Abstract
Dental caries (tooth decay) is the most common chronic disease, worldwide, affecting most children and adults. Though dental caries is highly heritable, few caries-related genes have been discovered. We investigated whether 18 genetic variants in the group of nonamelogenin enamel matrix genes (AMBN, ENAM, TUFT1, and TFIP11) were associated with dental caries experience in 13 age- and race-stratified samples from six parent studies (N=3,600). Linear regression was used to model genetic associations and test gene-byfluoride interaction effects for two sources of fluoride: daily tooth brushing and home water fluoride concentration. Meta-analysis was used to combine results across five child and eight adult samples. We observed the statistically significant association of rs2337359 upstream of TUFT1 with dental caries experience via meta-analysis across adult samples (p<0.002) and the suggestive association for multiple variants in TFIP11 across child samples (p<0.05). Moreover, we discovered two genetic variants (rs2337359 upstream of TUFT1 and missense rs7439186 in AMBN) involved in gene-by-fluoride interactions. For each interaction, participants with the risk allele/genotype exhibited greater dental caries experience only if they were not exposed to the source of fluoride. Altogether, these results confirm that variation in enamel matrix genes contributes to individual differences in dental caries liability, and demonstrate that the effects of these genes may be moderated by protective fluoride exposures. In short, genes may exert greater influence on dental caries in unprotected environments, or equivalently, the protective effects of fluoride may obviate the effects of genetic risk alleles.
Keywords: tooth decay, genetic association, ameloblastin, enamelin, tuftelin, tuftelin-interacting protein
INTRODUCTION
Dental caries affects a majority of children and adults worldwide, and represents a major public health problem due to the fact that disease burden and associated concomitants (pain, tooth loss, trouble learning, eating, and sleeping, days of missed school/work, emergency room visits) are concentrated in vulnerable populations such as racial and ethnic minorities, and those living in rural areas and in poverty. In this regard, preventive measures such as fluoride exposure through tooth brushing and community water fluoridation are beneficial (Centers for Disease Control and Prevention Division of Oral Health (2013) Community Water Fluoridation, Using Fluoride to Prevent and Control Tooth Decay in the United States (www.cdc.gov/fluoridation/factsheets/fl_caries.htm)). However, fluoride exposures alone are insufficient to prevent tooth decay in some individuals. Numerous factors are thought to influence susceptibility to dental caries, and chief among these is host genetics. Heritability estimates for dental caries experience range from 30-70% (Boraas et al. 1988; Bretz et al. 2005; Conry et al. 1993; Shaffer et al. 2012a; Shaffer et al. 2013; Shaffer et al. 2012b; Wang et al. 2010), with host genes hypothesized to influence risk of dental caries by affecting tooth and enamel development, defense against cariogenic bacteria, dietary preferences, and protective features of the oral environment, including saliva composition and flow rate. Though few specific caries-related genes have been identified and rigorously validated, those that have been most extensively studied are the family of extracellular enamel matrix genes.
Enamel matrix genes code the proteinaceous content of tooth enamel, and mutations in these and other genes regulating their function are known to cause inherited enamel dysplasia, amelogenesis imperfecta. While the non-amelogenin enamel matrix proteins (enamelin, ameloblastin, and tuftelin) cumulatively comprise only a fraction of the protein component of developing enamel, mutations in the genes coding these proteins, particularly mutations in ENAM, are the most common cause of amelogenesis imperfecta, indicating a vital role in enamel development. Previous candidate gene studies have explored whether these enamel matrix and related genes are associated with dental caries (Deeley et al. 2008; Gasse et al. 2013; Jeremias et al. 2013; Kang et al. 2011; Olszowski et al. 2012; Patir et al. 2008; Shimizu et al. 2012; Slayton et al. 2005). From these studies, the most consistent genetic associations have been observed between TUFT1 and dental caries experience (Deeley et al. 2008; Patir et al. 2008; Shimizu et al. 2012; Stanley et al. 2014). Results for the other non-amelogenin enamel matrix genes, however, have been inconsistent. Individual studies have suggested that variants in AMBN (Patir et al. 2008; Shimizu et al. 2012), ENAM (Jeremias et al. 2013; Patir et al. 2008; Shimizu et al. 2012), and TFIP11 (Jeremias et al. 2013; Shaffer et al. 2011) influence dental caries, but the majority of studies have not (Deeley et al. 2008; Olszowski et al. 2012; Patir et al. 2008; Shimizu et al. 2012; Slayton et al. 2005; Wang et al. 2012b). Among the many possible explanations for the inconsistencies observed across studies is heterogeneity with regard to environmental exposures, such as fluoride, that profoundly impact risk of dental caries. In light of the current uncertainty regarding the effects of non-amelogenin enamel matrix genes, the purpose of this study was to investigate their effects on dental caries susceptibility in children and adults, and to determine whether their effects are moderated by fluoride exposures. We hypothesize that variants in AMBN, ENAM, TUFT1, and/or TFIP11 affect dental caries experience, and that genetic susceptibilities are especially pronounced in individuals lacking adequate protective exposure to fluoride.
MATERIALS AND METHODS
Samples and data collection
We included samples from six parent studies in this investigation: The Center for Oral Health Research in Appalachia, cohort 1 (COHRA1; N=1,769 (Polk et al. 2008)), Iowa Head Start (IHS; N=64 (Slayton et al. 2005)) Study, Iowa Fluoride Study (IFS; N=136 (Wang et al. 2012b)), Dental Strategies Concentrating on Risk Evaluation (Dental SCORE; N=502 (Aiyer et al. 2007a; Aiyer et al. 2007b)) the Dental Registry and DNA Repository (DRDR; N=875 (Wang et al. 2012a)), and the Center for Education and Drug Abuse Research (CEDAR; N=241 (Vanyukov et al. 2004)). Details regarding study design and participant recruitment for each parent study have been previously reported (Stanley et al. 2014) and are summarized in the Supplemental Material. Phenotype assessment protocols were similar across studies. All participants underwent an intra-oral examination by a dentist or research dental hygienist to assess dental caries experience, from which traditional caries indices, DMFT and dft, were generated. DMFT was defined as the number of decayed, missing due to decay, or restored (filled) teeth of the permanent dentition, excluding third molars. Likewise, dft was defined as the number of decayed or restored teeth of the primary dentition. Note, missing primary teeth did not contribute to dft scores due to the difficulty in determining the cause of missingness. Phenotype assessments were reliable and reproducible (ICC for COHRA1 was >0.99 for inter-examiners reliability and 0.86-0.99 for intra-examiner reliability (Polk et al. 2008; Wendell et al. 2010)). Data on two sources of fluoride exposure, home water source fluoride concentration and tooth brushing behavior, were collected in the COHRA1 and IFS samples. Home water source fluoride concentration (ppm) was measured using fluoride ion-specific electrodes of home water samples provided by participants. Daily tooth brushing frequency was assessed via questionnaire. All study protocols for participant recruitment and data collection were approved by all pertinent Universities’ Institutional Review Boards.
Genotypes
Genotyping for a custom panel of single nucleotide polymorphisms (SNPs) was performed by the Center for Inherited Disease Research (CIDR) at Johns-Hopkins University using the Illumina GoldenGate platform (San Diego, USA). Whereas the majority of the panel was chosen to follow-up putative associations from a number of GWAS scans, we also included SNPs such as those in and near enamel matrix genes, based on our interest in strong candidate genes. For this study, we interrogated 18 SNPs distributed across four non-amelogenin enamel matrix genes: AMBN, ENAM, TFIP11, and TUFT1 (see Table 1). These genes were chosen because of their known roles in amelogenesis and implication in Mendelian enamel defects. SNPs in these genes were chosen according to several criteria, including their high minor allele frequencies, ability to capture much of the gene-level variation in as few SNPs as possible, low linkage disequilibrium (i.e., correlation) with each other, as well as genotyping constraints such as compatibility with the GoldenGate platform, and mutual compatibility with other genotyped SNPs on the custom panel. Details regarding the design and quality of the genotyping panel are available elsewhere (Stanley et al. 2014).
Table 1.
Genetic variants in enamel matrix genes
| gene | SNP | chromosome | positiona | MAFb | base change | location / functionality |
|---|---|---|---|---|---|---|
| AMBN | rs17149026 | 4 | 71440940 | 0.0901 | G-T | upstream |
| rs17733915 | 4 | 71447093 | 0.4477 | C-T | upstream | |
| rs7439186 | 4 | 71469604 | 0.0802 | A-G | missense (Ala-Val) | |
| ENAM | rs1967376 | 4 | 71501744 | 0.0877 | C-T | intron |
| rs12640848c | 4 | 71506412 | 0.3729 | A-G | intron | |
| TFIP11 | rs17402286 | 22 | 26894879 | 0.0383 | A-G | synonymous (Leu-Leu) |
| rs6005060 | 22 | 26895736 | 0.0924 | A-T | intron | |
| rs713900 | 22 | 26898242 | 0.0726 | A-G | intron | |
| rs134134 | 22 | 26898891 | 0.1308 | C-T | intron | |
| rs134135 | 22 | 26898962 | 0.1589 | C-G | intron | |
| rs2097470 | 22 | 26904965 | 0.1245 | C-T | intron | |
| rs134145 | 22 | 26909750 | 0.3727 | A-G | upstream | |
| TUFT1 | rs2337359 | 1 | 151495796 | 0.1854 | C-T | upstream |
| rs1045298 | 1 | 151510825 | 0.1370 | C-T | upstream | |
| rs10158855 | 1 | 151515654 | 0.4212 | G-T | intron | |
| rs17640579 | 1 | 151521933 | 0.2228 | A-G | intron | |
| rs16833391 | 1 | 151547253 | 0.1068 | C-T | inton | |
| rs12749d | 1 | 151555741 | 0.1944 | C-T | 3′UTR |
based on Build 37
MAF = minor allele frequency in the COHRA1 sample
this SNP was independently genotyped and tested for association with dental caries experience in a larger sample of IFS subjects (Wang et al. 2012b); the previous study used different genotyping technology and a different statistical approach; a subset of the participants from the previous study was included in the present study
this SNP was independently genotyped and tested for association with dental caries in an independent sample of IHS participants (Slayton et al. 2005)
Statistical analysis
From the six parent studies, 13 race- and age-stratified samples were available (Table 2). Analyses of dental caries experience were performed separately in each sample and were limited to children 3-12 years of age for the primary dentition (dft) and adults ≥18 years of age for the permanent dentition (DMFT). One exception was the CEDAR sample, which included adolescents >15 years and for the purposes of this study was considered an adult sample. Analyses were also limited to self-reported non-Hispanic whites and blacks (analyzed separately) in order to obviate potential biases due to population stratification. Linear regression was used to test for genetic association between dft/DMFT and each SNP (under the additive genetic model) while simultaneously adjusting for the effects of age and sex. Regression models were inspected for influential points. Note, some samples were small after stratifying by race and age, therefore limited in their value; results for these samples should be interpreted with caution. Tests of association in black samples also were also adjusted for the first two principal components of ancestry to guard against confounding due to admixture/population structure. Adjustments for ancestry were not performed in whites due to lack of population structure at the level that can be adequately measured by our custom panel. Tests of genetic main effects and genetic ancestry modeling were performed in PLINK (Purcell et al. 2007). Evidence of association was combined across samples using Stouffer's inverse variance weighted method of meta-analysis, the most appropriate method given the likely non-random heterogeneity among the cohorts, as implemented in METAL (Willer et al. 2010). Meta-analyses were performed for whites-only, and whites and blacks combined.
Table 2.
Characteristics of the study samples: mean (range) or percentage, %
| sample | N | female sex | age, years | caries prevalencea | dft/DMFTb | fluoridated water (%) | daily tooth brushing | tooth brushing per day |
|---|---|---|---|---|---|---|---|---|
| children | ||||||||
| COHRA1 whites | 608 | 46.7 | 7.3 (3.0-12.0) | 55.4 | 2.3 (0-17) | 60.2 | 92.8 | 1.6 (0-4) |
| COHRA1 blacks | 81 | 46.9 | 7.6 (3.2-11.8) | 49.4 | 1.8 (0-8) | 86.8 | 95.1 | 1.6 (0-3) |
| IHS whites | 41 | 58.5 | 4.1 (3.2-5.3) | 80.5 | 6.3 (0-20) | - | - | - |
| IHS blacks | 23 | 52.2 | 4.3 (3.4-5.6) | 82.6 | 5.7 (0-17) | - | - | - |
| IFS whites | 136 | 48.5 | 5.2 (4.4-6.8) | 37.8 | 1.2 (0-16) | 73.6 | 88.6 | 1.4 (0-3) |
| adults | ||||||||
| COHRA1 whites | 994 | 62.8 | 34.3 (18.0-75.0) | 96.5 | 10.5 (0-28) | 58.8 | 89.3 | 1.5 (0-2) |
| COHRA1 blacks | 86 | 70.9 | 36.2 (18.2-60.8) | 94.2 | 9.3 (9-28) | 88.0 | 92.3 | 1.5 (0-2) |
| Dental SCORE whites | 277 | 63.2 | 64.0 (48.0-78.0) | 100.0 | 16.4 (2-28) | - | - | - |
| Dental SCORE blacks | 225 | 72.9 | 61.6 (47.0-79.0) | 100.0 | 14.8 (1-28) | - | - | - |
| DRDR whites | 702 | 50.0 | 43.0 (18.0-74.8) | 97.9 | 16.6 (0-28) | - | - | - |
| DRDR blacks | 173 | 57.8 | 44.5 (18.0-74.4) | 98.3 | 16.5 (0-28) | - | - | - |
| CEDAR whites | 173 | 31.2 | 20.4 (15.7-28.6) | 82.1 | 5.4 (0-21) | - | - | - |
| CEDAR blacks | 68 | 44.3 | 20.2 (15.6-27.8) | 88.6 | 6.4 (0-16) | - | - | - |
caries prevalence was defined as dfs ≥ 1 in children or DMFT ≥ 1 in adults
dft was the measure of caries experience of the primary dentition in child samples; DMFT was the measure of caries experience of the permanent dentition in adult samples
SNP-by-fluoride interaction effects were modeled using linear regression while adjusting for age, sex, (principal components of ancestry in blacks,) and SNP and fluoride main effects. Two dichotomous fluoride exposures were considered: home water source fluoride concentration (less than vs. greater than 0.7 ppm) and daily tooth brushing (once or more vs. less than once per day). For interaction models, heterozygotes and rare homozygotes were combined (i.e., the dominant genetic model) for SNPs with minor allele frequencies less than 25% to avoid modeling strata represented by few participants. Interaction effects between the two fluoride exposures were also modeled (in the absence of SNP effects). All interaction models and descriptive statistics were generated in R (R Foundation for Statistical Computing, Vienna, AU). Given the multiple comparisons, we used the threshold of p-values less than 0.003 to declare statistical significance for genetic associations, which corresponds to the Bonferroni correction for 18 SNPs. P-values less than 0.05 were considered “suggestive” for SNP associations.
RESULTS
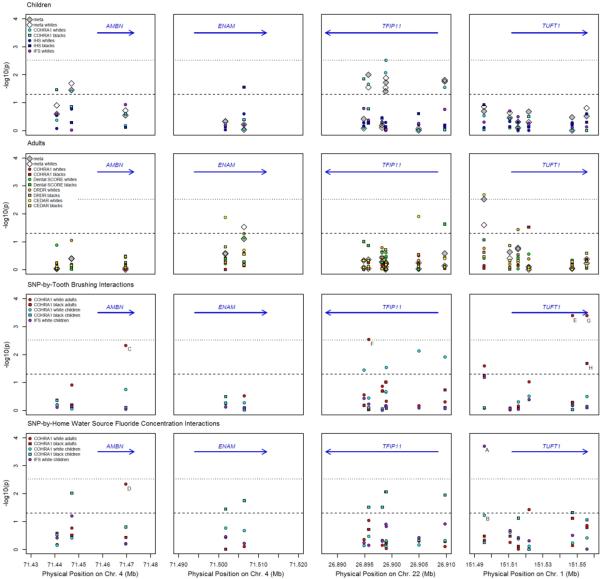
Characteristics of the 13 samples are shown in Table 2. Considerable variation in dental caries experience was observed, which was expected given the differences in age and demography across the samples. Figure 1 shows the results of tests of genetic association for four non-amelogenin enamel matrix genes: AMBN, ENAM, TFIP11, and TUFT1. Negative log10-transformed p-values are plotted against physical positions of each SNP such that SNPs showing significant evidence of association occur at –log10(p)=2.52 or higher on the plot (indicated by the dotted line), and SNPs showing suggestive association occur above –log10(p)=1.30 (indicated by the dashed line). Full association results are available in the Supplemental Table.
Fig 1.
Genetic association in enamel matrix genes (columns from left to right): AMBN, ENAM, TFIP11, and TUFT1. Rows (top to bottom) represent (first row) main effects in child samples, (second row) main effects in adult samples, (third row) SNP-by-tooth brushing interaction effects for daily brushing, and (fourth row) SNP-by-fluoride concentration interaction effects for home water source. Negative log10-transformed p-values are shown for association tests in all samples: COHRA1 children (cyan), IHS (blue), IFS (purple), COHRA1 adults (red), Dental SCORE (green), DRDR (orange), and CEDAR (yellow). Circles represent white samples and squares represents black samples. White diamonds represent meta-analysis across all white samples and gray diamonds represent meta-analysis across all black and white samples, combined. The dashed lines are displayed at p-value = 0.05. The blue arrows represent the physical location and direction of genes. Points labeled A to H correspond to the interactions shown in panels A to H, respectively, of Figure 3.
SNP-wise tests of genetic association (Figure 1, first and second rows) were performed separately for each sample, and meta-analyzed across child samples and across adult samples. In children (Figure 1, first row), a significant association was observed for SNP rs134135 in TFIP11 (p=0.003) for COHRA1 white children. No meta-analyses in children (across either the three white or all five child samples) showed significant associations. Suggestive associations (p<0.05) were observed for COHRA1 black and white children with SNP rs17733915 in AMBN and with several SNPs in TFIP11. Meta-analyses of these SNPs were also suggestive.
In adults, a significant association was observed for SNP rs2337359 upstream of TUFT1 in the CEDAR white sample (p=0.002; Figure 1, second row). This SNP was also significant in the meta-analysis across all eight adult samples (p=0.003). ENAM SNP rs12640848 showed suggestive association (p=0.02) via meta-analysis across white adult samples. Individual samples showed suggestive associations (p<0.05) for some SNPs (i.e., rs1967376 and rs2097470 in CEDAR whites, rs134145 in Dental SCORE blacks, rs10158855 in DRDR whites, and rs17640579 in COHRA1 blacks), though meta-analyses of these SNPs were not significant. No SNPs showed significant association or compelling suggestive association in both children and adults.
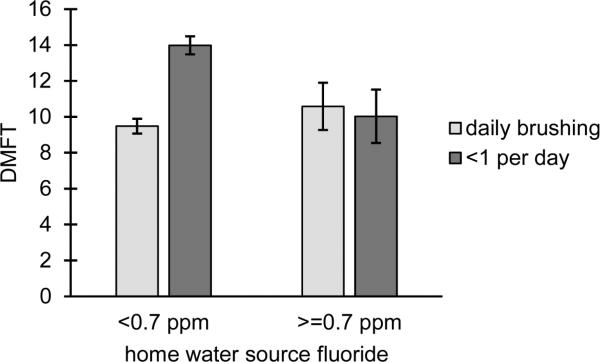
In addition to tests of SNP main effects, we also tested the effects of two fluoride exposures (daily tooth brushing and home water source fluoride concentration) in the COHRA1 and IFS samples, for which fluoride data were available. Daily brushing was significantly protective in COHRA1 white adults (equating to 2.1 fewer carious teeth in daily brushers compared to non-brushers; p=0.002). Though showing a similar trend in COHRA1 black adults, daily brushing was not significantly associated with dental caries experience in any other cohorts (data not shown). Likewise, home water fluoride concentration, alone, did not show significant effects on dental caries experience (data not shown). Interestingly, the interaction between daily tooth brushing and home water fluoride concentration was statistically significant (p=0.02) in COHRA1 white adults, with participants exposed to fluoride either through daily tooth brushing, fluoridated home water, or both, exhibiting fewer carious teeth than individuals who were not protected by either source of fluoride (Figure 2). Note, few participants in other samples (COHRA1 black adults, COHRA1 children, IFS) lacked both fluoride exposures, and thus the fluoride-by-fluoride interaction effect could not be tested.
Fig 2.
Interaction between tooth brushing and home water source fluoride concentration in COHRA1 white adults indicates that participants not exposed to either source of fluoride have more carious teeth than participants exposed to one or both sources of fluoride (p=0.02)
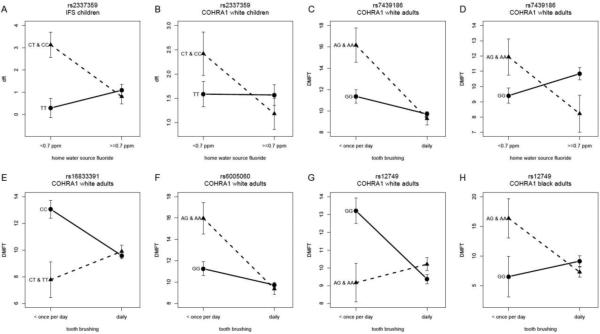
Given the previous studies suggesting the presence of interactions between enamel matrix genes and fluoride exposures (Kang et al. 2011; Shaffer et al. 2011), we tested for such interaction effects in the COHRA1 and IFS samples (Figure 1, third and fourth rows). Four significant, and several suggestive interactions were observed. The most significant interaction was observed for SNP rs2337359, upstream of TUFT1, which showed an interaction with home water fluoride in IFS (p=0.0002). Though not statistically significant, this interaction trend was also observed in COHRA1 white children (p=0.06). In both samples, increased dental caries was observed for participants carrying the C allele who were not exposed to the fluoride source (Figure 3A-B).
Fig 3.
(A-B) Interactions between rs2337359 and home water source fluoride concentration in (A) IFS and (B) COHRA1 white children indicate that participants with CT and CC genotypes exhibit greater dental caries experience if not exposed to the fluoride source. (C-D) Interactions between rs7439186 and (C) tooth brushing and (D) home water source fluoride concentration in COHRA1 white adults indicates that participants with the A allele exhibit greater dental caries experience if not exposed to the fluoride source. (E-H) Interactions between SNPs (E) rs16833391, (F) rs6005060, and (G-H) rs12749 and tooth brushing in COHRA1 white (E-G) and black (H) adults show greater differences between genotype groups in participants not exposed to the fluoride source. Note, for all the interaction models the extreme strata (i.e., rarer genotype group in the unexposed environment) comprise at least 15 participants except for the extreme strata in (H) which comprises 3 participants.
In adults, compelling suggestive interactions were observed between the AMBN missense SNP rs7439186 and both tooth brushing (p=0.005; Figure 3C) and home water fluoride concentration (p=0.004; Figure 3D) in COHRA1 white adults. For both interactions, participants with the A allele had greater dental caries experience if not exposed to fluoride sources. The two fluoride exposures are not correlated in COHRA1 white adults (Pearson's R=0.01; p=0.81), and therefore, the tests each provide independent evidence of interaction between this SNP and fluoride exposure.
Another significant interaction was observed between TFIP11 SNP rs6005060 and tooth brushing in COHRA1 white adults (p=0.003, Figure 3F), though this relationship was not observed for other samples or for the other fluoride exposure. Two SNPs in TUFT1 showed significant interactions with tooth brushing frequency in COHRA1 white adults (rs16833391, p=0.0004, Figure 3E, and rs12749, p=0.0004; Figure 3G). A similar suggestive trend was observed in COHRA1 black adults for rs12749 (p=0.02; Figure 3H), although the sample size was small. As with the other interactions, the genetic effect was greater in the participants not exposed to sources of fluoride. However, the risk alleles differed between whites and blacks.
DISCUSSION
Given their role in the developing tooth enamel, non-amelogenin enamel matrix genes are sensible candidate genes for dental caries. Previous genetic association studies of AMBN, ENAM, TFIP11, and TUFT1 have been performed, albeit with inconsistent findings (Deeley et al. 2008; Jeremias et al. 2013; Kang et al. 2011; Olszowski et al. 2012; Shimizu et al. 2012; Slayton et al. 2005; Wang et al. 2012b). Here we report associations in the largest study of these genes, to date, comprising 13 sub-samples from six parent projects, with nearly 3,600 participants in total. Significant associations were observed for SNPs in TFIP11 and TUFT1, each showing evidence of association across meta-analyses of children or adults. These results reinforce the notion that genetic variation in TUFT1 and TFIP11 influence susceptibility to dental caries.
Echoing the previously-published body of literature, we also observed heterogeneity across the samples, including associations that were specific to individual samples. Several explanations for these inconsistencies are possible, including differences in power to detect association across the samples, false discoveries, genetic heterogeneity (especially between racial groups), and differences in phenotype assessments, ages, dentitions, demography, and risk profiles across the samples. To some extent, each of these explanations is likely to have impacted our results.
We investigated two sources of fluoride, tooth brushing and home water source, and showed evidence of a statistical interaction between them in that individuals lacking both sources had greater dental caries experience than individuals protected by one fluoride source or the other (or both). This result reiterates the important protective role of fluoride, and also suggests that having at least some regular fluoride exposure is more important than having multiple types of exposure. Also, we were particularly interested in exploring the hypothesis that differences in environmental fluoride exposures, which profoundly impact risk of dental caries, may account for the lack of replication across samples, a conundrum observed both in this study and in the literature as a whole. In particular, we hypothesized that the comparatively smaller genetic effects may be masked by fluoride exposures, and therefore observable in some environments (and some studies), but not others.
Two SNP-by-fluoride interactions showed strong evidence with consistency of risk alleles/genotypes across two independent fluoride sources (missense rs7439186 in AMBN), or across two independent samples (rs2337359 upstream of TUFT1). Other significant SNP-by-fluoride interactions showed inconsistencies across fluoride sources and/or samples, but nevertheless exhibited greater genetic effects in the unprotected environments. Altogether, these results confirm that the effects of genetic variants in enamel matrix genes may be moderated by fluoride exposures. In short, genetic factors may exert greater influence on dental caries in unprotected environments, or equivalently, the protective effects of fluoride may obviate the effects of genetic risk alleles. These results support our hypothesis that genes may play a larger role in dental caries experience in participants who are not otherwise protected by their exposure to fluoride.
Despite the significant associations and gene-by-fluoride interactions observed in this study, our understanding of how genetic factors involved in enamel development contribute to dental caries susceptibility is far from complete. Other genes may also influence normal variation in enamel. These include AMELX (which accounts for most of the proteinaceous component during tooth enamel development but is only responsible for a small fraction of cases of Mendelian amelogenesis imperfecta), DSPP (a dentin matrix protein important for the biomineralization of dentin, the tissue that supports the enamel), MMP20 and KLK4 (proteases that degrade organic matter during enamel maturation), and others. More work is need to fully understand how variation in genes involved in amelogenesis influence subsequent risk of dental caries, and whether and how these variants interact with environmental exposures.
Overall, our results fit in with a broader notion that the impact of environmental exposures varies based on an individual's genome. While moderate fluoride exposure is recommended for everyone, there may be other environmental exposures that benefit specific subsets of the population, such as antimicrobial or pH-buffering rinses, or prescription strength topical fluoride. Understanding the gene-by-environment interactions contributing to dental caries susceptibility may enable tailored prevention strategies, such as these, in the near future under the personalized medicine model. Furthermore, understanding the genetic contributors to variation in dental caries risk may lead to new avenues of prevention, early identification of high-risk patients, and/or new targets for therapy.
Supplementary Material
ACKNOWLEDGEMENTS
We would like to express our deep gratitude to the participants and research teams of the six parent studies whose contributions made this work possible. This work was supported by the following National Institutes of Health grants: U01-DE018903, R01-DE014899, R01-DE009551, R01-DE012101, R01-DE018914, P50-DA005605, and R01-DA019157, as well as the National Science Foundation/Department of Defense grant DBI-1263020. The Dental Registry and DNA Repository is supported by the University of Pittsburgh School of Dental Medicine. The Dental SCORE sample is partially supported by Commonwealth of Pennsylvania Department of Health grant ME-02-384.
Footnotes
CONFLICTS OF INTEREST
The authors declare no conflicts of interest.
REFERENCES
- Aiyer AN, Kip KE, Marroquin OC, Mulukutla SR, Edmundowicz D, Reis SE. Racial differences in coronary artery calcification are not attributed to differences in lipoprotein particle sizes: the Heart Strategies Concentrating on Risk Evaluation (Heart SCORE) Study American heart journal. 2007a;153:328–334. doi: 10.1016/j.ahj.2006.11.002. doi:10.1016/j.ahj.2006.11.002. [DOI] [PubMed] [Google Scholar]
- Aiyer AN, Kip KE, Mulukutla SR, Marroquin OC, Hipps L, Jr., Reis SE. Predictors of significant short-term increases in blood pressure in a community-based population. The American journal of medicine. 2007b;120:960–967. doi: 10.1016/j.amjmed.2007.06.021. doi:10.1016/j.amjmed.2007.06.021. [DOI] [PubMed] [Google Scholar]
- Boraas JC, Messer LB, Till MJ. A genetic contribution to dental caries, occlusion, and morphology as demonstrated by twins reared apart. J Dent Res. 1988;67:1150–1155. doi: 10.1177/00220345880670090201. [DOI] [PubMed] [Google Scholar]
- Bretz WA, et al. Longitudinal analysis of heritability for dental caries traits. J Dent Res. 2005;84:1047–1051. doi: 10.1177/154405910508401115. doi:84/11/1047 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Centers for Disease Control and Prevention Division of Oral Health Community Water Fluoridation, Using Fluoride to Prevent and Control Tooth Decay in the United States. 2013 ( www.cdc.gov/fluoridation/factsheets/fl_caries.htm)
- Conry JP, Messer LB, Boraas JC, Aeppli DP, Bouchard TJ., Jr Dental caries and treatment characteristics in human twins reared apart. Arch Oral Biol. 1993;38:937–943. doi: 10.1016/0003-9969(93)90106-v. doi:0003-9969(93)90106-V [pii] [DOI] [PubMed] [Google Scholar]
- Deeley K, Letra A, Rose EK, Brandon CA, Resick JM, Marazita ML, Vieira AR. Possible association of amelogenin to high caries experience in a Guatemalan-Mayan population. Caries Res. 2008;42:8–13. doi: 10.1159/000111744. doi:000111744 [pii] 10.1159/000111744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasse B, et al. Common SNPs of AmelogeninX (AMELX) and dental caries susceptibility. J Dent Res. 2013;92:418–424. doi: 10.1177/0022034513482941. doi:10.1177/0022034513482941. [DOI] [PubMed] [Google Scholar]
- Jeremias F, et al. Genes expressed in dental enamel development are associated with molar-incisor hypomineralization. Arch Oral Biol. 2013;58:1434–1442. doi: 10.1016/j.archoralbio.2013.05.005. doi:10.1016/j.archoralbio.2013.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang SW, Yoon I, Lee HW, Cho J. Association between AMELX polymorphisms and dental caries in Koreans. Oral diseases. 2011;17:399–406. doi: 10.1111/j.1601-0825.2010.01766.x. doi:10.1111/j.1601-0825.2010.01766.x. [DOI] [PubMed] [Google Scholar]
- Olszowski T, Adler G, Janiszewska-Olszowska J, Safranow K, Kaczmarczyk M. MBL2, MASP2, AMELX, and ENAM gene polymorphisms and dental caries in Polish children. Oral diseases. 2012;18:389–395. doi: 10.1111/j.1601-0825.2011.01887.x. doi:10.1111/j.1601-0825.2011.01887.x. [DOI] [PubMed] [Google Scholar]
- Patir A, Seymen F, Yildirim M, Deeley K, Cooper ME, Marazita ML, Vieira AR. Enamel formation genes are associated with high caries experience in Turkish children. Caries Res. 2008;42:394–400. doi: 10.1159/000154785. doi:10.1159/000154785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polk DE, Weyant RJ, Crout RJ, McNeil DW, Tarter RE, Thomas JG, Marazita ML. Study protocol of the Center for Oral Health Research in Appalachia (COHRA) etiology study. BMC Oral Health. 2008;8:18. doi: 10.1186/1472-6831-8-18. doi:1472-6831-8-18 [pii] 10.1186/1472-6831-8-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell S, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. doi:S0002-9297(07)61352-4 [pii] 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaffer JR, et al. Heritable patterns of tooth decay in the permanent dentition: principal components and factor analyses. BMC Oral Health. 2012a;12:7. doi: 10.1186/1472-6831-12-7. doi:10.1186/1472-6831-12-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaffer JR, et al. Clustering tooth surfaces into biologically informative caries outcomes. J Dent Res. 2013;92:32–37. doi: 10.1177/0022034512463241. doi:10.1177/0022034512463241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaffer JR, et al. Genetic susceptibility to dental caries on pit and fissure and smooth surfaces. Caries Res. 2012b;46:38–46. doi: 10.1159/000335099. doi:10.1159/000335099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaffer JR, et al. Genome-wide association scan for childhood caries implicates novel genes. J Dent Res. 2011;90:1457–1462. doi: 10.1177/0022034511422910. doi:0022034511422910 [pii] 10.1177/0022034511422910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu T, et al. Enamel formation genes influence enamel microhardness before and after cariogenic challenge. PloS one. 2012;7:e45022. doi: 10.1371/journal.pone.0045022. doi:10.1371/journal.pone.0045022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slayton RL, Cooper ME, Marazita ML. Tuftelin, mutans streptococci, and dental caries susceptibility. J Dent Res. 2005;84:711–714. doi: 10.1177/154405910508400805. doi:84/8/711 [pii] [DOI] [PubMed] [Google Scholar]
- Stanley BO, et al. Genetic Association of MPPED2 and ACTN2 with Dental Caries. J Dent Res. 2014 doi: 10.1177/0022034514534688. doi:10.1177/0022034514534688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanyukov MM, Maher BS, Devlin B, Tarter RE, Kirillova GP, Yu LM, Ferrell RE. Haplotypes of the monoamine oxidase genes and the risk for substance use disorders American journal of medical genetics Part B. Neuropsychiatric genetics : the official publication of the International Society of Psychiatric Genetics. 2004;125B:120–125. doi: 10.1002/ajmg.b.20105. doi:10.1002/ajmg.b.20105. [DOI] [PubMed] [Google Scholar]
- Wang X, et al. Genes and Their Effects on Dental Caries May Differ between Primary and Permanent Dentitions. Caries Res. 2010;44:277–284. doi: 10.1159/000314676. doi:000314676 [pii] 10.1159/000314676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, et al. Genome-wide association scan of dental caries in the permanent dentition. BMC Oral Health. 2012a;12:57. doi: 10.1186/1472-6831-12-57. doi:10.1186/1472-6831-12-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, et al. Genetic and environmental factors associated with dental caries in children: the Iowa Fluoride Study. Caries Res. 2012b;46:177–184. doi: 10.1159/000337282. doi:10.1159/000337282 000337282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wendell S, et al. Taste genes associated with dental caries. J Dent Res. 2010;89:1198–1202. doi: 10.1177/0022034510381502. doi:0022034510381502 [pii] 10.1177/0022034510381502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willer CJ, Li Y, Abecasis GR. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics. 2010;26:2190–2191. doi: 10.1093/bioinformatics/btq340. doi:10.1093/bioinformatics/btq340. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.