Abstract
Aging is the predominant risk factor for neurodegenerative diseases. One key phenotype as the brain ages is an aberrant innate immune response characterized by proinflammation. However, the molecular mechanisms underlying aging-associated proinflammation are poorly defined. Whether chronic inflammation plays a causal role in cognitive decline in aging and neurodegeneration has not been established. Here we report a mechanistic link between chronic inflammation and aging microglia and a causal role of aging microglia in neurodegenerative cognitive deficits. We showed that SIRT1 is reduced with the aging of microglia and that microglial SIRT1 deficiency has a causative role in aging- or tau-mediated memory deficits via IL-1β upregulation in mice. Interestingly, the selective activation of IL-1β transcription by SIRT1 deficiency is likely mediated through hypomethylating the specific CpG sites on IL-1β proximal promoter. In humans, hypomethylation of IL-1β is strongly associated with chronological age and with elevated IL-1β transcription. Our findings reveal a novel epigenetic mechanism in aging microglia that contributes to cognitive deficits in aging and neurodegenerative diseases.
Keywords: epigenetic, innate immunity, interleukin, memory deficits, neuroinflammation, NF-κ B
Introduction
Aging is associated with aberrant inflammatory responses in human brains (Lu et al., 2004; Cribbs et al., 2012). Specifically, basal levels of proinflammation cytokines are elevated with aging (Sierra et al., 2007), whereas those of anti-inflammatory cytokines are reduced (Ye and Johnson, 1999). In addition, other genes involved in innate immune responses, such as the complement pathway, toll-like receptor signaling, and inflammasome activation, are also upregulated as the brain ages (Cribbs et al., 2012). Age is the predominant risk factor for neurodegenerative diseases, including frontotemporal dementia (FTD). These diseases are also characterized by chronic inflammation (Lucin and Wyss-Coray, 2009; Glass et al., 2010; Perry et al., 2010). However, whether chronic inflammation is an epiphenomenon or plays a causal role in cognitive decline in aging and neurodegeneration is unknown.
Microglia are the key mediators of innate immunity in brain (Ransohoff and Perry, 2009). During aging, levels of markers normally in activated microglia, including MHC II antigens, CD11b, CD14, and pattern recognition receptors, are elevated (Perry et al., 1993; Ogura et al., 1994; Letiembre et al., 2007). However, unlike the overactivated microglia induced by acute injury, microglia in aging brain are dystrophic, senescent, and associated with decreased ability to mount a normal response to injury (Damani et al., 2011; Hart et al., 2012). Microglia also appear to have telomere shortening and decreased telomerase activity with aging (Flanary and Streit, 2005) and in Alzheimer disease (AD) brains (Flanary et al., 2007). However, little is known about the molecular mechanisms underlying aging-induced microglial dysfunction.
Sirtuin 1 (SIRT1), a member of the sirtuin family, plays an important role in key cellular processes, including senescence/aging and inflammation (Gan and Mucke, 2008; Libert and Guarente, 2013). SIRT1 deacetylates intracellular targets, including transcription factors, signaling molecules, and histones (Michan and Sinclair, 2007; Zhang et al., 2011). SIRT1 suppresses inflammatory responses via deacetylation of RelA/p65 (Yeung et al., 2004; Chen et al., 2005; Rajendrasozhan et al., 2008; Schug et al., 2010). In cultured primary cortical cultures, overexpression of SIRT1 in microglia protected against Aβ toxicity, most likely by inhibiting NF-κB signaling (Chen et al., 2005). SIRT1 could also protect against cellular senescence by inactivating NF-κB (Rovillain et al., 2011; Tilstra et al., 2012) or deacetylating the FOXO3 transcription factor (Yao et al., 2012).
Here we showed that SIRT1 levels in microglia exhibit an age-dependent decline, and microglial SIRT1 deficiency exacerbated aging, or tau-associated cognitive deficits. The extent of aging- or tau-mediated memory function negatively correlated with levels of IL-1β transcripts, which are selectively regulated by SIRT1. To dissect the underlying molecular mechanisms, we established cellular models of myeloid lineage by transforming bone marrow-derived macrophage progenitor cells isolated from SIRT1F/F or LysM-Cre/SIRT1F/F mice. We found that an epigenetic mechanism involving hypomethylation of specific CpG sites on IL-1β underlies its selective upregulation in SIRT1-deficient microglia/myeloid cells. Notably, selective hypomethylation of IL-1β in aging humans and in demented patients with tauopathy strongly support validity of epigenetic regulation of IL-1β. We unraveled a novel epigenetic mechanism that modulates inflammatory pathways in aging and neurodegeneration.
Materials and Methods
Mice.
To delete floxed exon 4 of SIRT1 in the microglia, SIRT1F/F mice were crossed with LysM-Cre mice (Clausen et al., 1999; Cheng et al., 2003). The first cross yielded LysM-Cre/SIRT1F/+ mice, which were crossed with SIRT1F/F to obtain SIRT1F/F or LysM-Cre/SIRT1F/F mice. To delete the floxed exon 4 of microglial SIRT1 in the FTD mice, SIRT1F/F mice were crossed with hTau-P301S mice (Yoshiyama et al., 2007). The resulting hTau/SIRT1F/F mice were subsequently crossed with LysM-Cre/SIRT1F/F mice to obtain four genotypes: hTau/SIRT1F/F, hTau/LysM-Cre/SIRT1F/F, SIRT1F/F, and LysM-Cre/SIRT1F/F. Both male and female mice were used in the study. The number of male or female mice used in each study is given in the respective figure legends. All animal procedures were performed under University of California, San Francisco, Institutional Animal Care and Use Committee approved guidelines.
Isolation of adult microglia.
Adult microglia were isolated from SIRT1F/F or LysM-Cre/SIRT1F/F mice as described previously (Hickman et al., 2008) with some modifications. Mice were deeply anesthetized and perfused with ice-cold PBS without Ca2+ and Mg2+. Brains were placed in a culture dish with enzyme solution: RPMI 1640 containing 2 mm l-glutamine (Mediatech), 3 U/ml dispase, and 0.2% collagenase type 3 (Worthington Biochemicals), and then minced with razor blades and incubated for 30 min at 37°C, followed by 15 min of additional incubation with DNase I (Roche Applied Science). The enzymes were inactivated by adding PBS without Ca2+ and Mg2+ but with 2 mm EDTA and 2% fetal bovine serum (FBS), and the digested brains were triturated with a 20 ml pipette and passed through 70 μm filter. After centrifugation, cells were incubated with antimyelin microbeads (Miltenyi Biotech) in MACS buffer (2% FBS in PBS) for 15 min at 4°C. Cells were washed and centrifuged at 300 × g at 18°C for 7 min. Cells were resuspended with MACS buffer and passed through an autoMACS Pro Separator (Miltenyi Biotech). Myelin-negative fractions were collected and further processed by incubating with anti-CD11b microbeads at 4°C for 15 min. After washing, the resuspended myelin-negative fraction was passed through an autoMACS Pro Separator, and then the CD11b-positive population was separated from CD11b-negative population. Cells were centrifuged, and the pellets were lysed with RNA lysis buffer (Qiagen) for microarray or quantitative RT-PCR.
Microarray and WGCNA.
Total RNA was sent to the UCLA Neuroscience Genomics Core (http://www.semel.ucla.edu/ungc) and quantified using a Ribogreen Assay. Quality was assessed using an Agilent Bioanalyzer (Agilent Technologies). Amplified and labeled samples were then hybridized to Illumina MouseRef-8 v2.0 Expression BeadChips arrays. Detailed code for the microarray analysis can be found in Coppola (2011). Briefly, raw expression data were imported into R (http://www.r-project.org/), and the quality was checked by examining the interarray Pearson correlation and clustering based on the top variant genes using Bioconductor packages (http://www.bioconductor.org/). After quantile normalization, differential expression analysis was performed using the limma package (Smyth, 2004). After linear model fitting, a Bayesian estimate of differential expression was calculated, and the significance threshold was set at p < 0.005. Microarray data have been deposited in the NCBI Gene Expression Omnibus database (GEO; www.ncbi.nlm.nih.gov/geo) with the GEO series accession number GSE56452. Network analysis was performed as described previously (Oldham et al., 2008), using the WGCNA package in R. Briefly, correlation coefficients were constructed between expression levels of genes, and a connectivity measure (topological overlap; TO) was calculated for each gene by summing the connection strength with other genes. Genes were then clustered based on their TO, and groups of coexpressed genes (modules) were identified. Each module was assigned a color, and the first principal component of a module (module eigengene) was extracted from the module and considered to be representative of the gene expression profiles in a module. Phenotypic traits of interest were then regressed on the eigengene to examine whether there was a significant relationship between the module and the trait. Pathway analysis was performed by using the Ingenuity Pathways Analysis software (Ingenuity Systems, www.ingenuity.com).
Sample cohort of human IL-1β methylation data.
The cohort consisted of 122 patients with FTD, nine patients with FTD with motor neuron disease (MND), 43 patients with progressive supranuclear palsy (PSP), and 335 nondemented controls. The Illumina HumanMethylation 450K was used to assay methylation of DNA extracted from peripheral whole blood in three batches. For a 167 subjects, including a subset of 33 with FTD, four with FTD/MND, 11 with PSP, 90 nondemented controls, and an additional 29 with AD, mRNA expression in peripheral whole blood was quantified using the Illumina HumanHT-12 v4.0 Gene Expression BeadChip. The data from this study were accepted for publication (Li et al., 2014) and will be available in the GEO repository (GSE accession no. GSE53740).
Novel place recognition.
Mice were transferred to the testing room and acclimated for at least 1 h. On the first day, mice were habituated to the chamber for 15 min. On the second day, mice were placed in same chamber, exposed to two identical objects, and allowed to explore the objects and the chamber for 10 min as training. After 4 h, one of the objects was moved to a new location, and the mice were allowed to explore the objects and the chamber for 10 min. Time spent exploring each object was recorded for data analysis. The recognition index was obtained as the percentage of time spent with the object in a new location or in an old location.
Passive avoidance.
Passive avoidance was examined as described previously (Cho et al., 2011). Mice were initially placed in the lighted chamber. After 15 s, a sliding door to the dark chamber was opened, and the latency to enter the dark chamber was recorded. The door was closed as soon as all four paws of the mouse were inside the dark chamber, and an electric shock of 0.3 mA (duration, 2 s) was delivered. Retention was tested 24 and 144 h later.
Quantitative RT-PCR.
Real-time reverse transcription (RT)-PCR was done as described previously (Cho et al., 2011). The amount of gene expression was calculated as the difference (ΔCT) between the CT value of the sample for the target gene and the CT value of that sample for the endogenous control GAPDH [ΔCT = CT(target gene) − CT(GAPDH)]. The relative amount of gene expression for each target gene was determined by 2−ΔCT and expressed as the fold-change compared with a control.
Generation of immortalized bone marrow-derived macrophages.
Bone marrow cells were obtained from 1.8-month-old SIRT1F/F or LysM-Cre/SIRT1F/F mice. To immortalize myeloid cells, Ficoll-purified progenitors were infected with ER-Hoxb8 retrovirus and maintained in GM-CSF medium with estrogen as described previously (Wang et al., 2006; see Fig. 4A). For the experiment, macrophages that were differentiated from ER-Hoxb8 GM-CSF myeloid progenitor cells were used after culturing for 5 or 7 d in the culture medium without estrogen. Cells were treated with 5 μm 5-aza-2′-deoxycytidine (5-aza-CdR) at differentiation day 3 for 2 d. Due to short half-life, fresh drug was added every 24 h by replacing the medium.
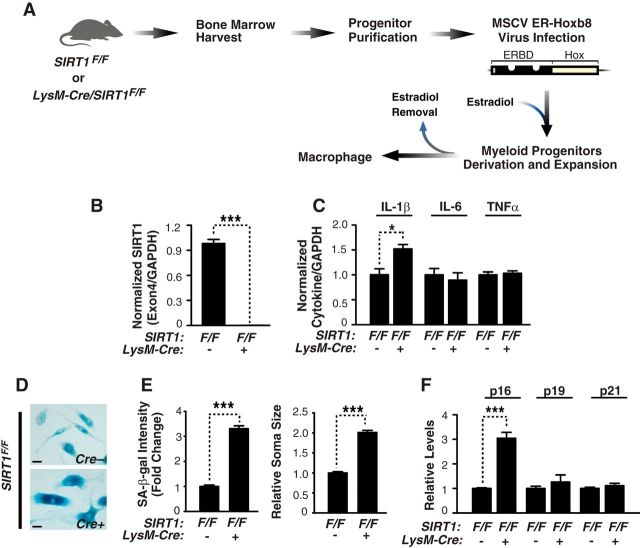
Figure 4.
SIRT1 deficiency elevates IL-1β transcription and accelerates senescence in myeloid cells. A, Flow chart describing the process of generating SIRT1-deficient ER-Hoxb8 macrophages. Bone marrow cells were isolated from 1.8-month-old SIRT1F/F or LysM-Cre/SIRT1F/F mice. In the presence of estrogen, ER-Hoxb8-infected macrophage-committed progenitor cells were immortalized. Upon estrogen withdrawal, progenitors were differentiated into macrophages. ERBD, Estrogen-binding domain. B, Quantifying exon 4-containing SIRT1 mRNA in SIRT1F/F or LysM-Cre/SIRT1F/F macrophages by qRT-PCR. n = 5/genotype;***p < 0.001, unpaired Student's t test. C, Quantifying IL-1β, IL-6, or TNFα by qRT-PCR; n = 4 from two experiments; *p < 0.05, unpaired Student's t test. D, Representative picture of senescence associated (SA)-β-gal staining in the SIRT1F/F or LysM-Cre/SIRT1F/F macrophages. Scale bars, 25 μm. E, Quantification of SA-β-gal staining intensity normalized by cell number or soma area; n = 6 independent sample, >450 cells (staining intensity) or >150 cells (soma area) were quantified for each independent sample; ***p < 0.001, unpaired Student's t test. F, Quantification of p16Ink4a, p19Arf, or p21Cip1 by qRT-PCR; n = 4 from two independent experiments; ***p < 0.001, unpaired Student's t test. Values are mean ± SEM (B, C, E, F).
SA-βgal staining.
Cells were stained with β-galactosidase as described previously (Debacq-Chainiaux et al., 2009). Cells were washed with PBS, fixed for 5 min at room temperature with 2% formaldehyde/0.2% glutaraldehyde and then stained for 16 h at 37°C with fresh β-galactosidase staining solution: 40 mm citric acid/sodium phosphate buffer, pH 6.0, 5 mm potassium ferricyanide (Sigma-Aldrich), 5 mm potassium ferrocyanide (Sigma-Aldrich), 150 mm NaCl, 2 mm MgCl2, and 1 mg/ml Xgal in distilled water. Images were obtained with Leica DM5000B, and staining intensity and soma area were measured with ImageJ (National Institutes of Health).
Pyrosequencing.
DNA methylation analysis was performed by EpigenDx. Briefly, 500 ng of sample DNA was treated with a proprietary bisulfite salt solution. Cells were resuspended in 40 μl of 1× proteinase K digestion buffer and digested with proteinase K (40 μg) at 50°C for 30 min. Cell debris were pelleted by centrifugation at 14,000 × g for 10 min and 20 μl was used in the bisulfite conversion reaction. DNA was diluted to 45 μl, and then 5 μl of 3N NaOH was added, followed by 30 min incubation at 42°C to denature the DNA. One-hundred microliters of bisulfite salt solution was added to the DNA and incubated for 14 h at 50°C. Bisulfite-treated DNA was purified using Zymogen DNA columns and was eluted with 20 μl of T1E0.2, pH 8.0, and 1 μl was used for each PCR. PCR was performed with 0.2 μm of each primer, and one of the PCR primers was biotinylated to purify the final PCR product with Sepharose beads. The PCR product was bound to streptavidin sepharose HP (GE Healthcare), and the sepharose beads containing the immobilized PCR product were purified, washed, and denatured with a 0.2 m NaOH solution and rewashed using the Pyrosequencing Vacuum Prep Tool (Qiagen), as recommended by the manufacturer. Then 0.2 μm pyrosequencing primer was annealed to the purified single-stranded PCR product. PCR products (10 μl) were sequenced by Pyrosequencing PSQ96 HS System (Biotage AB), following the manufacturer's instructions (Qiagen). The methylation status of each locus was analyzed individually as a T/C SNP with QCpG software (Qiagen).
FACS analysis.
CD11b+ cells isolated by magnetic beads were centrifuged and resuspended in PBS containing 5% FBS/0.5% BSA/0.1% NaN3. Cells were incubated for 15 min with anti-CD16/CD32 monoclonal antibodies (1:200, BD PharMingen) to block Fc receptors on microglia. Cells were then stained with allophycocyanin (APC)-labeled anti-CD11b (1:100, Biolegend) or APC-labeled isotype control (1:100, Biolegend) for 30 min on ice, followed by fixation with 2% PFA. Fluorescence intensity was measured using a FACSCalibur (BD Biosciences) flow cytometer.
Immunohistochemistry.
Perfusion, fixation, sectioning of mice brains were done as described previously (Cho et al., 2011). Sections were incubated with mouse rat anti-CD68 (1:200, Serotec). Expression of specific proteins was visualized with the avidin-biotin-peroxidase complex from the VECTASTAIN ABC Elite kit (Vector Laboratories). Intensity of staining was measured using ImageJ (National Institutes of Health). Three coronal sections were analyzed per mouse, and the average of the individual measurements was used to do the statistics.
DNMT activity assay.
Nuclear extracts from SIRT1F/F or LysM-Cre/SIRT1F/F, or 5 μm 5-aza-CdR-treated SIRT1F/F cells were incubated with DNA methyltransferase (DNMT) substrate with methyl donor per manufacturer's instruction (Epigentek). Methylated DNA was recognized with an anti-5-methylcytosine antibody and measured by optical density.
Western blot analysis.
Western blotting was done as described previously (Cho et al., 2011). After blocking, membranes were probed with a mouse monoclonal anti-SIRT1 (1:1000, Cell Signaling Technology), anti-human Tau (1:20,000; Thermo Scientific), and anti-GAPDH (1:10,000; Millipore) overnight at 4°C. Horseradish peroxidase-conjugated goat anti-mouse secondary antibody (1:2000, EMD Chemicals) was used to detect the primary antibodies.
Primary mouse culture.
Primary cultures were established from SIRT1F/F or LysM-Cre/SIRT1F/F mice on postnatal day 0 or 1. For neuronal culture, purified cells were plated at 160,000 cells/ml in neurobasal medium supplemented with B27 (Invitrogen) on poly-ornithine (PO)-coated plates. To generate mixed culture, cells were plated in medium containing DMEM, 10% FBS, 0.5 mm GlutaMAX, and B16 conditioned media. For primary microglial culture, mixed cultures were maintained in DMEM medium mentioned above for 2 weeks with changing the medium at day 5. After 2 weeks of culture, flasks were gently shaken, and the microglia containing supernatants were harvested. After microglia were shaken off, astrocytes were detached using trypsin-EDTA and plated onto PO-coated dish to obtain astrocyte culture.
IL-1β cytokine immunogenicity assay.
Cortical brain lysates were prepared and centrifuged to pellet debris. The supernatant was used to measure the protein concentration of IL-1β using a high-sensitivity multiplexed immunoassay (Meso-Scale Discovery) or traditional enzyme-linked immunosorbent assay kit (Abcam) per the manufacturer's instructions.
Results
Reducing SIRT1 levels in microglia in aging mice alters the IL-1R pathway
SIRT1, a class III deacetylase, is implicated in anti-aging and anti-inflammation (Chen et al., 2005; Gan and Mucke, 2008; Libert and Guarente, 2013). As microglia are key mediators of innate immunity in brain, we determined whether SIRT1 levels were regulated in microglia during aging. We used CD11b magnetic bead selection to isolate microglia to high purity (>97%) after myelin depletion (Hickman et al., 2008; Williamson et al., 2011) from young (2- to 3-month-old), middle-aged (13- to 15-month-old), and old (20- to 26-month-old) mice. By qRT-PCR, SIRT1 mRNA levels were reduced in microglia from middle-aged mice and reduced further in 20- to 26-month-old mice (Fig. 1A).
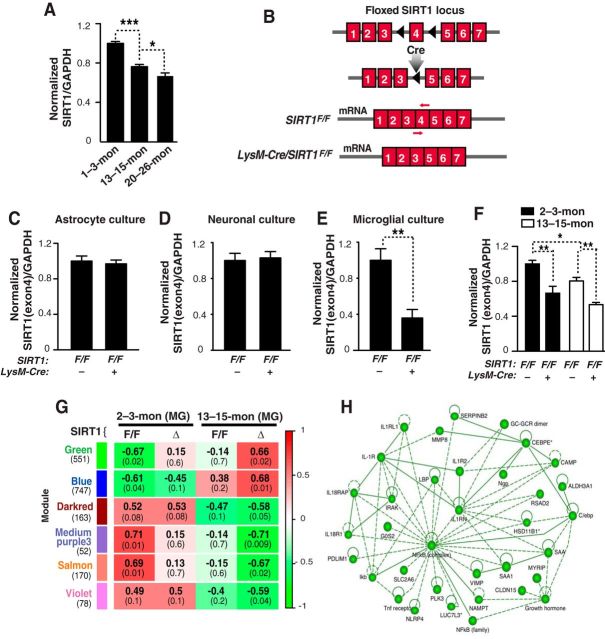
Figure 1.
Reducing SIRT1 levels in microglia in aging mice alters the IL-1R pathway. A, SIRT1 mRNA quantified by qRT-PCR in adult microglia from mice of the indicated months (mon) of age; n = 3–6 sample/genotype; four brains pooled for each sample; *p < 0.05; ***p < 0.001, one-way ANOVA with Newman–Keuls multiple-comparison test. B, Schematic diagram illustrating the loxP/LysM-Cre-mediated deletion of floxed exon 4 in SIRT1F/F gene. A primer set targets the exon 4 of SIRT1 (red arrows). C–E, Quantification of exon 4 containing SIRT1 mRNA in primary cultures of astrocytes, neurons or microglia. Astrocyte culture; n = 8–9/genotype, neuronal culture; n = 3–4/genotype, and microglia culture; n = 5–7/genotype; **p < 0.01, unpaired Student's t test. F, Quantifying SIRT1 mRNA in the SIRT1F/F or LysM-Cre/SIRT1F/F adult microglia; n = 3–4 sample/genotype, three brains pooled for each sample; *p < 0.05; **p < 0.01, one-way ANOVA with Newman–Keuls multiple-comparison test. G, Weighted gene coexpression network analysis. Each row corresponds to a module (more precisely, module eigengene), and each column corresponds to a trait. In each cell, the upper number indicates the correlation coefficient between the eigengene and a trait. The number in parentheses indicates the p value. Cells are color-coded based on the strength and direction of correlation, according to the scale on the right. H, Ingenuity Pathway Analysis of green module genes. MG, Microglia. Values are mean ± SEM (A, C–F).
Next we genetically reduced SIRT1 levels in microglia of young and middle-aged mice. SIRT1F/F mice were crossed with LysM-Cre mice to delete floxed exon 4, which is the functional domain of SIRT1 (Fig. 1B; Clausen et al., 1999; Cheng et al., 2003). To determine whether LysM-Cre leads to selective deletion in microglia, we quantified exon 4-containing SIRT1 levels in different brain cells derived from SIRT1F/F mice in the presence or absence of LysM-Cre. LysM-Cre expression did not affect levels of exon 4-containng SIRT1 in cultured astroglia or neurons, but reduced those in microglia significantly (Fig. 1C–E). In isolated adult microglia, LysM-Cre expression led to a significant reduction, not ablation, of microglial SIRT1 in young and middle-aged mice. The lowest microglial SIRT1 levels were in middle-aged SIRT1F/F mice that express LysM-Cre (Fig. 1F).
To identify gene networks that are affected by microglial SIRT1 reduction with aging, we performed weighted gene coexpression network analyses (WGCNA), which was developed to identify clusters (modules) of genes that were highly correlated, based on gene expression profile (Zhang and Horvath, 2005). Among 32 modules identified, six had significant correlation with middle-aged LysM-Cre/SIRT1F/F mice with lowest microglial SIRT1. Of interest is the Green module (551 genes), in which the first component correlated positively with gene expression in middle-aged ΔSIRT1 (LysM-Cre/SIRT1F/F) microglia (with lowest SIRT1), but negatively with young SIRT1F/F microglia (with highest SIRT1; Fig. 1G). Ingenuity Pathway Analyses of the Green module showed that one of the top networks is enriched with NF-κB-related genes and genes in the interleukin-1 receptor (IL-1R) superfamily, including type I and II IL-1Rs (IL-1R1 and IL-1R2), IL-1 receptor-like 1 (IL1RL1), IL-18 receptor 1 (IL18R1), and IL-18 receptor accessory protein (IL18RAP; Fig. 1H; Kakkar and Lee, 2008). Thus, SIRT1 deficiency in aging microglia is associated with upregulated IL-1R-related pathways.
Microglial SIRT1 deficiency elevates IL-1β transcription selectively and exacerbates memory deficits in aging
In adult microglia isolated from young, middle-aged, and aged mice, levels of IL-1β mRNA were significantly elevated (Fig. 2A). The increase in IL-1β mRNA was associated with the reduction in microglial SIRT1 (n = 9, r = −0.6276, p = 0.02, Pearson correlation). In brain lysates, levels of IL-1β protein, measured with ELISA, were also significantly higher in old mice than those in young mice (Fig. 2B,C). Not surprisingly, a strong positive correlation was observed between levels of IL-1β mRNA and protein across different ages (n = 27, r = 0.4425, p = 0.02, Pearson's correlation). Moreover, reducing SIRT1 levels in microglia elevated levels of IL-1β without affecting levels of IL-6 or TNFα, supporting the selective regulation of IL-1β transcription by SIRT1 in vivo (Fig. 2D).
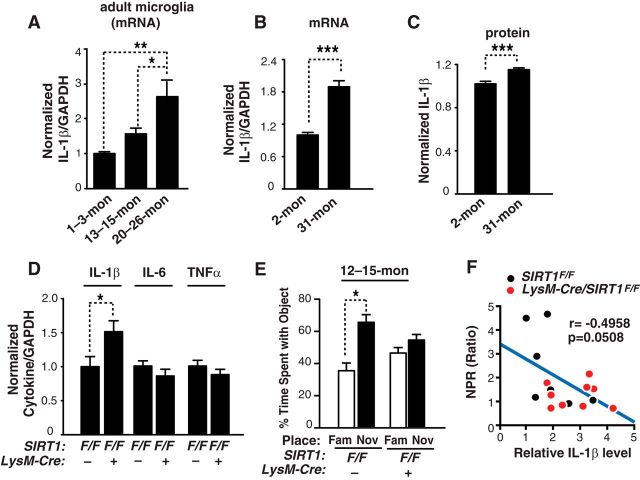
Figure 2.
Microglial SIRT1 deficiency elevates IL-1β transcription selectively and exacerbates memory deficits in aging. A, Quantification of IL-1β mRNA in the adult microglia from mice of different ages; n = 3–6 sample/genotype, four brains pooled for each sample; *p < 0.05; **p < 0.01, one-way ANOVA with Newman–Keuls multiple-comparison test. B, C, Quantifying IL-1β mRNA (B) and protein in the cortices of 2- and 31-month-old mice; n = 9–18 mice/genotype, males were used; ***p < 0.001, unpaired Student's t test. D, Quantification of IL-1β, IL-6 and TNFα mRNA in the cortices of 12- to 15-month-old SIRT1F/F or LysM-Cre/SIRT1F/F mice; n = 9–10 mice/genotype. SIRT1F/F: seven males and two females; LysM-Cre/SIRT1F/F: five males and five females; *p < 0.05, unpaired Student's t test. E, Measuring spatial memory with a novel place recognition test. SIRT1F/F or LysM-Cre/SIRT1F/F mice were exposed to two identical objects (objects A and B) during training. Four hours later, spatial memory was measured as the time spent with object moved to the novel place (NOV) versus that in the familiar place (FAM); n = 8–9 mice/genotype. SIRT1F/F: five males and three females; LysM-Cre/SIRT1F/F: four males and five females; *p < 0.05, paired Student's t test. F, Levels of brain IL-1β negatively correlated with performance in the novel place recognition test in 12- to15-month-old SIRT1F/F and LysM-Cre/SIRT1F/F mice; n = 7–9 mice/genotype. SIRT1F/F: four males and three females; LysM-Cre/SIRT1F/F: four males and five females; Pearson's correlation. Values are mean ± SEM (A–E).
IL-1β has been implicated in the impairment of hippocampal-dependent memory in rodents (Rachal Pugh et al., 2001; Barrientos et al., 2002; Shaftel et al., 2008). We then determined whether microglial SIRT1 deficiency adversely affects cognitive function in aging mice. A novel place recognition task was used to measure hippocampal-dependent spatial memory. At 12 to 15 months of age, wild-type mice (SIRT1F/F) showed a significant preference for the object in a new location, indicating normal spatial memory (Fig. 2E). However, mice with reduced levels of microglial SIRT1 (LysM-Cre/SIRT1F/F) exhibited no such preference, suggesting that spatial memory was impaired in these mice (Fig. 2E). Interestingly, the worsened spatial memory induced by microglial SIRT1 deficiency exhibited a modest association with higher levels of IL-1β (Fig. 2F).
Microglial SIRT1 deficiency elevates IL-1β transcription selectively and exacerbates memory deficits in human P301S tau mice
Aging is a major risk factor for neurodegenerative diseases, including FTD. We hypothesize that age-associated changes on microglia play a causal role in cognitive deficits associated with FTD. Mutations in the microtubule-binding protein tau cause FTD that is pathologically characterized by an accumulation of neurofibrillary tangles (FTLD-tau). PS19 mice, which overexpresses human tau with familial FTLD-linked mutation P301S, exhibit age-dependent synaptic loss and microglial activation preceding tangle formation (Yoshiyama et al., 2007). They also exhibit age-dependent memory deficits. In the brains, SIRT1 protein levels were significantly reduced in hTau mice compared with those of NTG mice (Fig. 3A). In contrast, expression of hTau significantly elevated levels of IL-1β mRNA and protein (Fig. 3B,C), which exhibited strong positive correlation (n = 19, r = 0.7223, p = 0.0005, Pearson's correlation). To genetically reduce levels of microglial SIRT1, LysM-Cre/SIRT1F/F mice were crossed with PS19 mice. We found that reducing microglial SIRT1 markedly elevated levels of IL-1β mRNA (Fig. 3D). The effects of microglial SIRT1 reduction on IL-6 were modest and not statistically significant (Fig. 3E). Overall microglial activation induced by tau expression, as measured with CD68 immunoreactivity, was not altered by microglial SIRT1 deficiency (Fig. 3F,G). In a similar vein, TNFα transcription was strongly upregulated by hTau-P301S expression whereas reducing microglial SIRT1 levels had no effect on TNFα transcription (Fig. 3H). These findings further established that IL-1β is selectively upregulated by microglial SIRT1 deficiency in tau-mediated neurodegeneration.
Figure 3.
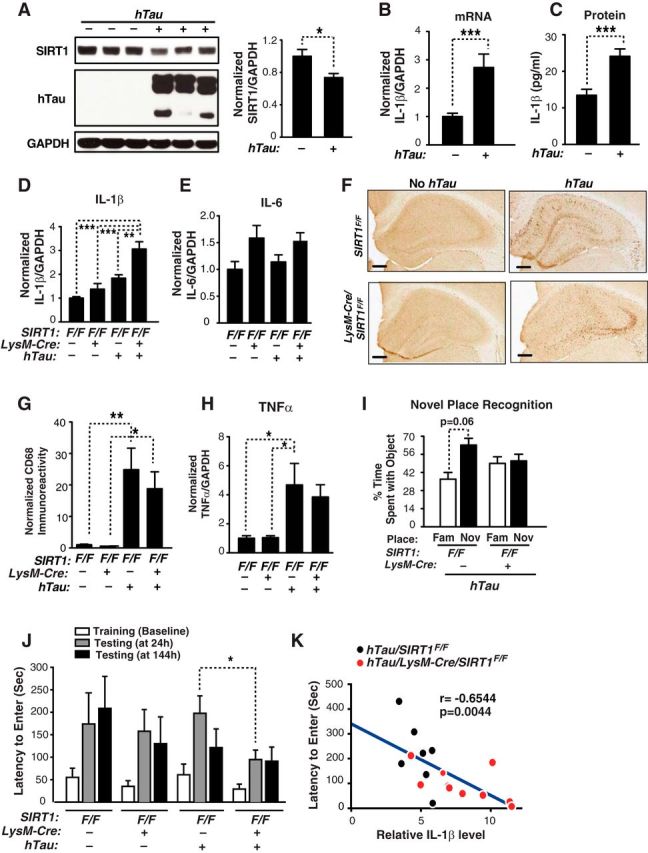
Microglial SIRT1 deficiency elevates IL-1β transcription selectively and exacerbates memory deficits in human TauP301S (PS19) mice. A, Representative Western blot and quantification of SIRT1 in cortical brain lysates of NTG and hTau mice that are 7 to 10 months of age; n = 6–7 mice/genotype; NTG: three males and three females; hTau: three males and four females; *p < 0.05, unpaired Student's t test. Quantifying IL-1β mRNA (B) and IL-1β protein (C) in the cortices of 7- to 9-month-old NTG and hTau mice; n = 8–11 mice/genotype; NTG: 4 males and 7 females; hTau: 4 males and 4 females; ***p < 0.001, unpaired Student's t test. MSD (Meso-Scale Discovery) immunoassay was used to quantifying IL-1β protein levels. D, E, Quantifying IL-1β and IL-6 mRNA in the cortices of 7- to 10-month-old SIRT1F/F, hTau/SIRT1F/F, LysM-Cre/SIRT1F/F or hTau/LysM-Cre/SIRT1F/F mice; n = 6–10 mice/genotype; SIRT1F/F: three (IL-6) or four (IL-1β) males and three females; LysM-Cre/SIRT1F/F: three males and five (IL-6) or six (IL-1β) females; hTau/SIRT1F/F: three males and four females; hTau/LysM-Cre/SIRT1F/F: three males and seven females; **p < 0.01; ***p < 0.001, one-way ANOVA with Tukey's multiple-comparison test. F, Representative pictures of CD68 immunoreactivity in the hippocampus of 7- to 10-month-old SIRT1F/F, LysM-Cre/SIRT1F/F, hTau/SIRT1F/F, hTau/LysM-Cre/SIRT1F/F mice. Scale bars, 500 μm. G, Quantification of CD68 immunoreactivity; n = 7–10/genotype; SIRT1F/F: four males and three females; LysM-Cre/SIRT1F/F: three males and six females; hTau/SIRT1F/F: three males and four females; hTau/LysM-Cre/SIRT1F/F: three males and seven females; *p < 0.05; **p < 0.01, one-way ANOVA with Tukey's multiple-comparison test. H, Quantifying TNFα mRNA in the cortices of 7- to 10-month-old SIRT1F/F, hTau/SIRT1F/F, LysM-Cre/SIRT1F/F or hTau/LysM-Cre/SIRT1F/F mice; n = 7–10 mice/genotype; SIRT1F/F: four males and three females; LysM-Cre/SIRT1F/F: three males and six females; hTau/SIRT1F/F: three males and four females; hTau/LysM-Cre/SIRT1F/F: three males and seven females; *p < 0.05, one-way ANOVA with Tukey's multiple-comparison test. I, Measuring spatial memory with a novel place recognition test. Tau/SIRT1F/F or Tau/LysM-Cre/SIRT1F/F mice were exposed to two identical objects (objects A and B) during training. Four hours later, spatial memory was measured as the time spent with object moved to the novel place (NOV) versus that in the familiar place (FAM). Six to 11 months of age; n = 5–11 mice/genotype; paired Student's t test. J, Microglial SIRT1 deficiency exacerbates tau-mediated memory deficits in passive avoidance test. Tests were performed 24 and 144 h after training; n = 7–10 mice/genotype; 4- to 7 months of age; SIRT1F/F: four males and three females; LysM-Cre/SIRT1F/F: three males and six females; hTau/SIRT1F/F: three males and four females; hTau/LysM-Cre/SIRT1F/F: three males and seven females; *p < 0.05, unpaired Student's t test. K, Levels of IL-1β in hTau transgenic mice (Tau/SIRT1F/F, Tau/LysM-Cre/SIRT1F/F) negatively correlated with the passive avoidance test; n = 7–10 mice/genotype, Pearson's correlation. Values are mean ± SEM (A–E, G–J).
Tau-mediated memory deficits were first assessed with a novel place recognition task similar to the aging cohort. Microglial SIRT1 intact hTau mice (hTau/SIRT1F/F) showed a trend toward preference for the object in a new location (Fig. 3I). However, hTau mice with reduced levels of microglial SIRT1 (hTau/LysM-Cre/SIRT1F/F) exhibited no such preference, suggesting that spatial memory was impaired in these mice (Fig. 3I). To further assess tau-mediated cognitive deficits, we performed passive avoidance test, which is commonly used to assess cognitive function. During the training session, mice readily entered the dark chamber from the lighted chamber due to an innate preference for darkness. No significant difference was observed between groups during the training (Fig. 3J). In a memory retention test measured 24 h after the shock, microglial SIRT1 deficiency led to a significantly shorter delay in hTau mice. By 144 h, hTau mice with or without intact SIRT1 were impaired (Fig. 3J). These findings suggest that tau-mediated memory deficits were exacerbated by microglial SIRT1 deficiency. Notably, a strong correlation between elevated levels of IL-1β and poor memory in passive avoidance test was observed in hTau mice with intact or deficient microglial SIRT1 (Fig. 3K). Thus, in independent mouse models of aging and neurodegeneration, microglial SIRT1 deficiency selectively upregulates IL-1β, which is associated with tau-mediated cognitive deficits.
SIRT1 deficiency elevates IL-1β transcription and accelerates senescence in myeloid cells
To dissect the mechanisms underlying the regulation of IL-1β by SIRT1, we established a myeloid model of SIRT1 deficiency. Bone-marrow progenitor cells from LysM-Cre/SIRT1F/F mice were infected with virus expressing estrogen-regulated Hoxb8 (ER-Hoxb8) and differentiated into macrophages (Wang et al., 2006; Fig. 4A). Unlike the partial deletion in brain, LysM-Cre expression led to complete SIRT1 ablation in macrophages (Fig. 4B). SIRT1 deletion elevated levels of IL-1β, but not those of IL-6 or TNFα, indicating that SIRT1 regulates IL-1β transcription selectively in cells of myeloid lineage in a cell-autonomous manner (Fig. 4C). SIRT1 deletion also led to increased senescence-associated β-galactosidase staining, an enlarged soma area, and increased p16Ink4a expression, consistent with previous studies (Brooks and Gu, 2009; Yao et al., 2012; Fig. 4D–F). Our findings support cell-autonomous regulation of cellular senescence and IL-1β transcription by SIRT1 in cells of myeloid lineage (macrophage/microglia).
SIRT1 deficiency increases IL-1β transcription via downregulating DNMT activity and hypomethylating IL-1β proximal promoter
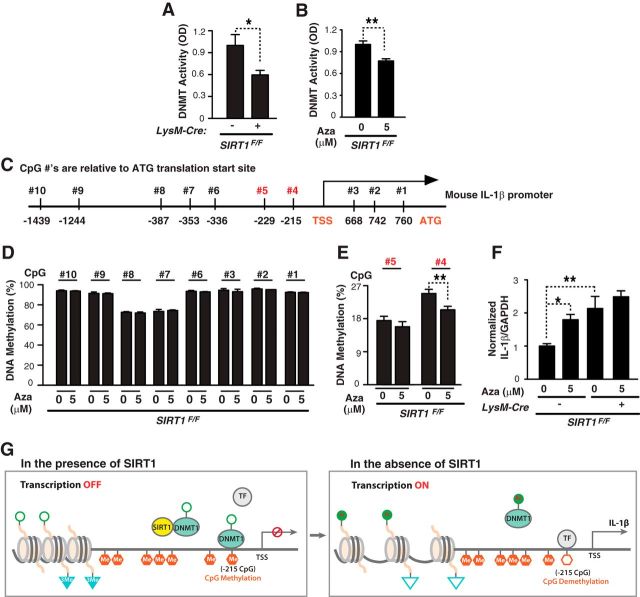
DNA methylation, catalyzed by DNA methyltransferases (DNMTs), is an epigenetic mechanism that underlies transcriptional suppression of various factors, including cytokines (Hashimoto et al., 2009; Hartnett and Egan, 2012; Jones, 2012). SIRT1 could activate DNMT1, a key DNMT, and enhance methylation via deacetylation (Peng et al., 2011). We hypothesize that SIRT1 deficiency inactivates DNMTs, leading to hypomethylation of IL-1β promoter and transcriptional activation of IL-1β. SIRT1 depletion lowered DNMT activity in myeloid cells (Fig. 5A). DNMT activity in myeloid cells can be inhibited by 5-aza-CdR (Fig. 5B). To determine how DNMT inhibition affects the methylation status of IL-1β promoter, we examined >10 CpGs for the region −1439 to the ATG translation start site (Fig. 5C). Most CpGs, except nos. 4 and 5, were heavily methylated and not affected by DNMT inhibition (Fig. 5D). In contrast, the percentage of DNA methylation at no. 4 (215 bp upstream from the transcriptional start site) was reduced by DNMT inhibition (Fig. 5E). Moreover, DNMT inhibition elevated IL-1β transcription in wild-type myeloid cells, but not in those lacking SIRT1 (Fig. 5F). Thus, SIRT1 deficiency, which lowers DNMT activity, occludes DNMT inhibition-induced increase in IL-1β. Our findings support a model in which SIRT1 deficiency inhibits DNMT activity and reduces methylation of the proximal promoter of IL-1β, leading to IL-1β transcriptional upregulation (Fig. 5G).
Figure 5.
SIRT1 deficiency increases IL-1β transcription via downregulating DNMT activity and hypomethylating IL-1β proximal promoter. A, B, Nuclear extracts from SIRT1F/F or LysM-Cre/SIRT1F/F or 5 μm 5-aza-CdR-treated SIRT1F/F cells were incubated with DNMT substrate with methyl donor. Methylated DNA was recognized with anti-5-methylcytosine antibody and measured by optical density; n = 4–5 samples/condition from two experiments; *p < 0.05; **p < 0.01, unpaired Student's t test. C, Mouse IL-1β promoter. Vertical lines indicate CpG sites. CpG sites −1439 bp to translation start site in the IL-1β promoter were analyzed. D, E, Percentage of IL-1β methylation at 10 CpG sites with or without 5 μm 5-aza-CdR for 48 h;. n = 4–10/samples from two experiments; **p < 0.01, unpaired Student's t test. F, SIRT1F/F or LysM-Cre/SIRT1F/F cells were treated with or without 5 μm 5-aza-CdR for 48 h. Using qRT-PCR, IL-1β mRNA were measured; n = 4–5 samples/condition from two experiments; *p < 0.05; **p < 0.01, one-way ANOVA with Tukey's multiple-comparison test. G, Potential mechanism of microglial SIRT1-mediated regulation of IL-1β transcript. With SIRT1, DNMT1 is deacetylated and activated. Activated DNMT1 methylates CpG at −215bp, recruits corepressor complex, and prevents the transcription factors (TFs) from binding to the promoter, and suppresses IL-1β transcription. Without SIRT1, DNMT1 remains acetylated and its activity is inhibited, leading to hypomethylation of IL-1β promoter at −215 bp, allowing TF binding. Values are mean ± SEM (A, B, D–F).
IL-1β CpG hypomethylation is associated with increased IL-1β expression
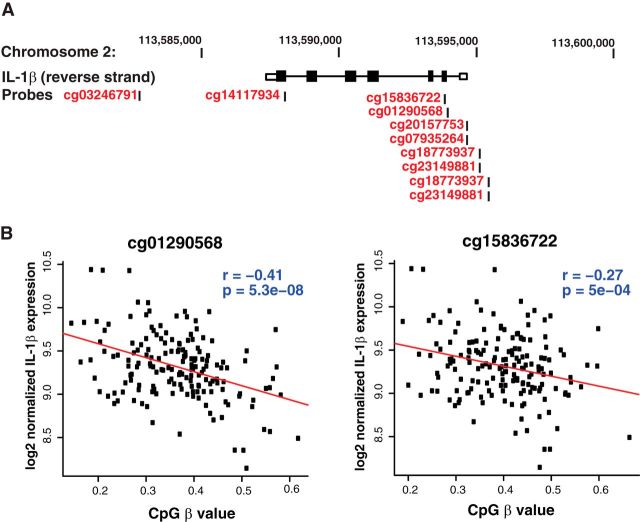
To validate the relationship between IL-1β hypomethylation with IL-1β expression in humans, we used the Illumina HumanMethylation 450K to assay methylation of human blood samples from 167 subjects, in which levels of IL-1β mRNA were obtained using Illumina HumanHT-12 v4.0 Gene Expression BeadChip. Ten probes were found near the IL-1β gene (Fig. 6A). Consistent with the finding that hypomethylation of IL-1β upregulates its expression in mouse myeloid cells, methylation of two CpGs (cg01290568 and cg15836722) negatively correlated with IL-1β expression in humans (Fig. 6B). This suggests that, in humans, IL-1β transcript levels are regulated by methylation.
Figure 6.
IL-1β CpG hypomethylation is associated with increased IL-1β expression. A, Schematic diagram of the human IL-1β gene on chromosome 2 and 10 CpG sites covered by the Illumina HumanMethylation 450K array. Coordinates on chromosome 2 are based on hg19/GRCh37. Boxes represent exons, whereas the connecting lines represent introns; filled boxes correspond to coding sequence, and unfilled boxes correspond to UTR (untranslated region). B, Illumina HumanMethylation 450K was used to assay methylation of DNA from peripheral whole blood. mRNA expression in peripheral whole blood was quantified with the Illumina HumanHT-12 v4.0 Gene Expression BeadChip. Methylation levels at cg01290568 and cg15836722 negatively correlated with expression of IL-1β in 167 whole-blood samples.
Two IL-1β CpG sites are hypomethylated in normal aging and in demented patients with tauopathy
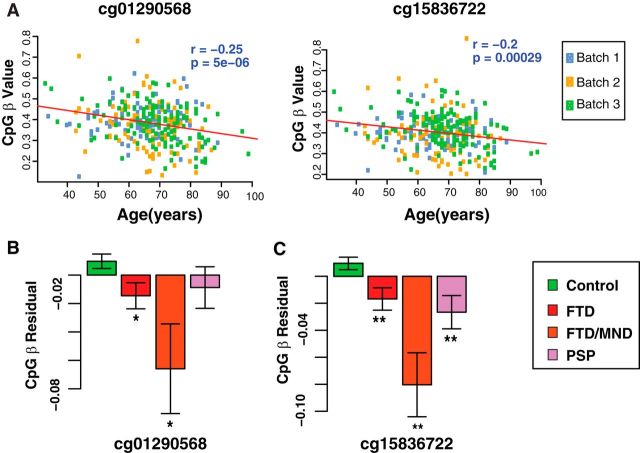
Because microglial SIRT1 was decreased with the aging of microglia and lack of SIRT1 decreased DNA methylation in myeloid cells (Figs. 1A, 5E), we hypothesize that inverse relationship exists between IL-1β methylation and aging in humans. The Illumina HumanMethylation 450K was used to assay methylation in three batches of human blood samples. Methylation of cg01290568 and cg15836722 was inversely correlated with chronological age, after correction for multiple comparisons (Fig. 7A). To account for a possible batch effect, samples were colored by array batch, and no confounding effect was observed. We compared IL-1β methylation in dementia patients and nondemented controls. To exclude the possible confounding effect of chronological age in this analysis, the residual of the methylation level after regression for age was used. Two CpG sites (cg01290568 and cg15836722) that were hypomethylated with normal aging were also hypomethylated in demented patients (Fig. 7B). The hypomethylation appears to be highly selective as no other CpG sites were affected by aging or tau-mediated dementia (data not shown).
Figure 7.
Two IL-1β CpG sites are hypomethylated in normal aging and in demented patients with tauopathy. A, Methylation at cg01290568 and cg15836722 correlated with chronological age of 335 nondemented controls assayed by Illumina HumanMethylation 450K array. Data for each CpG were fitted to a linear model (red line). Samples were colored according to array batch. B, C, Average methylation residual at cg01290568 or cg15836722 in nondemented controls (n = 335) and patients with FTD (n = 122), FTD/MND (n = 9), and PSP (n = 43). Two-sided t test was used to compare dementia patients versus nondemented controls. Values are mean ± SEM (B, C).
Discussion
In the current study, we provided direct evidence that alterations in aging microglia play a causative role in cognitive decline in normal aging and in FTD-related neurodegenerative diseases, and identified downregulation of SIRT1 in aging microglia as a key mediator. Genetic reduction of microglial SIRT1 selectively activated IL-1β transcription and exacerbated cognitive decline in aging and in hTau (PS19) transgenic mouse models. Elevated levels of IL-1β were associated with aging- or tau-mediated memory deficit. We showed that SIRT1 deficiency inhibits DNMT activity, resulting in hypomethylation of the proximal promoter of IL-1β and a selective increase in IL-1β transcription. Notably, in humans, hypomethylation of IL-1β is strongly associated with chronological age and with elevated IL-1β transcription. These results support a novel epigenetic mechanism in aging microglia that contributes to cognitive deficits in aging and neurodegeneration.
Recent studies identified molecular signatures that distinguish microglia from macrophages or young microglia from old microglia or resting microglia from microglia in neurodegeneration (Chiu et al., 2013; Hickman et al., 2013; Butovsky et al., 2014; Raj et al., 2014). Our findings are consistent with previous findings that IL-1β transcript levels were increased in microglia isolated from old mice compared with those from young mice (Hickman et al., 2013). Levels of IL-1β mRNA were also significantly higher in microglia isolated from SOD1G293A mutated ALS mouse model compared with those from wild-type SOD1 mice (Chiu et al., 2013). Interestingly, we found that SIRT1 exerts a remarkable selective and potent regulation of IL-1β by on the transcriptional level. Active forms of IL-1β are regulated on multiple levels, including transcriptional, translational, and processing. IL-1β is synthesized as a proprotein that is proteolytically processed to its active form by caspase 1 predominantly (Thornberry et al., 1992). Using WGCNA, the top network in the module that positively correlates with aging and microglial SIRT1 deficiency features IL-1β-related pathways. Moreover, in mice expressing hTau, LysM-Cre-mediated reduction of SIRT1 led to a significant increase of IL-1β without altering transcription of IL-6 or TNFα. Similarly, in myeloid cells, deleting SIRT1 elevated IL-1β selectively without affecting IL-6 or TNFα.
How does SIRT1 deficiency enhance IL-1β transcription selectively? The selective regulation of IL-1β by SIRT1 deficiency could be, at least partially, explained by an epigenetic mechanism of methylation of CpGs located in the proximal promoter of IL-1β, whereas most CpGs on the promoter are not affected. The highly selective epigenetic regulation of specific CpG sites is not uncommon. Depending on position of CpG on the promoter, some sites have more important roles in regulating gene transcription than others (Hashimoto et al., 2009). Although expression of TNFα is not affected by SIRT1 deficiency, it can be regulated by the methylation status of the proximal promoter region (Pieper et al., 2008; Gowers et al., 2011). One possible mechanism underlying the differential effects of SIRT1 on IL-1β versus other cytokines could be explained by the recruitment of distinct DNMTs onto different promoters. For example, although SIRT1 deacetylates DNMT1 in the catalytic region to activate its DNA methyltransferase activity (Peng et al., 2011), the activities of other DNMTs, which could be involved in regulating methylation status of TNFα promoters, may not be regulated by SIRT1.
Our finding that transcriptional regulation of IL-1β through methylation in mouse myeloid cells is remarkably consistent with findings in human normal aging and demented patients with tauopathy. We found one specific CpG site, 215 bp upstream of transcription start site (TSS), whose methylation status affects IL-1β transcript levels. When we aligned mouse and human IL-1β sequences, there was no CpG site in human where mouse CpG is selectively regulated. However, in humans, there are two CpGs (cg01290568 and cg15836722) within the IL-1β gene body near the TSS whose methylation negatively correlates with aging and dementia. This suggests selective regulation of specific CpGs at IL-1β gene in both mice and humans. Microglial SIRT1 levels were decreased with the aging of microglia and deficiency of microglial SIRT1 led to hypomethylation. In line with the findings in mouse, two IL-1β CpG sites were hypomethylated in normal aging and dementia. Most studies show a negative correlation between DNA methylation and expression, but some recent studies show a positive association between gene expression and methylation at the gene body (Jones, 2012). Our findings show that, in mice and humans, hypomethylation of IL-1β at specific CpG sites is associated with its increased expression.
Our study established a critical protective role of microglial SIRT1 in vivo for the first time. Microglial SIRT1 deficiency exacerbates memory deficits in aging and in tau-mediated memory deficits. In dissecting the underlying mechanism, we explored genes regulated by SIRT1 deficiency in aging brain. The IL-1R-related pathway emerged as a key pathway. Indeed, we confirmed that SIRT1 deficiency elevates levels of IL-1β in cultured myeloid cells, aging brain, and brains that express high levels of hTau. The importance of excessive IL-1β in hippocampal-dependent deficits was established in previous studies. Intracerebral ventricular infusion of IL-1β into rat brain led to impaired contextual fear conditioning but not auditory-cue fear conditioning (Rachal Pugh et al., 2001). In contrast, pretreatment with IL-1ra to block IL-1R signaling abolished LPS-induced memory deficits in contextual fear conditioning task (Terrando et al., 2010). Our current study showed that elevated levels of IL-1β induced by SIRT1 deficiency correlated strongly with aging and, especially, with tau-mediated, cognitive deficits. Our findings are consistent with an earlier study, which showed that blocking IL-1β signaling with an antibody against IL-1R alleviated cognitive deficits in the Morris water maze and passive avoidance task in 3XTG-AD mice that express mutant tau and hAPP (Kitazawa et al., 2011).
In summary, we showed that SIRT1 levels in microglia exhibit an age-dependent decline, and microglial SIRT1 deficiency is causative in cognitive decline in normal aging and in FTD-related neurodegenerative diseases. Aging-induced SIRT1 deficiency in microglia could initiate epigenetic alterations on IL-1β, leading to its enhanced expression that is associated with impairments in memory and related cognitive decline. Strategies that target epigenetic alterations and/or IL-1β-related pathways in aging microglia could offer new therapeutic avenue in neurodegenerative diseases.
Footnotes
This work was supported by NIH Grants 1R01AG036884 and R01AG030207 to L.G., 1F31NS084556 to J.A.C.; S.D. Bechtel Jr Foundation to L.G.; a grant from Consortium for Frontotemporal dementia (L.G., G.C.); the NINDS Informatics Center for Neurogenetics and Neurogenomics (P30 NS062691); and the Gladstone Institutes received support from a National Center for Research Resources Grant RR18928. We thank Dr Mariel Finucane for advice on statistical analyses, Dr Ravi Ponnusamy for advice on behavioral tests, Dr Robert V. Farese Jr for advice on multiplexed immunoassay, Gary Howard for editorial review, John Carroll and Giovanni Maki for graphics assistance, and Latrice Goss for administrative assistance.
The authors declare no competing financial interests.
References
- Barrientos RM, Higgins EA, Sprunger DB, Watkins LR, Rudy JW, Maier SF. Memory for context is impaired by a post context exposure injection of interleukin-1 beta into dorsal hippocampus. Behav Brain Res. 2002;134:291–298. doi: 10.1016/S0166-4328(02)00043-8. [DOI] [PubMed] [Google Scholar]
- Brooks CL, Gu W. How does SIRT1 affect metabolism, senescence and cancer? Nat Rev Cancer. 2009;9:123–128. doi: 10.1038/nrc2562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butovsky O, Jedrychowski MP, Moore CS, Cialic R, Lanser AJ, Gabriely G, Koeglsperger T, Dake B, Wu PM, Doykan CE, Fanek Z, Liu L, Chen Z, Rothstein JD, Ransohoff RM, Gygi SP, Antel JP, Weiner HL. Identification of a unique TGF-beta-dependent molecular and functional signature in microglia. Nat Neurosci. 2014;17:131–143. doi: 10.1038/nn.3599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Zhou Y, Mueller-Steiner S, Chen LF, Kwon H, Yi S, Mucke L, Gan L. SIRT1 protects against microglia-dependent amyloid-β toxicity through inhibiting NF-κB signaling. J Biol Chem. 2005;280:40364–40374. doi: 10.1074/jbc.M509329200. [DOI] [PubMed] [Google Scholar]
- Cheng HL, Mostoslavsky R, Saito S, Manis JP, Gu Y, Patel P, Bronson R, Appella E, Alt FW, Chua KF. Developmental defects and p53 hyperacetylation in Sir2 homolog (SIRT1)-deficient mice. Proc Natl Acad Sci U S A. 2003;100:10794–10799. doi: 10.1073/pnas.1934713100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu IM, Morimoto ET, Goodarzi H, Liao JT, O'Keeffe S, Phatnani HP, Muratet M, Carroll MC, Levy S, Tavazoie S, Myers RM, Maniatis T. A neurodegeneration-specific gene-expression signature of acutely isolated microglia from an amyotrophic lateral sclerosis mouse model. Cell Reports. 2013;4:385–401. doi: 10.1016/j.celrep.2013.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho SH, Sun B, Zhou Y, Kauppinen TM, Halabisky B, Wes P, Ransohoff RM, Gan L. CX3CR1 protein signaling modulates microglial activation and protects against plaque-independent cognitive deficits in a mouse model of Alzheimer disease. J Biol Chem. 2011;286:32713–32722. doi: 10.1074/jbc.M111.254268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clausen BE, Burkhardt C, Reith W, Renkawitz R, Förster I. Conditional gene targeting in macrophages and granulocytes using LysMcre mice. Transgenic Res. 1999;8:265–277. doi: 10.1023/A:1008942828960. [DOI] [PubMed] [Google Scholar]
- Coppola G. Designing, performing, and interpreting a microarray-based gene expression study. Methods Mol Biol. 2011;793:417–439. doi: 10.1007/978-1-61779-328-8_28. [DOI] [PubMed] [Google Scholar]
- Cribbs DH, Berchtold NC, Perreau V, Coleman PD, Rogers J, Tenner AJ, Cotman CW. Extensive innate immune gene activation accompanies brain aging, increasing vulnerability to cognitive decline and neurodegeneration: a microarray study. J Neuroinflammation. 2012;9:179. doi: 10.1186/1742-2094-9-179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damani MR, Zhao L, Fontainhas AM, Amaral J, Fariss RN, Wong WT. Age-related alterations in the dynamic behavior of microglia. Aging Cell. 2011;10:263–276. doi: 10.1111/j.1474-9726.2010.00660.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debacq-Chainiaux F, Erusalimsky JD, Campisi J, Toussaint O. Protocols to detect senescence-associated beta-galactosidase (SA-betagal) activity, a biomarker of senescent cells in culture and in vivo. Nat Protoc. 2009;4:1798–1806. doi: 10.1038/nprot.2009.191. [DOI] [PubMed] [Google Scholar]
- Flanary BE, Streit WJ. Effects of axotomy on telomere length, telomerase activity, and protein in activated microglia. J Neurosci Res. 2005;82:160–171. doi: 10.1002/jnr.20636. [DOI] [PubMed] [Google Scholar]
- Flanary BE, Sammons NW, Nguyen C, Walker D, Streit WJ. Evidence that aging and amyloid promote microglial cell senescence. Rejuvenation Res. 2007;10:61–74. doi: 10.1089/rej.2006.9096. [DOI] [PubMed] [Google Scholar]
- Gan L, Mucke L. Paths of convergence: sirtuins in aging and neurodegeneration. Neuron. 2008;58:10–14. doi: 10.1016/j.neuron.2008.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glass CK, Saijo K, Winner B, Marchetto MC, Gage FH. Mechanisms underlying inflammation in neurodegeneration. Cell. 2010;140:918–934. doi: 10.1016/j.cell.2010.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gowers IR, Walters K, Kiss-Toth E, Read RC, Duff GW, Wilson AG. Age-related loss of CpG methylation in the tumour necrosis factor promoter. Cytokine. 2011;56:792–797. doi: 10.1016/j.cyto.2011.09.009. [DOI] [PubMed] [Google Scholar]
- Hart AD, Wyttenbach A, Perry VH, Teeling JL. Age related changes in microglial phenotype vary between CNS regions: grey versus white matter differences. Brain Behav Immun. 2012;26:754–765. doi: 10.1016/j.bbi.2011.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartnett L, Egan LJ. Inflammation, DNA methylation and colitis-associated cancer. Carcinogenesis. 2012;33:723–731. doi: 10.1093/carcin/bgs006. [DOI] [PubMed] [Google Scholar]
- Hashimoto K, Oreffo RO, Gibson MB, Goldring MB, Roach HI. DNA demethylation at specific CpG sites in the IL1B promoter in response to inflammatory cytokines in human articular chondrocytes. Arthritis Rheum. 2009;60:3303–3313. doi: 10.1002/art.24882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hickman SE, Allison EK, El Khoury J. Microglial dysfunction and defective β-amyloid clearance pathways in aging Alzheimer's disease mice. J Neurosci. 2008;28:8354–8360. doi: 10.1523/JNEUROSCI.0616-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hickman SE, Kingery ND, Ohsumi TK, Borowsky ML, Wang LC, Means TK, El Khoury J. The microglial sensome revealed by direct RNA sequencing. Nat Neurosci. 2013;16:1896–1905. doi: 10.1038/nn.3554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones PA. Functions of DNA methylation: islands, start sites, gene bodies and beyond. Nat Rev Genet. 2012;13:484–492. doi: 10.1038/nrg3230. [DOI] [PubMed] [Google Scholar]
- Kakkar R, Lee RT. The IL-33/ST2 pathway: therapeutic target and novel biomarker. Nat Rev Drug Discov. 2008;7:827–840. doi: 10.1038/nrd2660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitazawa M, Cheng D, Tsukamoto MR, Koike MA, Wes PD, Vasilevko V, Cribbs DH, LaFerla FM. Blocking IL-1 signaling rescues cognition, attenuates tau pathology, and restores neuronal beta-catenin pathway function in an Alzheimer's disease model. J Immunol. 2011;187:6539–6549. doi: 10.4049/jimmunol.1100620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letiembre M, Hao W, Liu Y, Walter S, Mihaljevic I, Rivest S, Hartmann T, Fassbender K. Innate immune receptor expression in normal brain aging. Neuroscience. 2007;146:248–254. doi: 10.1016/j.neuroscience.2007.01.004. [DOI] [PubMed] [Google Scholar]
- Li Y, Chen JA, Sears RL, Gao F, Klein ED, Karydas A, Geschwind MD, Rosen HJ, Boxer AL, Guo W, Pellegrini M, Horvath S, Miller BL, Geschwind DH, Coppola G. An epigenetic signature in peripheral blood associated with the haplotype on 17q21.31, a risk factor for neurodegenerative tauopathy. PLoS Genet. 2014;10:e1004211. doi: 10.1371/journal.pgen.1004211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Libert S, Guarente L. Metabolic and neuropsychiatric effects of calorie restriction and sirtuins. Annu Rev Physiol. 2013;75:669–684. doi: 10.1146/annurev-physiol-030212-183800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu T, Pan Y, Kao SY, Li C, Kohane I, Chan J, Yankner BA. Gene regulation and DNA damage in the ageing human brain. Nature. 2004;429:883–891. doi: 10.1038/nature02661. [DOI] [PubMed] [Google Scholar]
- Lucin KM, Wyss-Coray T. Immune activation in brain aging and neurodegeneration: too much or too little? Neuron. 2009;64:110–122. doi: 10.1016/j.neuron.2009.08.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michan S, Sinclair D. Sirtuins in mammals: insights into their biological function. Biochem J. 2007;404:1–13. doi: 10.1042/BJ20070140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogura K, Ogawa M, Yoshida M. Effects of ageing on microglia in the normal rat brain: immunohistochemical observations. Neuroreport. 1994;5:1224–1226. doi: 10.1097/00001756-199406020-00016. [DOI] [PubMed] [Google Scholar]
- Oldham MC, Konopka G, Iwamoto K, Langfelder P, Kato T, Horvath S, Geschwind DH. Functional organization of the transcriptome in human brain. Nat Neurosci. 2008;11:1271–1282. doi: 10.1038/nn.2207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng L, Yuan Z, Ling H, Fukasawa K, Robertson K, Olashaw N, Koomen J, Chen J, Lane WS, Seto E. SIRT1 deacetylates the DNA methyltransferase 1 (DNMT1) protein and alters its activities. Mol Cell Biol. 2011;31:4720–4734. doi: 10.1128/MCB.06147-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry VH, Matyszak MK, Fearn S. Altered antigen expression of microglia in the aged rodent CNS. Glia. 1993;7:60–67. doi: 10.1002/glia.440070111. [DOI] [PubMed] [Google Scholar]
- Perry VH, Nicoll JA, Holmes C. Microglia in neurodegenerative disease. Nat Rev Neurol. 2010;6:193–201. doi: 10.1038/nrneurol.2010.17. [DOI] [PubMed] [Google Scholar]
- Pieper HC, Evert BO, Kaut O, Riederer PF, Waha A, Wüllner U. Different methylation of the TNF-alpha promoter in cortex and substantia nigra: implications for selective neuronal vulnerability. Neurobiol Dis. 2008;32:521–527. doi: 10.1016/j.nbd.2008.09.010. [DOI] [PubMed] [Google Scholar]
- Rachal Pugh C, Fleshner M, Watkins LR, Maier SF, Rudy JW. The immune system and memory consolidation: a role for the cytokine IL-1beta. Neurosci Biobehav Rev. 2001;25:29–41. doi: 10.1016/S0149-7634(00)00048-8. [DOI] [PubMed] [Google Scholar]
- Raj DD, Jaarsma D, Holtman IR, Olah M, Ferreira FM, Schaafsma W, Brouwer N, Meijer MM, de Waard MC, van der Pluijm I, Brandt R, Kreft KL, Laman JD, de Haan G, Biber KP, Hoeijmakers JH, Eggen BJ, Boddeke HW. Priming of microglia in a DNA-repair deficient model of accelerated aging. Neurobiol Aging. 2014;35:2147–2160. doi: 10.1016/j.neurobiolaging.2014.03.025. [DOI] [PubMed] [Google Scholar]
- Rajendrasozhan S, Yang SR, Kinnula VL, Rahman I. SIRT1, an antiinflammatory and antiaging protein, is decreased in lungs of patients with chronic obstructive pulmonary disease. Am J Respir Crit Care Med. 2008;177:861–870. doi: 10.1164/rccm.200708-1269OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ransohoff RM, Perry VH. Microglial physiology: unique stimuli, specialized responses. Annu Rev Immunol. 2009;27:119–145. doi: 10.1146/annurev.immunol.021908.132528. [DOI] [PubMed] [Google Scholar]
- Rovillain E, Mansfield L, Caetano C, Alvarez-Fernandez M, Caballero OL, Medema RH, Hummerich H, Jat PS. Activation of nuclear factor-kappa B signalling promotes cellular senescence. Oncogene. 2011;30:2356–2366. doi: 10.1038/onc.2010.611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schug TT, Xu Q, Gao H, Peres-da-Silva A, Draper DW, Fessler MB, Purushotham A, Li X. Myeloid deletion of SIRT1 induces inflammatory signaling in response to environmental stress. Mol Cell Biol. 2010;30:4712–4721. doi: 10.1128/MCB.00657-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaftel SS, Griffin WS, O'Banion MK. The role of interleukin-1 in neuroinflammation and Alzheimer disease: an evolving perspective. J Neuroinflammation. 2008;5:7. doi: 10.1186/1742-2094-5-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sierra A, Gottfried-Blackmore AC, McEwen BS, Bulloch K. Microglia derived from aging mice exhibit an altered inflammatory profile. Glia. 2007;55:412–424. doi: 10.1002/glia.20468. [DOI] [PubMed] [Google Scholar]
- Smyth GK. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol. 2004;3:Article3. doi: 10.2202/1544-6115.1027. [DOI] [PubMed] [Google Scholar]
- Terrando N, Rei Fidalgo A, Vizcaychipi M, Cibelli M, Ma D, Monaco C, Feldmann M, Maze M. The impact of IL-1 modulation on the development of lipopolysaccharide-induced cognitive dysfunction. Crit Care. 2010;14:R88. doi: 10.1186/cc9019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thornberry NA, Bull HG, Calaycay JR, Chapman KT, Howard AD, Kostura MJ, Miller DK, Molineaux SM, Weidner JR, Aunins J. A novel heterodimeric cysteine protease is required for interleukin-1 beta processing in monocytes. Nature. 1992;356:768–774. doi: 10.1038/356768a0. [DOI] [PubMed] [Google Scholar]
- Tilstra JS, Robinson AR, Wang J, Gregg SQ, Clauson CL, Reay DP, Nasto LA, St Croix CM, Usas A, Vo N, Huard J, Clemens PR, Stolz DB, Guttridge DC, Watkins SC, Garinis GA, Wang Y, Niedernhofer LJ, Robbins PD. NF-kappaB inhibition delays DNA damage-induced senescence and aging in mice. J Clin Invest. 2012;122:2601–2612. doi: 10.1172/JCI45785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang GG, Calvo KR, Pasillas MP, Sykes DB, Häcker H, Kamps MP. Quantitative production of macrophages or neutrophils ex vivo using conditional Hoxb8. Nat Methods. 2006;3:287–293. doi: 10.1038/nmeth865. [DOI] [PubMed] [Google Scholar]
- Williamson LL, Sholar PW, Mistry RS, Smith SH, Bilbo SD. Microglia and memory: modulation by early-life infection. J Neurosci. 2011;31:15511–15521. doi: 10.1523/JNEUROSCI.3688-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao H, Chung S, Hwang JW, Rajendrasozhan S, Sundar IK, Dean DA, McBurney MW, Guarente L, Gu W, Rönty M, Kinnula VL, Rahman I. SIRT1 protects against emphysema via FOXO3-mediated reduction of premature senescence in mice. J Clin Invest. 2012;122:2032–2045. doi: 10.1172/JCI60132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye SM, Johnson RW. Increased interleukin-6 expression by microglia from brain of aged mice. J Neuroimmunol. 1999;93:139–148. doi: 10.1016/S0165-5728(98)00217-3. [DOI] [PubMed] [Google Scholar]
- Yeung F, Hoberg JE, Ramsey CS, Keller MD, Jones DR, Frye RA, Mayo MW. Modulation of NF-κB-dependent transcription and cell survival by the SIRT1 deacetylase. EMBO J. 2004;23:2369–2380. doi: 10.1038/sj.emboj.7600244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshiyama Y, Higuchi M, Zhang B, Huang SM, Iwata N, Saido TC, Maeda J, Suhara T, Trojanowski JQ, Lee VM. Synapse loss and microglial activation precede tangles in a P301S tauopathy mouse model. Neuron. 2007;53:337–351. doi: 10.1016/j.neuron.2007.01.010. [DOI] [PubMed] [Google Scholar]
- Zhang B, Horvath S. A general framework for weighted gene coexpression network analysis. Stat Appl Genet Mol Biol. 2005;4:Article17. doi: 10.2202/1544-6115.1128. [DOI] [PubMed] [Google Scholar]
- Zhang F, Wang S, Gan L, Vosler PS, Gao Y, Zigmond MJ, Chen J. Protective effects and mechanisms of sirtuins in the nervous system. Prog Neurobiol. 2011;95:373–395. doi: 10.1016/j.pneurobio.2011.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]