Abstract
Objective
To define the genetic landscape of amyotrophic lateral sclerosis (ALS) and assess the contribution of possible oligogenic inheritance, we aimed to comprehensively sequence 17 known ALS genes in 391 ALS patients from the United States.
Methods
Targeted pooled-sample sequencing was used to identify variants in 17 ALS genes. Fragment size analysis was used to define ATXN2 and C9ORF72 expansion sizes. Genotype-phenotype correlations were made with individual variants and total burden of variants. Rare variant associations for risk of ALS were investigated at both the single variant and gene level.
Results
64.3% of familial and 27.8% of sporadic subjects carried potentially pathogenic novel or rare coding variants identified by sequencing or an expanded repeat in C9ORF72 or ATXN2. 3.8% of subjects had variants in more than one ALS gene, and these individuals had disease onset ten years earlier (p=0.0046) than subjects with variants in a single gene. The number of potentially pathogenic coding variants did not influence disease duration or site of onset.
Interpretation
Rare and potentially pathogenic variants in known ALS genes are present in over 25% of apparently sporadic and 64% of familial patients, significantly higher than previous reports using less comprehensive sequencing approaches. A significant number of subjects carried variants in more than one gene, which influenced the age of symptom onset and supports oligogenic inheritance as relevant to disease pathogenesis.
INTRODUCTION
Amyotrophic lateral sclerosis (ALS) is caused by degeneration of upper and lower motor neurons which results in progressive paralysis and ultimately death. As the most common motor neuron disease, the incidence of ALS is 0.44–3.2/100,000 person years1 and data from the National ALS Registry demonstrates a prevalence of 3.9/100,000 cases in the US.2 5–10% of ALS patients have a family history of the disease (FALS)3–5 and the genetic analysis of these FALS pedigrees has fueled the discovery of more than 20 ALS genes-some with high-penetrance and others with lower penetrance or tentative associations to disease (reviewed in Harms and Baloh, Andersen and Al-Chalabi).6,7 Mutations in many of these genes are also found in patients without a family history of ALS (sporadic ALS, or SALS), with high-penetrance mutations found in ~10%.8–14 Recently, the heritability of SALS has been estimated to be 12–21% from genome-wide association studies15,16 and as high as 61% in twin studies17 suggesting additional genetic influences on ALS risk remain to be identified.
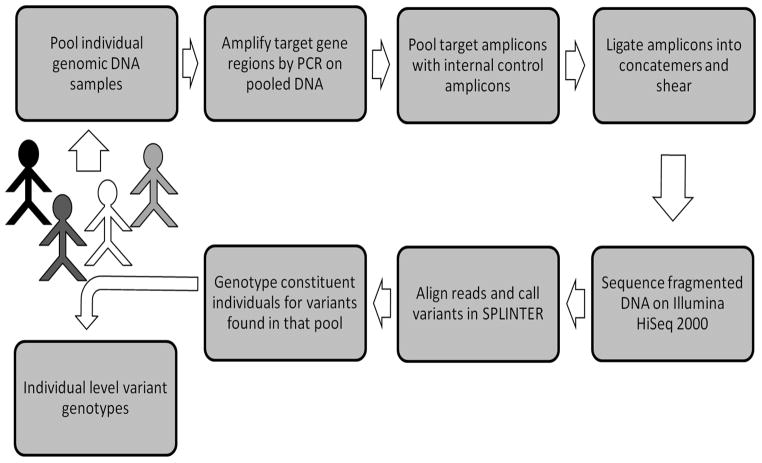
The emergence of next-generation sequencing techniques has driven down sequencing costs and made it feasible for studies to abandon sequential candidate gene sequencing in favor of analyzing larger numbers of genes simultaneously. One of the more powerful and cost-effective sequencing techniques for screening moderate number of genes in medium sized cohorts is termed pooled-sample or pooled-DNA sequencing (Figure 1).18 In this method, DNA samples from multiple patients are pooled prior to PCR amplification of target regions. PCR products are then combined and sequenced en masse using short-read/next-generation sequencing platforms.18 Analysis programs such as SPLINTER utilize statistical algorithms to identify potential variants with high sensitivity, and are capable of detecting single alleles in pools of up to 500 individuals.19 Pooled-sample sequencing therefore overcomes the resource and time-intensive drawbacks of traditional Sanger sequencing approaches at a fraction of the cost.18,19
Figure 1.
Schema of pooled-sample sequencing workflow
As a result of next-generation sequencing advances, studies have begun addressing the relative contributions of individual genes in ALS subjects with and without family histories, revealing significant heterogeneity between populations.8–12,20 Furthermore, screening multiple ALS genes in parallel has also uncovered a number of patients that carry potentially pathogenic variants in more than one known ALS gene.12 The unexpected frequency of this phenomenon has raised the hypothesis that some fraction of apparently sporadic ALS8,12 could be caused by the co-occurrence of two or more genetic variants with additive or synergistic deleterious effects. Each variant alone could be tolerated but when combined with a second variant would exceed the threshold required for neurodegeneration. Although several papers have reported cases with multiple variants in ALS genes, no effect on phenotype or disease manifestations has been noted.9,12
We have used pooled-sample sequencing as the major technique to analyze 17 ALS-associated genes in 391 ALS subjects from a United States clinic-based cohort. In creating the most comprehensively-sequenced North American ALS cohort to date, this study measures the burden of rare and novel variants in known ALS genes and defines the frequency of potential oligogenic cases.
METHODS
Subjects
Between 2005 and 2011, patients diagnosed with ALS at the Washington University Neuromuscular Disease Center in St. Louis, Missouri (WUSM) or at the Virginia Mason Medical Center (VMMC) were systematically asked to participate in genetic studies. All subjects provided informed and written consent for clinical-genetic correlation studies of ALS that had been approved by institutional ethics review boards. At WUSM, subjects with or without a family history of ALS were included, while only sporadic cases were enrolled at VMMC. All subjects had been evaluated by neuromuscular specialists and diagnosed with probable or definite ALS according to El Escorial criteria.21 A subset of included subjects (mostly with FALS) also underwent sequencing for one or more ALS genes at commercial reference laboratories, which identified 6 subjects with SOD1 or TARDBP mutations.
Genetic investigations
Sequencing of ALS-associated genes
All coding exons and 20 flanking bases of SOD1, FUS, TARDBP, ANG, OPTN, VCP, VAPB, DAO, DCTN1, FIG4, SETX, TAF15, EWSR1, UBQLN2, and SQSTM1 were sequenced in our cohort using the pooled-sample method as previously described in detail and schematized in Figure 1.18,22 Genomic DNA was extracted from whole blood or saliva of individual subjects according to standard protocols. Double-stranded DNA was carefully quantified by fluorimetry based on SYBR gold fluorescence. Pooled-sample gDNA pools were then created by combining equimolar amounts of DNA from multiple individuals: two pools containing 21 samples each were used to validate the method, while the remaining samples were divided into 8 pools of 30–50 samples each.
Primer pairs for all coding exons and at least 20bp of flanking sequence were designed using Primer3 (http://biotools.umassmed.edu/bioapps/primer3_www.cgi) and the RefSeq gene annotations found in GRCh37/hg19 (accession numbers NM_000454.4, NM_004960.3, NM_007375.3, NM_001145.4, NM_001008211.1, NM_007126.3, NM_004738.4, NM_001917.4, NM_004082.4, NM_014845.5, NM_015046.5, NM_139215.1, NM_013986.3, NM_013444.3, and NM_003900.4). Primer sequences are available upon request. Amplicons from each pool were sequenced on one lane of HiSeq2000 (Illumina), with single-end 42bp reads. UBQLN2 and SQSTM1 were reported after initial sequencing was underway and all subjects were sequenced as part of 6 pools across two lanes of Illumina HiSeq2000. Exon 1 of SQSTM1 was not sufficiently covered using pooled-sample methods and required Sanger sequencing of each individual subject. Mutations in PFN1 were reported after analysis was already underway so this gene was not assessed.23
In total, 144 PCR amplicons were required to amplify 193 exons of the 15 genes analyzed by pooled-sample sequencing. Twelve lanes of next-generation sequencing yielded 1.2 billion total reads (~3 million per subject) to produce a coverage depth exceeding 67× per allele for all amplicons across all pools. Most amplicons showed considerably higher coverage (range: 67–987, median=474.76, IQR=355.84–568.92).
Sensitivity for single alleles (i.e. heterozygous variants present in a single individual within a given pool) was 98% (100% in 12 of 16 pools and 92% in the remainder), as determined by the detection rate of positive control singleton variants. We also compared pooled-sample results to whole-exome data for 35 subjects and found no missed variants in targeted genes. Finally, we detected all six previously-found mutations with the correctly assigned singleton frequencies.
To assess the false positive rate at the low allele frequencies in which we were interested, we performed validation genotyping of 99 allele calls for 67 non-synonymous or splice-site variants that were either rare (<1% minor allele frequency) or absent in population databases. 13 of 99 calls (8 SNPs, including 4 SNPs that were identified and validated in other pools) were not validated by subsequent genotyping, resulting in a false-positive rate of 13%. The false-positives included 5 calls that were true in other pools.
After filtering and validation, 65 rare or novel coding variants were identified (63 by pooled-sample sequencing and two by direct sequencing of exon 1 of SQSTM1). Variants were identified in all sequenced genes except for UBQLN2.
Bioinformatic analysis
Sequence alignment and variant calling were performed using “short indel prediction by large deviation inference and nonlinear true frequency estimation by recursion” (SPLINTER).19 The SPLINTER program generates an error model based on the negative control for each run. The error model is used to calculate a p-value for each SNP that is detected. SPLINTER calculates the p-value cutoff that has the highest sensitivity and specificity to distinguish true variants in the positive control vector and uses the ratio of sequencing reads with and without variant nucleotides to estimate the frequency of a given variant within a pool.
All variants called by SPLINTER were filtered for variants within exons or the 10 flanking bases and then visually inspected using Integrated Genomics Viewer(IGV)24,25 after realignment to Hg19 using Novoalign (http://www.novocraft.com) and SAMtools.26 Variants were annotated using SeattleSeq (http://sngs.washington.edu/SeattleSeqAnnotation131/), SIFT (http://sift.jcvi.org/), MutationTaster (http://www.mutationtaster.org/), and PolyPhen2 (http://genetics.bwh.harvard.edu/pph2/). The effect of splice-site mutations was predicted by Human Splicing Finder (http://www.umd.be/HSF). Population frequencies for each variant were determined in dbSNP, the 1000 Genomes Project, and the NHLBI Exome Sequencing Project version ESP6500 exome variant server (ESP6500) (http://ESP6500.gs.washington.edu/ESP6500/ [1 Dec 2013]).
Variant validation and classification
Novel and rare (<1% MAF in ESP6500) non-synonymous and splicing variants that passed visual inspection were genotyped in individual DNA samples by either Sequenom or Sanger sequencing to both validate the variant and determine which subject(s) carry them. Validated variants were assigned to four categories based on their presence in the ALS literature and frequencies in population databases. Category 1 variants have been previously reported in ALS patients but are absent from population databases. Category 2 variants have been reported in ALS patients but are present in population databases. Category 3 variants are novel (i.e. they have not been reported in ALS patients or population databases). Category 4 variants have not been reported in ALS patients but are present in population databases. Pathogenicity prediction algorithms were not utilized for category assignments because of their poor track-record in predicting disease-causing mutations.27,28 All four categories of variants were considered to be potentially pathogenic mutations.
C9ORF72 repeat expansion detection
All subjects were also screened for C9ORF72 repeat expansions using standard repeat-primed PCR.29 Many subjects in this study overlapped with those screened for our previous publication on the topic22, but are again included here to demonstrate the relative frequencies of individual genes in the cohort and to investigate patients for multiple mutations.
ATXN2 repeat size
The CAG repeat region was amplified using primers 5′ FAM-CCC CGC CCG GCG TGC GAG CCG GTG TAT G 3′ and 5′ CGG GCT TGC GGA CAT TGG 3′. PCR was performed with PhusionHigh-Fidelity PCR Master Mix with HF Buffer (New England BioLabs) with cycles as follows: 30 seconds 98°C, 35 cycles (10 seconds 98°C, 30 seconds 72°C), and 2 minutes 72°C. Repeat lengths were determined by fluorescent capillary gel electrophoresis. While intermediate-length alleles were originally considered to be 27–33 repeats30, subsequent meta-analysis has shown 29 repeats to be the optimal cutoff to distinguish ALS subjects from controls.31 Therefore we considered repeat sizes of 29–33 to be of intermediate length.
Statistical Analysis
Disease characteristics were compared in subjects possessing different numbers of mutations in ALS genes using R v3.0.1-Wilcoxon rank-sum tests were used to assess age of onset and survival, with Fisher’s exact tests were used to analyze site of symptom onset.
To identify rare and novel SNPs that might be over-represented in sporadic ALS subjects, we used Fisher’s exact tests to compare each candidate SNV’s allele frequency in sporadic ALS versus controls. By genotyping variants across a range of frequencies, we found that SPLINTER predicted frequencies and genotyped frequencies were highly correlated (r2=0.9596) as in prior studies19 (data not shown). Therefore SPLINTER-predicted frequencies were used for ALS SNPs that were not genotyped. We included only SALS samples with self-reported non-Hispanic white backgrounds (n=309) and used subjects with European ancestry in ESP6500 and the 1000 Genomes Project (1000genomes.org)32 as our control population(n=4679). Variants were selected for replication based on p-values and potential functional significance. Selected variants were genotyped in 552 ALS cases and 464 neurologically normal controls from the Coriell DNA repository. Tests were performed in R v3.0.1.
Gene-based tests comparing the burden of rare variants in cases compared to controls were performed using SKAT-O.33 We included the same individuals as were used for single-variant testing. Only missense and nonsense variants with MAF<0.1 in the control cohort were included in analysis. We used a Bonferonni correction to account for multiple testing (α=0.0036). UBQLN2 was excluded from analysis since SKAT does not handle data from the X-chromosome.
RESULTS
Subject Characteristics
Demographic and disease characteristics for the 391 sequenced subjects with ALS are shown in Table 1. Age at onset, site of first symptom, and overall disease survival were similar to other population-based and referral center-based cohorts (reviewed in Harms and Baloh).6 42 subjects (10.7%) had a family history of ALS, which is comparable to other ALS referral center-based cohorts.
Table 1.
Subject demographics and disease characteristics.
| Total ALS cases | 391 |
| Subjects with family history of ALS | 10.7% |
| Self-reported Caucasian (%)a | 93.1% |
| Male sex (%) | 57% |
| Limb Onset (%)b | 69.2% |
| Age at Onset (mean, stdev)c | 59.7±12.8 |
| Age at Onset (range) | 14–85 |
| Survival in months (mean, stdev)d | 41.3±27.7 |
| Survival in months (median) | 34 |
| Survival in months (range) | 7–147 |
Data for ethnicity,
site of onset, and
age at onset was missing for 14, 5, and 4 subjects respectively.
Survival data was available for 172 subjects.
Variant Identification and Classification
Coverage of targeted bases was ≥130 fold for each subject. Sensitivity for detecting a variant present as a single allele within the pool of normal alleles averaged 98% (100% in 12 of 16 pools and 92% in the remainder). Based on validation genotyping, 13% of variants were determined to be false-positives.
In total, we found 65 rare or novel coding or splice-site variants (Table 2). One-third of these (n=23) were previously reported in ALS patients. Ten of these ALS-associated variants were not found in population databases of genetic variation (Category 1) and review of the literature showed that all of them are well-established causal mutations. The remainder of variants previously reported in ALS (n=13) were found to be present in population databases (Category 2). With the exception of SOD1 D91A (where causality is clear), these variants lacked conclusive evidence of causation in the literature. Two-thirds (n=42) of variants we identified have not been previously reported in ALS, including 17 that are absent from population databases (Category 3) and 25 that are rare in population databases (Category 4).
Table 2.
Novel and rare coding variants identified in ALS genes.
| Gene Name | Genomic locationa | dbSNP IDb | Predicted cDNA changec | Predicted protein changec | Allele Countsd | ||||
|---|---|---|---|---|---|---|---|---|---|
| FALS | SALS | Globale | Populationf | ||||||
| Expansions | ATXN2 | 12:112036785 | rs193922927 | c.532_534CAG | Q188(29–33) | 1/84 | 11/698* | 19/2982 | 19/2982 |
| C9ORF72 | 9:27573539 | - | 14/84* | 21/698 | 11/7598 | 11/7598 | |||
Category 1:
|
FUS | 16:31202739 | rs121909668 | c.1561C>G | R521G | 1/84 | 0/698 | 0/15190 | 0/9358 |
| FUS | 16:31202752 | - | c.1574C>T | P525L | 0/84* | 1/698 | 0/15190 | 0/9358 | |
| SOD1 | 21:33032096 | rs121912442 | c.14C>T | A5V | 1/84* | 1/698 | 0/15190 | 0/9358 | |
| SOD1 | 21:33036142 | rs121912431 | c.112G>A | G38R | 1/84* | 0/698 | 0/15190 | 0/9358 | |
| SOD1 | 21:33038821 | - | c.229G>T | D77Y | 1/84 | 0/698 | 0/15190 | 0/9358 | |
| SOD1 | 21:33039600 | - | c.269C>T | A90V | 0/84 | 1/698 | 0/15190 | 0/9358 | |
| SOD1 | 21:33039672 | rs121912441 | c.341T>C | I114T | 1/84 | 1/698 | 0/15190 | 0/9358 | |
| TARDBP | 1:11082325 | rs80356719 | c.859G>A | G287S | 0/84 | 1/698* | 0/15190 | 0/9358 | |
| TARDBP | 1:11082409 | rs80356726 | c.943G>A | A315T | 1/84 | 0/698 | 0/15190 | 0/9358 | |
| VCP | 9:35065360 | rs121909329 | c.464G>A | R155H | 1/84 | 0/698 | 0/15190 | 0/9358 | |
Category 2:
|
ANG | 14:21161845 | rs121909536 | c.122A>T | K41I | 0/84 | 1/698* | 27/15190 | 23/9358 |
| ANG | 14:21162130 | rs121909543 | c.407C>T | P136L | 1/84* | 0/698 | 1/15190 | 1/9358 | |
| DCTN1 | 2:74588717 | rs72466496 | c.3746C>T | T1249I | 2/84* | 1/698* | 44/15190 | 39/9358 | |
| DCTN1 | 2:74592252 | rs72659383 | c.3146G>A | R1049Q1 | 0/84 | 1/698* | 22/15190 | 21/9358 | |
| FIG4 | 6:110036336 | rs121908287 | c.122T>C | I41T2 | 0/84 | 1/698 | 17/15190 | 16/9358 | |
| OPTN | 10:13166053 | rs142812715 | c.941A>T | Q314L | 0/84 | 1/698 | 3/15190 | 3/9358 | |
| SETX | 9:135140020 | rs151117904 | c.7640T>C | I2547T | 2/84 | 8(1hom)/698* | 76/15190 | 71/9358 | |
| SETX | 9:135202325 | rs112089123 | c.4660T>G | C1554G | 0/84 | 6/698* | 47/15190 | 40/9358 | |
| SOD1 | 21:33039603 | rs80265967 | c.272A>C | D91A | 1/84 | 2(1hom)/698 | 9/15190 | 9/9358 | |
| SQSTM1 | 5:179248034 | rs200396166 | c.98T>T | A33V | 0/84 | 2/698 | 6/15190 | 6/9358 | |
| SQSTM1 | 5:179251013 | rs145056421 | c.457G>A | V153I | 1/84* | 0/698 | 9/15190 | 9/9358 | |
| SQSTM1 | 5:179252184 | rs11548633 | c.712A>G | K238E | 0/84 | 5/698* | 41/15190 | 32/9358 | |
| TAF15 | 17:34171525 | rs200175347 | c.1222C>T | R408C | 0/84* | 1/698 | 2/15190 | 2/9358 | |
Category 3:
|
DAO | 12:109278977 | - | c.194+1G>A | Splice donor | 0/84 | 1/698 | 0/15190 | 0/9358 |
| DCTN1 | 2:74588653 | - | c.3810C>A | H1270Q | 0/84 | 1/698* | 0/15190 | 0/9358 | |
| DCTN1 | 2:74590527 | - | c.3239C>T | S1080F | 0/84 | 1/698 | 0/15190 | 0/4898† | |
| EWSR1 | 22:29682932 | - | c.620C>G | T207S | 0/84 | 1/698 | 0/15190 | 0/9358 | |
| FIG4 | 6:110087935 | - | c.1588_1589delTT | F530Ter | 0/84 | 1/698 | 0/15190 | 0/9358 | |
| FUS | 16:31202282 | - | c.1394-2delA | Splice site | 1/84 | 0/698 | 0/15190 | 0/9358 | |
| OPTN | 10:13160964 | - | c.703C>T | Q235Ter | 0/84 | 1/698 | 0/15190 | 0/9358 | |
| SETX | 9:135202223 | - | c.4762G>A | A1588T | 0/84 | 1/698 | 0/15190 | 0/572§ | |
| SETX | 9:135203632 | - | c.3353C>A | T1118K | 0/84 | 1/698 | 0/15190 | 0/9358 | |
| SETX | 9:135206694 | - | c.980A>T | E327V | 0/84 | 1/698 | 0/15190 | 0/9358 | |
| SETX | 9:135210013 | - | c.820A>G | M274V | 0/84 | 1/698* | 0/15190 | 0/9358 | |
| SETX | 9:135211743 | - | c.658A>C | K220Q | 0/84 | 1/698 | 0/15190 | 0/9358 | |
| SETX | 9:135211898 | - | c.503G>A | R168Q | 0/84 | 1/698* | 0/15190 | 0/9358 | |
| SETX | 9:135224775 | - | c.41C>T | T14I | 0/84 | 1/698 | 0/15190 | 0/9358 | |
| SOD1 | 21:33038791 | - | c.199C>G | P67A | 1/84* | 0/698 | 0/15190 | 0/9358 | |
| SQSTM1 | 5:179248079 | - | c.143T>T | L48P | 0/84 | 1/698 | 0/15190 | 0/9358 | |
| TARDBP | 1:11082589 | - | c.1123A>G | S375G | 0/84 | 1/698 | 0/15190 | 0/9358 | |
Category 4:
|
ANG | 14:21161973 | rs17560 | c.250A>G | K84E | 0/84 | 1/698 | 70/15190 | 69/4898† |
| DAO | 12:109294259 | rs4262766 | c.992G>A | G331E | 0/84 | 1/698 | 4/15190 | 0/9358 | |
| DAO | 12:109294301 | rs143732132 | c.1034C>T | S345F | 1/84* | 0/698 | 3/15190 | 3/9358 | |
| DCTN1 | 2:74593101 | rs145130328 | c.2805C>G | I935M | 0/84 | 1/698 | 4/15190 | 0/362‡ | |
| DCTN1 | 2:74598723 | rs55862001 | c.586A>G | I196V | 1/84 | 4/698* | 77/15190 | 70/9358 | |
| DCTN1 | 2:74604801 | rs374419252 | c.332C>G | S111C | 0/84 | 1/698 | 1/15190 | 0/9358 | |
| EWSR1 | 22:29682919 | rs144503053 | c.607T>A | S203T | 0/84 | 1/698 | 1/15190 | 1/9358 | |
| FIG4 | 6:110081543 | rs142463699 | c.1228A>C | T410P | 0/84 | 1/698 | 1/15190 | 0/362‡ | |
| FIG4 | 6:110107636 | rs143531641 | c.2080A>G | M694V | 0/84 | 1/698* | 3/15190 | 3/9358 | |
| FIG4 | 6:110113852 | rs375414729 | c.2444T>C | F815S | 0/84 | 1/698 | 1/15190 | 1/9358 | |
| FUS | 16:31201719 | rs186547381 | c.1292C>T | P431L | 0/84 | 1/698 | 3/15190 | 2/9358 | |
| FUS | 16:31202343 | rs201772423 | c.1453C>T | R485W | 0/84 | 1/698* | 1/15190 | 1/9358 | |
| SETX | 9:135140063 | rs202121071 | c.7597C>T | H2533Y | 1/84 | 0/698 | 1/15190 | 1/9358 | |
| SETX | 9:135147182 | rs150673589 | c.7114G>A | D2372N | 0/84 | 1/698 | 19/15190 | 6/362‡ | |
| SETX | 9:135202120 | rs140781535 | c.4865C>T | P1622L | 0/84 | 1/698 | 1/15190 | 0/9358 | |
| SETX | 9:135204004 | rs149546633 | c.2981A>G | D994G | 0/84 | 1/698 | 31/15190 | 0/9358 | |
| SETX | 9:135204235 | rs376022544 | c.2750T>C | M917T | 0/84 | 1/698 | 1/15190 | 1/9358 | |
| SETX | 9:135205116 | rs139200312 | c.1869A>C | E623D | 0/84 | 1/698 | 3/15190 | 3/9358 | |
| SETX | 9:135205594 | rs200614765 | c.1391C>T | S464L | 0/84 | 1/698 | 11/15190 | 6/9358 | |
| SETX | 9:135206706 | rs372193033 | c.968G>A | S323N | 0/84 | 2/698* | 1/15190 | 1/9358 | |
| SETX | 9:135218103 | rs145438764 | c.472T>G | L158V | 0/84 | 1/698 | 53/15190 | 48/9358 | |
| SQSTM1 | 5:179263547 | rs201239306 | c.1277C>T | A426V | 0/84 | 1/698 | 1/15190 | 0/9358 | |
| TAF15 | 17:34171358 | rs140268553 | c.1163G>A | R388H | 0/84 | 1/698 | 7/15190 | 7/9358 | |
| VAPB | 20:57014075 | rs146459055 | c.390T>G | D130E | 0/84 | 1/698 | 11/15190 | 11/9358 | |
| VAPB | 20:57016076 | rs143144050 | c.510G>A | M170I | 0/84 | 5/698* | 19/15190 | 18/9358 | |
Rare was considered a global minor allele frequency <1%.
GRCh37/hg19
dbSNP138
cDNA location and predicted protein changes refer to isoforms listed in Methods.
Allele counts are listed as alternate alleles found/total alleles assayed.
For all but the ATXN2 and C9ORF72 repeats, global allele counts were calculated from all subjects in the 1000 Genomes and NHLBI Exome Sequencing Projects. Global allele counts for C9ORF72 repeat expansions were derived from37 while intermediate CAG repeats in ATXN2 were derived from 30,31.
Population allele count refers to the population most closely matching that of the ALS subjects(s) carrying the variant. Unless indicated by a symbol, this is European ancestry (EA subjects from ESP6500 and 1000genomesEUR). Symbols used to denote other populations:
African American (AA subjects from ESP6500+1000genomesAFR);
Hispanic (1000genomesAMR)
Asian (1000genomesASN)
indicates at least one subject carrying that specific variant also carried another variant(s) in an analyzed ALS gene.
Prevalence of variants in ALS genes
We considered all 65 rare and novel variants identified by sequencing to be potentially pathogenic. 83 subjects (21.2% overall, 35.7% in FALS and 19.5% in SALS) carried at least one of these variants. The C9ORF72 repeat expansion (found in 8.7% of subjects, n=34) and ATXN2 intermediate-length CAG repeats (found in 3.1% of subjects, n=12) were also considered to be potentially pathogenic. In total, 124 subjects (31.7% overall; 64.3% across all categories of FALS, 27.8% in SALS) carried one or more of these potentially pathogenic variants (Supplementary Table 1), a higher number than reported in many prior studies.8–12,14
The proportion of subjects carrying a potentially pathogenic variant was heavily influenced by the strength of evidence for familial transmission of ALS (Table 3), with the highest rate of variant discovery in definite FALS (81.6%). The frequency of variants declined with less evidence for transmission, but even 27.8% of sporadic/simplex subjects were carriers
Table 3.
Prevalence of variants in ALS genes by family categorization
| Sporadic | All FALS | Definite | Probable A | Probable B | Possible | |
|---|---|---|---|---|---|---|
| Total Subjects | 349 | 42 | 16 | 3 | 18 | 5 |
| ANG | 2 | 1 | 1 | - | - | - |
| ATXN2 | 11 | 1 | - | - | 1 | - |
| C9ORF72 | 21 | 14 | 7 | 1 | 6 | - |
| DAO | 2 | 1 | 1 | - | - | - |
| DCTN1 | 10 | 3 | 2 | - | - | 1 |
| EWSR1 | 2 | - | - | - | - | - |
| FIG4 | 5 | - | - | - | - | - |
| FUS | 3 | 2 | 1 | - | - | 1 |
| OPTN | 2 | - | - | - | - | - |
| SETX | 29 | 3 | 1 | - | 2 | - |
| SOD1 | 4 | 6 | 4 | - | 2 | - |
| SQSTM1 | 9 | 1 | - | - | 1 | - |
| TAF15 | 2 | - | - | - | - | - |
| TARDBP | 2 | 1 | 1 | - | - | - |
| UBQLN2 | - | - | - | - | - | - |
| VAPB | 6 | - | - | - | - | - |
| VCP | - | 1 | - | 1 | - | - |
| Total Variants | 110 | 34 | 18 | 2 | 12 | 2 |
| Subjects with any variant | 97 | 27 | 13 | 2 | 10 | 2 |
| % Subjects with variants | 27.8% | 64.3% | 81.3% | 66.7% | 55.6% | 40.0% |
Definite FALS (38% of families): at least two first- or second-degree relatives with ALS;
Probable A FALS (7% of families): one first-degree relative with ALS;
Probable B FALS (43% of families): one second-degree relative with ALS;
Possible FALS (12% of families): one distant relative with ALS;
Sporadic ALS (89% of entire cohort): all subjects not meeting criteria for any FALS category.
We identified 4 sporadic subjects with potentially recessive causes of their ALS (Table 4). One subject was homozygous for SOD1 p.D91A (D90A), while three others carried two mutations in SETX. One subject tested homozygous for SETX p.I2547T, but we did not exclude the possibility of a deletion on one allele. The two additional subjects could each be compound heterozygotes comprised of a rare variant (p.C1554G or p.I2547T with 0.3% and 0.5% MAF in population database respectively) and a novel variant (p.R168Q or p.T14I respectively). The subject carrying p.I2547T and p.T14I was also heterozygous for TAF15 p.R408C which has previously been reported in a subject with SALS.34 Due to the absence of additional family members for segregation or tissue for cDNA sequencing, we were unable to determine if these SETX variants are in cis or trans. Because recessive mutations in SETX are associated with ataxia-ocular apraxia type 2 (OMIM 606002) and SETX-associated ALS is dominantly inherited, we reviewed the medical records of these 3 individuals. All three showed typical ALS disease course without clinically apparent eye movement abnormalities or ataxia.
Table 4.
Subjects with multiple variants in ALS genes
| Patient Type | Variant 1 | Variant 2 | Variant 3 | Possible model |
|---|---|---|---|---|
| Sporadic | SOD1(p.D91A) | SOD1(p.D91A) | - | Homozygous recessive |
| Sporadic | SETX(p.I2547T) | SETX(p.I2547T) | - | Homozygous recessive |
| Sporadic | SETX(p.C1554G) | SETX(p.R168Q) | - | Potential compound het |
| Familial | SOD1(p.A5V) | DAO(p.S345F) | - | Oligogenic |
| Familial | SOD1(p.P67A) | SETX(p.I2547T) | - | Oligogenic |
| Familial | C9ORF72 | DCTN1(p.I196V) | - | Oligogenic |
| Familial | C9ORF72 | SQSTM1(p.V153I) | - | Oligogenic |
| Familial | C9ORF72 | SETX(p.I2547T) | - | Oligogenic |
| Sporadic | ANG(p.K41I) | VAPB(p.M170I) | - | Oligogenic |
| Sporadic | ATXN2(22/31) | SQSTM1(p.K238E) | - | Oligogenic |
| Sporadic | FUS(p.R485W) | SETX(p.I2547T) | - | Oligogenic |
| Sporadic | DCTN1(p.R1049Q) | SETX(p.S323N) | - | Oligogenic |
| Sporadic | FUS(p.P525L) | ATXN2(23/31) | - | Oligogenic |
| Sporadic | TARDBP(p.G287S) | VAPB(p.M170I) | - | Oligogenic |
| Familial | SOD1(p.G38R) | ANG(p.P136L) | DCTN1(p.T1249I) | Oligogenic |
| Sporadic | ATXN2(22/32) | DCTN1(p.T1249I) | SETX(p.M274V) | Oligogenic |
| Sporadic | TAF15(p.R408C) | SETX(p.I2547T) | SETX(p.T14I) | Oligogenic, potential compound het |
| Sporadic | SETX(p.C1554G) | DCTN1(p.H1270Q) | FIG4(p.M694V) | Oligogenic |
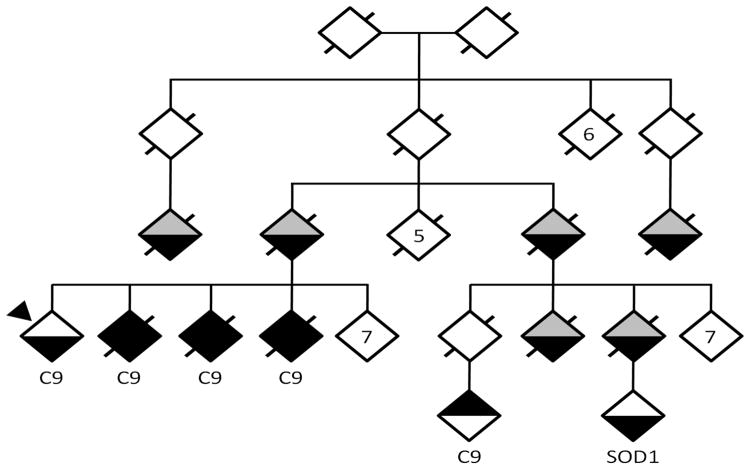
We also identified a pedigree with FALS with independently-segregating causative mutations (Figure 2). The proband, three affected siblings and a first cousin once-removed all tested positive for the C9ORF72 repeat expansion. Another second first cousin once-removed was diagnosed with ALS at another center test but tested negative for the expansion, including by Southern blot (data not shown). Instead, this individual was found to carry a heterozygous SOD1 p.D91A mutation detected by pooled-sample sequencing.
Figure 2. Segregation of distinct mutations in a FALS pedigree.
WUNM0026 has been de-identified, with exclusion of some unaffected branches. If a diamond represents more than one individual it is indicated by a number in the diamond. The upper portion of each diamond denotes presence or absence of frontotemporal dementia while the lower portion denotes presence or absence of ALS. White = unaffected; black=affected; gray=unknown. Slashes denote deceased individuals. Proband is marked with an arrow-head. Those carrying C9ORF72 expansions labeled as C9, and the single individual who carried the SOD p.D91A variant is marked as SOD1.
Prevalence of potential oligogenic subjects
We assessed the number of genes with potentially pathogenic rare variants in each individual. Fifteen subjects (3.8% overall, 14% in FALS, 2.6% in SALS) harbored potentially pathogenic variants in at least two ALS genes: 11 with variants in two ALS genes, while 4 had variants in three genes each (Table 4).
Six potentially oligogenic subjects had a family history of ALS subjects and in all cases one of their variants was either the C9ORF72 repeat expansion or a missense variant in SOD1 in combination with additional rare or novel variant(s), several of which have also been previously reported in ALS subjects. Interestingly, one FALS proband carried 3 variants, each of which has previously been reported as pathogenic: SOD1 p.G38R, ANG p.P136L, and DCTN1 p.T1249I.
Nine apparently sporadic subjects had variants in multiple genes (Table 4), but only two were well-established ALS mutations: TARDBP p.G287S was found in combination with VAPB p.M170I while a subject with juvenile-onset ALS carried a de novo FUS p.P525L mutation with a paternally-inherited intermediate-sized CAG expansion in ATXN2. Two SALS patients carried multiple ALS-associated variants that are rare in population databases (ANG p.K41I with VAPB p.M170I and TAF15 p.R408C with SETX p.I2547T and SETX p.T14I).
Correlation of variant genes with disease characteristics
In an oligogenic model of disease, the additive or synergistic effects of multiple variants can influence not only the risk of developing disease, but also phenotypic manifestations of the disease. Age at symptom onset was significantly earlier in cases carrying variants in multiple genes (median=46, IQR=39–61) compared to all other subjects (median=61, IQR=51–70, p=0.0046) and when compared to cases with mutations in just one gene (median=60, IQR=48–60, p=0.017). Even when the subject with juvenile onset was removed a difference of 10 years earlier remained (median=50.5, IQR=40.25–61.5, p=0.013 against all others and p=0.041 against other single-gene variant carriers). Furthermore, there was a weak, but statistically significant negative correlation between age of onset and the number of genes with variants (spearman’s rho=−0.11, p=0.024). The number of ALS genes with variants did not influence disease duration or site of onset in our cohort.
Rare variants as modifiers of ALS risk
We also used our sequencing results to search for single variants in known ALS genes that increased or decreased ALS risk. To do so, we analyzed all coding variants found in our ALS cohort and also present in population databases (n=61, with 47 having a population MAF <1%). Three SNPs in SETX (rs1183768, rs543573, and rs2296871) were in perfect linkage disequilibrium and were considered to be one signal represented by rs2296871. We included only ALS subjects of European ancestry and compared to controls of European ancestry from ESP6500 and the 1000 Genomes Project. SPLINTER-predicted allele frequencies were used for common variants that were not confirmed by genotyping in ALS subjects. Using a Bonferonni-corrected significance level of 8.2×10−4, 3 variants were significantly more common in our ALS discovery cohort (rs3739927 and rs882709 in SETX, and rs41311143 in EWSR1). To follow up, we genotyped these 3 SNPs and 28 additional candidate variants in a validation cohort of 552 sporadic ALS cases and 464 controls from Coriell reference panels. However, none of the 31 tested variants showed a significant association with ALS in either direction (Supplementary Table 2).
We also asked whether the burden of rare coding variants in any of the tested ALS genes was higher in sporadic subjects compared to controls using SKAT (Table 5).33 After correcting for multiple tests (α=3.57×10−3), SOD1 was the only gene that showed a significant association (p=1.59×10−5) while TARDBP and VAPB approached statistical significance (p=5.57×10−3 and p=5.99×10−3 respectively).
Table 5.
Gene-based rare variant association tests
| Gene | P-value | # Markers |
|---|---|---|
| SOD1 | 1.59×10−5 | 4 |
| TARDBP | 5.57×10−3 | 10 |
| VAPB | 5.99×10−3 | 8 |
| SQSTM1 | 0.126 | 39 |
| SETX | 0.165 | 125 |
| FUS | 0.323 | 25 |
| DAO | 0.425 | 26 |
| DCTN1 | 0.443 | 58 |
| EWSR1 | 0.450 | 21 |
| ANG | 0.487 | 9 |
| TAF15 | 0.573 | 34 |
| VCP | 0.693 | 5 |
| OPTN | 0.765 | 20 |
| FIG4 | 0.863 | 32 |
Association tests were performed with SKAT using the optimal.adj method and the default linear, weighted kernel, with significance level=3.57×10−3. Only coding variants with minor allele frequencies <1% were included in the analysis. Only subjects of European Ancestry were used from our cohort and controls (self-declared Caucasians compared to EA in ESP6500 and 1000genomesEUR).
DISCUSSION
Rapid progress toward defining the genetic landscape of ALS has been fueled by the emergence of next-generation sequencing. In this study, we used the efficiency and power of pooled-sample sequencing to investigate the frequency of pathogenic and potentially-pathogenic variants in known ALS genes in a large cohort of US patients. Our approach produced highly accurate sequence data for 15 known genes in a time, sample, and cost-efficient manner. We estimated that this study required 83% less DNA per subject and cost 10% of performing the equivalent study by traditional Sanger sequencing. In doing so, we have generated the most comprehensively sequenced North American cohort to date.
In this group of subjects we identified 27 novel variants (i.e. not found in databases of variation) and an additional 38 that are very rare in control populations. Not surprisingly, the highest rate of variant detection occurred in families with the strongest ALS histories: we found explanatory mutations in 80% of these pedigrees. This rate is higher than many previous reports of all FALS8–14 and partially stems from our use of a strict definition of familial ALS favoring pedigrees with clearly dominant transmission patterns that undoubtedly enrich for Mendelian genes. Our elevated variant detection rate is also influenced by the large number genes analyzed in each family. Because our cohort was a clinic-based, we cannot address whether differences in populations are also involved.
Although the frequency of variant detection in our sporadic ALS subjects was lower than in familial ALS, it was still 28%. This is considerably higher than other studies 8–12,14, likely due in part to the large number of genes we sequenced. In support of this, we note that the frequency of variants in commonly sequenced genes (e.g. C9ORF72, SOD1, TARDBP) was within previously reported ranges. To directly compare our findings with a similar study of an Irish population9, we limited both data sets to genes shared between the two studies and only included novel variants (i.e. not seen in any population database). The total number of subjects found with at least one potentially pathogenic mutation was 16.4% in this study compared to 12.8% in the Irish population. This difference is not statistically significant (p=0.12) and was driven by the absence of SOD1 mutations in the Irish cohort. These broad differences in populations need to be given appropriate consideration when genetic testing or counseling is being provided to patients.
Based on previous reports of oligogenic inheritance in ALS, we looked for subjects with potentially pathogenic variants in more than one ALS gene. We found mutations in at least two ALS genes in 3.8% of our subjects (14% in FALS, 2.6% in SALS). This rate is higher than in prior reports, but direct comparisons are prevented by differences in i) which genes were sequenced, ii) how complete variant ascertainment was, iii) relative numbers of familial and sporadic cases, and iv) which variants were considered to be potentially pathogenic.9,12 In most cases, one of the identified variants is a known mutation with clearly established pathogenicity, however many of the additional variants are of unknown significance. It is possible that these additional variants co-occur with pathogenic mutations by chance. However, the fact that subjects with potentially pathogenic variants in more than one gene had disease onset 10 years earlier than other subjects supports a model of ALS where the additive or synergistic effects of multiple defective genes increases risk and influences disease phenotype.
This study evaluated known ALS genes only. With many efforts underway to generate exome and genome-wide variant data on large numbers of ALS patients, these types of interactions should become easier to detect and validate. These large datasets should also allow unbiased searches for new ALS genes using rare variant burden testing. As a test of this principle, we asked whether rare variant burden testing would identify any of the known ALS genes we sequenced. In our modestly-sized cohort we demonstrated a significant association for SOD1 and suggestive associations for TARDBP and VAPB. We also noticed an abundance of variants in the SETX gene, an intriguing finding that was also evident in a prior study.9 These findings predict that well-powered genome-wide studies will identify new ALS genes.
Our study also highlights important lessons regarding mutation screening in ALS. First, a significant number of individuals will harbor more than one potentially pathogenic mutation. This fact dramatically influences estimates of transmission risk and even prognosis. Therefore, comprehensive screening of known genes is preferable to single-gene testing and made more cost-effective by next-generation approaches to sequencing. Second, as our pedigree with independently segregating SOD1 and C9ORF72 mutations demonstrates, even once a causative mutation has been identified in a pedigree, each affected individual should be sequenced for confirmation. Third, despite the frequency with which our study found variants in ALS subjects, 36% of FALS and 74% of SALS subjects had no variants in any of 17 ALS genes we analyzed. Efforts are therefore needed to identify additional genes influencing ALS risk.
Finally, we note that many of the novel and rare variants identified by this study and others are of unknown significance and will require further study to validate a possible contribution to ALS pathogenesis. The complexity of determining pathogenicity of variants is highlighted by the 13 variants we identified that had been previously associated with ALS but have since been found in control databases at rates higher than expected for moderate or high penetrance mutations. Although these variants could represent mutations with reduced penetrance, or the presence of pre-symptomatic individuals in control populations, they most likely result from including limited controls in the original studies. In fact, many variants previously reported as pathogenic for ALS and other diseases are now found in the 1000 Genomes Project or the Exome Sequencing Project at frequencies exceeding those expected for moderately or highly penetrant mutations.35 To prevent the literature from becoming confused with disease-associated variants that are not pathogenic, we support increased attention to variants are reported in disease populations, including the creation of levels of genetic evidence for pathogenicity as recently proposed.36
Supplementary Material
Acknowledgments
This work was supported by NIH grants K08-NS075094 (Dr Harms), R01-NS069669 (Dr Baloh) and the Genetics Epidemiology Training grant 5-T32-HL-83822-5 (Ms. Cady). We thank Ryan T. Libby (Virginia Mason Medical Center) and Shaughn Bell (Cedars Sinai Medical Center) for assistance with DNA preparation; Francesco Valliana for help in implementing SPLINTER; Joseph Giacalone for providing pooled sample sequencing controls; Carlos Cruchaga for providing custom scripts to analyze coverage for SPLINTER sequencing, and Carolyn Hilliard (St. Louis University School of Medicine) for generating ATXN2 genotypes. We thank the Washington University Sequenom Core and the Protein and Nucleic Acids Laboratory. We thank the Genome Technology Access Center in the Department of Genetics at Washington University School of Medicine for help with genomic analysis. The Center is partially supported by NCI Cancer Center Support Grant #P30 CA91842 to the Siteman Cancer Center and by ICTS/CTSA Grant# UL1 TR000448 from the National Center for Research Resources (NCRR), a component of the National Institutes of Health (NIH), and NIH Roadmap for Medical Research. This publication was made possible by grant UL1 RR024992 from the National Center for Research Resources, a component of the NIH, and NIH Roadmap for Medical Research. This publication is solely the responsibility of the authors and does not necessarily represent the official view of NCRR or NIH.
Footnotes
AUTHOR CONTRIBUTIONS:
Janet Cady contributed to the study design, data acquisition, analysis, and manuscript preparation. Peggy Allred, Taha Bali, Alan Pestronk, and Timothy M. Miller contributed to data acquisition and analysis. Robi Mitra and Alison Goate contributed to study design and data analysis. John Ravits contributed to study design. Robert H. Baloh and Matthew B. Harms contributed to study design, data acquisition, data analysis and manuscript preparation.
POTENTIAL CONFLICTS OF INTEREST:
Matthew Harms reports NIH grant funding related to the submitted work. All other authors have nothing to disclose related to the submitted work. Outside the submitted work, authors Janet Cady, Peggy Allred, Taha Bali, Alan Pestronk, John Ravits, and Robert H. Baloh report nothing to disclose. Alison Goate reports grants from NIA during the conduct of the study; personal fees from Finnegan HC, personal fees from Cognition Therapeutics, personal fees from Finnegan HC, personal fees from Dickstein Shapiro, grants from Genentech, grants from Pfizer, grants from Astra Zeneca, personal fees from Genentech, personal fees from Amgen and patent US20070258898. Timothy M. Miller reports grants and personal fees from Biogen Idec, non-financial support from Regulus Therapeutics, grants and personal fees from Isis Pharmaceuticals, and patent PCT/US2012/060597 licensed to C2N Diagnostics. Rob Mitra reports NIH grants. Matthew B. Harms reports personal fees from Genzyme, grants from Millstone Family Foundation, Biogen Idec, the Hope Center for Neurological Disorders, and the McDonnell Center.
References
- 1.Abhinav K, Stanton B, Johnston C, et al. Amyotrophic lateral sclerosis in SouthEast England: a population-based study. The South-East England register for amyotrophic lateral sclerosis (SEALS Registry) Neuroepidemiology. 2007;29(1–2):44–48. doi: 10.1159/000108917. [DOI] [PubMed] [Google Scholar]
- 2.Mehta P, Antao V, Kaye W, et al. Prevalence of amyotrophic lateral sclerosis - United States, 2010–2011. Morb Mortal Wkly Rep Surveill Summ Wash DC 2002. 2014;63 (Suppl 7):1–14. [PubMed] [Google Scholar]
- 3.Gros-Louis F, Gaspar C, Rouleau GA. Genetics of familial and sporadic amyotrophic lateral sclerosis. Biochim Biophys Acta. 2006;1762(11–12):956–972. doi: 10.1016/j.bbadis.2006.01.004. [DOI] [PubMed] [Google Scholar]
- 4.Kurland LT, Mulder DW. Epidemiologic investigations of amyotrophic lateral sclerosis. 2. Familial aggregations indicative of dominant inheritance. I. Neurology. 1955;5(3):182–196. doi: 10.1212/wnl.5.3.182. [DOI] [PubMed] [Google Scholar]
- 5.Kurland LT, Mulder DW. Epidemiologic investigations of amyotrophic lateral sclerosis. 2. Familial aggregations indicative of dominant inheritance II. Neurology. 1955;5(4):249–268. doi: 10.1212/wnl.5.4.249. [DOI] [PubMed] [Google Scholar]
- 6.Harms MB, Baloh RH. Clinical neurogenetics: amyotrophic lateral sclerosis. Neurol Clin. 2013;31(4):929–950. doi: 10.1016/j.ncl.2013.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Andersen PM, Al-Chalabi A. Clinical genetics of amyotrophic lateral sclerosis: what do we really know? Nat Rev Neurol. 2011;7(11):603–615. doi: 10.1038/nrneurol.2011.150. [DOI] [PubMed] [Google Scholar]
- 8.Lattante S, Conte A, Zollino M, et al. Contribution of major amyotrophic lateral sclerosis genes to the etiology of sporadic disease. Neurology. 2012;79(1):66–72. doi: 10.1212/WNL.0b013e31825dceca. [DOI] [PubMed] [Google Scholar]
- 9.Kenna KP, McLaughlin RL, Byrne S, et al. Delineating the genetic heterogeneity of ALS using targeted high-throughput sequencing. J Med Genet. 2013;50(11):776–783. doi: 10.1136/jmedgenet-2013-101795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chiò A, Calvo A, Mazzini L, et al. Extensive genetics of ALS: a population-based study in Italy. Neurology. 2012;79(19):1983–1989. doi: 10.1212/WNL.0b013e3182735d36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kwon M-J, Baek W, Ki C-S, et al. Screening of the SOD1, FUS, TARDBP, ANG, and OPTN mutations in Korean patients with familial and sporadic ALS. Neurobiol Aging. 2012;33(5):1017, e17–23. doi: 10.1016/j.neurobiolaging.2011.12.003. [DOI] [PubMed] [Google Scholar]
- 12.Van Blitterswijk M, van Es MA, Hennekam EAM, et al. Evidence for an oligogenic basis of amyotrophic lateral sclerosis. Hum Mol Genet. 2012;21(17):3776–3784. doi: 10.1093/hmg/dds199. [DOI] [PubMed] [Google Scholar]
- 13.Conte A, Lattante S, Luigetti M, et al. Classification of familial amyotrophic lateral sclerosis by family history: effects on frequency of genes mutation. J Neurol Neurosurg Psychiatry. 2012;83(12):1201–1203. doi: 10.1136/jnnp-2012-302897. [DOI] [PubMed] [Google Scholar]
- 14.Renton AE, Chiò A, Traynor BJ. State of play in amyotrophic lateral sclerosis genetics. Nat Neurosci. 2014;17(1):17–23. doi: 10.1038/nn.3584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fogh I, Ratti A, Gellera C, et al. A genome-wide association meta-analysis identifies a novel locus at 17q11.2 associated with sporadic amyotrophic lateral sclerosis. Hum Mol Genet. 2014;23(8):2220–2231. doi: 10.1093/hmg/ddt587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Keller MF, Ferrucci L, Singleton AB, et al. Genome-Wide Analysis of the Heritability of Amyotrophic Lateral Sclerosis. JAMA Neurol. 2014;71(9):1123. doi: 10.1001/jamaneurol.2014.1184. cited 2014 Oct 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Al-Chalabi A, Fang F, Hanby MF, et al. An estimate of amyotrophic lateral sclerosis heritability using twin data. J Neurol Neurosurg Psychiatry. 2010;81(12):1324–1326. doi: 10.1136/jnnp.2010.207464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Druley TE, Vallania FLM, Wegner DJ, et al. Quantification of rare allelic variants from pooled genomic DNA. Nat Methods. 2009;6(4):263–265. doi: 10.1038/nmeth.1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vallania FLM, Druley TE, Ramos E, et al. High-throughput discovery of rare insertions and deletions in large cohorts. Genome Res. 2010;20(12):1711–1718. doi: 10.1101/gr.109157.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Brown JA, Min J, Staropoli JF, et al. SOD1, ANG, TARDBP and FUS mutations in amyotrophic lateral sclerosis: a United States clinical testing lab experience. Amyotroph Lateral Scler Off Publ World Fed Neurol Res Group Mot Neuron Dis. 2012;13(2):217–222. doi: 10.3109/17482968.2011.643899. [DOI] [PubMed] [Google Scholar]
- 21.Brooks BR. El Escorial World Federation of Neurology criteria for the diagnosis of amyotrophic lateral sclerosis. Subcommittee on Motor Neuron Diseases/Amyotrophic Lateral Sclerosis of the World Federation of Neurology Research Group on Neuromuscular Diseases and the El Escorial “Clinical limits of amyotrophic lateral sclerosis” workshop contributors. J Neurol Sci. 1994;124 (Suppl):96–107. doi: 10.1016/0022-510x(94)90191-0. [DOI] [PubMed] [Google Scholar]
- 22.Harms MB, Cady J, Zaidman C, et al. Lack of C9ORF72 coding mutations supports a gain of function for repeat expansions in amyotrophic lateral sclerosis. Neurobiol Aging. 2013;34(9):2234, e13–19. doi: 10.1016/j.neurobiolaging.2013.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wu C-H, Fallini C, Ticozzi N, et al. Mutations in the profilin 1 gene cause familial amyotrophic lateral sclerosis. Nature. 2012;488(7412):499–503. doi: 10.1038/nature11280. cited 2014 Jul 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Robinson JT, Thorvaldsdóttir H, Winckler W, et al. Integrative genomics viewer. Nat Biotechnol. 2011;29(1):24–26. doi: 10.1038/nbt.1754. cited 2013 Jun 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Thorvaldsdottir H, Robinson JT, Mesirov JP. Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Brief Bioinform. 2012;14(2):178–192. doi: 10.1093/bib/bbs017. cited 2013 Jun 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li H, Handsaker B, Wysoker A, et al. The Sequence Alignment/Map format and SAMtools. Bioinforma Oxf Engl. 2009;25(16):2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Thusberg J, Olatubosun A, Vihinen M. Performance of mutation pathogenicity prediction methods on missense variants. Hum Mutat. 2011;32(4):358–368. doi: 10.1002/humu.21445. cited 2014 Jun 20. [DOI] [PubMed] [Google Scholar]
- 28.Flanagan SE, Patch A-M, Ellard S. Using SIFT and PolyPhen to predict loss-of-function and gain-of-function mutations. Genet Test Mol Biomark. 2010;14(4):533–537. doi: 10.1089/gtmb.2010.0036. [DOI] [PubMed] [Google Scholar]
- 29.DeJesus-Hernandez M, Mackenzie IR, Boeve BF, et al. Expanded GGGGCC hexanucleotide repeat in noncoding region of C9ORF72 causes chromosome 9p-linked FTD and ALS. Neuron. 2011;72(2):245–256. doi: 10.1016/j.neuron.2011.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Elden AC, Kim H-J, Hart MP, et al. Ataxin-2 intermediate-length polyglutamine expansions are associated with increased risk for ALS. Nature. 2010;466(7310):1069–1075. doi: 10.1038/nature09320. cited 2014 Jun 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Van Damme P, Veldink JH, van Blitterswijk M, et al. Expanded ATXN2 CAG repeat size in ALS identifies genetic overlap between ALS and SCA2. Neurology. 2011;76(24):2066–2072. doi: 10.1212/WNL.0b013e31821f445b. [DOI] [PubMed] [Google Scholar]
- 32.1000 Genomes Project Consortium. Abecasis GR, Auton A, et al. An integrated map of genetic variation from 1,092 human genomes. Nature. 2012;491(7422):56–65. doi: 10.1038/nature11632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wu MC, Lee S, Cai T, et al. Rare-variant association testing for sequencing data with the sequence kernel association test. Am J Hum Genet. 2011;89(1):82–93. doi: 10.1016/j.ajhg.2011.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Couthouis J, Hart MP, Shorter J, et al. A yeast functional screen predicts new candidate ALS disease genes. Proc Natl Acad Sci U S A. 2011;108(52):20881–20890. doi: 10.1073/pnas.1109434108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Xue Y, Chen Y, Ayub Q, et al. Deleterious- and disease-allele prevalence in healthy individuals: insights from current predictions, mutation databases, and population-scale resequencing. Am J Hum Genet. 2012;91(6):1022–1032. doi: 10.1016/j.ajhg.2012.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.MacArthur DG, Manolio TA, Dimmock DP, et al. Guidelines for investigating causality of sequence variants in human disease. Nature. 2014;508(7497):469–476. doi: 10.1038/nature13127. cited 2014 Jul 25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Beck J, Poulter M, Hensman D, et al. Large C9orf72 Hexanucleotide Repeat Expansions Are Seen in Multiple Neurodegenerative Syndromes and Are More Frequent Than Expected in the UK Population. Am J Hum Genet. 2013;92(3):345–353. doi: 10.1016/j.ajhg.2013.01.011. cited 2014 Jun 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Byrne S, Bede P, Elamin M, et al. Proposed criteria for familial amyotrophic lateral sclerosis. Amyotroph Lateral Scler Off Publ World Fed Neurol Res Group Mot Neuron Dis. 2011;12(3):157–159. doi: 10.3109/17482968.2010.545420. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.