Abstract
Clearance of apoptotic cells (efferocytosis) is achieved through phagocytosis by professional or amateur phagocytes. It is critical for tissue homeostasis and remodeling in all animals. Failure in this process can contribute to the development of inflammatory autoimmune or neurodegenerative diseases. We previously found that the PALL-SCF E3-Ubiquitin ligase complex promotes apoptotic cell clearance, yet it remained unclear as to how it did so. Here, we show that the F-Box protein PALL interacts with phosphorylated Ribosomal protein S6 (RpS6) to promote its ubiquitylation and proteasomal degradation. This leads to RAC2 GTPase up-regulation and activation and F-actin remodeling that promotes efferocytosis. We further show that the specific role of PALL in efferocytosis is driven by its apoptotic cell-induced nuclear export. Finding a role for RpS6 in negatively regulating efferocytosis provides the opportunity to develop new strategies to regulate this process.
INTRODUCTION
Innate immunity is the first line of host defense of all animals; it limits infection by clearing pathogens (Modlin, 2012). Humoral responses in innate immunity have been extensively studied (Shishido et al., 2012), but its cellular responses are still poorly understood. This includes phagocytosis, the process by which specialized cells, the phagocytes, recognize, engulf and digest pathogens, tissue debris and necrotic cells during infection and in wound healing (Brown, 1995). Phagocytosis is also critical for tissue homeostasis, as an estimated 50-70 billions of our cells die daily by apoptosis (Han and Ravichandran, 2011). Professional and amateur phagocytes rapidly clear these apoptotic cells (ACs), a process known as efferocytosis (Henson et al., 2001). Failure to clear ACs can promote neurodegeneration and autoimmunity (Fuller and Van Eldik, 2008; Munoz et al., 2010). It has also been associated with age-related macular retinal degeneration and with systemic lupus erythematosus (Finnemann et al., 2002; Hanayama et al., 2006). A thorough understanding of phagocytosis is essential to design new drugs to fight infection, and to prevent and/or treat neurodegenerative and autoimmune diseases.
Drosophila is an ideal model organism in which to study innate immune responses including phagocytosis (Ferrandon et al., 2007). It has macrophage-like cells (Tepass et al., 1994), and Drosophila Schneider S2 cells behave like macrophages and are amenable to RNAi and biochemistry (Schneider, 1972). Using genetic and genome-wide RNAi screens, we previously identified several molecules required for efferocytosis, including Pallbearer (PALL), an F-box protein that acts within a SkpA/dCullin-1/F-Box (SCF) complex (Silva et al., 2007). This PALL-SCF complex functions as an E3-Ubiquitin ligase to promote efficient AC clearance (Silva et al., 2007).
E3-Ubiquitin ligases are involved in a variety of biological processes where the F-Box protein is generally responsible for the binding specificity of the substrate to be degraded via the proteasome (Deshaies, 1999). Many F-box proteins have specific protein-protein interaction domains such as Leucine-rich repeats, WD-40 repeats, Sec7 or others, which have facilitated the identification of their substrate(s). However, many other F-Box proteins do not have such domains, making it more difficult to identify their substrates. PALL belongs to this sub-class and what its substrate(s) for degradation may be in efferocytosis is not yet known.
Here we have used both biochemistry and genetics to find the PALL substrate(s). We show that PALL interacts with the ribosomal protein S6 (RpS6) and that this interaction depends on the phosphorylation of RpS6. Treatment of S2 cells with the proteasome inhibitor MG132 results in accumulation of poly-ubiquitylated RpS6 thus revealing a role for the Ubiquitin-Proteasome pathway in the regulation of RpS6 levels. The poly-ubiquitylation and degradation of RpS6 are PALL-dependent, as we observed less poly-ubiquitylated forms of RpS6 in pall RNAi-treated S2 cells. RNAi of RpS6 enhances AC engulfment in S2 cells, and conversely, overexpressing RpS6 in embryonic macrophages partially inhibits AC clearance in vivo. Mutation of RpS6 suppresses the AC clearance defect phenotype of a pall mutant allele. We have found that pall and RpS6 mutant macrophages have opposite F-actin phenotypes, with F-actin accumulation in pall mutants and diminished F-actin in RpS6 mutants. Also, pall and RpS6 RNAi-treated S2 cells have opposite staining phenotypes for RAC. RAC regulates actin cytoskeleton during AC clearance in C. elegans and mammalian systems (Gumienny et al., 2001; Kinchen et al., 2005), and RAC2 is required for efferocytosis in Drosophila S2 cells (Cuttell et al., 2008). Cells treated with pall RNAi have less total and active RAC2, while RpS6 RNAi-treated S2 cells have more. Importantly, over-expressing RAC2 in pall mutant macrophages rescued their phagocytosis defect. Thus, we propose that the PALL/SCF complex promotes the proteasomal degradation of RpS6, which acts as a negative regulator of efferocytosis; this degradation leads to F-actin cytoskeleton remodeling via the up-regulation and activation of RAC2 thereby promoting phagocytosis. Finally, we show that PALL is not required for phagocytosis of bacteria and that its specificity in efferocytosis is driven by its AC-induced nuclear export.
RESULTS
PALL physically interacts with RpS6
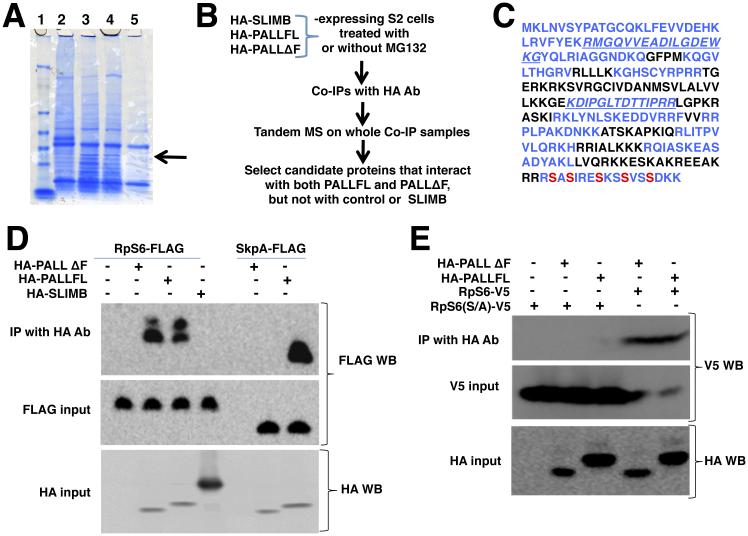
We previously proposed that PALL promotes efferocytosis by promoting ubiquitylation and proteasomal degradation of one or more phosphorylated substrates (Silva et al., 2007). To identify the PALL substrate(s), we established three stable S2 cell lines that express HA-tagged full length PALL (HA-PALLFL), its F-Box deleted version (HA-PALLΔF), or the F-box protein SLIMB (HA-SLIMB), which plays a role in innate immunity but not in efferocytosis (Khush et al., 2002; Silva et al., 2007). We used protein extracts of these stable cell lines in immunoprecipitations (IPs) with HA antibody (Ab) and carried out comparative analyses of the IPs on SDS-PAGE. We found one endogenous protein of around 26kDa in co-IPs with both HA-PALLFL and HA-PALLΔf, but not with HA-SLIMB. Using MALDI-TOF mass spectrometry (MS), we identified this interactor as the Ribosomal protein S6 (RpS6), a component of the 40S subunit of the ribosome (Fig. 1A). Although ribosomal proteins are abundant and often pulled non-specifically in IPs, RpS6 was the only ribosomal protein being consistently pulled down with HA-PALLFL and HA-PALLΔf but not with HA-SLIMB in three other independent IPs subjected to comparative Tandem MS (Fig. 1B). RpS6 sequences recovered in the MS are indicated in blue in Fig. 1C. Treatment of S2 cells with cycloheximide (CHX), an inhibitor of protein translation, did not significantly enhance or reduce S2 cells ability to clear ACs, arguing that the role of RpS6 in efferocytosis is independent of its role as a component of the small ribosomal sub-unit in the context of protein synthesis (Fig. S1A). Deleting the F-Box domain of PALL prevents its interaction with SkpA ((Silva et al., 2007) and Fig. 1D), eliminating the possibility that RpS6 is binding to the other components of the PALL-SCF complex. We further confirmed the interaction between PALL and RpS6 in co-transfections in S2 cells using HA-PALLFL and RpS6-FLAG tagged constructs (Fig. 1D), as well as using HA-PALLFL and HA-PALLΔF and RpS6-V5 tagged constructs (Fig. 1E and Fig. S1B). Thus RpS6 interacts with PALL and is a candidate substrate of PALL.
Figure 1. PALL physically interacts with phosphorylated RpS6.
(A) SDS-PAGE and coomassie blue staining of HA Ab immunoprecipitates of HA-PALLFL, HA-PALLΔF or HA-SLIMB-expressing S2 cells extracts. RpS6 identified by differential band cut-out and MALDI-TOF is indicated by an arrow. Lane 1, protein ladder; Lane 2, HA-SLIMB (unrelated F-box); Lane 3, HA-PALLFL; Lane 4, HA-PALLΔF (F-box deleted PALL); Lane 5, control S2 cells. (B) Flow chart of IPs and Tandem MS with HA-SLIMB (control irrelevant F-Box protein), HA-PALLFL and HA-PALLΔF (F-box deleted PALL) stable S2 cells. (C) Primary amino-acid sequence of endogenous RpS6. In blue are sequences found by Tandem MS. Phosphorylated S are indicated in red. (D) IPs with HA Ab of crude protein extracts from transiently transfected S2 cells expressing RpS6-FLAG or SkpA-FLAG and HA-SLIMB, HA-PALLFL, or HA-PALLΔF. RpS6-FLAG immunoprecipitates with HA-PALLFL and HA-PALLΔF but not with HA-SLIMB. As a control, SkpA-FLAG immunoprecipitates with HA-PALLFL but not HA-PALLΔF. Inputs are given by WB detection on protein extracts of RpS6-FLAG and SkpA-FLAG, or of HA-SLIMB, HA-PALLFL, and HA-PALLΔF expressing cells with FLAG and HA Abs, respectively (see also Fig. S1B)(E) IPs with HA Ab of protein extracts from S2 cells expressing RpS6-V5 or its RpS6(S/A)-V5 mutant with HA-PALLFL or HA-PALLΔF, followed by V5 Ab WB. The PALL/RpS6 interaction is lost when RpS6 is mutated at its S phosphorylation sites. See also Fig. S1.
RpS6 phosphorylation is required for its interaction with PALL
RpS6 is a component of the 40S subunit of the ribosome and the substrate of several mammalian Serine/Threonine protein kinases that can phosphorylate RpS6 at Serines 233, 235, 239, 242 and 245 (S indicated in red in Fig. 1C), all of which are highly conserved (Radimerski et al., 2000). One of the RpS6 peptide identified by MS included a carboxy-terminal peptide that contains the conserved S, as highlighted in blue in Fig. 1C. Substrates of F-Box proteins are generally phosphorylated prior to specifically interacting with their respective F-Box proteins. To address whether RpS6 phosphorylation at its conserved S is a prerequisite for its interaction with PALL, we generated a V5-tagged mutated construct of RpS6 under the control of the metallothionin (MT)-inducible promoter where all five S were mutated into Alanines (A)(MT-RpS6(S/A)-V5). We co-transfected this construct into S2 cells with either HA-PALLFL and HA-PALLΔF constructs, carried out IPs with HA Ab, and probed for RpS6(S/A)-V5 using V5 Ab on western blots (WBs). RpS6(S/A)-V5 could no longer co-immunoprecipitate with either HA-PALLFL or HA-PALLΔF (Fig. 1E). Therefore, S phosphorylation of RpS6 is required for its interaction with PALL.
PALL promotes poly-ubiquitylation and proteasomal degradation of RpS6
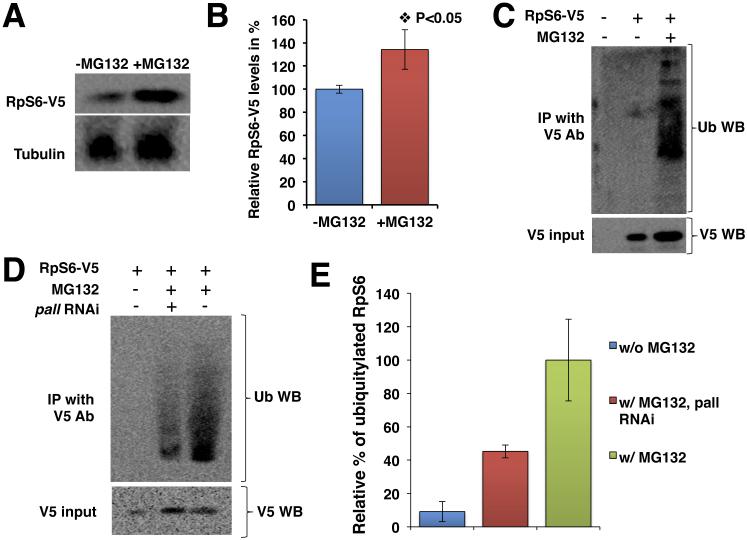
The interaction of phosphorylated RpS6 with PALL suggested that RpS6 is a target of the PALL-SCF complex for poly-ubiquitylation and proteasomal degradation. This hypothesis was further strengthened by our observation that the input level of expression of the mutated form of RpS6, RpS6(S/A)-V5 in S2 cells was greater than that of RpS6-V5 in similar co-transfection conditions, as well as in the sole presence of endogenous PALL (see V5 input in Fig. 1E). This result argues that mutating the S phosphorylation sites of RpS6 into A, which abolishes RpS6 interaction with PALL, prevents its ubiquitylation and degradation via the proteasome. To test this, we asked whether RpS6 could accumulate when inhibiting the proteasome. We overexpressed RpS6-V5 in HA-PALLFL stable S2 cells in the presence or absence of the proteasome inhibitor MG132 and quantified the level of RpS6 protein in WBs. RpS6-V5 accumulated in the presence of MG132 (Fig. 2A), as its expression level significantly increased by 34.2±17.1% (P value <0.05) when compared to that in absence of MG132 (Fig. 2B). We could not evaluate the levels of phosphorylated RpS6, as commercial antibodies against mammalian phosphorylated RpS6 do not cross-react with Drosophila RpS6.
Figure 2. RpS6 is a substrate of PALL for poly-ubiquitylation and proteasomal degradation.
(A) HA-PALLFL stable S2 cells transfected with RpS6-V5 and treated with or without MG132, followed by WB with V5 Ab. The Tubulin WB serves as a loading control. (B) Graph summarizing the quantification of (A). Bars represent the relative percentage when compared to untreated (-MG132) cells ± standard errors from the mean (SEM) of three independent experiments. (C) HA-PALLFL stable S2 cells transfected with RpS6-V5 and Act5C-Ub constructs and treated with or without MG132. V5 Ab immunoprecipitates of RpS6 blotted with Ub Ab to detect poly-ubiquitylated forms of RpS6. (D) Same experiments as in (C) but with or without pre-treating the S2 cells with pall RNAi prior to MG132 treatment and IP with V5 Ab and WB with Ub Ab. In C-D, the input of RpS6-V5 protein is shown by WB of crude cell extracts with V5 Ab. (E) Graph showing the relative quantification of the mean % ± SEM of ubiquitylated RpS6 in S2 cells without MG132 (w/o MG132) and pall-RNAi cells in the presence of MG132 (w/MG132 pall RNAi) when compared to control S2 cells with MG132 (w/MG132).
To further address whether RpS6 is a substrate of PALL-SCF for ubiquitylation, we overexpressed RpS6-V5 with an Act5C-Ubiquitin (Act5C-Ub) construct in HA-PALLFL stable S2 cells in presence or absence of the proteasome inhibitor MG132. We immunoprecipitated RpS6-V5 with V5 Ab and performed a WB with Ubiquitin (Ub) Ab to assess whether RpS6 could be ubiquitylated (Fig. 2C). We detected several poly-ubiquitylated forms of RpS6 in the presence of MG132, but not in its absence (Fig. 2C). These results conclusively demonstrate that RpS6 is a substrate for poly-ubiquitylation.
We then asked whether ubiquitylation of RpS6 was PALL-dependent. For this, we repeated the above experiments in pall RNAi-treated S2 cells in the presence of MG132. We found that the level of detectable poly-ubiquitylated RpS6 was reduced by almost 55% when compared to MG132-treated control S2 cells (Fig. 2D and E). As in figure 2A, the level of RpS6-V5 expression was higher in MG132-treated cells than in untreated control S2 cells (see V5 input WB in Fig. 2D). Altogether, these results demonstrate a previously unappreciated role for the Ubiquitin-Proteasome pathway in the regulation of RpS6 protein levels. Thus PALL is required for specific interaction of RpS6 to the PALL-SCF complex and subsequent RpS6 poly-ubiquitylation and proteasomal degradation.
An RpS6 loss-of-function mutation suppresses the loss-of-function phenotype of pall
To further assess whether RpS6 acts as a substrate for poly-ubiquitylation and proteasomal degradation via the PALL-SCF complex in vivo, we asked whether RpS6 and pall might genetically interact. Because a loss-of-function of pall is expected to result in RpS6 accumulation, we asked whether a loss-of-function of RpS6 might suppress the loss-of-function phenotype of pall, i.e. restore AC clearance in the pall null allele. Previously characterized pall alleles were not single mutant of pall but also affected neighboring genes (Silva et al., 2007). Thus, we generated a pallko null allele by homologous recombination (Fig. S2A, B and B”). As anticipated, pallko macrophages have a defect in phagocytosis of ACs in vivo when compared to wild-type macrophages (Fig. S2C). Of note is that pallko does not appear to have any obvious growth-related phenotypes at any developmental stages. Although we did not look extensively for these types of phenotypes at the cellular level, the phosphorylation of RpS6 in the context of efferocytosis is also independent of the Target of Rapamycin (TOR) and S6 kinases pathway (HX and NCF, unpublished data).
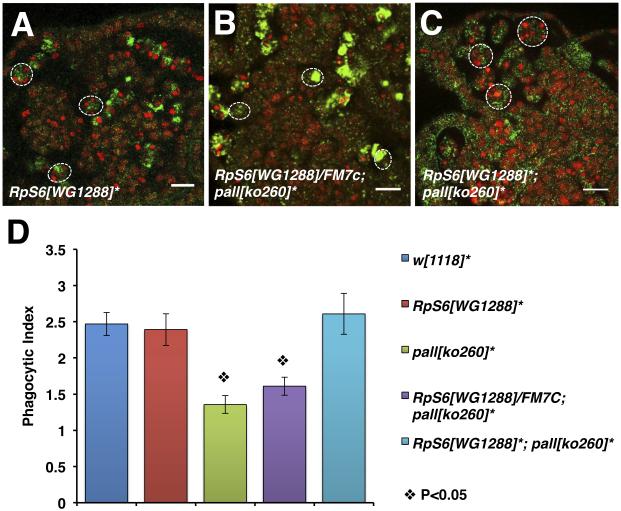
We then crossed our pallko to a previously characterized strong hypomorphic loss-of-function allele of RpS6, RpS6WG1288 (Watson et al., 1992), and assessed the phagocytosis phenotypes and phagocytic indices (PIs) of homozygous pallko macrophages that were either heterozygous or homozygous for RpS6WG1288 by CRQ Ab and 7-AAD double staining and confocal microscopy (Silva et al., 2007)(Fig. 3). Double homozygous mutant macrophages were capable of engulfing ACs (Fig. 3C) with a phagocytic index (PI) of 2.61±0.28 similar to that of control macrophages (Fig. 3D) and of RpS6WG1288 homozygous macrophages (Fig. 3A) with PIs of 2.47±0.16 and 2.4±0.22, respectively (Fig. 3D)(p values>0.05). Thus, loss-of-function of RpS6 rescued the phagocytosis phenotype defect of homozygous pallko mutant macrophages, which had a PI of 1.35±0.12, while mutating one copy of RpS6 did not significantly affect the homozygous pallko mutant phenotype (Fig. 3B and D)(p values<0.05 when compared to control and >0.05 when compared to pallko). Altogether, these results demonstrate that RpS6 and pall genetically interact and act in the same pathway in vivo where RpS6 acts downstream of pall as its substrate for poly-ubiquitylation and proteasomal degradation.
Figure 3. A LOF mutation of RpS6 suppresses the LOF mutation phenotype of pall in vivo.
(A-C) Confocal micrographs of RpS6WG1288 homozygous embryos, RpS6WG1288/+; pallko260/pallko260 embryos, and RpS6WG1288; pallko260 double homozygous embryos stained with the CRQ Ab (green) and 7-AAD (red). Dotted white circles outline single macrophages. Scale bars correspond to 10 μm. (D) Graph showing the mean PIs ± SEM for each genotype. * denotes homozygous alleles. See also Fig. S2.
RpS6 acts as a negative regulator of efferocytosis
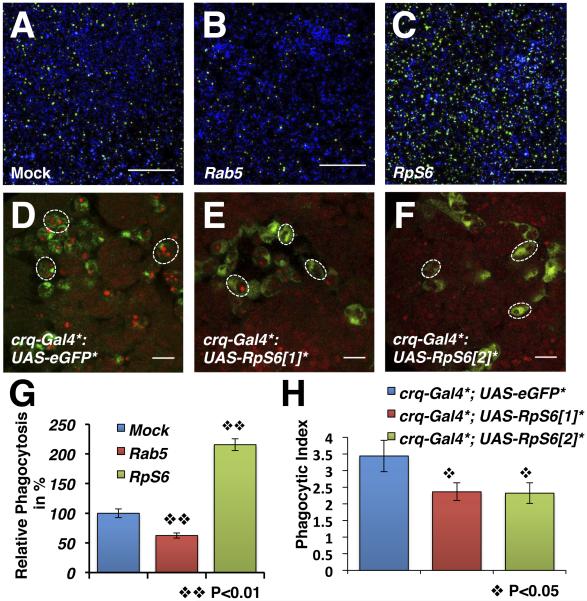
RpS6 is a substrate of PALL for proteasomal degradation to promote AC clearance, thus arguing that RpS6 acts in macrophages as a negative regulator of this process. In a genome-wide RNAi screen for genes required in AC clearance in S2 cells, we found that RpS6 RNAi-treatment of S2 cells with both DRSC18712 and DRSC25010 could enhance phagocytosis of ACs (http://www.flyrnai.org, Fig. 4). Compared with mock-treated S2 cells (Fig. 4A), RpS6 RNAi-treated S2 cells with DRSC25010 were over 2 fold more phagocytic when given a same amount of ACs (Fig. 4C and G). RNAi-treatments of two other components of the small ribosome subunit such as RpS10b (the knock-down of which enhances phagocytosis of Listeria monocytogenes (Agaisse et al., 2005)) and RpS2 served as controls and did not significantly affect the S2 cells ability to engulf ACs (Fig. S3A and B).
Figure 4. RpS6 acts as a negative regulator of AC clearance.
(A-C) Phagocytosis of ACs by mock- (A), Rab5 RNAi- (B) or RpS6 RNAi- (C) treated S2 cells. Live cells are blue, engulfed FITC-labeled ACs are green. Scale bars represent 200 μm. (D-F) Merged confocal images of yw; +; crq-Gal4, UAS-eGFP (wild-type reference)(D), and yw; UAST-RpS6; crq-Gal4, UAS-eGFP macrophages with two independent transgenic UAST-RpS6 lines [1] and [2] in E and F, respectively. ACs are stained with 7-AAD (red), GFP-expressing macrophages appear green. Scale bars represent 10 μm. (G) Graph summarizing the quantification of A-C with rab5-RNAi S2 cells as a control. Bars represent the relative phagocytosis when compared to mock-treated S2 cells ± SEM of three independent experiments with duplicated wells. (H) Graph showing the mean PIs ± SEM for each genotype in D-F. See also Fig. S3.
We further tested whether RpS6 could act as an inhibitor of efferocytosis in vivo by driving the expression of two independent UAS-RpS6 transgenic lines under the control of the macrophage-specific crq-Gal4 transgene (Fig. 4D-F and H). Overexpressing RpS6 with either UAS-RpS6 lines resulted in >30% decrease in phagocytosis by embryonic macrophages in vivo (compare Fig. 4E and F to Fig. 4D) with PIs of 2.36±0.27 and 2.32±0.31 versus 3.44±0.47 in crq-gal4 control (p<0.05)(see Fig. 4H). The macrophage-specific over-expression of other components of the small ribosome sub-units, including RpS2 and RpS10b did not affect their phagocytic ability (Fig. S3C). Thus, RpS6 acts as a negative regulator of efferocytosis, which further fits with its role as a PALL substrate for poly-ubiquitylation and proteasomal degradation, thereby promoting efficient engulfment of ACs. Furthermore, this role for RpS6 appears independent of its ribosomal function.
PALL and RpS6 regulate F-actin cytoskeleton rearrangement in efferocytosis
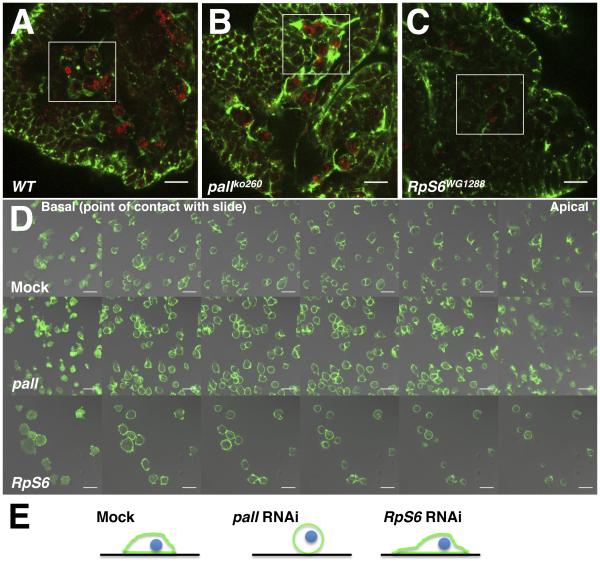
How do PALL and RpS6 act in regulating AC clearance? F-actin cytoskeleton rearrangement is an important hallmark of phagocytosis (Allen and Aderem, 1996). Recently, phosphorylated RpS6 was attributed a role in regulating F-actin organization and junctional protein recruitment at the blood-testis barrier during spermatogenesis, where increased level of phosphorylated RpS6 disrupted the barrier and co-localized with disorganized F-actin (Mok et al., 2012). Thus, we next asked whether pall facilitates AC clearance by promoting RpS6 poly-ubiquitylation and degradation, and subsequent F-actin remodeling. We first assessed the F-actin phenotype of mock- and pall RNAi-treated S2 cells (Fig. S4A and B, respectively) by phalloidin staining. The F-actin immunostaining increased by 73±6.4% in pall RNAi-treated S2 cells when compared to the mock-treated cells (Fig. S4C). The pall mRNA expression was down by about 75% in pall RNAi-treated S2 cells when compared to mock-treated cells (Fig. S4D). To ask whether pall regulates F-actin in vivo, we further assessed the actin phenotype of pallko macrophages. As for Phalloidin staining in pall RNAi-treated S2 cells, F-actin immunostaining increased in pallko macrophages (Fig. 5B) when compared to that of wild-type macrophages (Fig. 5A). In contrast, F-actin immunostaining was weaker in RpS6WG1288 mutant macrophages (Fig. 5C) when compared to wild type macrophages (Fig. 5A), thus having the anticipated opposite phenotype. Similar results were obtained with Phalloidin staining of pall and RpS6 mutant macrophages in vivo (data not shown). These data argue that PALL and RpS6 play a role in regulating F-actin cytoskeleton in Drosophila macrophages in vivo. Furthermore, pall RNAi-treated S2 cells were more rounded than mock-treated cells with F-actin distributed evenly around their cell cortex, while F-actin staining was weaker in RpS6-RNAi cells and more localized at their basal membrane (i.e. at their point of contact with the glass slide)(Fig. 5D and E). RpS6 RNAi-treated cells spread onto glass slides and made more membrane ruffles than mock-treated cells (Fig. 5D and E). Tubulin and Actin monomers expression levels in pall- or RpS6- RNAi cells were comparable to that of mock-treated cells (Fig. 6A). Thus PALL and RpS6 regulate F-actin cytoskeleton rearrangement in vivo and in S2 cells.
Figure 5. PALL promotes Actin remodeling during AC clearance.
(A-C) Confocal micrographs of Actin (green) and CRQ (red) immunostaining of yw embryos (wild-type (WT) control in A), pallko260 (B) and RpS6WG1288 (C) mutant embryos. (D) Confocal micrographs of Mock-treated, pall- or RpS6-RNAi S2 cells stained with phalloidin. Z-stack images through the cells were collected 1.74 μm apart (basal membrane is in contact with the glass slide). Scale bars correspond to 10 μm. (E) Schematic drawings summarize F-actin staining (green) distribution and cell shape for mock-treated, and pall- and RpS6-RNAi S2 cells seen in D. Green lines and blue circles represent Actin and nuclei, respectively. Black lines represent the slides. See also Fig. S4.
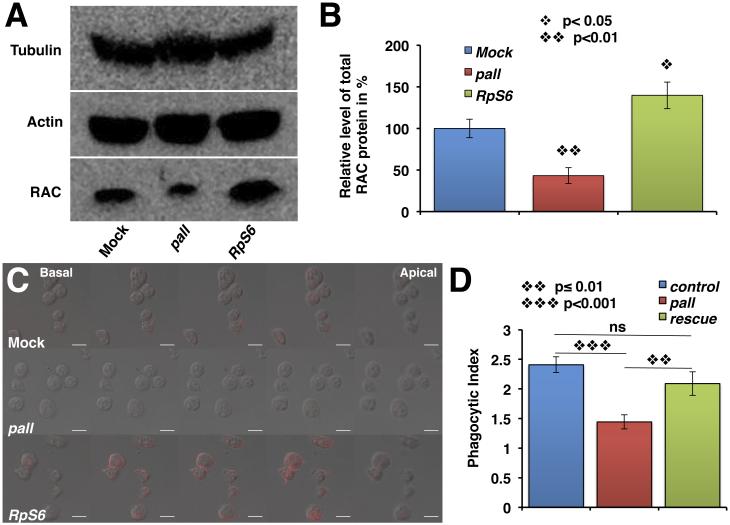
Figure 6. RpS6 targeting by PALL promotes RAC up-regulation and activation.
(A) WB of protein extracts from mock-treated, pall- or RpS6-RNAi S2 cells probed with Tubulin and monomeric Actin (internal controls), and mammalian RAC1 Abs. (B) Graph showing the % ± SEM of RAC protein expression in pall- and RpS6-RNAi S2 cells relative to mock-treated cells (control normalized at 100%) based on three separate experiments. (C) Confocal micrographs of mock-treated, pall- and RpS6-RNAi S2 cells stained with active RAC Ab. Z-stack images through the cells were collected at 1.74 μm intervals. Scale bars correspond to 10 μm. (D) Graph showing the mean PIs ± SEM of crq-gal4 control-, crq-gal4;pallko mutant- and pallko-macrophages that overexpress UAS-Rac2 under the control of crq-gal4. See also Fig. S5.
PALL and RpS6 regulate Drosophila RAC2 level and activity in efferocytosis
The small GTPase RAC has been shown to play an important role in AC clearance in both C. elegans and mammalian systems (Gumienny et al., 2001; Kinchen et al., 2005) by promoting Actin remodeling. In a genome-wide RNAi screen for genes required for AC clearance by S2 cells, we have found a role for the small GTPase RAC2, but not for the other RAC family members, RAC1 and MTL (Mig-2 like)(Cuttell et al., 2008). We asked whether PALL and RpS6 might regulate RAC levels and/or activity in S2 cells. On WB of total protein extracts from RNAi-treated S2 cells, we observed that there was approximately 57% less and 40% more total RAC in pall-RNAi and RpS6-RNAi cells, respectively, than in mock-treated cells (Fig. 6A and B). We then assessed the localization of RAC and active RAC by immunostaining in these cells. Mammalian RAC1 Ab stainings, which cross-reacts with Drosophila RAC1 and 2 (23A8, Thermo Scientific), confirmed our WB findings, as RpS6 and pall RNAi-treated S2 cells exhibited increased and decreased immunostainings, respectively, when compared to mock-treated S2 cells (Fig. S5A). To further ask whether RAC levels resulted in similar active RAC levels in the cells, we also stained these RNAi-treated cells with a monoclonal Ab directed against active mammalian RAC1. This Ab cross-reacted with Drosophila RAC2 specifically (Fig. S5B) and labeled the membrane of all treated cells. Again, pall and RpS6 RNAi-treated S2 cells showed reduced and increased stainings, respectively, when compared to mock-treated S2 cells (Fig. 6C). We then asked whether we could rescue the phagocytosis defect of pall by overexpressing the Drosophila UAS-Rac2 transgene in pallko mutant under the control of the macrophage-specific crq-gal4 driver. As previously for pallko, crq-gal4;pallko homozygous macrophages had a significantly diminished PI of 1.44±0.12 when compared to UAS-Rac2/+;crq-gal4/+;pallko/+ heterozygous control macrophages (PI=2.41±0.13; p<0.001), while pallko rescue macrophages that overexpressed UAS-Rac2 under the control of crq-gal4 (UAS-Rac2/+;crq-gal4/+;pallko) had a PI of 2.09±0.19 that was statistically significantly rescued compared to that of crq-gal4;pallko homozygous macrophages (p=0.01) but not statistically different from the UAS-Rac2/+;crq-gal4/+;pallko/+ heterozygous control (p=0.3)(Fig. 6D and Fig. S5C-E). Thus PALL allows for the degradation of RpS6 via the proteasome that leads to the up-regulation and activation of RAC2 and F-actin re-arrangement to promote efferocytosis in vivo.
Nuclear translocation of PALL in response to ACs confers its phagocytic specificity
Considering the role of PALL upstream of RAC2 activation in efferocytosis, and because bacteria are cleared by Drosophila hemocytes via RAC2-mediated phagocytosis (Avet-Rochex et al., 2007), we next asked whether pallko mutants were susceptible to infection. Survival rates to Gram negative Erwinia carotovora carotovora 15 (Ecc15) and Gram positive Enterococcus faecalis (Ef)(Fig. S6A), as well as their bacterial loads (Fig. S6B) were similar in pallko mutant flies and control flies. Injections of Alexa488 (green) E. coli in pallko (Fig. S6C’) and their control flies (Fig. S6C) or of Alexa 488 S. aureus (data not shown), or of their pH-sensitive pHrodo-red equivalent (data not shown and Fig. S6D-D’) did not reveal any defect in their uptake (Fig. S6C-C’), or in their phagosomal degradation (Fig. S6D-D’). The pallko flies survived to E. coli (Fig. S6E) and S. aureus (Fig. S6F) septic injuries equally well to their controls, and the loss of pall did not exacerbate the phenotype of mutant flies of the IMD pathway that are susceptible to a low dose of infection by E. coli (Fig. S6E). Thus pall is not required for bacteria killing or phagocytosis and plays a specific role in efferocytosis.
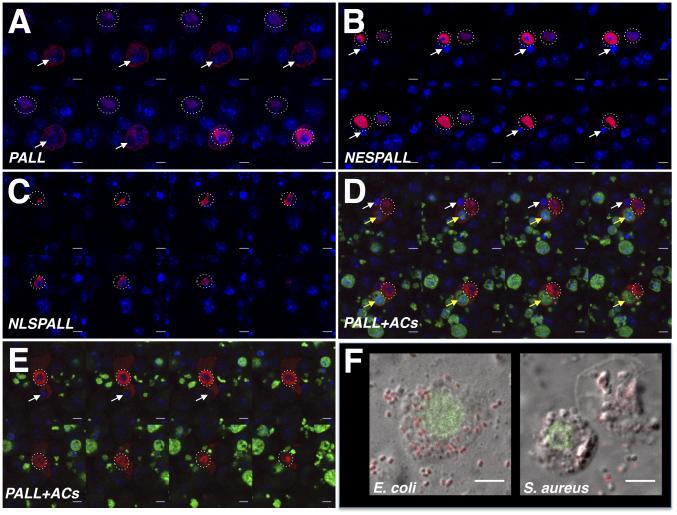
Having previously found that ACs regulate the expression level of CRQ, which localizes at the phagosomal membranes during AC clearance and participates in efferocytosis (Franc et al., 1996; Franc et al., 1999), we asked whether ACs could regulate PALL expression and/or its localization. PALL has two potential nuclear export signals (NES) at its L amino-acids 6 and 10, as predicted by the NetNES 1.1 server (la Cour et al., 2004)(Fig. S6G). We independently expressed in S2 cells a wild-type HA-tagged PALL (HA-PALLFL, Fig. S6H and Supp. Mov.1) and versions of PALL in which the L of its NES were mutated into A (HA-NESPALL)(Fig. S6H and Supp. Mov.2) or in which the NES was replaced by a nuclear localization signal (NLS)(HA-NLSPALL)(Fig. S6H and Supp. Mov.3). After staining these cells with a HA Ab and either DAPI or DRAQ5 DNA dyes, we found that both HA-NESPALL (Fig. 7B) and HA-NLSPALL (Fig. 7C) were strictly localized to the nuclei of all transfected S2 cells, as anticipated. However, while HA-PALLFL was expressed in the nuclei of most transfected S2 cells, it was also found to localize both in the nuclei and cytoplasm of all transfected S2 cells that were bound to/engulfing and/or had fully ingested ACs (Fig. 7A and Supp. Mov.1). In contrast, mutated HA-NESPALL never translocated even when these cells engulfed endogenous ACs (see white arrow in Fig. 7B and Supp. Mov.2; of note is that endogenous PALL is present in these cells), neither did mutated HA-NLSPALL (Fig. 7C and Supp. Mov.3). Moreover, HA-PALLFL remained nuclear in S2 cells that had bound or fully ingested gram negative (E. coli) or gram positive (S. aureus) bacteria (Fig. 7F). Thus the NES is required for the translocation of PALL from the nuclei to the cytoplasm of S2 cells in response to ACs, and this translocation is dependent on the binding/recognition of ACs (Fig. 7D and E). These results demonstrate that ACs play an active role in regulating PALL localization that promotes efferocytosis and confers PALL its specificity in this process.
Figure 7. Regulation of PALL localization by ACs confers its specificity to efferocytosis.
(A-E) Z-stack confocal images through DRAQ5- (A-C) or DAPI- (D-E)(in blue) and HA Ab (in red) double-stained S2 cells expressing various forms of PALL. (A) A HA-PALLFL expressing S2 cell that has engulfed an endogenous AC (white arrow) shows both nuclear (dotted circles surround cell nuclei of interest) and cytoplasm HA-PALLFL protein expression, while a cell that has not engulfed nor bound to AC shows HA-PALLFL strictly in its nucleus. (B) HA-NESPALL expressing S2 cells where the L residues of the NES of PALL were mutated into A that prevents PALL nuclear export even when cells have engulfed endogenous ACs. (C) HA-NLSPALL expressing S2 cells where the NES was replaced by a NLS show its nuclear localization. (D-E) A HA-PALLFL expressing S2 cell that has engulfed an endogenous AC (white arrow) and a FITC-labeled AC (yellow arrow) (D), and a HA-PALLFL expressing S2 cell that is engulfing an endogenous AC (white arrow)(E) showing both nuclear (dotted circle around nucleus) and cytoplasmic localization of HA-PALLFL. (F) Nuclear localization of PALL on confocal micrographs of HA Ab-stained (green) HA-PALLFL-expressing S2 cells that have engulfed E. coli and S. aureus bacteria. Scale bars are 5 μm. See also Fig. S6.
DISCUSSION
The PALL-SCF complex and proteasomal degradation are required for efficient AC engulfment (Silva et al., 2007), however, it was not clear how this promoted efferocytosis. Here we provide new insights into the molecular mechanisms by which PALL promotes efferocytosis: upon recognition and binding of ACs, PALL translocates from the nucleus to the cytoplasm where it forms a PALL-SCF complex and specifically interacts with and targets phosphorylated RpS6 to poly-ubiquitylation and degradation via the 26S proteasome that results in increased level and activity of RAC2, followed by actin remodeling to promote efferocytosis.
RpS6, a component of the small 40S subunit of the ribosome, has highly conserved S that are phosphorylated by S6 and RSK kinases following a wide variety of stimuli (Meyuhas, 2008). RpS6 has long been thought to be required for various functions including global protein synthesis, the translation of mRNAs containing 5’ terminal oligopyrimidine tract (TOP mRNAs), cell size and proliferation, and glucose homeostasis (Meyuhas, 2008). However, RpS6P−/− knock-in mice in which all five S of RpS6 were mutated into A, show a 2.5 fold increase rather than a decrease in global protein synthesis, and have a similar rate of translational activation of TOP mRNAs (Ruvinsky et al., 2005). RpS6P−/− mice have similar weights to wild-type mice, although some of their cells are small due to impaired growth as they undergo compensatory proliferation (Ruvinsky et al., 2005). RpS6P−/− mice also have reduced glucose disposal capacity due to a 2-fold reduction in secretion of circulating and pancreatic insulin by small β-cells (Ruvinsky et al., 2005). In at least one study, intravenous injection of mice with rat breast carcinoma cells results in poorly phagocytic alveolar macrophages with reduced cell-autonomous glucose oxidation that leads to tumor metastasis (Gudewicz and Saba, 1977). It will be interesting to ask whether reduced glucose oxidation in these alveolar macrophages correlates with reduced glucose homeostasis due to defective RpS6 regulation.
Drosophila RpS6 appears to play immune-specific roles, as RpS6 mutants have overgrown lymph glands (the larval hematopietic organs), abnormal proliferation and differentiation of enlarged hemocytes, and melanotic masses caused by erroneous encapsulation of larval tissues (Stewart and Denell, 1993; Watson et al., 1992). These phenotypes suggest that loosing RpS6 results in hemocyte hyperactivation. This is consistent with our data showing that RpS6-RNAi S2 cells are more phagocytic. We did not, however, observe any increased efferocytosis by macrophages in RpS6 mutant embryos in vivo. This is likely due to the limited amount of ACs present in the embryo compared to the excess of ACs used in our S2 cell assay. While RpS6 acts as a negative regulator of efferocytosis, other proteins of the ribosome small sub-unit, including RpS2 and RpS10b, do not participate in this process, as RNAi in S2 cells or in vivo overexpression experiments did not reveal any significant effect on efferocytosis (Fig. S3). Yet another ribosomal protein RpS17 appears to act as a positive regulator of efferocytosis, as RNAi for RpS17 in S2 cells results in these cells being less efficient at engulfing ACs (HX and NCF, unpublished data). Further experiments will be required to determine the precise role of RpS17 in AC clearance. However, based on these experiments, we propose that RpS6 acts in efferocytosis independently of its ribosomal function. Interestingly, mammalian RpS6 can interact with proteins outside the ribosome, including the heat-shock protein 90, the alphavirus non-structural protein and the death-associated protein kinase (DAPK)(Meyuhas, 2008) and is likely to also have functions that are independent of the translational machinery.
The nuclear export of PALL in response to ACs, but not to bacteria, is particularly important, as it confers a specific role for RpS6 in efferocytosis. Indeed, RpS6 RNAi in S2 cells results in an increase in phagocytosis of the gram positive bacteria Listeria monocytogenes (Agaisse et al., 2005), although the specific role of RpS6 in this system is not yet known. We have carried out similar RpS6 RNAi in S2 cells with new validation amplicons that have no target effects and exposed those S2 cells to fluorescent E. coli and S. aureus bacteria. We have also found that knocking down RpS6 enhanced phagocytosis of both E. coli and S. aureus (HX and NCF, unpublished data). Thus RpS6 acts as a negative regulator of bacterial clearance, although the mechanism by which RpS6 is regulated during this process is not known. Finally, RpS6 RNAi in S2 cells also confers tolerance to Drosophila C virus infection, although it does so by acting in the context of its ribosomal function together with other ribosomal proteins (Cherry et al., 2005). Thus RpS6 can play many roles in Drosophila immunity either directly or indirectly, via its ribosomal function or more specific functions that will merit further investigation.
Phosphorylated RpS6 regulates F-actin organization and junctional protein recruitment at the blood-testis barrier during spermatogenesis, where increased levels of phosphorylated RpS6 disrupt the barrier and co-localize with disorganized F-actin (Mok et al., 2012). F-actin rearrangement in phagocytosis is regulated by small GTPases of the RAC family. Our results demonstrate a negative role for Drosophila RpS6 in AC clearance and that proteasomal degradation of phosphorylated RpS6 by PALL-SCF leads to F-actin remodeling via the up-regulation and activation of RAC2. RAC2 is the only GTPase required for efferocytosis in S2 cells (Cuttell et al., 2008), and over-expressing RAC2 in pallko mutant macrophages rescued their phagocytosis defect, thus linking PALL and RAC2 functions in vivo. Of further note, myoblasts of RpS6P−/− mice have less contractile proteins, including myosins (Ruvinsky et al., 2009), suggesting that phosphorylation of RpS6 (and perhaps its degradation) is required to regulate myosin expression. Together these observations further argue in favor of a role for RpS6 in the regulation of specific proteins in both mouse and Drosophila, rather than a role in global protein synthesis. Whether RpS6 phosphorylation and proteasomal degradation in Drosophila macrophages or in other cells result in increased levels of proteins other than RAC2 to promote efferocytosis or other RpS6-mediated functions will need further investigation.
How the degradation of phosphorylated RpS6 results in higher levels and activation of RAC2 remains to be determined. Recent studies, however, have highlighted a role for specific E3 ligases, including SCFFBXL19 and HACE1, in regulating mammalian RAC1 protein levels in cell migration and tumorigenesis (Torrino et al., 2011; Zhao et al., 2013). The small GTPase RhoA is also regulated by SCFFBXL19 as well as another E3 ubiquitin ligase Smurf1 in cell migration (Wei et al., 2013). In this instance, Synaptopodin, an actin-associated protein, can compete with Smurf1 for binding to RhoA thereby preventing its degradation (Asanuma et al., 2006). One possibility is that the accumulation in the pall mutant of phosphorylated RpS6 may displace such an inhibitory protein, thereby allowing a specific E3-ubiquitin ligase to bind to and target RAC2 to poly-ubiquitylation and proteasomal degradation. In contrast, in wild type context, the PALL-SCF mediated ubiquitylation and degradation of phosphorylated RpS6 may prevent the poly-ubiquitylation and degradation of RAC2 by allowing binding of a specific inhibitory protein to the RAC2-specific E3-ubiquitin ligase, resulting in elevated levels and activity of RAC2. Of note is that Smurf1 can also regulate a guanine nucleotide exchange factor (GEF) for yet another small GTPase CDC42 (Yamaguchi et al., 2008). Thus the regulation of RpS6 may also lead to activation of RAC2 by indirectly regulating RAC2-specific GEFs or GAPs.
Future studies of the role of Drosophila RpS6 and the regulation of its phosphorylation and levels are likely to provide new insights into the role and regulation of mammalian RpS6 that may have implications for the development of treatments to fight autoimmune and neurodegenerative diseases. Moreover, high levels of phosphorylated RpS6 correlate with tumor progression and shorter survival prognostic in patients with oesophageal squamous cell carcinoma (Chaisuparat et al., 2012). Whether RpS6 plays a role in AC clearance in mammals remains to be determined, but a lack of engulfment of cancer cells by poorly phagocytic macrophages with high levels of phosphorylated RpS6 may favor the development of this cancer. Thus, a better understanding of the phosphorylation and regulation of RpS6 levels and its role in phagocytosis will likely provide us with drug targets to fight cancer progression.
EXPERIMENTAL PROCEDURES
Fly Strains
Most fly stocks were from the Bloomington Drosophila stock center. RpS6WG1288 and S6kIIign-Δ58-1 flies were provided by Leonie M. Quinn (University of Melbourne, Parkville, Australia) and by F. Rob Jackson (Tufts University School of Medicine, Boston, MA, USA), respectively. UAS transgenic fly lines were generated at BestGene Inc (Chino Hill, CA, USA) or supplied by FLYORF. Fly stocks were maintained at 25°C in Superfly incubators (Genesee, Inc.) on standard medium.
Plasmid Constructs (see supplemental procedures)
Generation of HA-PALLFL, HA-PALLΔF and HA-SLIMB stable S2 cell lines
S2 cells were transfected with Effectene (QIAGEN) according to the supplier’s protocol using 950 ng of HA-PALLFL, HA-PALLΔF or HA-SLIMB (N-terminus tagged constructs in pMT-N-3xHA tag vector) DNA and 50 ng of the pHygro vector DNA. Cells were resuspended in complete media with 300 μg/ml of Hygromycin B to select clones that had integrated pHygro DNA. Clones were tested for integration of pMT constructs and expression of HA-tagged proteins by WB following standard protocols after treating the cells with 0.7 mM of CuSO4 to induce expression under the control of the Metallothionin (MT) promoter.
Immunoprecipitations and Tandem MS analysis
PALL, PALLΔF or SLIMB expression was induced with 0.7 mM CuSO4. Cells were treated with 25 μM MG132 for 6 hrs (Sigma), harvested at 48 and 72 hrs post-CuSO4 treatment, and lysed in 50 mM Tris HCl (pH 7.4), 150 mM NaCl, 1 mM EDTA, 1 % NP40 with Complete EDTA-free protease inhibitor (Roche). Lysate were centrifuged at 10,000 g for 30 min, and supernatants were added to EZ view HA beads (Sigma) overnight at 4°C. After several washes, proteins were eluted in 0.2 M Glycine (pH 2.5), neutralized with ammonium bicarbonate, impurities removed by Trichloro-acetic acid (TCA) precipitation and samples were subjected to Tandem MS analysis (see supplemental procedures).
Transient Transfections and Immunoprecipitations
S2 cells were transfected with Effectene (QIAGEN) according to the supplier’s protocol. Expression of tagged versions of RpS6, PALL and SkpA were induced with CuSO4 at 24 hr and the cells harvested and lysed at 48 to 72 hr after transfection. Lysates were centrifuged at 10,000 g for 30 min and supernatants incubated with EZview HA beads or anti-V5 agarose affinity gel (Sigma). Purified samples were resolved by 4-15% gradient SDS PAGE (Biorad). In WBs, Abs were used at the following dilutions: rat anti-HA (Roche) and mouse anti-FLAG (Sigma), 1:2,000; mouse anti-V5 (Invitrogen), 1:5,000. Anti Rat or mouse HRP-coupled secondary Abs (Jackson Laboratories) were used at a 1:10,000 dilution followed by ECL detection following supplier’s protocol (Pierce).
Stability and ubiquitylation assays
For the stability assay, pAc5.1/V5-HisA-RpS6 was transiently transfected into HA-PALLFL stable S2 cell line with Effectene (QIAGEN) according to the supplier’s instructions. The cells were then treated with CuSO4 at 24 hr post-transfection followed by MG132 (Sigma) at a concentration of 50 μM for 4h. For the ubiquitylation assay, pAc5.1/V5-HisA-RpS6 and pAc5.1/UB were transiently transfected into HA-PALLFL stable S2 cell line with Effectene (QIAGEN). PALL expression was induced with CuSO4 at 24 hr post-transfection. The cells were then treated with MG132 (Sigma) at a concentration of 50 μM for 4h, lysed and subjected to IPs and WBs as described above. The Ub (P4D1) mouse monoclonal Ab (Santa Cruz Biotechnology) was used at a 1:200 dilution.
In vivo phagocytosis assay, phagocytosis index quantification, RNA interference and S2 cell phagocytosis assays, and their statistical analyses were performed as in (Cuttell et al., 2008; Silva et al., 2007).
Statistical analyses
Statistical p values were derived using ANOVA (for PIs) or the student T-test and are indicated in the text and figures.
Ends-out gene targeting
Donor targeting constructs were built by insertion of two 3-kb regions (upstream and downstream of the pall target gene) into two multiple cloning sites of the targeting vector pXH87. These constructs were then injected into w1118 embryos using established methods to obtain transgenics (Bestgene Inc.). Ends-out gene targeting procedure was performed as described in (Chen et al., 2009). To induce double-stranded DNA breaks, heat shock was performed at 38°C for 90 min at day 3 after egg-laying. Target gene-specific PCR amplification was performed using primers corresponding to a sequence within the pall gene to verify homozygous knockouts (no PCR product). crq primer sets were used as a control (see supplemental procedures).
Actin staining and RAC immunostainings were performed according to standard protocols (seesupplemental procedures).
RAC quantification
RAC WBs were performed after 3 days of RNAi treatment of S2 cells, as described above. Cells were lysed in in 50 mM Tris HCl (pH 7.4), 0.1 % SDS, 200 mM NaCl, 5 mM MgCl2, 1 mM DTT, 1 % NP40, 10% Glycerol with Complete EDTA-free protease inhibitor (Roche) and subjected to WBs. Mouse RAC1 monoclonal Ab (23A8, Thermo Scientific), mouse Actin Ab (A-2066; Sigma) and rabbit alpha-Tubulin Ab (11H10, Cell Signaling) were used at 1:1,000. Secondary anti-Mouse HRP-coupled Abs (Jackson Laboratories) were used at 1:10,000 followed by ECL detection.
Supplementary Material
ACKNOWLEDGMENTS
We thank Dr. Xun Huang (Institute of Genetics and Developmental Biology, Chinese Academy of Sciences, Beijing, China) for the pXH87 plasmid, Dr. Jin Jiang (UT Southwestern Medical Center, Dallas, USA) for the pUAST-UB construct, Dr. Leonie M. Quinn (University of Melbourne, Parkville, Australia), Dr. F. Rob Jackson (Tufts University School of Medicine, Boston, MA, USA) and the Bloomington Stock Center and FLYORF for fly strains. We are grateful to Claude Desplan, Sergio Grinstein and Céline DerMardirossian for critically reading this manuscript, and to Céline DerMardirossian for valuable advice on RAC and actin stainings. HX, HW, ES, JT, AG, NB, NCF conceived, designed and performed the experiments, analyzed and interpreted the data and wrote the manuscript. This is manuscript #24072 from The Scripps Research Institute.
FUNDING STATEMENT
The work was funded by a program leader-track grant from the Medical Research Council to NCF and core support to the MRC Cell Biology Unit (NCF, ES), an Irvington Institute post-doctoral fellowship of the Cancer Research Institute to HX, a TSRI-Advanced Discovery Institute grant provided by Novartis and a National Institute of Health grant of the National Institute of Allergy and Infectious Diseases R01 AI093687-01A1 to NCF, and a Research Resource grant P41 RR011823 of the National Institute of Health to JRY. Stocks from the Bloomington Drosophila Stock Center (NIH P40OD018537) were used in this study.
Footnotes
PALL degrades RpS6 to boost efferocytosis via RAC
The authors declare having no financial interest related to this work.
REFERENCES
- Agaisse H, Burrack LS, Philips JA, Rubin EJ, Perrimon N, Higgins DE. Genome-wide RNAi screen for host factors required for intracellular bacterial infection. Science. 2005;309:1248–1251. doi: 10.1126/science.1116008. [DOI] [PubMed] [Google Scholar]
- Allen LA, Aderem A. Mechanisms of phagocytosis. Curr Opin Immunol. 1996;8:36–40. doi: 10.1016/s0952-7915(96)80102-6. [DOI] [PubMed] [Google Scholar]
- Asanuma K, Yanagida-Asanuma E, Faul C, Tomino Y, Kim K, Mundel P. Synaptopodin orchestrates actin organization and cell motility via regulation of RhoA signalling. Nat Cell Biol. 2006;8:485–491. doi: 10.1038/ncb1400. [DOI] [PubMed] [Google Scholar]
- Avet-Rochex A, Perrin J, Bergeret E, Fauvarque MO. Rac2 is a major actor of Drosophila resistance to Pseudomonas aeruginosa acting in phagocytic cells. Genes Cells. 2007;12:1193–1204. doi: 10.1111/j.1365-2443.2007.01121.x. [DOI] [PubMed] [Google Scholar]
- Brown EJ. Phagocytosis. Bioessays. 1995;17:109–117. doi: 10.1002/bies.950170206. [DOI] [PubMed] [Google Scholar]
- Chaisuparat R, Rojanawatsirivej S, Yodsanga S. Ribosomal Protein S6 Phosphorylation is Associated with Epithelial Dysplasia and Squamous Cell Carcinoma of the Oral Cavity. Pathol Oncol Res. 2012 doi: 10.1007/s12253-012-9568-y. [DOI] [PubMed] [Google Scholar]
- Chen H, Ma Z, Liu Z, Tian Y, Xiang Y, Wang C, Scott MP, Huang X. Case studies of ends-out gene targeting in Drosophila. Genesis. 2009;47:305–308. doi: 10.1002/dvg.20501. [DOI] [PubMed] [Google Scholar]
- Cherry S, Doukas T, Armknecht S, Whelan S, Wang H, Sarnow P, Perrimon N. Genome-wide RNAi screen reveals a specific sensitivity of IRES-containing RNA viruses to host translation inhibition. Genes Dev. 2005;19:445–452. doi: 10.1101/gad.1267905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuttell L, Vaughan A, Silva E, Escaron CJ, Lavine M, Van Goethem E, Eid JP, Quirin M, Franc NC. Undertaker, a Drosophila Junctophilin, links Draper-mediated phagocytosis and calcium homeostasis. Cell. 2008;135:524–534. doi: 10.1016/j.cell.2008.08.033. [DOI] [PubMed] [Google Scholar]
- Deshaies RJ. SCF and Cullin/Ring H2-based ubiquitin ligases. Annu Rev Cell Dev Biol. 1999;15:435–467. doi: 10.1146/annurev.cellbio.15.1.435. [DOI] [PubMed] [Google Scholar]
- Ferrandon D, Imler JL, Hetru C, Hoffmann JA. The Drosophila systemic immune response: sensing and signalling during bacterial and fungal infections. Nat Rev Immunol. 2007;7:862–874. doi: 10.1038/nri2194. [DOI] [PubMed] [Google Scholar]
- Finnemann SC, Leung LW, Rodriguez-Boulan E. The lipofuscin component A2E selectively inhibits phagolysosomal degradation of photoreceptor phospholipid by the retinal pigment epithelium. Proc Natl Acad Sci U S A. 2002;99:3842–3847. doi: 10.1073/pnas.052025899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franc NC, Dimarcq JL, Lagueux M, Hoffmann J, Ezekowitz RA. Croquemort, a novel Drosophila hemocyte/macrophage receptor that recognizes apoptotic cells. Immunity. 1996;4:431–443. doi: 10.1016/s1074-7613(00)80410-0. [DOI] [PubMed] [Google Scholar]
- Franc NC, Heitzler P, Ezekowitz RA, White K. Requirement for croquemort in phagocytosis of apoptotic cells in Drosophila. Science. 1999;284:1991–1994. doi: 10.1126/science.284.5422.1991. [DOI] [PubMed] [Google Scholar]
- Fuller AD, Van Eldik LJ. MFG-E8 regulates microglial phagocytosis of apoptotic neurons. J Neuroimmune Pharmacol. 2008;3:246–256. doi: 10.1007/s11481-008-9118-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gudewicz PW, Saba TM. Inhibition of phagocytosis and glucose metabolism of alveolar macrophages during pulmonary tumour growth. Br J Cancer. 1977;36:670–677. doi: 10.1038/bjc.1977.249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gumienny TL, Brugnera E, Tosello-Trampont AC, Kinchen JM, Haney LB, Nishiwaki K, Walk SF, Nemergut ME, Macara IG, Francis R, et al. CED-12/ELMO, a novel member of the CrkII/Dock180/Rac pathway, is required for phagocytosis and cell migration. Cell. 2001;107:27–41. doi: 10.1016/s0092-8674(01)00520-7. [DOI] [PubMed] [Google Scholar]
- Han CZ, Ravichandran KS. Metabolic connections during apoptotic cell engulfment. Cell. 2011;147:1442–1445. doi: 10.1016/j.cell.2011.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanayama R, Miyasaka K, Nakaya M, Nagata S. MFG-E8-dependent clearance of apoptotic cells, and autoimmunity caused by its failure. Curr Dir Autoimmun. 2006;9:162–172. doi: 10.1159/000090780. [DOI] [PubMed] [Google Scholar]
- Henson PM, Bratton DL, Fadok VA. Apoptotic cell removal. Curr Biol. 2001;11:R795–805. doi: 10.1016/s0960-9822(01)00474-2. [DOI] [PubMed] [Google Scholar]
- Khush RS, Cornwell WD, Uram JN, Lemaitre B. A ubiquitin-proteasome pathway represses the Drosophila immune deficiency signaling cascade. Curr Biol. 2002;12:1728–1737. doi: 10.1016/s0960-9822(02)01214-9. [DOI] [PubMed] [Google Scholar]
- Kinchen JM, Cabello J, Klingele D, Wong K, Feichtinger R, Schnabel H, Schnabel R, Hengartner MO. Two pathways converge at CED-10 to mediate actin rearrangement and corpse removal in C. elegans. Nature. 2005;434:93–99. doi: 10.1038/nature03263. [DOI] [PubMed] [Google Scholar]
- la Cour T, Kiemer L, Molgaard A, Gupta R, Skriver K, Brunak S. Analysis and prediction of leucine-rich nuclear export signals. Protein Eng Des Sel. 2004;17:527–536. doi: 10.1093/protein/gzh062. [DOI] [PubMed] [Google Scholar]
- Meyuhas O. Physiological roles of ribosomal protein S6: one of its kind. Int Rev Cell Mol Biol. 2008;268:1–37. doi: 10.1016/S1937-6448(08)00801-0. [DOI] [PubMed] [Google Scholar]
- Modlin RL. Innate immunity: ignored for decades, but not forgotten. J Invest Dermatol. 2012;132:882–886. doi: 10.1038/jid.2011.373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mok KW, Mruk DD, Silvestrini B, Cheng CY. rpS6 Regulates blood-testis barrier dynamics by affecting F-actin organization and protein recruitment. Endocrinology. 2012;153:5036–5048. doi: 10.1210/en.2012-1665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munoz LE, Janko C, Schulze C, Schorn C, Sarter K, Schett G, Herrmann M. Autoimmunity and chronic inflammation - two clearance-related steps in the etiopathogenesis of SLE. Autoimmun Rev. 2010;10:38–42. doi: 10.1016/j.autrev.2010.08.015. [DOI] [PubMed] [Google Scholar]
- Radimerski T, Mini T, Schneider U, Wettenhall RE, Thomas G, Jeno P. Identification of insulin-induced sites of ribosomal protein S6 phosphorylation in Drosophila melanogaster. Biochemistry. 2000;39:5766–5774. doi: 10.1021/bi9927484. [DOI] [PubMed] [Google Scholar]
- Ruvinsky I, Katz M, Dreazen A, Gielchinsky Y, Saada A, Freedman N, Mishani E, Zimmerman G, Kasir J, Meyuhas O. Mice deficient in ribosomal protein S6 phosphorylation suffer from muscle weakness that reflects a growth defect and energy deficit. PLoS One. 2009;4:e5618. doi: 10.1371/journal.pone.0005618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruvinsky I, Sharon N, Lerer T, Cohen H, Stolovich-Rain M, Nir T, Dor Y, Zisman P, Meyuhas O. Ribosomal protein S6 phosphorylation is a determinant of cell size and glucose homeostasis. Genes Dev. 2005;19:2199–2211. doi: 10.1101/gad.351605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider I. Cell lines derived from late embryonic stages of Drosophila melanogaster. J Embryol Exp Morphol. 1972;27:353–365. [PubMed] [Google Scholar]
- Shishido SN, Varahan S, Yuan K, Li X, Fleming SD. Humoral innate immune response and disease. Clin Immunol. 2012;144:142–158. doi: 10.1016/j.clim.2012.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva E, Au-Yeung HW, Van Goethem E, Burden J, Franc NC. Requirement for a Drosophila E3-ubiquitin ligase in phagocytosis of apoptotic cells. Immunity. 2007;27:585–596. doi: 10.1016/j.immuni.2007.08.016. [DOI] [PubMed] [Google Scholar]
- Stewart MJ, Denell R. Mutations in the Drosophila gene encoding ribosomal protein S6 cause tissue overgrowth. Mol Cell Biol. 1993;13:2524–2535. doi: 10.1128/mcb.13.4.2524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tepass U, Fessler LI, Aziz A, Hartenstein V. Embryonic origin of hemocytes and their relationship to cell death in Drosophila. Development. 1994;120:1829–1837. doi: 10.1242/dev.120.7.1829. [DOI] [PubMed] [Google Scholar]
- Torrino S, Visvikis O, Doye A, Boyer L, Stefani C, Munro P, Bertoglio J, Gacon G, Mettouchi A, Lemichez E. The E3 ubiquitin-ligase HACE1 catalyzes the ubiquitylation of active Rac1. Dev Cell. 2011;21:959–965. doi: 10.1016/j.devcel.2011.08.015. [DOI] [PubMed] [Google Scholar]
- Watson KL, Konrad KD, Woods DF, Bryant PJ. Drosophila homolog of the human S6 ribosomal protein is required for tumor suppression in the hematopoietic system. Proc Natl Acad Sci U S A. 1992;89:11302–11306. doi: 10.1073/pnas.89.23.11302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei J, Mialki RK, Dong S, Khoo A, Mallampalli RK, Zhao Y, Zhao J. A new mechanism of RhoA ubiquitination and degradation: roles of SCF(FBXL19) E3 ligase and Erk2. Biochim Biophys Acta. 2013;1833:2757–2764. doi: 10.1016/j.bbamcr.2013.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi K, Ohara O, Ando A, Nagase T. Smurf1 directly targets hPEM-2, a GEF for Cdc42, via a novel combination of protein interaction modules in the ubiquitin-proteasome pathway. Biol Chem. 2008;389:405–413. doi: 10.1515/BC.2008.036. [DOI] [PubMed] [Google Scholar]
- Zhao J, Mialki RK, Wei J, Coon TA, Zou C, Chen BB, Mallampalli RK, Zhao Y. SCF E3 ligase F-box protein complex SCF(FBXL19) regulates cell migration by mediating Rac1 ubiquitination and degradation. Faseb J. 2013;27:2611–2619. doi: 10.1096/fj.12-223099. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.