Abstract
The cellulose microfibril has more subtlety than is commonly recognized. Details of its structure may influence how matrix polysaccharides interact with its distinctive hydrophobic and hydrophilic surfaces to form a strong yet extensible structure. Recent advances in this field include the first structures of bacterial and plant cellulose synthases and revised estimates of microfibril structure, reduced from 36 to 18 chains. New results also indicate that cellulose interactions with xyloglucan are more limited than commonly believed, whereas pectin-cellulose interactions are more prevalent. Computational results indicate that xyloglucan binds tightest to the hydrophobic surface of cellulose microfibrils. Wall extensibility may be controlled at limited regions (“biomechanical hotspots”) where cellulose-cellulose contacts are made, potentially mediated by trace amounts of xyloglucan.
Introduction
Our concepts of plant cell wall structure, its synthesis and its dynamics are rapidly changing. Partly this is a result of accelerated identification of genes underlying the synthesis of cell wall components, e.g. [1-6], some of which are formidably complex multi-domain structures such as a proteoglycan covalently linked to pectin and xylan chains [7*]. Gene identification facilitates the identification of mutants, sometimes with phenotypes that surprise us, either because of lack of phenotype when current models demand one, or because of unanticipated phenotypes. Surprises lead to new insights about how a system functions. Gene identification has also led to fluorescent versions of wall-related proteins, allowing researchers to track their location and movement by confocal microscopy. This has elegantly revealed the dynamics of cellulose synthesis and provided a new means to identify components associated with the cellulose synthesis complex (CSC) [8, 9*,10]. Our ability to monitor specific matrix polysaccharides is more limited. While antibodies and carbohydrate binding modules have proved informative [11,12], epitope masking can be problematic [13]. Click labeling of pectins has given insights into pectin secretion and its aftermath [14*] while fluorescent monolignols enable new ways to monitor the process of lignification [15*,16].
Another key contributor to recent progress is use of advanced physical methods to probe whole cell walls. Defining the covalent structure of isolated cell wall components and the underlying genes is only the beginning of a molecular understanding of cell wall structure and its dynamic properties. We need to elucidate the interactions among wall components at a level of detail that X-ray crystallography can provide, but because walls lack crystalline order, other physical methods have to be used, sometimes in combination with computational approaches, e.g. [17*]. The goal is to build better cell wall models that go beyond merely illustrating prevailing concepts. Useful models provide testable predictions that lead to further insights into cell wall structure.
The following minireview summarizes selected recent advances in the construction of the primary cell wall. Regrettably, space limitations precluded discussion of many important advances, such as recent insights into the construction of the Casperian strip [18**-21], paradoxical results linking pectin demethylation with wall softening during morphogenesis at the shoot apical meristem [22-26], and new insights into lignin polymerization [27,28*], to name but a few recent developments related to cell wall biology.
Cellulose microfibril structure
Cellulose was first isolated and named nearly two centuries ago by Anselme Payen. It has vast economic value in the form of paper, textiles, wood products, polymers, animal feed, and biomass for energy uses, so it may come as a surprise to many biologists to learn that structural features of the cellulose microfibril, as well as its mechanism of synthesis, remain subjects of continuing research and debate. The cellulose microfibril is composed of numerous linear β1,4-linked glucan chains synthesized in parallel by protein complexes embedded in the plasma membrane. The chains are packed into an ordered microfibril of indefinite length and uncertain cross sectional area and shape. These geometrical features are important determinants of the physical properties of cellulose and its interaction with matrix components (Figure 1). For instance, the cellulose microfibril has distinct hydrophobic and hydrophilic surfaces which are thought to bind xyloglucan [29*], xylan [30*], and lignin [31] with different affinities and which are attacked by lytic enzymes and chemical treatments in different ways. Hopes to understand the molecular architecture of the cell wall and to develop realistic computational models of it depend on these microfibril features.
Figure 1.
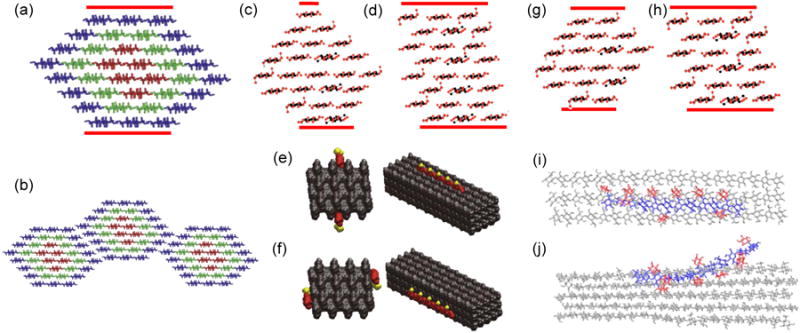
Potential shapes of cellulose microfibrils in cross section and impact on interactions with other wall components. Top row: potential cross sections of 36-, 24- and 18-chain microfibrils. The hydrophobic surface in each structure is indicated with the red lines. (a) Common depiction of a 36-chain microfibril cross section in a hexagonal shape. The colors represent chain mobility, with internal residues (red) more rigid than surface residues (blue). (b) According to Ding et al. [32,77], microfibrils may associate laterally via their hydrophilic surfaces. Others postulate preferential association via the hydrophobic surfaces. Two versions of a microfibril with 24-chain cross section are shown (c) in diamond shape or (d) in rectangular shape. Note how shape affects the proportion of hydrophilic and hydrophilic surfaces. (e) Busse-Wicher et al. [30*] illustrate two ways in which acetylated xylan (red residues with yellow acetyl groups) may bind to the hydrophilic surface (shown in two views) or (f) to the hydrophobic surface (likewise shown in two views). The two models in (e) and (f) make use of the 24-chain rectangular microfibril shown in (d). In (e) the xylan fits into the grooves of the hydrophilic surface of the rectangular microfibril, with the evenly-spaced acetyl groups exposed to the outside. This model would not work for the diamond-shaped structure – a striking example of how cellulose packing may affect interaction with matrix polysaccharides. Two versions of 18-chain cross sections are shown in (g) and in (h). The interaction of xyloglucan with the hydrophobic and hydrophilic surface of cellulose are illustrated in (i) and (j), based on MDS by Zhao et al. [29*]. Image credits: (a) and (b) adapted from Ding et al. [32], (i) and (j) from Zhao et al. [29], copyright Springer Verlag, used with permission. (c), (d), (g), (h) adapted from Fernandes et al. [33], copyright Proceedings of the National Academy of Sciences, used with permission. (e) and (f) from Busse-Wicher et al. [30*], copyright John Wiley & Sons, used with permission.
In recent years the microfibril has most often been represented as a hexagonal arrangement of 36 chains, e.g. [17*,32], but this is partly a rough guess based on estimates of microfibril diameter and partly speculation derived from the hexameric appearance of the ‘particle rosettes’ (the CSC) seen in TEM replica images of freeze-fractured plasma membranes. Like the Babylonian predilection for base-60 numerology (which is why an hour is divided into 60 minutes), the number of chains in the cellulose microfibril is often cited as 6 × 6, an idea stemming from the hexameric appearance of CSCs, the seductive appeal of a ‘hexamer of hexamers’ concept, and the limited resolution of traditional methods for giving precise chain numbers in a microfibril of ∼3 nm width. Technical improvements have steadily decreased published estimates of microfibril size and application of advanced physical methods, combined with modeling, have recently led to yet smaller estimates, in the range of 18-24 chains [33]. Thomas et al. [34*] used wide-angle x-ray scattering (WAXS), small-angle neutron scattering (SANS), solid-state nuclear magnetic resonance (ssNMR), polarized FTIR and other methods to examine cellulose isolated from celery collenchyma. They concluded that 18-24 chain models were the likeliest fit to the results, with a slight nod towards a 24-chain model. They also found evidence for microfibril aggregation. The experimental results are also compatible with chain numbers in between 18 and 24 (e.g. 20 or 21), but the appearance of hexameric rosettes biases models towards chain numbers that are multiples of six. Newman et al. [35*] analyzed cellulose from primary cell walls of mung bean by synchrotron WAXS. The results, when combined with computational diffractograms and published ssNMR data, best fit an 18-chain model in which microfibrils assumed a variety of cross sectional shapes and occasionally ‘twinned’, meaning regions of two microfibrils coalesced. These twinned regions may be the aggregated regions noted by Thomas et al. [34] and are perhaps related to the ‘biomechanical hotspots’ or regions of microfibril contact in primary cell walls, discussed below.
One consequence of an 18-chain microfibril is that the individual particles in the hexameric rosette could contain as few as three cellulose synthases (CESAs). This would neatly fit genetic results indicating that concurrent expression of three different CESA genes is normally needed for cellulose synthesis and, to add icing on the cake, it would provide a simple mechanism for self assembly of CESAs into a trimeric complex (=one particle) and then into rosettes (Figure 2) [35*].
Figure 2.
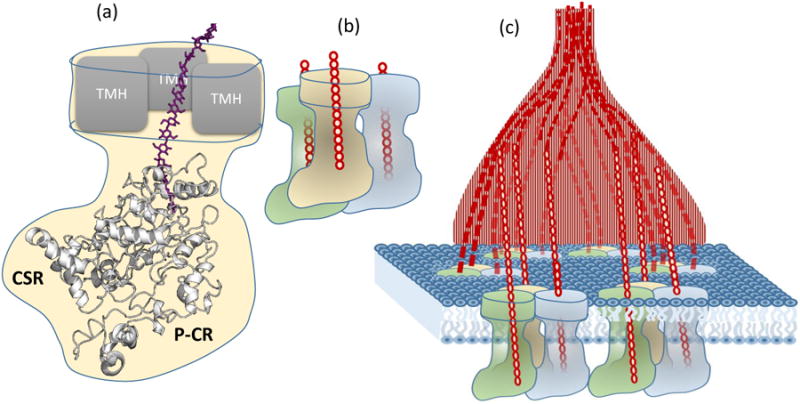
Updated models of plant CESAs and CSCs. (a) A computational model of a plant CESA catalytic domain with P-CR and CSR regions (light grey). The glucan chain (purple) is from the homologous Rhodobacter structure. The location of the transmembrane helices (TMH) is represented with grey boxes. (b) Three CESAs, encoded by three different genes, may interact to form a trimeric particle, which in turn may assemble into a hexameric rosette, depicted in (c). The glucan chains are represented in red. Image (a) is adapted from Slaubaugh et al. [37] and is courtesy of Jonathan Davis.
Cellulose microfibril synthesis & guidance
Major advances in understanding the mechanism of cellulose chain polymerization were recently achieved with the astonishing solution of a Rhodobacter cellulose synthase by X-ray crystallography [36**] and computational modeling of the catalytic domain of plant CESAs [37, 38*,39]. The Rhodobacter structure, composed of two proteins, includes the catalytic domain, the transmembrane domain and the intra-protein tunnel that provides a low-energy pathway for translocating the growing glucan chain to the external membrane surface from the cytoplasmic side, where the catalytic site transfers a glucose residue from UDP-glucose to the reducing end of the glucan. Electron density compatible with an 18-residue glucan was found within the protein tunnel – a very sweet result, as it enabled structural details of the translocation mechanism to be discerned. Compared with the long and troubled history of unstable and inactive CESA preparations from plant tissues, it is a remarkable and fortunate result that the two proteins making the Rhodobacter complex proved sufficient for cellulose synthesis in vitro [40]. Based on the structure, a plausible catalytic scheme was presented in which the newly-added glucose residue would rotate 180° between each step in the synthesis, shaping the chain into a linear two-fold helix for translocation through the tunnel. In plants multiple chains coalesce outside the plasma membrane to form a microfibril, but Rhodobacter is not known to form microfibrils. This protein structure lays to rest previous debate about the number of active sites needed for glucan polymerization [41]; one is evidently sufficient.
Prior to publication of the Rhodobacter structure, Sethaphong et al. [38*] developed a computational model of the catalytic domain of a plant CESA (Figure 2a). Despite low sequence similarity, the two protein folds showed structural congruence, indicating functional and evolutionary similarity. Several CESA missense mutations were mapped to protein regions close to the catalytic site. Unlike bacterial homologs, plant CESAs also contain distinctive regions named P-CR and CSR that may participate in formation of the multimeric CSC. These regions were modeled as loops external to the catalytic fold, forming potential interfaces between CESAs. Using protein threading based on the Rhodobacter structure, Olek et al. [39] likewise developed a CESA model, but their approach yielded wildly varying PCR and CSR structures. They reported successful recombinant expression of a CESA catalytic domain that formed homodimers in vitro. SAXS data were interpreted to mean the monomeric protein formed a warped boomerang-shaped structure with the catalytic domain in the middle and the P-CR and CSR domains at opposite poles, differing from the prediction of Sethaphong et al. [38]. Further experimental work will be needed to decide which of these predicted structures is closer to reality and whether CESAs indeed form a homodimer within functional CSCs, as predicted by Olek et al. [39]. A dimer-based structure would almost certainly mean six CESAs per particle or 36 CESAs per rosette. If 18 is the correct number of chains in the microfibril as produced by a single CSC, then half the CESAs would be inactive at any given instant. There is evidence that microfibril diameter may vary developmentally and across species [35,42-44], so a CSC with an ability to synthesize microfibrils of variable chain number would provide a creative solution to what is otherwise a vexing problem: how to account for variable microfibril diameters with a common CSC structure? Perhaps with a 36-cylinder engine that idles some cylinders. An alternative possibility is that microfibril diameters vary by other, post-synthetic mechanisms, e.g. by trimming of nascent microfibrils by endoglucanases [9,10,45] or by microfibril aggregation.
To understand the initial stages of microfibril formation (largely terra incognita), Haigler et al. [42] combined molecular dynamics simulation (MDS) with freeze-fracture TEM of Zinnia mesophyll cells transdifferentiating into tracheary elements. MDS was limited to 6 chains and did not include water in the simulation, so it must be regarded as a limited first step in approaching this problem. The fibrils were highly disordered, probably as a result of the limited number of chains used for the simulation. Other work indicates that 6 chains may be too few to form a stable structure resembling native cellulose [46]. One novel result to emerge from the MDS was the accumulation of noncrystallized glucan chains at the outer surface of the CSC prior to formation of a protofibril. TEM images showed structures interpreted as nascent microfibrils, with diameters varying from <2 to >5 nm and at their base were swollen regions interpreted as pools of disorganized glucans. The authors point out that such pools would buffer between the crystallization process and the concerted polymerization process by the 18-36 CESAs that contribute to the microfibril. It is unclear how a pool of disorganized glucan would spontaneously form the precise crystalline form (known as cellulose Iβ) that is dominant in plant cell walls. Crystallization may be a spontaneous process [44] or possibly it is a guided process in which cellulose-binding proteins such as CHITANASE-LIKE1 [47] and BRITTLE CULM1 [48] facilitate microfibril assembly.
Although cellulose is often described as a crystalline structure, primary wall cellulose has low crystallinity. This stems in part from the thinness of the microfibril and the conformational disorder of surface chains, which are more dynamic and flexible than the more-constrained internal chains, and in part from sloppy packing of internal chains [34]. The internal disorder may arise from twisting of the microfibril and the resulting need to periodically relieve the internal stress [46]. It may also arise from entrapment of xyloglucan within the microfibril – an attractive idea that dates from the 1980's [49] but still has only circumstantial support. A recent study confirmed the selective effect of xyloglucan on the crystallinity of cellulose formed by Gluconoacetobacter [50]. This study detected crystalline cellulose by SFG (sum frequency generation) spectroscopy, a nonlinear optical method that can provide information about the crystallinity and meso-scale ordering of microfibrils in intact cell walls [51]. Because SFG is selective for crystalline cellulose, whole cell walls may be measured without the need to extract lignin, hemicelluloses and other wall materials that interfere with most common physical methods of cell wall analysis. However, primary cell walls typically give rather weak SFG signals, probably because of low crystallinity and high dispersion of cellulose microfibrils compared with the dense, parallel packing of cellulose in secondary cell walls.
Cellulose alignment in the wall
Microfibril orientation has long been considered a major determinant of the directionality of cell growth [52*-54], and microtubules have been implicated in microfibril alignment since Green's seminal study showing that microtubule manipulations affect cellulose orientation [55], but the mechanism has been uncertain. Recent advances have identified a physical linker between CESA and cortical microtubules: it is CSI1 [56,57*], a large protein (2151 amino acids) that binds to CESA and to microtubules. Given its large size and multiple domains, CSI1 may have other functions as well [58]. Growth of the csi1 mutant is reduced and cells in the stem become twisted [59]. Although microtubule guidance is lost in the csi1 mutant, the CESA trajectories in the plasma membrane do not become random, but tend toward a transverse orientation, suggesting another, as yet unknown, guidance system may operate in the absence of functional CSI1.
Mechanical coupling of cellulose microfibrils by xyloglucan and pectins?
Since the early 1970's xyloglucan has been considered an essential structural component whose metabolism is central to concepts of wall loosening and induction of growth by auxin and other hormones [49]. Its putative role in wall structure and mechanics is emphasized in depictions of cell walls as nearly parallel arrays of cellulose microfibrils tethered together by long, extended xyloglucan chains that coat the cellulose surface (to prevent cellulose-cellulose contacts) and mechanically link adjacent microfibrils together. This tethered network model, which has held sway for more than two decades, depicts several untested aspects of cell wall architecture. Its validity was seriously shaken by the discovery that an Arabidopsis double mutant (xxt1/xxt2) lacked detectable xyloglucan, yet displayed only relatively minor growth reduction [60]. XXT1 and XXT2 are xylosyl transferases that add xylose side chains to the glucan backbone of xyloglucan. The minor phenotype was shocking because xyloglucan was considered a central and essential structural element of primary walls. Hypocotyls from the xxt1/xxt2 line were ∼30% weaker than WT (measured as breaking strength or as stiffness). Yet despite this mechanically weaker condition, the cell walls were less extensible in creep assays [61]. Cell walls from the mutant displayed reduced sensitivity to wall loosening by α-expansins, accounting for the reduced growth and extensibility (creep) of the cell walls. Moreover the walls of the mutant were more sensitive to loosening by treatments that loosened pectins and xylans, indicating that other components of the matrix assumed a larger mechanical role in the xyloglucan-deficient walls. This mutant has prompted a reappraisal of the role of xyloglucan in primary wall structure.
Analyses of WT Arabidopsis cell walls by multidimensional ssNMR indicate that only a small proportion of the cellulose surface is in contact with xyloglucan [62,63], in contrast to the common view that most of the cellulose surface is coated with xyloglucan and prevents direct cellulose-cellulose contacts [49]. Instead of xyloglucan, pectic sugars (most likely from rhamnogalacturonan-I) were close to the cellulose surface (within spin diffusion distance, ∼1 nm). These and other results have led to the proposal that pectins serve as mechanical tethers between microfibrils [62,64], in parallel with xyloglucan. One difficulty with this notion is that the acidic pectic polymers do not bind appreciably to cellulose surfaces in vitro [65], so they would seem to make feeble tethers. Moreover, treatments that lyse or solubilize pectins cause negligible wall loosening in WT Arabidopsis cell wall, as measured by induction of cell wall creep [61]. The pectin-cellulose proximity detected by ssNMR might result from molecular crowding within the cell wall. Alternatively, the neutral pectin side chains of rhamnogalacturonan-I (i.e. galactans and arabinans), which display intermediate binding affinity to cellulose in vitro [65,66], might draw rhamnogalacturonan-1 close to cellulose. However, such direct interactions have not yet been demonstrated by ssNMR or other methods. Thus the hypothesis of mechanical tethering by pectins needs further testing.
Pectins are the most dynamic (mobile) polymers in the wall and their hydrophilic character likely helps to reduce direct cellulose-cellulose contacts, lubricating microfibril motions as the cell wall expands. Based on ssNMR spin diffusion, ∼50% of the cellulose surface makes contact with pectin [67]. Partial extraction of pectin from Arabidopsis cell walls rigidified the remaining matrix polysaccharides [67] and greatly slowed magnetic spin transfer from water to cellulose [68]. The latter result was attributed to water stabilization by pectins (spin transfer is reduced by rapid motions of water). These results are consistent with a picture of cellulose intimately coated by pectins (about half of its surface area, perhaps only by weak interactions) and limited coating by xyloglucan.
Unsolved issues include the location of the xyloglucan (it comprises ∼20% of the Arabidopsis wall) and why, with its high affinity for cellulose surfaces, it does not coat more of the cellulose surfaces. One possibility is that xyloglucan assumes a random coil shape [69], rather than the extended conformation commonly depicted. Upon secretion, a coiled xyloglucan may immediately bind to the first bare cellulose surface it contacts, resulting in limited contact with cellulose, but extensive contact with the mobile pectins, consistent with ssNMR results [63]. In-vitro experiments indicate xyloglucan binding to cellulose is irreversible, so an anchored xyloglucan chain might have little ability to reposition itself, particularly in view of dense crowding by pectins and other components in the wall.
These considerations highlight the fact that some of our conventional ideas about cell wall structure and assembly are influenced by the results of in-vitro binding experiments in which dilute solutions of polymers are allowed to bind to cellulose surfaces. In contrast, the living cell wall is assembled in organized layers (lamellae) in a confined space where polymer crowding, entanglements, entrapment, competing interactions, and enzymatic modifications may greatly influence the interactions among wall components.
Xyloglucan tethering, direct cellulose contacts and biomechanical hotspots
The concept of microfibril tethering by xyloglucan was tested by an enzymatic approach in which cell walls were clamped in a constant-force extensometer and treated with a series of GH12 endoglucanases with differing substrate specificities [70*]. Xyloglucan-specific endoglucanase digested much of the xyloglucan from the wall, but did not weaken the walls. Only enzymes able to cut both xyloglucan and cellulose were effective in loosening the wall. Remarkably, a mixture of cellulose-specific and xyloglucan-specific endoglucanases did not loosen the cell wall. The proposed explanation of these enigmatic results is the ‘biomechanical hotspot’ concept: that wall extension is controlled at limited sites of close contact between cellulose microfibrils, mediated by xyloglucan chains. Xyloglucans may be intertwined with cellulose chains in these inaccessible sites, forming an amalgam that requires an enzyme with both cellulase and xyloglucanase activity to digest. Alternatively xyloglucan in these sites may be rendered inaccessible by forming a tight monomolecular junction between two or more cellulose microfibrils. Only enzymes that could digest both cellulose and xyloglucan would be able to progressively digest the intermingled or appressed xyloglucan-cellulose chains that bonded the microfibrils together. This concept was tested in a computational model in which xyloglucan was configured as a bonding agent between two cellulose microfibrils [29*]. The result showed that a tight sandwich-like structure was formed between the hydrophobic surfaces of microfibrils. This junction was energetically stable and strong enough to withstand substantial force when the microfibrils were pulled apart, strong enough to withstand the tensile forces generated by cell turgor pressure.
Cells walls lacking xyloglucan showed diminished creep responses to endoglucanases and to expansins as well, leading to the suggestion that these ‘biomechanical hotspots’ are also targets of expansin action [70]. Expansins are wall-loosening proteins that induce cell wall extension without lytic breakdown of the wall components [71]. Expansin binding sites in complex Arabidopsis walls were recently characterized by ssNMR [72*] with use of method called dynamic nuclear polarization to increase sensitivity. Interpretation was aided by use of expansin mutants with altered binding to cellulose and pectin. The results showed the effective target to be cellulose with a slightly smaller NMR shift compared with bulk cellulose, indicating a slightly different configuration of the internal chains. The expansin binding site was also in close proximity to xyloglucan. This structure is remarkably similar to the proposed site of endoglucanases-catalyzed wall loosening, i.e. the biomechanical hotspots.
According to the hotspot hypothesis, cellulose microfibrils are linked with one another via direct load-bearing junctions, mediated by intimate bonding by xyloglucan in some scenarios. This idea is at odds with common depictions of primary cell walls which show well-spaced microfibrils kept apart by matrix polysaccharides. The view that microfibrils do not make direct contact may stem from an early depiction of the primary cell wall by Frey-Wyssling [73]. This aspect of wall structure was incorporated into the influential molecular model of cell walls of sycamore suspension cell cultures by Keegstra et al. [74] and was also included into most subsequent models. In contrast to these depictions of wall structure, many high-resolution micrographs of primary cell walls show aggregation or bundling of microfibrils. In some cases it might be argued that microfibril aggregation occurs because of extraction or dehydration of the sample in preparation for imaging [75], but in a recent study such technical problems were avoided by use of atomic force microscopy (AFM) to image the newly-deposited surface of unextracted, never-dried walls under water [76]. This AFM-based method enhances microfibril visibility, perhaps because the AFM tip passes through a surface layer of hydrated pectins which obscure EM-based methods. Microfibrils were seen to merge into and out of short junctions where they come into close contact with one another (Figure 3). Are such junctions the biomechanical hotspots described above? We do not know yet. The effective scale of this network is in 100-500 nm range, whereas the molecular depictions of cell wall structure are at a much smaller scale, 10-50 nm range. The hotspot concept brings the implication that wall extensibility is controlled by limited, specific junctions between microfibrils rather than the bulk viscoelasticity of the matrix. Such a mechanism could provide cells with a finer means for dynamic control of their growth.
Figure 3.
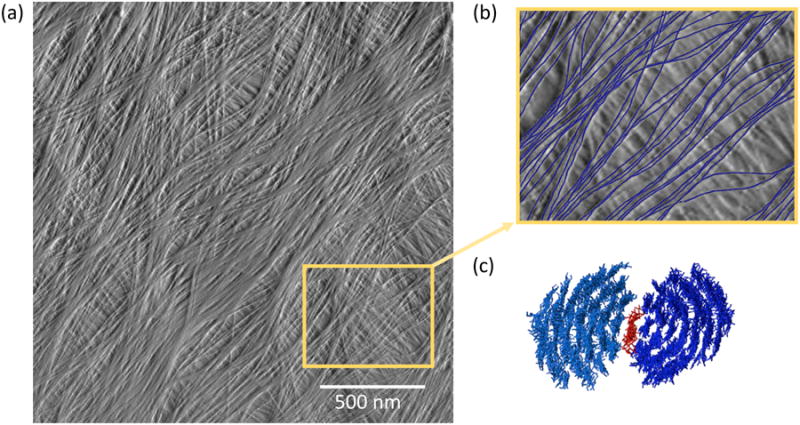
(a) Multilayered arrangement of cellulose microfibrils at the cell wall surface from onion scale, visualized by atomic force microscopy under water, without extraction or drying of the sample. (b) A close up from (a), with the microfibrils in the surface layer drawn in blue. Note that microfibrils merge into and out of regions of close contract. (c) One potential arrangement in which xyloglucan (red) serves as a monomolecular adhesive between the hydrophobic surfaces of two cellulose microfibrils (blue). Image credit: (a) is from Zhang et al. [76], (c) is from Zhao et al. [29*], both copyright Springer Verlag and used with permission.
This concept of wall mechanics raises numerous questions. How are the biomechanical hotspots formed? Do cells have specific molecular mechanisms to control the density and location of such structures, or are they formed by a stochastic process requiring ‘accidental’ co-localization of nascent microfibrils in proximity with appropriate matrix components? Are hotspots destroyed in the course of cell wall extension? Are they regenerated? What kinds of microfibril motions are limited by the hotspots, i.e. lateral separation (unzipping) of microfibrils or slippage (sliding) of microfibrils? Many growing cell walls are assemblages of multiple lamellae, each made up of a monolayer of microfibrils approximately oriented in the same direction, but with different orientations in each lamella. Cell wall expansion in such a structure likely entails a combination of lateral separation and sliding of microfibrils in the different lamellae, potentially with different structures limiting each type of microfibril movement.
Some of the concepts discussed in this review are summarized in Figure 4, which is a colorized and ornamented version of the AFM image shown in Figure 3b. Microfibrils in the surface-most lamella are traced in blue. Regions where microfibrils make close contact with one another are given red highlights (potential biomechanical hotspots). Because matrix polymers are not well visualized in this AFM image, they were added at plausible sites based on their abundance, likely conformation and in-muro interactions as detected by NMR studies (green=xyloglucans; yellow=pectins), but these features of the figure must be considered as speculative placeholders until additional data help to refine their location and conformations.
Figure 4.
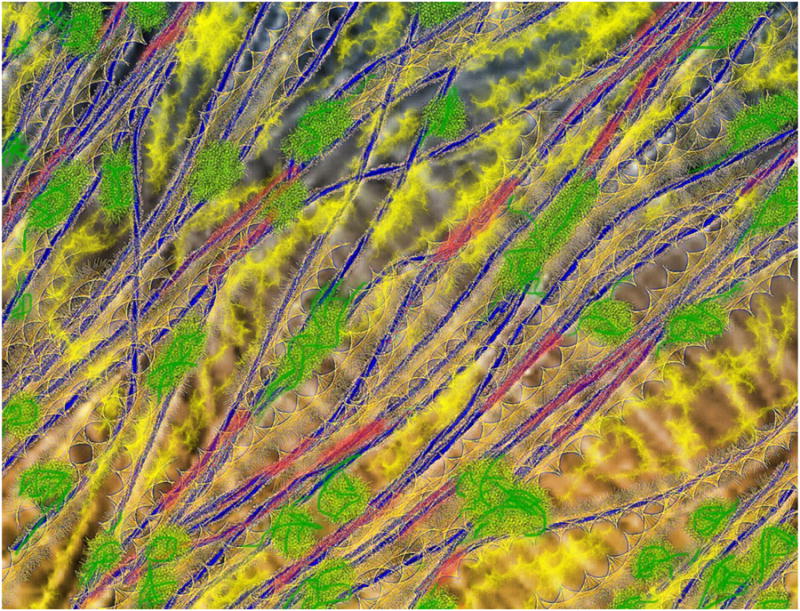
Artistic depiction of the cell wall, based on the microfibril arrangement shown in Figure 3(b). Cellulose microfibrils were traced in blue. Red regions indicate potential biomechanical hotspots where two or more microfibrils merge into close contact. Highly dispersed, mobile pectins are represented in yellow (different textures to indicate different pectic domains) whereas xyloglucans are shown as green coiled structures anchored to microfibril surfaces at limited locations.
Conclusions
Concepts of the identity, spatial distribution and scale of the load-bearing elements that restrict expansion of the cell wall are continuing to evolve. Relatively limited contact points between cellulose microfibrils may be key sites of wall loosening, and their creation may originate with the formation of the cellulose microfibril and its distinctive interaction with matrix polymers on hydrophobic and hydrophilic surfaces and noncrystalline regions. Molecular models of cell walls need to incorporate these physical and biomechanical aspects of cell wall architecture and to undergo critical testing and refinement by a combination of approaches in order to bring the next advance in our understanding of mechanisms by which cells control and limit cell wall expansion.
Highlights.
Cellulose interactions with the matrix depends on microfibril shape.
Physical data point to a microfibril made of 18 to 24 chains, not 36 chains.
Recent structures for cellulose synthases yield insights into glucan polymerization.
Revised roles for xyloglucan and pectin in wall structure are emerging.
Direct contact between cellulose microfibrils may control wall extensibility.
Acknowledgments
This work is supported as part of The Center for Lignocellulose Structure and Formation, an Energy Frontier Research Center funded by the U.S. Department of Energy, Office of Science, Office of Basic Energy Sciences under Award Number DE-SC0001090.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References and recommended reading
- 1.Atmodjo MA, Sakuragi Y, Zhu X, Burrell AJ, Mohanty SS, Atwood JA, 3rd, Orlando R, Scheller HV, Mohnen D. Galacturonosyltransferase (GAUT)1 and GAUT7 are the core of a plant cell wall pectin biosynthetic homogalacturonan:galacturonosyltransferase complex. Proc Natl Acad Sci U S A. 2011;108:20225–20230. doi: 10.1073/pnas.1112816108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bromley JR, Busse-Wicher M, Tryfona T, Mortimer JC, Zhang Z, Brown DM, Dupree P. GUX1 and GUX2 glucuronyltransferases decorate distinct domains of glucuronoxylan with different substitution patterns. Plant J. 2013;74:423–434. doi: 10.1111/tpj.12135. [DOI] [PubMed] [Google Scholar]
- 3.Schultink A, Cheng K, Park YB, Cosgrove DJ, Pauly M. The identification of two arabinosyltransferases from tomato reveals functional equivalency of xyloglucan side chain substituents. Plant Physiol. 2013;163:86–94. doi: 10.1104/pp.113.221788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.de Souza A, Hull PA, Gille S, Pauly M. Identification and functional characterization of the distinct plant pectin esterases PAE8 and PAE9 and their deletion mutants. Planta. 2014 doi: 10.1007/s00425-014-2139-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zabotina OA, Avci U, Cavalier D, Pattathil S, Chou YH, Eberhard S, Danhof L, Keegstra K, Hahn MG. Mutations in multiple XXT genes of Arabidopsis reveal the complexity of xyloglucan biosynthesis. Plant Physiol. 2012;159:1367–1384. doi: 10.1104/pp.112.198119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hijazi M, Velasquez SM, Jamet E, Estevez JM, Albenne C. An update on post-translational modifications of hydroxyproline-rich glycoproteins: toward a model highlighting their contribution to plant cell wall architecture. Front Plant Sci. 2014;5:395. doi: 10.3389/fpls.2014.00395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7*.Tan L, Eberhard S, Pattathil S, Warder C, Glushka J, Yuan C, Hao Z, Zhu X, Avci U, Miller JS, et al. An arabidopsis cell wall proteoglycan consists of pectin and arabinoxylan covalently linked to an arabinogalactan protein. Plant Cell. 2013;25:270–287. doi: 10.1105/tpc.112.107334. This tour-de-force biochemical characterization shows that an arabinogalactan protein is covalently linked to pectin and xylan domains and postulates that it might cross link cell walls. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Miart F, Desprez T, Biot E, Morin H, Belcram K, Hofte H, Gonneau M, Vernhettes S. Spatio-temporal analysis of cellulose synthesis during cell plate formation in Arabidopsis. Plant J. 2013 doi: 10.1111/tpj.12362. [DOI] [PubMed] [Google Scholar]
- 9*.Vain T, Crowell EF, Timpano H, Biot E, Desprez T, Mansoori N, Trindade LM, Pagant S, Robert S, Hofte H, et al. The cellulase KORRIGAN is part of the cellulose synthase complex. Plant Physiol. 2014;165:1521–1532. doi: 10.1104/pp.114.241216. The KOR endoglucanase is associated, perhaps transiently, with the CSC and influences its intracellular trafficking. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lei L, Zhang T, Strasser R, Lee CM, Gonneau M, Mach L, Vernhettes S, Kim SH, Cosgrove DJ, Li S, et al. The jiaoyao1 mutant is an allele of korrigan1 that abolishes endoglucanase activity and affects the organization of both cellulose microfibrils and microtubules in Arabidopsis. Plant Cell. 2014;26:2601–2616. doi: 10.1105/tpc.114.126193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McCann MC, Knox JP. Plant cell wall biology: Polysaccharides in architectural and developmental contexts. Annual Plant Reviews. 2011;41:343–366. [Google Scholar]
- 12.Gilbert HJ, Knox JP, Boraston AB. Advances in understanding the molecular basis of plant cell wall polysaccharide recognition by carbohydrate-binding modules. Curr Opin Struct Biol. 2013;23:669–677. doi: 10.1016/j.sbi.2013.05.005. [DOI] [PubMed] [Google Scholar]
- 13.Xue J, Bosch M, Knox JP. Heterogeneity and glycan masking of cell wall microstructures in the stems of Miscanthus × giganteus, and its parents M. sinensis and M. sacchariflorus. PLoS One. 2013;8:e82114. doi: 10.1371/journal.pone.0082114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14*.Anderson CT, Wallace IS, Somerville CR. Metabolic click-labeling with a fucose analog reveals pectin delivery, architecture, and dynamics in Arabidopsis cell walls. Proc Natl Acad Sci U S A. 2012;109:1329–1334. doi: 10.1073/pnas.1120429109. Pulse labeling of pectins by ‘click’ chemistry enabled the delivery of pectins to the cell wall and their subsequent spread to be visualized with confocal microscropy. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15*.Tobimatsu Y, Wagner A, Donaldson L, Mitra P, Niculaes C, Dima O, Kim JI, Anderson N, Loque D, Boerjan W, et al. Visualization of plant cell wall lignification using fluorescence-tagged monolignols. Plant J. 2013;76:357–366. doi: 10.1111/tpj.12299. This technical development provides a new way to monitor lignin polymerization. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tobimatsu Y, Wouwer DVd, Allen E, Kumpf R, Vanholme B, Boerjan W, Ralph J. A click chemistry strategy for visualization of plant cell wall lignification. Chemical Communications. 2014;50:12262–12265. doi: 10.1039/c4cc04692g. [DOI] [PubMed] [Google Scholar]
- 17*.Langan P, Petridis L, O'Neill HM, Pingali SV, Foston M, Nishiyama Y, Schulz R, Lindner B, Hanson BL, Harton S, et al. Common processes drive the thermochemical pretreatment of lignocellulosic biomass. Green Chemistry. 2014;16:63–68. This study combines physical analysis with computational approaches to understand the restructuring of secondary cell walls upon thermochemical treatment to reduce recalcitrance for biofuel production. The discussion emphasizes the role of water in cell wall structure and present attractive images, based on computational analsis, of possible cellulose-matrix interactions. [Google Scholar]
- 18**.Lee Y, Rubio MC, Alassimone J, Geldner N. A mechanism for localized lignin deposition in the endodermis. Cell. 2013;153:402–412. doi: 10.1016/j.cell.2013.02.045. The Casperian strip is a narrow hydrophobic strip of cell wall in the endodermis. This study shows how membrane NADPH oxidases are brought together with cell wall peroxidases, via the action of membrane proteins (CASPs), to cause the local formation of lignin (not suberin, as textbooks state) [DOI] [PubMed] [Google Scholar]
- 19.Hosmani PS, Kamiya T, Danku J, Naseer S, Geldner N, Guerinot ML, Salt DE. Dirigent domain-containing protein is part of the machinery required for formation of the lignin-based Casparian strip in the root. Proc Natl Acad Sci U S A. 2013;110:14498–14503. doi: 10.1073/pnas.1308412110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Geldner N. The endodermis. Annu Rev Plant Biol. 2013;64:531–558. doi: 10.1146/annurev-arplant-050312-120050. [DOI] [PubMed] [Google Scholar]
- 21.Naseer S, Lee Y, Lapierre C, Franke R, Nawrath C, Geldner N. Casparian strip diffusion barrier in Arabidopsis is made of a lignin polymer without suberin. Proc Natl Acad Sci U S A. 2012;109:10101–10106. doi: 10.1073/pnas.1205726109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Braybrook SA, Peaucelle A. Mechano-chemical aspects of organ formation in Arabidopsis thaliana: the relationship between auxin and pectin. PLoS One. 2013;8:e57813. doi: 10.1371/journal.pone.0057813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wolf S, Mravec J, Greiner S, Mouille G, Hofte H. Plant cell wall homeostasis is mediated by brassinosteroid feedback signaling. Curr Biol. 2012;22:1732–1737. doi: 10.1016/j.cub.2012.07.036. [DOI] [PubMed] [Google Scholar]
- 24.Peaucelle A, Braybrook SA, Le Guillou L, Bron E, Kuhlemeier C, Hofte H. Pectin-induced changes in cell wall mechanics underlie organ initiation in Arabidopsis. Curr Biol. 2011;21:1720–1726. doi: 10.1016/j.cub.2011.08.057. [DOI] [PubMed] [Google Scholar]
- 25.Routier-Kierzkowska AL, Weber A, Kochova P, Felekis D, Nelson BJ, Kuhlemeier C, Smith RS. Cellular force microscopy for in vivo measurements of plant tissue mechanics. Plant Physiol. 2012;158:1514–1522. doi: 10.1104/pp.111.191460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kierzkowski D, Nakayama N, Routier-Kierzkowska AL, Weber A, Bayer E, Schorderet M, Reinhardt D, Kuhlemeier C, Smith RS. Elastic domains regulate growth and organogenesis in the plant shoot apical meristem. Science. 2012;335:1096–1099. doi: 10.1126/science.1213100. [DOI] [PubMed] [Google Scholar]
- 27.Wilkerson CG, Mansfield SD, Lu F, Withers S, Park JY, Karlen SD, Gonzales-Vigil E, Padmakshan D, Unda F, Rencoret J, et al. Monolignol ferulate transferase introduces chemically labile linkages into the lignin backbone. Science. 2014;344:90–93. doi: 10.1126/science.1250161. [DOI] [PubMed] [Google Scholar]
- 28*.Bonawitz ND, Kim JI, Tobimatsu Y, Ciesielski PN, Anderson NA, Ximenes E, Maeda J, Ralph J, Donohoe BS, Ladisch M, et al. Disruption of Mediator rescues the stunted growth of a lignin-deficient Arabidopsis mutant. Nature. 2014 doi: 10.1038/nature13084. A common phenotype of genetic lesions that disrupt lignin formation is a rather sick plant. This study shows that many symptoms of such sickness can be rescued by mutation of a MED transcriptional complex, which normally regulates phenylpropanoid homeostasis. [DOI] [PubMed] [Google Scholar]
- 29*.Zhao Z, Crespi VH, Kubicki JD, Cosgrove DJ, Zhong L. Molecular dynamics simulation study of xyloglucan adsorption on cellulose surfaces: effects of surface hydrophobicity and side-chain variation. Cellulose. 2014;21:1025–1039. Xyloglucan binds much more tightly to the hydrophobic surface of cellulose microfibrils and through this surface can bind microfibrils together through a strong, monomolecular layer that dehydrates the cellulose and xyloglucan surfaces. [Google Scholar]
- 30*.Busse-Wicher M, Gomes TC, Tryfona T, Nikolovski N, Stott K, Grantham NJ, Bolam DN, Skaf MS, Dupree P. The pattern of xylan acetylation suggests xylan may interact with cellulose microfibrils as a twofold helical screw in the secondary plant cell wall of Arabidopsis thaliana. Plant J. 2014;79:492–506. doi: 10.1111/tpj.12575. Biochemical analysis of xylans is combined with computational analysis to create new concepts about the potential interactions of xylans with cellulose surfaces. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Besombes S, Mazeau K. The cellulose/lignin assembly assessed by molecular modeling. Part 2: Seeking for evidence of organization of lignin molecules at the interface with cellulose. Plant Physiol Biochem. 2005;43:277–286. doi: 10.1016/j.plaphy.2005.02.004. [DOI] [PubMed] [Google Scholar]
- 32.Ding SY, Zhao S, Zeng Y. Size, shape, and arrangement of native cellulose fibrils in maize cell walls. Cellulose. 2014;21:863–871. [Google Scholar]
- 33.Fernandes AN, Thomas LH, Altaner CM, Callow P, Forsyth VT, Apperley DC, Kennedy CJ, Jarvis MC. Nanostructure of cellulose microfibrils in spruce wood. Proc Natl Acad Sci U S A. 2011;108:E1195–1203. doi: 10.1073/pnas.1108942108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34*.Thomas LH, Forsyth VT, Sturcova A, Kennedy CJ, May RP, Altaner CM, Apperley DC, Wess TJ, Jarvis MC. Structure of cellulose microfibrils in primary cell walls from collenchyma. Plant Physiol. 2013;161:465–476. doi: 10.1104/pp.112.206359. Like the study by Fernandes et al. (2011), this study combines several physical techniques to assess cellulose microfibril structure, coming to the conclusion that 18- to 24-chain microfibrils provide best fits to the data. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35*.Newman RH, Hill SJ, Harris PJ. Wide-angle x-ray scattering and solid-state nuclear magnetic resonance data combined to test models for cellulose microfibrils in mung bean cell walls. Plant Physiol. 2013;163:1558–1567. doi: 10.1104/pp.113.228262. Physical analyses shows that a 36-chain model of cellulose microfibril provides a poorer fit to the data than an 18-chain model. This consequences of this idea are extended to the structure of the CSC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36**.Morgan JL, Strumillo J, Zimmer J. Crystallographic snapshot of cellulose synthesis and membrane translocation. Nature. 2013;493:181–186. doi: 10.1038/nature11744. A landmark advance in our understanding of how cellulose is synthesized. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Slabaugh E, Davis JK, Haigler CH, Yingling YG, Zimmer J. Cellulose synthases: new insights from crystallography and modeling. Trends Plant Sci. 2014;19:99–106. doi: 10.1016/j.tplants.2013.09.009. [DOI] [PubMed] [Google Scholar]
- 38*.Sethaphong L, Haigler CH, Kubicki JD, Zimmer J, Bonetta D, DeBolt S, Yingling YG. Tertiary model of a plant cellulose synthase. Proc Natl Acad Sci U S A. 2013;110:7512–7517. doi: 10.1073/pnas.1301027110. This computational study provides the first detailed structral model of the catalytic domain of CESA and offers ideas about how the CSC may be constructed. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Olek AT, Rayon C, Makowski L, Kim HR, Ciesielski P, Badger J, Paul LN, Ghosh S, Kihara D, Crowley M, et al. The structure of the catalytic domain of a plant cellulose synthase and its assembly into dimers. Plant Cell. 2014;26:2996–3009. doi: 10.1105/tpc.114.126862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Omadjela O, Narahari A, Strumillo J, Melida H, Mazur O, Bulone V, Zimmer J. BcsA and BcsB form the catalytically active core of bacterial cellulose synthase sufficient for in vitro cellulose synthesis. Proc Natl Acad Sci U S A. 2013;110:17856–17861. doi: 10.1073/pnas.1314063110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Carpita NC. Update on mechanisms of plant cell wall biosynthesis: how plants make cellulose and other (1->4)-beta-D-glycans. Plant Physiology. 2011;155:171–184. doi: 10.1104/pp.110.163360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Haigler CH, Grimson MJ, Gervais J, Le Moigne N, Hofte H, Monasse B, Navard P. Molecular modeling and imaging of initial stages of cellulose fibril assembly: evidence for a disordered intermediate stage. PLoS One. 2014;9:e93981. doi: 10.1371/journal.pone.0093981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jarvis MC. Cellulose biosynthesis: counting the chains. Plant Physiol. 2013;163:1485–1486. doi: 10.1104/pp.113.231092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Guerriero G, Fugelstad J, Bulone V. What do we really know about cellulose biosynthesis in higher plants? J Integrative Plant Biology. 2010;52:161–175. doi: 10.1111/j.1744-7909.2010.00935.x. [DOI] [PubMed] [Google Scholar]
- 45.Park YW, Tominaga R, Sugiyama J, Furuta Y, Tanimoto E, Samejima M, Sakai F, Hayashi T. Enhancement of growth by expression of poplar cellulase in Arabidopsis thaliana. Plant Journal. 2003;33:1099–1106. doi: 10.1046/j.1365-313x.2003.01696.x. [DOI] [PubMed] [Google Scholar]
- 46.Zhao Z, Shklyaev OE, Nili A, Mohamed MNA, Kubicki JD, Crespi VH, Zhong LH. Cellulose microfibril twist, mechanics, and implication for cellulose biosynthesis. J Physical Chemistry A. 2013;117:2580–2589. doi: 10.1021/jp3089929. [DOI] [PubMed] [Google Scholar]
- 47.Sanchez-Rodriguez C, Bauer S, Hematy K, Saxe F, Ibanez AB, Vodermaier V, Konlechner C, Sampathkumar A, Ruggeberg M, Aichinger E, et al. Chitinase-like1/pom-pom1 and its homolog CTL2 are glucan-interacting proteins important for cellulose biosynthesis in Arabidopsis. Plant Cell. 2012;24:589–607. doi: 10.1105/tpc.111.094672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Liu L, Shang-Guan K, Zhang B, Liu X, Yan M, Zhang L, Shi Y, Zhang M, Qian Q, Li J, et al. Brittle Culm1, a COBRA-like protein, functions in cellulose assembly through binding cellulose microfibrils. PLoS Genet. 2013;9:e1003704. doi: 10.1371/journal.pgen.1003704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hayashi T. Xyloglucans in the primary cell wall. Annual Review of Plant Physiology and Plant Molecular Biology. 1989;40:139–168. [Google Scholar]
- 50.Park YB, Lee CM, Kafle K, Park S, Cosgrove DJ, Kim SH. Effects of plant cell wall matrix polysaccharides on bacterial cellulose structure studied with vibrational sum frequency generation spectroscopy and X-ray diffraction. Biomacromolecules. 2014;15:2718–2724. doi: 10.1021/bm500567v. [DOI] [PubMed] [Google Scholar]
- 51.Park YB, Lee CM, Koo BW, Park S, Cosgrove DJ, Kim SH. Monitoring meso-scale ordering of cellulose in intact plant cell walls using sum frequency generation spectroscopy. Plant Physiology. 2013;163:907–913. doi: 10.1104/pp.113.225235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52*.Baskin TI, Jensen OE. On the role of stress anisotropy in the growth of stems. J Exp Bot. 2013;64:4697–4707. doi: 10.1093/jxb/ert176. This excellent review shows that multiple subtle effects must be considered in the mechanical analysis of stem growth. Similar issues no doubt arise in the growth of other multicellular organs. [DOI] [PubMed] [Google Scholar]
- 53.Bashline L, Lei L, Li S, Gu Y. Cell wall, cytoskeleton, and cell expansion in higher plants. Mol Plant. 2014;7:586–600. doi: 10.1093/mp/ssu018. [DOI] [PubMed] [Google Scholar]
- 54.Ivakov A, Persson S. Plant cell shape: modulators and measurements. Front Plant Sci. 2013;4:439. doi: 10.3389/fpls.2013.00439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Green PB. Mechanism for plant cellular morphogenesis. Science. 1962;138:1404–&. doi: 10.1126/science.138.3548.1404. [DOI] [PubMed] [Google Scholar]
- 56.Bringmann M, Li E, Sampathkumar A, Kocabek T, Hauser MT, Persson S. POM-POM2/cellulose synthase interacting1 is essential for the functional association of cellulose synthase and microtubules in Arabidopsis. Plant Cell. 2012;24:163–177. doi: 10.1105/tpc.111.093575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57*.Li S, Lei L, Somerville CR, Gu Y. Cellulose synthase interactive protein 1 (CSI1) links microtubules and cellulose synthase complexes. Proc Natl Acad Sci U S A. 2012;109:185–190. doi: 10.1073/pnas.1118560109. This large protein (CSI1) mediates microtubule guidance of CSC movement in the plasma membrane. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gu Y, Kaplinsky N, Bringmann M, Cobb A, Carroll A, Sampathkumar A, Baskin TI, Persson S, Somerville CR. Identification of a cellulose synthase-associated protein required for cellulose biosynthesis. Proc Natl Acad Sci U S A. 2010;107:12866–12871. doi: 10.1073/pnas.1007092107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Landrein B, Lathe R, Bringmann M, Vouillot C, Ivakov A, Boudaoud A, Persson S, Hamant O. Impaired cellulose synthase guidance leads to stem torsion and twists phyllotactic patterns in Arabidopsis. Curr Biol. 2013;23:895–900. doi: 10.1016/j.cub.2013.04.013. [DOI] [PubMed] [Google Scholar]
- 60.Cavalier DM, Lerouxel O, Neumetzler L, Yamauchi K, Reinecke A, Freshour G, Zabotina OA, Hahn MG, Burgert I, Pauly M, et al. Disrupting two Arabidopsis thaliana xylosyltransferase genes results in plants deficient in xyloglucan, a major primary cell wall component. Plant Cell. 2008;20:1519–1537. doi: 10.1105/tpc.108.059873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Park YB, Cosgrove DJ. Changes in cell wall biomechanical properties in the xyloglucan-deficient xxt1/xxt2 mutant of Arabidopsis. Plant Physiology. 2012;158:465–475. doi: 10.1104/pp.111.189779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wang T, Zabotina O, Hong M. Pectin-cellulose interactions in the Arabidopsis primary cell wall from two-dimensional magic-angle-spinning solid-state nuclear magnetic resonance. Biochemistry. 2012;51:9846–9856. doi: 10.1021/bi3015532. [DOI] [PubMed] [Google Scholar]
- 63.Dick-Perez M, Zhang Y, Hayes J, Salazar A, Zabotina OA, Hong M. Structure and interactions of plant cell-wall polysaccharides by two- and three-dimensional magic-angle-spinning solid-state NMR. Biochemistry. 2011;50:989–1000. doi: 10.1021/bi101795q. [DOI] [PubMed] [Google Scholar]
- 64.Peaucelle A, Braybrook S, Hofte H. Cell wall mechanics and growth control in plants: the role of pectins revisited. Front Plant Sci. 2012;3:121. doi: 10.3389/fpls.2012.00121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zykwinska A, Thibault JF, Ralet MC. Competitive binding of pectin and xyloglucan with primary cell wall cellulose. Carbohydrate Polymers. 2008;74:957–961. [Google Scholar]
- 66.Zykwinska A, Thibault JF, Ralet MC. Organization of pectic arabinan and galactan side chains in association with cellulose microfibrils in primary cell walls and related models envisaged. J Exp Botany. 2007;58:1795–1802. doi: 10.1093/jxb/erm037. [DOI] [PubMed] [Google Scholar]
- 67.Dick-Perez M, Wang T, Salazar A, Zabotina OA, Hong M. Multidimensional solid-state NMR studies of the structure and dynamics of pectic polysaccharides in uniformly 13C-labeled Arabidopsis primary cell walls. Magn Reson Chem. 2012;50:539–550. doi: 10.1002/mrc.3836. [DOI] [PubMed] [Google Scholar]
- 68.White PB, Wang T, Park YB, Cosgrove DJ, Hong M. Water-polysaccharide interactions in the primary cell wall of Arabidopsis thaliana from polarization transfer solid-state NMR. J Am Chem Soc. 2014;136:10399–10409. doi: 10.1021/ja504108h. [DOI] [PubMed] [Google Scholar]
- 69.Muller F, Manet S, Jean B, Chambat G, Boue F, Heux L, Cousin F. SANS measurements of semiflexible xyloglucan polysaccharide chains in water reveal their self-avoiding statistics. Biomacromolecules. 2011;12:3330–3336. doi: 10.1021/bm200881x. [DOI] [PubMed] [Google Scholar]
- 70*.Park YB, Cosgrove DJ. A revised architecture of primary cell walls based on biomechanical changes induced by substrate-specific endoglucanases. Plant Physiol. 2012;158:1933–1943. doi: 10.1104/pp.111.192880. The tethered network hypothesis is put to the test by mechanical assays combined with cell wall disgestion by related endoglucanases with differering substrate specificities. The tethering concept fails the test, but the results point to a mechanical role for a limited subset of enzyme-inaccessible xyloglucans. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sampedro J, Cosgrove DJ. The expansin superfamily. Genome Biology. 2005;6:242. doi: 10.1186/gb-2005-6-12-242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72*.Wang T, Park YB, Caporini MA, Rosay M, Zhong L, Cosgrove DJ, Hong M. Sensitivity-enhanced solid-state NMR detection of expansin's target in plant cell walls. Proc Natl Acad Sci U S A. 2013;110:16444–16449. doi: 10.1073/pnas.1316290110. This study measures the 13C NMR spectrum of polysaccharides within spin diffusion distance (a few nm) of expansin bound to complex cell walls. The results show that expansin targets a form of cellulose with a slightly different NMR shift than bulk cellulose. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Frey-Wyssling A. The fine structure of cellulose microfibrils. Science. 1954;119:80–82. doi: 10.1126/science.119.3081.80. [DOI] [PubMed] [Google Scholar]
- 74.Keegstra K, Talmadge KW, Bauer WD, Albersheim P. The structure of plant cell walls. III. A model of the walls of suspension-cultured sycamore cells based on the interconnections of the macromolecular components. Plant Physiology. 1973;51:188–196. doi: 10.1104/pp.51.1.188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.McCann MC, Wells B, Roberts K. Direct visualization of cross-links in the primary plant cell wall. J Cell Sci. 1990;96:323–334. [Google Scholar]
- 76.Zhang T, Mahgsoudy-Louyeh S, Tittmann B, Cosgrove DJ. Visualization of the nanoscale pattern of recently-deposited cellulose microfibrils and matrix materials in never-dried primary walls of the onion epidermis. Cellulose. 2014;21:853–862. [Google Scholar]
- 77.Ding SY, Liu YS, Zeng Y, Himmel ME, Baker JO, Bayer EA. How does plant cell wall nanoscale architecture correlate with enzymatic digestibility? Science. 2012;338:1055–1060. doi: 10.1126/science.1227491. [DOI] [PubMed] [Google Scholar]


