Abstract
Semantic web technologies offer an approach to data integration and sharing, even for resources developed independently or broadly distributed across the web. This approach is particularly suitable for scientific domains that profit from large amounts of data that reside in the public domain and that have to be exploited in combination. Translational medicine is such a domain, which in addition has to integrate private data from the clinical domain with proprietary data from the pharmaceutical domain. In this survey, we present the results of our analysis of translational medicine solutions that follow a semantic web approach. We assessed these solutions in terms of their target medical use case; the resources covered to achieve their objectives; and their use of existing semantic web resources for the purposes of data sharing, data interoperability and knowledge discovery. The semantic web technologies seem to fulfill their role in facilitating the integration and exploration of data from disparate sources, but it is also clear that simply using them is not enough. It is fundamental to reuse resources, to define mappings between resources, to share data and knowledge. All these aspects allow the instantiation of translational medicine at the semantic web-scale, thus resulting in a network of solutions that can share resources for a faster transfer of new scientific results into the clinical practice. The envisioned network of translational medicine solutions is on its way, but it still requires resolving the challenges of sharing protected data and of integrating semantic-driven technologies into the clinical practice.
Keywords: semantic web, translational medicine, data integration, data sharing, data interoperability, knowledge discovery
INTRODUCTION
Biomedical research has evolved into a data-intensive science, where prodigious amounts of data can be collected from disparate resources at any time [1]. However, the value of data can only be leveraged through its analysis, which ultimately results in the acquisition of knowledge. In domains such as translational medicine, where multiple types of data are involved, often from different sources and in different formats, data integration and interoperability are key requirements for an efficient data analysis.
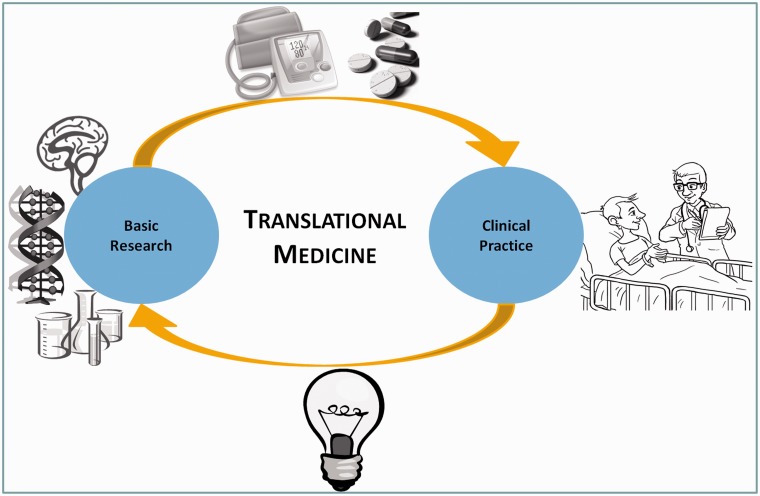
Translational medicine focuses on the improvement of human health by bridging the gap between basic science research and clinical practice [2–4]. This bridging is done at two distinct levels: at the level of basic science research, translating it into new devices or treatments (‘from the bench to the bedside’); and at the level of clinical practice, transferring the new treatments into the daily routine (Figure 1) [4,5]. Additionally, knowledge in translational medicine can also flow in the contrary direction, resulting in the initiation of new basic research based on the clinical observations of a disease development. Included in the first bridging level is genomic medicine, which consists in exploring the molecular genetics knowledge of diseases and translating it into personalized treatments with more beneficial treatment responses and with reduced undesired effects [6]. For example, a clinician may analyze a patient’s mutations to explain observed drug side effects or may retrieve the list of biomarkers and their functions that have been associated with a specific cancer type.
Figure 1:
Knowledge workflow in translational medicine. Translational medicine improves the knowledge on human diseases by translating basic science research results into new exams, devices and treatments, which are then incorporated into the clinical practice. It also explores the knowledge collected during patient care to identify new research topics and topics that need further research.
It is unquestionable that translational medicine is a multidisciplinary research domain that relies both on public and protected data. Public data include resources such as medical guidelines, scientific literature and biomedical databases, whereas protected data are composed of private patient data and proprietary data from pharmaceutical and publishing companies. Translational medicine thus requires appropriate technologies for the interpretation of distributed and disparate data resources, and it is easy to conceive that such a large scale endeavor will require a versatile infrastructure that preserves data semantics at all integration levels.
Using the semantic web for data integration
The need for data integration and data interoperability has a long-standing history. The Committee on Models for Biomedical Research proposed in 1985 a structured and integrated view of biology to cope with the available data [7]. Ten years later, in 1995, Davidson et al. questioned the feasibility of data integration, since the resulting data structure has to follow changes in the data itself and individual research groups fail to comply with the integration structure [8]. In 2007, the challenges identified for data integration in genomic medicine were the lack of clinical data sources; the privacy issues linked to clinical data; the inherent complexity of medical records; and finally, the lack of data representation standards in the clinical domain [6]. These selected examples clearly show that data integration remains an open research area and that its complexity escalates with the increase in number of heterogeneous domains to be integrated.
The World Wide Web is the key information channel for the communication of public data, particularly for the scientific community, since it allows the fast publication of methods, results and opinions, and it is easily reached by virtually anyone anywhere. This information channel fulfills the requirements for efficient data exchange between scientific communities and data repositories, and thus should also be explored in translational medicine for optimal progress. However, its usefulness in this context is counterbalanced by the lack of data standards across domains, of explicit data representations, and of interoperability of the data resources, which hinder the sharing of data between the biomedical and the clinical domains [9].
Tim Berners-Lee et al. proposed the vision of the semantic web, where the web of documents is replaced by the web of data, thus allowing the manipulation of data over disparate domains and solving most of the problems previously stated for data integration [10]. The manipulation of data is achieved by substituting the links connecting web pages (i.e. the documents) with links connecting the data elements themselves and adding semantics to them all. The data elements in the web thus represent real-world entities and the links between data elements embody the logical relations between those entities. When independent applications share this representation of the reality, interoperability and effective data integration across knowledge domains are achieved. The semantic web thus becomes the framework for data integration at the web-scale, independent from the knowledge domain and focused on the semantics and context of the data. The result is a network of linked data that can be exploited by computers: by following the links between data elements, jumping from data set to data set; by querying the whole network, and thus providing an answer based on otherwise independent data sets; and by reasoning over the data, based on its formal representation, thus identifying new implicit connections between data elements [11].
The semantic web reaches beyond data integration toward data sharing across institutions, and makes data integration and interoperability a standard feature instead of a requirement. If built on this infrastructure, many of the technical challenges faced by translational medicine are thus prevented. However, it is important to bear in mind that, as happens with other technologies, the semantic web is inherently constrained by the complexity of the domain of knowledge.
In this work, we analyzed how the semantic web and its technologies have been used in the translational medicine domain. In particular, we analyzed which technologies are more often exploited and how they are used. For that purpose, we analyzed 11 noncommercial systems integrating genetic and medical data, developed from 2007 to 2013. These systems are presented in terms of the medical context in which they were developed, the resources that were embedded, their compliance with the semantic web principles and, finally, the extent to which the new knowledge can reach the everyday clinical practice.
SEMANTIC WEB RESOURCES FOR TRANSLATIONAL MEDICINE
Combining resources from public and private repositories, either in an open infrastructure or in a clinical environment, requires data representation standards, semantic normalization and ultimately data sharing (with appropriate access control policies). The infrastructure of the World Wide Web can be exploited to this end, but it has to be focused on the semantic representation of data and on the interoperability of data, or, in other words, it has to become a semantic web.
Technological standards in the semantic web
Over the past decade, the semantic web community, and in particular the World Wide Web Consortium (W3C), has been developing a set of core technologies to realize the vision of the semantic web. Some of these technologies have since become de facto standards, and have brought the semantic web to life [12,13].
The Resource Description Framework (RDF) is a standard language for data representation and interchange on the Web [14]. It uses the Universal Resource Identifier (URI) to identify each data element represented [15]. The basic structure of RDF is the triple, a statement composed of a subject connected with an object through a predicate, similar to narrative statements in English (e.g. ‘HomoSapiens isA mammal.’, ‘Dopamin treats ParkinsonSyndrome.’). Since either of these elements can be part of different statements, data in RDF are best visualized through a directed graph, where the nodes represent the subjects and objects, and the arcs represent the predicates (or relations).
The RDB to RDF Mapping Language (R2RML, in which RDB stands for relational database) is a language that expresses customized mappings from relational databases to RDF data sets [16]. As such, it assists in the integration of data from relational databases by exporting it in RDF.
Owing to its basic and simple format, RDF restricts the representation of data to low levels of expressiveness (e.g. it does not allow the union of concepts, the definition of hierarchic relations between concepts or the definition of cardinality in nonhierarchical relations). To overcome this limitation, two other technologies have been proposed: the RDF Schema (RDFS), a specification language for data properties based on RDF; and the Web Ontology Language (OWL), a language to formally define semantics, which also enables reasoning based on Description Logics [17–19]. Both formal languages extend RDF and enable the inference of new knowledge. As a result, knowledge can be shared and at the same time assessed for formal semantic consistency.
SPARQL, a self-referencing acronym for SPARQL Protocol and RDF Query Language, is a query language to access RDF data [20]. Since RDF data may be distributed over disparate data sources (including data stores exporting RDF from non-RDF relational databases), SPARQL has to retrieve data from all these resources. Due to the graph structure of RDF, SPARQL queries are transformed into graph pattern searches that rely only on the knowledge about the relations between concepts but not on a particular data model. SPARQL is also able to query RDFS and OWL provided that the graph pattern matching of the SPARQL query is defined with semantic entailment relations instead of the explicit graph structures [21]. Although other query languages exist for RDF (e.g. RDQL [22]), the availability of a SPARQL end point (i.e. an interface that provides access to a data set through SPARQL queries) guarantees the independence from software and implementation specifications.
Although necessary, these standards are not sufficient for the implementation of the web of data. This can be achieved with the representation of domain knowledge with ontologies and with the semantic characterization of links between resources.
Domain knowledge representation
Semantic interoperability is a key requirement in the realization of the semantic web and it is mainly achieved through the generation of resources that reliably represent the abstraction of real-world objects and their interactions. These representations exist in the form of ontologies and controlled vocabularies in general. An ontology is ‘an explicit specification of a conceptualization’ that provides a means to formally describe domain knowledge in a structured manner [23]. If an ontology is accepted as a reference by the community (e.g. the Gene Ontology and the SNOMED-CT), its representation of the reality becomes a standard, and data integration is facilitated [24,25]. This is true even if different abstraction levels are provided from unrelated data sets, since the hierarchical structure of ontologies supports the identification of a common ancestor for any two related concepts, by traversing the ontology graph [26].
The representation of ontologies in RDFS or OWL provides additional advantages, namely, novel interpretations of the existing data against the ontological knowledge enabled by the mapping of data elements in RDF representation (‘instances’) to the ontological concepts (‘classes’ or ‘types’); and more detailed semantic comparisons of concepts that exploit the expressiveness of these formats [27].
The Open Biomedical Ontologies (OBO) format also exists for ontology representation, although it is not a standard semantic web technology [28]. Due to its popularity in the health care and life sciences domains, extensive work has been done in the conversion of ontologies in this format to OWL [29–31].
Linking data
Mappings between resources are another key element in the semantic web, enabling interlinked structured data according to the principles defined by Tim Berners-Lee: (i) use Uniform Resource Identifier(s) (URIs) as names for things; (ii) use resolvable URIs (e.g. based on the HTTP protocol) so that those names can be looked up (either by people or machines); (iii) provide useful information for lookup through the URI, using the standards (e.g. RDF, SPARQL); and (iv) include links to other URIs, so that they can discover more things [32–34]. The URI can then be used to define any real-world entity (or ‘thing’), be it an object or an abstract concept [35].
Examples of real-world entities in the biomedical domain are diseases, drugs, facts related to genes and protein functions, patient symptoms, biological measurements and family history. Ideally, each individual entity should have only one URI, so that every application points to the same source, regardless of its domain. This means that if the entity is altered in the original source, all applications pointing to it will be automatically updated. Additionally, the correct definition of URIs ensures that mappings between resources do not lead to semantic inconsistencies.
The links established between resources can be defined both at instance-level (i.e. between data instances) and at schema-level (i.e. between concepts or properties defined in different vocabularies). Heath and Bizer state the existence of three important types of instance-level links: ‘Vocabulary Links’ that map an instance to the definition of the vocabulary concept used to represent it; ‘Identity Links’, used to indicate when two instances with different URIs refer to the same real-world entity (defined in OWL through the property ‘sameAs’); and ‘Relationship Links’ that map an instance in a data set to related things in other data sets (e.g. people to places) [36]. There are also three types of links that can be defined at schema-level: ‘Equivalence Links’ (similar to the identity links at instance-level) used to indicate when two concepts are equivalent and therefore have the same set of instances (owl:equivalentClass) or when two properties represent the same relationship (owl:equivalentProperty); ‘Hierarchical Links’ that define a hierarchical relation between concepts (defined in RDFS as subClassOf) or between properties (rdfs:subPropertyOf); and ‘Relationship Links’, which can be used to relate concepts from different data sets through any definable relation (e.g. a concept ‘Gene’ in one vocabulary can be related through the property associatedTo to a concept ‘Disease’ in another vocabulary).
Exploring linked data
If data providers follow the principles of publishing and interlinking structured data on the web as indicated above, including the definition of mappings, data will be integrated as in a large-scale database, forming a Linked Open Data Cloud. The integrated resources can then be explored by crawling or on-the-fly exploration, through query federation or a virtual knowledge broker [37,38]. Crawling the web means traversing the links between resources in advance, to reduce the response time of queries during run-time. However, it may lead to the retrieval of outdated data. On-the-fly exploration means accessing the data only during run-time, which ensures the data are always up-to-date, but may lead to longer waiting periods. Query federation consists in sending queries, or portions of complex queries, to a fixed set of resources (e.g. FeDeRate [37]). Although this is the most advantageous approach due to the flexibility of query formulations, it presents the same limitations as data federations, namely, the low performance of complex queries when considering a large number of data sources. Finally, the virtual knowledge broker exploits distributed data resources and makes use of the semantic data representation to deliver a coherent view to the end users, with the possibility of being instantiated in different locations [38].
The Linking Open Data project, under the tutelage of the W3C Semantic Web Education and Outreach Interest Group, is one key distribution channel for the publishing of data sets in the web using the semantic web standard language RDF and the definition of links connecting them [36,39]. Currently (as of August 2013), 337 data sets are available from disparate domains such as geography, governance and life sciences [40,41]. The latter includes examples such as the Gene Ontology, PubMed and UniProt [24,42,43].
The notion of open data is based on the free usage and redistribution of data. The arguments supporting the openness of data are based on the fact that government and scientific data are financed by public taxes and therefore should be publicly available. In the particular case of the translational medicine domain, the notions of linked data and linked open data are markedly distinct and present a fundamental limitation in achieving data integration.
SOLUTIONS FOR TRANSLATIONAL MEDICINE
According to our analysis, 11 systems have been reported in the scientific literature that present translational medicine solutions dealing with medical conditions as disparate as cardiovascular diseases, cancer and diabetes.
Three systems focused on the cardiovascular system: one on the identification and prioritization of candidate genes for cardiovascular diseases; another one on genetic association studies for hypercholesterolemia; and the third one also addressing association studies but for cerebrovascular diseases [44–46].
Two systems targeted cancer and its causes: one exploring genetic association studies for cervical cancer (Association Studies aSsisted by Inference and Semantic Technologies (ASSIST)); and the other one identifying personalized treatments for colon cancer patients (MATCH) [47,48].
Two other systems targeted type 2 diabetes mellitus: one focused on the understanding of its causes to discover novel treatment hypotheses (Semantic Enrichment of the Scientific Literature (SESL)); and the other one on genetic association studies [49,50]. The latter covered hypothyroidism in addition to type 2 diabetes.
Each of the remaining four solutions tackled different biomedical tasks: neuroscience research (Receptor Explorer); the repurposing of drugs; Traditional Chinese Medicine (TCM); and congenital muscular dystrophy [37,51–53].
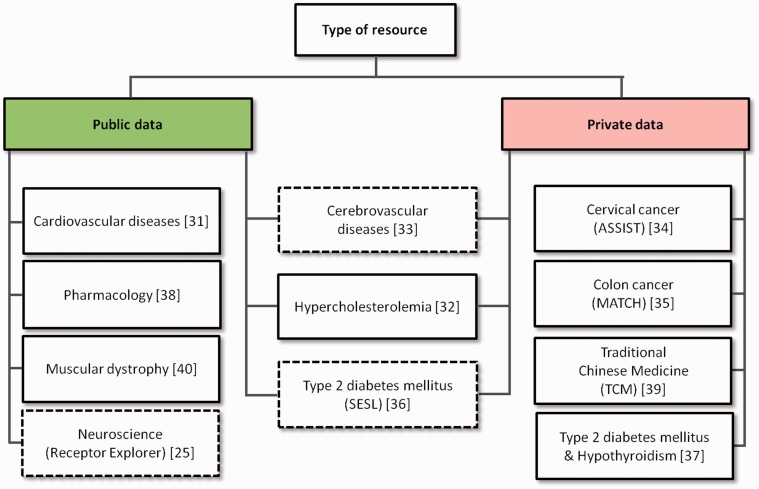
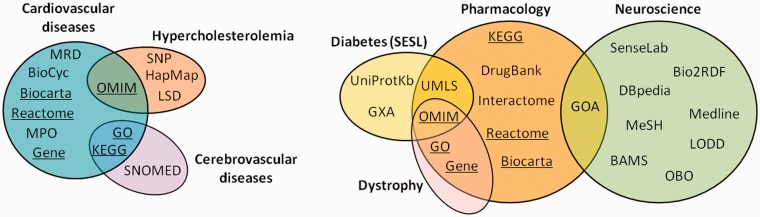
Seven of the 11 translational medicine systems surveyed integrate public resources (see Figure 2) but four of them consider only private data. Figure 3 shows the distribution of public resources integrated in each system.
Figure 2:
The type of data used by the 11 translational medicine systems surveyed. Four systems use solely public data, three integrate both public and private data and four use only private data. Receptor Explorer, the cerebrovascular diseases system and SESL (represented with dashed borders) are the only systems that provide open access to their integrated resources.
Figure 3:
Public resources integrated by the translational medicine systems surveyed. The resources shown on the left are those integrated by the three systems targeting the cardiovascular system, whereas the resources shown on the right side are those integrated by the remaining four systems. The resources integrated in the cardiovascular system subdomain that were also considered in at least one of the other subdomains are underlined. MRD—Mental Retardation Database; MPO—Mammalian Phenotype Ontology; Gene—NCBI Gene Database; OMIM—Online Mendelian Inheritance in Man; GO—Gene Ontology; KEGG—Kyoto Enclycopedia of Genes and Genomes; SNP—SNP Database; LSD—Locus-Specific Databases; GXA—Gene Expression Atlas; UMLS—Unified Medical Language System; GOA—Gene Ontology Annotation; OBO—Open Biomedical Ontologies; LODD—Linked Open Drug Data; BAMS—Brain Architecture Management System; MeSH—Medical Subject Headings.
Exploitation of semantic web resources
As previously pointed out, exploiting the semantic web to its full potential requires four key constructs: (i) structured (and ideally shared) knowledge representations; (ii) mappings between resources; (iii) data sharing; and (iv) use of semantic web technology standards in the previous three constructs.
To evaluate how the translational medicine systems exploited the semantic web and its technologies, we assessed them in view of three fundamental parameters for data integration: (i) degree of data sharing, (ii) data interoperability and (iii) knowledge discovery.
Data integration requires primarily data sharing, which in translational medicine can be achieved with public resources (e.g. gene and protein data) and/or private repositories (e.g. patient data). Conversely, the integration of data from different sources can also lead to data sharing if the resources are then made available to a wider audience. It is important to note that sharing data means that the data are accessible by third-party members having the appropriate access rights, but not necessarily accessible by the general public.
Data interoperability is achieved with the support of semantic web technologies and resources, through the use of the technological standards (e.g. RDF, URIs, RDFS and OWL), the linking of data and the representation of domain knowledge with controlled vocabularies.
Finally, data integration can lead to knowledge discovery by enabling the exploration of a potentially unlimited set of resources covering different knowledge domains, from which new associations can be discovered and previously hypothesized associations can be validated. In the semantic web context, knowledge discovery is founded on the use of the standards (e.g. RDFS and OWL) and the exploration of available linked data resources at a web-scale [11].
Data sharing
All of the translational medicine systems surveyed took advantage of shared data from public or private resources to achieve data integration (Figure 2). However, out of the seven systems that integrate public data, only three shared their data after integration: Receptor Explorer (neuroscience context), SESL (type 2 diabetes mellitus context) and the cerebrovascular diseases system (all three systems are represented with dashed borders in Figure 2). Receptor Explorer integrates public resources, some of which are maintained in their original location (e.g. DBpedia), and exposes them both as linked data and through a SPARQL end point. SESL, on the other hand, integrates both public and proprietary resources in a local triple store, exposing them through the links established with Wikipedia and a SPARQL end point. However, it requires specific access rights for accessing parts of the scientific literature. The SESL portal functions as a virtual knowledge broker [38]. The cerebrovascular diseases system works as a bridge (or share point) for resources from different institutions, but does not disclose the data to the general public.
The four systems that integrate exclusively private data (see Figure 2) function as nonpublic share points in the same way as the cerebrovascular diseases system.
The remaining four systems do not explicitly state that the integrated data or resultant knowledge is shared in any manner, and thus are assumed to instantiate a local and closed translational medicine solution available only to the directly involved parties.
Data interoperability
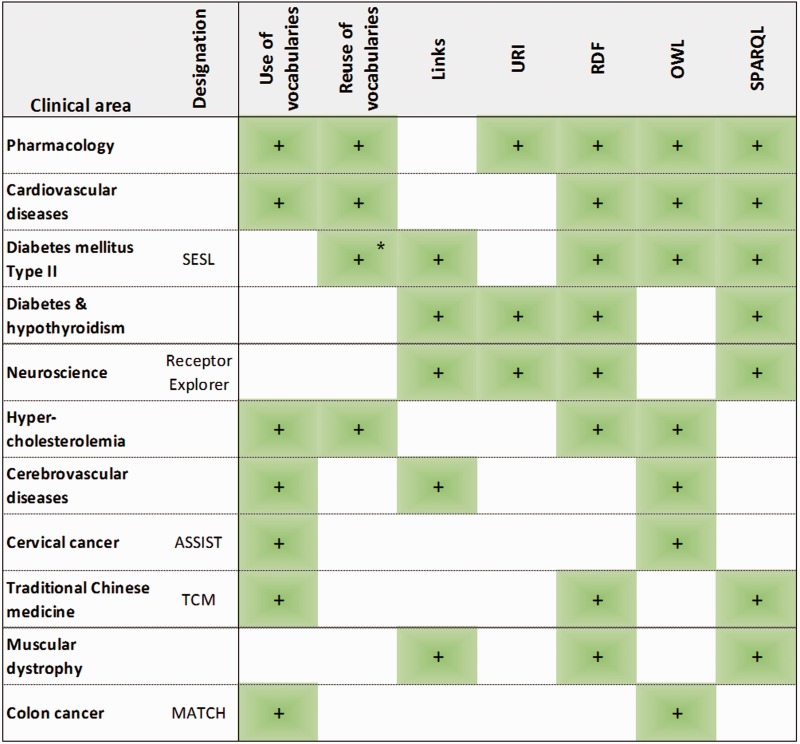
All systems incorporate semantic web technologies enabling data interoperability, which include the representation of domain knowledge with controlled vocabularies, links between resources and the use of the semantic web standards (see Figure 4).
Figure 4:
Technical description of the translational medicine systems surveyed. This figure shows the use of controlled vocabularies for knowledge representation, as well as their reuse for knowledge representation and data annotation (marked with *). Furthermore, it shows the definition of mappings between resources, the consideration of URIs, and lists the use of three semantic web standard technologies: RDF, OWL and SPARQL. All this information is indicated for all the translational medicine systems discussed.
Seven of the 11 systems used controlled vocabularies to represent their domain knowledge: the TCM system adopting the RDFS language, and the other six systems adopting OWL. From these seven systems, three reused existing vocabularies, whereas the other four developed their own. SESL reused existing controlled vocabularies only for data annotation. Regarding the implementation of links, Receptor Explorer and the muscular dystrophy system defined links between data resources, the cerebrovascular diseases system and the diabetes/hypothyroidism system defined links between data resources and controlled vocabularies, and SESL defined both types of links.
Among the standard technologies, RDF, OWL and SPARQL are the most common (see Figure 4), with only three systems not using RDF: cerebrovascular diseases, cervical and colon cancer. RDFS is only adopted by the TCM system and R2RML by the diabetes/hypothyroidism system. Only three systems (diabetes/hypothyroidism, pharmacology and Receptor Explorer) use URIs, even though their advantages were praised by several of the authors of the remaining systems. These three systems use locally defined URIs to represent the integrated data elements, but Receptor Explorer provides open access to the resources, thus making their URIs tractable and exploitable by third parties.
Knowledge discovery
Exploring a set of integrated resources by following existing mappings is a straightforward form of knowledge discovery. A more complex form involves inference mechanisms that uncover knowledge that does not have a previous explicit representation. Both approaches contribute to either formulate new hypotheses or refine and validate existing ones, which can lead to new research ideas and eventually to new treatments for individual patients.
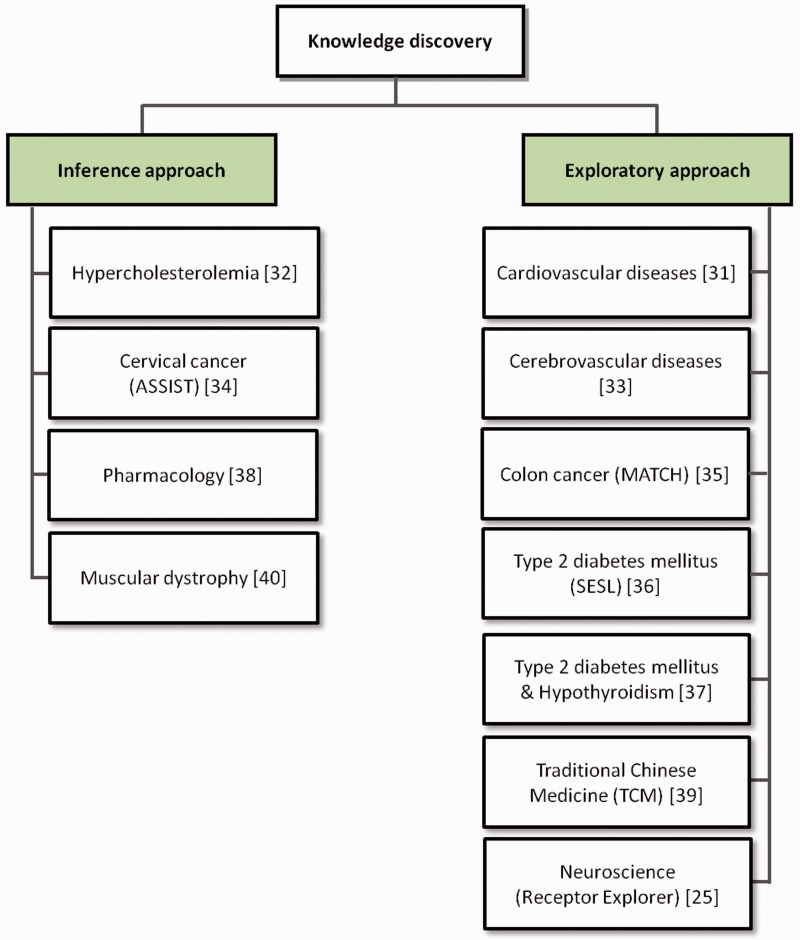
All surveyed translational medicine systems perform knowledge discovery, with seven following the exploratory approach and the remaining four the inference approach (see Figure 5). Among the seven systems that follow the exploratory approach, four use RDFS/OWL ontologies for knowledge representation, which means that they do not exploit the reasoning potential of those languages. Of the systems exploring inference, the muscular dystrophy system defined custom rules over RDF instead of using either RDFS or OWL, whereas the pharmacology system defined custom rules over RDF despite using OWL, owing to the fact that their chosen triple store did not support inference over OWL.
Figure 5:
The knowledge discovery approaches followed by the translational medicine systems surveyed. All of the systems performed knowledge discovery over their integrated resources: some exploiting inference, and the others following an exploratory approach.
Receptor Explorer is an example of a system implementing the exploratory approach to knowledge discovery. In this system, a knowledge base was created that aggregates the Neurocommons knowledge base and the data sets generated by the W3C’s Linking Open Drug Data task force [37,54,55]. The Neurocommons contains biomedical databases and ontologies such as the OBO and parts of the SenseLab Neurobiology databases, while the Linking Open Drug Data sets include data concerning clinical trials and disease–gene associations [56,57]. In addition to these locally stored data sets, Receptor Explorer integrates data from resources maintained at their original location, namely DBpedia, Bio2RDF and the Linked Clinical Trials project [58–60]. Through this pipeline of resources, it is possible to select a neural receptor, obtain its description, the genes involved in it, as well as publications and clinical trials involving the receptor.
The pharmacology system, on the other hand, is an example of a system implementing knowledge discovery through inference. The resources it integrates include DrugBank, Unified Medical Language System, Kyoto Enclycopedia of Genes and Genomes, National Center for Biotechnology Information's Entrez Gene database (from which Gene Ontology annotations were extracted for human genes) and Online Mendelian Inheritance in Man [61–65]. The authors present a good example of how knowledge discovery through inference enables the identification of a connection (until then undefined) between a drug approved for the treatment of hypertension and a connective tissue disorder. The identification of this connection was only possible owing to the use of the data inferred from the Gene Ontology.
IS SEMANTIC WEB TECHNOLOGY ENABLING TRANSLATIONAL MEDICINE?
Delivering solutions from the ‘bench to the bedside’ and incorporating them into the health care practice requires that the data flow from research in molecular biology, genetics and pharmacology into the clinical domain and in reverse. Within this flow of data and knowledge, research on the molecular mechanisms of diseases and drugs can be translated more quickly into novel treatment approaches, and conversely, observations about patients can lead to novel hypotheses and experimental conditions. The setup for this exchange requires not only the integration of data between the intervening research communities, but also the adaptation of data for safe and potentially unrestricted use by both communities.
Given the number of intervening parties in the translational medicine setting, data integration is fundamental for the evolution of this domain of knowledge. As we have shown, the semantic web has the potential to assist in many of the difficulties posed by the integration of data from disparate sources, as four of its underlying principles accelerate data integration and its exploration:
Represent data and knowledge with technologies that serve as a standard across the entire community.
Define mappings between resources.
Provide access to the resources so they can be integrated.
Share the effort of resource integration among data providers and data users.
The analysis of the translational medicine systems presented in the previous sections provides an overview of how the semantic web resources are being exploited in this domain of knowledge. It shows that most translational medicine systems adhere in earnest to the first principles described above, with RDF for data structuring, formal semantics and exploratory knowledge discovery among the features most commonly used. However, many systems neglect or ignore the remaining three principles.
By itself, the use of standard semantic web technologies does not fulfill the semantic web vision. For example, of the seven surveyed systems that use controlled vocabularies developed in OWL or RDFS for knowledge representation, only three reuse existing controlled vocabularies. The other three systems have created their own vocabularies, opting for a representation of the domain knowledge not shared by other researchers. Despite using standard semantic web technologies, these systems do not promote interoperability between applications and thus fall short of the semantic web vision.
The definition of mappings between resources is also critical in this context, as mappings facilitate the access to the resources that have them, increase the interoperability between applications that use these resources and increase the impact of these resources in the knowledge discovery process. Despite the clear advantages of using mappings, only five of the surveyed systems exploit them.
The definition of mappings is evidently linked to the third and fourth principles above: on the one hand, one form of access that can be given to resources is precisely through mappings between local resources and external resources; on the other hand, when open access is not an issue, the effort of creating mappings need not be supported exclusively by the data provider, as it can be divided with the users.
Resource sharing and integration are paramount to realize the semantic web vision, as without them, translational medicine systems can only be implemented on a local scale. Sharing integrated public resources that were not originally in a semantic web representation is particularly valuable, as it promotes distributed efforts in data integration. However, most of the current translational medicine systems are more focused on gathering and exploiting accessible resources than on making them accessible. Despite incorporating public resources, the majority of the systems surveyed do not share their data after integration, and thus forego the opportunity to contribute to translational medicine on the web-scale.
Nevertheless, the importance of sharing resources, reusing existing vocabularies and defining links between resources is not oblivious to all translational medicine systems, as some have done the base work for the future integration with one another through the resources they integrate. This is evident on Figure 3, which shows several resources that are integrated by more than one system. In addition to those resources, the cardiovascular system and the pharmacology system reuse the SNOMED-CT and the NCI Thesaurus, resources that were mapped to the cerebrovascular diseases system and to the diabetes/hypothyroidism system, respectively [25,66].
Relevance of semantic web technologies for translational medicine solutions
Translational medicine implementations can vary in focus, being centered on a disease or an organ, studying specific functions such as immune responses and reproduction, and performing analyses at the level of cellular processes or epidemiological phenomena. In our vision of translational medicine, each individual solution, independently of its focus, is an integral part of an interoperable and collaborative network of solutions. In this network, each solution can consume and contribute with data and knowledge, be it resources or new scientific findings, which are useful for the other solutions. The semantic web can assist in this endeavor but only if the solutions respect the four principles presented above, since by doing so the solutions become an integral part of the translational medicine network. The nonconnected remaining solutions are separate islets that, although contributing to the advancement of their specific medical focus, will not be contributing to the advancement of translational medicine as a whole.
With the exception of the cardiovascular diseases system and the pharmacology system, all the systems surveyed are totally independent, developed by different groups of researchers and without explicit connections between them. However, as discussed above, owing to the use of the same standard technologies, the integration and the reuse of the same resources and the definition of mappings with external resources, these systems currently have the potential for a seamless integration. If this integration were achieved, the knowledge obtained in one system could eventually be exploited in another. Some of the relations between systems are fairly direct, as is the case with the three systems targeting the cardiovascular system: new candidate genes in cardiovascular diseases might direct the genetic association studies in hypercholesterolemia or cerebrovascular diseases. Another example is that of the diabetes/hypothyroidism system that can identify genetic associations in diabetes that might assist SESL in identifying new treatments. Finally, the pharmacology system might identify connections between drugs and diseases that highlight unknown metabolic pathways affected in other diseases, which can be any of those targeted by the other systems.
The transition from isolated single-focused solutions based on semantic web technologies to translational medicine in the semantic web is necessary and there are already signs that it will happen. Such signs include the translational medicine examples that integrate data from several medical institutions (e.g. cerebrovascular diseases and colon cancer), and the work developed by the Committee on A Framework for Developing a New Taxonomy of Disease, which aims at developing a new taxonomy of disease integrating data from biomedical research, the public health and the health care delivery domains [67]. Sharing our network vision of translational medicine, the Committee intends to explore the creation of a knowledge network to connect all the participants, and through which new knowledge will be contributed back to the community.
Nonetheless, there are two issues that are essential for the transition to occur: the sharing of private data, and the incorporation of semantic web technologies in the clinical practice.
The integration of private data is essential for the instantiation of translational medicine, in particular in the subdomain of genomic medicine. However, while public data can be easily integrated into private data, the reverse is not true. Sharing private data, be it proprietary or patient data, is a sensitive issue that cannot be addressed without the implementation of proper security measures. Surprisingly enough, of the translational medicine systems surveyed, all those that integrate solely private data share their data and knowledge among several participating institutions, whereas most of the systems integrating public data do not.
The restrictions to data sharing imposed by private data are not limited to the questions of what data can be shared and with whom, but must also contemplate the questions of liability, how to acknowledge and keep track of the data owner, and how to judge the reliability of the data. These questions lead to the definition and specification of data provenance and access control policies (i.e. access authorization). The tractability of data provenance, namely the source of the data and how it was obtained, is fundamental to assess the reliability of the data sources from which new knowledge is extracted [68]. This is particularly important when relying on automatic applications that need to make decisions even when faced with contradictory and confusing pieces of information. A candidate recommendation for data provenance specification proposed by the W3C is the provenance data model (PROV-DM) [69].
No less important than the origin of the data, is who can have access to it, and specifically who can use it and alter it. This is achievable through the definition of the referred access control policies, which can be defined at several levels, namely, identification, authentication, authorization, confidentiality and accountability. An example of a tested access control model is the one implemented in the Simple Sloppy Semantic Database (S3DB) Core [70,71]. This model uses operators to define relationships between the system users and the entities stored in the system. These operators allow the definition of an access relationship between every user and every instance of the system, but they also allow the propagation of relationships from more generic entities to more specific entities (e.g. from classes to subclasses), thus reducing drastically the amount of access control data that needs to be stored. Nonetheless, these propagation models rapidly became inefficient owing to the increase in the amount of data stored. The propagation of the control relationships to all entities becomes prohibitive for large data sets, especially if they are rapidly evolving and the control relationships have to be continuously updated.
The incorporation of semantic web technologies in the clinical practice is another important issue, since these technologies are not sufficient by themselves to translate newly discovered medical tests or treatments into the patient care. Despite the advantages presented by these technologies, from the clinical perspective, the overheads associated with their implementation still exceed the benefits ensuing from the shared data. The solution for this issue is the establishment of partnerships between academia and health care providers (or other private entities). The key for the success of such partnerships lies in the already wide acceptance of the semantic web technologies in academia, and—on the other side—in the reduction of implementation overheads for the health care providers. A promising example of a partnership between academia and private entities such as pharmaceutical companies is the Open PHACTS project [72]. Funded by the Innovative Medicines Initiative, the goal of this project is to create an open space of shared data, knowledge and resources built with semantic web technologies, to stimulate drug discovery research [73].
CONCLUSIONS
The semantic web and its resources play a fundamental role in data integration, data interoperability and knowledge discovery. All these features have the potential to drive translational medicine forward, helping to deliver its goals of a better understanding of diseases and tailored treatments ‘from the bench to the bedside’, and of an efficient incorporation of these results in the everyday clinical practice, grounded on the knowledge gathered by all the intervening parties.
This article has surveyed a number of systems that make use of semantic web technologies to enable translational medicine. These systems successfully integrate public and private resources, represent their domain knowledge with controlled vocabularies and extract knowledge either by exploring the integrated resources or by performing inference over them. The semantic web technologies play an essential role by facilitating all these steps in the data exploration process, thus assisting in the concretization of translational medicine. However, only some of the solutions establish connections with external resources, hence improving their interoperability with other systems. Additionally, only some of the systems aim beyond tackling a medical problem, toward sharing the new knowledge with other researchers, be they partner institutions or the general public. Overall, the potential of the semantic web and its technologies is markedly untapped.
In our view of translational medicine, individual solutions tackling specific medical use cases should participate in a network of solutions that facilitates the flow of data and knowledge. Nevertheless, the creation of such network faces challenges, namely those posed by the share of private data and the overheads associated with the incorporation of semantic web technologies into the clinical practice. The solution for the first challenge lies with the semantic web community, which must address the technical issues related with the tracking of data and knowledge ownership and reliability, the control of data users and editors and the correct creation and maintenance of URIs. The solution for the second challenge can be achieved by sharing the costs associated with the implementation of semantic web solutions between the academia and the clinical practice, through the establishment of partnerships. It is also through these partnerships that new translational medicine results can be incorporated more quickly in the patient care.
The advent of high-throughput sequencing and genotyping technologies offers unprecedented opportunities for the development of new translational medicine applications, such as in the identification of mutations relevant for diagnosis and therapy. Thus, translational medicine solutions are bound to multiply as increasingly more data becomes available. Exploiting semantic web technologies in these solutions from the beginning offers the invaluable opportunity of creating systems intrinsically wired for interoperability.
It is our belief that individual solutions serve translational medicine best by embracing the vision of the semantic web, ideally through a close collaboration between data producers, data engineers and data consumers.
Key Points.
Translational medicine focuses on the understanding of human diseases through the exploration of heterogeneous data sources, ranging from public databases to proprietary pharmacology research data and patients’ private medical information.
The semantic web is a medium of excellence for the development of solutions that depend on the integration of data from heterogeneous sources, being thus extremely useful for translational medicine applications.
In addition to a set of technologies, the semantic web is also a set of principles on how to use those technologies, how to represent domain knowledge with controlled vocabularies and how to link resources and also share them so that they can be explored by others
The majority of the translational medicine systems surveyed use the semantic web technologies in a local-scale solution, with little regard to the use of existing vocabularies, the linking of resources or the share of data and knowledge with other researchers
To create a network of translational medicine solutions and maximize the potential of the semantic web, both technical and legal aspects regarding the secure share of data need to be solved and emphasis should be put in the creation of partnerships between academia and health care providers to better distribute both data and individual expertise
Acknowledgements
The authors would like to thank Miguel Gabriel for drawing the clinical practice illustration in Figure 1, Doctor Daniel Faria for his assistance in revising the final manuscript and the reviewers for their helpful insights and suggestions.
Biographies
Catia M. Machado is a PhD student at the Department of Informatics of the Faculty of Sciences and at INESC-ID, of the University of Lisbon. Her research interests are data representation and integration, in particular with semantic web technologies, knowledge discovery and translational medicine.
Dietrich Rebholz-Schuhmann (PhD) is ‘Oberassistent’ (similar to Associate Professor) with the University of Zürich, Department of Computational Linguistics. His research interests are biomedical literature and data analysis, data integration and knowledge discovery.
Ana Teresa Freitas (PhD) is an Associate Professor with the Technical University of Lisbon, Department of Computer Science and Engineering and the head of the group Knowledge Discovery and Bioinformatics at INESC-ID. Her research interests are in the areas of Computational Biology, Human genetics, Algorithms and Data Mining.
Francisco M. Couto (PhD) is an Assistant Professor at the Department of Informatics of Faculty of Sciences (University of Lisbon). He is a Senior Researcher of LASIGE where he coordinates the Biomedical Informatics research line. His research interests are in the areas of Text and Data Mining, Information Retrieval and Extraction, Ontologies and Bioinformatics.
FUNDING
Fundação para a Ciência e a Tecnologia through the Multi-annual Funding Program, the doctoral grant SFRH/BD/65257/2009, the projects SOMER (PTDC/EIA-EIA/119119/2010) and PEst-OE/EEI/LA0021/2013.
References
- 1.Hey T, Tansley S, Tolle K. The Fourth Paradigm: Data-Intensive Scientific Discovery. Washington: Microsoft Research; 2009. [Google Scholar]
- 2.Webb CP, Pass HI. Translation research: from accurate diagnosis to appropriate treatment. J Transl Med. 2004;2:35. doi: 10.1186/1479-5876-2-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Albani S, Prakken B. The advancement of translational medicine—from regional challenges to global solutions. Nat Med. 2009;15:1006–9. doi: 10.1038/nm0909-1006. [DOI] [PubMed] [Google Scholar]
- 4.Woolf SH. The meaning of translational research and why it matters. JAMA. 2008;299:211–3. doi: 10.1001/jama.2007.26. [DOI] [PubMed] [Google Scholar]
- 5.Wang W. Global health and translational medicine new drivers for medicine and medical sciences. J Med Med Sci. 2012;3:126–7. [Google Scholar]
- 6.Louie B, Mork P, Martin-Sanchez F, et al. Data integration and genomic medicine. J Biomed Inform. 2007;40:5–16. doi: 10.1016/j.jbi.2006.02.007. [DOI] [PubMed] [Google Scholar]
- 7.Committee on Models for Biomedical Reasearch. Models for Biomedical Research: A New Perspective. Washington, D.C.: National Academy Press; 1985. [Google Scholar]
- 8.Davidson SB, Overton C, Buneman P. Challenges in integrating biological data sources. J Comput Biol. 1995;2:557–72. doi: 10.1089/cmb.1995.2.557. [DOI] [PubMed] [Google Scholar]
- 9.Sagotsky JA, Zhang L, Wang Z, et al. Life sciences and the web: a new era for collaboration. Mol Syst Biol. 2008;4:201. doi: 10.1038/msb.2008.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Berners-Lee T, Hendler J, Lassila O. The Semantic Web. Sci Am. 2001;284:34–43. [Google Scholar]
- 11.Rebholz-Schuhmann D, Oellrich A, Hoehndorf R. Text-mining solutions for biomedical research: enabling integrative biology. Nat Rev Genet. 2012;13:829–39. doi: 10.1038/nrg3337. [DOI] [PubMed] [Google Scholar]
- 12.World Wide Web Consortium. http://www.w3.org/ (9 July 2013, date last accessed) [Google Scholar]
- 13.Semantic Web Standards. http://www.w3.org/standards/semanticweb/ (9 July 2013, date last accessed) [Google Scholar]
- 14.RDF Primer—W3C Recommendation. http://www.w3.org/TR/2004/REC-rdf-primer-20040210/ (9 July 2013, date last accessed) [Google Scholar]
- 15. Universal Resource Identifier (URI) current status. http://www.w3.org/standards/techs/uri#w3c_all (9 July 2013, date last accessed)
- 16.R2RML: RDB to RDF Mapping Language. http://www.w3.org/TR/2012/REC-r2rml-20120927/ (9 July 2013, date last accessed) [Google Scholar]
- 17.RDF Vocabulary Description Language 1.0: RDF Schema. http://www.w3.org/TR/rdf-schema/ (9 July 2013, date last accessed) [Google Scholar]
- 18. OWL Web Ontology Language Current Status. http://www.w3.org/standards/techs/owl#w3c all (9 July 2013, date last accessed)
- 19.Baader F, Calvanese D, McGuinness D, et al., editors. The Description Logic Handbook: Theory, Implementation, and Applications. New York: Cambridge University Press; 2003. [Google Scholar]
- 20. SPARQL Current Status. http://www.w3.org/standards/techs/sparql#w3c all (9 July 2013, date last accessed)
- 21.SPARQL 1.1 Entailment Regimes. http://www.w3.org/TR/2013/REC-sparql11-entailment-20130321/ (31 August 2013, date last accessed) [Google Scholar]
- 22.RDQL—A Query Language for RDF. http://www.w3.org/Submission/RDQL/ (9 July 2013, date last accessed) [Google Scholar]
- 23.Gruber TR. A translation approach to portable ontology specifications. Knowl Acquis. 1993;5:199–220. [Google Scholar]
- 24.Ashburner M, Ball CA, Blake JA, et al. Gene Ontology: tool for the unification of biology. Nat Genet. 2000;25:25–9. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Systematized Nomenclature of Medicine-Clinical Terms (SNOMED) http://www.ihtsdo.org/snomed-ct/ (9 July 2013, date last accessed) [Google Scholar]
- 26.Stein LD. Integrating biological databases. Nat Rev Genet. 2003;4:337–45. doi: 10.1038/nrg1065. [DOI] [PubMed] [Google Scholar]
- 27.Couto F, Pinto H. The next generation of similarity measures that fully explore the semantics in biomedical ontologies. J Bioinform Comput Biol. 2013;11:1–12. doi: 10.1142/S0219720013710017. [DOI] [PubMed] [Google Scholar]
- 28.The OBO Flat File Format Specification, version 1.2. http://www.geneontology.org/GO.format.obo-1_2.shtml (9 July 2013, date last accessed) [Google Scholar]
- 29.OBO Flat File Format Syntax and Semantics and Mapping to OWL Web Ontology Language. http://www.cs.man.ac.uk/∼horrocks/obo/ (6 October 2013, date last accessed) [Google Scholar]
- 30.OboInOwl - Mapping OBO to OWL. http://www.bioontology.org/wiki/index.php/OboInOwl:Main_Page (6 October 2013, date last accessed) [Google Scholar]
- 31.Tirmizi SH, Aitken S, Moreira DA, et al. Mapping between the OBO and OWL ontology languages. J Biomed Semantics. 2011;2: S3. doi: 10.1186/2041-1480-2-S1-S3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Linked Data - Design Issues. http://www.w3.org/DesignIssues/LinkedData.html (9 July 2013, date last accessed) [Google Scholar]
- 33.Best Practices for Publishing Linked Data. https://dvcs.w3.org/hg/gld/raw-file/default/bp/index.html (9 July 2013, date last accessed) [Google Scholar]
- 34.Linked Data Glossary. http://www.w3.org/TR/2013/NOTE-ld-glossary-20130627/ (9 July 2013, date last accessed) [Google Scholar]
- 35.Dodds L, Davis I. 2012. Linked Data Patterns: A pattern catalogue for modelling, publishing, and consuming Linked Data. http://patterns.dataincubator.org/book/ (31 August 2013, date last accessed) [Google Scholar]
- 36.Heath T, Bizer C. Linked Data: Evolving the Web into a Global Data Space. (1st edition). Synthesis Lectures on the Semantic Web: Theory and Technology; 1:1,1–136. Morgan & Claypool, 2011. [Google Scholar]
- 37.Cheung K, Frost H, Marshall M, et al. A journey to semantic web query federation in the life sciences. BMC Bioinformatics. 2009;10:S10. doi: 10.1186/1471-2105-10-S10-S10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Harrow I, Filsell W, Woollard P, et al. Towards virtual knowledge broker services for semantic integration of life science literature and data sources. Drug Discov Today. 2013;18:428–34. doi: 10.1016/j.drudis.2012.11.012. [DOI] [PubMed] [Google Scholar]
- 39.Linking Open Data by the W3C Semantic Web Education and Outreach Interest Group. http://www.w3.org/wiki/SweoIG/TaskForces/CommunityProjects/LinkingOpenData (9 July 2013, date last accessed) [Google Scholar]
- 40.Linking Open Data Cloud. http://datahub.io/group/lodcloud (19 August 2013, date last accessed) [Google Scholar]
- 41.State of the LOD Cloud. http://www4.wiwiss.fu-berlin.de/lodcloud/state/ (19 August 2013, date last accessed) [Google Scholar]
- 42.PubMed. http://www.ncbi.nlm.nih.gov/pubmed/ (9 July 2013, date last accessed) [Google Scholar]
- 43.The UniProt Consortium. The Universal Protein Resource (UniProt) Nucleic Acids Res. 2007;35:D193–7. doi: 10.1093/nar/gkl929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gudivada RC, Qu XA, Chen J, et al. Identifying disease-causal genes using Semantic Web-based representation of integrated genomic and phenomic knowledge. J Biomed Inform. 2008;41:717–29. doi: 10.1016/j.jbi.2008.07.004. [DOI] [PubMed] [Google Scholar]
- 45.Coulet A, Smail-Tabbone M, Benlian P, et al. Ontology-guided data preparation for discovering genotype-phenotype relationships. BMC Bioinformatics. 2008;9:S3. doi: 10.1186/1471-2105-9-S4-S3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Colombo G, Merico D, Boncoraglio G, et al. An ontological modeling approach to cerebrovascular disease studies: the NEUROWEB case. J Biomed Inform. 2010;43:469–84. doi: 10.1016/j.jbi.2009.12.005. [DOI] [PubMed] [Google Scholar]
- 47.Agorastos T, Koutkias V, Falelakis M, et al. Semantic integration of cervical cancer data repositories to facilitate multicenter association studies: the ASSIST approach. Cancer Inform. 2009;8:31–44. doi: 10.4137/cin.s963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Siddiqi J, Akhgar B, Gruzdz A, et al. Fifth International Conference on Information Technology: New Generations. IEEE Press; 2008. Automated Diagnosis System to Support Colon Cancer Treatment: MATCH; pp. 201–5. [Google Scholar]
- 49.Rebholz-Schuhmann D, Grabmuller C, Kavaliauskas S, et al. Semantic integration of gene-disease associations for Type 2 Diabetes mellitus from literature and 2 biomedical data resources. Drug Discov Today. 2013 doi: 10.1016/j.drudis.2013.10.024. [DOI] [PubMed] [Google Scholar]
- 50.Pathak J, Kiefer RC, Bielinski SJ, et al. Applying semantic web technologies for phenome-wide scan using an electronic health record linked Biobank. J Biomed Semant. 2012;3:10. doi: 10.1186/2041-1480-3-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Qu X, Gudivada R, Jegga A, et al. Bioinformatics and Biomedicine Workshops. IEEE International Conference; 2007. Semantic Web-based data representation and reasoning applied to disease mechanism and pharmacology; pp. 131–43. [Google Scholar]
- 52.Chen H, Mao Y, Zheng X, et al. Towards semantic e-science for traditional Chinese medicine. BMC Bioinformatics. 2007;8:S6. doi: 10.1186/1471-2105-8-S3-S6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sahoo SS, Zeng K, Bodenreider O, et al. From “glycosyltransferase” to “congenital muscular dystrophy”: integrating knowledge from NCBI Entrez Gene and the Gene Ontology. Studies in Health Technology and Informatics. Vol. 129 MEDINFO 2007. Amsterdam, Netherlands: IOS Press, 2007, p.1260–4. [PMC free article] [PubMed] [Google Scholar]
- 54.Ruttenberg A, Rees JA, Samwald M, et al. Life sciences on the Semantic Web: the Neurocommons and beyond. Brief Bioinform. 2009;10:193–204. doi: 10.1093/bib/bbp004. [DOI] [PubMed] [Google Scholar]
- 55.Semantic web Health Care and Life Sciences Interest Group/Linking Open Drug Data. http://www.w3.org/wiki/HCLSIG/LODD (6 September 2013, date last accessed) [Google Scholar]
- 56.The Open Biological and Biomedical Ontologies. http://www.obofoundry.org/ (6 September 2013, date last accessed) [Google Scholar]
- 57. SenseLab Neurobiology databases. http://senselab.med.yale.edu/ (6 September 2013, date last accessed)
- 58.DBpedia. http://dbpedia.org/About (6 September 2013, date last accessed) [Google Scholar]
- 59.Belleau F, Nolin M, Tourigny N, et al. Bio2RDF: towards a mashup to build bioinformatics knowledge systems. J Biomed Inf. 2008;41:706–16. doi: 10.1016/j.jbi.2008.03.004. [DOI] [PubMed] [Google Scholar]
- 60.Linked Clinical Trials (LinkedCT) project. http://linkedct.org/about/ (6 September 2013, date last accessed) [Google Scholar]
- 61.Wishart DS, Knox C, Guo AC, et al. DrugBank: a comprehensive resource for in silico drug discovery and exploration. Nucleic Acids Res. 2006;34:D668–72. doi: 10.1093/nar/gkj067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bodenreider O. The Unified Medical Language System (UMLS): integrating biomedical terminology. Nucleic Acids Res. 2004;32:D267–70. doi: 10.1093/nar/gkh061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kanehisa M, Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Maglott D, Ostell J, Pruitt KD, et al. Entrez Gene: gene-centered information at NCBI. Nucleic Acids Res. 2005;33:D54–8. doi: 10.1093/nar/gki031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hamosh A, Scott AF, Amberger JS, et al. Online Mendelian Inheritance in Man (OMIM), a knowledgebase of human genes and genetic disorders. Nucleic Acids Res. 2005;33:D514–7. doi: 10.1093/nar/gki033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sioutos N, Coronado S, Haber MW, et al. NCI Thesaurus: a semantic model integrating cancer-related clinical and molecular information. J Biomed Inform. 2007;40:30–43. doi: 10.1016/j.jbi.2006.02.013. [DOI] [PubMed] [Google Scholar]
- 67.Committee on A Framework for Developing a New Taxonomy of Disease. Toward a Precision Medicine: Building a Knowledge Network for Biomedical Research and a New Taxonomy of Disease. Washington, D.C.: National Academy Press; 2011. [PubMed] [Google Scholar]
- 68.Sahoo SS, Nguyen V, Bodenreider O, et al. A unified framework for managing provenance information in translational research. BMC Bioinformatics. 2011;12:461. doi: 10.1186/1471-2105-12-461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. PROV-DM: The PROV Data Model. http://www.w3.org/TR/2012/CR-prov-dm-20121211/ (9 July 2013, date last accessed)
- 70.Almeida JS, Deus HF, Maass W. S3DB core: a framework for RDF generation and management in bioinformatics infrastructures. BMC Bioinformatics. 2010;11:387. doi: 10.1186/1471-2105-11-387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Deus HF, Correa MC, Stanislaus R, et al. S3QL: a distributed domain specific language for controlled semantic integration of life sciences data. BMC Bioinformatics. 2011;12:285. doi: 10.1186/1471-2105-12-285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Williams AJ, Harland L, Groth P, et al. Open PHACTS: semantic interoperability for drug discovery. Drug Discov Today. 2012;17:1188–98. doi: 10.1016/j.drudis.2012.05.016. [DOI] [PubMed] [Google Scholar]
- 73.The Innovative Medicines Initiative. http://www.imi.europa.eu/ (9 July 2013, date last accessed) [Google Scholar]