normal (Bottom) specimens. The gates delineate PD1hi and PD1lo cells. D, co-localization of CD163 and PD-L1 expression in invasive front (left panel) and TIL/Stroma (right panel) areas of a representative set of MSS (lower panels) and MSI (upper panel) CRC specimens were assessed by IHC; x20 magnification. Scale bars, 100μM. Red stars indicate the tumor stroma. E, PD-L1 expression scores in 7 MSS (blue) and 7 MSI (red) CRC specimens (average of 5 hpf per sample). F, MFC analysis of PD-L1 expression on MSI CRC-infiltrating myeloid cells. Dot plots represent the expression of myeloid associated markers on CD11b+HLA-DR−/low cells. Infiltrating myeloid cells were characterized as CD15−CD14+CD33+CD11c+ cells. PD-L1 expression (dark gray) is overlaid with corresponding isotype control (light gray).
Finally, we analyzed PD-L1 protein expression by IHC. MSI tumors demonstrated much higher PD-L1 expression than MSS tumors (Fig. 4D). Surprisingly, in contrast to other cancers such as melanoma, renal cancer and lung cancer (19), there was virtually no discernable PD-L1 expression on tumor cells of MSI tumors by IHC; rather, co-staining with CD163 demonstrated that the majority of PD-L1 expression was by myeloid cells. There were large numbers of PD-L1+ myeloid cells at the invasive front and in the stroma and some were intercalated between epithelial cells in the tumor nests of MSI specimens (Fig. 4D). Histological scoring of PDL1+ cells in TIL and the invasive front regions confirmed the high expression of PD-L1 in MSI tumors (Fig. 4E). Omitting the MSS outlier patient (high CD8 infiltration and IFN-γ production), that also demonstrated high PD-L1 expression, we found that the difference between MSS and MSI specimens reached statistical significance in the TIL area (p<0.05, Mann Whitney U test). MFC analysis performed on freshly dissociated MSI tumors confirmed high levels of PD-L1 expression on viable CD11b+HLA-DRloCD15−CD14+CD33+ myeloid cells (Fig. 4F). Because most human cancers appear to up-regulate PD-L1 as an adaptive response to IFN-γ (17), the lack of clear PD-L1 expression on tumor cells in the MSI specimens as assayed by IHC was unexpected, particularly given the high IFN-γ level in these tumors. Of note, while PD-L1 was up-regulated on a number of both MSI and MSS colon tumor cell lines after incubation with IFN-γ, this up-regulation was, in general, much less than the one observed in melanoma cell lines (Fig. S4). Interestingly, cell lines with the weakest PD-L1 induction also showed weak MHC II induction after IFN-γ treatment, suggesting that colon tumors may have relatively dampened Stat1 signaling.
MSI tumors have a much higher mutational load (and thus potentially more neoantigens) than MSS tumors, and this could be related to the presence of the vigorous immune microenvironment in this genotypic subset of CRC. We therefore wondered whether the highly infiltrated MSS tumor described above had more mutations than expected for an MSS tumor. To address this question, we performed exome sequencing on the microdissected tumor of this individual. We found only 49 nonsynonymous, somatic mutations in this tumor, a number well within the typical range for MSS CRC and much less than in MSI tumors (20). Additionally, no mutations in POLE and POLD1 genes were identified; tumors with mutations in these genes also contain a high mutational load despite having stable microsatellites (21). Thus, in this individual tumor, the prominent T cell infiltration was not due to an unusually high mutational load (Table S2).
Discussion
Direct analysis of the immune microenvironment of tumors is emerging as the most important means of understanding the relationship between patients’ immune systems and their cancer (1). These analyses are bearing fruit in informing prognosis and guiding immunotherapy, particularly using antibody blockade of immune checkpoints. We demonstrate here that the immune microenvironment of DNA-repair deficient MSI CRC contains a strong Th1 and CTL component not found in the vast majority of DNA repair sufficient MSS tumors, in keeping with previous reports of high T cell infiltration in MSI tumors (13-16). More importantly, we address a vexing question in the field: why are human tumors, which are apparently strongly immunogenic, as judged by histopathologic criteria, not rejected by the host? The numbers of infiltrating activated lymphocytes and CTLs in some tumors are huge, in some cases larger than the number of neoplastic cells. The answer to this question was found through the analysis of immune checkpoints. Multiple key immune inhibitory ligands, receptors and metabolic enzymes, including those that are IFN-γ inducible (PD-L1, IDO), were highly up-regulated in MSI tumors relative to MSS (Fig. 2). We suggest that MSI represents a classic example of adaptive resistance in which an active immune Th1/CTL microenvironment results in compensatory induction of checkpoints that protect the tumor from killing (11).
The mechanistic basis for the link between MSI genetic status and a high Th1/CTL microenvironment in CRC is not known. However, one important factor may be the increased number of tumor “neoantigens” in DNA repair-deficient tumors created by the high mutational load, typically between 10 and 50 times that of DNA repair-sufficient tumors (14, 16). The notion is that mutation-generated neoantigens are truly tumor specific and may not induce immune tolerance to the same extent as self-antigens, even those up-regulated in tumors due to epigenetic dysregulation (22). Ultimately, an altered amino acid due to a coding mutation is only relevant as a tumor neoantigen for T cells if it can be processed and presented on self-MHC. Exomic sequencing and application of a multifactorial algorithm to the mutations in the MSS with an active Th1/Tc1 microenvironment may predict a number of potential neoepitopes capable of being presented on the tumor's HLA class I alleles (23). The multiple variables involved in antigen processing and presentation suggest that while mutational density may correlate with neoantigenicity on a population level, individual tumors with lower mutational load can none-the-less generate good T cell neoepitopes if the mutations are appropriately positioned. Additionally, many other mechanisms besides neoantigenicity (i.e. production of immune regulatory cytokines and chemokines by the tumor and non-transformed stromal cells) likely contribute to the character of the immune microenvironment.
Among the many immune inhibitory ligand-receptor pairs (checkpoints) and metabolic enzymes discovered to date, CTLA-4, PD-1, PD-L1, LAG-3 and IDO are of particular interest because inhibitory antibodies or drugs (in the case of IDO) are currently in active clinical testing to enhance therapeutic anti-tumor responses. Of note, all of these were highly elevated in all three compartments (TIL, stroma and invasive front) of MSI tumors. IHC of PD-1, PD-L1 and LAG-3 confirmed the gene expression findings and demonstrated that MSS tumors and their infiltrating and invasive front lymphocytes (with the exception of the outlier) express very little of these checkpoint molecules. Recent findings point to the elevated expression of these checkpoints in tumors (particularly PD-L1 and IDO) as an adaptive response to cytokines such as IFN-γ that are part-and-parcel of an active intratumoral immune response (24, 25). This mechanism appears to be operative in MSI tumors. What is strikingly different in MSI CRC relative to melanoma, renal or lung cancer is that there is little PD-L1 expressed by tumor cells; rather it is expressed predominantly by infiltrating myeloid cells. The modest up-regulation of PD-L1 by CRC cell lines in response to in vitro incubation with IFN-γ suggests that adaptive PD-L1 expression in CRC is predominantly at the level of infiltrating myeloid cells, though low levels of PDL1 could be expressed by tumor cells in situ that is below the detection level of IHC. Regardless, a very important prediction of our findings on differential expression of immune inhibitory ligands, receptors and metabolic enzymes is that MSI tumors will respond to checkpoint blockade with agents such as anti-PD-1 or anti-PD-L1 antibodies, while MSS tumors will be much less responsive. Indeed, based on these findings, 2 clinical trials testing anti-PD-1 antibodies in selected MSI patients have been initiated (NCT01876511 and NCT02060188).
Materials and Methods
Patient selection, tumor samples and cell lines
Tumor tissues were collected at Johns Hopkins Hospital (Baltimore, MD, USA) from patients with primary sporadic CRC and free of prior chemotherapy. Demographic, pathologic (tumor location, TNM grade) and genetic status are detailed in Table S1. This study was approved by the Johns Hopkins Institutional Review Board. All samples were obtained in accordance with the Health Insurance and Accountability Act.
Assessment of microsatellite instability (MSI) was done using the length of a panel of microsatellite markers in the tumor and a normal reference (either normal mucosa or germline) by using fragment analysis of PCR products labeled with fluorescent dyes. Fragment analysis determined the expression level of the proteins in charge of maintaining the integrity of microsatellite tracts. Differences in the length of 2 or more markers (the standard Bethesda panel uses 5 markers) was indicative of MSI status. Eleven patients tested as MSI positive and fourteen patients as MSS (Table S1).
Analysis of mutations on the MSS outlier sample was performed using DNA exomic sequencing from formalin-fixed, paraffin-embedded (FFPE) tissues. Identification of tumor-specific mutations involved comparison with normal matched cells derived from FFPE sections that contained no tumor. Genomic DNA libraries were prepared following Illumina's suggested protocol with small modification. Briefly, the ends of sheared genomic DNA fragments are blunted and Illumina adaptors are ligated. The fragment library was amplified by 15 cycles of PCR. Human exome capture is performed following a modified protocol from Agilent's SureSelect Paired-End Version 2.0 Human Exome Kit (Agilent, Santa Clare, CA). The genomic library is hybridized to the SureSelect probes. After washes, the genomic regions captured by the probes are eluted. The captured region covers 38 Mb of exomic sequences. Following PCR amplification of 6 cycles the library is hybridized to an Illumina flow cell and eventually sequenced according to Illumina's protocols. Mismatched bases were identified as a mutation based on rigorous criteria described previously. Mutations were further confirmed to not be variants present in the normal population according to the NCBI dbSNP database.
For analysis of cell lines, three microsatellite instable (SW48, HCT116 and DLD1) and 3 chromosomal instable (HT29, Caco-2 and SW480) human CRC cell lines were used. These cell lines were obtained from the Genetic resources core facility (GRCF) - Institute of Genetic Medicine - Johns Hopkins School of Medicine, which purchased these cells from the authenticated ATCC (American Type Culture Collection) repository. The cell lines were maintained in culture in 10% FCS DMEM supplemented with L-Glutamine, non-essential amino acids, sodium pyruvate and antibiotics.
Immunohistochemistry
FFPE specimens were cut into 5-μm sections and mounted on glass slides. For each specimen, CD3 (clone PS1, Leica Biosystems, Buffalo Grove, IL), CD4 (clone Sp35, Ventana, Tuscon, AZ), CD8 (clone C8144B, Cell Marque, Rocklin, CA), Foxp3 (236A/E7, Abcam, Cambridge, MA), CD163 (clone Novacastra10D6, Leica Biosystems) stainings were performed according to standard protocols. IHC for PD-L1 (clone 5H1), PD-1 (clone M3) stainings were performed according to a protocol previously reported (24). Positive cells were counted in 5 HPF (0.028 mm/HPF; Olympus BX41) by two blinded pathologists (R.A.A and M.C) in three distinct tumor areas denominated TIL, tumor stroma and Invasive Front. LAG3 IHC was performed using clone 17B4 (LS Bio, Seattle, WA).
Laser capture microdissection and Taqman quantitative RT-PCR
5um FFPE and H&E stained tissue sections were used for LCM procedure using the Leica LMD 7000 system (Leica, Buffalo Grove, Il). For each patient, tissues were microdissected from the 3 previously defined areas and directly collected in tissue lysis buffer for RNA extraction. RNA was isolated following the manufacturer instructions (High pure paraffin kit, Roche). RNA was converted to cDNA using high capacity RNA to cDNA kit (Life Technologies, Grand Island, NY) and a step of preamplification was first done using a pool of primers and preamplification master mix kit (Life technologies, ) followed by Taqman RT-PCR (list of primers in Table S3). Data are expressed as 2−ΔCt where ΔCt represents Ctgene-Ctctrl. For our calculation Ctctrl is the average of Ct for 2 ubiquitous genes (GAPDH, GUSB). When undetectable, a value of 40 was assigned as Ct.
Tumor processing and flow cytometry
Tumor specimens were collected and freshly dissociated using enzymatic cocktail (0.1% DNaseI and Liberase 400u/ml, Roche). Leukocytes were enriched by Percoll density gradient (GE healthcare, US) and cells were banked in liquid nitrogen until further analysis. MFC was performed with a LSR II cytometer (BD Biosciences). Samples were stained with anti-CD45 (2D1, BD Biosciences) anti-CD3 (UCHT1, BD Biosciences), anti-CD4 (OKT4, BD Bioscience), anti-CD8 (RPA-T8, BD Biosciences), anti-PD-1 (EH12.1, BD Biosciences), anti-Foxp3 (259D/C7, BD Biosciences). Cytokine intracellular staining (ICS) for IFN-γ (B27, BD Biosciences) and IL-17 (SCPL1362, BD Biosciences) was performed following 3 hr -in vitro stimulation in the presence of stimulation cocktail (PMA+ionomycin; Ebioscience) and GolgiStop (Monensin; BD Biosciences) according to the manufacturer instructions. Data were analyzed using DIVA 6.1 Software (BD Biosciences). Myeloid cells were stained using anti-CD45, anti-CD11b (ICRF44, BD Biosciences), anti-DR (G46.6, BD Biosciences), anti-CD15 (HI98, BD Biosciences), anti-CD14 (M5E2, BD Biosciences), anti-CD33 (WM53, BD Biosciences), anti-CD11c (B-Ly6, BD Biosciences) and anti-PD-L1 (29E.2A3, Biolegend, San Diego, CA). Cell lines were cultured for 3 days in the presence or absence of IFN-γ (500IU/ml). Cells were stained with DR and B7-H1/PD-L1 mAb prior to analysis by flow cytometry.
Statistical analysis
IHC scoring in each of the three histological areas (i.e., TIL, stroma and invasive front) and MFC data were summarized using scatter plots and compared between MSI and MSS patients using means and non-parametric Mann Whitney U test. For the gene expression analysis, scatter plots and geometric means were used to characterize MSI/MSS patient groups for each of the three locations. Genes were grouped by lineage and/or function (Th1/Tc1, CTL, Th17, Treg, Pro-inflammation, Metabolism) and to distinguish which groups of genes were differentially expressed based on MSS/MSI status, a re-sampling based permutation test was conducted based on the maximum Wilcoxon Mann Whitney test statistic within the gene group. Individual gene expression was also compared across MSI/MSS status using the Wilcoxon-Mann-Whitney test. All tests are descriptive and no multiplicity adjustment was considered. Statistical analysis were performed using the R statistical package (version 2.15.1)
Supplementary Material
Significance.
The findings reported in this article are the first to demonstrate a link between a genetically defined subtype of cancer and its corresponding expression of immune checkpoints in the tumor microenvironment. The mismatch repair defective subset of CRC selectively up-regulates at least 5 checkpoint molecules that are targets of inhibitors currently being clinically tested.
Acknowledgements
We would like to thank K. Judge and C. Blair for her excellent management and coordination of CRC patient recruitment and specimen banking. We acknowledge Robert Scharpf (Department of Biostatistics, Johns Hopkins University School of Medicine) for his helpful expertise in genomic data analysis.
Financial Support: This work was supported by National institutes of Health through P50 CA062924 (GI SPORE to FH), RO1 CA151393 (CLS and DMP), K08 DK087856 (ECW), 5T32CA126607-05 (EMH), P30 DK089502 (GI Core), P30 CA006973 (SKCCC core). Additional support was provided by GSRRIG-015 (American Society of Colon and Rectal Surgeons to EMH), the Mérieux Foundation (CLS and DMP), a Stand Up To Cancer–Cancer Research Institute Cancer Immunology Dream Team Translational Research Grant SU2C-AACR-DT1012 (Stand Up To Cancer is a program of the Entertainment Industry Foundation administered by the American Association for Cancer Research), a Melanoma Research Alliance award (to DMP), the Commonwealth Foundation (KWK, NP, and BV), The Virginia and D.K. Ludwig Fund for Cancer Research and the Ludwig Institute (KWK and BV).
Abbreviations list
- ARG
arginase
- CRC
colorectal cancer
- CTL
cytotoxic T lymphocytes
- FFPE
formalin-fixed paraffin-embedded
- H&E
hematoxylin and eosin
- IDO
indolamine 2’3’ dioxygenase
- IHC
immunohistochemistry
- LCM
laser capture microdissection
- MSI
microsatellite instable
- MSS
microsatellite stable
- MFC
multiparameter flow cytometry
- Th
T helper
- TIL
tumor-infiltrating lymphocyte
- TME
tumor microenvironment
- Treg
regulatory T cells
Footnotes
Conflict of Interest Disclosure: The authors disclose no potential conflicts of interest
References
- 1.Fridman WH, Pages F, Sautes-Fridman C, Galon J. The immune contexture in human tumours: impact on clinical outcome. Nat Rev Cancer. 2012;12:298–306. doi: 10.1038/nrc3245. [DOI] [PubMed] [Google Scholar]
- 2.Galon J, Costes A, Sanchez-Cabo F, Kirilovsky A, Mlecnik B, Lagorce-Pages C, et al. Type, density, and location of immune cells within human colorectal tumors predict clinical outcome. Science. 2006;313:1960–4. doi: 10.1126/science.1129139. [DOI] [PubMed] [Google Scholar]
- 3.Tosolini M, Kirilovsky A, Mlecnik B, Fredriksen T, Mauger S, Bindea G, et al. Clinical impact of different classes of infiltrating T cytotoxic and helper cells (Th1, th2, treg, th17) in patients with colorectal cancer. Cancer Res. 2011;71:1263–71. doi: 10.1158/0008-5472.CAN-10-2907. [DOI] [PubMed] [Google Scholar]
- 4.Shankaran V, Ikeda H, Bruce AT, White JM, Swanson PE, Old LJ, et al. IFNgamma and lymphocytes prevent primary tumour development and shape tumour immunogenicity. Nature. 2001;410:1107–11. doi: 10.1038/35074122. [DOI] [PubMed] [Google Scholar]
- 5.Wilke CM, Kryczek I, Wei S, Zhao E, Wu K, Wang G, et al. Th17 cells in cancer: help or hindrance? Carcinogenesis. 2011;32:643–9. doi: 10.1093/carcin/bgr019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Topalian SL, Hodi FS, Brahmer JR, Gettinger SN, Smith DC, McDermott DF, et al. Safety, activity, and immune correlates of anti-PD-1 antibody in cancer. N Engl J Med. 2012;366:2443–54. doi: 10.1056/NEJMoa1200690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brahmer JR, Tykodi SS, Chow LQ, Hwu WJ, Topalian SL, Hwu P, et al. Safety and activity of anti-PDL1 antibody in patients with advanced cancer. N Engl J Med. 2012;366:2455–65. doi: 10.1056/NEJMoa1200694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Topalian SL, Sznol M, McDermott DF, Kluger HM, Carvajal RD, Sharfman WH, et al. Survival, durable tumor remission, and long-term safety in patients with advanced melanoma receiving nivolumab. J Clin Oncol. 2014;32:1020–30. doi: 10.1200/JCO.2013.53.0105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hamid O, Robert C, Daud A, Hodi FS, Hwu WJ, Kefford R, et al. Safety and tumor responses with lambrolizumab (anti-PD-1) in melanoma. N Engl J Med. 2013;369:134–44. doi: 10.1056/NEJMoa1305133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hirano F, Kaneko K, Tamura H, Dong H, Wang S, Ichikawa M, et al. Blockade of B7-H1 and PD-1 by monoclonal antibodies potentiates cancer therapeutic immunity. Cancer Res. 2005;65:1089–96. [PubMed] [Google Scholar]
- 11.Pardoll DM. The blockade of immune checkpoints in cancer immunotherapy. Nat Rev Cancer. 2012;12:252–64. doi: 10.1038/nrc3239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Boland CR, Goel A. Microsatellite instability in colorectal cancer. Gastroenterology. 2010;138:2073–87. e3. doi: 10.1053/j.gastro.2009.12.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kim H, Jen J, Vogelstein B, Hamilton SR. Clinical and pathological characteristics of sporadic colorectal carcinomas with DNA replication errors in microsatellite sequences. Am J Pathol. 1994;145:148–56. [PMC free article] [PubMed] [Google Scholar]
- 14.Smyrk TC, Watson P, Kaul K, Lynch HT. Tumor-infiltrating lymphocytes are a marker for microsatellite instability in colorectal carcinoma. Cancer. 2001;91:2417–22. [PubMed] [Google Scholar]
- 15.Dolcetti R, Viel A, Doglioni C, Russo A, Guidoboni M, Capozzi E, et al. High prevalence of activated intraepithelial cytotoxic T lymphocytes and increased neoplastic cell apoptosis in colorectal carcinomas with microsatellite instability. Am J Pathol. 1999;154:1805–13. doi: 10.1016/S0002-9440(10)65436-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Phillips SM, Banerjea A, Feakins R, Li SR, Bustin SA, Dorudi S. Tumour-infiltrating lymphocytes in colorectal cancer with microsatellite instability are activated and cytotoxic. Br J Surg. 2004;91:469–75. doi: 10.1002/bjs.4472. [DOI] [PubMed] [Google Scholar]
- 17.Dong H, Strome SE, Salomao DR, Tamura H, Hirano F, Flies DB, et al. Tumor-associated B7-H1 promotes T-cell apoptosis: a potential mechanism of immune evasion. Nat Med. 2002;8:793–800. doi: 10.1038/nm730. [DOI] [PubMed] [Google Scholar]
- 18.Kao C, Oestreich KJ, Paley MA, Crawford A, Angelosanto JM, Ali MA, et al. Transcription factor T-bet represses expression of the inhibitory receptor PD-1 and sustains virus-specific CD8+ T cell responses during chronic infection. Nat Immunol. 2011;12:663–71. doi: 10.1038/ni.2046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Taube JM, Klein A, Brahmer JR, Xu H, Pan X, Kim JH, et al. Association of PD-1, PD-1 Ligands, and Other Features of the Tumor Immune Microenvironment with Response to Anti-PD-1 Therapy. Clin Cancer Res. 2014;20:5064–74. doi: 10.1158/1078-0432.CCR-13-3271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vogelstein B, Papadopoulos N, Velculescu VE, Zhou S, Diaz LA, Jr., Kinzler KW. Cancer genome landscapes. Science. 2013;339:1546–58. doi: 10.1126/science.1235122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Palles C, Cazier JB, Howarth KM, Domingo E, Jones AM, Broderick P, et al. Germline mutations affecting the proofreading domains of POLE and POLD1 predispose to colorectal adenomas and carcinomas. Nat Genet. 2013;45:136–44. doi: 10.1038/ng.2503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Segal NH, Parsons DW, Peggs KS, Velculescu V, Kinzler KW, Vogelstein B, et al. Epitope landscape in breast and colorectal cancer. Cancer Res. 2008;68:889–92. doi: 10.1158/0008-5472.CAN-07-3095. [DOI] [PubMed] [Google Scholar]
- 23.van Rooij N, van Buuren MM, Philips D, Velds A, Toebes M, Heemskerk B, et al. Tumor exome analysis reveals neoantigen-specific T-cell reactivity in an ipilimumab-responsive melanoma. J Clin Oncol. 2013;31:e439–42. doi: 10.1200/JCO.2012.47.7521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Taube JM, Anders RA, Young GD, Xu H, Sharma R, McMiller TL, et al. Colocalization of inflammatory response with B7-h1 expression in human melanocytic lesions supports an adaptive resistance mechanism of immune escape. Sci Transl Med. 2012;4:127ra37. doi: 10.1126/scitranslmed.3003689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Spranger S, Spaapen RM, Zha Y, Williams J, Meng Y, Ha TT, et al. Up-regulation of PD-L1, IDO, and T(regs) in the melanoma tumor microenvironment is driven by CD8(+) T cells. Sci Transl Med. 2013;5:200ra116. doi: 10.1126/scitranslmed.3006504. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.