Abstract
Lotus japonicus was shown to contain the two nitrile glucosides rhodiocyanoside A and rhodiocyanoside D as well as the cyanogenic glucosides linamarin and lotaustralin. The content of cyanogenic and nitrile glucosides in L. japonicus depends on plant developmental stage and tissue. The cyanide potential is highest in young seedlings and in apical leaves of mature plants. Roots and seeds are acyanogenic. Biosynthetic studies using radioisotopes demonstrated that lotaustralin, rhodiocyanoside A, and rhodiocyanoside D are derived from the amino acid l-Ile, whereas linamarin is derived from Val. In silico homology searches identified two cytochromes P450 designated CYP79D3 and CYP79D4 in L. japonicus. The two cytochromes P450 are 94% identical at the amino acid level and both catalyze the conversion of Val and Ile to the corresponding aldoximes in biosynthesis of cyanogenic glucosides and nitrile glucosides in L. japonicus. CYP79D3 and CYP79D4 are differentially expressed. CYP79D3 is exclusively expressed in aerial parts and CYP79D4 in roots. Recombinantly expressed CYP79D3 and CYP79D4 in yeast cells showed higher catalytic efficiency with l-Ile as substrate than with l-Val, in agreement with lotaustralin and rhodiocyanoside A and D being the major cyanogenic and nitrile glucosides in L. japonicus. Ectopic expression of CYP79D2 from cassava (Manihot esculenta Crantz.) in L. japonicus resulted in a 5- to 20-fold increase of linamarin content, whereas the relative amounts of lotaustralin and rhodiocyanoside A/D were unaltered.
Cyanogenic glucosides are widely distributed in the plant kingdom. They are present in more than 2,650 different plant species derived from about 550 genera and more than 130 families (Seigler, 1991) and these include ferns, angiosperms, and gymnosperms. Cyanogenic glucosides are β-glucosides of α-hydroxynitriles. When the subcellular structures of plant tissue containing cyanogenic glucosides are disrupted, e.g. by chewing insects, the cyanogenic glucosides are degraded by β-glucosidases and α-hydroxynitrilases. This results in concomitant release of toxic hydrogen cyanide, Glc, and an aldehyde or ketone. This binary system—two sets of components that separately are chemically inert—provides plants with an immediate chemical defense response to herbivores and pathogens that cause tissue damage (Møller and Seigler, 1999). Accordingly, cyanogenic glucosides are classified as phytoanticipins (VanEtten et al., 1994). Another suggested role for cyanogenic glucosides is as nitrogen storage compounds (Forslund and Jonsson, 1997; Busk and Møller, 2002).
Cyanogenic glucosides are derived from the protein amino acids l-Val, l-Ile, l-Leu, l-Phe, or l-Tyr and from the nonprotein amino acid cyclopentenyl-Gly. The Val and Ile derived cyanogenic glucosides linamarin and lotaustralin usually cooccur and are widespread in nature (Seigler, 1975; Møller and Seigler, 1999). The biosynthetic pathway for cyanogenic glucosides has been extensively studied in a number of plant species including sorghum (Sorghum bicolor), cassava (Manihot esculenta), seaside arrow grass (Triglochin maritimum), and barley (Hordeum vulgare; Koch et al., 1995; Jones et al., 1999; Nielsen and Møller, 1999; Andersen et al., 2000; Nielsen et al., 2002). The synthesis is catalyzed by two membrane bound multifunctional cytochromes P450 and by a soluble UDP-Glc (UDPG)-glucosyltransferase (Fig. 1). A cytochrome P450 belonging to the CYP79 family catalyses the first committed step, the conversion of amino acids to the corresponding oximes. The first CYP79 enzyme that was isolated and characterized was CYP79A1 from sorghum involved in biosynthesis of Tyr derived dhurrin (Halkier and Møller, 1989; Koch et al., 1992; Sibbesen et al., 1994; Koch et al., 1995; Sibbesen et al., 1995). Subsequently, CYP79s from seaside arrow grass (Nielsen and Møller, 1999) and cassava (Andersen et al., 2000) have been characterized. The oxime is converted to a cyanohydrin by the second cytochrome P450 and subsequently glucosylated by a UDPG-glucosyltransferase to produce the cyanogenic glucosides (Fig. 1). Hitherto, postoxime enzymes have exclusively been cloned from sorghum. The sorghum postoxime enzymes are designated CYP71E1 (Bak et al., 1998) and UGT85B1 (Jones et al., 1999).
Figure 1.
Biosynthetic pathways for the cyanogenic glucosides linamarin and lotaustralin, and proposed pathways of biosynthesis of the nitrile glucosides, rhodiocyanoside A and D in L. japonicus.
Nitrile glucosides have in a few cases been shown to cooccur with cyanogenic glucosides (Lechtenberg et al., 1996; Nielsen et al., 2002). As cyanogenic glucosides, nitrile glucosides are β-glucosides of hydroxy nitriles. However, in nitrile glucosides, the hydroxyl and nitrile groups are not linked to the same carbon atom of the aglycone. Accordingly, hydrolysis of nitrile glucosides by β-glucosidases does not result in HCN release. Rhodiocyanoside A, B, and D, as well as sarmentosine, sarmentosine epoxide, and isosarmentosine are nitrile glucosides that based on their structural characteristics may be envisioned to be derived from Ile (Lechtenberg and Nahrstedt, 1999). The corresponding nitrile and epoxide glucosides that likewise might be formed from Val have not been discovered. While the biosynthetic pathway for cyanogenic glucosides has been elucidated and some biological roles have been identified, no experimental data are available on how nitrile glucosides are synthesized and what their biological function might be. In addition to cyanogenic and nitrile glucosides, glucosides of cyanogenic glucosides, often referred to as diglucosides, have been identified in cyanogenic plants and hypothesized to be transport forms of cyanogenic glucosides in the so-called linustatin pathway (Selmar et al., 1988).
Lotus corniculatus has previously been reported to contain the cyanogenic glucosides lotaustralin and linamarin (Jones, 1977). Lotus japonicus is being introduced as a model plant to investigate the interaction between symbiotic nitrogen fixing bacteria and legumes, supported by a number of genomic research programs (VandenBosch and Stacey, 2003). L. japonicus holds many of the traits that provide an excellent genetic model system. It is diploid, has a small genome (approximately 450 Mb), a short generation time, is self-compatible, relatively easy to transform by Agrobacterium tumefaciens, mutants are available, and micro-arrays are being produced (Handberg and Stougaard, 1992; Asamizu et al., 2000; Nakamura et al., 2002; Perry et al., 2003). In addition, cDNA and genomic sequencing is in progress at the Kazusa DNA Research Institute (http://www.kazusa.or.jp/en/database.html).
None of the previously studied cyanogenic glucoside producing plant species offers such overall advantages. Accordingly, we want to introduce L. japonicus as the genetic model system to study cyanogenic glucosides in higher plants. In this paper, we analyze the presence of cyanogenic glucosides in dependence of plant development. We document that the two nitrile glucosides, rhodiocyanoside A and D, are also formed, and that they are synthesized from Ile. We show that two independent biosynthetic pathways appear to operate in roots and aerial parts of L. japonicus, each supported by a separate CYP79 enzyme. The ability to modify composition as well as site of accumulation of cyanogenic glucosides in L. japonicus is demonstrated by the ectopic expression of an orthologous cytochrome P450 from cassava, CYP79D2, which processes different catalytic properties compared to L. japonicus CYP79D3 and CYP79D4.
RESULTS
Distribution of the Cyanogenic and Nitrile Glucosides in L. japonicus
The presence and absence of cyanogenic glucosides, cyanogenic diglucosides, and nitrile glucosides in various L. japonicus tissues was analyzed by liquid chromatography-mass spectrometry-selected ion monitoring (LC-MS-SIM). Glucosides readily form adducts with sodium ions. Sodium adducts of the cyanogenic glucosides linamarin (rt = 4.5 min) and lotaustralin (rt = 14.32 min) and of the nitrile glucosides, rhodiocyanoside D (rt = 8.5 min) and rhodiocyanoside A (rt = 9.5 min), were detected, albeit only in aerial tissues (Fig. 2). The identity of the compounds was confirmed by NMR spectrometry (data not shown). In roots, minute amounts of lotaustralin were occasionally detected. The nitrile glucosides rhodiocyanoside A and D were never detected in roots. The glucosides of linamarin and lotaustralin, denoted linustatin and neolinostatin, respectively, were not detected in any of the tissues analyzed. In L. japonicus aerial tissues, rhodiocyanoside A and lotaustralin are two major components, while linamarin and rhodiocyanoside D are minor components (Fig. 2). These four constituents cooccur in the different aerial tissues of L. japonicus, and the ratio between the components is relatively constant, except in cotyledons where the proportion of linamarin is higher (Fig. 2).
Figure 2.
The percentage content of the cyanogenic and nitrile glucosides in L. japonicus tissues. Except for flowers, the tissues were obtained from 18-d-old hydroponically grown plants. Quantification is based on peak areas from LC-MS-SIM chromatograms.
When cyanogenic glucosides are degraded by β-glucosidases they release stoichiometric amounts of HCN, which can be detected colorimetrically (e.g. Forslund and Jonsson, 1997). Nitrile glucosides such as rhodiocyanoside A and D are also hydrolyzed by β-glucosidase activity, but do not release HCN. Accordingly, nitrile glucosides are not detected by the colorimetric assay for HCN potential. However, as cyanogenic and nitrile glucosides accumulate to the same relative amounts in all tissues examined (Fig. 2), the HCN potential provides a fast and convenient assay to follow the presence and absence of cyanogenic and nitrile glucosides during plant development and in different tissues (Fig. 3). L. japonicus leaves were found to contain an endogenous β-glucosidase activity that, upon disruption of the leaf tissue by freeze/thawing cycles, was able to hydrolyze linamarin and lotaustralin. Full cleavage of the cyanogenic glucoside content was accomplished by incubation of the frozen/thawed material for 30 min, as evidenced by no increased cyanide release upon external addition of an excess of β-glucosidase in the form of crude cassava linamarase.
Figure 3.
Cyanogenic glucosides in L. japonicus in dependence of plant age and tissue expressed as μmol HCN per plant in young plants (A), as μmol HCN per plant during the entire growth cycle (B), as nmol HCN per milligram FW (C), and as nmol HCN per milligram FW in different tissue of 53-d-old plant (D). The error bars represent sd from three replicates at each time point.
Cyanide potential was detectable in L. japonicus seedlings 2 d after germination (Fig. 3, A–C). The cyanide potential per shoot increased with plant age and size (Fig. 3, A and B), demonstrating that cyanogenic glucosides are de novo synthesized throughout the life cycle of the plant. The cyanide potential increased rapidly in the developing seedling per gram fresh weight (FW) and reached the highest level, 10 nmol HCN (mg FW)−1, 4 d after germination (Fig. 3C). Thereafter, cyanide potential per milligram FW declined with plant age. The peaks in cyanide potential at day 4 and day 11 (Fig. 3, A and C) correspond to the time points where the cotyledons are developed and the first leaf becomes visible, and when the first leaf is fully developed and the second leaf is visible, respectively. Cyanide potential was never detected in seeds or in roots (Fig. 3D).
Biosynthesis of Cyanogenic Glucosides and Nitrile Glucosides
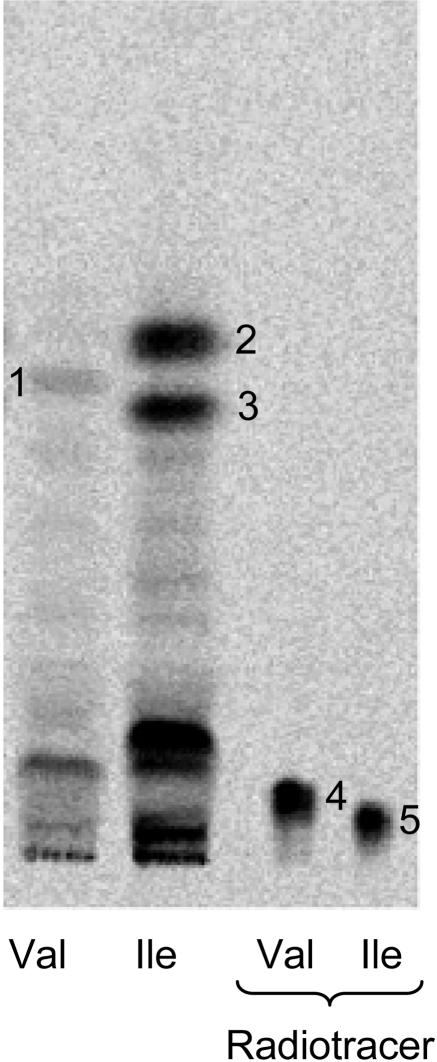
In vivo synthesis of linamarin from l-Val and of lotaustralin and rhodiocyanoside A as well as D from l-Ile was shown by administration of radiolabeled amino acids to detached apical leaves followed by analysis of the products by radio thin-layer chromatography (TLC; Fig. 4). The radiolabeled products were subsequently eluted from the TLC plate and identified by LC-MS as linamarin (1), lotaustralin (2), and rhodiocyanoside A and D (3). Rhodiocyanoside A and D comigrated in the solvent TLC system applied, but could be separated by LC-MS (see below, Fig. 9). In general, the total amount of l-[14C]Val taken up by the leaflet was considerably lower compared to the total amount of l-[14C]Ile taken up. The low uptake of l-Val in comparison to l-Ile is accompanied by a relatively low conversion of absorbed tracer into linamarin in comparison to the conversion of l-Ile into lotaustralin and rhodiocyanoside A and D. The remaining radiolabeled bands reflect l-Val and l-Ile metabolism, not related to biosynthesis of cyanogenic glucosides or nitrile glucosides.
Figure 4.
Biosynthesis of cyanogenic glucosides and nitrile glucosides in L. japonicus monitored by radio TLC after administration of radiolabeled l-[U-14C]Val and l-[U-14C]Ile to detached apical leaves. After incubation overnight, metabolites were extracted with boiling methanol and analyzed by TLC. The identity of the radiolabeled products 1, 2, and 3 as confirmed by LS-MS-SIM analysis of the eluted bands. 1, linamarin; 2, lotaustralin; 3, rhodiocyanoside A/D; 4, l-[U-14C]Val tracer; and 5, l-[U-14C]Ile tracer. The radiolabeled components that are not identified by a number represent general metabolites of l-Val and l-Ile in L. japonicus apical leaves.
Figure 9.
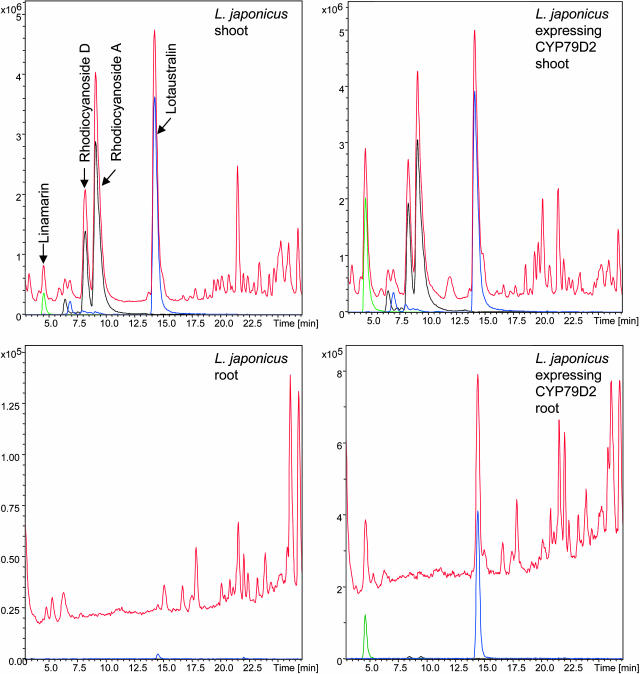
Analysis by LC-MS-SIM of cyanogenic glucosides and nitrile glucosides in shoots and roots of wild-type L. japonicus and a transgenic line expressing cassava CYP79D2 (35S::CYP79D2#5). Total ion chromatogram and extracted ion chromatogram (EIC) of specific [M+ Na]+ adducts are presented. Linamarin (EIC, 270 [green], rt = 4.5 min), rhodiocyanoside D (EIC 282 [black] rt = 8.5 min), rhodiocyanoside A (EIC, 282 [black] rt = 9.5 min), and lotaustralin (EIC 284, [blue] rt = 14.3 min). To visualize the minute amounts of cyanogenic glucosides and nitrile glucosides in roots, different scales are applied for shoots and roots.
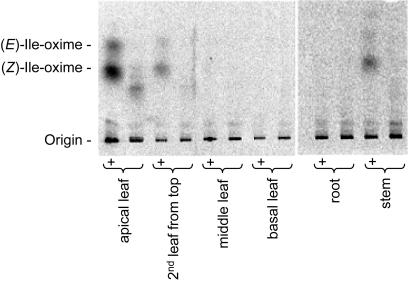
According to the general scheme for synthesis of cyanogenic glucosides, l-Ile and l-Val are converted to the corresponding aldoximes by a microsomal cytochrome P450 enzyme belonging to the CYP79 family. The aldoximes are subsequently converted by a second microsomal cytochrome P450 to the corresponding cyanohydrins that are finally glucosylated by a soluble UDPG-glucosyltransferase (Fig. 1). These putative cytochrome P450 activities were studied using isolated microsomes. The microsomal production of radiolabeled (E) and (Z)-2-methylbutanal oxime (Ile-ox) and (E) and (Z)-2-methylpropanal oxime (Val-ox) from l-[14C]Ile and l-[14C]Val reflects the catalytic activity of the first cytochrome P450 and was dependent on NADPH as expected for a cytochrome P450 catalyzed reaction. Results obtained using l-[14C]Ile as substrate are shown in (Fig. 5). Biosynthesis of cyanogenic glucosides is highly channeled. To facilitate detection of the radiolabeled aldoximes formed, an excess of either unlabeled Ile-ox or Val-ox was added to the reaction mixture as a trap. The aldoxime producing activity of the microsomes was highest in apical leaves and no activity was detected in roots and older leaves throughout the plant development (Fig. 5). The microsomal enzyme system showed a strong preference for l-Ile as a substrate but also accepted l-Val as a substrate (Fig. 6A). Metabolism of l-Leu, l-Phe, or l-Tyr to the corresponding oximes was not detectable in consistence with the absence of cyanogenic glucosides derived from these amino acids (data not shown). The ability of the microsomal enzyme system to metabolize oximes was analyzed by measurements of HCN release from the spontaneous decomposition of the cyanohydrins formed by the action of CYP71E1 ortholog(s) present in the microsomes. As opposed to the l-Val and l-Ile metabolizing system, the putative aldoxime metabolizing CYP71E1 ortholog(s) showed overall less substrate specificity. Thus, all four aldoximes tested were metabolized, although Ile-ox and Val-ox were preferred over Phe-ox and Tyr-ox (Fig. 6B).
Figure 5.
The ability of different tissues of L. japonicus to metabolizeL-[U-14C]Ile measured using microsomes isolated from 53-d-old plants and monitored by radio TLC. The data presented are based on the use of equivalent amounts of plant material from the individual tissues for preparation of microsomes. In lanes marked with +, an excess of unlabeled aldoxime was added as a trap to prevent further metabolism of enzymatically formed radiolabeled oxime by CYP71E1 orthologs present in the microsomal enzyme system. To enable visualization of the low oxime metabolizing activity in stems, the TLC to the right has been developed with a higher sensitivity than the TLC to the left.
Figure 6.
Substrate specificity of L. japonicus microsomes with respect to metabolism of amino acids (section A) and oximes (section B). Microsomes isolated from 3-d-old seedlings were used for the experiments. Radiolabeled l-Val and l-Ile were administered as indicated (section A). After incubation, the reaction mixtures were extracted with ethyl acetate, and analyzed by TLC. NADPH and an excess of unlabeled oximes were added to each reaction. Metabolism of oximes (section B) was quantified as HCN release (see “Material and Methods”). Aliquots (15 μL) of microsomes from the same preparation were used for all experiments.
L. japonicus Contains Two CYP79 Genes
To identify the l-Val and l-Ile metabolizing cytochrome P450 in L. japonicus, a bioinformatics approach was undertaken. In silico homology searches of L. japonicus expressed sequence tags (ESTs; http://www.kazusa.or.jp/en/plant/lotus/EST/) using sorghum CYP79A1, cassava CYP79D1, and CYP79D2 identified several overlapping ESTs of which AV406380 turned out to be full length. A genomic L. japonicus library was subsequently screened by PCR and two new CYP79D subfamily members were identified that both mapped to the short arm of chromosome 3. The two L. japonicus CYP79s were designated CYP79D3 and CYP79D4 by the P450 nomenclature committee (http://drnelson.utmem.edu/CytochromeP450.html). CYP79D3 and CYP79D4 share 94% sequence identity at the amino acid level, whereas the promoter regions showed very low degree of sequence identity. The position of the intron in CYP79D3 and CYP79D4 was identical and corresponded to the highly conserved phase 0 intron found in most A-type P450s (Paquette et al., 2000; Werck-Reichhart et al., 2002). The intron length was 1,986 bp for CYP79D3 and 489 bp for CYP79D4. CYP79D3 and CYP79D4 showed approximately 55% sequence identity to cassava CYP79D1 and CYP79D2 at the amino acid level. Whereas several ESTs were identified for CYP79D3 in the public databases, none could be identified for CYP79D4.
CYP79D3 and CYP79D4 Are Differentially Expressed
To determine if the differences in biosynthetic activity and accumulation of cyanogenic glucosides and nitrile glucosides in different tissues of L. japonicus were due to different mRNA levels of the biosynthetic enzymes, the amounts of CYP79D3 and CYP79D4 transcript were estimated by reverse transcription (RT)-PCR (Burleigh, 2001). Amplification of actin cDNA was used to ensure that an equal amount of cDNA was used in each RT-PCR reaction. Amplification of CYP98 cDNA was used as a positive control, since it is involved in lignification and is known to be expressed in all tissues (Schoch et al., 2001). RT-PCR was performed on mRNA isolated from various tissues from 21-d-old plants. CYP79D3 and CYP79D4 are nearly identical sequences, and the two primer sets work equally well on genomic DNA (data not shown). Of the tissues analyzed, the amount of CYP79D3 transcript was highest in the apical leaves and was also detected in the second leaf from the top and in stem tissue (Fig. 7). No CYP79D3 transcript was detected in older leaves and roots. Conversely, CYP79D4 transcripts were exclusively detected in root tissue. The control amplification of CYP98 cDNA shows a lower expression in the developing leaves as compared to the other tissues, in agreement with an involvement in lignin biosynthesis (Schoch et al., 2001).
Figure 7.
Gene expression of CYP79D3 and CYP79D4 in different tissues of 21-d-old seedlings. mRNA was isolated from different tissues and quantified by RT-PCR with specific primers against CYP79D3, CYP79D4, CYP98, and actin.
Characterization of Recombinant CYP79D3 and CYP79D4
CYP79D3 and CYP79D4 were functionally expressed in a yeast (Saccharomyces cerevisiae) strain system that enables coexpression of the Arabidopsis P450 reductase, ATR1, to facilitate delivery of electrons from NADPH (Pompon et al., 1996). Yeast microsomes containing either recombinant CYP79D3 or CYP79D4 metabolized both l-Ile and l-Val to the corresponding aldoxime (Table I). No aldoximes were detected when l-Tyr, l-Phe, l-Leu, l-Trp, l-Met, and l-Pro were used as substrates in agreement with the data obtained with microsomes isolated from L. japonicus (data not shown). For both CYP79D3 and CYP79D4, a higher activity was observed with l-Ile as substrate compared with l-Val. These data are in accordance with the results obtained with L. japonicus microsomes. The Km and Kcat values for both enzymes favor metabolism of l-Ile rather than of l-Val with the catalytic efficiency (Kcat/Km) being 6-fold higher with l-Ile than with l-Val as substrate (Table I).
Table I.
Catalytic parameters of recombinant L. japonicus CYP79D3 and CYP79D4 compared to cassava CYP79D1
| Enzyme | Km (Ile) | Km (Val) | Kcat (Ile) | Kcat (Val) | Kcat/Km (Ile) | Kcat/Km (Val) |
|---|---|---|---|---|---|---|
| mm | mm | min−1 | min−1 | min−1 mm−1 | min−1 mm−1 | |
| CYP79D3 | 1.8 ± 0.6 | 2.6 ± 0.5 | 220 ± 50 | 50 ± 6 | 122 | 19 |
| CYP79D4 | 0.70 ± 0.8 | 1.7 ± 0.4 | 120 ± 6 | 44 ± 5 | 171 | 26 |
| CYP79D1a | 1.3–1.7 | 1.2–2.2 | 2.3–6.4 | 4.3–9.7 | 1.3–5.0 | 0.6–8.0 |
Metabolic Engineering of L. japonicus
To study whether the ratio between linamarin and lotaustralin is determined by the availability of the amino acid precursors or the catalytic efficiencies of the enzyme systems, the cassava ortholog CYP79D2 was ectopically expressed in L. japonicus. Twenty-two individual transformants were selected and the ratio between linamarin and lotaustralin was determined by LC-MS-SIM (Fig. 8). Of the 22 lines tested, the line designated 35S::CYP79D2#5 was selected for further analysis as it contained a single insert as monitored by segregation analysis, and because it contained the highest ratio of linamarin to lotaustralin content. The line appeared morphometrically indistinguishable from wild type. In line 35S::CYP79D2#5, the cyanide potential was 16 nmol cyanide/FW (mg)−1, approximately twice as high as in wild-type plants. While the linamarin content was increased approximately20-fold, the lotaustralin content was only slightly increased. Accordingly, the increase in cyanide potential is mainly the consequence of increased linamarin content. The ratio of rhodiocyanoside A and D to lotaustralin was unaltered in leaves (Fig. 9). In roots expressing cassava CYP79D2, linamarin and lotaustralin could be detected although in much smaller quantities than in green tissue (Fig. 9). In wild-type roots, the level of lotaustralin and especially linamarin was very low and in most cases below detection limit (Fig. 9).
Figure 8.
Linamarin to lotaustralin ratio in 22 independent 35S::CYP79D2 L. japonicus lines, in two wild-type L. japonicus plants, and cassava apical leaves. The transgenic lines are designated D1-D20, #5 and #6. Cyanogenic glucosides were extracted with methanol from apical leaves of primary transformants and analyzed by LC-MS-SIM.
DISCUSSION
Cyanogenic and Nitrile Glucosides in L. japonicus
Most Lotus species are cyanogenic (Gebrehiwot and Beuselinck, 2001). This is thought to reflect the presence of the two cyanogenic glucosides linamarin and lotaustralin that have been shown to be present in Lotus tenuis and Lotus arabicus (Abrol and Conn, 1966; Jones, 1977). In both these species, linamarin content is generally twice as high as the content of lotaustralin (Abrol and Conn, 1966). In line with these previous studies, we find that L. japonicus contains linamarin and lotaustralin. In contrast to Lotus species previously examined, lotaustralin is the main cyanogenic glucoside in L. japonicus, being approximately 10 times higher than the linamarin content (Fig. 2). In addition, to our surprise, L. japonicus was found to contain the two nitrile glucosides rhodiocyanoside A and D (Fig. 2). In aerial parts of L. japonicus, the content of rhodiocyanoside A is similar to the content of lotaustralin, whereas rhodiocyanoside D and linamarin are present in minor amounts (Fig. 2). Because rhodiocyanoside A and D are not glucosides of α-hydroxynitriles, these compounds do not give rise to release of hydrogen cyanide upon degradation with β-glucosidase and would escape direct detection in survey screens based on cyanide release. LC-MS-SIM analyses of methanol extracts of Lotus corniculatus species did not reveal presence of rhodiocyanosides (Zagrobelny et al., 2004). Accordingly, L. japonicus is the first Lotus species found to produce rhodiocyanosides. Rhodiocyanoside A and D were first isolated and their structures elucidated from underground parts of Rhodiola quadrifida (Pall.) Fisch. et Mey and from Rhodiola sacra (Prain ex Hamet) S. H. Fu. (Yoshikawa et al., 1995, 1997), respectively. In a third Rhodiola species, rhodiocyanoside A and lotaustralin were shown to cooccur (Yoshikawa et al., 1996). Rhodiocyanoside A and D as well as lotaustralin are known in Chinese natural medicine as antiallergic constituents that exhibit inhibitory activity on histamine release (Yoshikawa et al., 1995, 1996, 1997). The biological role of rhodiocyanoside A and D in plants is not known.
The total content of lotaustralin and linamarin in L. japonicus as measured per plant increases during plant growth and development (Fig. 3). The highest cyanide potential per gram FW is reached at day four when the cotyledons are fully developed and the first true leaves start to emerge (Fig. 3, A and B). During the entire life cycle of L. japonicus, the cyanide potential is highest in newly formed tissues, e.g. cotyledons and primary leaves at the seedling stage and apical leaves and flowers at later developmental stages. No cyanide potential was detected in roots or in dry seeds at any time during development. On few occasions, e.g. as shown in Figure 9, very low levels of lotaustralin were detectable in roots of L. japonicus by LC-MS-SIM analysis, which is much more sensitive than measurements of cyanide potential. High amounts of cyanogenic glucosides in actively growing tissues and absence or presence of only minute amounts of cyanogenic glucosides in roots and seeds has earlier been reported in other Lotus species (Gebrehiwot and Beuselinck, 2001) as well as in sorghum and barley (Erb et al., 1981; Halkier et al., 1988; Halkier and Møller, 1989; Forslund and Jonsson, 1997).
The overall tissue distribution of lotaustralin and linamarin (Fig. 3) in comparison to the microsomal enzyme activity (Fig. 5) shows that young growing tissues like seedlings, apical leaves, and stems are major sites of synthesis and storage. The presence of only tiny amounts of cyanogenic glucosides and the apparent lack of rhodiocyanosides A and D in roots indicate that in L. japonicus, neither cyanogenic glucosides nor nitrile glucosides are transported over long distances within the plant. Diglucosides have been proposed as long distance transport forms of cyanogenic glucosides in Hevea braziliensis (Selmar et al., 1988). In L. japonicus, the diglucosides linustatin and neolinustatin were not detectable by LC-MS-SIM. L. japonicus thus differs from cassava, where the diglucosides linustatin and neolinustatin were detectable (Lykkesfeldt and Møller, 1994) and where high amounts of linamarin and lotaustralin accumulate in tubers although the majority of the cyanogenic glucosides are synthesized in the aerial parts of the plant (Koch et al., 1992; Du et al., 1995).
Biosynthesis of Cyanogenic and Nitrile Glucosides in L. japonicus
The general biosynthetic pathway for cyanogenic glucosides involves two membrane-bound cytochrome P450s and a soluble UDPG-glucosyl transferase (Jones et al., 2000). In this study, we identified CYP79D3 and CYP79D4 as involved in cyanogenic glucoside synthesis in L. japonicus. CYP79D3 is exclusively expressed in aerial parts of L. japonicus and CYP79D4 in the roots (Fig. 7). CYP79D3 and CYP79D4 have essentially the same catalytic parameters (Table I), but compared to the expression level of CYP79D3 in apical parts, the expression level of CYP79D4 in roots appears to be lower. Overall, the expression level of CYP79D3 correlates well with the biosynthetic activity (Figs. 5 and 7), indicating that the activity of CYP79D3 is regulated at the transcriptional level. In the monocotyledonous plant sorghum, transcriptional regulation of CYP79s has been studied and it was shown that mRNA levels correlate both with biosynthetic activity and with cyanogenic glucoside content (Busk and Møller, 2002).
Microsomal preparations obtained from 3-d-old seedlings of L. japonicus as well as recombinantly expressed CYP79D3 and CYP79D4 were found to exclusively metabolize l-Ile and l-Val, the two expected precursors for lotaustralin and linamarin. This result is in agreement with previous observations that the CYP79 enzyme(s) defines the substrate specificity of the entire pathway (Koch et al., 1992; Kahn et al., 1999; Andersen et al., 2000; Busk and Møller, 2002). CYP79D3 and CYP79D4 both catalyze the conversion of l-Ile into the corresponding oxime with a catalytic efficiency (Kcat/Km) 6 times greater than for l-Val. Most likely this explains why lotaustralin is the predominant cyanogenic glucoside in L. japonicus, although differences in turnover rates and differences in availability of amino acid precursors may also be important factors. However, the transgenic lines ectopically expressing cassava CYP79D2 accumulate up to 20-fold more linamarin than wild-type plants. This implies that the ratio of linamarin to lotaustralin is mainly determined by the catalytic properties of CYP79D3 and CYP79D4 and not by free amino acid availability.
The biosynthetic pathway for rhodiocyanoside A and D has to our knowledge not previously been studied. We demonstrate using radiolabeling that rhodiocyanoside A and D are derived from Ile (Fig. 4). The relative content of lotaustralin, rhodiocyanoside A, and rhodiocyanoside D remains nearly constant in aerial tissues (Fig. 2). This suggests that lotaustralin and the two rhodiocyanosides are derived from the same pool of Ile-ox (Fig. 1).
Catalytic Properties of CYP79 Determine the Profile of Cyanogenic Glucosides in Planta
CYP79D4 is specifically expressed in root tissue and has a lower Km value for l-Ile and l-Val when compared to the corresponding Km values of CYP79D3 present in the aerial parts of the plant. The relative lower Km values of CYP79D4 compared to CYP79D3 for l-Ile and l-Val do not correlate with a higher level of lotaustralin and linamarin in the root. Accordingly, the low levels of linamarin and lotaustralin in roots most likely reflect a relatively lower expression level of CYP79D4 (Fig. 7) or lower levels of the substrates l-Val and l-Ile. Another possibility is the presence of CYP79 inhibitors in roots. This possibility was ruled out by demonstrating that CYP79D2 from cassava is active in L. japonicus roots (Fig. 9). The high Km value of both CYP79D3 and CYP79D4 for l-Val compared to l-Ile may reflect the low abundance of linamarin in comparison to lotaustralin in all tissue examined.
Cassava is allotetraploid and contains two CYP79 copies, CYP79D1 and CYP79D2, that both metabolize l-Val and l-Ile (Andersen et al., 2000). The catalytic properties of CYP79D1 have been elucidated using recombinant CYP79D1 expressed in Pichia pastoris. CYP79D1 has a Km of 1.3 to 1.7 mm for l-Ile and a Km of 1.2 to 2.2 mm for l-Val (Andersen et al., 2000), which is in the same range as for CYP79D3 and CYP79D4 in L. japonicus (Table I). The rate of aldoxime formation in the cassava study was determined using a TLC assay (Andersen et al., 2000), which inevitably leads to evaporation of the radiolabeled aldoximes despite administration of a large excess of unlabeled carrier aldoximes. Therefore, the Kcat values determined for the cassava enzyme cannot be directly compared to those in our study, where the TLC assay was replaced by a more accurate assay based on phase separation followed by liquid scintillation counting, to eliminate the risk of aldoxime loss due to evaporation. The catalytic efficiencies (Kcat/Km) for the cassava enzyme CYP79D1 for l-Ile and l-Val were determined as 4.8 min−1 μm−1 and 4.4 min−1 μm−1, respectively. As Val-ox is more volatile than Ile-ox, the catalytic efficiencies toward especially l-Val may in fact have been underestimated. This is in contrast to the catalytic efficiency of the CYP79 orthologs in L. japonicus where l-Ile is a 6-times better substrate than l-Val.
To study the impact of free amino acid availability versus the catalytic properties of the CYP79s on the type and amounts of cyanogenic glucosides that accumulate in L. japonicus, the CYP79D2 ortholog from cassava was ectopically expressed in L. japonicus. The transgenic plants showed up to 20-fold increase in linamarin content in shoots and a small increase in lotaustralin content (Figs. 8 and 9). Wild-type L. japonicus accumulate approximately 10 times more lotaustralin than linamarin (Fig. 2), while in the highest expresser the lotaustralin level is only approximately 1.5-fold higher than the linamarin level (Fig. 8). The observed change in linamarin to lotaustralin ratio is in accordance with the reported catalytic properties of the cassava ortholog CYP79D2 (Andersen et al., 2000; Table I) in that the catalytic efficiencies of the cassava orthologs toward l-Val and l-Ile are similar. Accordingly, the ratio of linamarin to lotaustralin in the best transgenic lines approaches the ratio found in cassava leaves (Fig. 8). The capability to increase the level of linamarin 20-fold in L. japonicus demonstrates that the level of free l-Val is not the limiting factor, and that the activities of the postoxime enzymes in the pathway easily can accommodate an increased flux of intermediates. Furthermore, the expression of cassava CYP79D2 in roots significantly increases the level of linamarin as well as lotaustralin, and demonstrates that L. japonicus roots have the capacity to synthesize linamarin and lotaustralin, but not rhodiocyanoside A and D (Fig. 9). This substantiates that the low levels of linamarin and lotaustralin in roots are not due to the presence of inhibitors, but are a reflection of substrate availability, catalytic properties of the enzyme system, and expression levels.
Ectopic expression of CYP79s has been used to alter the profiles of glucosinolates (Mikkelsen et al., 2002). The ability to modify the glucosinolate profile by ectopic expression of CYP79s from cyanogenic plants relies on the ability of CYP79s to create a metabolic cross talk with the postoxime biosynthetic enzymes in the glucosinolate pathway (Bak et al., 1999; Mikkelsen et al., 2002). Among the 22 transgenic L. japonicus plants obtained that express CYP79D2 from cassava, the highest expresser 35S::CYP79D2#5 accumulates up to 2.8% dry weight as cyanogenic glucosides. In comparison, ectopic expression of CYP79D2 in Arabidopsis resulted in the accumulation of up to 8.2 nmol of Val and Ile derived glucosinolates per milligram dry weight corresponding to approximately 0.3% dry weight (Mikkelsen and Halkier, 2003). When CYP79A1 from sorghum was ectopically expressed in Arabidopsis using the same 35S promoter, l-Tyr derived glucosinolates accumulated up to 52 nmol per milligram dry weight corresponding to approximately 2.3% dry weight (Bak et al., 1999; Petersen et al., 2001; Mikkelsen et al., 2002). In comparison, transgenic Arabidopsis expressing the entire biosynthetic pathway for the Tyr derived cyanogenic glucoside dhurrin driven by 35S promoters accumulate up to 4% dhurrin (Tattersall et al., 2001). An explanation for the differences observed in accumulation of novel glucosinolates or cyanogenic glucosides, could be due to insertional effects of the transgenes. However, as only the best producing lines out of between 10 and 35 in the mentioned studies are compared, position effects are likely be minimal. A more likely explanation for the different product levels observed in transgenic plants expressing CYP79A1 and CYP79D2 could be differences in the catalytic efficiencies of CYP79A1 versus CYP79D2. The catalytic efficiency for the substrate Tyr is approximately 9.5 min−1 mm−1 for CYP79A1 (Km: 0.21 mm; Kcat: 200 min−1; Sibbesen et al., 1995). Although the reported values for the catalytic efficiency of CYP79D1 (Table I; Andersen et al., 2000) may not be accurate due to the assay employed in that study, it is clear that the Km for CYP79A1 is lower than for CYP79D1. This implies that CYP79A1 is a more effective enzyme than CYP79D2. Another important factor in plants may be the availability of substrates. In Arabidopsis and L. japonicus leaves, Ile, Val, and Tyr accumulate to relatively low levels in comparison to other amino acids (our unpublished data). The observed higher accumulation of Tyr derived products may also reflect that Tyr is derived from the shikimate pathway, a pathway that accommodates up to 20% of the flux of carbon fixed in plants, and which primarily is regulated by feed-back inhibition to facilitate adjustment to metabolic alterations.
Accordingly, our results substantiate the conclusion that the substrate specificity and efficiency of the CYP79 catalyzed step exerts quantitative and qualitative control of the flux through the pathway.
Two Pathways Are Operating in L. japonicus
Ectopic expression of CYP79D2 in L. japonicus resulted in up to 20-fold increase in linamarin in shoots, whereas the relative amounts of lotaustralin, rhodiocyanoside A, and rhodiocyanoside D were almost unaltered (Fig. 9). Low amounts of linamarin and lotaustralin were also detectable in transgenic roots, but were accompanied by minute amounts of rhodiocyanosides A and D (Fig. 9). The most obvious explanation for this is the operation of two independent pathways in aerial parts of the plant and in roots. In favor of this hypothesis is the existence of two differentially expressed CYP79D paralogs, CYP79D3 and CYP79D4, of which CYP79D4 is specifically expressed in roots and CYP79D3 in the aerial parts (Fig. 7). The operation of two independent pathways would imply the existence of at least two CYP71E1 orthologs in L. japonicus. The CYP71E1 ortholog expressed in aerial parts of the plants would then be able to convert Val-ox and Ile-ox into the aglycones of linamarin and those of lotaustralin and rhodiocyanosides A and D, respectively. The CYP71E1 ortholog expressed in roots would primarily convert Val-ox and Ile-ox into the aglycons of linamarin and lotaustralin, respectively, i.e. primarily catalyze hydroxylation of the α-carbon of the nitrile intermediates (Fig. 1). The three enzymes CYP79A1, CYP71E1, and UGT85B1 that catalyze dhurrin synthesis in sorghum have recently been shown to form a metabolon (K.A. Nielsen, D.B. Tattersall, P.R. Jones, and B.L. Møller, personal communication). We propose, as observed in sorghum, that the two independent pathways in aerial parts and roots are organized into two separate metabolons to facilitate a high degree of channeling to avoid liberation of reactive and unstable intermediates into the cytosol.
Previously, the entire pathway for synthesis of the cyanogenic glucoside dhurrin has been introduced into Arabidopsis by genetic engineering (Tattersall et al., 2001). Arabidopsis does not naturally produce cyanogenic glucosides but glucosinolates. Because the biosynthetic pathways for cyanogenic glucosides and glucosinolates share aldoxime as common intermediates, introduction of CYP79A1 into Arabidopsis results in simultaneous accumulation of the Tyr-derived glucosinolate p-hydroxybenzylglucosinolate (Bak et al., 1999, 2000). In addition, the endogenous β-glucosidase activity toward cyanogenic glucosides is insignificant in Arabidopsis, rendering cyanide release from the transgenic plants very slow (Tattersall et al., 2001). Such experimental difficulties are not encountered when using L. japonicus as a model system to study cyanogenic glucosides in higher plants. We have characterized the first committed step in biosynthesis of cyanogenic glucosides and nitrile glucosides in L. japonicus. By ectopic expression of an orthologous cassava CYP79, the feasibility of altering the ratio of linamarin to lotaustralin in L. japonicus has been documented. The ability to metabolically engineer L. japonicus paves the way for production of novel cyanogenic glucosides in L. japonicus as well as for generation of acyanogenic L. japonicus plants. Such transgenic plants will be used to study the function of cyanogenic glucosides and nitrile glucosides such as rhodiocyanoside A and D as defense compounds and in other environmental interactions. The biological reason for the apparent existence of two pathways and the apparent lack of rhodiocyanoside A and D in roots in L. japonicus is currently not understood. The use of transgenic L. japonicus plants with altered cyanogenic glucoside and nitrile glucoside profiles will hopefully help to clarify these issues.
MATERIALS AND METHODS
Culture of Plants
Seeds from Lotus japonicus GIFU B-129-S9 were scarified by a short treatment with sandpaper and surface sterilized in 2% hypochlorite with 0.02% Tween 20 (shaking, 20 min). The seeds were washed in 6 shifts of autoclaved water and germinated for 1 week on water saturated filter paper (20°C, photosynthetic flux of 100–120 μmol photons m−2 s−1, and a light dark regime of 16/8 h). All plant material was grown hydroponically with the seedlings mounted with foam in Eppendorf tubes from which the bottom had been cut off. The Eppendorf tubes were then mounted in the lid of a 4-L high density polyethylene container filled with aerated nutrient solution (Husted et al., 2002), and placed in a greenhouse at 24°C fitted with extra light bulbs ensuring a minimum photosynthetic flux of 100 to 120 μmol photons m−2 s−1 and a 16/8 light dark regime. The nutrient solution was changed once per week. Plant tissue used for the different analyses was harvested into liquid nitrogen and stored at −80° until use.
Liquid Chromatography-Mass Spectrometry
To determine the content of individual cyanogenic glucosides as well as nitrile glucosides, L. japonicus plant material was extracted in 85% (v/v) boiling methanol. The solvent was evaporated by lyophilization and the residue dissolved in water and extracted three times with n-pentane. Aliquots of the aqueous phase were subjected to LC-MS-SIM analysis using an HP1100 LC coupled to a Bruker Esquire-LC ion trap mass spectrometer (Bruker Instruments, Billerica, MA). A Waters Xterra MS C18 column (Waters, Milford, MA) was used and the LC conditions were as described in (Nielsen et al., 2002). The mass spectrometer was run in positive ion mode. The results were analyzed by Bruker Daltonics DataAnalysis version 3.0. Retention time for cyanogenic and nitrile glucosides: linamarin, 4.5 min; rhodiocyanoside D, 8.5 min; rhodiocyanoside A, 9.5 min; and lotaustralin, 14.3 min. Linamarin was verified based on identical mass, retention times, and fragmentation pattern compared to an authentic standard. Lotaustralin and rhodiocyanoside A and D were isolated by preparative LC and their identity verified by NMR-spectrometry.
Cyanide Determination
To determine the total content of linamarin and lotaustralin, plant material was harvested into liquid nitrogen, homogenized, and extracted (10 min) in 85% (v/v) boiling methanol. After removal of solvent by evaporation and phase separation with sterile water/n-pentane (1:10, v/v), the water phase was collected. Assay mixtures (total volume: 200 μL) contained aliquots (1–10 μL) of the water phase, crude cassava (Manihot esculenta Crantz.) linamarase extract, (as a source of β-glucosidase; 10 μL), and 50 mm Tricine (pH 7.9). After incubation (1 h, 30°C) the cyanide content was quantified colorimetrically (triplicates; Forslund and Jonsson, 1997). To monitor the presence of endogenous β-glucosidase activity, a second cyanide determination method was used in which L. japonicus plant material in 50 mm Tricine (pH 7.9) was taken through three freeze-thaw cycles and incubated (1 h, 30°C) followed by quantification of the cyanide released as described above.
Biosynthetic Activity of L. japonicus Microsomes
Microsomes were prepared from 3-d-old L. japonicus seedlings (0.1–0.5 g) as described for barley (Nielsen et al., 2002). Biosynthetic activity of the microsomes was determined in assay mixtures (total volume: 25 μL) containing 15 μL of microsomes, 0.5 μCi (300 mCi mmol−1) l-[ U-14C]amino acids (l-Val, l-Ile, l-Leu, l-Tyr, or l-Phe; Amersham Biosciences, Uppsala), 1 mm NADPH, and 3.3 mm Ile-ox or Val-ox in 50 mm Tricine (pH 7.9). After incubation (30°C, 30 min) and extraction with ethyl acetate (2 volumes), the ethyl acetate phase was applied to a TLC sheet (Silica Gel 60 F254 sheets, Merck, Rahway, NJ) that was developed in dichloromethane/ethyl acetate (85:15 v/v). Radiolabeled products were visualized using a STORM 840 phosphor imager (Molecular Dynamics, Sunnyvale, CA). The ability of L. japonicus microsomes to metabolize oximes derived from l-Val, l-Ile, l-Tyr, or l-Phe was determined in assay mixtures (total volume: 200 μL) containing 15 μL microsomes, 10 mm substrate, and 1 mm NADPH in 50 mm Tricine (pH 7.9). After incubation (30°C, 30 min), the assay mixtures were frozen in liquid nitrogen, and 40 μL of 6 m NaOH added. Cyanide content was determined colorimetrically (Forslund and Jonsson, 1997).
l-Val and l-Ile as Glucoside Precursors in L. japonicus
Detached leaflets of 2-week-old L. japonicus seedlings were mounted in Eppendorf tubes containing 5 μL l-[U-14C]Ile or l-[U-14C]Val (0.5 μCi, 300 mCi mmol−1). Water (45 μL) was administrated to the leaves after the tracer had been taken up. After incubation for 24 h in closed Eppendorf tubes, the leaves were extracted with 85% (v/v) boiling methanol (5 min). The extracts were applied to TLC plates (Silica Gel 60 F254, Merck), and developed in ethyl acetate/acetone/chloroform/methanol/water (40:30:12:10:8, v/v/v/v/v). The presence of radiolabeled products was monitored as described above.
Isolation and Expression of CYP79D3 and CYP79D4 in Yeast
PCR fragments corresponding to the open reading frame of CYP79D3 and CYP79D4 were isolated by a PCR approach, using the EST AV 406380 as template for CYP79D3 and the BAC clone BMO 53279 from Kazusa DNA Research Institute as template for CYP79D4. The two CYP79D4 exons were combined by overlap extension PCR. BamHI/KpnI and BamHI/SacI sites were introduced by PCR and used to ligate CYP79D3 and CYP79D4, respectively, into the pYeDP60 vector (Pompon et al., 1996). PCR (total volume: 50 μL) were performed in Pfu polymerase PCR buffer including 200 μm dNTPs, 1.5 mm MgCl2, 10 pmol of each primer, and 1.25 units of Pfu polymerase (Stratagene). The PCR program was as follows: 96°C for 3 min followed by 5 cycles (94°C for 30 s, 50°C for 30 s, and 72°C for 2.5 min), 25 cycles (94°C for 30 s, 60°C for 30 s, and 72°C for 2.5 min), and a final cycle of 72°C for 5 min. The expression constructs were verified by sequencing and introduced into yeast (Saccharomyces cerevisiae) WAT11 cells (Pompon et al., 1996) by the LiCl method (Schiestl and Gietz, 1989). Yeast microsomes were isolated (Pompon et al., 1996), and total P450 content was measured spectrophotometrically (Omura and Sato, 1964).
Analysis of Recombinant CYP79D3 and CYP79D4
To determine the substrate specificity of CYP79D3 and CYP79D4, yeast microsomes (10 pmol CYP79) harboring recombinant CYP79D3 or CYP79D4 were incubated (30 min, 28°C, gentle agitation (350 rpm) with l-[U-14C]labeled Val, Ile, Leu, Tyr, Phe, Met, Trp, or Pro, (Amersham Biosciences) in assay mixtures containing NADPH (1 mm), oxime (3.3 mm) and Tricine (10 mm, pH 7.5). Reaction mixtures to which no NADPH was added and reaction mixtures containing microsomes prepared from WAT11 transformed with empty vector served as negative controls. After incubation, reaction mixtures were analyzed as described above for the plant microsomes. Kcat and Km values with l-Ile or l-Val as substrates were determined using assay mixtures (total volume of 30 μL) containing microsomes (6.5 μL; 9.32 pmol of CYP79D3 or 11.35 pmol of CYP79D4), substrate (50 μm to 2 mm), NADPH (1 mm), and Ile-ox (3.3 mm) or Val-ox (3.3 mm) in 50 mm Tricine (pH 7.5). After incubation (5 min, 28°C) reactions were stopped by addition of ethyl acetate (50 μL). The organic phase was collected and its radioactive content determined by liquid scintillation counting. All measurements were carried out in duplicates. Radioactivity detected in the ethyl acetate phase in assay mixtures containing void vector were subtracted as background. Sigma Plot 2001 and EnzymeKinetics 1.10 (SPSS, Chicago) were used to calculate Kcat and Km values.
RT-PCR
Poly(A) RNA was isolated from 21-d-old hydroponically grown plants using Micro Poly-A-Pure (Ambion, Austin, TX) according to the manufacturer's instructions. The poly(A) RNA was treated (30 min, 37°C) with RNase free DNase I (RNasine, Stratagene, La Jolla, CA) followed by phenol-chloroform extraction and precipitation with ethanol to remove traces of contaminating DNA. Poly(A) RNA (0.5 μg) from each tissue was reverse transcribed using SuperScript II reverse transcriptase (Invitrogen, Carlsbad, CA) following the manufacturer's instructions. Random hexamers [200 ng, Oligo dN (pd(N)6); Amersham Biosciences] were used to prime the reaction. Reverse-transcribed cDNA samples were diluted 20 times with water and 1 μL was PCR-amplified. PCR reactions (total volume 50 μL) were performed in PCR buffer (Invitrogen) containing 200 μm dNTPs, 1.5 mm MgCl2, 10 pmol of forward and reverse primers, and 2.5 units of Taq DNA polymerase (Invitrogen). The PCR programs were as follows: 94°C for 2 min followed by 26 cycles (actin homolog, CYP98A3 homolog, and CYP79D3), or 32 cycles (CYP79D4) of: 94°C for 30 s, 60°C for 30 s, and 72°C for 30 s. The number of cycles chosen for these gene specific primer sets was set to enable a semiquantitative comparison between samples. Using sequences in GenBank, primer pairs were designed to amplify transcribed portions of: L. japonicus putative actin homolog (EST no. AU089544) forward: 5′-CTT TTA ATA CCC CCG CTA TGT ATG-3′ and reverse 5′-GGT GGT AAA AGA ATA ACC ACG TTC-3′; CYP79D3 (GenBank accession no. AY599895): forward 5′-ACC AAG ATT GGC TGC AGA ACT C-3′ and reverse 5′-TTC AAC AAC TGC ATG GCT TTA CAA-3′; CYP79D4 (GenBank accession no. AY599896): forward 5′-CGA TCT TAC CCA GTC CAG TGA T-3′ and reverse 5′-CAC ATG CGT GAG TAA CAT CAA AC-3′; L. japonicus CYP98A3 homolog (EST no. AV417871): forward 5′-CTC TCA CCT CCA AAT TCT CCA C-3′ and reverse 5′-CCT TCT CCT TAA GAA CCT CCT TG-3′. The specificity and efficiency of each primer pair was established by amplification from L. japonicus genomic DNA and confirmed by DNA sequencing. PCR products were analyzed by gel electrophoresis on 1% (w/v) agarose gels. Bands were visualized by ethidium bromide staining and quantified using a Gel Doc 2000 Transilluminator (Bio-Rad, Hercules, CA). All experiments were repeated using independent tissue samples. Amplification of actin cDNA was used to control that similar amounts of cDNA from each sample were used in the PCR experiments.
Transgenic L. japonicus Expressing Cassava CYP79D2
The cassava CYP79D2 cDNA containing the 35S promoter and polyadenylation site was excised from pPZP221 cauliflower mosaic virus 35S::CYP79D2 constructs (Mikkelsen and Halkier, 2003) using HindIII and ligated into the corresponding HindIII site in pPZP111 (Hajdukiewicz et al., 1994) to confer resistance to G-418 disulphate, and the final construct was introduced into Agrobacterium tumefaciens by electroporation (Shen and Forde, 1989). L. japonicus was transformed using hypocotyls (Handberg and Stougaard, 1992). The explants were grown at 25°C with light dark regime of 16/8 h. Transformants were selected using the gentamycin analog G-418 (25 mg L−1) and tested for expression of the neomycin phophotransferase NPT II protein using the NPT II ELISA kit (Patho Screen NPTII, Agdia, Elkhart, IN). Independent transgenic lines were selected as evidenced by the expression of NPTII product and segregation analysis on G-418 disulfate. Cuttings from the in vitro grown transformants were grown hydroponically as described above.
Sequence data from this article have been deposited with the EMBL/GenBank data libraries under accession numbers AY599895 and AY599896.
Acknowledgments
Professor Birger Lindberg Møller and Dr. David Tattersall are thanked for helpful discussions. Dr. Michael Dalgaard Mikkelsen is thanked for providing the pPZP221 35S::CYP79D2 construct. Anne Vinther Rasmussen and Mika Zagrobelny are thanked for critically reading the manuscript, Steen Malmmose for taking care of the hydroponically grown L. japonicus plants, and Winnie Dam and Mai-Britt Eicke for skilful technical assistance. Dr. Jens Stougaard provided seeds of L. japonicus and Dr. Kirsten Jørgensen provided crude cassava linamarase.
This work was supported by the Danish National Research Foundation by a grant to the Center for Molecular Plant Physiology (PlaCe), by the Danish Agricultural and Veterinary Research Council (grant no. 23–02–0095), and by the Swedish Research Council for Environment, Agricultural Sciences and Spatial Planning (FORMAS; grant no. 672.0825/0096 to K.F.).
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.103.038059.
References
- Abrol YP, Conn EE (1966) Studies of cyanide metabolism in Lotus arabicus L. and Lotus tenuis L. Phytochemistry 5: 237–242 [Google Scholar]
- Andersen MD, Busk PK, Svendsen I, Møller BL (2000) Cytochromes P-450 from cassava (Manihot esculenta Crantz) catalyzing the first steps in the biosynthesis of the cyanogenic glucosides linamarin and lotaustralin: cloning, functional expression in Pichia pastoris, and substrate specificity of the isolated recombinant enzymes. J Biol Chem 275: 1966–1975 [DOI] [PubMed] [Google Scholar]
- Asamizu E, Nakamura Y, Sato S, Tabata S (2000) Generation of 7137 non-redundant expressed sequence tags from a legume, Lotus japonicus. DNA Res 7: 127–130 [DOI] [PubMed] [Google Scholar]
- Bak S, Kahn RA, Nielsen HL, Moller BL, Halkier BA (1998) Cloning of three A-type cytochromes p450, CYP71E1, CYP98, and CYP99 from Sorghum bicolor (L.) Moench by a PCR approach and identification by expression in Escherichia coli of CYP71E1 as a multifunctional cytochrome p450 in the biosynthesis of the cyanogenic glucoside dhurrin. Plant Mol Biol 36: 393–405 [DOI] [PubMed] [Google Scholar]
- Bak S, Olsen CE, Halkier BA, Møller BL (2000) Transgenic tobacco and Arabidopsis plants expressing the two multifunctional sorghum cytochrome P450 enzymes, CYP79A1 and CYP71E1, are cyanogenic and accumulate metabolites derived from intermediates in dhurrin biosynthesis. Plant Physiol 123: 1437–1448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bak S, Olsen CE, Petersen BL, Møller BL, Halkier BA (1999) Metabolic engineering of p-hydroxybenzylglucosinolate in Arabidopsis by expression of the cyanogenic CYP79A1 from Sorghum bicolor. Plant J 20: 663–671 [DOI] [PubMed] [Google Scholar]
- Burleigh SH (2001) Relative quantitative RT-PCR to study the expression of plant nutrient transporters in arbuscular mycorrhizas. Plant Sci 160: 899–904 [DOI] [PubMed] [Google Scholar]
- Busk PK, Møller BL (2002) Dhurrin synthesis in sorghum is regulated at the transcriptional level and induced by nitrogen fertilization in older plants. Plant Physiol 129: 1222–1231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du LC, Bokanga M, Møller BL, Halkier BA (1995) The biosynthesis of cyanogenic glucosides in roots of cassava. Phytochemistry 39: 323–326 [Google Scholar]
- Erb N, Zinsmeister HD, Nahrstedt A (1981) Die cyanogenen glukoside von triticum, secale und sorghum. Planta Med 41: 84–89 [DOI] [PubMed] [Google Scholar]
- Forslund K, Jonsson L (1997) Cyanogenic glycosides and their metabolic enzymes in barley, in relation to nitrogen levels. Physiol Plant 101: 367–372 [Google Scholar]
- Gebrehiwot L, Beuselinck PR (2001) Seasonal variations in hydrogen cyanide concentration of three Lotus species. Agron J 93: 603–608 [Google Scholar]
- Hajdukiewicz P, Svab Z, Maliga P (1994) The small, versatile pPZP family of Agrobacterium binary vectors for plant transformation. Plant Mol Biol 25: 989–994 [DOI] [PubMed] [Google Scholar]
- Halkier BA, Møller BL (1989) Biosynthesis of the cyanogenic glucoside dhurrin in seedlings of Sorghum bicolor (L.) Moench and partial-purification of the enzyme-system involved. Plant Physiol 90: 1552–1559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halkier BA, Scheller HV, Møller BL (1988) Cyanogenic glucosides: the biosynthetic-pathway and the enzyme-system involved. Ciba F Symp 140: 49–66 [DOI] [PubMed] [Google Scholar]
- Handberg K, Stougaard J (1992) Lotus japonicus, an autogamous, diploid legume species for classical and molecular-genetics. Plant J 2: 487–496 [Google Scholar]
- Husted S, Mattsson M, Møllers C, Wallbraun M, Schjoerring JK (2002) Photorespiratory NH4+ production in leaves of wild-type and glutamine synthetase 2 antisense oilseed rape. Plant Physiol 130: 989–998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones D (1977) Polymorphism of cyanogenesis in Lotus corniculatus L. VII. The distribution of cyanogenic form in western-Europe. Heredity 39: 27–44 [Google Scholar]
- Jones PR, Andersen MD, Nielsen JS, Hoj PB, Moller BL (2000) The biosynthesis, degradation, transport and possible function of cyanogenic glucosides. Recent Adv Phytochem 34: 191–247 [Google Scholar]
- Jones PR, Møller BL, Høj PB (1999) The UDP-glucose: p-hydroxymandelonitrile-O-glucosyltransferase that catalyzes the last step in synthesis of the cyanogenic glucoside dhurrin in Sorghum bicolor: isolation, cloning, heterologous expression, and substrate specificity. J Biol Chem 274: 35483–35491 [DOI] [PubMed] [Google Scholar]
- Kahn RA, Fahrendorf T, Halkier BA, Møller BL (1999) Substrate specificity of the cytochrome P450 enzymes CYP79A1 and CYP71E1 involved in the biosynthesis of the cyanogenic glucoside dhurrin in Sorghum bicolor (L.) Moench. Arch Biochem Biophys 363: 9–18 [DOI] [PubMed] [Google Scholar]
- Koch B, Nielsen VS, Halkier BA, Olsen CE, Møller BL (1992) The biosynthesis of cyanogenic glucosides in seedlings of cassava (Manihot esculenta Crantz). Arch Biochem Biophys 292: 141–150 [DOI] [PubMed] [Google Scholar]
- Koch BM, Sibbesen O, Halkier BA, Svendsen I, Møller BL (1995) The primary sequence of cytochrome P450tyr, the multifunctional N-hydroxylase catalyzing the conversion of L-tyrosine to P-hydroxyphenylacetaldehyde oxime in the biosynthesis of the cyanogenic glucoside dhurrin in Sorghum bicolor (L.) Moench. Arch Biochem Biophys 323: 177–186 [DOI] [PubMed] [Google Scholar]
- Lechtenberg M, Nahrstedt A (1999) Cyanogenic glycosides. In R Ikan, ed, Naturally Occurring Glycosides. John Wiley & Sons, New York
- Lechtenberg M, Nahrstedt A, Fronczek FR (1996) Leucine-derived nitrile glucosides in the rosaceae and their systematic significance. Phytochemistry 41: 779–785 [Google Scholar]
- Lykkesfeldt J, Møller BL (1994) Cyanogenic glycosides in cassava, Manihot esculenta Crantz. Acta Chem Scand 48: 178–180 [Google Scholar]
- Mikkelsen MD, Halkier BA (2003) Metabolic engineering of valine- and isoleucine-derived glucosinolates in Arabidopsis expressing CYP79D2 from cassava. Plant Physiol 131: 773–779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikkelsen MD, Petersen BL, Olsen CE, Halkier BA (2002) Biosynthesis and metabolic engineering of glucosinolates. Amino Acids 22: 279–295 [DOI] [PubMed] [Google Scholar]
- Møller BL, Seigler DS (1999). In BK Singh, ed, Plant Amino Acids, Biochemistry and Biotechnology. Marcel Dekker, New York, pp 563–609
- Nakamura Y, Kaneko T, Asamizu E, Kato T, Sato S, Tabata S (2002) Structural analysis of a Lotus japonicus genome. II. Sequence features and mapping of sixty-five TAC clones which cover the 6.5-Mb regions of the genome. DNA Res 9: 63–70 [DOI] [PubMed] [Google Scholar]
- Nielsen JS, Møller BL (1999) Biosynthesis of cyanogenic glucosides in Triglochin maritima and the involvement of cytochrome P450 enzymes. Arch Biochem Biophys 368: 121–130 [DOI] [PubMed] [Google Scholar]
- Nielsen KA, Olsen CE, Pontoppidan K, Møller BL (2002) Leucine-derived cyano glucosides in barley. Plant Physiol 129: 1066–1075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omura T, Sato R (1964) The carbon monoxide-binding pigment of liver microsomes. I. Evidence for its hemoprotein nature. J Biol Chem 239: 2370–2378 [PubMed] [Google Scholar]
- Paquette SM, Bak S, Feyereisen R (2000) Intron-exon organization and phylogeny in a large superfamily, the paralogous cytochrome P450 genes of Arabidopsis thaliana. DNA Cell Biol 19: 307–317 [DOI] [PubMed] [Google Scholar]
- Perry JA, Wang TL, Welham TJ, Gardner S, Pike JM, Yoshida S, Parniske M (2003) A TILLING reverse genetics tool and a web-accessible collection of mutants of the legume Lotus japonicus. Plant Physiol 131: 866–871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen BL, Andreasson E, Bak S, Agerbirk N, Halkier BA (2001) Characterization of transgenic Arabidopsis thaliana with metabolically engineered high levels of p-hydroxybenzylglucosinolate. Planta 212: 612–618 [DOI] [PubMed] [Google Scholar]
- Pompon D, Louerat B, Bronine A, Urban P (1996) Yeast expression of animal and plant P450s in optimized redox environments. Methods Enzymol 272: 51–64 [DOI] [PubMed] [Google Scholar]
- Schoch G, Goepfert S, Morant M, Hehn A, Meyer D, Ullmann P, Werck-Reichhart D (2001) CYP98A3 from Arabidopsis thaliana is a 3′-hydroxylase of phenolic esters, a missing link in the phenylpropanoid pathway. J Biol Chem 276: 36566–36574 [DOI] [PubMed] [Google Scholar]
- Seigler D (1991) Cyanide and cyanogenic glucosides. In GA Rosenthal, MR Berenbaum, eds, Herbivores: Their Interaction with Secondary Plant Metabolites. Academic Press, San Diego, pp 35–77
- Seigler DS (1975) Isolation and characterization of naturally occurring cyanogenic compounds. Phytochemistry 14: 9–29 [Google Scholar]
- Selmar D, Lieberei R, Biehl B (1988) Mobilization and utilization of cyanogenic glycosides: the linustatin pathway. Plant Physiol 86: 711–716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiestl RH, Gietz RD (1989) High efficiency transformation of intact yeast cells using single stranded nucleic acids as a carrier. Curr Genet 16: 339–346 [DOI] [PubMed] [Google Scholar]
- Shen WJ, Forde BG (1989) Efficient transformation of Agrobacterium spp. by high voltage electroporation. Nucleic Acids Res 17: 8385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sibbesen O, Koch B, Halkier BA, Møller BL (1994) Isolation of the heme-thiolate enzyme cytochrome P-450 (Tyr), which catalyzes the committed step in the biosynthesis of the cyanogenic glucoside dhurrin in Sorghum bicolor (L.) Moench. Proc Natl Acad Sci USA 91: 9740–9744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sibbesen O, Koch B, Halkier BA, Møller BL (1995) Cytochrome P-450 (Tyr) is a multifunctional heme-thiolate enzyme catalyzing the conversion of L-tyrosine to P-hydroxyphenylacetaldehyde oxime in the biosynthesis of the cyanogenic glucoside dhurrin in Sorghum bicolor (L.) Moench. J Biol Chem 270: 3506–3511 [DOI] [PubMed] [Google Scholar]
- Tattersall DB, Bak S, Jones PR, Olsen CE, Nielsen JK, Hansen ML, Høj PB, Møller BL (2001) Resistance to an herbivore through engineered cyanogenic glucoside synthesis. Science 293: 1826–1828 [DOI] [PubMed] [Google Scholar]
- VandenBosch K, Stacey G (2003) Summaries of legume genomics projects from around the globe. Community resources for crops and models. Plant Physiol 131: 840–865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- VanEtten HD, Mansfield JW, Bailey JA, Farmer EE (1994) Two classed of plant antibiotics: phytoalexins versus phytoanticipins. Plant Cell 6: 1191–1192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werck-Reichhart D, Bak S, Paquette S (2002) Cytochromes P450. In The Arabidopsis Book. American Society of Plant Biologists, Rockville, MD, pp 1–29 [DOI] [PMC free article] [PubMed]
- Yoshikawa M, Shimada H, Horikawa S, Murakami T, Shimoda H, Yamahara J, Matsuda H (1997) Bioactive constituents of Chinese natural medicines. 4. Rhodiolae Radix. 2. On the histamine release inhibitors from the underground part of Rhodiola sacra (Prain ex Hamet) S.H. Fu (Crassulaceae): chemical structures of rhodiocyanoside D and sacranosides A and B. Chem Pharm Bull (Tokyo) 45: 1498–1503 [DOI] [PubMed] [Google Scholar]
- Yoshikawa M, Shimada H, Shimoda H, Matsuda H, Yamahara J, Murakami N (1995) Rhodiocyanoside-A and Rhodiocyanoside-B, new antiallergic cyanoglycosides from Chinese natural medicine Si-Li-Hong-Jing-Tian, the underground part of Rhodiola quadrifida (Pall) Fisch Et Mey. Chem Pharm Bull (Tokyo) 43: 1245–1247 [DOI] [PubMed] [Google Scholar]
- Yoshikawa M, Shimada H, Shimoda H, Murakami N, Yamahara J, Matsuda H (1996) Bioactive constituents of Chinese natural medicines. 2. Rhodiolae radix. 1. Chemical structures and antiallergic activity of rhodiocyanosides A and B from the underground part of Rhodiola quadrifida (Pall) Fisch et MEY (Crassulaceae). Chem Pharm Bull (Tokyo) 44: 2086–2091 [DOI] [PubMed] [Google Scholar]
- Zagrobelny M, Bak S, Rasmussen AV, Jørgensen B, Naumann CM, Møller BL (2004) Cyanogenic glucosides and plant-insect interactions. Phytochemistry 65: 293–306 [DOI] [PubMed] [Google Scholar]