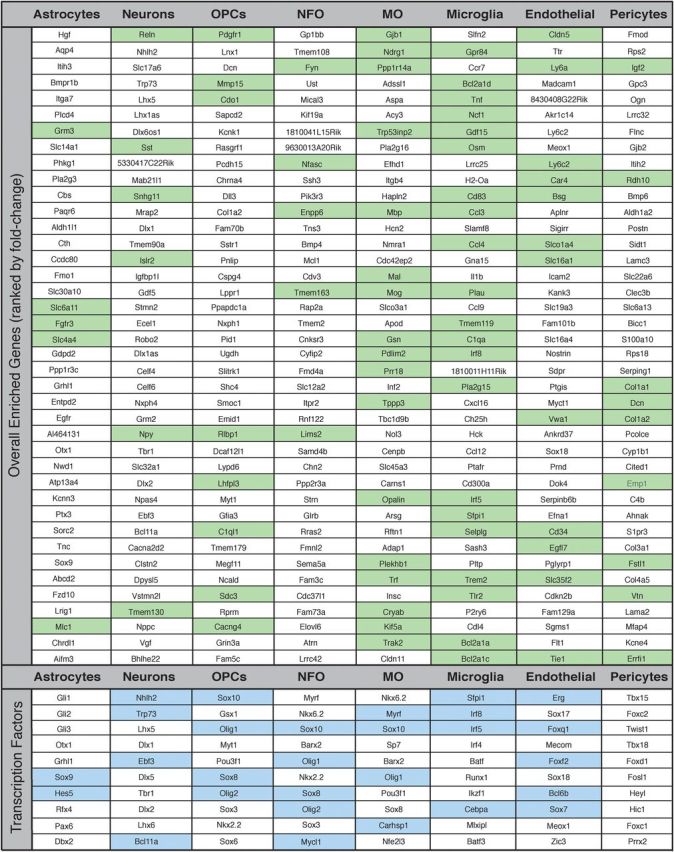
In the article “An RNA-Sequencing Transcriptome and Splicing Database of Glia, Neurons, and Vascular Cells of the Cerebral Cortex” by Ye Zhang, Kenian Chen, Steven A. Sloan, Mariko L. Bennett, Anja R. Scholze, Sean O'Keeffe, Hemali P. Phatnani, Paolo Guarnieri, Christine Caneda, Nadine Ruderisch, Shuyun Deng, Shane A. Liddelow, Chaolin Zhang, Richard Daneman, Tom Maniatis, Ben A. Barres, and Jian Qian Wu, which appeared on pages 11929–11947 of the September 3, 2014 issue, an oversight led to the omission of recognizing two authors, Dr. Kenian Chen and Dr. Jian Qian Wu, who contributed toward the writing of the paper. The Author Contribution footnote should have read “Author contributions: Y.Z., S.A.S., M.L.B., R.D., T.M., B.A.B., and J.Q.W. designed research; Y.Z., S.A.S., M.L.B., A.R.S., C.C., N.R., and S.D. performed research; S.A.L. and C.Z. contributed unpublished reagents/analytic tools; Y.Z., K.C., S.A.S., M.L.B., S.O., H.P.P., and P.G. analyzed data; Y.Z., K.C., S.A.S., B.A.B., and J.Q.W. wrote the paper. Also, note that a few typos have been noted in some gene names in Tables 2 and 3, and in Figure 4. The author contributions, Tables 2 and 3 and Figure 4 have been corrected on the online PDF version. The corrected tables and figure are shown below.
Table 2.
Cell type-enriched ligands and receptors
| Astrocyte | Neuron | OPC | NFO | MO | Microglia | Endothelial | Pericyte | |
|---|---|---|---|---|---|---|---|---|
| Enriched transmembrane receptors | Ptprz1 | Gpc1 | Pdgfra | Gpr17 | Efnb3 | Csf1r | Tfrc | Pdgfrb |
| Ednrb | Ptprn | Gpr17 | Sema4d | Gpr37 | Cd83 | Pglyrp1 | Colec12 | |
| S1pr1 | Caly | Itgav | Plxnb3 | Sema4d | Tyrobp | H2-D1 | Sfrp1 | |
| Fgfr3 | Grin1 | Omg | Ddr1 | Lpar1 | Ccrl2 | Eltd1 | S1pr3 | |
| Gabbr1 | Ptprn2 | Gfra1 | Efnb3 | Ddr1 | B2m | Kdr | Abcc9 | |
| Tnfrsf19 | Gria1 | Gria3 | Gpr37 | Omg | Trem2 | Eng | Rarres2 | |
| Vcam1 | Cnr1 | Sstr1 | Lpar1 | Ephb1 | Sirpa | Tie1 | Pdgfrl | |
| Adcyap1r1 | Opcml | Il1rap | Pdgfra | Gprc5b | Fcer1g | Cav1 | Mrc2 | |
| Gria2 | Stx1b | Adora1 | Omg | S1pr5 | Cd14 | Flt1 | Fas | |
| F3 | Cxadr | Lypd1 | Ephb1 | Gpr17 | Icam1 | Fcgrt | Ednra | |
| Grm3 | Nptxr | Gria4 | Sema5a | Plxnb3 | Cx3cr1 | Sema7a | Lepr | |
| Dag1 | Grm2 | Chrna4 | Il1rap | Gpr62 | Lag3 | Lsr | Osmr | |
| Plxnb1 | Robo2 | Sema5a | S1pr5 | Il1rap | Gpr56 | Acvrl1 | Gprc5a | |
| Ntsr2 | Kit | Calcrl | Prkcz | Prkcz | Itgam | Tek | Ifitm1 | |
| Fgfr1 | Gabrb3 | Gabra3 | Erbb3 | Itgb4 | Fcgr3 | Gpr116 | Ddr2 | |
| Ptch1 | Gabrg2 | Grin3a | Gpr62 | Erbb3 | P2ry12 | Fzd6 | Scarf2 | |
| Fgfr2 | Sarm1 | Oprl1 | Ptpre | Sema5a | P2ry13 | Ptprb | Tgfbr3 | |
| Gpr19 | Gabra2 | Grik4 | Grik3 | Tnfrsf12a | Aplnr | Fzd5 | ||
| Itga6 | Darc | Plxnb3 | Grik2 | H2-K1 | Ptprg | Npy1r | ||
| Gabbr2 | Celsr3 | Grik3 | Casr | Gpr183 | Plxnd1 | Celsr1 | ||
| Enriched ligands | Sparcl1 | Reln | Matn4 | Gsn | Trf | Cst3 | Sparc | Igf2 |
| Cpe | Sst | Scrg1 | Lgi3 | Gsn | C1qa | Sepp1 | Vtn | |
| Cyr61 | Npy | Olfm2 | Scrg1 | Apod | Ccl4 | Pltp | Cxcl12 | |
| Mfge8 | Olfm1 | Vcan | Enpp6 | Lgi3 | Ccl3 | Igfbp7 | Col4a1 | |
| Clu | Dkk3 | Emid1 | Matn4 | Metrn | C1qb | Spock2 | Col1a2 | |
| Htra1 | Ccl27a | Tnr | Tnr | Endod1 | C1qc | Ctla2a | Bgn | |
| Igfbp2 | Cx3cl1 | Nxph1 | Ddr1 | Adamts4 | Selpig | Pglyrp1 | Dcn | |
| Vegfa | Cck | Timp4 | Adamts4 | Cntn2 | Ctsb | Col4a1 | Ptgds | |
| Scg3 | Vgf | Spon1 | Metrn | Enpp6 | B2m | Egfl7 | Cxcl1 | |
| Ncan | Vstm2l | Igsf21 | Fam3c | Hapln2 | Gdf15 | AU021092 | Col1a1 | |
| Pla2g7 | Chgb | Gsn | C1ql1 | Il1rap | Olfm3 | Srgn | Fstl1 | |
| Fjx1 | Scg2 | Fam5c | Vcan | Erbb3 | Tnf | Fn1 | Col3a1 | |
| Timp3 | C1qtnf4 | Qpct | Timp4 | Slpi | Pla2g15 | Kdr | Mdk | |
| Il18 | Cxadr | C1ql3 | Il1rap | Klk6 | Tcn2 | Apln | Igfbp5 | |
| Btbd17 | Col6a2 | Smoc1 | Col11a2 | Col11a2 | Ly86 | Wfdc1 | Serpinf1 | |
| Itih3 | Resp18 | Gpc5 | Bmp4 | Matn4 | Plod1 | Angptl4 | Nbl1 | |
| Hapln1 | Vstm2a | Il1rap | Elfn2 | Dlk2 | Il1a | Htra3 | Nid2 | |
| Lcat | Car11 | Dscam | Dlx2 | Il23a | Tgfb1 | Smpdl3a | Islr | |
| Chrdl1 | Igfbpl1 | Chga | Spon1 | Wnt3 | Lgals9 | Lama4 | Ptx3 | |
| Pla2g3 | Nppc | Nptx2 | Dscam | Npb | Ccl2 | Emcn | Vasn |
Table 3.
Cell type-specific splicing events
| Gene | Coverage | dI | p value | FDR | |
|---|---|---|---|---|---|
| Astrocyte | Fyn | 771 | −0.27 | 1.15 E-176 | 1.95 E-174 |
| Prom1 | 251 | 0.86 | 1.12 E-133 | 4.59 E-131 | |
| Ncam1 | 3576 | 0.24 | 1.63 E-132 | 5.93 E-130 | |
| Ptprf | 480 | −0.58 | 1.03 E-120 | 2.98 E-118 | |
| Srgap3 | 308 | 0.69 | 1.99 E-84 | 1.44 E-82 | |
| Kif1a | 180 | 0.75 | 3.12 E-66 | 1.76 E-64 | |
| Ptk2 | 134 | −0.47 | 1.33 E-63 | 6.73 E-62 | |
| Camk2g | 903 | −0.44 | 5.75 E-56 | 2.43 E-54 | |
| Mapk8 | 270 | −0.55 | 6.98 E-37 | 2.36 E-35 | |
| Pkm2 | 1112 | −0.23 | 2.85 E-25 | 6.58 E-24 | |
| Neurons | Agrn | 907 | −0.5 | <1 E-300 | <1 E-300 |
| App | 5181 | 0.2 | <1 E-300 | <1 E-300 | |
| Atp6v0a1 | 1815 | −0.66 | <1 E-300 | <1 E-300 | |
| Clta | 3032 | −0.81 | <1 E-300 | <1 E-300 | |
| Dync1i2 | 1618 | −0.82 | <1 E-300 | <1 E-300 | |
| Nfasc | 821 | −0.94 | <1 E-300 | <1 E-300 | |
| Rab6 | 3058 | 0.34 | <1 E-300 | <1 E-300 | |
| Mtss1 | 1244 | −0.6 | 5.19 E-216 | 7.02 E-214 | |
| Srgap3 | 834 | 0.62 | 1.69 E-132 | 1.52 E-130 | |
| Lrp8 | 328 | −0.75 | 3.23 E-119 | 2.51 E-117 | |
| Oligodendrocytes | Phldb1 | 2947 | −0.58 | <1 E-300 | <1 E-300 |
| Aplp2 | 1358 | 0.48 | <1 E-300 | <1 E-300 | |
| Capzb | 1350 | −0.58 | 5.97 E-263 | 3.54 E-260 | |
| Add1 | 1515 | −0.51 | 1.41 E-231 | 5.95 E-229 | |
| Mpzl1 | 1165 | 0.45 | 4.37 E-227 | 3.15 E-223 | |
| Cldnd1 | 1187 | −0.6 | 2.49 E-209 | 8.2 E-207 | |
| Enpp2 | 1550 | 0.23 | 1.63 E-209 | 2.35 E-191 | |
| H2afy | 320 | 0.43 | 1.31 E-35 | 5.11 E-34 | |
| Mtss1 | 181 | 0.44 | 1.58 E-24 | 4.54 E-23 | |
| Snap25 | 100 | 0.39 | 8.8 E-22 | 2.1 E-20 | |
| Microglia | Clstn1 | 588 | 0.91 | <1 E-300 | <1 E-300 |
| H13 | 1263 | 0.2 | 6.2 E-296 | 3.94 E-283 | |
| Sema4d | 768 | 0.6 | 1.19 E-282 | 6.79 E-280 | |
| App | 705 | −0.6 | 5.33 E-240 | 2.04 E-237 | |
| Add1 | 509 | 0.57 | 9.97 E-188 | 3.17 E-185 | |
| Lass5 | 408 | 0.65 | 2.49 E-174 | 7.12 E-172 | |
| Rapgef1 | 355 | 0.45 | 2.22 E-173 | 5.79 E-171 | |
| Fmnl1 | 493 | 0.44 | 1.32 E-158 | 3.15 E-156 | |
| Fez2 | 853 | 0.36 | 1.61 E-139 | 3.08 E-137 | |
| Fyn | 131 | 0.68 | 7.07 E-89 | 9.31 E-87 | |
| Endothelial | Adam15 | 893 | 0.7 | <1 E-300 | <1 E-300 |
| Mcf2l | 629 | 0.74 | <1 E-300 | <1 E-300 | |
| Palm | 942 | −0.65 | <1 E-300 | <1 E-300 | |
| Ablim1 | 1025 | 0.47 | <1 E-300 | <1 E-300 | |
| Mprip | 3292 | −0.51 | <1 E-300 | <1 E-300 | |
| Actn4 | 1805 | −0.31 | <1 E-300 | <1 E-300 | |
| Ktn1 | 809 | −0.7 | 3.84 E-226 | 1.02 E-223 | |
| Arhgef1 | 865 | 0.36 | 2.23 E-219 | 5.66 E-217 | |
| Eif4h | 1199 | 0.46 | 1.14 E-197 | 2.29 E-195 | |
| Pkp4 | 577 | 0.63 | 1.68 E-195 | 3.26 E-193 |
Figure 4.