Abstract
Background and Aims
A greater understanding of cholestatic disease is necessary to advance diagnostic tools and therapeutic options for conditions such as primary biliary cirrhosis (PBC) and primary sclerosing cholangitis (PSC). The purpose of this study was to determine and compare the serum metabolomes of patients with PBC (n=18) or PSC (n=21) and healthy controls (n=10) and to identify metabolites that may differentiate these two cholestatic diseases.
Methods and Results
Using a mass spectrometry-based, non-targeted biochemical profiling approach we identified 420 serum metabolites, 101 that differed significantly (p≤0.05) between PBC and control groups, 115 that differed significantly between PSC and control groups, and 56 that differed significantly between PSC and PBC groups. Random forest classification analysis was able to distinguish patients with PBC or PSC with 95% accuracy with selected biochemicals reflective of protein and amino acid metabolism identified as the major contributors. Metabolites related to bile acid metabolism, lipid metabolism, inflammation, and oxidative stress/lipid peroxidation were also identified as differing significantly when comparing the disease groups and controls, with some of these pathways differentially affected in the PBC and PSC groups.
Conclusion
In this study identified novel metabolic changes associated with cholestatic disease that were both consistent and different between PBC and PSC. Validation studies in larger patient cohorts are required to determine the utility of these biochemical markers for diagnosis and therapeutic monitoring of patients with PBC and PSC.
Introduction
Impaired hepatic production and excretion of bile, resulting in dramatic accumulation of circulating bile acids, form the basis for cholestatic liver disease. Toxins/drugs, gall stones, local tumors, genetic alterations, autoimmune disorders, and pregnancy, amongst other etiologies, may contribute to reductions in bile flow associated with cholestasis (1–3). Response to hepatobiliary injury ranges from proliferation of cholangiocytes and hepatocytes to development of periductular fibrosis, biliary fibrosis, and cirrhosis (4), ultimately giving rise to end-stage liver disease and the need for liver transplantation. In general, pathogenic mechanisms of cholestatic disease are poorly understood; therefore, clinical management of these complex disorders remains quite limited (5).
Two similar but distinct cholestatic diseases include primary biliary cirrhosis (PBC) and primary sclerosing cholangitis (PSC). PBC is an autoimmune disease in which progressive lymphocytic inflammation leads to destruction of intrahepatic bile ducts, and is predominantly observed in asymptomatic, middle-aged women (6). PSC is characterized by chronic inflammation of the biliary epithelium resulting in destruction of intra- and extrahepatic bile ducts, and is typically diagnosed in middle-aged males with a high preponderance of concurrent inflammatory bowel disease (7). Currently, differential diagnosis of PBC and PSC is sometimes complicated by overlapping clinical, serological, histological, and radiological findings and the overall lack of standardized diagnostic and treatment criteria (8). In addition, ursodeoxycholic acid (UDCA) is currently the only available therapy for cholestatic liver disease but several new investigational drugs are under evaluation for either PBC (Obeticholic acid, NCT01473524) or PSC (simtuzumab, NCT01672853). With the regulatory approval and eventual availability of these agents, a definitive diagnosis will be critical. Moreover, identification of a metabolic profile that would identify responders versus non-responders may provide an opportunity to personalize use of these therapeutic products.
Due to their downstream location within the biological spectrum, small molecules (ranging from 99–1000 Da) represent integration of various upstream processes (i.e., genetics, transcriptomics, and proteomics) and are often very closely related to phenotype (9). In order to gain insight into the pathogenesis of PBC and PSC and identify potential biomarkers to distinguish these two related disorders, we utilized a screening approach to profile the global metabolome (10) of serum samples from patients with PBC or PBC and healthy controls. Aside from a previous report in which 17 serum bile acids were profiled in serum from PBC and PSC patients (11), this is the first study to define and compare the global serum metabolomes of patients with PBC and PSC and to explore utilization of small molecule profiles to differentiate between the two groups.
Patients and Methods
Patients
Serum samples were obtained from a local blood bank (healthy controls) or from an ongoing liver bio bank at Indiana University. Liver bio bank was approved by the Indiana University Institutional Review Board and all participants signed an informed consent that allows archived serum and liver tissue samples to be used for future research purposes. Serum samples were prepared immediately after their acquisition and were frozen at −80°C until the metabolomic studies were conducted.
Metabolic profiling
Sample preparation
The non-targeted metabolic profiling platform employed for this analysis combined three independent platforms: ultrahigh performance liquid chromatography/tandem mass spectrometry (UHPLC/MS/MS) optimized for basic species, UHPLC/MS/MS optimized for acidic species, and gas chromatography/mass spectrometry (GC/MS). Samples were processed essentially as described previously (10, 12). For each sample, 100µL of serum was used for analyses. Using an automated liquid handler (Hamilton LabStar, Salt Lake City, UT), protein was precipitated with methanol that contained four standards to report on extraction efficiency. The resulting supernatant was split into equal aliquots for analysis on the three platforms. Aliquots, dried under nitrogen and vacuum-desiccated, were subsequently either reconstituted in 50µL 0.1% formic acid in water (acidic conditions) or in 50µL 6.5mM ammonium bicarbonate in water, pH 8 (basic conditions) for the two UHPLC/MS/MS analyses or derivatized to a final volume of 50µL for GC/MS analysis using equal parts bistrimethyl-silyl-trifluoroacetamide and solvent mixture acetonitrile:dichloromethane:cyclohexane (5:4:1) with 5% triethylamine at 60°C for one hour. In addition, three types of controls were analyzed in concert with the experimental samples: aliquots of a well-characterized human plasma pool served as technical replicates throughout the data set, extracted water samples served as process blanks, and a cocktail of standards spiked into every analyzed sample allowed instrument performance monitoring. Experimental samples and controls were randomized across two platform run days.
LC/MS/MS and GC/MS
For UHLC/MS/MS analysis, aliquots were separated using a Waters Acquity UPLC (Waters, Millford, MA) and analyzed using an LTQ mass spectrometer (Thermo Fisher Scientific, Inc., Waltham, MA) which consisted of an electrospray ionization (ESI) source and linear ion-trap (LIT) mass analyzer. The MS instrument scanned 99–1000 m/z and alternated between MS and MS2 scans using dynamic exclusion with approximately 6 scans per second. Derivatized samples for GC/MS were separated on a 5% phenyldimethyl silicone column with helium as the carrier gas and a temperature ramp from 60°C to 340°C and then analyzed on a Thermo-Finnigan Trace DSQ MS (Thermo Fisher Scientific, Inc.) operated at unit mass resolving power with electron impact ionization and a 50–750 atomic mass unit scan range.
Metabolite identification
Metabolites were identified by automated comparison of the ion features in the experimental samples to a reference library of chemical standard entries that included retention time, molecular weight (m/z), preferred adducts, and in-source fragments as well as associated MS spectra, and were curated by visual inspection for quality control using software developed at Metabolon (13). At the time of this writing, more than 4000 commercially-available purified standard compounds were included in the reference library.
Statistical analysis
For statistical analyses and data display purposes, any missing values were assumed to be below the limits of detection and these values were imputed with the compound minimum (minimum value imputation). Statistical analysis of log-transformed data was performed using “R” (http://cran.r-project.org/), which is a freely available, open-source software package. Welch’s two-sample t-tests were used to compare differences among groups. Classification by principal component analysis and unsupervised hierarchal clustering was carried out using Array Studio (V5.0). Classification by the unbiased and supervised random forest technique, providing an estimate of the ability to classify individuals, was carried out between groups using the R-package “RandomForest.” Briefly, a set of classification trees, based on continual sampling of the experimental units and compounds, was created and each observation was classified based on the majority votes from all classification trees. Identified drug metabolites contained within the xenobiotic pathway were not included in the RF classification scheme. P-values ≤0.05 were considered statistically significant and p-values <0.10 were reported as trends. Multiple comparisons were accounted for by estimating the false discovery rate (FDR) using q-values (14).
Results
Patient characteristics
A total of 49 subjects were included in this serum metabolic profiling study. Characteristics of the 18 patients with PBC, 21 patients with PSC, and 10 healthy control subjects, including demographics and liver biochemistries, are summarized in Table 1. Consistent with the established gender predispositions, the PBC group was predominantly female (94%) while the PSC group was predominantly male (76%). Additional characteristics such as age, Caucasian race, and body mass index (BMI) were generally well-matched across the cholestatic disease and healthy control groups. Statistical comparisons of liver biochemistries and additional biochemical parameters revealed significantly higher (p-value<0.05) levels of alkaline phosphatase and serum total protein in the PBC and PSC groups as compared to healthy controls, with non-significant elevations in alanine aminotransferase (ALT), aspartate aminotransferase (AST), and total bilirubin. Additional parameters such as serum albumin and serum creatinine were not different between the liver disease and healthy control groups, and the international normalized ratio (INR) and model for end-stage liver disease (MELD) scores were not different when comparing the PBC and PSC groups. It is important to note that the majority of PBC and PSC patients were receiving UDCA therapy (78% and 76%, respectively), and that a small subset of cholestatic patients were also taking anti-hyperlipidemic medications (33% of the PBC group and 10% of the PSC group). Administration of these medications has the potential to affect metabolic pathways highlighted in this report, including bile acid, cholesterol, and lipid metabolism.
Table 1.
Clinical characteristics and liver biochemistries of study participants.
| Primary Biliary Cirrhosis (n=18) |
Primary Sclerosing Cholangitis (n=21) |
Control (n=10) |
|
|---|---|---|---|
| Demographics | |||
| Age (years) | 49 ± 10 | 52 ± 16 | 45 ± 15 |
| Female (%) | 94 | 24 | 40 |
| Caucasian (%) | 89 | 95 | 90 |
| BMI (kg/m2) | 30 ± 7 | 34 ± 8 | 30 ± 6 |
| Hypertension (%) | 28 | 19 | N/A |
| Diabetes (%) | 22 | 5 | N/A |
| IBD (%) | 0 | 71 | N/A |
| Medications | |||
| Anti-hyperlipidimics (%) | 33 | 10 | N/A |
| Ursodeoxycholic acid (%) | 78 | 76 | N/A |
| Liver Biochemistries | |||
| ALT (U/L) | 96 ± 154 | 65 ± 48 | 17 ± 7 |
| AST (U/L) | 136 ± 327 | 57 ± 28 | 23 ± 3 |
| Alk P (U/L)¶ | 231 ± 117 | 249 ± 234 | 58 ± 15 |
| Total Bilirubin (mg/dL) | 1.5 ± 2.3 | 1.4 ± 1.1 | 0.5 ± 0.2 |
| Other parameters | |||
| Serum albumin (mg/dL) | 3.7 ± 0.6 | 3.6 ± 0.5 | 4.0 ± 2.3 |
| Serum total protein (mg/dL) ¶ | 7.5 ± 1.4 | 7.6 ± 0.7 | 6.3 ± 0.3 |
| INR | 1.0 ± 0.1 | 1.2 ± 0.5 | N/A |
| Serum creatinine (mg/dL) | 0.8 ± 0.2 | 0.9 ± 0.3 | 0.9 ± 0.1 |
| MELD score | 8.1 ± 2.8 | 10.6 ± 3.8 | N/A |
All values are expressed in mean ± standard deviation unless otherwise mentioned.
Abbreviations: BMI, body mass index; IBD, inflammatory bowel disease; ALT, alanine aminotransferase, AST, aspartate aminotransferase; Alk P, alkaline phosphatase; INR, international normalized ratio; MELD, model for end-stage liver disease; N/A, not applicable.
p-value<0.05
Metabolic Profiling
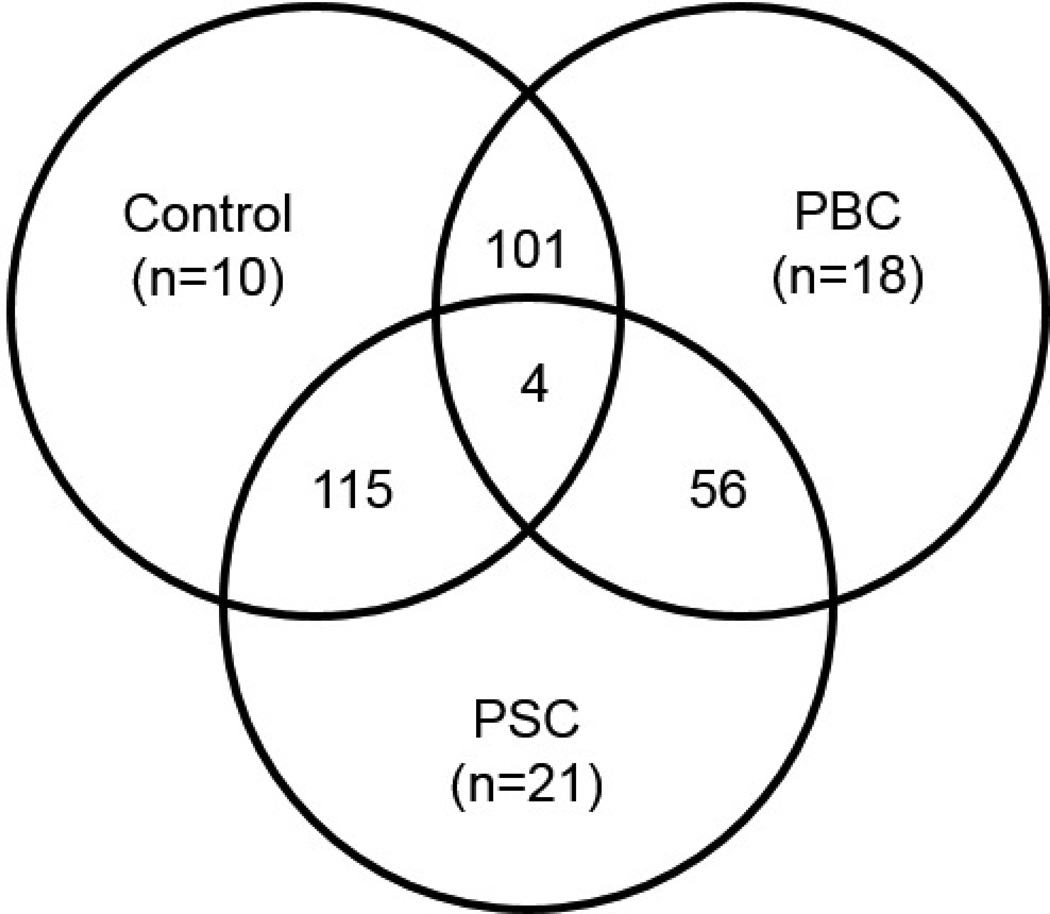
Findings from the global serum metabolomic analysis are summarized in Figure 1 and a list of all identified metabolites, grouped by super- and sub pathways, their changes when comparing among groups, and p- and q-values are provided as Supplementary Table 1. Overall, 420 metabolites were identified and, of these, 101 differed significantly (p≤0.05) when comparing PBC and control groups, 115 differed significantly when comparing PSC and control groups, 56 differed significantly when comparing PSC and PBC groups, and 4 differed significantly across all three comparisons (Figure 1). Directionality of these changes (i.e., biochemicals that were increased or decreased) is reported in Table 2. In addition to significant changes, trending (0.05<p<0.10) changes were also considered and a summary of the numbers and direction of these changes is included in Table 2. Although the number of subjects taking lipid-lowering medications was too low for meaningful comparisons, patients with PBC or PSC who were taking UDCA were compared to those not receiving therapy. In general, the number of statistically significant changes when comparing patients receiving UDCA to those not taking UDCA within the PBC (Δ=23) and PSC (Δ=29) groups was roughly equivalent to the number of changes that would be expected by random chance alone (5 changes for every 100 metabolites). Results from these comparisons are included within Supplementary Table 1.
Figure 1.
Summary of significant biochemical changes (out of 420 identified metabolites) when comparing among groups. Numbers of significant changes (p≤0.05) when comparing across the PBC, PSC, and healthy control groups, in addition to the numbers of subjects within each of the study groups, are displayed.
Table 2.
Summary of significant and trending biochemical changes observed when comparing serum from patients with primary biliary cirrhosis (PBC; n=18), primary sclerosing cholangitis (PSC; n=21), and healthy controls (n=10). The directionality of changes is also indicated in red (increase) or green (decrease).
|
PBC Control |
PSC Control |
PSC PBC |
|
|---|---|---|---|
| Total biochemicals p≤0.05 | 101 | 115 | 56 |
| Biochemicals (↑↓) |
61|40 | 67|48 | 19|37 |
| Total biochemicals 0.05<p<0.10 | 40 | 30 | 29 |
| Biochemicals (↑↓) |
23|17 | 16|14 | 16|13 |
Classification Analyses
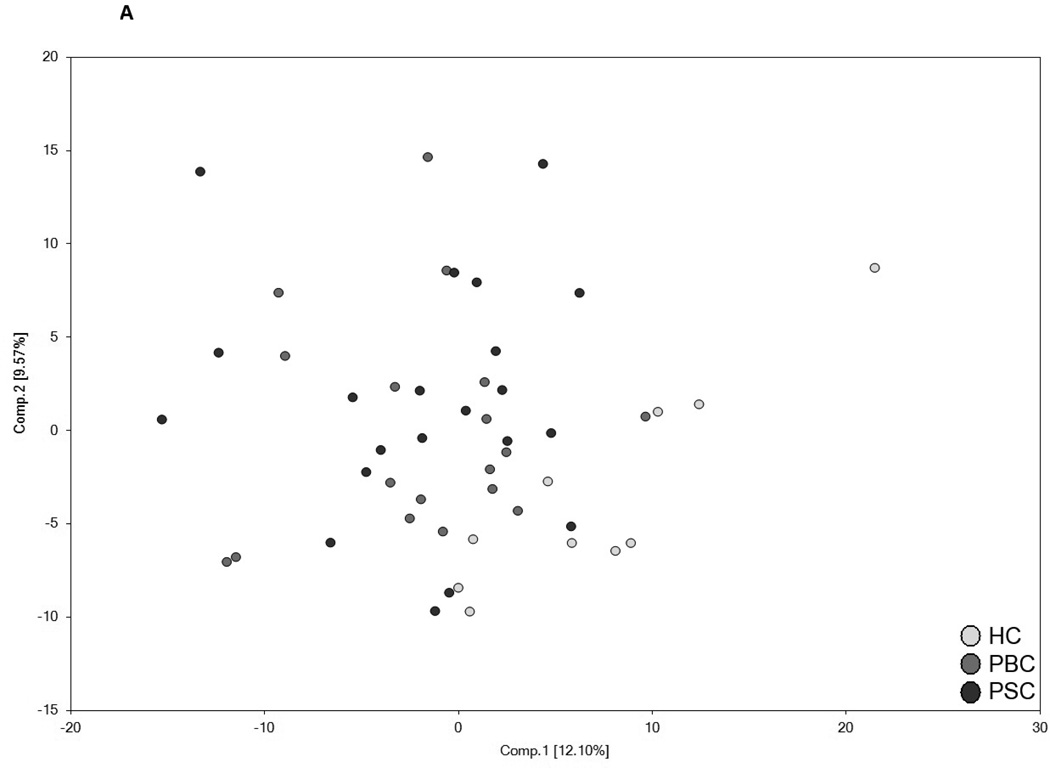
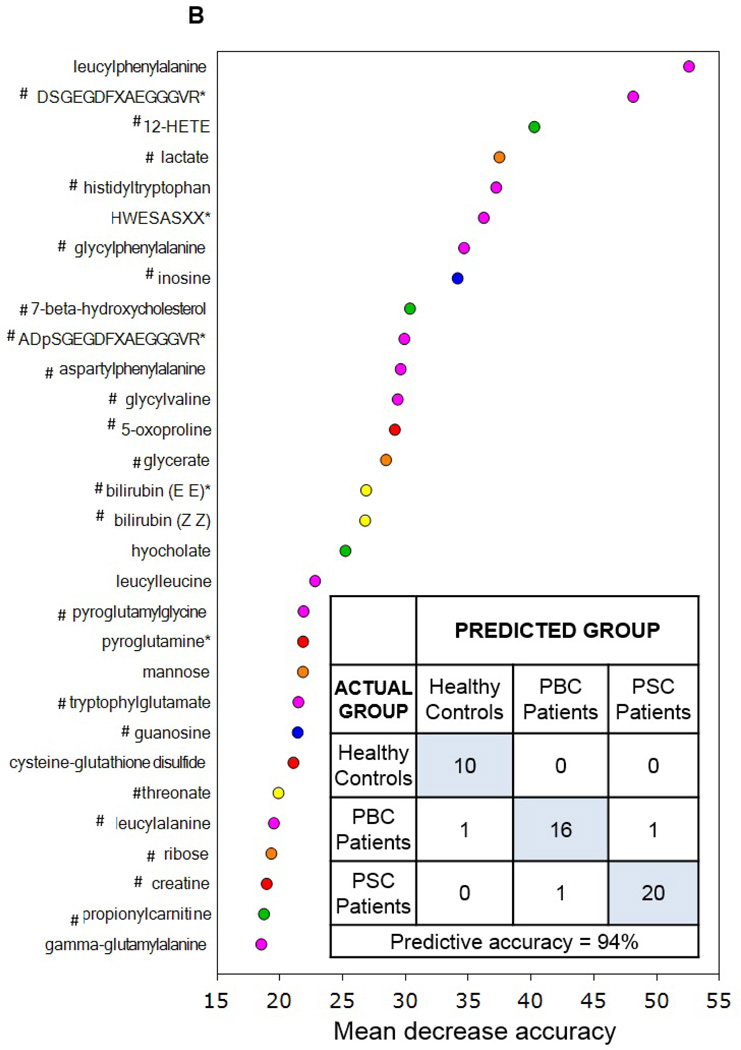
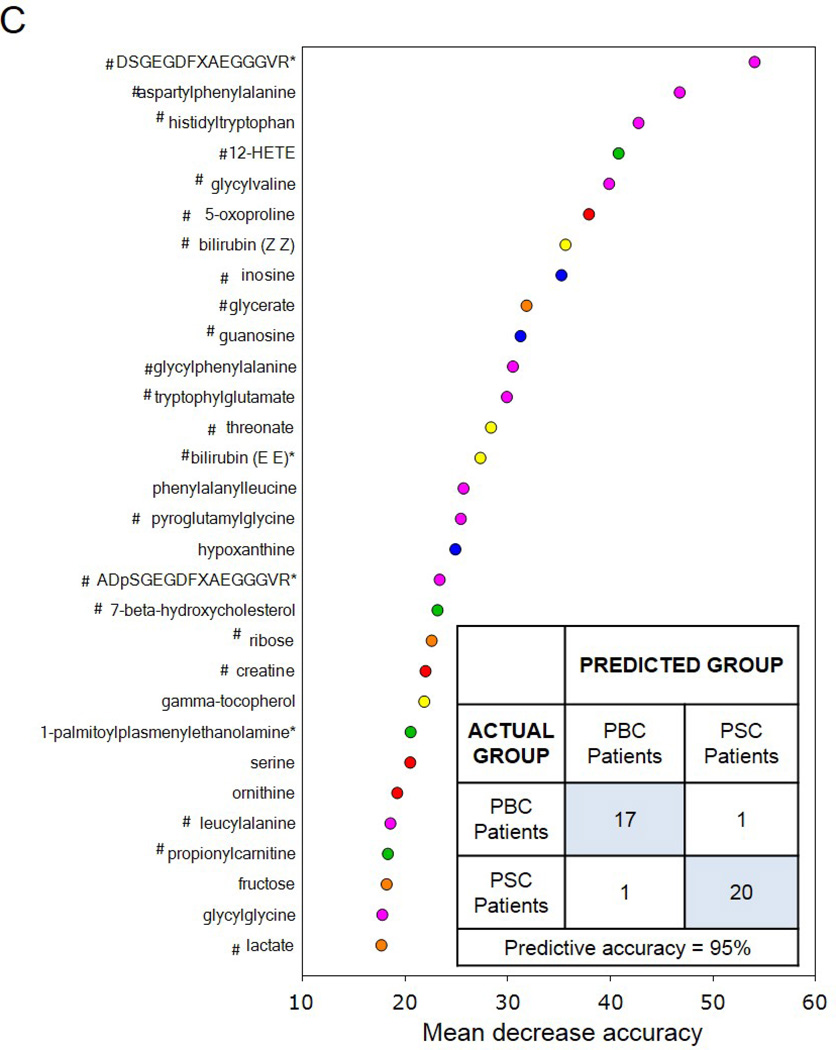
Utilization of global metabolic profiles for classification of control subjects and patients by disease type was determined by principal component analysis (PCA), random forest analysis (RF), and unsupervised hierarchal clustering. As shown in Figure 2A, results from the PCA demonstrate that control subjects were quite distinguishable from those with cholestatic disease, but that a substantial amount of overlap was present when differentiating patients with PBC and PSC. However, control subjects and patient groups were readily identified by RF analysis, which gave an overall predictive accuracy of 94% when differentiating between all three study groups (Figure 2B). Similarly, classification of only the PBC and PSC groups by RF gave an identical overall predictive accuracy of 95% as well (Figure 2C). RF produces a list of biochemicals ranked by their importance to the classification scheme, and the top 30 metabolites identified for each analysis are displayed in Figures 2B and 2C. Metabolites involved in amino acid/protein metabolism (including fibrinogen cleavage peptides), lipid metabolism (including eicosanoids, sterol metabolism, and bile acids), glucose metabolism, nucleotide metabolism, and xenobiotics were identified as contributing to the separation of groups and many of these biochemicals are discussed in more detail in this report. Of note, 22 common biochemicals were included in both RF importance plots. As shown in Supplementary Figure 1, unsupervised hierarchal clustering resulted in limited grouping of healthy control samples with extensive overlap observed between the two liver disease groups.
Figure 2.
Utilization of global metabolic profiles for classification of control subjects and patients by disease type by principal component analysis (PCA) and random forest analysis (RF). Results from the PCA (Panel A) demonstrated that healthy control subjects (HC; clear circles) were quite distinguishable from those with cholestatic disease, but that a substantial amount of overlap was present when differentiating patients with PBC (light grey circles) and PSC (dark grey circles). Classification of all three study groups (Panel B) or the two liver disease groups (Panel C) by RF gave overall predictive accuracies of 94% and 95% (inset confusion matrices), respectively, and 22 common metabolites (designated by#) were identified within the biochemical importance plots generated from the RF analyses. Importance plots display both rank of metabolites according to the contribution of each to the classification scheme (mean decrease accuracy) and biochemical pathway identifications (red=amino acid, orange=carbohydrate/energy, yellow=cofactors and vitamins, green=lipid, blue=nucleotide, and pink=peptide).
Pathway Analyses
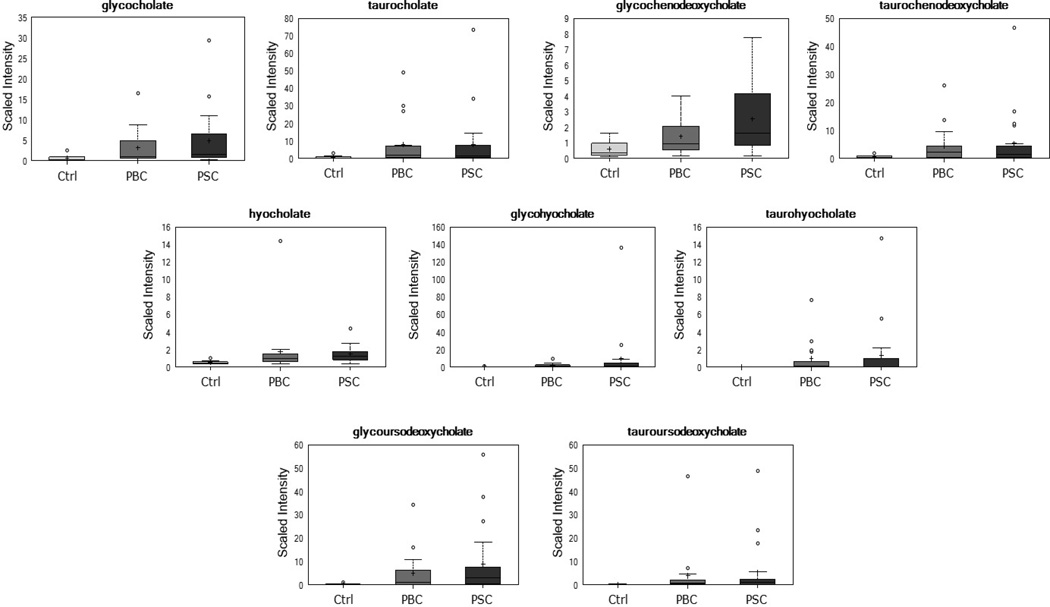
Further consideration of significant biochemical changes identified several metabolic pathways that were differentially affected in patients with PBC or PSC as compared to healthy controls. As shown in Figure 3, significant elevations in several bile acids were observed in the liver disease groups as indicated by both p≤0.05 and q≤0.10 when comparing the PBC or PSC groups and healthy controls. For example, higher levels of the conjugated primary bile acids glycocholate, taurocholate, glycochenodeoxycholate, and taurochenodeoxycholate were present in patients with PBC or PSC. Similarly, significant elevations were noted in the unusual trihydroxy bile acid hyocholate, as well as its conjugated derivatives, glycohyocholate and taurohyocholate, in patients with liver disease. Finally, while the secondary bile acid deoxycholate and its glycine- and taurine-conjugated derivatives were not different when comparing patients with liver disease and controls, greater than 23-fold elevations were observed in glycoursodeoxycholate and tauroursodeoxycholate in the PBC and PSC groups. Although UDCA is synthesized endogenously, these large increases in PBC and PSC patients also reflect exogenous administration of this bile acid as a treatment for cholestatic disease. Indeed, higher levels of glycoursodeoxycholate and tauroursodeoxycholate were identified in PBC and PSC patients taking UDCA as compared to those not receiving this therapy. As bile acids are the primary route for excretion of cholesterol from the body, it is also important to note the significant elevation (p≤0.05 and q≤0.10) in circulating cholesterol in both PBC and PSC patients as compared to controls.
Figure 3.
Differential bile acid levels in patients with PBC or PSC as compared to healthy controls. Boxplots convey the spread of the data with the middle 50% of the data represented by shaded box and the maximum and minimum values (unless there are extreme values) shown by the whiskers. Extreme values are defined as those that exceed, in either direction, 1.5 times the interquartile range. The solid bar across the box represents the median value while the + shows the mean. For each biochemical, data are median scaled with the median value across all samples set equal to 1.0; thus, the y-axis reflects scaled intensity for each metabolite. All displayed bile acids were significantly greater (as indicated by both p≤0.05 and q≤0.10) in the PBC and PSC groups as compared to healthy controls.
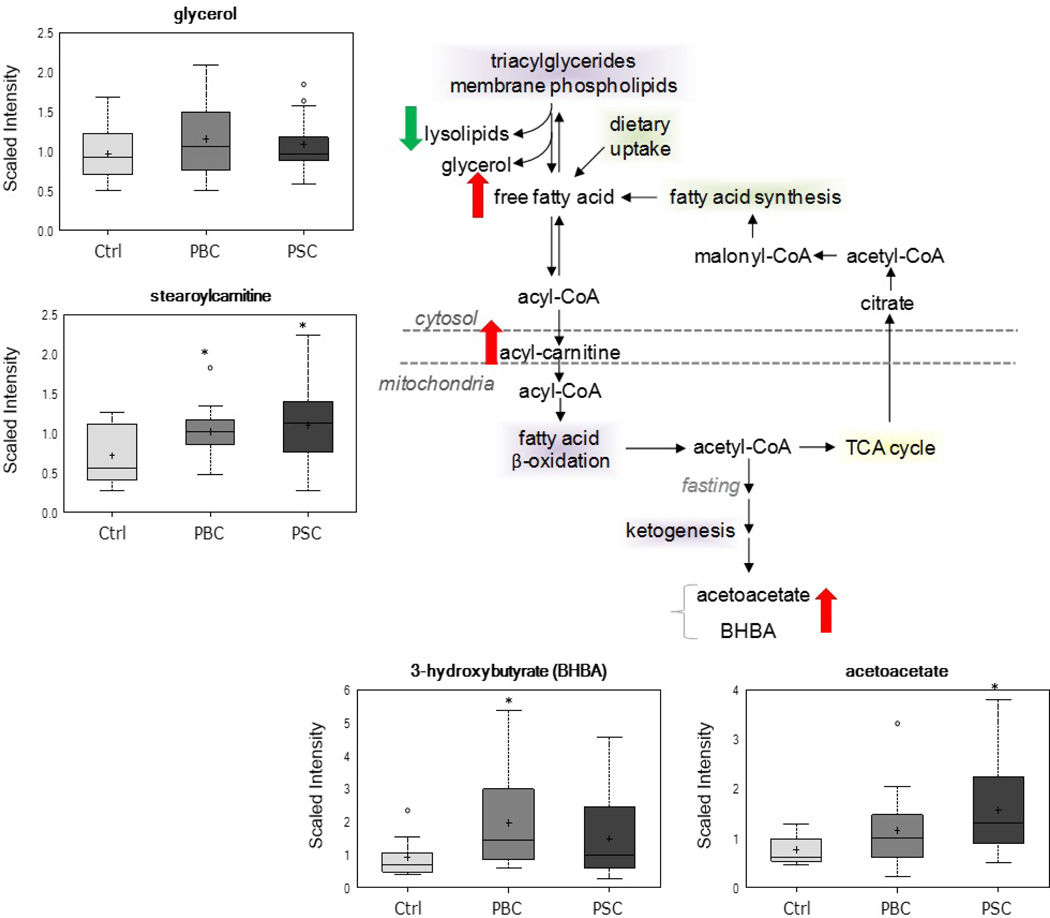
Alterations in biochemicals related to lipid metabolism were also observed in patients with cholestatic disease as compared to healthy controls (Figure 4). Despite unchanged levels of the lipolysis marker glycerol, consistent elevations (p≤0.05 and q≤0.10) in many free fatty acids, including the essential fatty acids linoleate (18:2n6) and linolenate (18:3n3 or 6) and the long-chain unsaturated fatty acids palmitoleate (16:1n7) and oleate (18:1n9), were noted in the PBC and PSC groups. As summarized in Table 3, both the number of trending (0.05<p<0.10) or significant (p≤0.05) changes as well as the magnitude of those changes was slightly more pronounced in patients with PBC as compared to PSC . In addition to changes in free fatty acids, significant elevations in the ketone bodies acetoacetate and 3-hydroxybutyrate (BHBA), as well as several acylcarnitines, including stearoylcarnitine, were also noted in the PBC and PSC groups, respectively, as compared to controls. It is important to note that administration of UDCA had a profound effect on circulating free fatty acid and ketone body levels, but this was only observed within the PSC group (Supplementary Table 1).
Figure 4.
Changes in metabolites related to lipid metabolism in the PBC and PSC groups. In addition to increased serum free fatty acid levels (Table 3), significant (p≤0.05) elevations in ketone bodies and acylcarnitines were also observed in the cholestatic disease groups as compared to controls. *p≤0.05 vs. control
Table 3.
Alterations in free fatty acids when comparing patients with primary biliary cirrhosis (PBC), primary sclerosing cholangitis (PSC), and healthy controls. Trending (0.05<p<0.10) or significant (p≤0.05) elevations are denoted by pink and red shading, respectively, while trending or significant reductions are indicated by light green and dark green shading, respectively.
| Sub Pathway | Biochemical Name |
PBC HC |
PSC HC |
PSC PBC |
|---|---|---|---|---|
| Medium Chain Fatty Acid | caproate (6:0) | 1.26 | 1.12 | 0.89 |
| 10-undecenoate (11:1n1) | 1.32 | 1.65 | 1.25 | |
| 5-dodecenoate (12:1n7) | 1.75 | 1.78 | 1.02 | |
| Long Chain Fatty Acid | myristoleate (14:1n5) | 2.32 | 2.29 | 0.99 |
| pentadecanoate (15:0) | 1.28 | 1.2 | 0.94 | |
| palmitate (16:0) | 1.31 | 1.28 | 0.98 | |
| palmitoleate (16:1n7) | 2.17 | 2.01 | 0.93 | |
| margarate (17:0) | 1.37 | 1.23 | 0.9 | |
| 10-heptadecenoate (17:1n7) | 2.12 | 1.81 | 0.85 | |
| oleate (18:1n9) | 1.84 | 1.82 | 0.99 | |
| Cis-vaccenate (18:1n7) | 1.79 | 1.5 | 0.84 | |
| nonadecanoate (19:0) | 1.33 | 1.14 | 0.86 | |
| 10-nonadecenoate (19:1n9) | 1.87 | 1.7 | 0.91 | |
| eicosenoate (20:1n9 or 11) | 1.97 | 1.65 | 0.84 | |
| Polyunsaturated Fatty Acid (n3 and n6) | stearidonate (18:4n3) | 1.54 | 1.63 | 1.06 |
| docosapentaenoate (n3 DPA; 22:5n3) | 1.45 | 1.37 | 0.94 | |
| docosahexaenoate (DHA; 22:6n3) | 1.27 | 1.38 | 1.09 | |
| linoleate (18:2n6) | 1.69 | 1.66 | 0.99 | |
| linolenate [alpha or gamma; (18:3n3 or 6)] | 1.74 | 1.84 | 1.06 | |
| docosapentaenoate (n6 DPA; 22:5n6) | 1.67 | 1.49 | 0.89 | |
| docosadienoate (22:2n6) | 1.73 | 1.54 | 0.89 | |
| dihomo-linoleate (20:2n6) | 1.55 | 1.54 | 0.99 |
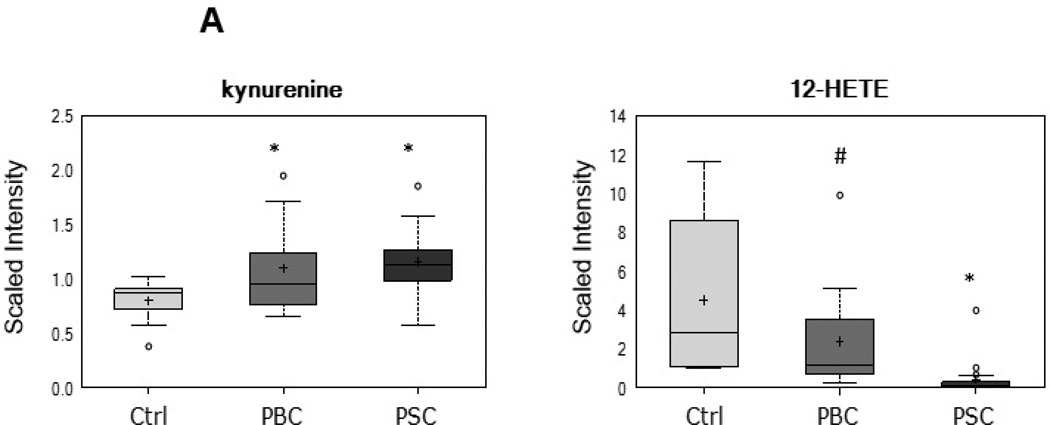
As displayed in Figure 5, several metabolites that serve as biochemical markers of inflammation and oxidative stress were altered in patients with PBC or PSC as compared to controls. For example, serum levels of the tryptophan degradation product kynurenine and the arachidonate-derived eicosanoid 12-hydroxyeicosatetraenoic acid (HETE) were altered in the PBC and PSC groups, suggestive of an inflammatory signature in these patients. Similarly, increased levels of the linoleate-derived product of lipid peroxidation 13-hydroxyoctadecadienoic acid (HODE) + 9-HODE, the marker of oxidative stress and pro-apoptotic metabolite 7β-hydroxycholesterol, and the free radical scavenger biliverdin were observed in PBC patients and increased levels of dimethylarginine (SDMA + ADMA) and biliverdin, along with altered antioxidant (vitamin C and vitamin E) metabolism, was observed in PSC patients.
Figure 5.
Alterations in biochemical markers of inflammation and oxidative stress in patients with PBC or PSC. Metabolites related to inflammation (Panel A) and oxidative stress (Panel B) that exhibited trending (0.05<p<0.10) or significant (p≤0.05) changes when comparing PBC and/or PSC patients to controls are displayed. *p≤0.05 vs. control; #0.05<p<0.10 vs. control
Finally, substantial changes with regard to protein and amino acid metabolism were observed in patients with PBC and PSC (Table 4). In particular, levels of several free amino acids (serine, aspartate, glutamate, phenylalanine, and ornithine) and 9 dipeptides were significantly (p≤0.05) lower in patients with PSC as compared to PBC. Moreover, significant elevations in three fibrinogen cleavage peptides, produced when fibrinogen is cleaved by thrombin for blood clot formation, were also observed in the PSC as compared to PBC group.
Table 4.
Differential alterations in free amino acids, dipeptides, and fibrinogen cleavage peptides when comparing patients with primary biliary cirrhosis (PBC), primary sclerosing cholangitis (PSC), and healthy controls. Trending (0.05<p<0.10) or significant (p≤0.05) elevations are denoted by pink and red shading, respectively, while trending or significant reductions are indicated by light green and dark green shading, respectively.
| Sub Pathway | Biochemical Name |
PBC HC |
PSC HC |
PSC PBC |
|---|---|---|---|---|
| Free Amino Acid | serine | 1.11 | 0.84 | 0.76 |
| aspartate | 1.28 | 0.89 | 0.69 | |
| glutamate | 1.44 | 0.98 | 0.68 | |
| histidine | 0.99 | 0.91 | 0.91 | |
| phenylalanine | 1.2 | 1.06 | 0.88 | |
| ornithine | 1.13 | 0.68 | 0.61 | |
| Dipeptide | aspartylphenylalanine | 1.45 | 0.71 | 0.49 |
| cyclo(phe-phe) | 1.08 | 0.69 | 0.64 | |
| glycylglycine | 1.12 | 1.64 | 1.47 | |
| glycylphenylalanine | 0.73 | 0.42 | 0.58 | |
| glycylvaline | 2.98 | 0.55 | 0.18 | |
| histidyltryptophan | 1.67 | 0.53 | 0.32 | |
| leucylalanine | 0.62 | 1.03 | 1.67 | |
| leucylleucine | 0.66 | 0.39 | 0.59 | |
| leucylphenylalanine | 0.37 | 0.3 | 0.82 | |
| phenylalanylleucine | 1.47 | 0.81 | 0.55 | |
| phenylalanylphenylalanine | 1.63 | 0.94 | 0.58 | |
| phenylalanylserine | 1.33 | 2.07 | 1.56 | |
| phenylalanyltryptophan | 0.69 | 0.55 | 0.8 | |
| pyroglutamylglycine | 1.7 | 1.1 | 0.65 | |
| tryptophylglutamate | 1.96 | 0.9 | 0.46 | |
| valylarginine | 0.89 | 1.37 | 1.53 | |
| Fibrinogen Cleavage Peptide | ADSGEGDFXAEGGGVR* | 1.45 | 1.9 | 1.32 |
| DSGEGDFXAEGGGVR* | 0.99 | 3.86 | 3.9 | |
| ADpSGEGDFXAEGGGVR* | 0.38 | 0.91 | 2.42 |
Discussion
Although metabolomic profiling has been used extensively for biomarker identification and informational studies involving many other liver diseases, particularly hepatocellular carcinoma (15) and drug-induced liver injury (16), to the best of our knowledge this study is the first to investigate the global serum metabolome of PBC and PSC. Here, we observed important differences in the serum metabolic profiles of patients with PBC or PSC as compared to healthy controls and, as a result, potential biomarker candidates for differentiating PBC from PSC were also identified. While clinical diagnosis of PBC is relatively straightforward, PSC presents a greater challenge without the use of endoscopic retrograde cholangiopancreatography (ERCP) or magnetic resonance cholangiopancreatography (MRCP). In addition, a serological hallmark that is present in PBC but not PSC includes the antimitochondrial antibody (AMA), an autoantibody that is found in 90–95% of PBC patients and less than 1% of normal controls (17–19). In the current study, differences in the serum metabolome were identified when comparing PBC and PSC patients and classification analyses demonstrated that, in general, the two disease states were quite distinguishable. In particular, predictive accuracy of the RF analyses, including classification of the PBC and PSC groups only, was high at 94–95% and analysis of the biochemical importance plots generated from both RF analyses revealed that metabolites reflective of protein metabolism (i.e., dipeptides and fibrinogen cleavage peptides) appear to have the greatest power in differentiating groups. Additional differentially-altered pathways with metabolites able to distinguish among groups were also identified when comparing patients with PBC or PSC and these findings are discussed in more detail below.
By definition, circulating levels of bile acids are increased in cholestatic disorders such as PBC and PSC as a result of biliary injury and hepatocyte accumulation of bile acids (3). In the current study, elevations in unconjugated, conjugated, and sulfated bile acids were observed in both liver disease groups, with minor changes in secondary bile acids derived from the gut microbiome. This pattern of change is mostly in agreement with the previous report by Trottier et al. demonstrating that the pathology of PBC and PSC mainly affects primary bile acids (11). One finding of particular interest included a significant elevation in serum hyocholate, an unusual trihydroxy bile acid synthesized by hydroxylation of chenodeoxycholate (20), in both PBC and PSC patients as compared to controls. Hyocholate has been found to be elevated in urine from bile acid-loaded adults (20) and children with Byler disease (21) and may therefore act as a biomarker of cholestasis and the ability of the liver to metabolize bile acids to their secretory forms. Indeed, hyocholate was the only bile acid included within the random forest importance plot as contributing to the separation of the control and liver disease groups.
Also of interest with regard to bile acid metabolism, circulating levels of cholesterol were significantly elevated in serum from both PBC and PSC patients. Perturbations in cholesterol homeostasis have been commonly associated with chronic cholestatic diseases (22). Changes in serum cholesterol are related to altered bile acid production through mechanism(s) that involve, at least to some extent, genetic, transcriptional, translational, and posttranslational modifications of hepatic cytochrome P450 7A1 (CYP7A1) expression/activity (23–25). On a related note, serum levels of the oxidized cholesterol derivatives 7-α-hydrocholesterol (bile acid precursor) and 7-β-hydroxycholesterol (marker of lipid oxidation and cytotoxic, pro-apoptotic lipid mediator) and the steroid hormone cortisol were elevated in only the PBC group. Although 7-α-hydroxycholesterol is not an accurate systemic indicator of hepatic bile acid synthesis (26), and cortisol can be derived from exogenous administration, these differential changes may be indicative of greater lipid peroxidation and potential abnormalities of the pituitary-adrenal axis in patients with PBC. These key differences observed when comparing the two cholestatic groups may point to specific metabolic alterations that contribute to the pathogenesis of PBC but not PSC.
While abnormalities in lipoprotein and cholesterol metabolism have been well-described in patients with cholestatic disease (22), investigation into changes in circulating free fatty acids and hepatic β-oxidation is quite limited (27). In the current study, although consistent elevations in serum free fatty acids were observed in both PBC and PSC patients as compared to controls, increases were slightly more pronounced in the PBC as compared to the PSC group. This finding may be attributable to the fact that PSC patients receiving UDCA therapy (76% of the PSC group) exhibited lower levels of circulating free fatty acids as compared to those not taking UDCA. As serum levels of the lipolysis marker glycerol were unchanged in the PBC and PSC groups, the observed changes in free fatty acids may be reflective of alterations in hepatic lipid metabolism as opposed to increased release from adipose tissue stores. Indeed, similar elevations in the ketone bodies acetoacetate and BHBA, along with modest accumulation of several medium- and long-chain acylcarnitines, were observed in both liver disease groups despite lower circulating levels of ketone bodies in PSC patients taking UDCA as compared to those not receiving therapy. Ketones are synthesized within the mitochondrial matrix from acetyl-CoA in a pathway that shares several steps with the cholesterol biosynthetic pathway. Therefore, these changes may be indicative of a general increase in activity of the mevalonate and ketogenesis pathways in both liver disease groups. However, mitochondrial dysfunction is known to play a key role in cholestatic disease (28) and is typically associated with impaired fatty acid β-oxidation. Thus, accumulation of acylcarnitines and ketone bodies in patients with PBC and PSC may instead be suggestive of reduced fatty acid oxidation and ketogenesis, resulting in increased circulating free fatty acids as well as decreased peripheral utilization of ketones by target tissues such as brain and cardiac muscle. Support for this hypothesis comes from previous studies demonstrating a central role for impaired ketogenesis in altered hepatic lipid metabolism observed in cholestatic rats following bile duct ligation (29, 30).
Inflammation and oxidative stress are known to contribute to the pathogenesis of cholestatic diseases including PBC and PSC (7, 28, 31). In the current study, a similar inflammatory signature was observed in both the PBC and PSC groups, with elevations in the tryptophan-derived metabolite kynurenine and reductions in the lipooxygenase-derived eicosanoid 12-HETE. The majority of tryptophan is degraded in the liver by the enzyme tryptophan dioxygenase (TDO), but this amino acid can also be metabolized by the highly inducible enzyme indoleamine 2,3-dioxygenase (IDO) that is stimulated by pro-inflammatory cytokines such as interferon-γ and tumor necrosis factor-α (32). Arachidonate-derived products of the lipid-peroxidating enzyme 12/15-lipoxygenase, including 12-HETE, generally possess anti-inflammatory properties to counteract pro-inflammatory responses (33). Therefore, higher levels of kynurenine and reductions in 12-HETE are suggestive of a systemic inflammatory signature that is similar in patients with PBC and PSC. Conversely, while elevations in various biochemical markers of oxidative stress were observed in both the PBC and PSC groups, differential changes in specific metabolites reflective of an oxidative environment were noted when comparing the two groups. For example, the oxysterol 7β-hydroxycholesterol, which is formed by autooxidation of cholesterol and is a specific marker of endogenous oxidative stress and lipid peroxidation, and the lipoxygenase-derived metabolite 13-HODE + 9-HODE, which also serves as a lipid peroxidation marker, were only elevated in the PBC group. In contrast, levels of asymmetric dimethylarginine (ADMA), which are elevated when oxidative stress increases the activity of arginine methylating enzymes (i.e., protein arginine N-methyltransferases) and decreases the activity of the ADMA-degrading enzyme dimethylarginine dimethylaminohydrolase (34), were increased only in patients with PSC. These findings suggest that while greater oxidative stress is present in both cholestatic diseases, the source(s) and pathways behind this change may differ between PBC and PSC.
Differential changes in metabolites related to protein and amino acid metabolism were also observed when comparing patients with PBC and PSC and several of these were identified as key biochemicals with regard to the RF classification scheme. In particular, levels of many free amino acids and dipeptides were lower in the PSC as compared to the PBC group. Reports in the literature corroborate findings from the current study demonstrating perturbations in protein and amino acid metabolism in cholestatic disease, including differential changes in the plasma amino acid profile of patients with PBC and PSC (35). For example, administration of alanine (significantly reduced in both the PBC and PSC groups in the current study) was shown to reduce bilirubin levels and improve symptoms in three patients with end-stage PBC (36), while reductions in circulating tyrosine levels (which were not significantly altered in the current study) may be related to greater fatigue and reduced quality of life in patients with PBC (35). In addition, increased protein degradation and inefficient protein synthesis are among the mechanisms associated with malnutrition in cholestatic liver disease (37), although it is important to note that in the current study levels of serum total protein were significantly greater in the PBC and PSC groups as compared to healthy controls . Together with the current study, findings from these previous investigations highlight an important role for changes in protein and amino acid metabolism in the cholestatic disease process.
Despite the small number of subjects profiled in this descriptive study and the inclusion of patients taking UDCA and/or anti-hyperlipidemic medications, novel perturbations in the global serum metabolome were identified in patients with PBC and PSC and this information was successfully utilized to classify cholestatic patients and healthy controls. In addition to common changes in patients with cholestatic disease, findings from this study also identified alterations in lipid metabolism, oxidative stress/lipid peroxidation, stress hormones, and protein/amino acid metabolism that differed when comparing the PBC and PSC groups. These differential changes may not only have diagnostic value with regard to identifying patients with PBC versus PSC, but also provide insight into differential metabolic pathways that contribute to the pathogenesis of these similar conditions. Moreover, identification of a novel therapeutic effect of UDCA on the serum free fatty acid profile was limited to the PSC group and is worthy of additional investigation. Metabolic profiling of an independent set of serum samples from cholestatic patients and healthy controls will be necessary to validate findings from this pilot study and determine the true diagnostic utility of biomarker candidates identified in the current study.
Supplementary Material
Keypoints.
The current study compares the global serum metabolomes of patients with PBC and PSC.
Metabolites reflective of protein metabolism appear to have the greatest power in differentiating between PBC and PSC.
Inflammatory signature (elevations in the tryptophan-derived metabolite kynurenine and reductions in the lipooxygenase-derived eicosanoid 12-HETE) was observed in both the PBC and PSC.
Levels of several free amino acids (serine, aspartate, glutamate, phenylalanine, and ornithine) and 9 dipeptides were significantly (p≤0.05) lower in patients with PSC as compared to PBC.
Acknowledgments
Financial Support: This work is in part supported by NIH K24 DK069290A (NC)
Abbreviations
- PBC
Primary Biliary Cirrhosis
- PSC
Primary Sclerosing Cholangitis
- UDCA
Ursodeoxycholic acid
- Da
Dalton
- UHPLC
Ultrahigh performance liquid chromatography
- MS
Mass spectrometry
- GC
Gas chromatography
- FDR
False discovery rate
- ALT
Alanine aminotransferase
- AST
Aspartate aminotransferase
- INR
International normalized ratio
- MELD
Model for end-stage liver disease
- HETE
Hydroxyeicosatetraenoic acid
- HODE
Hydroxyoctadecadienoic acid
- DMA
Dimethylarginine
- ERCP
Endoscopic retrograde cholangiopancreatography
- MRCP
Magnetic resonance cholangiopancreatography
- AMA
Antimitochondrial antibody
- CYP
Cytochrome P450
- TDA
tryptophan dioxygenase
- IDO
indoleamine 2,3-dioxygenase;
Footnotes
Disclosures
LNB and JW are employees of Metabolon, Inc. and, as such, have affiliations with or financial involvement with Metabolon, Inc. The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
Dr. Chalasani has consultant agreements and research grants from several pharmaceutical companies but none represent a potential conflict for this paper.
Dr. Vuppalanchi serves as a member of the speaker’s bureau, research grants from pharma and serves as a consultant but none represent a potential conflict for this paper.
Ms. Megan Comerford has no financial conflicts of interests to declare.
References
- 1.Woolbright BL, Jaeschke H. Novel insight into mechanisms of cholestatic liver injury. World J Gastroenterol. 2012;18:4985–4993. doi: 10.3748/wjg.v18.i36.4985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kim WR, Ludwig J, Lindor KD. Variant forms of cholestatic diseases involving small bile ducts in adults. Am J Gastroenterol. 2000;95:1130–1138. doi: 10.1111/j.1572-0241.2000.01999.x. [DOI] [PubMed] [Google Scholar]
- 3.Poupon R, Chazouilleres O, Poupon RE. Chronic cholestatic diseases. J Hepatol. 2000;32:129–140. doi: 10.1016/s0168-8278(00)80421-3. [DOI] [PubMed] [Google Scholar]
- 4.Hirschfield GM, Heathcote EJ, Gershwin ME. Pathogenesis of cholestatic liver disease and therapeutic approaches. Gastroenterology. 2010;139:1481–1496. doi: 10.1053/j.gastro.2010.09.004. [DOI] [PubMed] [Google Scholar]
- 5.Hirschfield GM, Chapman RW, Karlsen TH, Lammert F, Lazaridis KN, Mason AL. The genetics of complex cholestatic disorders. Gastroenterology. 2013;144:1357–1374. doi: 10.1053/j.gastro.2013.03.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Czul F, Peyton A, Levy C. Primary biliary cirrhosis: therapeutic advances. Clin Liver Dis. 2013;17:229–242. doi: 10.1016/j.cld.2012.12.003. [DOI] [PubMed] [Google Scholar]
- 7.Eaton JE, Talwalkar JA, Lazaridis KN, Gores GJ, Lindor KD. Pathogenesis of Primary Sclerosing Cholangitis and Advances in Diagnosis and Management. Gastroenterology. 2013 doi: 10.1053/j.gastro.2013.06.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Trivedi PJ, Hirschfield GM. Review article: overlap syndromes and autoimmune liver disease. Aliment Pharmacol Ther. 2012;36:517–533. doi: 10.1111/j.1365-2036.2012.05223.x. [DOI] [PubMed] [Google Scholar]
- 9.Eckhart AD, Beebe K, Milburn M. Metabolomics as a key integrator for “omic” advancement of personalized medicine and future therapies. Clin Transl Sci. 2012;5:285–288. doi: 10.1111/j.1752-8062.2011.00388.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Evans AM, DeHaven CD, Barrett T, Mitchell M, Milgram E. Integrated, nontargeted ultrahigh performance liquid chromatography/electrospray ionization tandem mass spectrometry platform for the identification and relative quantification of the small-molecule complement of biological systems. Anal Chem. 2009;81:6656–6667. doi: 10.1021/ac901536h. [DOI] [PubMed] [Google Scholar]
- 11.Trottier J, Bialek A, Caron P, Straka RJ, Heathcote J, Milkiewicz P, et al. Metabolomic profiling of 17 bile acids in serum from patients with primary biliary cirrhosis and primary sclerosing cholangitis: a pilot study. Dig Liver Dis. 2012;44:303–310. doi: 10.1016/j.dld.2011.10.025. [DOI] [PubMed] [Google Scholar]
- 12.Ohta T, Masutomi N, Tsutsui N, Sakairi T, Mitchell M, Milburn MV, et al. Untargeted metabolomic profiling as an evaluative tool of fenofibrate-induced toxicology in Fischer 344 male rats. Toxicol Pathol. 2009;37:521–535. doi: 10.1177/0192623309336152. [DOI] [PubMed] [Google Scholar]
- 13.Dehaven CD, Evans AM, Dai H, Lawton KA. Organization of GC/MS and LC/MS metabolomics data into chemical libraries. J Cheminform. 2010;2:9. doi: 10.1186/1758-2946-2-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Storey JD, Tibshirani R. Statistical significance for genomewide studies. Proc Natl Acad Sci U S A. 2003;100:9440–9445. doi: 10.1073/pnas.1530509100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang X, Zhang A, Sun H. Power of metabolomics in diagnosis and biomarker discovery of hepatocellular carcinoma. Hepatology. 2013;57:2072–2077. doi: 10.1002/hep.26130. [DOI] [PubMed] [Google Scholar]
- 16.Hawkins MT, Lewis JH. Latest advances in predicting DILI in human subjects: focus on biomarkers. Expert Opin Drug Metab Toxicol. 2012;8:1521–1530. doi: 10.1517/17425255.2012.724060. [DOI] [PubMed] [Google Scholar]
- 17.Lindor KD, Gershwin ME, Poupon R, Kaplan M, Bergasa NV, Heathcote EJ. American Association for Study of Liver D. Primary biliary cirrhosis. Hepatology. 2009;50:291–308. doi: 10.1002/hep.22906. [DOI] [PubMed] [Google Scholar]
- 18.Kaplan MM, Gershwin ME. Primary biliary cirrhosis. N Engl J Med. 2005;353:1261–1273. doi: 10.1056/NEJMra043898. [DOI] [PubMed] [Google Scholar]
- 19.Gershwin ME, Mackay IR, Sturgess A, Coppel RL. Identification and specificity of a cDNA encoding the 70 kd mitochondrial antigen recognized in primary biliary cirrhosis. J Immunol. 1987;138:3525–3531. [PubMed] [Google Scholar]
- 20.Nakashima T, Sano A, Seto Y, Nakajima T, Shima T, Sakamoto Y, et al. Unusual trihydroxy bile acids in the urine of patients treated with chenodeoxycholate, ursodeoxycholate or rifampicin and those with cirrhosis. Hepatology. 1990;11:255–260. doi: 10.1002/hep.1840110215. [DOI] [PubMed] [Google Scholar]
- 21.Jacquemin E, Dumont M, Bernard O, Erlinger S, Hadchouel M. Evidence for defective primary bile acid secretion in children with progressive familial intrahepatic cholestasis (Byler disease) Eur J Pediatr. 1994;153:424–428. doi: 10.1007/BF01983406. [DOI] [PubMed] [Google Scholar]
- 22.Kowdley KV. Lipids and lipid-activated vitamins in chronic cholestatic diseases. Clin Liver Dis. 1998;2:373–389. x. doi: 10.1016/s1089-3261(05)70013-1. [DOI] [PubMed] [Google Scholar]
- 23.Bertolotti M, Carulli L, Concari M, Martella P, Loria P, Tagliafico E, et al. Suppression of bile acid synthesis, but not of hepatic cholesterol 7alpha-hydroxylase expression, by obstructive cholestasis in humans. Hepatology. 2001;34:234–242. doi: 10.1053/jhep.2001.25958. [DOI] [PubMed] [Google Scholar]
- 24.Inamine T, Higa S, Noguchi F, Kondo S, Omagari K, Yatsuhashi H, et al. Association of genes involved in bile acid synthesis with the progression of primary biliary cirrhosis in Japanese patients. J Gastroenterol. 2013 doi: 10.1007/s00535-012-0730-9. [DOI] [PubMed] [Google Scholar]
- 25.Sauter G, Berr F, Beuers U, Fischer S, Paumgartner G. Serum concentrations of 7alpha-hydroxy-4-cholesten-3-one reflect bile acid synthesis in humans. Hepatology. 1996;24:123–126. doi: 10.1053/jhep.1996.v24.pm0008707250. [DOI] [PubMed] [Google Scholar]
- 26.Hahn C, Reichel C, von Bergmann K. Serum concentration of 7 alpha-hydroxycholesterol as an indicator of bile acid synthesis in humans. J Lipid Res. 1995;36:2059–2066. [PubMed] [Google Scholar]
- 27.Mortiaux A, Dawson AM. Plasma free fatty acid in liver disease. Gut. 1961;2:304–309. doi: 10.1136/gut.2.4.304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Arduini A, Serviddio G, Tormos AM, Monsalve M, Sastre J. Mitochondrial dysfunction in cholestatic liver diseases. Front Biosci (Elite Ed) 2012;4:2233–2252. doi: 10.2741/539. [DOI] [PubMed] [Google Scholar]
- 29.Lang C, Schafer M, Serra D, Hegardt F, Krahenbuhl L, Krahenbuhl S. Impaired hepatic fatty acid oxidation in rats with short-term cholestasis: characterization and mechanism. J Lipid Res. 2001;42:22–30. [PubMed] [Google Scholar]
- 30.Lang C, Berardi S, Schafer M, Serra D, Hegardt FG, Krahenbuhl L, Krahenbuhl S. Impaired ketogenesis is a major mechanism for disturbed hepatic fatty acid metabolism in rats with long-term cholestasis and after relief of biliary obstruction. J Hepatol. 2002;37:564–571. doi: 10.1016/s0168-8278(02)00248-9. [DOI] [PubMed] [Google Scholar]
- 31.Hirschfield GM, Gershwin ME. The immunobiology and pathophysiology of primary biliary cirrhosis. Annu Rev Pathol. 2013;8:303–330. doi: 10.1146/annurev-pathol-020712-164014. [DOI] [PubMed] [Google Scholar]
- 32.Dantzer R, O’Connor JC, Freund GG, Johnson RW, Kelley KW. From inflammation to sickness and depression: when the immune system subjugates the brain. Nat Rev Neurosci. 2008;9:46–56. doi: 10.1038/nrn2297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Conrad DJ. The arachidonate 12/15 lipoxygenases. A review of tissue expression and biologic function. Clin Rev Allergy Immunol. 1999;17:71–89. doi: 10.1007/BF02737598. [DOI] [PubMed] [Google Scholar]
- 34.Sydow K, Munzel T. ADMA and oxidative stress. Atheroscler Suppl. 2003;4:41–51. doi: 10.1016/s1567-5688(03)00033-3. [DOI] [PubMed] [Google Scholar]
- 35.ter Borg PC, Fekkes D, Vrolijk JM, van Buuren HR. The relation between plasma tyrosine concentration and fatigue in primary biliary cirrhosis and primary sclerosing cholangitis. BMC Gastroenterol. 2005;5:11. doi: 10.1186/1471-230X-5-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nishiguchi S, Habu D, Kubo S, Shiomi S, Tatsumi N, Tamori A, et al. Effects of alanine in patients with advanced primary biliary cirrhosis: preliminary report. Hepatol Res. 2003;25:8–13. doi: 10.1016/s1386-6346(02)00169-9. [DOI] [PubMed] [Google Scholar]
- 37.Munoz SJ. Nutritional therapies in liver disease. Semin Liver Dis. 1991;11:278–291. doi: 10.1055/s-2008-1040446. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.