Abstract
Dihydroneopterin aldolase (EC 4.1.2.25) is one of the enzymes of folate synthesis that remains to be cloned and characterized from plants. This enzyme catalyzes conversion of 7,8-dihydroneopterin (DHN) to 6-hydroxymethyl-7,8-dihydropterin, and is encoded by the folB gene in Escherichia coli. The E. coli FolB protein also mediates epimerization of DHN to 7,8-dihydromonapterin. Searches of the Arabidopsis genome detected three genes encoding substantially diverged FolB homologs (AtFolB1–3, sharing 57%–73% identity), for which cDNAs were isolated. A fourth cDNA specifying a FolB-like protein (LeFolB1) was obtained from tomato (Lycopersicon esculentum) by reverse transcription-PCR. When overproduced in E. coli, recombinant AtFolB1, AtFolB2, and LeFolB1 proteins all had both dihydroneopterin aldolase and epimerase activities, and carried out the aldol cleavage reaction on the epimerization product, 7,8-dihydromonapterin, as well as on DHN. AtFolB3, however, could not be expressed in active form. Size exclusion chromatography indicated that the plant enzyme is an octamer, like the bacterial enzyme. Quantifying expression of the Arabidopsis genes by real-time reverse transcription-PCR showed that AtFolB1 and AtFolB2 messages occur at low levels throughout the plant, whereas the AtFolB3 mRNA was detected only in siliques and only with an extremely low abundance. Sequence comparisons and phylogenetic analysis of FolB homologs from 16 plants indicated that their N-terminal regions are highly variable, and that most species have a small number of FolB genes that diverged after separation of the lineages leading to families. The substantial divergence of FolB homologs in Arabidopsis and other plants suggests that some of them may act on substrates other than DHN.
Folates are essential cofactors for various metabolic reactions involving one-carbon units. Bacteria, fungi, and plants synthesize folates de novo, but mammals and other higher animals lack a complete folate synthesis pathway and so need a dietary supply (Green et al., 1996; Cossins and Chen, 1997; Rébeillé and Douce, 1999). For humans, this supply comes mainly from plant foods (de Bree et al., 1997; Scott et al., 2000). Since inadequate folate intake is a worldwide problem in human nutrition, raising folate levels in crops is a target for metabolic engineering (Scott et al., 2000; Bouis, 2002).
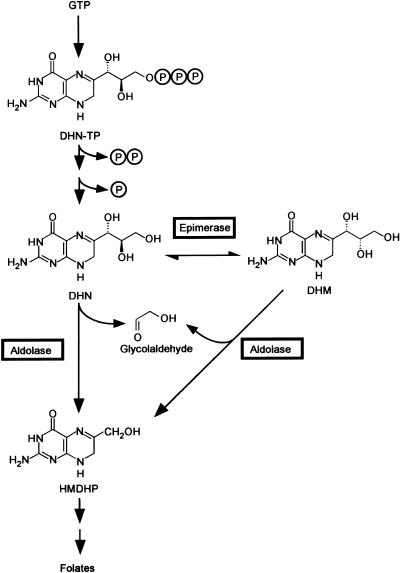
To engineer folate levels, it is important to know the steps in the folate synthesis pathway and to identify the corresponding enzymes and genes. Biochemical and genomics approaches have shown that plants have essentially the same pathway as Escherichia coli and have defined enzymes and genes for 7 of its 9 or 10 specific steps (Ravanel et al., 2001; Basset et al., 2002; Hanson and Gregory, 2002; Basset et al., 2004). One plant enzyme yet to be cloned and characterized is dihydroneopterin aldolase, which catalyzes the cleavage of the side chain of 7,8-dihydroneopterin (DHN), yielding 6-hydroxymethyl-7,8-dihydropterin (HMDHP) and glycolaldehyde (Fig. 1).
Figure 1.
The pteridine branch of the folate synthesis pathway. The aldolase and epimerase reactions catalyzed by E. coli DHN aldolase (FolB) are boxed. The phosphate groups of DHN triphosphate are removed in two steps. It is unclear whether the first step (elimination of pyrophosphate) is chemical or enzymatic (Suzuki and Brown, 1974; De Saizieu et al., 1995); the second step can be mediated by various nonspecific phosphomonoesterases (Suzuki and Brown, 1974). DHN-TP, 7,8-dihydroneopterin triphosphate; DHN, 7,8-dihydroneopterin; DHM, 7,8-dihydromonapterin; HMDHP, 6-hydroxymethyl-7,8-dihydropterin.
DHN aldolases have been cloned and characterized from several prokaryotes and one eukaryote, the fungus Pneumocystis carinii. The E. coli, Haemophilus influenzae, Staphylococcus aureus, and Synechocystis sp. enzymes are short, monofunctional proteins (118-150 residues) that form homooctamers (Haußmann et al., 1998; Hennig et al., 1998; Lee et al., 1999). Besides mediating the aldolase reaction, E. coli DHN aldolase (FolB) efficiently catalyzes epimerization of the 2′ carbon of the DHN side chain to give 7,8-dihydromonapterin (DHM), and cleavage of DHM to HMDHP (Fig. 1). The H. influenzae enzyme also shows these activities, but weakly (Haußmann et al., 1998). In Streptococcus pneumoniae, DHN aldolase is part of a bifunctional protein together with HMDHP pyrophosphokinase, the next enzyme in the folate pathway (Lopez and Lacks, 1993). P. carinii has a trifunctional protein comprising both these enzymes plus dihydropteroate synthase (Volpe et al., 1993).
Plants have been shown to contain small amounts of DHN and HMDHP (detected as their oxidized forms, neopterin and 6-hydroxymethylpterin) as well as molybdopterin (Iwai et al., 1976; Kohashi, 1980; Kohashi et al., 1980; Yoshida and Akino, 1980; Mendel and Hänsch, 2002). Very little else is known about pteridines in plants, there being just a few reports of pteridines such as monapterin, 6-carboxypterin, isoxanthopterin, and pterin (Sugiura and Goto, 1966; Kobayashi et al., 1967; Kohashi, 1980; Kohashi et al., 1980; Yoshida and Akino, 1980). Monapterin is most probably an enzymatic metabolite (or the oxidized form of one), but the rest are likely to be breakdown products of folates or folate precursors such as DHN. The lack of data on plant pteridines contrasts with the wealth of information for animals and bacteria, in which many pteridines are well documented; these include various pterin pigments, and tetrahydrobiopterin, the cofactor for certain hydroxylases and NO synthase (Forrest and Van Baalen, 1970; Thöny et al., 2000; Ziegler, 2003). Moreover, pteridine glycosides occur in many photosynthetic prokaryotes (e.g. Kang et al., 1998; Lee et al., 1999).
This study aimed to identify and characterize DHN aldolases from plants and to define their expression patterns. We focused on Arabidopsis and on tomato (Lycopersicon esculentum), which is our system for folate engineering. Having found that Arabidopsis has three diverged DHN aldolases, we used phylogenetic analysis to investigate the origin and potential significance of this divergence.
RESULTS
Identification of Plant FolB Homologs
Searches of the Arabidopsis genome identified three genes encoding short proteins with 31%–38% identity to the monofunctional DHN aldolases of E. coli (FolB), S. aureus, and Synechocystis. The products of these genes, At3g11750, At5g62980, and At3g21730, were designated AtFolB1, AtFolB2, and AtFolB3, respectively. Searches of plant genome and expressed sequence tag (EST) databases revealed DNA sequences encoding similar proteins from 14 other angiosperms, indicating that monofunctional FolB-like enzymes are probably ubiquitous in this group. As no tomato sequences were found in database searches, a full-length potato (Solanum tuberosum) EST (GenBank BQ512667) was used to design PCR primers to clone an orthologous tomato cDNA, specifying the protein LeFolB1.
Alignment of the Arabidopsis and tomato deduced protein sequences with those of prokaryotes revealed three features (Fig. 2). First is the conservation of residues inferred to be crucial from crystallographic and mutagenesis studies of the S. aureus enzyme (Hennig et al., 1998). Thus, Glu-22 and Lys-100 (numbers refer to the S. aureus sequence), both implicated in catalysis, are strictly conserved. Similarly, Ala-18, Val-48, Tyr-54, Leu-72, and Glu-74 are predicted to participate in substrate binding and are conserved or conservatively replaced. The second feature is that, unlike later enzymes in the folate pathway, the plant FolB proteins appear not to have mitochondrial targeting signals. Thus, their N termini are shorter than, or extend only a few residues beyond, that of Synechocystis, the longest prokaryote sequence. Furthermore, the N-terminal regions of the plant proteins lack the hallmark features of mitochondrial targeting peptides, i.e. enrichment in basic residues, no acidic residues, and the ability to form amphiphilic α-helices (Emanuelsson and von Heijne, 2001). It is, however, noteworthy that the two longer Arabidopsis proteins, AtFolB1 and AtFolB3, have an N-terminal exon (underlined in Fig. 2) that is missing from AtFolB2. The third feature is the substantial divergence among the Arabidopsis sequences, which share only 57%–73% identity.
Figure 2.
Alignment of the deduced amino acid sequences of Arabidopsis and tomato FolB homologs with the DHN aldolases of E. coli (FolB), S. aureus, and Synechocystis. Identical residues are shaded in black, similar residues in gray. Dashes are gaps introduced to maximize alignment. Underlining indicates an additional exon present in the AtFolB1 and AtFolB3 sequences. The arrow marks the position of an intron common to all three Arabidopsis sequences. Asterisks indicate conserved residues implicated in catalysis or pteridine binding in the S. aureus enzyme (Hennig et al., 1998). EcFolB, E. coli FolB (GenBank accession number P31055); SaDHNA, S. aureus DHN aldolase (GenBank P56740); SsDHNA, Synechocystis sp. PCC 6803 DHN aldolase (GenBank S76177); AtFolB1, gene At3g11750; AtFolB2, gene At5g62980; AtFolB3, gene At3g21730; LeFolB1, tomato DHN aldolase (GenBank AY422466).
Aldolase and Epimerase Activities of Plant FolB Homologs
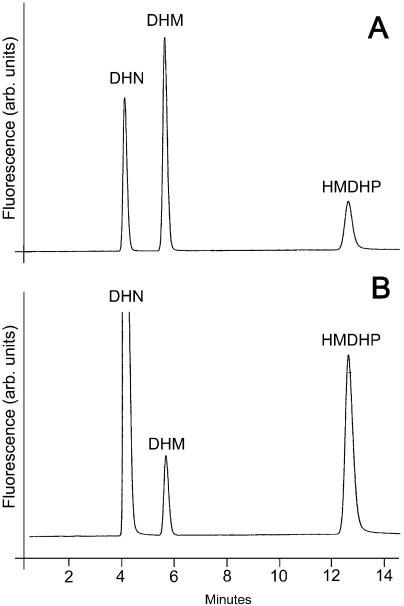
The Arabidopsis AtFolB1 and AtFolB2 proteins and the tomato LeFolB1 protein were overexpressed in E. coli, purified, and tested for aldolase and epimerase activities using DHN or DHM as substrate. All except AtFolB1 were strongly expressed (approximately 5% to 20% of total protein) in soluble form. To improve AtFolB1 expression, we deleted residues 1 to 20 and replaced Gly-21 by a start codon. This construct, designated as AtFolB1-ΔE, was expressed at high level (approximately 20% of total protein) and yielded active enzyme. Despite the substantial differences in their amino acid sequences, all three proteins had both aldolase and epimerase activities with DHN or DHM (Table I). Figure 3 shows a representative HPLC chromatogram of reaction products for the AtFolB1-ΔE enzyme acting on DHN. (The pteridines were oxidized to their fluorescent aromatic forms prior to chromatography.) Note the peak corresponding to HMDHP, the product of the aldolase reaction, and the smaller peak corresponding to DHM from the epimerase reaction. The Vmax values for the DHN and DHM aldolase reactions were similar. Although these values are approximately 10-fold lower than those reported by Haußmann et al. (1998) for the E. coli or H. influenzae enzymes, the difference can be ascribed, at least in part, to assay temperature (37°C for the bacterial enzymes versus 30°C for the plant enzymes), because when assayed at 30°C, the activity of E. coli FolB was near those of the plant proteins (V. Illarionova, unpublished data). The Vmax values for the DHN and DHM epimerase reactions were more variable but were similar to or greater than the corresponding values for the bacterial enzymes. The plant enzymes thus have relatively higher epimerase activities than their bacterial counterparts.
Table I.
Enzymatic activities of recombinant Arabidopsis and tomato FolB homologs
| Enzyme | Substrate | Product | Reaction Type | Reaction Rates |
|---|---|---|---|---|
| μmol min−1 mg −1 protein | ||||
| AtFolB1-ΔE | DHN | HMDHP | Cleavage | 0.32 |
| DHM | Epimerization | 0.20 | ||
| DHM | HMDHP | Cleavage | 0.31 | |
| DHN | Epimerization | 0.08 | ||
| AtFolB2 | DHN | HMDHP | Cleavage | 0.20 |
| DHM | Epimerization | 0.26 | ||
| DHM | HMDHP | Cleavage | 0.35 | |
| DHN | Epimerization | 0.20 | ||
| LeFolB1 | DHN | HMDHP | Cleavage | 0.28 |
| DHM | Epimerization | 0.08 | ||
| DHM | HMDHP | Cleavage | 0.27 | |
| DHN | Epimerization | 0.03 |
Enzymes were purified to homogeneity. Experiments were performed at 30°C. Data are mean Vmax values from at least two experiments.
Figure 3.
HPLC-fluorometric analysis of the products formed from DHN by recombinant AtFolB1-ΔE protein. A, Standard pteridines. Retention times: DHN, 4.2 min; DHM, 5.7 min; HMDHP, 12.8 min. B, Reaction mixture in which AtFolB1-ΔE protein was incubated with DHN for 2.5 min. Samples were treated with acidic I2/KI before HPLC separation to convert pteridines to their fully oxidized, fluorescent forms.
When expressed in E. coli using the procedures adopted for the other plant enzymes, the AtFolB3 protein made up approximately 25% of total protein, but was insoluble, and no recombinant aldolase or epimerase activity was detected in the soluble fraction. We tested a full-length construct (AtFolB3-WT) and a truncated variant lacking the N-terminal region (AtFolB3-ΔE). Various expression strategies using, for example, E. coli BL21-CodonPlus (DE3)/pRIL cells (which have plasmid-borne copies of tRNAs for codons that are rare in E. coli but common in plants) likewise failed to yield a soluble protein or any detectable recombinant DHN or DHM aldolase or epimerase activity.
Quaternary Structure of AtFolB1-ΔE, AtFolB2, and LeFolB1
The recombinant proteins were purified by chromatographic procedures to homogeneity as judged by SDS-PAGE and subjected to size exclusion chromatography to estimate native molecular mass. AtFolB1-ΔE, AtFolB2, and LeFolB1 had retention volumes close to that of the E. coli FolB protein, indicating that the plant enzymes, like their bacterial counterpart, are homooctamers (Table II).
Table II.
Size exclusion chromatography of native enzymes
| Enzyme | Subunit Mass | Retention Volume |
|---|---|---|
| kD | mL | |
| AtFolB1-ΔE | 14.1 | 198 |
| AtFolB2 | 13.6 | 198 |
| LeFolB1 | 14.6 | 199 |
| E. coli FolB | 13.6 | 206 |
Experiments were performed with a Superdex 200 column (2.6 × 60 cm) developed at a flow rate of 3 mL min−1 with 50 mm Tris-HCl, pH 7.8, containing 100 mm KCl.
Expression Patterns of Arabidopsis FolB Homologs
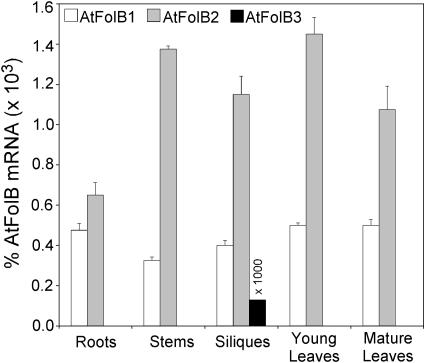
The expression patterns of the three Arabidopsis FolB genes were analyzed by real-time quantitative reverse transcription (RT)-PCR rather than northern blotting because EST data indicated that these genes are expressed at rather low levels, there being for all three genes together only five cognate sequences in the GenBank dbEST database of 1.9 × 105 Arabidopsis ESTs. The AtFolB1 and AtFolB2 mRNAs were expressed at low and roughly comparable levels in all organs examined, AtFolB2 being somewhat more abundant (Fig. 4). The mRNA levels measured for these two genes ranged from approximately 0.0003% to approximately 0.0015% of total RNA, which corresponds to an mRNA frequency of between 3 and 15 in 10,000, assuming mRNA to be approximately 1% of total RNA. In contrast, AtFolB3 mRNA was detected only in developing siliques, and at a level >1,000 times lower than the other two messages (Fig. 4).
Figure 4.
Quantification of AtFolB1, AtFolB2, and AtFolB3 mRNAs in Arabidopsis organs. Levels of mRNA were determined by real-time quantitative RT-PCR, using primers spanning two exons of each gene. Note that values for AtFolB3 are multiplied by 1,000. Roots were from hydroponically grown plants. Three independent RNA extracts were made of each organ, and triplicate mRNA determinations were made on each extract. Data are means of all nine determinations ±SE. Internal RNA standards were used to estimate recovery from RT-PCR of each sample; the data shown are corrected for recovery.
Phylogenetic Analysis
To analyze the diversity found among Arabidopsis FolB homologs, their sequences were aligned with 27 additional FolB-like sequences from tomato and 14 other angiosperms, and an unrooted phylogram was constructed using PHYLIP. The alignment (supplemental material 1, available at www.plantphysiol.org) revealed variation in the size and sequence of the N-terminal region, but not more than among the three Arabidopsis sequences (Fig. 2), which can therefore be considered representative.
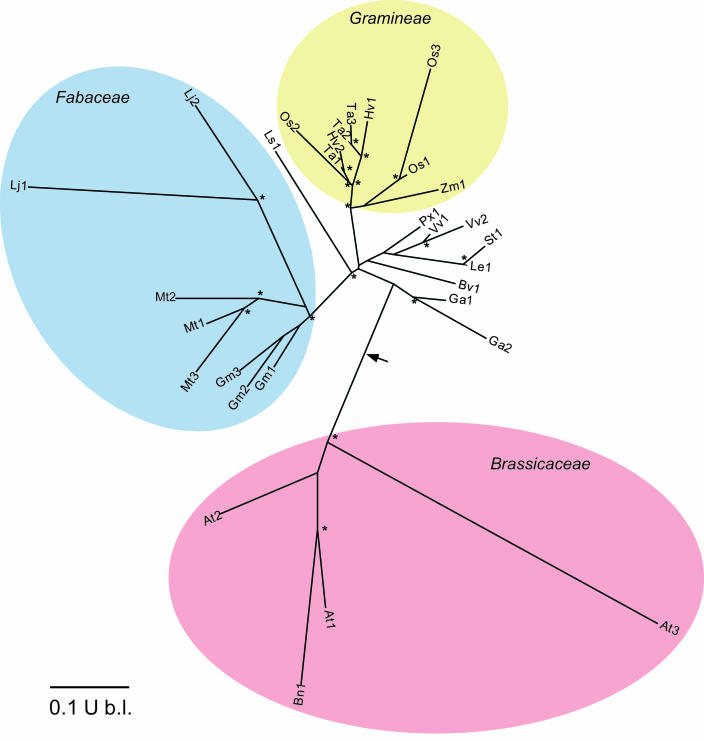
The phylogram (Fig. 5) illustrates four points. First, small FolB gene families occur in several species, of which most—like Arabidopsis—are diploids (rice [Oryza sativa], barley [Hordeum vulgare], Medicago truncatula, Lotus japonicus, and Gossypium arboreum), so that their multiple FolB genes are not simply homeologs (i.e. equivalent genes from different genomes). Second, sequences from members of the same families cluster together, indicating that the divergence occurred after separation of family lineages. In the legumes, the divergence apparently took place even later, in the lineages of individual genera. Third, there is a marked lack of correspondence between the broad FolB groups recovered and organismal phylogeny established using other genes (Soltis and Soltis, 2003; Soltis et al., 2000). Most notably, the grass family sequences cluster near those of eudicots, and the Brassicaceae sequences form an isolated clade. The isolation of the Brassicaceae becomes even more striking if the phylogram is rooted by including prokaryotic FolB sequences as an outgroup, for the root falls between the Brassicaceae cluster and all the others (Fig. 5, arrow). Last, the extent of divergence within the FolB genes of various taxa is unequal, the Arabidopsis genes being far more diverged than, for instance, the rice genes. Arabidopsis and rice can be validly compared because their FolB sequences are based on genomic as well as EST data, and we can thus be fairly sure that all their FolB homologs are included. This is not the case for the other 14 species, whose sequences come only from ESTs. These species may have other FolB genes that are not represented by ESTs. The divergences shown in the phylogram for these other species are thus minimum estimates.
Figure 5.
Unrooted phylogram generated using the neighbor-joining method and 1,000 bootstrap reiterations of 30 FolB-like sequences from 16 angiosperm species. Colored ovals show members of the Brassicaceae, Fabaceae, and Gramineae. Asterisks show branchpoints with bootstrap values >70%. The arrow indicates where the tree is rooted when prokaryotic FolB sequences are included as an outgroup in the analysis. Key to species: At, Arabidopsis thaliana; Bn, Brassica napus; Gm, Glycine max; Mt, Medicago truncatula; Lj, Lotus japonicus; Ls, Lactuca sativa; Os, Oryza sativa; Ta, Triticum aestivum; Hv, Hordeum vulgare; Zm, Zea mays; Px, Populus balsamifera × Populus deltoides; Vv, Vitis vinifera; St, Solanum tuberosum; Le, Lycopersicon esculentum; Bv, Beta vulgaris; Ga, Gossypium arboreum. The three sequences for rice (O. sativa) are based on genome data and at least one EST in each case. Sequences for other species are deduced from ESTs; all apparently encode complete proteins except for L. japonicus, where both sequences lack a few C-terminal residues.
DISCUSSION
This work, to our knowledge, is the first study of DHN aldolases from plants and is the first report of eukaryotic DHN aldolases that are not fused to other folate synthesis enzymes. Despite their relatively low overall amino acid identity with E. coli FolB and other prokaryotic DHN aldolases, the plant enzymes resemble their bacterial counterparts in quaternary structure (homooctamer) and in catalyzing aldolase and epimerase reactions using either DHN or DHM as substrate. The epimerase/aldolase activity ratios for both substrates are generally higher for the plant enzymes, where they vary from 0.11 to 1.3, compared to 0.007 to 0.16 for the E. coli and H. influenzae enzymes (Haußmann et al., 1998).
The metabolic significance of the epimerase reaction is unclear, but as this reaction occurs at a relatively high rate in vitro, it seems likely to be appreciable in planta. Consistent with this view, monapterin (the oxidized form of DHM) has been found in various plant tissues, typically at similar levels to neopterin (the oxidized form of DHN) (Kohashi, 1980; Kohashi et al., 1980). In this connection it is noteworthy that the reduced form of DHM, tetrahydromonapterin, is the main tetrahydropterin of E. coli (Ikemoto et al., 2002) and may act as a redox cofactor in bacteria (Guroff and Rhoads, 1969) in much the same way as tetrahydrobiopterin in animals. It is also noteworthy that plants have pterin-requiring enzymes (Yamamoto et al., 2001; Chandok et al., 2003), but the nature of the endogenous pterin cofactor remains an enigma because plants appear not to contain tetrahydrobiopterin (Kohashi et al., 1980). Tetrahydromonapterin, ultimately derived from the epimerase activity of DHN aldolase, is thus a plausible candidate for the missing plant cofactor.
In plants, the last four enzymes of folate synthesis—from HMDHP pyrophosphokinase onwards—occur in mitochondria and have typical mitochondrial targeting peptides (Rébeillé and Douce, 1999; Ravanel et al., 2001), whereas the enzyme that produces DHN triphosphate, GTP cyclohydrolase I, has no targeting peptide and is apparently cytosolic (Basset et al., 2002). As the Arabidopsis and tomato DHN aldolases also appear to lack targeting peptides, it is probable that DHN aldolase is likewise cytosolic. If so, it would follow that mitochondria import HMDHP.
The expression patterns of the three Arabidopsis genes indicate that AtFolB1 and AtFolB2 have a housekeeping function, as might be expected for enzymes in a pathway as vital as folate biosynthesis. AtFolB3 is different, being expressed only in siliques, and extremely weakly. Since the other two genes are strongly expressed in siliques it is conceivable that AtFolB3 is no longer physiologically important or maintained by selection, and that its gene has fallen virtually silent. The lack of detectable activity of recombinant AtFolB3 may be related to this. However, we cannot at this point exclude the possibilities that AtFolB3 is strongly expressed in a small subpopulation of the cells in siliques, or that it attacks a substrate other than DHN or DHM. We return to the latter idea below.
The Arabidopsis DHN aldolases share only moderate overall sequence identity (57%–73%) and vary most in the N-terminal region. Both of these features occur among the FolB homologs of other plants. Since DHN aldolase is the least conserved of the folate synthesis enzymes in bacteria (Bermingham and Derrick, 2002), the diversity among plant sequences may reflect rather relaxed structural requirements for activity in this enzyme. However, such diversity also suggests the possibility of functional differences. Some insight into this is provided by phylogenetic analysis, which indicates that DHN aldolase underwent a burst of evolution within the lineage leading to the Brassicaceae and within those leading to other families. The individual members of DHN aldolase families in living plant groups therefore presumably arose independently and quite recently, and are not orthologous to each other. While this pattern argues against ancient division of labor among these enzymes, it does not exclude such division having taken place at the family or genus level, as appears to have occurred among plant glucosyltransferases (Li et al., 2001; Lim et al., 2003). Many glucosyltransferases are specialized to attack different specific compounds but retain activity against certain core substrates. Given the dearth of information on plant pteridines, it is possible that something similar has taken place among DHN aldolases, i.e. that while retaining activity with DHN, they have evolved to act also on other pteridines that remain to be identified, and that may differ among plant taxa.
MATERIALS AND METHODS
Materials
DHN, DHM, and HMDHP were purchased from Schircks Laboratories (Jona, Switzerland). Oligonucleotides were from MWG (High Point, NC) or Interactiva (Ulm, Germany). Vent DNA polymerase was from New England Biolabs (Beverly, MA), T4 DNA ligase and SuperScript II reverse transcriptase were from Invitrogen (Carlsbad, CA), Pfu polymerase was from Stratagene (La Jolla, CA), and Taq polymerase was from Finnzyme (Epsoo, Finland). DNA fragments were purified with QIAquick PCR Purification Kits (Qiagen, Valencia, CA). A Nucleosil RP18 HPLC column (4 × 250 mm) was from Schambeck (Bad Honnef, Germany). Superdex 200, Q Sepharose Fast Flow, and Red Sepharose CL-6B were from Amersham Biosciences (Freiburg, Germany).
Plants and Growing Conditions
Arabidopsis plants (ecotype Columbia) were grown at 23°C in 12-h days (photosynthetic photon flux density 80 μE m−2 s−1) in potting soil and irrigated with water. Whole rosettes were harvested at the four-to-six-leaf stage (young leaves) and at the start of bolting (mature leaves). Stems and developing siliques were collected before flowering had ceased. When roots were required, plants were grown in hydroponic culture as described by Gibeaut et al. (1997).
cDNA Cloning and Sequence Analysis
Primers used to amplify cDNAs are listed in Table III. All cDNAs were verified by sequencing. To clone AtFolB2, total Arabidopsis RNA was isolated using RNeasy Plant Mini Kits (Qiagen, Valencia, CA) and reverse-transcribed using an oligo(dT) primer. The AtFolB2 open reading frame (ORF) was then amplified and cloned into pGEM-T Easy (Promega, Madison, WI). Using this construct as template, the ORF was reamplified using primers incorporating NdeI and XhoI sites (Table III); the amplicon was digested with these enzymes and cloned between the matching sites of the expression vector pET43.1a. The AtFolB1 ORF was amplified from a cDNA template. The amplicon was digested with EcoRI and BamHI and ligated between the matching sites of the expression vector pNCO113, yielding the plasmid pNCO-AtFolB1-WT. Plasmid pNCO-AtFolB1-ΔE (without N-terminal region) was constructed the same way except that a different forward primer was used (Table III). For AtFolB3, a full-length EST (GenBank BE529269) was obtained from the Arabidopsis Biological Resource Center. The ORF was amplified by PCR, and the amplicon was cloned into pNCO113 as above, yielding the plasmid pNCO-AtFolB3-WT. This was used as a template in a second PCR using a different forward primer (Table III) to remove the N-terminal region. The amplicon was cloned into pNCO113 as above, yielding pNCO-AtFolB3-ΔE. To clone LeFolB1, total RNA was prepared from pooled unripe and ripe tomato fruits as described (Hamilton et al., 1990), and mRNA was purified using the PolyATtract system (Promega). Single-stranded cDNA was prepared by RT and used as a template to amplify a 350-bp fragment of LeFolB1 using the primers 5′-GGAGACAACTTTCG-3′ (forward) and 5′-ATCTCGACACCYAAGTARTC-3′ (reverse), which were based on a potato DHN aldolase EST (GenBank BQ512667). The amplicon was ligated into pGEM-T Easy. The missing 5′ and 3′ ends were obtained using RACE systems from Invitrogen, and the following nested primers: for 5′-RACE, 5′-CCACATGAGGCTTTCCAACTT-3′ and 5′-CGAACAGCAGATACCTCTGGA-3′; for 3′-RACE, 5′-GGTGAAGCAAGAGGAAAGGAA-3′ and 5′-GGTCAGAAGTTCCTGGTAGAT-3′. The resulting fragments were cloned into pGEM-T Easy and sequenced. The full-length LeFolB1 cDNA was then amplified from the cDNA template using the primers shown in Table III. The amplicon was digested with NdeI and XhoI and cloned into pET-43.1a. Protein sequences were aligned using ClustalW. Phylogenetic analyses were made using PHYLIP at the Institut Pasteur server (bioweb.pasteur.fr).
Table III.
Sequences of primers used to clone Arabidopsis and tomato FolB cDNAs
| Construct | Forward Primer | Reverse Primer |
|---|---|---|
| AtFolB1-WT | ATAATAGAATTCATTAAAGAGGAGAAATTAACCATGCATAGCTCACTGGAGACCACG | }TATTATGGATCCTTAGTTCTTTGAACTAGTGTTTCGCTGTC |
| AtFolB1-ΔE | ATAATAGAATTCATTAAAGAGGAGAAATTAACCATGGACAAACTGATACTAAAAGGGTTG | |
| AtFolB2 | ATGGAGAAAGACATGGCAAT (for pGEM-T Easy) | TTATTCAGTTGCGCGATCTC (for pGEM-T Easy) |
| GGGAAACATATGGAGAAAGACATGGCAAT (for pET43.1a) | CCGCTCGAGTTATTCAGTTGCGCGATCTC (for pET43.1a) | |
| AtFolB3-WT | ATAATAGAATTCATTAAAGAGGAGAAATTAACTATGCATTCTTCTCTTAAGACGATG | }TATTATGGATCCTTAGTGATTTCGCTTCCTGAAAAG |
| AtFolB3-ΔE | ATAATAGAATTCATTAAAGAGGAGAAATTAACT-ATGGAGGATAAGCTTATACTGAGAGGGTTG | |
| LeFolB1 | CCAACCCATATGGACATGCCAAAAGGAGA | CCGACTCGAGCTAGTTTTGAACATCAAGA |
Primer sequences are in the 5′ → 3′ direction.
Protein Expression in Escherichia coli
The plasmids pNCO-AtFolB1-WT, pNCO-AtFolB1-ΔE, pNCO-AtFolB3-WT, and pNCO-AtFolB3-ΔE were transformed into E. coli XL-1-Blue cells (Bullock et al., 1987). Transformants were selected on Luria-Bertani plates supplemented with ampicillin (170 μg mL−1). The pNCO113-type plasmids reisolated from XL-1-Blue cells were transformed into E. coli M15 [pREP4] cells (Stüber et al., 1990) carrying the pREP4 repressor plasmid, which directs the synthesis of lac repressor protein. Kanamycin (15 μg mL−1) and ampicillin (170 μg mL−1) were added to secure the maintenance of both plasmids. Plasmids pNCO-AtFolB3-WT and pNCO-AtFolB3-ΔE reisolated from XL-1-Blue cells were also transformed into E. coli BL21-CodonPlus (DE3)/pRIL cells (Stratagene). Chloramphenicol (15 μg mL−1) and ampicillin (170 μg mL−1) were added to secure the maintenance of both plasmids. The pET-43.1a type plasmids coding for AtFolB2 and LeFolB1 were transformed into E. coli BL21(DE3) cells, and ampicillin (150 μg mL−1) was used for selection. Recombinant E. coli strains were grown in selective LB medium at 37°C with shaking overnight. Erlenmeyer flasks containing 500 mL of medium were then inoculated at a ratio of 1:50 and were incubated at 37°C with shaking until A600 reached 0.6, at which point isopropylthiogalactoside was added to a final concentration of 2 mm, and incubation was continued for 5 h. The cells were harvested by centrifugation, washed in 0.9% NaCl, and stored at −20°C. SDS-PAGE was performed with 16% polyacrylamide gels by published procedures (Laemmli, 1970). Molecular weight markers were from Sigma (Munich, Germany). Recombinant protein expression was estimated after Coomassie Blue staining.
Protein Purification
Recombinant E. coli FolB was purified as described previously (Haußmann et al., 1998). The endogenous DHN aldolase activity of the E. coli host strains used was negligible (approximately 1%) compared to that contributed by the recombinant plant enzymes, which were purified as follows. Frozen cell mass (6 g) was thawed in 35 mL of 50 mm Tris-HCl, pH 7.8. The suspension was sonicated and cleared by centrifugation. The supernatant was applied to a Q Sepharose Fast Flow column (90 mL) equilibrated with 50 mm Tris-HCl, pH 7.8. The column was developed with a linear gradient of 0–1.0 m KCl in 50 mm Tris-HCl, pH 7.8 (flow rate 1 mL min−1, total volume 500 mL). Active fractions were combined, concentrated by ultrafiltration, and applied to a Superdex 200 column (2.6 × 60 cm), which was developed with 50 mm Tris-HCl, pH 7.8, containing 100 mm KCl (flow rate 3 mL min−1). Active fractions were combined and loaded onto a column of Red Sepharose CL-6B (11 mL) equilibrated with 50 mm Tris-HCl, pH 7.8. The column was developed with a linear gradient of 0–1.0 m KCl (total volume 80 mL). Fractions were combined and concentrated by ultrafiltration. Protein concentration was monitored photometrically using the following absorbance coefficients: AtFolB1-ΔE, 9,530 m−1 cm−1; AtFolB2, 16,500 m−1 cm−1; LeFolB1, 13,490 m−1 cm−1 (280 nm). The enzymes were stored at −70°C.
Enzyme Assay
The assay used was based on published methods (Mathis and Brown, 1980; Haußmann et al., 1998). Assays contained 50 mm Tris-HCl, pH 7.8, 5 mm dithiothreitol, 200 μm DHN, and protein in a total volume of 150 μL. Incubation was at 37°C for 30 min. The reaction was stopped by adding 50 μL of 1 m HCl containing 1% (w/v) I2 and 2% (w/v) KI. After 5 min at room temperature, excess I2 was reduced by adding 20 μL of 3% (w/v) ascorbic acid. The samples were analyzed by reverse-phase HPLC using a Nucleosil RP18 column (4.6 × 250 mm) eluted with 0.5% (w/v) H3BO3, pH 4.7 (flow rate 2 mL min−1). The effluent was monitored fluorometrically (excitation 365 nm, emission 446 nm) using ChromGate HPLC software (Knauer, Berlin, Germany).
Steady-State Kinetic Analysis
Steady-state kinetic experiments were performed at 30°C. Reaction mixtures contained 50 mm Tris-HCl, pH 7.8, 5 mm dithiothreitol, a saturating concentration of DHN or DHM, and protein in a total volume of 2 mL. The reaction was started by adding enzyme. Aliquots (150 μL) were retrieved at 15-s intervals; the reaction was stopped and the products were analyzed as above. Vmax was determined by least square fit using the program Origin (Microcal, Northampton, MA).
Real-Time Quantitative RT-PCR
Total RNA was extracted from three samples of each tissue using RNeasy Plant Mini Kits, and treated with DNase (DNA-free Kit, Ambion, Austin, TX). Real-time quantitative RT-PCR was performed on 250 ng of RNA in 25-μL reactions using Taq-Man One-Step RT-PCR Master Mix Reagents (Applied Biosystems, Foster City, CA) and an Applied Biosystems GeneAmp 5700 sequence-detection system. The primers and Taq-Man probes (designed with Applied Biosystems Primer Express software) are described in Table IV. The fluorescent reporter dye 6-carboxyfluorescein and the quencher dye 6-carboxytetramethylrhodamine were bonded to the probes’ 5′ and 3′ ends, respectively. The amplicon length was 72 to 77 bp. For each gene, one of the primers spanned two exons, to avoid amplifying contaminating genomic DNA, and controls without reverse transcriptase were included to verify that no amplification of contaminating DNA was detectable. RT-PCR conditions were 48°C for 30 min, 95°C for 10 min, followed by 40 cycles of 95°C for 15 s and 60°C for 1 min. RNA standards were synthesized from cDNAs (see below); the standard curves were linear from 1.5 × 10−19 to 7.5 × 10−16 g. An internal standard of each RNA was added to each sample before the RT-PCR reaction to estimate recovery; recoveries were 54%–94%, except for AtFolB1 RNA in roots, where the value was 21%. A Ct threshold value was determined from amplification curves by selecting an optimal ΔRn (emission of the reporter dye over starting background fluorescence) in the exponential part of the plots.
Table IV.
Sequences of primers and probes for quantitative real-time RT-PCR
| Enzyme | Forward Primer | Reverse Primer | Probe |
|---|---|---|---|
| AtFolB1 | GATGCGTGGGTGAGTCTGAA | GCTGAAAATGTCGACGTAGCTAATT | AGGCTGGTGAATCAGACAACTTAGAAGATA |
| AtFolB2 | CAAAAAGGCTGGTCTATCAGACAA | TTCCTTTGCCACGCTGTAAA | TAGCTGATTCTGTCAGCTATGTCGAC |
| AtFolB3 | GCTTTGCTGATACTTTTAGGCTAGTG | CATCCGCAACCGTCTCGTA | AGAAAATTGTTGAAGGGCCACCAAGAAACCT |
Primer and probe sequences are in the 5′ → 3′ direction.
RNA Standard Synthesis and Quantification
The in vitro transcription (MAXIscript, Ambion) used 5.4 μm [5,6-3H]UTP or 3.1 μm [α-32P]UTP as limiting nucleotide. The efficiency of incorporation of [5,6-3H]UTP into RNA was 44%–89% as evaluated by TCA precipitation of an aliquot of the in vitro transcription product. The 3H- and 32P-labeled RNAs were run on a 5% (w/v) polyacrylamide gel (16 × 20 cm) containing 8 m urea. Each full-length [32P]RNA was localized by autoradiography, thus indicating the position of the corresponding [3H]RNA. The [3H]RNA band was excised, eluted overnight at 37°C , and quantified by scintillation counting. The overall [3H]RNA yield was 5%–15%.
Sequence data from this article have been deposited with the EMBL/GenBank data libraries under accession number AY422466.
Supplementary Material
Acknowledgments
We thank Dr. Douglas E. Soltis for advice on phylogenetic analysis and Michael J. Ziemak for preparing tomato RNA.
This work was supported in part by the National Science Foundation (grant no. MCB–0129944); by the Deutsche Forschungsgemeinschaft, the Fonds der Chemischen Industrie, and the Hans-Fischer-Gesellschaft; by an endowment from the C.V. Griffin, Sr. Foundation; and by the Florida Agricultural Experiment Station. Journal Series No. R-09930.
The online version of this article contains Web-only data.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.103.038430.
References
- Basset GJC, Quinlivan EP, Ravanel S, Rébeillé F, Nichols BP, Shinozaki K, Seki M, Adams-Phillips LC, Giovannoni JJ, Gregory JF III (2004) Folate synthesis in plants: the p-aminobenzoate branch is initiated by a bifunctional PabA-PabB protein that is targeted to plastids. Proc Natl Acad Sci USA 101: 1496–1501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basset G, Quinlivan EP, Ziemak MJ, Diaz De La Garza R, Fischer M, Schiffmann S, Bacher A, Gregory JF III, Hanson AD (2002) Folate synthesis in plants: the first step of the pterin branch is mediated by a unique bimodular GTP cyclohydrolase I. Proc Natl Acad Sci USA 99: 12489–12494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bermingham A, Derrick JP (2002) The folic acid biosynthesis pathway in bacteria: evaluation of potential for antibacterial drug discovery. Bioessays 24: 637–648 [DOI] [PubMed] [Google Scholar]
- Bouis HE (2002) Plant breeding: a new tool for fighting micronutrient malnutrition. J Nutr 132: 491S–494S [DOI] [PubMed] [Google Scholar]
- Bullock WO, Fernandez JM, Short JM (1987) XL1-Blue: a high efficiency plasmid transforming recA Escherichia coli strain with β-galactosidase selection. BioTechniques 5: 376–379 [Google Scholar]
- Chandok MR, Ytterberg AJ, van Wijk KJ, Klessig DF (2003) The pathogen-inducible nitric oxide synthase (iNOS) in plants is a variant of the P protein of the glycine decarboxylase complex. Cell 113: 469–482 [DOI] [PubMed] [Google Scholar]
- Cossins EA, Chen L (1997) Folates and one-carbon metabolism in plants and fungi. Phytochemistry 45: 437–452 [DOI] [PubMed] [Google Scholar]
- de Bree A, van Dusseldorp M, Brouwer IA, van het Hof KH, Steegers-Theunissen RPM (1997) Folate intake in Europe: recommended, actual and desired intake. Eur J Clin Nutr 51: 643–660 [DOI] [PubMed] [Google Scholar]
- De Saizieu A, Vankan P, van Loon AP (1995) Enzymic characterization of Bacillus subtilis GTP cyclohydrolase I. Evidence for a chemical dephosphorylation of dihydroneopterin triphosphate. Biochem J 306: 371–377 [PMC free article] [PubMed] [Google Scholar]
- Emanuelsson O, von Heijne G (2001) Prediction of organellar targeting signals. Biochim Biophys Acta 1541: 114–119 [DOI] [PubMed] [Google Scholar]
- Forrest HS, Van Baalen C (1970) Microbiology of unconjugated pteridines. Annu Rev Microbiol 24: 91–108 [DOI] [PubMed] [Google Scholar]
- Gibeaut DM, Hulett J, Cramer GR, Seemann JR (1997) Maximal biomass of Arabidopsis thaliana using a simple, low-maintenance hydroponic method and favorable environmental conditions. Plant Physiol 115: 317–319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green JC, Nichols BP, Matthews RG (1996) Folate biosynthesis, reduction, and polyglutamylation. In FC Neidhardt, ed, Escherichia coli and Salmonella: Cellular and Molecular Biology, Vol 1. ASM Press, Washington, DC, pp 665–673
- Guroff G, Rhoads CA (1969) Phenylalanine hydroxylation by Pseudomonas species (ATCC 11299a). Nature of the cofactor. J Biol Chem 244: 142–146 [PubMed] [Google Scholar]
- Hamilton AJ, Lycett GW, Grierson D (1990) Antisense gene that inhibits synthesis of the hormone ethylene in transgenic plants. Nature 346: 284–287 [Google Scholar]
- Hanson AD, Gregory JF III (2002) Synthesis and turnover of folates in plants. Curr Opin Plant Biol 5: 244–249 [DOI] [PubMed] [Google Scholar]
- Haußmann C, Rohdich F, Schmidt E, Bacher A, Richter G (1998) Biosynthesis of pteridines in Escherichia coli. Structural and mechanistic similarity of dihydroneopterin-triphosphate epimerase and dihydroneopterin aldolase. J Biol Chem 273: 17418–17424 [DOI] [PubMed] [Google Scholar]
- Hennig M, D'Arcy A, Hampele IC, Page MG, Oefner C, Dale GE (1998) Crystal structure and reaction mechanism of 7,8-dihydroneopterin aldolase from Staphylococcus aureus. Nat Struct Biol 5: 357–362 [DOI] [PubMed] [Google Scholar]
- Ikemoto K, Sugimoto T, Murata S, Tazawa M, Nomura T, Ichinose H, Nagatsu T (2002) (6R)-5,6,7,8-tetrahydro-L-monapterin from Escherichia coli, a novel natural unconjugated tetrahydropterin. Biol Chem 383: 325–330 [DOI] [PubMed] [Google Scholar]
- Iwai K, Bunno M, Kobashi M, Suzuki T (1976) Isolation and characterization of 6-hydroxymethylpterin as the Crithidia growth-promoting factor from spinach chloroplasts. Biochim Biophys Acta 444: 618–622 [DOI] [PubMed] [Google Scholar]
- Kang D, Kim S, Yim J (1998) Biosynthetic enzymes of tetrahydrolimipterin from green sulfur bacterium Chlorobium limicola. Pteridines 9: 69–84 [Google Scholar]
- Kobayashi K, Forrest HS, el-Emary M (1967) Isolation of 6-hydroxymethyllumazine (phosphodoxin?) and two other lumazine derivatives from spinach, and their synthesis. Arch Biochem Biophys 121: 220–223 [DOI] [PubMed] [Google Scholar]
- Kohashi M (1980) Isolation of six unconjugated pteridines from soybean (Glycine max L. Tsurunoko) seeds. J Biochem (Tokyo) 87: 1581–1586 [DOI] [PubMed] [Google Scholar]
- Kohashi M, Tomita K, Iwai K (1980) Analysis of unconjugated pterins in food resources and human urine. Agric Biol Chem 44: 2089–2094 [Google Scholar]
- Laemmli UK (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227: 680–685 [DOI] [PubMed] [Google Scholar]
- Lee SW, Lee HW, Chung HJ, Kim YA, Kim YJ, Hahn Y, Chung JH, Park YS (1999) Identification of the genes encoding enzymes involved in the early biosynthetic pathway of pteridines in Synechocystis sp. PCC 6803. FEMS Microbiol Lett 176: 169–176 [DOI] [PubMed] [Google Scholar]
- Li Y, Baldauf S, Lim EK, Bowles DJ (2001) Phylogenetic analysis of the UDP-glycosyltransferase multigene family of Arabidopsis thaliana. J Biol Chem 276: 4338–4343 [DOI] [PubMed] [Google Scholar]
- Lim EK, Baldauf S, Li Y, Elias L, Worrall D, Spencer SP, Jackson RG, Taguchi G, Ross J, Bowles DJ (2003) Evolution of substrate recognition across a multigene family of glycosyltransferases in Arabidopsis. Glycobiology 3: 139–145 [DOI] [PubMed] [Google Scholar]
- Lopez P, Lacks SA (1993) A bifunctional protein in the folate biosynthetic pathway of Streptococcus pneumoniae with dihydroneopterin aldolase and hydroxymethyldihydropterin pyrophosphokinase activities. J Bacteriol 175: 2214–2220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathis JB, Brown GM (1980) Dihydroneopterin aldolase from Escherichia coli. Methods Enzymol 66: 556–560 [DOI] [PubMed] [Google Scholar]
- Mendel RR, Hänsch R (2002) Molybdoenzymes and molybdenum cofactor in plants. J Exp Bot 53: 1689–1698 [DOI] [PubMed] [Google Scholar]
- Ravanel S, Cherest H, Jabrin S, Grunwald D, Surdin-Kerjan Y, Douce R, Rébeillé F (2001) Tetrahydrofolate biosynthesis in plants: molecular and functional characterization of dihydrofolate synthetase and three isoforms of folylpolyglutamate synthetase in Arabidopsis thaliana. Proc Natl Acad Sci USA 98: 15360–15365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rébeillé F, Douce R (1999) Folate synthesis and compartmentation in higher plants. In NJ Kruger, SA Hill, RG Ratcliffe, eds, Regulation of Primary Metabolic Pathways in Plants. Kluwer, Dordrecht, The Netherlands, pp 53–99
- Scott J, Rébeillé F, Fletcher J (2000) Folic acid and folates: the feasibility for nutritional enhancement in plant foods. J Sci Food Agric 80: 795–824 [Google Scholar]
- Soltis DE, Soltis PS (2003) The role of phylogenetics in comparative genetics. Plant Physiol 132: 1790–1800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soltis DE, Soltis PS, Chase MW, Mort ME, Albach DC, Zanis M, Savolainen V, Hahn WH, Hoot SB, Fay MF, et al. (2000) Angiosperm phylogeny inferred from a combined data set of 18S rDNA, rbcL and atpB sequences. Bot J Linn Soc 133: 381–461 [Google Scholar]
- Stüber D, Matile H, Garotta G (1990) System for high-level production in Escherichia coli and rapid purification of recombinant proteins: application to epitope mapping, preparation of antibodies and structure-function analysis. In I Lefkovits, P Pernis, eds, Immunological Methods, Vol 4. Academic Press, New York, pp 121–152
- Sugiura K, Goto M (1966) Isolation and identification of 6-hydroxymethyllumazine from spinach. J Biochem (Tokyo) 60: 335–337 [DOI] [PubMed] [Google Scholar]
- Suzuki Y, Brown GM (1974) The biosynthesis of folic acid. XII. Purification and properties of dihydroneopterin triphosphate pyrophosphohydrolase. J Biol Chem 249: 2405–2410 [PubMed] [Google Scholar]
- Thöny B, Auerbach G, Blau N (2000) Tetrahydrobiopterin biosynthesis, regeneration and functions. Biochem J 347: 1–16 [PMC free article] [PubMed] [Google Scholar]
- Volpe F, Ballantine SP, Delves CJ (1993) The multifunctional folic acid synthesis fas gene of Pneumocystis carinii encodes dihydroneopterin aldolase, hydroxymethyldihydropterin pyrophosphokinase and dihydropteroate synthase. Eur J Biochem 216: 449–458 [DOI] [PubMed] [Google Scholar]
- Yamamoto K, Kobayashi N, Yoshitama K, Teramoto S, Komamine A (2001) Isolation and purification of tyrosine hydroxylase from callus cultures of Portulaca grandiflora. Plant Cell Physiol 42: 969–975 [DOI] [PubMed] [Google Scholar]
- Yoshida T, Akino M (1980) Occurrence of unconjugated pterins in the higher plant, Stizolobium hassjoo. Experientia 36: 639–640 [Google Scholar]
- Ziegler I (2003) The pteridine pathway in zebrafish: regulation and specification during the determination of neural crest cell-fate. Pigment Cell Res 16: 172–182 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.