Abstract
Apoptolidin A has been described as among the top 0.1% most cell selective cytotoxic agents to be evaluated in the NCI 60 cell line panel. The molecular structure of apoptolidin A consists of a 20-membered macrolide with mono- and disaccharide moieties located at C9 and C27, respectively. In contrast to apoptolidin A, the aglycone (apoptolidinone) shows no cytotoxicity (>10 μM) when evaluated against several tumor cell lines. Apoptolidin H, the C27 deglycosylated analog of apoptolidin A, was produced by targeted glycosyl transferase gene deletion and displayed sub-micromolar activity against H292 lung carcinoma cells. Selective esterification of the C2′ hydroxyl group of apoptolidins A and H with 5-azidopentanoic acid afforded azido functionalized derivatives of potency equal to their parent macrolide. Azido apoptolidins readily underwent strain-promoted alkyne azido cycloaddition (SPAAC) reactions to provide access to fluorescent and biotin functionalized probes. Microscopy studies demonstrate apoptolidins A and H localize in the mitochondria of H292 human lung carcinoma cells.
Keywords: natural products, antitumor, polyketides, metabolism, chemical probe
The apoptolidins are macrocylic natural products produced by an actinomycete (Nocardiopsis sp. FU40) soil microbe by way of a type I polyketide synthase biosynthetic pathway.[1] Apoptolidin A (1) was reported to induce cell death in E1A transformed rat glia cells, a model cancer cell phenotype, while not affecting the growth of non-transformed glia cells.[2] The described selective cytotoxicity of apoptolidin A stimulated interest in its total synthesis and mechanism of induced cell death.[3] Salomon and Khosla employed a pharmacological approach to define the mechanism of cell death using LYas mouse lymphoma cells and concluded cell death proceeded by way of the mitochondria mediated apoptotic pathway (intrinsic pathway).[4] The same investigators suggested FOF1 ATPase as a potential target although inhibition potency (Ki = 4–5 μM) against yeast FOF1 ATPase in a biochemical assay did not correlate well with observed cytotoxicity in cell culture (EC50 0.2 μM) leaving open the possibilty of alternative cellular targets.
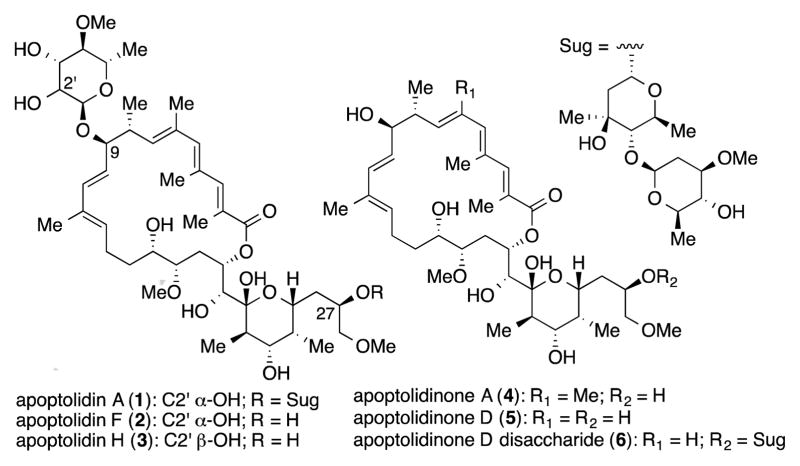
Following the reported isolation of apoptolidin A (1), other structural variants have been described either as minor microbial metabolites,[5] products of isomerization[6] or semi-[7] and total synthesis[8]. When evaluated for cytotoxicity against tumor cells, these apoptolidins reveal considerable tolerance of structural modifications within the macrolide core including deoxygenation [apoptolidins B and C][5c], demethylation [apoptolidin D][5a], and C2–C3 double bond isomerization [apoptolidin G][6c] without significant loss of cytotoxicity (sub-micromolar).
In contrast to structural changes within the core macrolides, removal of the deoxy sugars resulted in complete loss of activity with EC50 values of apoptolidinone A (4) and C (5) reported to be greater then 10 μmol against several tumor cell lines in cell viability assays.[8c,9] The observed loss in activity upon exhausitive deglycosylation of the core macrolide presented an opportunity to develop a series of apoptolidin derived probes to support mechanism of action studies. We report here methods to access apoptolidins of varying state of glycosylation (tri-, di-, mono and non-glycosylated) and preliminary studies on their use as cellular probes.
Apoptolidin A (1) is readily obtained by fermentation of the actinomycete Nocardiopsis sp. FU40 with a production of 50 – 100 mg per liter.[2,10] We previously described the identification and expression of the apoptolidin gene cluster that provided an opportunity to access glycovariants of apoptolidin A by targeted gene deletion.[10] Three genes encoding for glycosyl transferases (ApoGT1, ApoGT2 and ApoGT3) were identified. To date, targeted gene deletion of one of ApoGT2 via double crossover homologous recombination resulted in a Nocardiopsis variant producing a previously unreported glycovariant of apoptolidin A. In this case fermentation provided (50–100 mg per liter) of a new apoptolidin analog lacking the C27 disacharide and termed apoptolidin H (3). Nicolaou[8a] and Koert[8b] prepared 3 by total synthesis and Wender[5b] reported the isolation of a structurally related minor metabolite termed apoptolidin F (2) (<5 mg per liter) epimeric at C2′.[11]
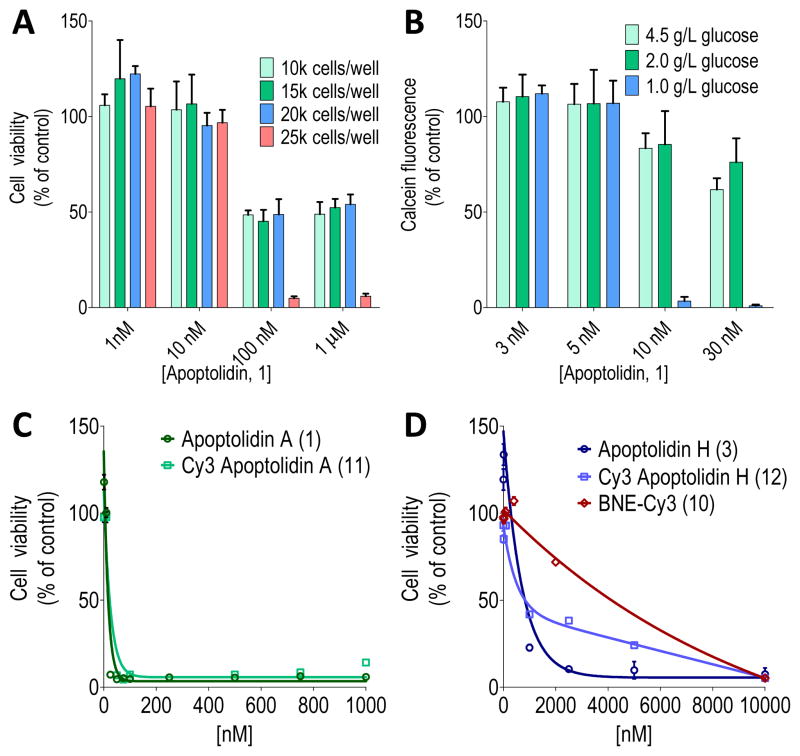
Employing a standard cell viability assay using H292 human lung cancer cells, apoptolidin A induced cell growth arrest without any indication of cell death. In this experiment, cells at ~20% confluency were treated with apoptolidin A and after 48 hours assayed for cell viability. Even treatment of cells with apopotolidin A for as long as 5 days resulted in only the observed antiproliferative effect but no loss of cell integrity. In contrast, cells grown to high confluency (~70%) prior to apoptolidin A treatment, resulted in >95% cell death after 4 days with a calculated EC50 of 20–30 nM. In order to standardize this assay, cells were systematically plated in a 96-well format (10, 15, 20 and 25 thousand cells per well), allowed to attach (16 hours), treated with apoptolidin A and assayed for viability after 4 days. As shown in Figure 2-A, 25,000 cells per well resulted in a reproducible cytotoxic effect (EC50 16 nM) against human lung (H292) as well as several other tumor cell lines (HCT116 colorectal, MDB MB321 breast, 1483 head and neck squamous). The results summarized in Figure 2-A also illustrate the antiproliferative activity of apoptolidin A (EC50 <100 nM) against lower confluency cells (10–20K cells). In separate experiments we observed that the cytotoxicity of apoptolidin A is potentiated by using cell culture media formulations of increasingly reduced glucose (Figure 2-B). Notably, such nutrient starvation conditions have been proposed to mimic poorly vascularized cells seen in solid tumors.[12] We hypothesize that high and low confluency cells differ in metabolic flux with low confluency cells primarily utilizing the Embden-Meyerhof glycolytic pathway (Warburg effect) and high-density cells using the more energetic oxidative phosphorylation (OXPHOS) manifold.[13] These results are in agreement with Salomon and co-workers results as they demonstrated glycolytic (apoptolidin unresponsive) cells were sensitized to apoptolidin by the addition of 2-deoxyglucose or oxamate, small molecules known to channel carbon flux from the Embden-Meyerhof to OXPHOS pathway.[4a]
Figure 2.
Analysis of apoptolidin toxicity in H292 cell line. A) Dependence of cytotoxic and cytostatic effects of apoptolidin A on cell confluency as measured by initial number of cells per well on a 96 well plate; B) Effect of cell growth media of varying glucose concentrations on apoptolidin A cytotoxicity; C) Cell viability assay for apoptolidin A (1) and Cy3 apoptolidin A (11); and D) apoptolidin H (3), Cy3 apoptolidin H (12) and BNE-Cy3 (10) evaluated using optimized conditions.
The macrolactones apoptolidinone A (4) and D (5), devoid of all three deoxy sugars common to apoptolidin A, were evaluated under the apoptolidin A sensitive high confluency H292 cell culture conditions and shown to be neither cytostatic or cytotoxic at concentrations of apoptolidinones to 10 μM.[8c] However apoptolidin H (3), bearing the C9 deoxy sugar, was cytotoxic against H292 cells under the same conditions with an EC50 of 810 nM (~50 times less potent then apoptolidin A) and apoptolidinone D disaccharide (6),[8c] lacking the C9 deoxysugar, (demonstrated an EC50 200 nM against the same cell line. Thus mono- or diglycoslyated variants of apoptolidin are sufficient to restore partial cell cytotoxicity. Finally, we note when assayed against yeast derived FOF1-ATPase apoptolidin A and H showed modest and comparable inhibition with Ki values of 4.9 and 13.7 μM, respectively, suggesting that the pharmacological importance of the C-27 disaccharide is in large part decoupled from the observed activity of apotolidins against FOF1-ATPase.
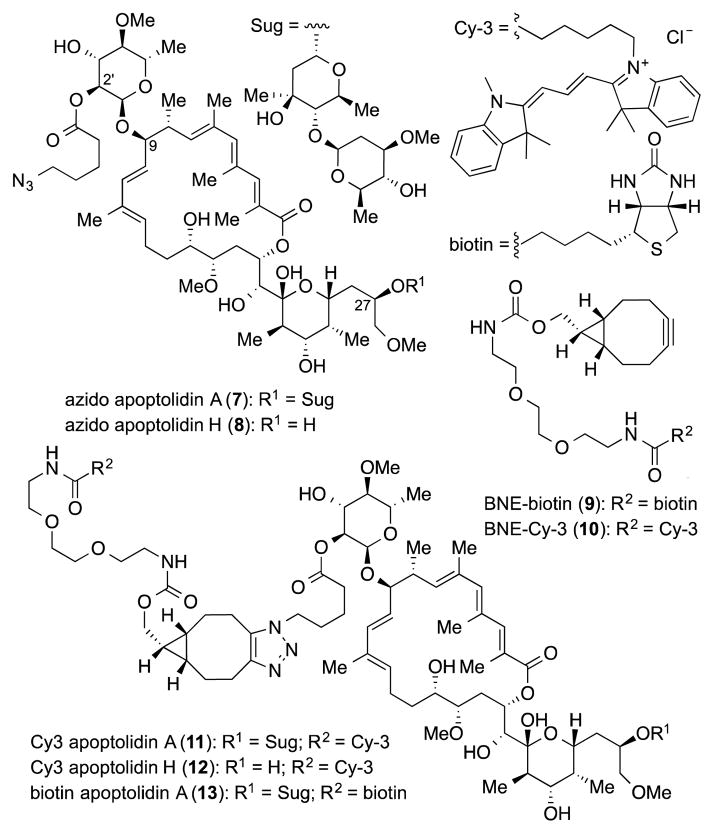
In order to initiate chemical probe studies we required the introduction of a azido functional group within the apoptolidin core to enable conjugation to either fluorescent or affinity tags using strain promoted alkyne azido cycloaddion (SPAAC) chemistry.[14] To this end we took advantage of a report by the Wender group describing the selective benzolylation of the C2′ hydroxyl group of the C9 sugar.[7a] And were pleased to observe selective acylation of the C2′ hydroxyl group of apoptolidins A and H using 5-azidopentanoic acid to afford azido derivatives 7 and 8 in 30–40% yield (Figure 3). Importantly, when evaluated in the cell viability assay, azido analogs 13 and 14 maintained activity comparable to their parent substrates (EC50 19 and 350 nM, respectively). As partner alkyne tags we selected cyanine dye biotin (9) and Cy-3 (10) PEG-tethered to click ready bicycle[6.1.0]nonynes.[15] Coupling of BNE-Cy-3 (10) with azido apoptolidins 7 and 8 proceeded smoothly in methanol at room temperature (4 h) to give fluorescently labeled apoptolidins 11 and 12 in 39 and 32% yield, respectively. In addition biotin-BNE (9) reacted with apoptolidin A under identical conditions to give biotinylated apoptolidin A (13) in 29% yield. Supporting their potential utility in functional imaging studies, conjugates 11–13 also maintained activity relative to their parent macrolides (11: EC50 22 nM and 12: EC50 820 nM, Figures 2-C and 2-D) when evaluated in the H292 cell assay, although biotin conjugate 13 dropped in activity to EC50 151 nM.
Figure 3.
Azido apoptolidins, strained alkyne tagging reagents and derived probes.
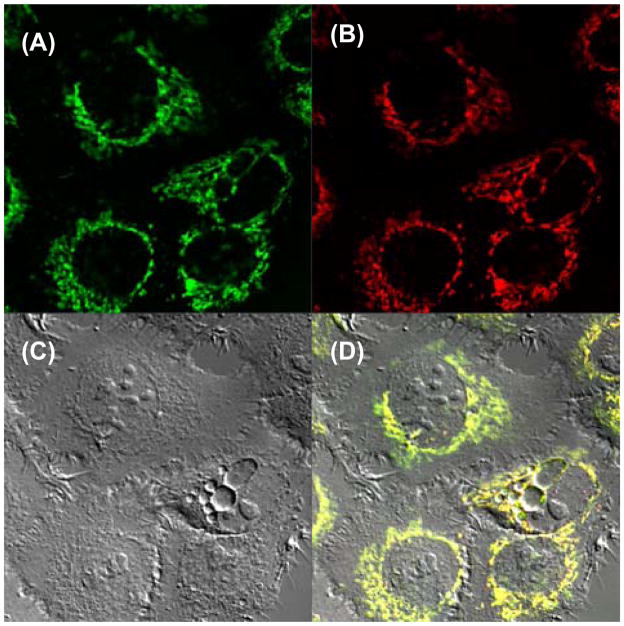
Confocal microscopy studies were conducted with Cy-3 apoptolidin conjugates 11 and 12 at concentrations of 200 nM applied to H292 human lung cancer cells. In these experiments compound treatment for 15 min was followed by a 60 min washout in order to dilute nonspecific binding. Cellular images of experiments using Cy3 apoptolidin A (11) are shown in Figure 4. Staining of washed cells with Mitotracker Green FM (Figure 4A) was conducted in order to evaluate whether 11 (Figure 4B) localized in the mitochondria. Inspection of the merged image (Figure 4D) confirmed colocalization of 11 with Mitotracker stain. Colocalization was further quantified by Costes’ analysis that showed excellent overlap with a Pearson’s coefficient of 0.89. An identical set of experiments using Cy-3 apoptolidin H (12) also demonstrated localization of 12 in the mitochondria.
Figure 4.
Costaining Cy3 apoptolidin A and MitoTracker in H292 cells. Fluorescence images of MitoTracker (A), Cy3 apoptolidin A (B), DIC image (C) and merged image (D) are shown.
We do note cationic dyes such as cyanine-3 tend to localize in the mitochondria,16 and bicyclononyne BNE-Cy-3 (10) localizes in the mitochondria but is not cytotoxic against H292 cells (EC50 4.6 μM) in the standard cell viability assay (see Figure 2-D). Significantly, regardless of the mechanism enabling localization of apoptolidin A analogs, mitochondrial localization did not reduce their activity. In order to more effectively judge whether the bioactivity enabled by glycosylation of the apoptolidins is due to enabling localization within the mitochondria we are now in the process of examining non-cationic dyes conjugated to apoptolidins and the non-toxic aglycone (apoptolidinone) in microscopy experiments.
The cell cytotoxicity and observed mitochondrial localization of Cy-3 conjugates 11 and 12 in human lung cancer cells H292 supports earlier conclusions of the apoptolidins act on a mitochondrial target. Further progress on defining the role of sugars to imparting activity against tumor cells and identification of cellular targets of the apoptolidins will be reported in due course.
Supplementary Material
Figure 1.
Members of the apoptolidin family of macrolides.
Acknowledgments
This research was supported by the National Institutes of Health (CA 059515). S. M. D. and D. C. E. acknowledges the support of the Vanderbilt Chemical Biology Interface (CBI) training program (T32 GM065086).
Footnotes
Supporting information for this article is given via a link at the end of the document.
Contributor Information
Dr. Sean M. DeGuire, Department of Chemistry, Vanderbilt University, Vanderbilt Institute of Chemical Biology, Nashville, TN 37232 (USA)
David C. Earl, Department of Chemistry, Vanderbilt University, Vanderbilt Institute of Chemical Biology, Nashville, TN 37232 (USA)
Dr. Yu Du, Department of Chemistry, Vanderbilt University, Vanderbilt Institute of Chemical Biology, Nashville, TN 37232 (USA)
Brenda A. Crews, Departments of Biochemistry and Chemistry, Vanderbilt University, Vanderbilt Institute of Chemical Biology, Nashville, TN 37232 (USA)
Dr. Aaron T. Jacobs, Departments of Biochemistry and Chemistry, Vanderbilt University, Vanderbilt Institute of Chemical Biology, Nashville, TN 37232 (USA)
Dr. Alessandro Ustione, Department of Molecular Physiology and Biophysics, Vanderbilt University Medical Center, Nashville, TN 37232 (USA)
Cristina Daniel, Departments of Biochemistry and Chemistry, Vanderbilt University, Vanderbilt Institute of Chemical Biology, Nashville, TN 37232 (USA).
Katherine Chong, Department of Chemistry, Vanderbilt University, Vanderbilt Institute of Chemical Biology, Nashville, TN 37232 (USA).
Prof. Lawrence J. Marnett, Departments of Biochemistry and Chemistry, Vanderbilt University, Vanderbilt Institute of Chemical Biology, Nashville, TN 37232 (USA)
Prof. David W. Piston, Department of Molecular Physiology and Biophysics, Vanderbilt University Medical Center, Nashville, TN 37232 (USA)
Prof. Brian O. Bachmann, Department of Chemistry, Vanderbilt University, Vanderbilt Institute of Chemical Biology, Nashville, TN 37232 (USA)
Prof. Gary A. Sulikowski, Department of Chemistry, Vanderbilt University, Vanderbilt Institute of Chemical Biology, Nashville, TN 37232 (USA).
References
- 1.Hayakawa Y, Kim JW, Adachi H, Shinya K, Fujita K, Seto H. J Am Chem Soc. 1998;120:3524–3525. [Google Scholar]
- 2.Kim JW, Adachi H, Shinya K, Hayakawa Y, Seto H. J Antibiot. 1997;50:628–630. doi: 10.7164/antibiotics.50.628. [DOI] [PubMed] [Google Scholar]
- 3.Review: Daniel PT, Koert U, Schuppan J. Angew Chem Int Ed. 2006;45:872–893. doi: 10.1002/anie.200502698.
- 4.a) Salomon A, Voehringer D, Herzenberg L, Khosla C. Proc Nat Acad Sci USA. 2000;97:14766–14771. doi: 10.1073/pnas.97.26.14766. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Salomon A, Voehringer D, Herzenberg L, Khosla C. Chemistry & Biology. 2001;8:71–80. doi: 10.1016/s1074-5521(00)00057-0. [DOI] [PubMed] [Google Scholar]
- 5.a) Wender PA, Longcore KE. Org Lett. 2007;9:691–694. doi: 10.1021/ol0630245. [DOI] [PubMed] [Google Scholar]; b) Wender PA, Longcore KE. Org Lett. 2009;11:5474–5477. doi: 10.1021/ol902308v. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Wender PA, Sukopp M, Longcore KEK. Org Lett. 2005;7:3025–3028. doi: 10.1021/ol051074o. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.a) Wender PA, Gulledge A, Jankowski O, Seto H. Org Lett. 2002;4:3819–3822. doi: 10.1021/ol0266222. [DOI] [PubMed] [Google Scholar]; b) Pennington J, Williams H, Salomon A, Sulikowski GA. Org Lett. 2002;4:3823–3825. doi: 10.1021/ol026829v. [DOI] [PubMed] [Google Scholar]; c) Bachmann BO, McNees R, Melancon BJ, Ghidu VP, Clark R, Crews BC, DeGuire SM, Marnett LJ, Sulikowski GA. Org Lett. 2010;12:2944–2947. doi: 10.1021/ol1009398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.a) Wender PA, Jankowski OD, Tabet EA, Seto H. Org Lett. 2003;5:487–490. doi: 10.1021/ol027366w. [DOI] [PubMed] [Google Scholar]; b) Lewis CA, Longcore KE, Miller SJ, Wender PA. J Nat Prod. 2009;72:1864–1869. doi: 10.1021/np9004932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.a) Nicolaou KC, Li Y, Sugita K, Monenchein H, Gutupalli P, Mitchell HJ, Fylaktakidou KC, Vourloumis D, Giannakakou P, O’Brate A. J Am Chem Soc. 2003;125:15433–15442. doi: 10.1021/ja0304953. [DOI] [PubMed] [Google Scholar]; b) Wehlan H, Dauber M, Fernaud MTM, Schuppan J, Keiper S, Mahrwald R, Garcia MEJ, Koert U. Chem Eur J. 2006;12:7378–7397. doi: 10.1002/chem.200600462. [DOI] [PubMed] [Google Scholar]; c) Ghidu VP, Wang J, Wu B, Liu Q, Jacobs AT, Marnett LJ, Sulikowski GA. J Org Chem. 2008;73:4949–4955. doi: 10.1021/jo800545r. [DOI] [PMC free article] [PubMed] [Google Scholar]; d) Ghidu VP, Ntai I, Wang J, Jacobs AT, Marnett LJ, Bachmann BO, Sulikowski GA. Org Lett. 2009;11:3032–3034. doi: 10.1021/ol901045v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schuppan J, Wehlan H, Keiper S, Koert U. Chem Eur J. 2006;12:7364–7377. doi: 10.1002/chem.200600461. [DOI] [PubMed] [Google Scholar]
- 10.Du Y, Derewacz DK, DeGuire SM, Teske J, Ravel J, Sulikowski GA. Tetrahedron. 2011;67:6568–6575. doi: 10.1016/j.tet.2011.05.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.The 13C NMR spectra of apoptolidin H and synthetic material reported by Koert (reference 8b) compare favorably (see Supporting information, Table S2). The 13C NMR spectra of apoptolidin H and F (reference 5b) differ.
- 12.Bereiter-Hahn J, Münnich A, Woiteneck P. Cell Struct Funct. 1998;23:85–93. doi: 10.1247/csf.23.85. [DOI] [PubMed] [Google Scholar]
- 13.a) Boxer MB, Jiang J-K, Vander Heiden MG, Shen M, Skoumbourdis AP, Southall N, Veith H, Leister W, Austin CP, Park HW, Inglese J, Cantley LC, Auld DS, Thomas CJ. J Med Chem. 2010;53:1048–1055. doi: 10.1021/jm901577g. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Jiang J-k, Boxer MB, Heiden MGV, Shen M, Skoumbourdis AP, Southall N, Veith H, Leister W, Austin CP, Park HW, Inglese J, Cantley LC, Auld DS, Thomas CJ. Bioorg Med Chem Lett. 2010;20:3387–3393. doi: 10.1016/j.bmcl.2010.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Anastasiou D, Yu Y, Israelsen WJ, Jiang J-k, Boxer MB, Hong BS, Tempel W, Dimov S, Shen M, Jha A, Yang H, Mattaini KR, Metallo CM, Fiske BP, Courtney KD, Malstrom S, Khan TM, Kung C, Skoumbourdis AP, Veith H, Southall N, Walsh MJ, Brimacombe KR, Leister W, Lunt SY, Johnson ZR, Yen KE, Kunii K, Davidson SM, Christofk HR, Austin CP, Inglese J, Harris MH, Asara JM, Stephanopoulos G, Salituro FG, Jin S, Dang L, Auld DS, Park H-W, Cantley LC, Thomas CJ, Vander Heiden MG. Nat Chem Biol. 2012;8:839–847. doi: 10.1038/nchembio.1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sletten EM, Bertozzi CR. Angew Chem Int Ed. 2009;48:6974–6998. doi: 10.1002/anie.200900942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dommerholt J, Schmidt S, Temming R, Hendriks LJA, Rutjes FPJT, Van Hest JCM, Lefeber DJ, Friedl P, Van Delft FL. Angew Chem Int Ed. 2010;49:9422–9425. doi: 10.1002/anie.201003761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kim YK, Ha HH, Lee JS, Bi X, Ahn YH, Hajar S, Lee JJ, Chang YT. J Am Chem Soc. 2010;132:576–579. doi: 10.1021/ja906862g. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.