Abstract
A variety of conventional methods allow the expression of multiple foreign proteins in plants by transgene stacking or pyramiding. However, most of these approaches have significant drawbacks. We describe a novel alternative, using a single transgene to coordinate expression of multiple proteins that are encoded as a polyprotein capable of dissociating into component proteins on translation. We demonstrate that this polyprotein system is compatible with the need to target proteins to a variety of subcellular locations, either cotranslationally or posttranslationally. It can also be used to coordinate the expression of selectable marker genes and effect genes or to link genes that are difficult to assay to reporter genes that are easily monitored. The unique features of this polyprotein system are based on the novel activity of the 2A peptide of Foot-and-mouth disease virus (FMDV) that acts cotranslationally to effect a dissociation of the polyprotein while allowing translation to continue. This polyprotein system has many applications both as a research tool and for metabolic engineering and protein factory applications of plant biotechnology.
Transgenic technology offers unrivaled possibilities for gaining valuable new insights into plant biology and also provides a novel route for crop improvement. To date, many different heterologous proteins have been introduced into plants, and the expression levels of resident enzymes have also been manipulated. An exciting prospect for the future is the possibility of introducing multiple heterologous proteins and, perhaps, entire new biochemical pathways into plants. However, despite rapid and continuing progress in this area, the vast majority of experiments reported to date involve the expression or manipulation of single genes and coordinate expression of multiple genes is still difficult to achieve. Pyramiding or stacking of transgenes is conventionally accomplished by crossing plants harboring different introduced genes, by successive retransformations or by cotransformation. Although these strategies have been used successfully (see Halpin and Boerjan, 2003), they cannot ensure coordinate expression of different transgenes—this has to be screened for in individual transgenics. Moreover, strategies that combine multiple unlinked transgenic loci that can subsequently segregate apart make breeding programs more complicated and expensive while repetition of, for example, regulatory sequences on different transgenes can compromise stable expression through homology-based silencing (for review, see Halpin et al., 2001).
We describe an alternative, novel strategy for coexpressing multiple proteins in plants by encoding them in a single open reading frame (ORF), in an artificial self-dissociating polyprotein. A short peptide of unique and novel function taken from the 2A region of Foot-and-mouth disease virus (FMDV) separates distinct coding sequences within the polyprotein. This 20-amino acid 2A peptide effects efficient cotranslational dissociation of the polyprotein into discrete protein products. Dissociation occurs at the carboxy terminus of the 2A sequence, most likely by disrupted peptide bond formation, a process that occurs cotranslationally within the ribosome without any requirement for cytosolic factors (de Felipe et al., 2003). This polyprotein system has been used for coexpression of two genes in a variety of eukaryotic systems (e.g. Roosien et al., 1990; Ryan and Drew, 1994; Suzuki et al., 2000; de Felipe et al., 2003), and preliminary work suggests that it can be used to express reporter genes in the cytoplasm of plant cells (Halpin et al., 1999). However, for most applications, this has limited value as many proteins need to be directed to specific subcellular locations in order to fulfill their metabolic functions. Such protein targeting occurs either cotranslationally (for proteins destined for the endoplasmic reticulum [ER], Golgi, vacuole, plasma membrane, or cell wall) or posttranslationally (for proteins destined for plastids, mitochondria, peroxisomes, and nucleus) and is mediated by distinct targeting signals encoded within the polypeptide sequence of proteins destined for these locations. In the current work we demonstrate that multiple proteins expressed in plant cells from a 2A-polyprotein can be correctly targeted to various subcellular locations via either cotranslational or posttranslational mechanisms. Even membrane proteins can be encoded and targeted in this way. All that is necessary is that each protein within the polyprotein contains its own independent targeting signals. Thus two proteins coencoded within a 2A-polyprotein can even be directed to different subcellular locations. Moreover, the polyprotein system allows the coordinated expression of three or more proteins from one polyprotein construct. The 2A-polyprotein system therefore offers a versatile, simple, one-step alternative to gene stacking for expressing combinations of two or more useful proteins in a variety of subcellular compartments in transgenic plants.
RESULTS
Synthesis of Multiple Polypeptides from One Promoter
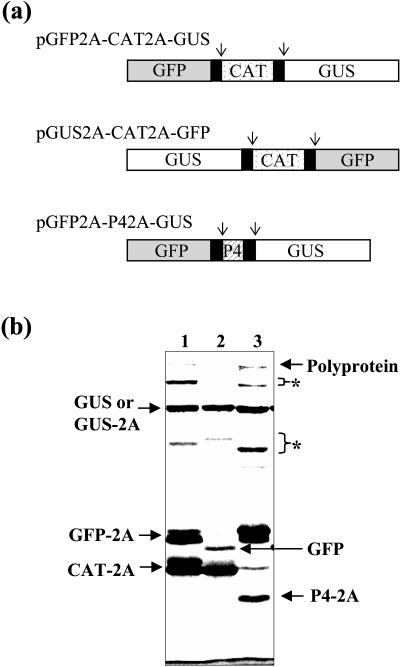
Clones for four different genes (β-glucuronidase, GUS; green fluorescent protein, GFP; chloramphenicol acetyltransferase, CAT; P4, a protein from Potato leaf roll virus) were used to make the various 2A-polyprotein constructs depicted in Figure 1a. In each construct, three different coding sequences were separated from each other by 2A and assembled together in a single ORF using PCR amplification methods as described in “Materials and Methods”. Each construct was used to program a TNT transcription-translation system, and the protein products of the translation are shown in Figure 1b.
Figure 1.
Expression of three genes from single polyprotein constructs. a, Three polyprotein constructs, each encoding a single ORF incorporating three different genes separated by FMDV 2A, were assembled and translated in a reticulocyte lysate in vitro. Arrows indicate the positions where the polyproteins would be expected to dissociate cotranslationally at the carboxy terminus of 2A. b, Products obtained when the polyprotein constructs are translated in vitro. Lane 1, Products of pGFP2A-CAT2A-GUS translation. Lane 2, Products of pGUS2A-CAT2A-GFP translation. Lane 3, products of pGFP2A-P42A-GUS translation.
For all constructs, dissociation at the C terminus of 2A apparently occurs efficiently during translation releasing the individual encoded polypeptides as the three predominant translation products. Thus translation of the construct pGFP2A-CAT2A-GUS yielded GFP2A, CAT2A, and GUS as the major polypeptide products, with minor amounts of GFP2A-CAT2A and CAT2A-GUS (Fig. 1b, lane 1; for clarity, minor products are just labeled with *). Similarly translation of pGUS2A-CAT2A-GFP yielded monomer products of GUS2A, CAT2A, and GFP almost exclusively (Fig. 1b, lane 2), while translation of pGFP2A-P42A-GUS yielded major products of GFP2A, P42A, and GUS with some dimer products and a small amount of the trimer polyprotein (Fig. 1b, lane 3). Thus 2A-polyproteins can be used to express three or more different polypeptides from a single promoter within a single construct or transgene.
Cotranslational Targeting to the ER
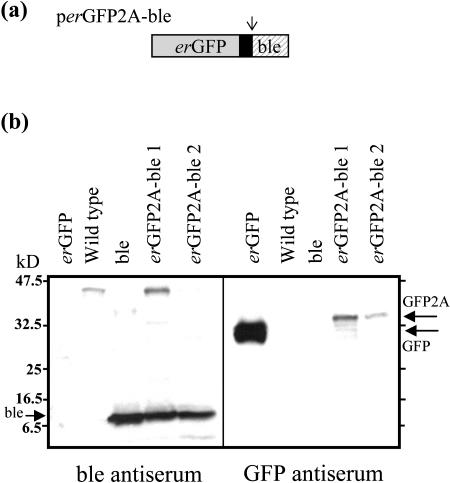
In order to determine whether proteins encoded at anterior positions within 2A-polyproteins could be cotranslationally targeted to the ER, a polyprotein construct was designed to express, from a single ORF, an ER-targeted version of GFP (erGFP) and a phleomycin resistance gene (ble) separated by 2A (Fig. 2a). This erGFP2A-ble polyprotein transgene was expressed in tobacco (Nicotiana tabacum cv Xanthi) plants from the cauliflower mosaic virus 35S promoter (CaMVp). After Agrobacterium tumefaciens-mediated transformation, regenerating plants were selected on medium containing phleomycin. A couple of phleomycin-resistant transgenics were selected at random, and leaf protein extracts were prepared for electrophoresis and western blotting. When probed with antiserum raised against the ble protein, extracts from both selected transgenics showed a major immunoreactive band of approximately 14 kD, the correct size for ble (Fig. 2b). This band was also present in a control transgenic that expressed ble alone (i.e. not from a polyprotein). Similarly, when probed with antiserum raised against GFP, extracts from both polyprotein-expressing plants showed a major band of 32 kD, the correct size for GFP2A (Fig. 2b). This band was slightly larger, due to the presence of the 2A peptide, than the band detected in a transgenic control plant expressing native GFP. These data indicate that the expected dissociation of the 2A-polyprotein to yield its component polypeptides has occurred during translation in plants. There was apparently little, if any, undissociated polyprotein present in either plant extract. A band of the expected size for the erGFP2A-ble polyprotein (approximately 46 kD) was present in one plant extract probed with ble antiserum but was also seen in the control untransformed plant extract (see Fig. 2b), so it is safe to assume it is not the polyprotein. In confirmation of this, the 46-kD band was not obvious in the other transformed plant extract or in either extract probed with GFP antiserum. Consequently, dissociation in planta of the GFP2A-ble polyprotein appears to have been highly efficient.
Figure 2.
Independent targeting of cytosolic and ER proteins expressed from a polyprotein construct. a, The cytosolic selectable marker protein ble and an ER-targeted version of GFP were separated by FMDV 2A and encoded as a polyprotein with a single ORF. The arrow indicates the position where the polyprotein would be expected to dissociate, on translation, at the carboxy terminus of 2A. b, Western blots of protein extracts from transgenic plants expressing the polyprotein erGFP2A-ble along with appropriate controls. Blots were probed with either antiserum raised against ble, or antiserum raised against GFP, as indicated. Extracts are from plants expressing an ER-targeted form of GFP (erGFP), from nontransgenic wild-type plants, from plants expressing the ble protein alone (ble), or from two randomly selected transgenics expressing the polyprotein (erGFP2A-ble 1 and erGFP2A-ble 2).
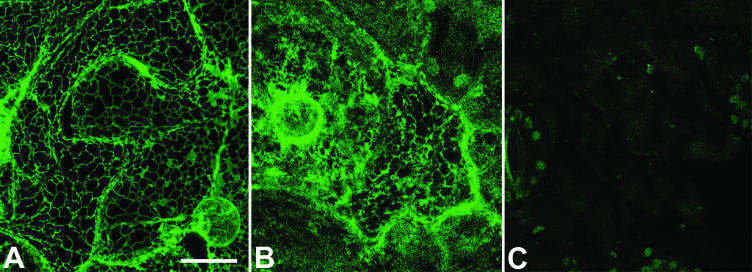
The western results and the effective phleomycin resistance of the transgenic plants indicate that ble was correctly localized in the cytoplasm on dissociation of the polyprotein. The localization of erGFP2A was investigated by visualizing leaf cells by confocal laser scanning microscopy. Figure 3 (middle panel) shows that in the erGFP2A-ble transgenic, fluorescence is present in a reticulate network throughout the cell. This pattern is completely distinct from the faint autofluorescence normally detected from untransformed tobacco cells (Fig. 3, right-hand panel). Moreover, the fluorescence pattern of erGFP2A is similar to that seen when a control construct used as an ER marker (i.e. erGFP), is expressed in tobacco cells (Fig. 3, left-hand panel). These results are therefore consistent with erGFP2A being correctly translocated into the plant endomembrane system. Thus erGFP2A and ble have both been faithfully localized to their respective different subcellular locations when expressed from a 2A-polyprotein.
Figure 3.
Localization of erGFP in the plant ER when expressed from an erGFP2A-ble polyprotein. A, Control plant expressing erGFP alone. B, Plant expressing erGFP2A-ble. C, Nontransformed control plant. Bar = 25 μm.
Targeting of an ER Membrane Protein
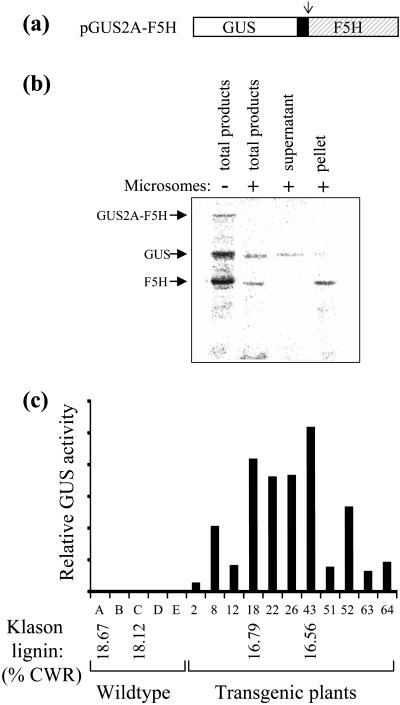
In order to determine whether membrane proteins can also be correctly targeted from within 2A-polyproteins, clones encoding GUS and Arabidopsis ferulate-5-hydroxylase (F5H), a plant cytochrome P450-dependent monooxygenase involved in lignin biosynthesis, were used to make the construct depicted in Figure 4a. F5H is predicted to be a membrane protein that is anchored, probably in the ER membrane, by a hydrophobic helix near its N terminus with most of the protein residing on the cytosolic face of the membrane (Chapple, 1998). Its N-terminal sequence has all the necessary features to mediate targeting to the ER. F5H was chosen for these experiments because of our interest in manipulating lignin biosynthesis. However, monitoring F5H expression in transgenic plants can be difficult as it cannot be easily assayed and there are no antibodies available for the Arabidopsis or tobacco proteins. By expressing it from a 2A-polyprotein, therefore, we hoped to be able to use GUS to report on F5H expression.
Figure 4.
Faithful targeting of a membrane protein and a cytosolic reporter protein expressed from a polyprotein construct. a, The cytosolic reporter protein, GUS, and a plant membrane protein, F5H, were separated by FMDV 2A and encoded as a polyprotein with a single ORF. The arrow indicates the position where the polyprotein would be expected to dissociate, on translation, at the carboxy terminus of 2A. b, Products obtained when the polyprotein construct was translated by TNT reticulocyte lysate in vitro in the absence (−) and presence (+) of microsome membranes. The left-hand lanes show total translation products. The right-hand lanes show the products present in the supernatant and in pelleted membranes after fractionation, by ultracentrifugation through a Suc cushion, of a translation reaction performed in the presence of microsomes. c, Protein extracts from plants transformed with pGUS2A-F5H and wild-type plants were assayed for GUS activity. Selected wild-type and transgenic plants were subjected to Klason lignin analysis to determine the effect of F5H expression on lignin content. Triplicate lignin measurements were made for each plant, and the mean Klason lignin is shown [as a % of extractive-free cell wall residue (CWR)]. Individual assay values varied from the mean by less than 2% (maximum sd = ±0.33).
Because of the difficulties of monitoring F5H in vivo, the GUS2A-F5H construct was first tested in vitro in a coupled transcription-translation system where reactions were performed both in the absence and presence of microsome membranes. Total translation products showed a similar pattern in each case (Fig. 4b), with major products of the expected sizes for GUS2A (69 kD) and F5H (59 kD). A small proportion of the GUS2A-F5H polyprotein was detected in the translation performed without microsomes. No polyprotein was obvious in the translation performed in the presence of microsomes, but this may just be because the translation efficiency was reduced overall, a common observation when using microsomes. Membrane-insertion of translated polypeptides was assessed by subsequently pelleting the microsomes through a Suc cushion by centrifugation at 100,000g. Supernatant and pellet fractions were collected, and translation products associated with each fraction were detected on a phosphorimager after electrophoresis. As illustrated in Figure 4b, F5H associated exclusively with the pelleted microsome membranes while GUS-2A was predominantly present in the soluble fraction. This indicates that expression of F5H from a posterior position in a 2A-polyprotein does not interfere with the cotranslational targeting and insertion of this membrane protein into ER-derived membranes.
The GUS2A-F5H polyprotein was cloned behind a vascular-specific promoter [the promoter of the Arabidopsis cinnamate 4-hydroxylase (C4H) gene] and expressed in transgenic tobacco (Nicotiana tabacum cv Samsun). A selection of regenerating transgenic plants was assayed for GUS activity. Figure 4c shows that there was a normal range of GUS expression levels in these plants. A couple of the highest GUS-expressing plants were selected for preliminary lignin analyses. Lignin content, when assayed by the Klason method, was found to be lower in these two transgenics than in corresponding wild-type plants (see Fig. 4c). This is consistent with previous results indicating that Arabidopsis F5H overexpression in tobacco results in reduced lignin content (Franke et al., 2000). A more comprehensive study on the lignin changes in this population of GUS2A-F5H-expressing plants will be published elsewhere (B.M. Askari and C. Halpin, unpublished data). However, the preliminary data presented here illustrate the advantages of using 2A-polyproteins to coexpress hard-to-assay proteins along with reporter genes—by screening our plants for GUS activity we were rapidly and easily able to detect plants with altered lignin biosynthesis due to F5H coexpression.
Posttranslational Targeting to Plastids
Many organelles, including plant plastids, acquire some of their component proteins posttranslationally. Nuclear-encoded plastid proteins are synthesized in the cytoplasm with N-terminal extension sequences, or transit peptides, responsible for targeting them to chloroplasts. On import into the chloroplast, the transit peptide is removed by chloroplast peptidases.
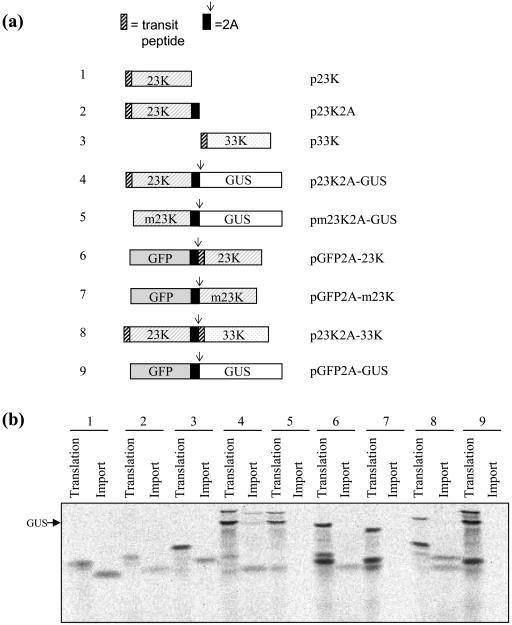
To investigate whether expressing proteins from 2A-polyproteins was consistent with the potential need to target them to plastids, a series of nine constructs was prepared (Fig. 5a) using two chloroplast protein genes (for the 23K and 33K polypeptides of the photosynthetic oxygen-evolving complex) along with reporter genes (GFP and GUS). Coding sequences were separated by 2A and assembled together in single ORFs. In most cases the 23K and 33K precursor polypeptides retained their native transit peptides for chloroplast targeting except for constructs 5 (pm23K2A-GUS) and 7 (pGFP2A-m23K) where the sequence for mature 23K has been used (denoted m23K).
Figure 5.
Expression and targeting of chloroplast proteins from polyprotein constructs. a, Polyprotein constructs encoding reporter proteins and plant chloroplast proteins were assembled along with control constructs. Arrows indicate the positions where the polyproteins would be expected to dissociate cotranslationally at the carboxy terminus of 2A. b, The constructs were translated in TNT wheat germ lysate in vitro and then incubated with isolated chloroplasts to determine whether the chloroplast proteins were competent for import by the organelles. Following import, reactions were treated with protease to remove protein that had not been imported. The results for each construct (identified by the numbers 1–9 by reference to a) are shown by a pair of lanes on the SDS-PAGE gel. The left-hand lane of each pair shows total translation products for that construct while the right-hand lane of each pair shows the proteins protected from protease after import into chloroplasts.
Each construct was translated in vitro in a transcription/translation reaction. After translation, the reactions were incubated with isolated pea (Pisum sativum) chloroplasts in a standard chloroplast import assay. After 25 min incubation, chloroplasts were collected and treated with protease to confirm true internalization of imported polypeptides. Figure 5b shows the results of this experiment. A pair of lanes is shown for each construct, one showing total translation products (left-hand lane of each pair) and the other showing the products imported into chloroplasts and protected from protease treatment (right-hand lane of each pair). Translations containing control plasmids p23K, p23K2A, p33K (constructs 1, 2, and 3) each showed a major polypeptide product of the expected Mr (i.e. 23 kD, 26 kD, and 33 kD, respectively). These products were efficiently imported into isolated chloroplasts, and the reduction in size of the imported polypeptides indicated concomitant removal of their native transit peptides by chloroplast peptidases (Fig. 5b). In contrast, translation directed by polyprotein constructs p23K2A-GUS, pm23K2A-GUS, pGFP2A-23K, pGFP2A-m23K, p23K2A-33K, and pGFP2A-GUS (constructs 4–9) showed three major translation products. The uppermost band in each case corresponds to the small proportion of polyprotein that remains undissociated after translation. The other major products in every case correspond to the individual proteins of the polyprotein, released by dissociation of the polyprotein at the carboxy terminus of 2A.
p23K2A-GUS (construct 4) yielded products of 23K2A and GUS on translation, and the 23K2A product alone was efficiently imported into chloroplasts with removal of the transit peptide. This indicates that proteins can be targeted to chloroplasts from the anterior position of 2A-polyproteins. Construct 5, pm23K2A-GUS, similarly yielded products of m23K2A and GUS but in this case, due to the lack of a transit peptide on the mature m23K polypeptide, no chloroplast import occurred. This result illustrates that chloroplast import is absolutely dependent on proteins possessing the relevant targeting signals. When the 23K was instead encoded in the posterior position within a2A-polyprotein as in constructs 6 and 7 (pGFP2A-23K and pGFP2A-m23K), it was again imported into chloroplasts only when it possessed a transit peptide (construct 6) and not when only the mature protein sequence was present (construct 7). These data indicate that proteins can be targeted posttranslationally and imported to plastids when expressed from either anterior or posterior positions within a2A-polyprotein if, and only if, they possess the appropriate targeting signal, a transit peptide. This was further confirmed by the behavior of the translation products (23K2A and 33K) of construct 8 (p23K2A-33K) which encoded chloroplast proteins in both positions. Both proteins were imported into chloroplasts and transit peptides removed. Conversely, when neither protein in the polyprotein contained a transit peptide, as in construct 9 (pGFP2A-GUS) neither product could be imported by the plastids.
DISCUSSION
In this study, we describe a versatile and simple strategy for introducing multiple proteins into plants that, moreover, allows coordinate expression from a single promoter and is compatible with targeting proteins to different subcellular compartments. Protein sequences are linked via the 20-amino acid FMDV 2A sequence within a single ORF in a polyprotein construct. On translation, the 2A peptide acts cotranslationally, while still on the ribosome, to effect a discontinuity in protein synthesis at its own C terminus such that a peptide bond is not formed yet translation continues. Thus the predominant products of translation of 2A-polyproteins of this type are the dissociated component proteins. Since 2A works rapidly during translation, it does not interfere with the targeting of component proteins to a variety of subcellular locations either cotranslationally or posttranslationally.
The translation of the 2A sequence on eukaryotic ribosomes is the absolute but sole requirement for dissociation of 2A-polyproteins. Thus 2A can be used a number of times in a polyprotein to effect efficient dissociation into multiple component polypeptides. We have shown this for three different polypeptides encoded in a variety of polyproteins such as pGFP2A-CAT2A-GUS, and 2A has also been used by others to express a concatamer of three CAT and one GUS polypeptides (Ma and Mitra, 2002). Thus, theoretically, at least, any number of polypeptides could be linked within 2A-polyprotein constructs to enable their rapid introduction into, and coordinate expression in, transgenic plants. This may alleviate some of the problems currently encountered when attempting to introduce whole new biochemical pathways or complex heteromeric proteins into plants. The development of golden rice, for example, is one of the most sophisticated exercises in plant metabolic engineering to date. This involved the cotransformation of three genes (plus a selectable marker gene) into rice to allow the grains to accumulate β-carotene (provitamin A), a development with potential to alleviate vitamin A deficiency in certain regions of the world (Ye et al., 2000). The nutritional benefits of golden rice could be further improved by additionally increasing resorbable iron which is necessary for efficient provitamin A absorption. It has been estimated that this may require introduction and expression of another three genes, a task that will be difficult and time-consuming using conventional technologies but that could be greatly simplified by the use of 2A-polyprotein technology.
Success in the golden rice story depended not only on the expression of multiple introduced proteins in rice endosperm, but on the ability to target those enzymes to the correct subcellular compartment for carotenoid biosynthesis, the plastid. The 2A-polyprotein system will only be truly valuable for plant metabolic engineering if its use is compatible with this need to target proteins to various subcellular locations. We have demonstrated here that two chloroplast proteins, the 23-kD and 33-kD polypeptides of the oxygen evolving complex of photosystem II, can be correctly targeted to, and imported into, chloroplasts when encoded within 2A-polyproteins. It did not matter whether the proteins occupied positions anterior or posterior to 2A in the polyprotein—so long as they contained the requisite transit peptide identifying them for chloroplast import, they were targeted correctly.
Like chloroplasts, the endomembrane system of plants is an important compartment that heterologous proteins might need to be targeted to, either for localization in the ER, Golgi or plasma membrane, or for secretion. Our data demonstrate that it is possible to target proteins, even membrane proteins, to the plant endomembrane system from anterior (as in the erGFP2A-ble construct) or posterior (as in the GUS2A-F5H construct) positions within a 2A-polyprotein. Similar work has confirmed the correct ER localization of proteins expressed from 2A-polyproteins in yeast (Saccharomyces cerevisiae) and has demonstrated that each polypeptide absolutely requires its own signal peptide for targeting into the ER (de Felipe et al., 2003). Our results therefore reinforce an accumulating body of data from other eukaryotic systems demonstrating the faithful ER-targeting of polypeptides expressed from 2A-polyproteins. Three independent groups have shown that 2A can be used to express a complex heterodimeric protein in mammalian or insect cells. When separated by 2A and expressed in COS or insect cells, the two polypeptides of interleukin-12 were correctly dissociated, translocated across the ER membrane, assembled, and secreted in a biologically active form (Chaplin et al., 1999; Kokuho et al., 1999; De Rose et al., 2000). These results indicate that 2A-polyproteins should be useful also for the production of complex, multimeric proteins in plant cells.
The cotranslational nature of 2A-mediated protein dissociation (de Felipe et al., 2003) distinguishes the 2A system from other polyprotein systems that have been used in plants. For example, several groups have assessed the value of a polyprotein system based on the proteolytic cleavage activity of NIa, a 48-kD protease of plant potyviruses (Marcos and Beachy, 1994; von Bodman et al., 1995), including an evaluation of the possibility of targeting polyprotein-encoded proteins to chloroplasts (Dasgupta et al., 1998). Constructs incorporating chloroplast-targeted proteins showed that, because cleavage by NIa occurs posttranslationally, it is possible for chloroplast localization to take place before complete processing of the polyprotein, resulting in mislocalization of incompletely processed polypeptides to chloroplasts (Dasgupta et al., 1998). This problem should be avoided by use of FMDV 2A since 2A-polyprotein dissociation occurs before nascent polypeptides are released from the ribosome and become free for chloroplast import.
Encoding proteins within 2A-polyproteins greatly simplifies the problems inherent with getting multiple proteins coordinately expressed in plants, but the system nevertheless has some limitations. Dissociation of 2A-polyproteins is thought to occur during their translation when peptide bond formation between the terminal Gly and Pro of the 2A sequence is disrupted (Ryan et al., 1999; Donnelly et al., 2001a). The protein anterior to 2A in the polyprotein therefore ends up with a C-terminal extension of 19 amino acids of 2A sequence, whereas the protein posterior to 2A has a single extra N-terminal Pro. Clearly, some proteins may be sensitive to a C-terminal extension that could inhibit activity or targeting or that might arouse regulatory concerns for products produced in this way. The N-terminal Pro is likely to be less of a problem since it will not destabilize proteins according to the N-end rule (Varshavsky, 1992), and it has also been shown to have no effect on ER targeting or translocation in yeast (de Felipe et al., 2003). We are currently testing constructs where the 2A sequence has been combined with other useful peptide motifs to produce polyprotein linkers that are completely removed after translation in order to overcome these concerns. Similarly, when translated in vitro, a proportion of each polyprotein remains undissociated and it will be important to ensure that, for in vivo applications, dissociation is as complete as possible. However, we have observed very little undissociated polyprotein when 2A-constructs are introduced into plants. Nevertheless work aimed at improving the efficiency of dissociation is ongoing as are experiments designed to quantify the exact relative stoichiometry of proteins coexpressed in this way.
2A-polyproteins have obvious biotechnological applications for expressing novel biochemical pathways or multimeric proteins rapidly in plants. Further benefits include the facts that expression of different proteins is coordinated from a single promoter and that the potential for transgene instability due to duplicated regulatory sequences is therefore reduced. However, our work also suggests how 2A-polyproteins might profitably be used to improve the design of transgenic experiments. For example, our erGFP2A-ble construct illustrates how an effect gene might be directly linked to a selectable marker gene via 2A, ensuring that all transformed individuals coming through selection will express the effect gene (provided the transgene is intact). This strategy has already been successfully used in animal cells (Precious et al., 1995). Likewise, our GUS2A-F5H construct illustrates how genes whose expression is difficult to detect, due to lack of appropriate probes or assays, can be easily monitored by linking them via 2A to a marker gene, such as GUS, that can report on their temporal, spatial, and quantitative expression in individual transgenics. In the light of the genomics revolution and the explosion in the discovery of new genes of no known function, this application for 2A-polyproteins may become increasingly important.
MATERIALS AND METHODS
Constructs
p23K2A-GUS, p23K2A, pm23K2A-GUS, pGFP2A-23K, p23K2A-33K, and pGFP2A-m23K
Primers: A, TTTTGGGCCCGAATTCGCCATGGCGTCCACCTCCTGC; B, TTTTATGCATCTATCTAGAGGTGGAGTCCAATCCCAGCAAAGGGTTTGGGATCGGCTTTTCCTGCGACGCTGAAGGATCC; C, TTTTGGGCCCGAATTCGCCATGGCCTACGGAGAAGCAGCC; D, TTTTGGGCCCGCCATGGCAGCGTCTCTCCAA; E, TTTTATGCATCTAGGTGGAGTCCAATCCCAGCAAAGGGTTTGGGATCGGCTTTCCGTTAGACTCGAGCTGCGC.
The wheat (Triticum aestivum) cDNA for the 23-kD polypeptide (James and Robinson, 1991) was used as template to produce p23K2A, p23K2A-GUS, pm23K2A-GUS, pGFP2A-23K and pGFP2A-m23K polyprotein constructs. Oligonucleotide primers were used in PCRs to introduce a C-terminal tag sequence and convenient N- and C-terminal restriction sites onto the precursor (primers A and B) or mature form (primers C and B) of the 23-kD protein. Primer B also removed the termination codon. Amplified products were digested with either (1) EcoRI and XbaI and used to replace GFP in pGFP2A-GUS (Donnelly et al., 2001a) to yield constructs p23K2A-GUS and p232A-GUS or (2) ApaI and NsiI and used to replace GUS in pGFP2A-GUS to yield constructs pGFP2A-23K and pGFP2A-m23K. p23K2A was made by digesting p23K2A-GUS with EcoRI and ApaI and cloning the fragment encoding 23K2A into pGEM11Zf+. A clone for the precursor of the 33-kD polypeptide (Meadows et al., 1991) was used as template to produce p23K2A-33K using primers D (introducing an ApaI site at the 5′ of the transit peptide) and E (incorporating the tag sequence, a stop codon, and an NsiI site) in PCR. The amplified product was cut (ApaI/NsiI) and used to replace GUS in p23K2A-GUS. To prepare plasmid p33K, the same ApaI-NsiI fragment was ligated into ApaI/NsiI-cut pGEM11Zf+.
pble and perGFP2A-ble
Primers: 5′GFP, CGGGATCC GATATCTCAACACAACATATACAAAACAAAC; 3′GFP, GGCATGCAAGCTCATCATGTTTGTATAGTT; 5′Shble, AGGGCCCATGGCCAAGTTGACCAGTGCCG; 3′Shble, GGCTGCAGGTCGACTCAGTCCTGCTCCTCGGCCACGAAG.
The cassette containing the CaMVp and nopaline synthase terminator from pJRI9 (Abbott et al., 2002) was moved into pPZP111 (Hajdukiewicz et al., 1994) as a HindIII-EcoRI fragment to obtain the binary vector pPZPC-A. The nopaline synthase terminator was moved from pJRI9 into pPZP111 (HindIII-PstI fragment) to obtain binary vector pPZPA. To make the p35S:Shble clone, the phleomycin-resistant gene (Shble) was released from pUT56 (Cayla, Toulouse, France) using EcoRV and BglII and inserted into pPZPC-A predigested with SmaI and BamHI. To make the p35S:ER-mGFP2A-ble clone, GFP was PCR-amplified from pRTL2 ER-mGFP using 5′-GFP and 3′-GFP primers. Vector pRTL2 ER-mGFP contains a modified version of GFP (mGFP5; Haseloff, 1999) with the TEV 5′ leader sequence and an ER-targeting sequence at its 5′ end. The GUS gene in pGUS-2A-GFP (Donnelly et al., 2001a) was replaced by the ER-mGFP PCR fragment using BamHI and SphI to obtain plasmid pER-mGFP-2A-GFP. The phleomycin resistance gene (Shble) was amplified from pUT56 (Cayla) using 5′-Shble and 3′-Shble primers. The second GFP in pER-mGFP-2A-GFP was replaced with the Shble PCR fragment digested with ApaI and PstI to obtain pER-mGFP-2A-Shble. This plasmid was cut with NdeI and SalI and fragment (3′)mGFP-2A- Shble isolated. pRTL2 ER-mGFP plasmid was cut with HincII and NdeI and the fragment 2 × 35S:ER-mGFP(5′) was isolated. The two fragments were ligated into pPZPA predigested with SmaI and SalI to obtain the binary vector pPZP2 × 35S:ER-mGFP-2A-Shble (denoted perGFP2A-ble in text).
pGUS2A-F5H
Primers: F5HA, GCGCGCGGGCCCATGGAGTCTTCTATATCAC; F5HB, GGCCGGCTGCAGTTAAAGAGCACAGATGAG.
The Arabidopsis F5H cDNA was PCR-amplified from pCC30 (gift from Clint Chapple, Purdue University, West Lafayette, IN) using Pfu DNA polymerase (Promega) and primers F5HA and F5HB which also attached ApaI and PstI restriction sites to 5′ and 3′ ends of the cDNA, respectively. The amplified product was cut with ApaI and PstI and used to replace GFP in pGUS2A-GFP. For plant transformation, the promoter-terminator cassette from pJR19 (Abbott et al., 2002) was isolated as an EcoRI/HindIII fragment and ligated into pMOG1006 cut with the same enzymes to make pMOG1006-35S. The 35S promoter was later replaced with the Arabidopsis C4H promoter (Bell-Lelong et al., 1997) to make pMOG-C4H. GUS2A-F5H was isolated as a SalI fragment and inserted into SalI-digested pMOG1006-35S or pMOG-C4H plasmids. Escherichia coli transformants were screened by colony hybridization, using a DIG-labeled F5H probe and the insert orientation of positive clones determined by PCR and restriction analysis.
pGFP2A-CAT2A-GUS, pGFP2A-P42A-GUS, and pGUS2A-CAT2A-GFP
Primers: Xbamyc, CGGCTCTAGAGAACAAAAACTCATCTCAGAAGAAGATCTGCAGCTGTTGAATTTTGAC; GUS2, TGATCAATTCCACAGTTTTC; HGUSBam, CGCGGGATCCATGCACCACCACCACCACCACTTA; XbaGUS, CGTCTAGATTGTTTGCCTCCCTG; ApaHis, CGCGGGGCCCCACCACCACCACCACCAC; P4Apa, CGCCGGGCCCTCAATGGTGGTGTACAACAAC; P4Bam, CGCCGGATCCACCATGTCAATGGTGGTGTAC; P4Xba, GCGCTCTAGATCCGCGCTTGATAAGTTTTGG; P4Xho, GCGCCTCGAGCTCCGCGCTTGATAAGTTTTGG; CATApa, CGCGGGGCCCGAGAAAAAAATCACTGC; CATBam, CGCGGGATCCACCATGGAGAAAAAAATCACTG; GFPXho, GCGCCTCGAGCCCCGG ACTTGTATAGT TC.
Using pSTA1 (Donnelly et al., 2001b) as template and primers Xbamyc and GUS2, a C-terminal c-myc tag was introduced onto GFP by PCR, yielding pcD23. The P4 gene in pSAB53 (Li et al., 2000) was amplified by PCR using primers P4Apa and P4Xho, and used to replace GUS in pcD23, yielding pcD24. GFP in pcD24 was replaced by GUS (PCR-amplified from pSTA1 using primers HGUSBam and XbaGUS) to obtain pcD34. pcD32 was constructed by replacing P4 in pcD34 with GFP (PCR-amplified from pSTA1 using primers ApaHis and GFPXho). GFP in pcD23 was replaced with CAT (PCR-amplified from pCAT2AGUS (Ryan and Drew, 1994) using primers CATBam and GUS2), to obtain pcD13. Similarly, GFP in pcD23 was replaced by P4 (PCR-amplified from oSAB53 using primers P4Bam and P4Xba) to obtain pcD43. CAT2A (PCR-amplified from pcD13 using primers CATApa and GUS2) was restricted with ApaI, and inserted into similarly cut pcD23 and pcD32, to obtain pcT213 and pcT312. Similarly, P42A (PCR-amplified from pcT213 using primers P4Apa and GUS2) was restricted with ApaI, and inserted into similarly cut pcD23 to obtain pcT243. Finally, the polyprotein sequences in pcT213, pcT312, and pcT243 were inserted as BamHI-XhoI fragments, into pcM, a vector derived from pcDNA3.1(+) with two modifications in the MCS region: (1) the c-myc sequence was inserted between the XhoI and XbaI sites; (2) the XbaI site was then eliminated. The resulting plasmids were named pGFP2A-CAT2A-GUS, pGFP2A-P42A-GUS, and pGUS2A-CAT2A-GFP.
Translation in Vitro
Coupled TNT transcription-translation wheat germ or TNT Quick Coupled Transcription/Translation system reticulocyte lysate (Promega) were programmed with plasmid DNA (100 ng−1 μg) and incubated in the presence of [35S]Met (Amersham, Buckinghamshire, UK) at 30°C for 60 to 90 min. Translation reactions were arrested by the addition of SDS loading buffer. Canine pancreatic microsomal membranes (Promega) were used according to supplier's instructions. Translation reactions were analyzed by SDS-PAGE and the distribution of radiolabel determined either by autoradiography or by phosphorimaging using a Fujix BAS 1000 (Fuji, Tokyo).
Chloroplast Preparation and Import Study
Intact chloroplasts were isolated from leaves of 9-d-old pea (Pisum sativum var. Feltham First) seedlings using differential centrifugation followed by Percoll gradient centrifugation as described by Bogsch et al. (1997). Chloroplasts were washed twice in import buffer, resuspended at 1 mg chlorophyll/mL, and used immediately. For import assays, chloroplasts (50 μg of chlorophyll) were preincubated in the presence of ATP for 10 min, then radiolabeled proteins were added and the mixture incubated for 25 min in the light. After import, chloroplasts were washed in 1 mL cold import buffer, pelleted, and resuspended in 120 μL import buffer containing thermolysin and CaCl2. The protease digestion was incubated on ice for 45 min and stopped by adding EDTA to 10 mm. Proteins were analyzed by SDS-PAGE (12.5%) and detected by autoradiography or phosphorimaging using a Fujix BAS 1000 (Fuji).
Plant Transformation
One-week etiolated seedlings (Nicotiana tabacum cv Xanthi) grown on vertical plates of Murashige and Skoog (MS) medium (4.41 g/L MS medium [Difco Laboratories, Detroit], 30 g/L Suc, 8 g/L bactoagar, pH 5.8) were inoculated with Agrobacterium tumefaciens LBA4404 strain harboring the binary vectors p35S:Shble or perGFP2A-ble and vacuum-infiltrated according to the method of Tinland et al. (1995). The inoculated plantlets were placed on MS plates with 0.1 mg/L naphthalene acetic acid and 1 mg/L 6-benzyl amino purine for 3 d, then on to MS plates with 0.1 mg/L naphthalene acetic acid, 1 mg/L 6-benzyl amino purine, 400 mg/L carbenacillin and 20 mg/L phleomycin. Shoots growing from calluses were planted into pots of MS media with 20 mg/L phleomycin for root induction. Plant transformation with pMOG-C4H:GUS2A-F5H was performed similarly except that Nicotiana tabacum cv Samsun NN was used.
Western Blots
Leaves of 1-month-old plants grown in MS media were used for protein extraction. Plant material was ground in liquid nitrogen, dissolved in Laemmli buffer, boiled, and centrifuged. Protein samples (30 μg) were separated by SDS-PAGE (10%–20%, Invitrogen, Carlsbad, CA) then blotted on to nitrocellulose membrane. Primary antibodies (anti-GFP, CLONTECH, Palo Alto, CA; anti-ShBle, Cayla) were used at 1/2,000 and 1/500 dilutions, respectively. Detection was carried out with the Phototope Western Blot detection kit (New England Biolabs, Beverly, MA).
Fluorescence Microscopy
Leaves of wild-type and transgenic tobacco plants (Nicotiana tabacum CV Samsun NN) expressing perGFP2A-ble were used for GFP detection on a Bio-Rad MRC 1000 confocal laser scanning microscope (Bio-Rad, Spectroscopy Group, Cambridge, MA) as described (Oparka et al., 1995).
Klason Lignin Analyses
Klason lignin analyses were performed according to previously described protocols (Lapierre et al., 1995).
Acknowledgments
We are grateful to Colin Robinson from the University of Warwick, (Coventry, UK) for providing p23 kD and p33 kD clones and for instruction in the chloroplast import assay. We thank Catherine Lapierre and her staff at Institut National de la Recherche Agronomique (INRA), Grignon, France, for performing Klason lignin analyses. We thank Leo Melchers, Syngenta, Research Triangle Park, North Carolina for pMOG1006, Jim Haseloff (University of Cambridge, Cambridge, UK) for an erGFP clone, and Clint Chapple (Purdue University, West Lafayette, IN) for a clone of Arabidopsis F5H.
This work was supported by research grants from the Biotechnology and Biological Sciences Research Council.
References
- Abbott JC, Barakate A, Pinçon G, Legrand M, Lapierre C, Mila I, Schuch W, Halpin C (2002) Simultaneous suppression of multiple genes by single transgenes. Downregulation of three unrelated lignin biosynthetic genes in tobacco. Plant Physiol 128: 844–853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell-Lelong DA, Cusumano JC, Meyer K, Chapple C (1997) Cinnamate-4-hydroxylase expression in Arabidopsis - regulation in response to development and the environment. Plant Physiol 113: 729–738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogsch E, Brink S, Robinson C (1997) Pathway specificity for a delta pH-dependent precursor thylakoid lumen protein is governed by a ‘Sec-avoidance’ motif in the transfer peptide and a ‘Sec-incompatible’ mature protein. EMBO J 16: 3851–3859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaplin PJ, Camon EB, Villarreal-Ramos B, Flint M, Ryan MD, Collins RA (1999) Production of interleukin-12 as a self-processing 2A polypeptide. J Interferon Cytokine Res 19: 235–241 [DOI] [PubMed] [Google Scholar]
- Chapple C (1998) Molecular-genetic analysis of plant cytochrome P450-dependent monooxygenases. Annu Rev Plant Physiol Plant Mol Biol 49: 311–343 [DOI] [PubMed] [Google Scholar]
- Dasgupta S, Collins GB, Hunt AG (1998) Co-ordinated expression of multiple enzymes in different subcellular compartments in plants. Plant J 16: 107–116 [DOI] [PubMed] [Google Scholar]
- de Felipe P, Hughes LE, Ryan MD, Brown JD (2003) Co-translational, intraribosomal cleavage of polypeptides by the foot-and-mouth disease virus 2A peptide. J Biol Chem 278: 11441–11448 [DOI] [PubMed] [Google Scholar]
- De Rose R, Scheerlinck JPY, Casey G, Wood PR, Tennent JM, Chaplin PJ (2000) Ovine interleukin-12: analysis of biologic function and species comparison. J Interferon Cytokine Res 20: 557–564 [DOI] [PubMed] [Google Scholar]
- Donnelly MLL, Hughes LE, Luke G, Mendoza H, ten Dam E, Gani D, Ryan MD (2001. b) The ‘cleavage’ activities of foot-and-mouth disease virus 2A site-directed mutants and naturally occurring ‘2A-like’ sequences. J Gen Virol 82: 1027–1041 [DOI] [PubMed] [Google Scholar]
- Donnelly MLL, Luke G, Mehrotra A, Li X, Hughes LE, Gani D, Ryan MD (2001. a) Analysis of the aphthovirus 2A/2B polyprotein ‘cleavage’ mechanism indicates not a proteolytic reaction, but a novel translational effect: a putative ribosomal ‘skip’. J Gen Virol 82: 1013–1025 [DOI] [PubMed] [Google Scholar]
- Franke R, McMichael CM, Meyer K, Shirley AM, Cusumano JC, Chapple C (2000) Modified lignin in tobacco and poplar plants over-expressing the Arabidopsis gene encoding ferulate 5-hydroxylase. Plant J 22: 223–234 [DOI] [PubMed] [Google Scholar]
- Hajdukiewicz P, Svab Z, Maliga P (1994) The small, versatile pPZP family of Agrobacterium binary vectors for plant transformation. Plant Mol Biol 25: 989–994 [DOI] [PubMed] [Google Scholar]
- Halpin C, Barakate A, Askari B, Abbott J, Ryan MD (2001) Enabling technologies for manipulating multiple genes on complex pathways. Plant Mol Biol 47: 295–310 [PubMed] [Google Scholar]
- Halpin C, Boerjan W (2003) Stacking transgenes in forest trees. Trends Plant Sci 8: 363–365 [DOI] [PubMed] [Google Scholar]
- Halpin C, Cooke SE, Barakate A, El Amrani A, Ryan MD (1999) Self-processing polyproteins - a system for co-ordinate expression of polyproteins in transgenic plants. Plant J 17: 453–459 [DOI] [PubMed] [Google Scholar]
- Haseloff J (1999) GFP variants for multispectral imaging of living cells. Methods Cell Biol 58: 139–151 [DOI] [PubMed] [Google Scholar]
- James HE, Robinson C (1991) Nucleotide sequence of cDNA encoding the precursor of the 23 kD protein of the photosynthetic oxygen-evolving complex from wheat. Plant Mol Biol 17: 179–182 [DOI] [PubMed] [Google Scholar]
- Kokuho T, Watanabe S, Yokomizo Y, Inumaru S (1999) Production of biologically active, heterodimeric porcine interleukin-12 using a monocistronic baculoviral expression system. Vet Immunol Immunopathol 72: 289–302 [DOI] [PubMed] [Google Scholar]
- Lapierre C, Pollet B, Rolando C (1995) New insight into the molecular architecture of hardwood lignins by chemical degradative methods. Res Chem Intermed 21: 397–412 [Google Scholar]
- Li X, Ryan MD, Lamb JW (2000) Potato leaf roll virus protein P1 contains a serine proteinase domain. J Gen Virol 81: 1857–1864 [DOI] [PubMed] [Google Scholar]
- Ma C, Mitra A (2002) Expressing multiple genes in a single open reading frame with the 2A region of foot-and-mouth disease virus as a linker. Mol Breed 9: 191–199 [Google Scholar]
- Marcos JF, Beachy RN (1994) In vitro characterization of a cassette to accumulate multiple proteins through synthesis of a self-processing polypeptide. Plant Mol Biol 24: 495–503 [DOI] [PubMed] [Google Scholar]
- Meadows JW, Hulford A, Raines CA, Robinson C (1991) Nucleotide sequence of a cDNA clone encoding the precursor of the 33 kDa protein of the oxygen-evolving complex from wheat. Plant Mol Biol 16: 1085–1087 [DOI] [PubMed] [Google Scholar]
- Oparka KJ, Roberts AG, Prior DAM, Chapman S, Baulcombe D, Santa Cruz S (1995) Imaging the green fluorescent protein in plants – viruses carry the torch. Protoplasma 189: 133–141 [Google Scholar]
- Precious B, Young DF, Bermingham A, Fearns R, Ryan MD, Randall RE (1995) Inducible expression of the P, V, and NP genes of the paramyxovirus simian virus 5 in cell lines and an examination of the NP-P and NP-V interactions. J Virol 69: 8001–8010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roosien J, Belsham GJ, Ryan MD, King AMQ, Vlak JM (1990) Synthesis of foot-and-mouth disease virus capsid proteins in insect cells using baculovirus expression vectors. J Gen Virol 71: 1703–1711 [DOI] [PubMed] [Google Scholar]
- Ryan MD, Donnelly MLL, Lewis A, Mehrotra AP, Wilkie J, Gani D (1999) A model for non-stoichiometric, co-translational protein scission in eukaryotic ribosomes. Bioorg Chem 27: 55–79 [Google Scholar]
- Ryan MD, Drew J (1994) Foot-and-mouth disease virus 2A ligopeptide mediated cleavage of an artificial polyprotein. EMBO J 134: 928–933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki N, Geletka LM, Nuss DL (2000) Essential and dispensable virus-encoded replication elements revealed by efforts to develop hypoviruses as gene expression vectors. J Virol 74: 7568–7577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tinland B, Schoumacher F, Gloeckler V, Bravo-Angel AM, Hohn B (1995) The Agrobacterium tumefaciens virulence D2 protein is responsible for precise integration of T-DNA into the plant genome. EMBO J 14: 3585–3595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varshavsky A (1992) The N-end rule. Cell 69: 725–735 [DOI] [PubMed] [Google Scholar]
- von Bodman S, Domier LL, Farrand SK (1995) Expression of multiple eukaryotic genes from a single promoter in Nicotiana. Biotechnology 13: 587–591 [DOI] [PubMed] [Google Scholar]
- Ye X, Al-Babili S, Klöti A, Zhang J, Lucca P, Beyer P, Potrykus I (2000) Engineering the provitamin A (β-carotene) biosynthetic pathway into (carotenoid-free) rice endosperm. Science 287: 303–305 [DOI] [PubMed] [Google Scholar]