Abstract
Yeasts have been used for thousands of years to make fermented foods and beverages, such as beer, wine, sake, and bread. However, the choice for a particular yeast strain or species for a specific industrial application is often based on historical, rather than scientific grounds. Moreover, new biotechnological yeast applications, such as the production of second-generation biofuels, confront yeast with environments and challenges that differ from those encountered in traditional food fermentations. Together, this implies that there are interesting opportunities to isolate or generate yeast variants that perform better than the currently used strains. Here, we discuss the different strategies of strain selection and improvement available for both conventional and nonconventional yeasts. Exploiting the existing natural diversity and using techniques such as mutagenesis, protoplast fusion, breeding, genome shuffling and directed evolution to generate artificial diversity, or the use of genetic modification strategies to alter traits in a more targeted way, have led to the selection of superior industrial yeasts. Furthermore, recent technological advances allowed the development of high-throughput techniques, such as ‘global transcription machinery engineering’ (gTME), to induce genetic variation, providing a new source of yeast genetic diversity.
Keywords: genetic engineering, metabolic engineering, Saccharomyces cerevisiae, GMO, non-Saccharomyces, evolutionary engineering
Introduction
Microorganisms, such as yeasts, bacteria, and algae, are key players in numerous industrial processes, ranging from the production of traditional fermented foods and beverages to recombinant proteins and other high-value molecules.
Many of these industrial processes rely heavily on the model yeast Saccharomyces cerevisiae. This yeast is traditionally used in the food industry for the production of alcoholic beverages, such as beer, wine, and sake, as well as for bread fermentation. More recently, S. cerevisiae has also been used in the bioethanol industry and for the production of heterologous compounds, such as human insulin, hepatitis vaccines, and human papillomavirus vaccines (Hou et al., 2012). Notwithstanding the fact that S. cerevisiae remains by far the most widely used industrial yeast species to date, other, so-called nonconventional yeasts, such as Scheffersomyces stipitis, Yarrowia lipolytica, Kluyveromyces lactis, and Dekkera bruxellensis, have also claimed their stake as valuable contributors to industrial fermentation processes.
Despite the intensive use of these and other yeasts in biotechnological applications and industrial fermentations, there is still significant room for improvement: industrial processes are rarely using the most suited or best-performing strain. This is because many industrial strains are currently used because of historical grounds, rather than being carefully selected for a specific application, and are therefore often suboptimal for their purposes. Additionally, demands for increased productivity, wider substrate range utilization, and production of nonconventional compounds in industry, as well as changing consumer preferences, lead to a great interest in further improving the currently used industrial strains and the selection or development of strains with novel properties.
This review aims to give a comprehensive overview of the different strategies that can be used to obtain strains with improved properties. The relevance and importance of these different techniques are illustrated with specific examples from the beer, wine, bread, and biofuel industry. Both the classical approaches of strain improvement and more recent techniques are discussed. In the first section, we discuss natural yeast diversity and the underlying genetic principles and also summarize how the existing natural diversity can be exploited to select strains with a suitable phenotype for a specific industrial application. In the second part, we show how this natural diversity can be further increased by artificially generating variants through mutagenesis, various hybridization methods, and/or directed evolution. Subsequently, the principles of genetic modification are explained, together with specific examples illustrating the applicability of this strategy to food-based fermentations and production of biofuels and high-value chemical compounds. In the final part, we outline several recent advances in genetic modification for the creation of phenotypic variation, illustrating how these cutting-edge techniques combine aspects of traditional technologies with genetic modification and how they could contribute to future improvement strategies for industrial yeasts. Although the major focus of this review is the improvement of the main fermentation workhorse S. cerevisiae and its close relatives, such as Saccharomyces pastorianus, we also highlight the advances that have been made with other, industrially relevant nonconventional yeasts.
Natural and artificial diversity
Introduction
There are multiple strategies developed that aim to provide suitable yeast strains for specific industrial goals. A deceivingly simple, yet very powerful way is to exploit the natural biodiversity by selecting a strain that performs best in a particular industrial process. Indeed, recent (meta)genomics studies indicate that the natural fungal biodiversity is enormous and largely unexplored, with the current industrial strains only representing a small fraction of the natural biodiversity (Liti et al., 2009; Wang et al., 2012b). This implies that nature possibly harbors multiple, as yet unknown species and strains that may prove superior for certain industrial fermentations. Even if many of these strains (for various reasons) turn out to be unsuitable for direct industrial implementation, they may possess certain industrially relevant characteristics. Specific strategies could allow transfer of these properties to industrial strains, thereby creating novel yeasts with extra beneficial features (Fig.1).
Figure 1.
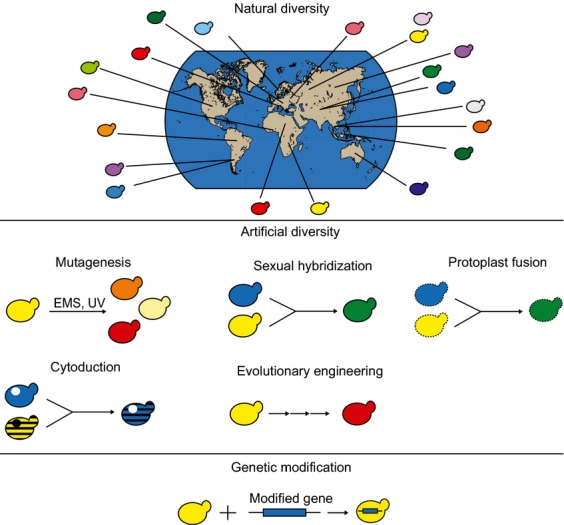
Overview of strategies to obtain superior industrial yeast strains. In order to select novel yeast strains for industrial applications, several strategies can be applied. First, the existing natural diversity can be explored by genotyping and phenotyping isolated feral strains or strains from yeast collections to select the most interesting variants. Apart from investigating naturally occurring yeasts, diversity can also be generated artificially. There are multiple strategies to induce genetic diversity in a single strain or shuffle the genomes of multiple strains. Strains resulting from these strategies are all considered non-genetically modified yeasts, implying that they can be freely used in industrial fermentations. These strategies will be further discussed in the section on ‘Generation of artificial diversity’. Lastly, strategies based on genetic engineering, where a recombinant piece of DNA is transformed in a target strain to confer a specific, industrially relevant phenotype to this strain, can be very efficient. However, this technique genetically modifies yeasts, currently limiting their use in food or beverage fermentations because of consumer concerns.
In addition to exploring naturally occurring yeast variants, several techniques allow researchers to further increase the diversity of yeasts by artificially generating variants, starting from feral or industrial strains. The applicability of these techniques depends on both the targeted phenotype and genetic background. The targeted phenotype is sometimes limited by its selectability (cf. infra), while the complex or understudied genetic background of certain yeasts can sometimes hamper their use in improvement strategies. For example, many industrial S. cerevisiae strains have a much more complex genetic architecture compared to laboratory strains, the latter being carefully bred and selected for sexual reproduction, optimal growth, and easy handling in the laboratory (e.g. no flocculation) (Mortimer & Johnston, 1986), while industrial strains often show aneuploidy and/or polyploidy, poor sporulation efficiency, unstable mating types, etc. Moreover, recent full-genome sequencing and large-scale phenotyping experiments underscore that these ‘tamed’ laboratory strains are not representative for the majority of industrial strains (Liti et al., 2009; Borneman et al., 2011; Warringer et al., 2011). Together, this implies that although most fundamental studies were performed on easy-to-use laboratory strains, such as S288c or EM93, many of these results cannot be simply extrapolated to industrial strains. Therefore, we also address the potential problems accompanying improvement strategies for genetically complex or understudied yeast strains and species. While this section only focuses on so-called non-genetically modified organisms (non-GMO) techniques to create artificial diversity, the last section will further discuss techniques based on recombinant DNA technologies, such as metabolic engineering and synthetic biology, which generate genetically modified (GM) yeasts.
Before we address examples of how natural or artificially generated diversity can provide industrially relevant strains, we first summarize the genetic mechanisms driving the genetic diversity in yeast. Next, different strategies to yield improved yeast strains are described, pros and cons are discussed, and the challenges associated with the selection of optimal strains are given.
Origins of yeast diversity
Yeasts represent a very diverse group of organisms, and even strains that are classified as the same species often show a high level of genetic divergence. The natural diversity of yeasts (between and even within species) has become very clear with the advent of next-generation sequencing technologies that enable in-depth characterization of the genetic variation (Dujon et al., 2004; Liti et al., 2009; Borneman et al., 2011; Wang et al., 2012b; Borneman et al., 2013; see section on ‘Natural yeast diversity and how it can be exploited in industry’).
Genetic variability can be generated by several different processes, including sexual reproduction (where the genomes of two parents are mixed and shuffled), changes in the DNA sequence such as point mutations (i.e. changes in single nucleotides), and InDels (i.e. insertion or deletion events of relatively short pieces of DNA), changes in ploidy level (where a whole genome, or large parts, is duplicated or lost), transposons (mobile genetic elements that can cause mutations by insertion in the genome), genetic recombination (where parts of the genome are re-organized; it can act on both homologous and nonhomologous loci), or acquisition of exogenic pieces of DNA by horizontal gene transfer (HGT) (Fig.2). Whereas each of these processes described above has been shown to occur in nature and therefore contribute to genetic diversity, the phenotypic outcome and importance of each of these processes is hard to estimate and often also depends on the exact environmental conditions.
Figure 2.
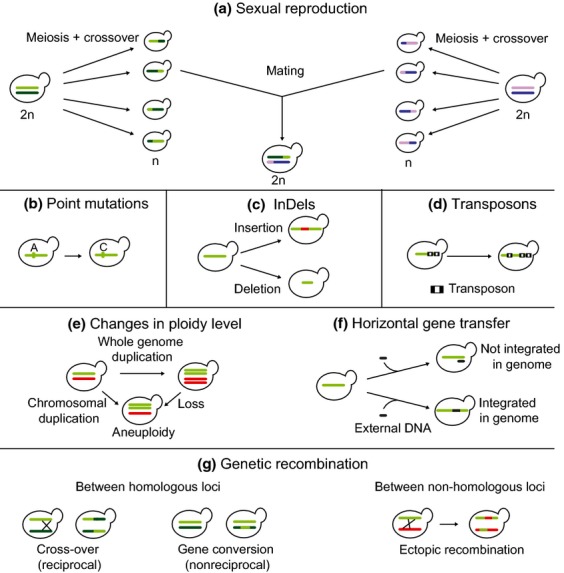
Origins of genetic variation in yeast. Genetic variation can be caused by several different mechanisms. For sake of simplicity, only one chromosome per yeast cell is displayed (green or purple). Different color shades represent homologous chromosomes. In (e), a second chromosome is represented in red. (a) Sexual reproduction: after sporulation and concomitant meiotic cross-over events in the parental strains (2n), genomes of two haploid (n) segregants can hybridize, a process called mating. (b) Point mutations: changes in single nucleotides. These mutations can be synonymous or nonsynonymous: synonymous mutations do not change the amino acid sequence, while nonsynonymous mutations do. Nonsynonymous mutations are therefore more likely to alter the phenotype. (c) InDels: insertion and deletion events of relatively short pieces of DNA. (d) Transposons: insertion of transposable elements in the genome. (e) Changes in ploidy level: the whole genome, or large parts, is duplicated or lost, which can result in poly- or aneuploidies. (f) Horizontal gene transfer: transfer of genes by means other than regular sexual reproduction. (g) Genetic recombination: reorganization of parts of the genome. It can act on both homologous (cross-over and gene conversion) and nonhomologous loci (ectopic recombination). Homologous recombination such as gene conversion (nonreciprocal transfer of genetic material between highly homologous genes) occurs relatively frequently and can sometimes give rise to novel or modified traits. Ectopic recombination events such as TY-promoted chromosomal translocations are more rare, but can drastically rearrange the genome, and even generate novel genes.
Sexual recombination is the single most important process that generates genetic diversity in higher eukaryotes such as animals and plants. Similarly, in yeasts with a sexual life cycle, such as Saccharomyces spp., sexual reproduction can reshuffle the genomes of different yeast strains, thereby altering their characteristics, and potentially even lead to the evolution of new species. Although sexual recombination is mostly thought of as a ‘natural’ process, mating also occurs between strains or species in industrial settings. A notable example is S. pastorianus, an interspecific hybrid of S. cerevisiae and Saccharomyces eubayanus, which may have originated in the fermentation tank of lager breweries (Libkind et al., 2011). The added advantage of cryotolerance (typical for S. eubayanus) may have led to its observed ability to carry out fermentations better at low temperatures than S. cerevisiae alone (Dunn & Sherlock, 2008).
In contrast to higher eukaryotes, yeasts such as Saccharomyces spp. also have the ability to reproduce asexually. Moreover, this vegetative asexual proliferation is a much more prevalent way of reproduction, with on average only one meiotic cycle for every 1000 mitotic divisions (Ruderfer et al., 2006; Tsai et al., 2008; Zörgö et al., 2012). During these asexual reproductive cycles, spontaneous mutations, such as point mutations, InDels, transposon insertions, and recombination events, can occur. The rate at which spontaneous mutations occur varies across the yeast genome, but has been estimated to be around 3.80 × 10−10 and 6.44 × 10−10/(bp × generation) when measured at two specific loci that allowed for phenotypic detection of mutations (Lang & Murray, 2008). However, spontaneous mutations occur much more frequently in so-called mutation hotspots such as subtelomeric regions and tandem repeats (Ellegren, 2004; Brown et al., 2010; Gemayel et al., 2010; Christiaens et al., 2012), where the mutation rates are often 10–100 000 times higher than average mutation rates in other parts of the genome (Gemayel et al., 2010). One particular category of spontaneous mutations which occurs often in these hotspots is genetic recombination. An interesting example of a gene family that is extremely prone to spontaneous mutations, because they are located subtelomerically and additionally contain tandem repeats, are the genes encoding flocculins (FLO genes). These flocculins are responsible for ‘flocculation’ of yeast cells, a trait of major importance in the wine, beer, and biofuel industry. The instability of the tandem repeats in these genes, which are causing expansion and contraction in the gene size, has allowed for the fast isolation of spontaneous mutants with altered flocculation characteristics (Hammond, 1996; Verstrepen et al., 2005; Ma et al., 2009). Additionally, the available arsenal of flocculins in Saccharomyces yeasts is hugely increased by the generation of chimeric FLO genes by ectopic recombination, which resulted in a huge diversity in the flocculation phenotype of industrial strains (Christiaens et al., 2012).
Although spontaneous mutations are one of the main driving forces behind evolution and enable organisms to adapt to specific ecological niches, most mutations are neutral or even deleterious (Drake, 1991). Most yeast species (including S. cerevisiae) generally exist in a diploid stage, which can mask the effect of (heterozygous) deleterious mutations. This can cause these mutations to accumulate during continuous asexual growth, leading to strains with a high mutational load. However, because of its complex sexual life cycle, yeast can filter out these deleterious mutations by a process called ‘genome renewal’ (Mortimer et al., 1994; Mortimer, 2000; Wang et al., 2012b). In essence, genome renewal results in the elimination of cells carrying deleterious recessive mutations from the population, while generating homozygous diploid cells in which these mutations are not present. Genome renewal is mainly described for indigenous Saccharomyces wine yeasts, which often show a high sporulation capacity and are homothallic (see Fig.3), two important prerequisites for genome renewal. Asexually growing cells (with a potentially high mutational load) will undergo meiosis and sporulate. After germinating, viable haploids will start reproducing asexually, and subsequently, mating can occur between neighboring ‘sister’ cells of the opposite mating type (a process called haplo-selfing), yielding a homozygous diploid inbred cell. In this way, the haploid stage filters out lethal recessive mutations and the subsequent haplo-selfing enables recessive, heterozygous mutations to become homozygous and thus influence the phenotype. Intratetrad mating (automixis) after sporulation will have the same effect (Katz Ezov et al., 2010). Together, this can improve cellular fitness and adaptability to the environment (Pretorius, 2000).
Figure 3.
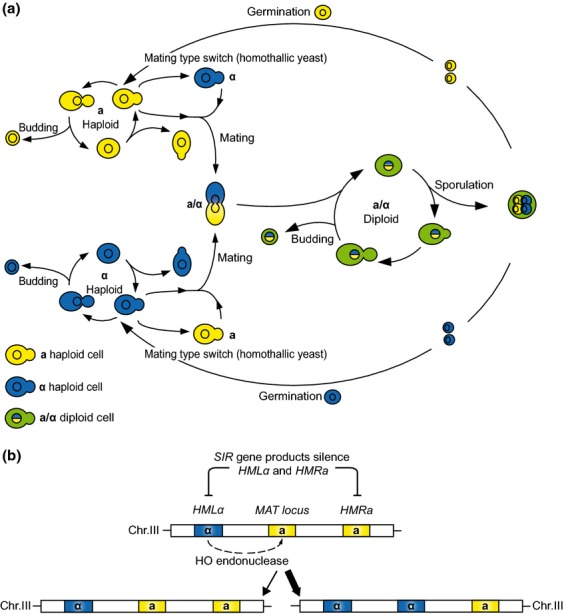
Life cycle of S. cerevisiae. Yeast cells can exist in both a haploid and diploid state. (a) Diploid cells are heterozygous for the mating type locus (mating type a/α), which makes diploids incapable of mating. Haploid cells have either mating type a or mating type α, making them capable of mating with a cell of the opposite mating type. In nutrient-rich conditions, both haploid and diploid cells can proliferate asexually by budding. When exposed to some nutrient-poor conditions, diploids can undergo sporulation (meiosis followed by spore formation), resulting in the conversion of a diploid cell into four haploid spores, two possessing mating type a and two having mating type α, which can germinate into haploid cells when conditions improve. In homothallic strains, the haploid derivatives can undergo a mating type switch (together with the mother cell), mediated by an endonuclease encoded by the HO gene. In this way, a mating type-switched cell can mate with neighboring sister cells of the opposite mating type, resulting in a homozygous (except for the MAT locus, which determines the mating type) diploid. In heterothallic strains, the HO gene is typically inactive and therefore haploid derivatives cannot switch mating type. (b) Mechanism of the mating type switch of homothallic strains. On chromosome III, the MAT locus is flanked by Hidden MAT Left and Right (HML and HMR, respectively), carrying a silenced copy of MATα and MATa, respectively. Homothallic strains contain the HO gene, a gene coding for an endonuclease that cleaves DNA specifically at the MAT locus. After breakdown of the MAT locus by exonucleases, a gene conversion event occurs, where HML or HMR is used as a template to repair the DNA strand. Because cells prefer to change their mating type, that is, a MATα cell will rather use HMR as a template and vice versa, mating-type switch occurs frequently.
In addition to sexual recombination and spontaneous mutations, the transfer of genetic material through asexual mechanisms, called horizontal gene transfer (HGT), can contribute to genetic diversity. HGT is rare in yeast, and the mechanisms underlying HGT are not yet elucidated, but natural transformation and conjugation have been proposed (Hall et al., 2005). An interesting example of how HGT altered industrially relevant phenotypes in yeast is described for the commonly used wine strain EC1118. Genetic analysis of EC1118 showed strong evidence for the HGT of three DNA regions, encompassing 34 genes involved in key wine fermentation functions (Novo et al., 2009). For example, FSY1, a gene coding for a high-affinity fructose/H+ symporter present in one of the regions, could confer a significant advantage during the wine fermentation process by enabling the yeast to efficiently utilize left-over fructose at the end of the fermentation (Galeote et al., 2010). Interestingly, two of these regions were acquired from non-Saccharomyces species, and Zygosaccharomyces bailii, a typical contaminant of wine fermentations, was identified as the donor of one region; indicating that HGT in yeast can cross genus boundaries.
Lastly, changes in ploidy level also occur and can have profound phenotypic effects. This is best described for the Saccharomyces lineage, which has undergone a whole-genome duplication (WGD) event about 100 million years ago. The most important consequences of this WGD are that an extra copy of the genome allowed a global rewiring of the yeast transcriptional network and gave the duplicated genes a chance to mutate and gain new or adapted functions compared to the original genes (reviewed by Piskur et al., 2006). Additionally, duplication of single genes pre- or post-WGD, such as the duplication of the alcohol dehydrogenase (ADH) gene approx. 80 million years ago, also contributed to the success of Saccharomyces species in the fermentation industry. In the case of ADH, reconstruction of the ancestral gene revealed that the encoding enzyme had a preference to convert acetaldehyde to ethanol and was therefore involved in the generation, and not the consumption, of ethanol (Thomson et al., 2005). Spontaneous duplication of this ancestral gene generated the genes encoding the Adh1 and Adh2 enzymes. These present-day enzymes show a different kinetic behavior, with Adh2 binding ethanol (its substrate) more strongly than Adh1. Duplication of the ancestral ADH gene as well as duplication of several other genes involved in ethanol metabolism, combined with the ability of Saccharomyces spp. to ferment glucose and accumulate ethanol even in the presence of oxygen (a phenomenon known as the Crabtree effect), resulted in the so-called make-accumulate-consume strategy of ethanol production, giving them a competitive advantage over other microorganisms during fermentation processes (Piskur et al., 2006).
Natural yeast diversity and how it can be exploited in industry
Although the basic principles underlying genetic variation are known (cf. supra), the extent of genetic biodiversity in Saccharomyces strains has only recently been elucidated. Two recent seminal papers provided a first comprehensive overview of the genetic architecture of feral (wild) and industrial Saccharomyces strains (Liti et al., 2009; Schacherer et al., 2009). Interestingly, genetic diversity within Saccharomyces paradoxus, a widespread feral species closely related to S. cerevisiae, can almost completely be explained by geographical origin. However, diversity within S. cerevisiae, the main yeast associated with human activity, was at least partly linked to its industrial application (Legras et al., 2007; Dunn & Sherlock, 2008; Liti et al., 2009; Schacherer et al., 2009).
Additionally, despite that it was generally believed that S. cerevisiae was a domesticated species with no truly natural strains existing, it was recently shown that diverged populations of wild S. cerevisiae exist independently of domesticated isolates (Fay & Benavides, 2005; Liti et al., 2009; Sicard & Legras, 2011; Wang et al., 2012b). Moreover, genetic analysis of these feral and industrial strains revealed that the genetic diversity within industrial strains is rather limited compared to the full spectrum of natural biodiversity. For example, while the nucleotide diversity (π, the average number of nucleotide differences per site between any two DNA sequences chosen randomly from the sample population) was calculated to be 0.56 × 10−3 in 14 representative wine yeasts, a more elaborate analysis of 138 strains, including both industrial and feral strains, revealed a sequence diversity that was more than one order of magnitude larger (7.27 × 10−3) (Wang et al., 2012b). It is important to note that this is truly remarkable, because the nucleotide diversity within Homo sapiens is estimated to be ‘only’ 1 × 10−3 (Jorde & Wooding, 2004). Furthermore, even the genetic diversity within geographically isolated feral S. cerevisiae populations can be substantial, illustrated by the fact that isolates from the primeval rainforests in Hainan, a tropical island in southern China, exhibited a magnitude of genetic diversity equivalent to the diversity of the complete human population (Wang et al., 2012b). Together, these observations suggest that the fermentation industry currently relies on only a very small fraction of the available genetic diversity of S. cerevisiae, ignoring a huge pool of unexplored (feral) strains. Although currently used strains have adapted well to the fermentation environment, this largely uncharted yeast pool might contain strains with characteristics potentially valuable for industrial applications.
To exploit this huge natural diversity, several teams have screened diverse feral yeast collections for industrially relevant traits (e.g. Pellegrini et al., 1999; Comitini et al., 2011). Additionally, isolation and analysis of feral or even contaminating yeast strains from niches similar (or identical) to a specific fermentation environment can yield interesting strains for starter cultures. Indeed, continuous evolution and adaptation of indigenous yeast strains to their environment have equipped these strains with phenotypes valuable for industry. For example, several research papers have described the isolation, selection, and incorporation of indigenous wine yeasts as starter cultures in the production of wine (Zagorc et al., 2001; Mannazzu et al., 2002; Lopes et al., 2007; Tosi et al., 2009; Capece et al., 2010; de Ullivarri et al., 2011; Scacco et al., 2012; Tristezza et al., 2012). Similarly, the application of indigenous yeast strains revolutionized the biofuel industry, where the initially used baker's strains were replaced by ‘contaminating’ strains, which were well adapted to the harsh fermentation environment (Basso et al., 1993, 2008; da Silva-Filho et al., 2005). Moreover, two recently isolated S. cerevisiae strains are now responsible for up to 70% of the total Brazilian biofuel production (Basso et al., 2008; Della-Bianca et al., 2012). Likewise, the identification of Dekkera bruxellensis as a contaminating yeast in a Swedish bioethanol production site has raised interest in applying this species as a starter culture, because this strain did not show a compromised fermentation efficiency and exhibited a more energy-efficient metabolism under oxygen limitation than the initially applied S. cerevisiae starter culture (Passoth et al., 2007; Blomqvist et al., 2010).
Generation of artificial diversity
Despite the immense wealth of natural yeast diversity, the extremely selective and specific conditions of industrial fermentations sometimes require (a combination of) phenotypic traits that might not be commonly encountered in nature. While the physiological behavior of feral yeasts is exclusively dedicated to survival and reproduction, most industrial fermentations require maximization of processes and characteristics that may not be beneficial in natural environments. Several techniques have therefore been developed to artificially increase the existing yeast diversity and generate variants that may perform better in industrial settings than the strains that are selected in natural environments.
Perhaps the most intuitive way to generate artificial diversity in yeasts is by (human-driven) sexual hybridization (also known as crossing or mating; Tables1 and S1). This practice is very similar to the common ‘selective breeding’ (or ‘artificial selection’) encountered in, for example, agriculture. This technique has been used by humans for thousands of years, for example by farmers who intuitively chose superior plants from their cultivations or animals from their stock to crossbreed in order to obtain crops or livestock with desired traits (Chambers et al., 2009; Steensels et al., 2012). Similarly, the close association of S. cerevisiae with human activities has led to the so-called domestication of this species, resulting in an organism that excels in its industrial task, but performs suboptimal in most other, more ‘natural’ environments (Fay & Benavides, 2005; Liti et al., 2009; Sicard & Legras, 2011). Moreover, human selection may even have given rise to new chimeric species in the Saccharomyces sensu stricto complex in industrial environments, such as the lager yeast S. pastorianus (Libkind et al., 2011). Although these processes occurred naturally (brewers, winemakers, or other craftsmen did not intentionally breed novel yeast strains), recent technological advances and the rapidly increasing knowledge about yeast physiology (e.g. the description of its sexual life cycle) paved the way for more targeted and large-scale approaches of yeast breeding, even beyond species barriers. This targeted breeding of yeast strains can now be used to create novel strains that combine different characteristics of the selected parents, or to optimize a single, often complex, trait, by crossing parents selected for the same phenotype. Using the latter approach, traits can theoretically be improved even beyond the phenotypic boundaries of the parental strains, a phenomenon called heterosis, or hybrid vigor (Lippman & Zamir, 2007). This phenomenon is occasionally encountered for certain traits in breeding experiments (Marullo et al., 2006; Timberlake et al., 2011), but the incidence is generally low (Zörgö et al., 2012). Later in this section, four approaches for strain improvement using sexual hybridization approaches will be discussed: direct mating, rare mating, mass mating, and genome shuffling.
Table 1.
Several key studies using hybridization techniques for the improvement of industrial yeasts. The phenotypes of interest from both parental strains are indicated. For a more extensive list, see Table S1
| Parental strain 1 | Parental strain 2 | Hybridization technique | Industrial application | References | |
|---|---|---|---|---|---|
| Intraspecific hybridization | |||||
| S. cerevisiae | x | S. cerevisiae | Protoplast fusion | Biofuel | Javadekar et al. (1995) |
| Fermentation performance | Flocculation | ||||
| S. cerevisiae | x | S. cerevisiae | Cytoduction | Beer | Hammond & Eckersley (1984) |
| Fermentation performance | Killer phenotype | ||||
| S. cerevisiae | x | S. cerevisiae | Rare mating | Bread | Oda & Ouchi (1990) |
| Fermentation performance | Fermentation performance | ||||
| S. cerevisiae | x | S. cerevisiae | Spore-to-cell mating | Biofuel | Benjaphokee et al. (2012) |
| Temperature tolerance | Ethanol tolerance | ||||
| Interspecific hybridization | |||||
| S. cerevisiae | x | S. cerevisiae (var. diastaticus) | Rare mating | Beer | Tubb et al. (1981) |
| Fermentation performance | Dextrin degradation | ||||
| S. cerevisiae | x | S. mikitae/S. paradoxus/S. kudriavzevii | Rare mating | Wine | Bellon et al. (2011, 2013) |
| Fermentation performance | Flavor profile | ||||
| S. cerevisiae | x | S. bayanus | Protoplast fusion | Biofuel | Choi et al. (2010) |
| Ethanol tolerance | Flocculation | ||||
| S. cerevisiae Fermentation performance | x | S. kudravzevii Low temperature | Spore-to-spore/rare mating/protoplast;fusion | Wine | Perez-Traves et al. (2012) |
| S. cerevisiae | x | S. bayanus | Spore-to-spore mating | Wine | Coloretti et al. (2006) |
| Flocculation | Fermentation performance | ||||
| Intergeneric hybridization | |||||
| S. cerevisiae | x | K. lactis | Protoplast fusion | Biofuel | Taya et al. (1984) |
| Fermentation performance | Lactose utilization | ||||
| S. cerevisiae Fermentation performance/ethanol tolerance | x | Sc. stipitis Xylose utilization | Protoplast fusion and genome shuffling | Biofuel | Zhang & Geng (2012) |
| S. cerevisiae | x | T. delbrueckii | Protoplast fusion | Polyol production/bakery | Lucca et al. (1999, 2002) |
| Fermentation performance | Osmotolerance | ||||
| S. cerevisiae | x | Sc. pombe | Protoplast fusion | Wine | Carrau et al. (1994) |
| Fermentation performance | Malic acid degradation | ||||
| S. cerevisiae (var. diastaticus) | x | Z. rouxii | Protoplast fusion | Bakery | Spencer et al. (1985) |
| Fermentation performance | Osmotolerance | ||||
K. lactis, Kluyveromyces lactis; S. bayanus, Saccharomyces bayanus; S. cerevisiae, Saccharomyces cerevisiae; S. cerevisiae (var. diastaticus), Saccharomyces cerevisiae (var. diastaticus); S. kudravzevii, Saccharomyces kudravzevii; S. mikitae, Saccharomyces mikitae; S. paradoxus, Saccharomyces paradoxus; Sc. pombe, Schizosaccharomyces pombe; Sc. stipitis, Scheffersomyces stipitis; T. delbrueckii, Torulaspora delbrueckii; Z. rouxii, Zygosaccharomyces rouxii.
However, because many industrial strains are, in contrary to most laboratory strains, polyploid or aneuploid and display a low sporulation efficiency and/or low spore viability (Mortimer et al., 1994; Codon et al., 1995), strain improvement by sexual hybridization is not always possible. Still, the unique features of yeast (such as its exceptional life cycle that combines sexual and asexual replication strategies (Fig.3) and their short generation time) facilitate several other strain improvement strategies. For example, a hybridization technique referred to as protoplast (or spheroplast) fusion was designed to hybridize cells asexually, thereby fully precluding the need for sporulation capacity of and sexual compatibility between both parental strains. Additionally, the transfer of non-Mendelian traits, caused by cytoplasmic factors such as dsRNA virus-like particles or prions, without compromising the genome structure of the parental cells, can be accomplished by cytoduction. These techniques, all aiming to combine traits from two parental strains by hybridization, are summarized in Fig.4. Other techniques, such as mutagenesis and directed evolution, use the asexual reproductive cycle of yeast and fully rely on random genetic mutations and strict selection procedures to isolate phenotypically improved variants. This section provides a comprehensive overview of all these techniques, which do not rely on recombinant DNA technology and for which resulting strains are thus considered non-GMO, and discusses their practical applicability, advantages, and limitations. Additionally, specific examples indicating how industrial fermentation processes have benefited from the resulting improved strains are given.
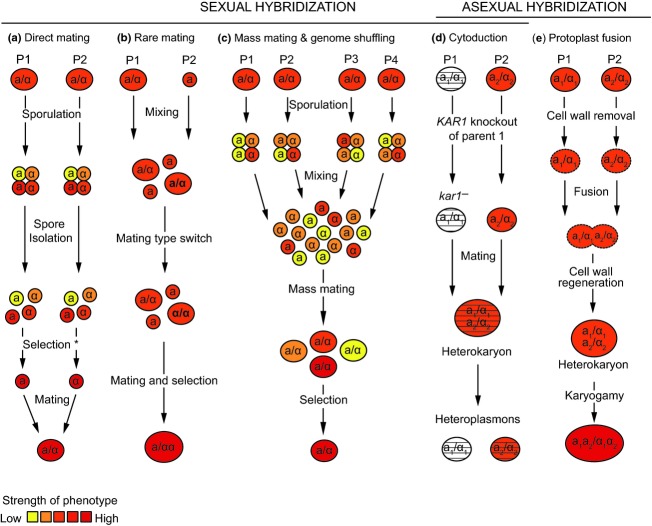
Figure 4.
Overview of different strain improvement techniques using hybridization. Sexual and asexual hybridization is a powerful technique to generate artificial diversity in yeast. Due to the sometimes complex genetics (ploidy, sporulation, …) of yeast, different techniques have been developed. Most techniques start from two parental (P) strains, selected for the target phenotype. The color scheme indicates the strength of the phenotype, for example red = strong ethanol tolerance, yellow = weak ethanol tolerance. In these examples, the parental strains are selected for the same phenotype, but combining different phenotypes of both parents is also possible. (a) In direct mating, two haploid cells or spores of opposite mating types are crossed. When the parental strains are both heterothallic, these haploids can be prescreened and selected, and cell-to-cell mating can be applied. When both or one of the parental strains is homothallic, spore-to-spore or spore-to-cell mating, respectively, can be used. In these latter cases, the selection step (indicated with *) cannot be applied. (b) In rare mating, strains are crossed without a sporulation step. This is possible because diploid yeasts occasionally (but rarely) undergo a homothallic mating-type switch, yielding an a/a or α/α diploid cell. These cells can subsequently hybridize with a haploid cell of the opposite mating type. It is important to note that rare mating is not limited to the development of triploid yeasts. For example, tetraploid hybrids can be obtained if P2 would be an a/a type yeast. (c) In mass mating, multiple parental strains, or a heterogeneous population of the same parental strain, can be used. After mass sporulation and mixing of the resulting spores, mass mating will occur. These rounds of mass sporulation and mass mating can be repeated multiple times, a process which is one way to perform so-called genome shuffling. In genome shuffling, the mass sporulation and mass mating steps can also be replaced by protoplast fusion. (d) Cytoduction can be used to transfer cytoplasmically inherited traits. First, the KAR1 gene of the parental strain containing the targeted cytoplasmic trait is deleted. Next, both parental strains are crossed (or fused by protoplast fusion), but because karyogamy is blocked, the heterokaryon segregates into cells containing a nucleus of only one parent but the cytoplasmic components of both parents (=heteroplasmons). With proper selection, this technique can also yield so-called disomic strains that contain the full chromosome complement of one parent plus one chromosome from the other parent. (e) In protoplast fusion, cells are asexually merged after cell wall removal in osmotically supportive medium. After cell wall regeneration, the formed transient heterokaryons may undergo karyogamy and form hybrids.
Direct mating
Direct mating is the most intuitive way of breeding organisms. Similar to selective breeding in agriculture, it consists of the crossing of two carefully selected parents possessing an interesting phenotype. In case of yeast strains, three distinct approaches exist: cell-to-cell, spore-to-cell, and spore-to-spore mating. The applicability of these approaches depends on the sexual cycle of the parental strains.
If both parental strains are heterothallic (Fig.3b), a prescreening of the stable vegetative haploid segregants of both parents can be carried out, after which the best haploid segregants can be selected for the hybridization experiment. This technique dates back to 1943, when it was described in a seminal paper by Lindegren & Lindegren (1943), and is now called ‘cell-to-cell’ mating. By simply mixing cell cultures of the two selected stable haploid parents and subsequent screening for diploid cells, hybrids can be isolated. The major advantage of cell-to-cell over mass and spore-to-spore mating (cf. infra) is that both haploid parents can be fully phenotyped prior to the breeding experiment (Lindegren, 1949), which increases the chance of yielding a superior hybrid. Additionally, no inbreeding can occur, because no cells of the opposite mating type from the same parental strain are present in the same experiment. Furthermore, the same parent can be used many times in a variety of different mating experiments and can theoretically be preserved indefinitely (Lindegren, 1949). Although there are some clear advantages to this approach, it is not used regularly to develop novel yeast hybrids. This is mainly due to the homothallic nature of most industrial and feral yeast strains, making them unsuited for this approach. However, it was recently described that several feral strains show a stable haploid mating type, due to a mutation in the HO endonuclease gene, a gene responsible for mating-type switching (Katz Ezov et al., 2010), making these strains fit for cell-to-cell mating experiments. In principle, also homothallic strains would be amenable to this approach after genetically disrupting the HO endonuclease gene (van Zyl et al., 1993; Walker et al., 2005; Blasco et al., 2011; Fig.3). However, this requires a genetic transformation, which implies that the resulting hybrid is classified as a GMO and is therefore subject to the GMO legislation. In addition, due to the heterozygous nature of many industrial strains, for some complex phenotypes that are difficult to measure, it can be laborious to identify a haploid descendant that exactly recapitulates the parental phenotype.
When one (‘spore-to-cell’ mating) or both (‘spore-to-spore’ mating) parental strains are homothallic, no stable haploid segregants can be obtained, and the additional prescreening step is not feasible. However, direct mating is still possible by placing two single spores of the strains to be hybridized close to one another on an agar surface, monitoring the hybridization event by microscopy, and isolating the developed zygotes (which can be formed if the spores are of the opposite mating type) using a micromanipulator. This approach is preferred when both parental strains are homothallic or when the hybridization efficiency of the two parental strains (outcrossing) is low compared to the hybridization efficiency of cells of the same parent (inbreeding) and no suitable hybrid selection markers are available to specifically isolate outbreds. The latter is, for example, the case in some interspecific crossing experiments, where the (however weak) pre- and postzygotic barriers (such as differences in germination timing or mating preferences) favor inbreeding (Maclean & Greig, 2008; Morales & Dujon, 2012; Murphy & Zeyl, 2012). In this case, spore-to-spore mating (instead of, e.g., mass mating) can be used to promote outcrossing. A major disadvantage of spore-to-spore compared to cell-to-cell mating is that the spores used in the experiment cannot be characterized prior to the mating and therefore might not display the desired phenotype of the parental strain, due to the segregation of causative alleles (Attfield & Bell, 2003). An additional step that can increase the frequency of developing hybrids with the desired phenotype is a phenotypic prescreening of the self-mated homozygous diploids formed after sporulation and tetrad dissection of the homothallic parental strains (Romano et al., 1985; Marullo et al., 2009), or use of a well-designed backcrossing scheme (Marullo et al., 2009).
Although time-consuming, direct mating has proven to be an effective way to obtain hybrids (Sipiczki, 2008); spore-to-spore and spore-to-cell mating are applied regularly to create novel, mainly interspecific hybrids for the fermentation industry (Table1). It has been used successfully to generate wine yeasts with improved cryotolerance, by crossing S. cerevisiae with cryotolerant species such as S. kudriavzevii or S. bayanus (Kishimoto, 1994; Zambonelli et al., 1997; Perez-Traves et al., 2012), or to introduce flocculation in a yeast strain for the production of sparkling wines (Coloretti et al., 2006). Recently, spore-to-cell mating was applied to develop thermotolerant (Marullo et al., 2009) and multistress-tolerant S. cerevisiae strains (Benjaphokee et al., 2012). Also, novel hybrids with improved characteristics developed by cell-to-cell mating have been reported. Hara et al. (1981) used this approach to construct cryotolerant wine yeasts able to produce killer toxins. Russell et al. (1983) described the use of cell-to-cell mating to eliminate the unwanted ‘phenolic off-flavor’ phenotype from brewer's yeast. This approach was also used to construct wine, bread, and beer yeasts with optimal fermentation characteristics (Gjermansen & Sigsgaard, 1981; Eschenbruch et al., 1982; Nakagawa & Ouchi, 1994; Marullo et al., 2006). More recently, it was used to combine specific phenotypes of ale and lager yeasts in order to improve stress resistance and fermentation performance (Garcia Sanchez et al., 2012).
Rare mating
As mentioned above, many natural and industrial yeasts show low sporulation efficiencies and/or low spore viability, hampering their use in direct mating (or mass mating, cf. infra) experiments. In these cases, rare mating can offer a way to obtain hybrids. Rare mating is based on the rare event that some cells in a diploid population can become homozygous for the mating-type locus (resulting in an a/a or α/α cell) and can subsequently be ‘force-mated’ with a cell of the opposite mating type (Gunge & Nakatomi, 1972), see also Figs3 and 4. Typically, in a rare mating experiment, dense cell suspensions of the parental strains are mixed, and subsequently outcrossed hybrids are isolated using a strong selection step. This selection is often achieved by using a respiratory-deficient and an auxotrophic parental strain, making rare hybrids easily selectable by their prototrophy and respiratory proficiency (Pretorius, 2000; Hammond, 2003; see ‘Selection of outcrossed hybrids’).
Although the frequencies of the mating-type switch and subsequent mating are usually very low (Gunge & Nakatomi, 1972; Hammond, 2003), rare mating has been used to study interspecific hybridization events (de Barros Lopes et al., 2002) as well as for the improvement of multiple yeast traits. Saccharomyces cerevisiae and S. cerevisiae (var. diastaticus) were crossed to develop yeasts able to ferment dextrins (low molecular weight carbohydrates, produced by the hydrolysis of starch) in order to produce low-calorie beers (Tubb et al., 1981). It has also been applied to construct cryotolerant wine yeasts (Perez-Traves et al., 2012), dextrin-fermenting and high ethanol-producing yeasts (Kim & Kim, 1996), and yeasts with higher leavening ability in dough fermentations (Oda & Ouchi, 1990). Recently, Bellon and coworkers used rare mating to construct triploid interspecific hybrids of S. cerevisiae and other Saccharomyces sensu stricto species (like Saccharomyces mikatae) to diversify the flavor profile of wines (Bellon et al., 2011, 2013).
Mass mating and genome shuffling
Because yeasts are such small organisms with short life cycles, it is possible to cultivate billions of individual cells and generate many crosses quickly and to execute consecutive rounds of crossing. These more evolved techniques, often referred to as ‘mass mating’ or ‘genome shuffling’, can significantly increase the throughput and thus success rate of the experiment. Mass mating is a technique in which large numbers of haploid yeast cells, often from different parental strains are mixed and allowed to randomly mate. Mass mating is a particularly useful improvement technique for homothallic strains, for strains that show low mating efficiency, or for the creation of interspecific hybrids if strong selective markers for outbreds are available (Kunicka-Styczynska & Rajkowska, 2011).
Mass mating has been used to generate industrial strains with improved characteristics. For instance, Higgins and coworkers used mass mating and selection to combine different properties of two types of bakery strains, namely high osmotolerance, which is typical for strains used in sweet dough, and good maltose utilization, a characteristic of strains used in unsugared dough. First, haploid segregants of seven industrial strains used in sweetened dough were mass-mated followed by selection for osmotolerance, and this procedure was repeated several times. Second, three strains used for unsugared dough fermentations were subjected to repeated cycles of mass mating and selection for growth on maltose. In a final step, the two enriched populations were sporulated and mass-mated, and strains capable of leavening both sweet and unsugared dough were recovered after additional rounds of mass mating and alternating selection for osmotolerance and growth on maltose (Higgins et al., 2001). Mass mating was also used to create new interspecific wine strains by crossing strains of S. cerevisiae and S. bayanus (Sato et al., 2002; Table1).
Conceptually related to mass mating, genome shuffling is one of the most recent techniques to improve complex phenotypes in microorganisms in a fast and relatively easy manner (see Gong et al., 2009). In a heterogeneous population, a cell displaying a specific phenotype might harbor beneficial mutations that differ from those present in another, phenotypically similar cell within this population. By applying repeated rounds of genetic recombination (either by protoplast fusion or by mass mating) and selection to this population, genome shuffling aims to combine many of these different beneficial mutations in the same cell, leading to additive or synergistic effects on the phenotype under study (Santos & Stephanopoulos, 2008).
Compared to other improvement techniques, genome shuffling has the advantage of exploiting the full genetic diversity in a population and makes it possible to combine useful mutations from many different individuals, while other hybridization methods, such as direct mating, typically involve only a limited number of haploid cells (Fig.4). Additionally, while classical methods of strain improvement often only select the best-performing mutant for the next round, genome shuffling exploits a much larger proportion of the diversity present in the population. The first convincing example of genome shuffling showed rapid improvement of tylosin production in the bacterium Streptomyces fradiae (Zhang et al., 2002). While it initially took 20 rounds of mutagenesis and selection to improve the tylosin titer ninefold, the same result was obtained by applying only two rounds of genome shuffling on a mutagenized population of the same starting strain, demonstrating the potential of this technique to take much larger leaps in the fitness landscape, enabling rapid improvement of a phenotype.
Most studies applying genome shuffling to yeast have focused on enhancing the tolerance to an industrially relevant stress factor and overall fermentation performance. To this end, variation is typically induced by mutagenizing a single strain. Mutants are then screened for the phenotype of interest, and cells showing phenotypic improvement are used as a starting population for multiple rounds of genome shuffling. After each round of genome shuffling, the severity of the stress is (usually) increased. In this way, both laboratory and industrial strains of S. cerevisiae have been improved for phenotypes such as ethanol tolerance, thermotolerance, acetic acid tolerance, and fermentation performance (Table2). Some recent studies also combine metabolic engineering with genome shuffling (Wang & Hou, 2010; Jingping et al., 2012; Tao et al., 2012; Wang et al., 2012a; Demeke et al., 2013). These approaches are promising to optimize strains for second-generation bioethanol production. Although the production of inferior, so-called crippled strains (cf. infra) is a potential disadvantage of genome shuffling because the prevalence of deleterious alleles may result in a majority of variants that perform better at the task they are selected for, but not other important traits, the first proof-of-principle use of a genome-shuffled S. cerevisiae strain in an industrial fermentation environment was recently published (Zheng et al., 2011a, b).
Table 2.
Studies using genome shuffling for the improvement of industrial yeasts. The phenotypes of interest from the parental strain(s) are indicated. The technique used to generate genetic variation and used for recombination is indicated
| Strain(s) | Phenotype | Technique variation | Recombination technique | Industrial application | Reference |
|---|---|---|---|---|---|
| S. cerevisiae (industrial haploid) | Ethanol tolerance, thermotolerance | UV mutagenesis | Protoplast fusion | Biofuel | Shi et al. (2009) |
| S. cerevisiae (laboratory diploid) | Ethanol tolerance | EMS mutagenesis | Mass mating | Biofuel | Hou (2009) |
| S. cerevisiae (industrial diploid) | VHG fermentation capacity | EMS mutagenesis | Mass mating | Biofuel/beer | Hou (2010) |
| S. cerevisiae (industrial diploid ale) | Wort and ethanol tolerance | EMS and UV mutagenesis | Mass mating | Beer | Wang & Hou (2010) |
| S. cerevisiae (industrial diploid biofuel) | Acetic acid tolerance | UV mutagenesis | Mass mating | Biofuel | Zheng et al. (2011a) |
| S. cerevisiae (industrial strains) | Multistress tolerance | Use of two strains | Mass mating | Biofuel | Zheng et al. (2011b) |
| S. cerevisiae (laboratory diploid) | VHG fermentation capacity | EMS mutagenesis | Mass mating | Biofuel/beer | Liu et al. (2011) |
| S. cerevisiae (industrial haploid strains) | Spent sulfite liquor tolerance | UV mutagenesis | Mass mating | Biofuel | Pinel et al. (2011) |
| S. cerevisiae (diploid soil isolate + GMO) | Ethanol production | Use of multiple strains | Protoplast fusion | Biofuel | Jingping et al. (2012) |
| S. cerevisiae (industrial strain) | Heat, acetic acid, and furfural tolerance | DES mutagenesis | Protoplast fusion | Biofuel | Lu et al. (2012) |
| S. cerevisiae (industrial strain) | VHG fermentation capacity | GMO strain | Mass mating | Biofuel | Tao et al. (2012) |
| S. cerevisiae (industrial strain) | VHG fermentation capacity | GMO strain and EMS/UV mutagenesis | Mass mating | Biofuel | Wang et al. (2012a) |
| S. cerevisiae (industrial near triploid) | VHG fermentation capacity | MCB mutagenesis | Mass mating | Biofuel | Zheng et al. (2013a) |
| S. cerevisiae (industrial near triploid) | VHG fermentation capacity and dessication tolerance | MCB mutagenesis | Mass mating | Biofuel | Zheng et al. (2013b) |
| S. cerevisiae (industrial strain) | Xylose fermentation | GMO strain and EMS mutagenesis | Mass mating | Biofuel | Demeke et al. (2013) |
| Z. rouxii (wild-type strain) | Salt tolerance | EMS mutagenesis | Protoplast fusion | Soy sauce | Cao et al. (2009) |
| H. anomala (wild-type strain) | Salt tolerance | EMS and UV mutagenesis | Protoplast fusion | Soy sauce | Cao et al. (2012) |
| Sc. stipitis (wild-type strain) | Spent sulfite liquor tolerance | UV mutagenesis | Mass mating | Biofuel | Bajwa et al. (2010) |
| C. krusei | Acetic acid tolerance | UV mutagenesis | Protoplast fusion | Biofuel | Wei et al. (2008) |
C. krusei, Candida krusei; H. anomala, Hansenula anomala; S. cerevisiae, Saccharomyces cerevisiae; Sc. stipitis, Scheffersomyces stipitis; Z. rouxii, Zygosaccharomyces rouxii; DES, diethylsulfate; EMS, ethyl methane sulfonate; GMO, genetically modified organism; UV, ultraviolet; MCB, methyl benzimidazole-2-yl-carbamate; VHG, very high gravity.
Genome shuffling has been applied to non-Saccharomyces yeasts as well. In high-salt soy sauce fermentations, salt-tolerant yeast strains are added for improved flavor. Cao et al. (2009) enhanced salt stress resistance and flavor formation of Zygosaccharomyces rouxii using three rounds of genome shuffling. In a similar approach, Cao et al. (2012) could increase the salt stress tolerance and soy sauce fermentation performance of Hansenula anomala. Bajwa et al. (2010) exploited the sexual cycle of the pentose-fermenting yeast Sc. stipitis to improve its tolerance to hardwood spent sulfite liquor. Lastly, the acetic acid tolerance of Candida krusei was improved using a protoplast fusion-based genome shuffling strategy (Wei et al., 2008).
Protoplast fusion
Although the rationale for protoplast fusion (often referred to as spheroplast fusion) is very similar to sexual hybridization (they both strive to combine positive traits of multiple parents in one hybrid strain), this technique can be used for strains that do not meet the requirements for sexual hybridization. This means that this technique is useful for strains that cannot sporulate, yield inviable spores, show unstable mating type or for strains that are incapable to mate with each other (Pretorius, 2000; Attfield & Bell, 2003), see also Fig.4. In this way, interspecific or even intergeneric crosses can be obtained. Because meiosis is not required, protoplast fusion can also be used to increase the ploidy of strains, which in some cases can increase cell productivity (Attfield & Bell, 2003).
In practice, protoplast fusion generally consists of three major steps: yeast cell wall degradation (generation of protoplasts), induction of hybridization, and cell wall regeneration. After hybridization, the parental nuclei temporarily coexist within a shared cytoplasm before (potentially) proceeding to karyogamy (Kavanagh & Whittaker, 1996). The success rate of the hybrid formation mainly depends on the taxonomic proximity of the strains and applied fusion protocol (Peberdy, 1980; Pina et al., 1986; Kavanagh & Whittaker, 1996; Attfield & Bell, 2003). Intraspecific fusion frequencies usually vary from 10−3 to 10−4, while for intergeneric fusions, it can be as low as 10−6 to 10−7 (Pina et al., 1986; Urano et al., 1993). Consequently, development of an optimal hybrid selection procedure is a crucial step in order to maximize the chance of achieving the desired genetic combination (further discussed in ‘Selection of outcrossed hybrids’).
An important disadvantage of using protoplast fusion as a strain improvement strategy is that many of the hybrids are mitotically unstable and chromosomal loss (resulting in aneuploidy) or dissociation into the parental strains often occurs (Pina et al., 1986; Attfield & Bell, 2003). Distantly related species are more prone to show this effect than closely related species (Morgan, 1983). Generally, protoplast fusion experiments result in hybrids containing the full genome of one parent, with a (few) extra chromosome(s) of the second parent (Yamazaki & Nonomura, 1994; Kavanagh & Whittaker, 1996). As a consequence, the phenotype and genotype of the resulting fused strain are very difficult to predict. The ratio at which both genomes are present in the hybrid might be a result of the fusion protocol employed, or the selection procedure used to isolate the hybrids (Kavanagh & Whittaker, 1996). Additionally, strains obtained by protoplast fusion are in some regions considered GMOs.
Although protoplast fusion is sometimes used in biotechnology to increase productivity of a strain by increasing the ploidy, it is mostly applied to combine characteristics from two parental strains. These parental strains can be from the same species, but often a Saccharomyces strain is combined with a nonconventional yeast displaying a specific trait, such as lactose utilization (Taya et al., 1984; Farahnak et al., 1986; Krishnamoorthy et al., 2010; Guo et al., 2012), temperature tolerance (Sakanaka et al., 1996), osmotolerance (Spencer et al., 1985; Loray et al., 1995; Lucca et al., 2002), starch degradation (Kishida et al., 1996), killer activity (Gunge & Sakaguchi, 1981), malic acid degradation (Carrau et al., 1994), or (hemi)cellulose hydrolysate utilization (Pina et al., 1986; Heluane et al., 1993; Table1).
Interestingly, protoplast fusion rarely yields nuclear hybrids (Chambers et al., 2009). While most research addressing protoplast fusion focuses on traits embedded in the genomic DNA, mitochondrial transfer without karyogamy occurs at a much higher frequency. Much like the strains resulting from cytoduction experiments (cf. infra), ‘cybrids’ can be defined as fusion products in which the cytoplasmic contents of the fusing protoplasts merge, without the concomitant fusion of, or exchange of genetic information between, the nuclei (Kavanagh & Whittaker, 1996). This phenomenon has been successfully used for the transfer of cytoplasmically inherited traits like the killer phenotype (Seki et al., 1985), respiratory competence (Richard et al., 1987; Kavanagh & Whittaker, 1996), or resistance to oligomycin (Matsuoka et al., 1982).
Cytoduction
Some factors underlying industrially relevant phenotypes are not embedded in the nuclear DNA, but are located in the mitochondrial DNA (e.g. several respiration-related genes) or present in the cytoplasm (e.g. killer plasmids). To selectively transfer these non-Mendelian traits from a donor to a recipient strain without disrupting the nuclear integrity of the recipient strain, a technique called cytoduction can be applied (Pretorius, 2000).
In cytoduction procedures, the donor strain (which contains the cytoplasmically transferable factor) carries a dysfunctional KAR1 gene. A kar1 mutant is defective in karyogamy (=nuclear fusion) after hybridization (Conde & Fink, 1976; Georgieva & Rothstein, 2002). As a consequence, through mating or protoplast fusion of donor and recipient strain, a zygote-like transient heterokaryon is formed which by subsequent mitotic divisions can bud off haploid heteroplasmons, containing only one genome but mixed cytoplasmic factors (Conde & Fink, 1976; Fig.4). These heteroplasmons, which still contain the full, undisrupted genome of the recipient strain but are supplemented with the (desired) cytoplasmic factors of the donor strain, have the desired combined phenotype. Occasionally, one or a few chromosomes of the second parent are transferred to the other nucleus, a process called ‘single-chromosome transfer’ (Nilsson-Tillgren et al., 1980; Dutcher, 1981). This results in ‘exceptional’ cytoductants that are sometimes used to examine individual chromosomes of industrial yeast strains in detail, for example chromosome III of lager yeasts (Nilsson-Tillgren et al., 1981; Kielland-Brandt et al., 1995).
Cytoduction is frequently used to obtain industrial strains with a positive killer phenotype, a trait encoded by a dsRNA virus-like particle (Ouchi et al., 1979; Young, 1983; Hammond & Eckersley, 1984; Seki et al., 1985; Yoshiuchi et al., 2000). Alternatively, it can be used to transfer flocculation characteristics (Barre et al., 1993), factors influencing carbon source utilization (Spencer et al., 1992) or yeast artificial chromosomes (YACs; Spencer et al., 1994). Cytoduction is also applied in fundamental research when studying amyloids (e.g. prions) in yeast (Saifitdinova et al., 2010; Wickner et al., 2012).
It is important to note that development of kar1 mutants does require genetic modification, which could hamper the use of this technique for industrial applications.
Mutagenesis
The in vivo induction of random mutations by chemical or physical mutagens and subsequent selection of phenotypically improved cells is one of the most widely used techniques to generate optimized microorganisms. One of the most impressive examples is the enormous increase, estimated to be more than three orders of magnitude, in penicillin production by Penicillium chrysogenum, which was achieved over a period of 60 years using multiple mutagenesis procedures (Demain, 2010). Over the last decades, mutagenesis has been applied to improve both monogenic and polygenic traits in a wide range of microorganisms (Giudici et al., 2005). Strain improvement using mutagens consists of two key steps: mutagenesis and screening. The screening is very similar to the screening procedures used in other procedures and will therefore be discussed in a separate paragraph (see section on ‘Selection of phenotypically improved cells’).
A typical mutagenesis experiment consists of overnight growth of the strain under study followed by the actual mutagenic treatment and a recovery step. Both the type of mutagen (different mutagens induce different types of mutations, see Table3) and the dose should be carefully selected or determined (see detailed reviews by Rowlands, 1984; Crook & Alper, 2012). However, it is often hard to predict which type of genetic alteration is required to improve a certain phenotype and hence which mutagen should be used. Therefore, it is advised to change the type of mutagen in a mutagenesis program consisting of multiple rounds, in order to sample as many different types of genetic changes as possible (Rowlands, 1984).
Table 3.
Mutagens often used in mutagenesis programs in yeast. Indicated are the mode of action of the mutagen and the resulting genetic alterations that it can induce. See for more information: Rowlands (1984) & Rubio-Texeira et al. (2010)
| Mutagen | Mode of action | Genetic alterations | |
|---|---|---|---|
| Physical | UV | Mitotic crossing over; mitotic gene conversion; pyrimidine dimers; hydroxylated bases; cross-linking DNA strands; reverse mutations | Frameshift mutations, base pair substitutions, transversions |
| Ionizing radiation | Single- and double-strand breaks in DNA; deamination and dehydroxylated bases | Point mutations | |
| Chemical | EMS | Alkylation | GC-AT transitions |
| MNNG | Alkylation, acts close to replication points | Transitions, transversions; clustered mutations |
EMS, ethyl methanesulfonate; MNNG, methylnitronitrosoguanidine; UV, ultraviolet.
As important as the type of mutagen is the mutagenic dose and exposure time. In general, a very low dose will yield a low proportion of mutants, making improved mutants hard to identify. Moreover, most mutants may only carry one or a few mutations, which reduces the chance to find improvements for which combinations of different mutations are needed. On the other hand, a high dose generates mutants that carry multiple mutations, of which many may be deleterious, leading to a large fraction of inferior or even unviable cells. Consequently, the optimal dose is the one that gives the largest proportion of beneficial mutants out of all cells that manage to survive; something which largely depends on the nature of the phenotype. Simple phenotypes that depend on one or a few mutations, like auxotrophy, typically show a monotonic dose–response curve, meaning that the fraction of desired mutants per survivor increases with increasing dose and reaches saturation at some point, after which the number of superior mutants may decline again because of the increasing proportion of individuals with deleterious mutations (Crook & Alper, 2012). For complex phenotypes, like the production of a compound/metabolite in a high concentration, that are influenced by many different genes, the ideal dose and the dose–response curve are harder to predict. Although a high dose will usually lead to fast phenotypic improvement, researchers usually prefer to use a low dosage. In this way, they avoid the accumulation of deleterious mutations and only increase the mutation rate when low killing rates do not yield improved mutants (Rowlands, 1983).
In both the wine and brewing industry, early studies applied random mutagenesis to generate mutants with improved industrial characteristics. In wine strain improvement, mutagenesis is often used to purge undesired monogenic traits (Giudici et al., 2005). Industrial yeast strains are (at least) diploid, implying that only dominant mutations can alter the phenotype directly. Therefore, haploid derivatives are preferred for mutagenesis programs (Pretorius, 2000). However, these yeasts are often homothallic, so no stable haploid cell cultures can be maintained for these strains. To circumvent this issue, mutagenesis can be applied to spores (instead of vegetative haploid cells) derived from such homothallic strains (Romano et al., 1983; Rous et al., 1983). After autodiploidization, these recessive mutations become homozygous and potentially influence the phenotype. However, it is not essential to use haploid strains or spores in a mutagenesis program. Mutagenesis of polyploid brewing yeasts yielded strains producing lower amount of the off-flavors diacetyl and H2S (Molzahn, 1977). In a recent study, the commercial diploid wine strain PDM was mutagenized directly using EMS, and mutants with reduced H2S production were obtained (Cordente et al., 2009). Also, sake strains in their natural ploidy were successfully mutagenized to obtain auxotrophic mutants (Hashimoto et al., 2005), which can be of great value in breeding and metabolic engineering strategies (Crook & Alper, 2012). Auxotrophic mutants were also developed by applying EMS mutagenesis to a haploid derivative of a commercial wine strain, in order to decrease higher alcohol production (Rous et al., 1983). Mobini-Dehkordi et al. (2008) used EMS mutagenesis to develop mutants with increased ethanol production. In a different study, EMS was used to develop mutants with increased dough fermentation capacity (Angelov et al., 1996).
Mutagenesis approaches are not limited to Saccharomyces yeasts. For instance, EMS mutagenesis has been used to increase astaxanthin production in Xanthophyllomyces dendrorhous (Phaffia rhodozyma; Brehm-Stecher & Johnson, 2012). Recently, EMS mutagenesis was successfully applied to improve the secretion of a heterologous protein in Ashbya gossypii (Ribeiro et al., 2013). Induced mutants of Sc. stipitis for improved lignocellulose fermentation were generated in various studies (Watanabe et al., 2010; Hughes et al., 2011), whereas multiple rounds of mutagenesis boosted the ethanol production of Kluyveromyces marxianus (Pang et al., 2010).
In recent studies, mutagenesis is often the first step to generate genetic variation in a population, after which genome shuffling of the best-performing mutants is applied to combine multiple beneficial mutations in the same cell, or the mutant population is subjected to directed evolution (cf. infra). Alternatively, strains obtained by genetic modification or breeding can also be further improved using random mutagenesis. For instance, Kumari & Pramanik (2012) subjected a hybrid between S. cerevisiae and Pachysolen tannophilus to multiple rounds of mutagenesis in order to increase its tolerance to high temperature, ethanol, and toxic compounds.
Directed evolution
Strain improvement through evolutionary engineering, a term first coined by Butler et al. (1996) and later also often referred to as adaptive, directed, or experimental evolution, relies on the basic principles of (natural and/or induced) genetic variation and subsequent selection acting on this variation. In general, a population of cells is grown under continuous selection for the phenotype of interest for many generations (cell divisions). Over time, random mutants will arise in this population. Directed evolution can also be combined with the use of mutagens and/or sexual hybridization within the evolving population(s) in order to increase the genetic and phenotypic variability that selection can act on. If a specific mutation (or mutations) endows a cell with a fitness advantage, this variant will be selected and enriched for in the population. Because of the short generation time and easy manipulation and cultivation of microorganisms in the laboratory, evolutionary engineering is a feasible route to generate yeast strains with improved phenotypes in a relatively fast fashion (Elena & Lenski, 2003; Buckling et al., 2009).
Different experimental setups can be used for growing cells under the desired selective conditions: batch culture, with serial passaging of cells, or a continuous culture system such as a chemostat or a turbidostat. In a chemostat, cells are kept at physiological steady state, and growth occurs at a constant rate. This constant growth rate is maintained by the continuous influx of a growth-limiting substrate, setting a fixed dilution rate (Dykhuizen & Hartl, 1983). In a turbidostat, there is continuous feedback between the inflow of the medium and the cell density of an exponentially growing culture, measured through, for example, an optical sensor (Bryson & Szybalski, 1952). In principle, it is also possible to evolve populations directly in the industrial setting where they are to be employed. It is important that the selection conditions match the industrial parameters as closely as possible in order to avoid ‘crippled’ strains that show improvement for the selected trait, but are inferior to the parent for other relevant traits (cf. infra). In fact, in applications where yeasts are continuously used for longer time periods, for example serial repitching of brewer's yeast in beer fermentations (Gibson et al., 2007), these populations are (unintentionally) being subjected to directed evolution, yielding strains with (sometimes positively) adjusted phenotypes. For example, at the end of the beer fermentation process, yeast cells sediment to the bottom of the fermentation tank. It has been shown that re-using cells from specific layers (near the top or bottom) of this pack of sedimented cells can influence the sedimentation behavior (including flocculation) in later fermentation rounds (Powell et al., 2004).
Directed evolution has proven to be a valuable tool to create yeast strains with specific, improved characteristics (Sauer, 2001). Examples of industrially relevant phenotypes improved through this strategy can be found in Table4. Several studies have investigated how yeast cells adapt to specific nutrient limitations, for example glucose, phosphate, or sulfate limitation (Paquin & Adams, 1983a, b; Dunham et al., 2002; Gresham et al., 2008). Because the main focus of these studies was to determine the exact genetic underpinnings of how cells adapt to a specific stress and also how reproducible the adaptation was, this research was performed with standard laboratory yeast strains that have only limited industrial relevance. However, several more applied studies have started from strains commonly used in the wine, beer, biofuel, and baking industry. The phenotypes targeted include resistance to individual stresses, such as high levels of acetate (Aarnio et al., 1991) or ethanol (Brown & Oliver, 1982; Dinh et al., 2008), osmotic stress (Ekberg et al., 2013), and high concentrations of metal ions such as copper (Adamo et al., 2012) and cobalt (Cakar et al., 2009), as well as (improved) utilization of alternative carbon sources such as xylose and arabinose (starting from metabolically engineered strains, see also below; Sonderegger & Sauer, 2003; Wisselink et al., 2009; Scalcinati et al., 2012; Demeke et al., 2013). However, in industrial settings, cells are often faced with a combination of different stresses: during brewing fermentations for example, cells encounter osmotic stress, high levels of ethanol, and nutrient deprivation (Gibson et al., 2007). By performing the selection steps under conditions resembling this harsh environment, researchers have succeeded to isolate multistress-tolerant lager strains with improved fermentation capacity in high-gravity wort (Blieck et al., 2007; Huuskonen et al., 2010). Using long-term batch culturing on gluconate, a carbon source poorly assimilated by S. cerevisiae, Cadière et al. evolved a commercially used wine strain with increased flux through the pentose phosphate pathway (Cadière et al., 2011). Evolved strains showed higher fermentation rates and increased aroma production compared to the parental strain in laboratory-scale fermentations. Interestingly, similar phenotypic improvements were observed when this evolved strain was used in pilot-scale fermentation trials (Cadière et al., 2012). Most evolutionary engineering studies have been performed using the model organism S. cerevisiae, but other yeast species have been subjected to evolutionary engineering as well. Examples include lager strains (S. pastorianus), subjected to sequential selection for tolerance to high ethanol levels and rapid growth at high osmolarity, to obtain strains with enhanced fermentation capacity under industrially used brewing conditions (Ekberg et al., 2013).
Table 4.
Studies using directed evolution for the improvement of industrial yeasts. The targeted phenotype, target species, and technique used to generate variation in the starting population are indicated
| Improved phenotype | Species | Starting population | Reference |
|---|---|---|---|
| Ethanol tolerance | S. uvarum | EMS-mutagenized and nonmutagenized | Brown & Oliver (1982) |
| S. cerevisiae | EMS-mutagenized and nonmutagenized | Dinh et al. (2008) | |
| S. cerevisiae | EMS-mutagenized and nonmutagenized | Stanley et al. (2010) | |
| Acetic acid tolerance | S. cerevisiae | Nonmodified | Aarnio et al. (1991) |
| Thermotolerance | S. cerevisiae | UV and EMS-mutagenized | Balakumar et al. (2001) |
| Copper resistance | S. cerevisiae | Nonmodified | Adamo et al. (2012) |
| C. humilis | Nonmodified | ||
| Cobalt resistance | S. cerevisiae | EMS mutagenized | Cakar et al. (2009) |
| Tolerance to inhibitors in lignocellulosic hydrolysates | S. cerevisiae | Nonmodified | Almario et al. (2013) |
| Freeze tolerance | S. cerevisiae | UV mutagenized | Teunissen et al. (2002) |
| Glycerol production | S. cerevisiae | Nonmodified | Kutyna et al. (2012) |
| Wine fermentation properties | S. cerevisiae | Metabolic engineered and EMS mutagenized | Cadière et al. (2011) |
| Flux through pentose phosphate pathway | S. cerevisiae | Metabolic engineered and EMS mutagenized | Cadière et al. (2011) |
| Fermentation under high-gravity conditions | S. pastorianus | EMS mutagenized | Huuskonen et al. (2010) |
| S. pastorianus | UV mutagenized | Blieck et al. (2007) | |
| Ethanol tolerance, growth, and high osmotic stress | S. pastorianus | EMS mutagenized | Ekberg et al. (2013) |
| Oxidative, freeze–thawing, high temperature, and ethanol stress | S. cerevisiae | EMS mutagenized | Cakar et al. (2005) |
| Utilization of glucose, xylose, and arabinose mix | S. cerevisiae | Metabolic engineered and EMS mutagenized | Wisselink et al. (2009) |
| Anaerobic growth on xylose | S. cerevisiae | Metabolic engineered and EMS mutagenized | Sonderegger & Sauer (2003) |
| Xylose fermentation | S. cerevisiae | Metabolic engineered | Shen et al. (2012) |
| S. cerevisiae | Metabolic engineered, EMS mutagenized, and genome shuffled | Demeke et al. (2013) |
C. humilis, Candida humilis; S. cerevisiae, Saccharomyces cerevisiae; S. pastorianus, Saccharomyces pastorianus; S. uvarum, Saccharomyces uvarum; EMS, ethyl methane sulfonate; UV, ultraviolet.
Directed evolution is most commonly used to fine-tune a specific phenotype that is already present in the starting strain but is not optimal yet. The initial population can be completely isogenic, using a single, often already commercially used, strain as starting point. To increase genetic variability of the starting population, researchers have also used UV- or EMS-mutagenized populations to initiate directed evolution strategies (see also paragraph on ‘Mutagenesis’ and Table4). Directed evolution can also be used to further optimize characteristics of (industrial) yeast strains created through other methods, such as metabolic engineering or genome shuffling. A prime example of a phenotype improved in such a way is fermentation of the pentose sugar xylose (Sonderegger & Sauer, 2003; Wisselink et al., 2009; Demeke et al., 2013). Although this sugar is present in high amounts in the lignocellulosic biomass that is frequently used for second-generation bioethanol production, S. cerevisiae is unable to metabolize xylose. Several studies have engineered this novel metabolic capacity into S. cerevisiae and subsequently used a directed evolution approach to further optimize xylose fermentation. Starting from a commercially used bioethanol strain, Demeke et al. (2013) applied metabolic engineering to introduce the necessary enzymes for xylose utilization, followed by mutagenesis and genome shuffling. Finally, the resulting strains were evolutionarily adapted by serial transfer in spruce hydrolysate containing D-xylose. One of the final clones isolated showed an increased fermentation performance on xylose and was also tolerant to growth inhibitors commonly found in lignocellulosic hydrolysates. Importantly, the evolved phenotype remained stable in the absence of selective pressure.
The advent of novel sequencing technologies has now also made it possible to sequence the complete genomes of evolved strains. Together with expression analysis of evolved clones, this can yield insight into how a specific phenotype is established. It can yield valuable information to guide other methods of strain improvement, such as reverse metabolic engineering.
Selection of phenotypically improved cells and outcrossed hybrids
A crucial factor determining the success rate of the improvement techniques mentioned above is the selectability of the targeted trait and, in case of hybridization experiments, of the outcrossed hybrids. Whereas some phenotypes, such as stress resistance, allow for a relatively easy high-throughput screening, selecting for so-called difficult phenotypes, such as general fermentation performance or flavor production, is much more labor-intensive and can limit the applicability of the strain improvement technique. Below, different approaches to select improved cells or hybrids from a heterogeneous population are discussed.
Selection of phenotypically improved cells
In all screens of natural or artificially created yeast diversity, the choice of the selection procedure is vital. Depending on the targeted phenotype, this selection step can be straightforward or extremely difficult and/or labor-intensive. Cells with improved tolerance for a specific stress factor can be readily selected from a pool of millions of different cells by applying this specific stress and selecting the surviving or growing cells. For example, cells with improved thermotolerance are easily isolated by mass growth on high temperatures (Steinmetz et al., 2002). Apart from selection procedures based on improved stress tolerance, the use of chemical analogs of nutrient sources or intermediary metabolites to select mutants with an altered metabolism is a widely used selection technique. The best-known examples are the isolation of mutants defective in URA3 or LYS2 using a selection medium supplemented with 5-fluoroorotic acid (5-FOA) or α-aminoadipic acid (α-AA), respectively (Zaret & Sherman, 1985; Boeke et al., 1987). 5-FOA and α-AA are analogs of intermediary metabolites in the biosynthesis of uracil and lysine, respectively, and each is converted to a toxic compound by URA3 (in the case of 5-FOA) and LYS2 (in the case of α-AA). Cells that have acquired a disruptive mutation in URA3 (or LYS2) will not produce and accumulate the toxic compound anymore and can thus grow on the supplemented medium. Mutants containing these so-called counter-selectable ‘auxotrophic’ markers can then be used in, for example, hybridization experiments (see section on ‘Generation of artificial diversity’). Other examples where chemical analogs can be used to select phenotypically improved variants are approaches to select cells lacking catabolite repression. These experiments used fermentation medium supplemented with nonmetabolizable glucose analogs, such as glucosamine (Hockney & Freeman, 1980) or 2-deoxyglucose (Jones et al., 1986). In addition, the growth medium used in these experiments contained maltose (or a different nonpreferred sugar) as the sole assimilable carbon source. Consequently, only cells in which the genes required for utilization of the nonpreferred sugar are not inhibited by the glucose analog are able to multiply. These mutants are of particular interest for the beer industry, where efficient co-fermentation of glucose and maltose, two important carbon sources in beer wort, can significantly shorten the fermentation time.
In other cases, it is difficult to directly select superior variants. However, the close association of certain easy and difficult phenotypes sometimes allows efficient selection for these latter phenotypes. For instance, genome shuffling studies often ultimately aim for improving general fermentation performance. However, because it would be too laborious to test each of the newly generated hybrids individually in small-scale fermentations (therefore making general fermentation performance a ‘difficult’ phenotype), researchers have found several ways to circumvent this issue. Firstly, it is possible to carry out a prescreening by subjecting the hybrid population to a severe stress by plating the cells on medium that contains for instance high ethanol or acetic acid levels, conditions encountered during fermentation. Next, only fast-growing colonies are tested individually in small-scale fermentations, and only superior hybrids are used for a next round of shuffling (Shi et al., 2009; Zheng et al., 2011a, b, 2013a, b; Tao et al., 2012). Other investigators have instead tried to first improve stress tolerance and found that hybrids generated after multiple rounds of genome shuffling and selection also showed increased general fermentation performance (Wei et al., 2008; Cao et al., 2009, 2010, 2012; Hou, 2009; Wang & Hou, 2010; Jingping et al., 2012; Lu et al., 2012; Wang et al., 2012a). Alternatively, some researchers inoculated their entire hybrid population in a very high-gravity fermentation and harvested cells for a next round of shuffling when the viability of the culture had considerably dropped, thereby enriching for the best adapted hybrids (Hou, 2010; Liu et al., 2011).
Similar to general fermentation performance, screening for cells with an improved flavor production profile can be extremely difficult, because differences in flavor production do not result in a clear and easily selectable fitness advantage for the yeast cell. Therefore, the main bottlenecks of strain improvement strategies for flavor production are the labor-intensive fermentation experiments of each individual hybrid or mutant, followed by measurements of the produced flavor compounds to select the individuals showing an improved flavor profile. To avoid this, several strategies that use chemicals that favor strains with a higher production of flavor (precursors) were developed. For example, an increased production of specific aromatic higher alcohols can be achieved by selecting strains with higher resistance to amino acid precursor analogs, such as 5,5,5-trifluoro-d,l-leucine, a leucine analog, 2-fluoro-l-tyrosine, a tyrosine analog, or p-fluoro-dl-phenylalanine, a phenylalanine analog (Fukuda et al., 1991). These analogs select for cells with decreased feedback inhibition of amino acid synthesis and increase the production of, respectively, isoamyl alcohol, tyrosol, and β-phenethyl alcohol. Cerulenin, an inhibitor of fatty acid synthesis, is used to select for increased fatty acid synthesis (Ichikawa et al., 2002; Vicente et al., 2006; de Souza et al., 2012), yielding strains with increased ethyl ester production. An increased production of isoamyl acetate (without increasing ethyl acetate production) was achieved by screening for resistance to pregnenolone. This steroid is detoxified by Atf2, an enzyme also responsible for isoamyl acetate, but not ethyl acetate, production (Kitagaki & Kitamoto, 2013). These chemicals can therefore be used to select cells with higher production of flavor compounds from a huge heterogeneous pool of cells, thereby facilitating high-throughput selection of this difficult phenotype.
Other techniques exploit the physiological differences (more specifically the tolerance to stress) of actively growing cells and cells in stationary phase. For example, the higher temperature sensitivity of exponentially growing cells compared to cells in stationary phase can be used to enrich for auxotrophic mutants in a heterozygous population (Walton et al., 1979). In this setup, a mutagenized (and thus heterogeneous) pool of cells in stationary phase, presumably containing some auxotrophic mutant cells, are transferred to fresh fermentation medium, counter-selecting for specific auxotrophies and inducing exponential growth of prototrophic cells. Next, exponentially growing cells are selectively killed by high temperatures, thereby enriching for auxotrophic mutants. Similarly, nystatin (instead of high temperatures) can be applied, because it acts selectively on actively growing cells (Snow, 1966; Sanchez et al., 1978).
Selection of outcrossed hybrids
In hybridization experiments, the hybridization efficiency is often very low (e.g. in protoplast fusion), or inbreeding is hugely favored (e.g. in interspecific mating experiments), highlighting the importance of a good screening strategy to select for outcrossed hybrids. Four main strategies are designed to isolate outcrossed hybrids. Firstly, both parental strains can be genetically transformed to introduce resistance against different antibiotics, so that outcrossed hybrids can be selected on medium supplemented with both antibiotics. To this end, plasmid-based approaches (Nakazawa et al., 1999), genomic insertion of resistance markers, and induction of antibiotic resistance using mutagenic agents (Putrament & Baranowska, 1973; Putrament et al., 1978) have been described. However, an important disadvantage of the first two strategies is that the resulting strains are GM strains, limiting their use in the food and beverage industries. Secondly, if each starting strain contains a different auxotrophy, outcrossed hybrids will be prototrophic and can be selected on appropriate selective medium. However, industrial strains are rarely auxotrophic, and screening for spontaneous or induced auxotrophic mutants is necessary (cf. infra). Thirdly, phenotypic complementation is a method in which each starting strain possesses a unique phenotype that can be easily selected by, for instance, cryotolerance or utilization of a specific carbon source, such as melibiose, in which case the hybrids can be selected for the presence of both phenotypes (Sato et al., 2002). Lastly, each strain can be reversibly stained with a different fluorescent dye, and after hybridization, fluorescence activated cell sorting (FACS) can be used to enrich for dual-stained cells (Bell et al., 1998). Alternatively, a combination of each of these strategies can be used.
Discussion – pros and cons of exploiting natural and artificial diversity
The existing natural diversity of yeast strains provides a rich, yet underexplored source of strains with industrial potential. Recent advances in next-generation sequencing technologies have allowed scientists to chart the diversity to an unprecedented level of detail. This revealed that the genetic diversity of currently employed industrial strains is relatively limited. Therefore, high-throughput screening of (natural) yeast collections or investigation of the phenotypic potential of indigenous strains might already yield yeasts with superior characteristics compared to the currently used strains. Moreover, additional (artificial) diversity can be generated through different strain improvement techniques like mutagenesis, sexual hybridization, protoplast fusion, and directed evolution. These strategies have successfully enriched the available biodiversity and yielded strains that valuably contributed to many different industrial fermentation processes. Perhaps the greatest promise lies in the combination of carefully selecting the best strains from the immense natural biodiversity, followed by a (combination of) technique(s) to further improve these natural yeasts in order to generate superior variants for industrial use.
In contrast to the genetic modification approaches described in the next section, one of the advantages of the approaches described above is that the resulting superior yeasts are mostly considered to be non-GMO, and food and beverages produced with these yeasts do not suffer from any problems with specific legislation and/or consumer acceptance. Moreover, they do not require an a priori knowledge of the genetic circuitry underlying the phenotype under study. In this way, complex polygenic phenotypes, such as ethanol tolerance, can be improved, without knowing exactly which genes need to be targeted. This allows scientists to use less well-described, nonconventional yeasts in improvement experiments. For example, efficient mutagenesis experiments targeting the basidiomycetous yeast P. rhodozyma have been performed for many decades (Brehm-Stecher & Johnson, 2012), while only very rudimentary genetic manipulation techniques are available for this organism (Lin et al., 2012). Additionally, detailed analysis of improved strains (whole-genome sequencing, expression analysis,…) can yield valuable insights into how a specific trait is established. This in turn can point to possible targets for genetic or metabolic engineering and can thus guide other methods of strain improvement.
On the other hand, these ‘non-GMO’ techniques also have important limitations. First and foremost, they only allow changing yeasts within the limitations of what is achievable through natural diversity, mutation, and crossing. Secondly, it is often difficult to select the few cells that show improved properties from the large and heterogeneous pool of cells. This prevents some industrially desired phenotypes, such as increased production of specific flavor compounds, for which a clear fitness advantage has not been discovered (yet), from being efficiently improved through these approaches. In these cases, selection has to be carried out on individual clones (e.g. in separate fermentation experiments), thereby severely limiting the pool of variants from which improved strains can be selected. In these cases, the best strategy to obtain improved variants is often the use of directed mating, rare mating, or protoplast fusion, where (in contrast to, e.g., mass mating or genome shuffling) parental strains can be thoroughly screened prior to the hybridization event. Thirdly, selection is often limited to the specific phenotype targeted, while other phenotypes are not under selection and may therefore deteriorate during the experiment. This can result in so-called crippled strains: strains performing only well for the targeted phenotype, but performing worse than the parental strain for other (industrially) important phenotypes that were not selected for. To circumvent this issue as much as possible, investigators have tried to evolve (in case of experimental evolution) or select (in case of the other approaches) strains or mutants under conditions very similar to the conditions in which the improved strain will be used in industry. However, practical limitations often prevent mimicking a full-scale industrial setting. Fourth, the complex ploidy of feral and industrial strains (which are often polyploid or even aneuploid) and their distinctive sexual life cycle (which is often characterized by poor sporulation efficiency, low spore viability, and/or homothallism) can severely hamper the success of improvement strategies such as sexual hybridization. Additionally, because of the lack of a sexual life cycle (or if it is not yet discovered), sexual hybridization experiments of several nonconventional yeasts, such as Candida stellata (an indigenous wine yeast with interesting oenological properties, cf. infra), are not possible.
Nonetheless, several of these disadvantages can be circumvented by carefully considering the genetic characteristics of the target strain(s) when one selects an improvement strategy, or by combining different techniques. For example, induction of auxotrophies by mutagenesis in industrial strains unable to sporulate (and thus unfit for use in sexual hybridization experiments) can provide mutants with appropriate selection markers for rare mating or protoplast fusion experiments. Alternatively, recombinant DNA technologies can be used, an approach that allows a more targeted strain improvement, which enables researchers to cross the borders of what is present in nature or achievable by mutations or hybridization. This approach will be further discussed in the next section.
Genetic modification
Introduction
The best documented approach of yeast improvement is genetic modification. Genetic modification comprises the controlled and precise modification of an organism's genome using recombinant DNA and other molecular techniques, in order to alter a trait of interest. Initial applications of genetic modification consisted of producing human proteins in bacteria and yeast for therapeutic treatments (e.g. Itakura et al., 1977). By modifying existing biochemical pathways, or even introducing complete heterologous pathways, a huge array of genotypic and phenotypic variability can be generated using recombinant DNA technology. Although this technique is already widely accepted in the pharmaceutical industry, the implementation of GMOs in the food or biofuel industry is still heavily debated.
Strain improvement by genetic modification differs from the non-GMO approaches described above in two important characteristics. Firstly, the DNA of the organism is changed in a specific, predetermined manner, without changing other parts of the genome. This implies that, in theory, one or several phenotypes can be changed, without negatively affecting other important characteristics. This reduces the chance of creating ‘crippled’ strains (cf. supra). Second, the technique is not limited to naturally occurring or randomly induced mutations. Modern technologies enable changing the DNA code in almost any possible way – including the transfer of DNA between different species, genera, or kingdoms, and the construction of completely artificial, man-made DNA fragments or even complete chromosomes and genomes (Gibson et al., 2008; Dymond et al., 2011). On the other hand, genetic modification also has several limitations compared to non-GMO techniques. Most importantly, genetic modification requires insight into the genetic alterations needed to obtain a desired phenotype. However, some of the newest applications of genetic modification circumvent this problem by, for example, applying ‘inverse metabolic engineering’ or by combining high-throughput and ‘semi-random’ genetic modification with selection techniques described above (e.g. ‘global transcription machinery engineering’). A second limitation is that genetic modification often involves optimization of specific cloning vectors and transformation protocols and tools for each organism.
Here, we summarize traditional as well as more recent and future approaches for genetically modifying industrial yeasts and discuss several recent industrially relevant outcomes. Although interesting, a historical overview and a discussion of the currently applied shuttle vectors and transformation protocols are only briefly mentioned, but are described extensively elsewhere (e.g. by Struhl, 1983; Gietz & Woods, 2001; Da Silva & Srikrishnan, 2012). The first part of this section will focus on Saccharomyces yeasts, while the genetic improvement of a few other, nonconventional yeasts with a well-established role in the fermentation industry is discussed separately.
Principles of genetic modification
Recombinant DNA technology
Whereas model bacteria like Escherichia coli are relatively easy to manipulate genetically because of their natural propensity to take up and incorporate foreign extracellular DNA, the uptake of foreign DNA into yeast is less efficient, and it was not until the seminal work by Hinnen, Hicks, and Fink in 1978 that yeast was genetically transformed efficiently (Hinnen et al., 1978). Since then, multiple different techniques and strategies have been developed to introduce recombinant DNA in yeast cells (reviewed by Gietz & Woods, 2001), and many different vectors (usually shuttle plasmids) and transformation protocols were designed to efficiently introduce DNA fragments.
There are two major ways to efficiently express foreign DNA in yeasts: using plasmids or fixed integration in the host's genome (or a combination of both). Several types of plasmid vectors exist, which allow variation of the copy number of the introduced DNA fragment (e.g. 10–40 copies per cell for ‘YEp’ vectors and 1–2 copies per cell for ‘YCp’ vectors; Clarke & Carbon, 1980; Christianson et al., 1992; Romanos et al., 1992). To ensure stability over multiple generations, these low-copy number ‘YCp’ vectors carry an origin of replication and a centromere sequence which allows for a high segregational stability of the plasmid under selective conditions. On the other hand, multicopy plasmids have limited segregational stability, which can lead to copy number variation between cells of the same population and thus also cell-to-cell expression heterogeneity.
Engineering of a yeast strain (e.g. for the production of a specific compound) often requires the introduction of multiple heterologous genes or genetic modifications. However, stably maintaining multiple plasmids in one cell, each with a different gene construct, can be difficult (Futcher & Carbon, 1986). Bidirectional promoter plasmid series as well as yeast artificial chromosomes exist for these purposes (Murray & Szostak, 1983; Miller et al., 1998; Li et al., 2008). However, the method of choice for introducing multiple genes is usually integration into the yeast genome. Issues with segregational stability and copy number control when plasmids are used for the introduction of heterologous genes, as well as the need for specific selective conditions during cell propagation to ensure plasmid maintenance and stability, make that a genetically stable engineered strain is usually created by integrating DNA into the yeast genome by homologous recombination. Vector- or PCR-based generated DNA fragments are generally used for insertion. Homologous recombination is highly efficient in S. cerevisiae: small flanking regions of homology (30–45 bp) are sufficient for targeted integration into the yeast genome (Manivasakam et al., 1995). Locations in the yeast genome that are often used for the integration of heterologous DNA fragments include rRNA genes (Lopes et al., 1989) and delta sequences (Sakai et al., 1990; Lee & Da Silva, 1997). When multiple genes need to be integrated, a dispersed nature of integration sites causes the inserted DNA fragments to be more stably maintained than when tandemly inserted (Da Silva & Srikrishnan, 2012).
As mentioned above, there are also several different transformation protocols described. While the initial transformation protocol (Hinnen et al., 1978) entailed the spheroplasting of yeast cells prior to transformation, protocols using intact cells are considered the golden standard today. The use of intact cells in transformation protocols was already mentioned in 1981. It was fine-tuned in a decisive paper by Ito et al. (1983) describing how alkali cations (such as Li+) in combination with polyethylene glycol (PEG) increased the efficiency of plasmid DNA uptake. In the next decades, small changes (including the addition of single-stranded carrier DNA or RNA) were made to the protocol, ultimately leading to the transformation method of choice for most yeast scientists today (Gietz & Woods, 2001). Alternative methods using electroporation or glass beads successfully increased the speed of the protocol, but are often less preferred due to higher equipment costs (electroporation) or a lower yield of transformants (glass beads; Gietz & Woods, 2001). For the introduction of mitochondrial genes, the most effective current protocol includes the use of biolistic methods, where yeast cells are bombarded with 0.5-μm gold or tungsten projectiles coated with DNA using compressed helium.
From genetic to metabolic engineering
In the 1980s and especially the 1990s, many research groups adopted the principles of recombinant DNA technology and started developing new, improved yeast strains for industrial processes using this technology (Dequin, 2001). Initially, these experiments were focusing on single genes and enzymes, aiming to manipulate genes directly involved in creating the product of interest (Tyo, 2008). This approach is mostly referred to as ‘genetic engineering’, although this term is often used in a broader context. In 1991, two seminal papers (Bailey, 1991; Stephanopoulos & Vallino, 1991) introduced a novel systematic approach of genetic modification where multiple genes were targeted, termed ‘metabolic engineering’. This technique uses directed modification of metabolic pathways and/or their regulation to optimize or establish the synthesis of various products (Ostergaard et al., 2000; Stephanopoulos, 2012). The main targets for modulation are enzymes, transporters, and regulatory proteins (Woolston et al., 2013). The opportunities of metabolic (and genetic) engineering include adjusting or fine-tuning the (1) gene expression level, (2) gene expression regulation, (3) in vivo protein/enzyme activity, and (4) protein subcellular location (Nevoigt, 2008). Key in this approach is the availability of detailed genetic information about the relevant pathways, enzymes, and their regulation. Using this information, a model on how these protein activities can be optimized to achieve the desired metabolic flux or phenotypic trait is built, and targeted genetic modifications using recombinant DNA technology are performed.
When aiming to optimize a specific trait by metabolic engineering, there are several important factors to keep in mind. First, high expression levels of heterologous genes are not always desirable because it can impose a significant metabolic burden on the cell (e.g. through the depletion of cofactors). Moreover, high enzyme levels do not necessarily correspond to the optimal level needed for production of a specific compound (see, for example, the article by Jin et al., 2003). Therefore, optimization of both the promoter and terminator region of the targeted gene(s) is crucial. Both constitutive and inducible promoters of varying strength are available for metabolic engineering of yeast, and several expression-enhancing terminators for S. cerevisiae were identified (Curran et al., 2013). Additionally, several methods to develop synthetic promoter libraries with promoter variants spanning a wide range of ‘activities’ have been developed (Jensen & Hammer, 1998; Jeppsson et al., 2003; Alper et al., 2005). This allows researchers to perform screenings with these libraries in order to fine-tune gene expression for their specific application. For example, error-prone PCR (see ‘Novel techniques’) was applied to the strong constitutive TEF1 promoter of S. cerevisiae, generating promoters with a strength ranging from 8% to 120% of the native TEF1 promoter (Alper et al., 2005; Nevoigt et al., 2006). A similar approach was used to generate a promoter library based on the constitutive GAP promoter for Komagataella (Pichia) pastoris (Qin et al., 2011).
Secondly, expression levels of multiple genes of a particular pathway often need to be optimized to balance metabolic flux within this pathway. For example, multiple gene promoter shuffling allows to find the optimal levels of expression of multiple genes at a time (Lu & Jeffries, 2007). In this approach, promoters of varying strength are fused to the genes of interest, these constructs are joined via ligation, and subsequently, a screen for the phenotype of interest is performed. A similar method, termed COMPACTER (customized optimization of metabolic pathways by combinatorial transcriptional engineering), was used for fine-tuning the expression of heterologous genes involved in xylose and cellobiose metabolization pathways introduced in S. cerevisiae (Du et al., 2012). This latter study also demonstrated that the optimal expression level strongly depends on the genetic background.
Thirdly, apart from regulation at the level of transcription, protein levels also depend on translational efficiency, and this in turn is highly influenced by codon usage. Codon optimization of heterologous genes, by changing codons to codons of highly expressed genes, such as glycolytic genes in the case of S. cerevisiae, can significantly increase protein levels (Wiedemann & Boles, 2008; Brat et al., 2012). Interestingly, even expression of endogenous genes can be increased through codon optimization (Brat et al., 2012).
Inverse metabolic engineering
As already indicated above, a crucial factor determining the applicability of metabolic engineering is the detailed knowledge and understanding of genetics underlying a certain trait or pathway. The lack of knowledge often impedes the success of metabolic engineering approaches, even for an intensively characterized model organism like S. cerevisiae. To circumvent this drawback, a novel approach of metabolic engineering called ‘inverse metabolic engineering’ was described (Bailey et al., 2002), conceptually identical to the more generally used term ‘reverse engineering’ (Oud et al., 2012). Roughly put, the rationale of inverse metabolic engineering is very similar to classical biochemical genetics, yet including newer techniques.
In this approach, the initial step consists of unraveling the genetics underlying an interesting, but genetically cryptic phenotype observed in a certain strain or condition. Next, this information is used to optimize a production strain. In practice, the first step can be achieved by screening a heterogeneous yeast collection for the phenotype of interest. The diversity in the collection can both be natural (e.g. a large collection of different industrial strains) or artificially created (e.g. by mutagenesis or genome shuffling). Next, the genotype–phenotype relationship of this trait must be unraveled. This can be accomplished by genetic mapping or genetic association analysis (reviewed by, e.g., Liti & Louis, 2012) or by using -omics technologies to analyze differences in gene, mRNA, protein, or metabolite abundance in different conditions or in different strains and link it to the observed phenotype. Lastly, this information is used to genetically modify a production strain.
Inverse metabolic engineering has already yielded various industrial strains (cf. infra; Bro et al., 2005; Blieck et al., 2007; Rossouw et al., 2008; Yoshida et al., 2008; Perez-Torrado et al., 2010; Duong et al., 2011). A clear example of how inverse metabolic engineering can be applied to target complex phenotypes is a study by Rossouw et al. (2008). By studying the transcriptome and exo-metabolome of several wine yeasts during fermentation conditions, a link between expression of certain genes (BAT1, AAD10, AAD14, and ACS1) and production of aroma compounds was identified and subsequently confirmed by overexpressing these genes in a commercial wine yeast (Rossouw et al., 2008). Similarly, Yoshida et al. (2008) identified genes involved in sulfite production of baker's yeast by combined analysis of the transcriptome and endo-metabolome, and successfully applied this knowledge to engineer a lager's yeast strain that produced high sulfite, but not excessive sulfide concentrations (Yoshida et al., 2008).
Synthetic biology
A relatively new and incredibly powerful strategy for creating novel industrial yeast strains relies on synthetic biology. Broadly, the goal of synthetic biology is the design and construction of new biological parts, devices, and systems [ranging from (parts of) single genes to completely new organisms], or re-designing existing, natural biological systems for useful purposes. Using individual genetic elements (sometimes referred to as ‘biobricks’), synthetic biology assembles more complex genetic systems such as metabolic pathways (Stephanopoulos, 2012). The term ‘synthetic biology’ comprises two different approaches: The first is the use of man-made molecules to mimic natural molecules with the goal of creating artificial life. The second is the use of natural molecules to assemble a system that acts unnaturally.
Although S. cerevisiae has many desirable characteristics for use in synthetic biology, the preferred model organism of synthetic biologists is E. coli (see, e.g., the articles by Fung et al., 2005; Levskaya et al., 2005). However, the pioneering work of the J. Craig Venture institute has shown that yeasts have the ability to assemble and maintain entire bacterial genomes, indicating that large synthetic networks could be constructed and implemented in the yeast's DNA arsenal (Gibson et al., 2008). Moreover, the Synthetic Yeast 2.0 (Sc2.0) project, a project seeking to reconstruct and redesign the full S. cerevisiae yeast genome, is progressing swiftly. This recently yielded the publication of the first partially synthetic eukaryotic chromosomes, synIXR, and semi-synVIL (Dymond et al., 2011).
Production of certain valuable compounds through complex pathways was already achieved in S. cerevisiae, for example the antimalarial drug precursor artemisinic acid (Ro et al., 2006). In this study, a mevalonate pathway, amorphadiene synthase (ADS), and a novel cytochrome P450 monooxygenase (CYP71AV1) from Artemisia annua were introduced and expressed in S. cerevisiae, yielding titers up to 100 mg L−1 of artemisinic acid when cultured in a simple growth medium.
Theoretically, the possibilities of synthetic biology are endless. However, it is unlikely that creating fully functional and optimal yeasts for industrial processes from scratch will occur in the very near future. Although it is often stated that the availability of biobricks will be the major bottleneck in advances in synthetic biology, a more fundamental challenge will likely be to completely map and design a functional cell and optimize it to the level of robust performance required for commercial operation (Stephanopoulos, 2012). That is why, so far, the majority of studies applying synthetic biology are still leaning more toward basic biology rather than applied research (Serrano, 2007; Dymond et al., 2011; Voordeckers et al., 2013).
GM Saccharomyces yeasts for the production of fermented foods
The use of GM yeasts in food fermentation processes is still controversial and heavily debated. Nonetheless, numerous research groups and companies are using genetic modification to alter industrial yeast properties. In this section, an overview of the most notable progress in industrial yeasts engineered with recombinant DNA technologies for the production of foods and beverages is given. Phenotypes that are often targeted in these strains can be broadly categorized into four different groups: (1) propagation, fermentation and storage efficiency, (2) sensorial quality of the end product, (3) health-related quality of the end product, and (4) microbial stability. It is important to note that some phenotypes show overlap between these different areas; for example, the production of a high concentration of glycerol can be both of sensorial importance and a health-driven approach to lower the ethanol content of beverages. Below, some examples of previous research studies in these areas are discussed; an extensive overview is given in Supporting Information, Table S2.
Propagation, fermentation, and storage efficiency
Environmental conditions during fermentation or propagation are often harsh for yeast cells, which are faced with, and must quickly respond to, fluctuations in dissolved oxygen concentration, pH, osmolarity, ethanol concentration, nutrient supply, and temperature (reviewed by Gibson et al., 2007). Hence, the development of more robust strains that are still capable of producing a high-quality end product is a prime focus in biotechnology (Kim et al., 1996; Perez-Torrado et al., 2002, 2010; Panadero et al., 2007; Kaino et al., 2008; Gomez-Pastor et al., 2012). For example, using an inverse metabolic engineering approach, Perez-Torrado et al. (2010) identified two genes, CAF19 and ORC2, important for osmotolerance. Overexpression of these genes exerted a positive impact on the leavening activity of baker's yeast. In a different study, Kaino et al. (2008) improved the freeze tolerance of baker's yeast by increasing the intracellular proline accumulation. Disrupting PUT1 (encoding a proline oxidase) and replacing the wild-type PRO1 (encoding a γ-glutamyl kinase) by a modified allele less sensitive to proline feedback inhibition significantly increased the freeze tolerance of commercial baker's yeast without influencing the fermentation ability.
The composition of the medium used for fermentation or propagation can be another cause of suboptimal yeast performance. Molasses for example, a cheap medium classically used for yeast propagation, is generally suboptimal for the generation of high amounts of biomass of Crabtree-positive yeasts, like S. cerevisiae, because these yeasts prefer a fermentative lifestyle when glucose is present, which leads to inefficient use of the substrate. Regulation of the central carbon flux to improve biomass yield has been a major target in strain improvement. For example, overexpression of HAP4, a transcriptional regulator of respiration-related genes, led to a redirection from fermentation to respiration flux and a concomitant increase in biomass production (Duenas-Sanchez et al., 2010). In many cases, multiple nonpreferred carbon sources, like maltose, maltotriose, galactose, fructose, and melibiose, or nonpreferred nitrogen sources, like proline and arginine, are present in the propagation or fermentation medium used in industrial applications. Efficient utilization of these nutrients can be achieved by neutralizing catabolite repression or modifying various regulators, enzymes, or transporters (Klein et al., 1996; Salmon & Barre, 1998; Higgins et al., 1999; Guillaume et al., 2007). For example, in the wine industry, stuck fermentations are sometimes caused by the inability of yeast to metabolize all fructose present in the wine must. By introducing a mutated HXT3 gene (coding for a hexose transporter), wine yeasts can become able to completely ferment grape must sugars (Guillaume et al., 2007). Similarly, introduction of a modified high-affinity specific proline permease gene (PUT4) in a lager yeast resulted in strains with an improved proline assimilation profile during lager fermentations (Omura et al., 2005). Furthermore, a beer yeast able to utilize dextrins, leading to the production of highly attenuated beers, was the first GM strain that was approved by the British government for commercial use (Hammond, 1995; cf. supra). However, due to the negative perception of GMOs by consumers (often fueled by specific organizations), this strain has never attained commercial success.
Lastly, yeast physiology can also be altered to facilitate downstream handling of the fermentation product. For example, flocculation facilitates the removal of the biomass and thus clarification of, for example, beer or sparkling wine. Related to this phenotype is ‘flotation’, which is the ability of yeast cells to trap CO2 bubbles in a fermenting liquid and form a film or vellum at the top of fermentation vessels, for example in traditional ales or flor sherry (Pretorius, 2000; Verstrepen et al., 2003a, b). Both traits are linked to expression of FLO genes, and modifying these genes (Ishida-Fujii et al., 1998) or coupling their expression to a suitable promoter (Verstrepen et al., 2001) can enable appropriate timing and intensity of yeast aggregation.
Sensorial quality
The distinctive aroma of wine, beer, sake, spirits, bread, and all other fermented foodstuffs is highly affected by many yeast-associated compounds, including esters, higher alcohols, ketones, phenolic compounds, sulfuric compounds, and terpenes. Consequently, increased or decreased production of these aroma-active compounds is a main focus of several studies. In addition, yeast-induced flavor stability has also received some attention, for example by increasing the production of antioxidants such as glutathione and sulfite, which delays staling of fermented beverages.
Although volatile esters and higher alcohols are only present in trace amounts in fermented beverages such as beer and wine, they are extremely important for the flavor profile (Verstrepen et al., 2003b). As already mentioned above, strains with improved flavor production cannot be easily selected from large pools of variants, and therefore, many improvement strategies targeting this phenotype, such as genome shuffling or directed evolution, require testing of individual clones. As a result, much research has focused on the use of genetic modification to produce strains with superior ester and higher alcohol profiles. The regulatory mechanisms and factors influencing the production of these secondary metabolites are still not completely understood, but fine-tuning expression of specific genes, like ATF1, BAT1, and EHT1, has proven to be a fruitful strategy for strain improvement (Hirata et al., 1992; Lilly et al., 2000, 2006; Verstrepen et al., 2003b; Blieck et al., 2007; Rossouw et al., 2008; Zhang et al., 2013).
Similarly, reduction of unwanted off-flavors, like diacetyl and dimethyl sulfide (DMS) in beer or H2S in wine or beer, is also an important target in strain improvement. Interestingly, some aroma-active compounds, like 4-vinyl guaiacol, a phenolic compound, are considered to be unwanted off-flavors in some fermented beverages (e.g. in most ale beers or sake), while in limited concentrations they are perceived positively in certain wines. As a result, some studies focus on eliminating phenolic compounds from yeast metabolism, while others try to promote or balance their production, for example by introducing heterologous genes encoding a phenolic acid decarboxylase from Bacillus subtilis and the p-coumaric acid decarboxylase from Lactobacillus plantarum. This approach leads to a significant increase in the formation of volatile phenols (Smit et al., 2003).
The potential of yeast strains to produce or utilize certain organic acids, and therefore influence the (volatile) acidity or rheology of the food, is another major focus for strain improvement. Obtaining a balanced acidity profile by fine-tuning production of citric, succinic, malic, lactic, and acetic acid, or allowing yeasts to perform malolactic wine fermentations, have all been studied in detail. ML01, a strain engineered for this latter purpose, has been approved for commercial wine production in a number of countries, including the USA, Canada, and Moldova (Chambers & Pretorius, 2010). This strain carries two chromosomally integrated heterologous genes – the Schizosaccharomyces pombe malate transporter gene (mae1) and the Oenococcus oeni malolactic enzyme gene (mleA) – regulated by the S. cerevisiae PGK1 promoter and terminator, and is able to simultaneously metabolize malic acid and perform an alcoholic fermentation (Husnik et al., 2006).
Health-related quality
There is an increasing interest to use genetic modification to improve the nutritional and/or health-promoting qualities of foods. For example, decreasing the concentration of ethyl carbamate, a suspected carcinogen that occurs in most fermented foods and beverages, is an important goal. In aging wines or sake, the concentration of ethyl carbamate, which is mainly formed by a spontaneous reaction of ethanol with urea, can be relatively high (Pretorius & Bauer, 2002; Dahabieh et al., 2010). By adjusting the urea catabolism of the yeast strain, ethyl carbamate concentrations can be significantly lowered (Kitamoto et al., 1991; Coulon et al., 2006; Dahabieh et al., 2010). The most successful resulting strain so far, called 522EC−, was approved for commercial use (Chambers & Pretorius, 2010). In 522EC−, DUR1,2 is constitutively expressed, causing the degradation of urea and thus minimizing the amount of urea released in the medium (Coulon et al., 2006).
Secondly, the production of health-promoting substances such as antioxidants and/or compounds that are believed to protect against DNA damage also becomes more and more important (Becker et al., 2003; Wang et al., 2011). Because S. cerevisiae does not produce most of these compounds naturally, heterologous expression of genes from other species is required. For instance, carotenoids, tetraterpenoids consisting of 40 carbon atoms, are of interest because they possess antioxidant activities, but can also be applied as coloring agents (see the review by Victor & Bhatia, 2012). Early studies reported β-carotene and lycopene production, although at low levels, in S. cerevisiae by overexpression of the bacterial genes involved in biosynthesis of carotenoids from episomal vectors (Yamano et al., 1994). Higher levels [5.9 mg g−1 (dry weight)] of β-carotene were achieved by chromosomal integration and overexpression of genes from the red yeast X. dendrorhous (Verwaal et al., 2007).
Terpenes are another class of molecules that, in addition to their contribution to the aroma of fermented foodstuffs and use as dyes, are of interest for nutritional and medical purposes. Various metabolic engineering strategies in S. cerevisiae have been adopted to express terpenes (described, for instance, by Liu et al., 2013). For example, Rico et al. (2010) engineered a wine yeast to produce linalool by the heterologous expression of the linalool synthase gene from Clarkia breweri. In addition, the titer could be doubled by overexpression of the native enzyme 3-hydroxy-3-methylglutaryl coenzyme A reductase, which formed the rate-limiting step in the endogenous mevalonate pathway.
Resveratrol is a compound that, apart from its antifungal characteristics, has been proposed to reduce the risk of cancer and coronary heart disease (Jang et al., 1997; Becker et al., 2003). Introducing a low-affinity, high-capacity E. coli arabinose transporter gene (araE) or introducing a mutated, codon-optimized tyrosine ammonia lyase gene from Rhodobacter sphaeroides into yeast significantly enhanced resveratrol production (Wang et al., 2011). In addition, Sydor et al. (2010) expressed the Arabidopsis thaliana gene coding for 4-coumaroyl-coenzyme A ligase and the Vitis vinifera gene for stilbene synthase in various strains and found that, together with using rich medium, an industrial sugarcane-fermenting yeast strain reached a resveratrol titer of 391 mg L−1.
Thirdly, due to the increasing demand for healthier foodstuffs, the reduction of ethanol and carbohydrates in alcoholic beverages, especially beer and wine, is of considerable commercial interest (Pretorius, 2000; Kutyna et al., 2010; Saerens et al., 2010). Reducing the level of (nonfermentable) dextrins in beer wort yields beers with less carbohydrates and thus a lower caloric content. The production of such ‘light’ or ‘diet’ beers can be achieved by adding exogenous enzymes (which is inefficient and expensive) or by developing yeast strains that can produce and secrete starch-decomposing enzymes such as α-amylase (Perry & Meaden, 1988; Steyn & Pretorius, 1991; Hammond, 1995; Randezgil et al., 1995; Marin et al., 2001). A dextrin-assimilating brewer's yeast, equipped with the S. cerevisiae (var. diastaticus) gene STA2, encoding an extracellular glucoamylase, is one of the few genetically engineered strains to receive official approval for commercial use from the British Government (Hammond, 1995; Dequin, 2001). However, as already mentioned above, negative perception of this GMO by consumers hampered its commercial success.
Next to low-calorie beverages, there is an increasing demand from both consumers and producers for fermented beverages with reduced alcohol content, but without the loss of product quality (Kutyna et al., 2010). It is argued that excessive alcohol levels in wines can compromise wine quality and cause health issues, while the costs to the consumer in countries where taxes are calculated based on ethanol content increase. Approaches implementing additional processing steps, like postfermentation removal of alcohol, or modified fermentation parameters, yielded only beverages with inferior quality and usually implicated extra processing costs. Numerous strategies, mainly based on targeting genes involved in glycerol production or regulating the redox balance, have been described to alter yeast carbon flux in order to reduce ethanol production. Due to the drastic nature of this metabolic reorganization, a balanced and well-considered combination of different strategies, for example as described in Cordier et al. (2007), will probably yield the most useful outcome. This strategy employs the combined effect of genes involved in glycerol production and transport (FPS1 and GPD1), conversion of DHAP to GAP in the glycolysis pathway (TPI), and the conversion of acetaldehyde to ethanol and acetic acid (ADH1 and ALD3).
Because of the high consumer concerns about safety and other health-related properties of foodstuffs, and the solutions GM yeasts can provide for these potential problems, it is plausible that the use of GMOs in this field might be the key to a more rational and balanced consumer attitude to GMOs in the field of foods and beverages and pave the way for broader applications.
Microbial stability
The quality of fermentation products largely depends on the microbial actors in the fermenter. Contamination during the fermentation usually has dramatic effects on the fermentation efficiency and/or sensorial quality of the end product. To prevent contamination by other microorganisms during fermentation, yeast strains with antimicrobial properties have been developed, either by hybridization or by genetic engineering. The most commonly applied strategy is to equip the yeast strain with a killer phenotype, such as K1 (Boone et al., 1990), which makes it capable of producing antifungal toxins, to which the strain itself is immune (Schmitt & Breinig, 2002). Sporadically, there are reports of strains with other engineered antifungal (Carstens et al., 2003) or antibacterial properties (Schoeman et al., 1999).
GM Saccharomyces yeasts for the production of biofuels
The modern biofuel industry confronts industrial microorganisms with specific challenges that differ from those encountered in many food fermentations. Whereas S. cerevisiae has been used for thousands of years in a wide range of food-related fermentations, the substrates used for these traditional fermentations differ significantly from the complex substrates and harsh conditions in second-generation biofuel production processes. Moreover, S. cerevisiae is now also used to produce a range of different biofuels such as butanol, which is not a natural fermentation product of the common brewer's yeast. Microorganisms that naturally utilize these complex substrates or produce these compounds, such as Sc. stipitis for xylose metabolism and Clostridium acetobutylicum for butanol production, come with disadvantages. These include a lack of knowledge on genetics and physiology, limited stress and/or alcohol tolerance, and the lack of a molecular toolbox to modify the organism's characteristics (Alper & Stephanopoulos, 2009). Metabolic and genetic engineering strategies have been and still are being developed to modify the classical fermentation workhorse S. cerevisiae in order to tackle these new challenges. A detailed overview of all metabolic engineering strategies for biofuel production falls outside the scope of this review, but in this section we briefly discuss the most important contributions of genetic modification to various characteristics relevant to the modern biofuel industry.
Substrate range
Saccharomyces cerevisiae efficiently ferments a range of hexose sugars (including the disaccharide maltose and the trisaccharide maltotriose) into ethanol, and it is therefore the organism of choice in first-generation bioethanol production. However, it cannot consume pentose sugars like xylose, the most abundant pentose in lignocellulose and the chief substrate for second-generation biofuels. Researchers have explored two main strategies to introduce and express genes from various pentose-utilizing microorganisms into S. cerevisiae, in order to make it capable of utilizing these nonpreferred sugars [see reviews by Van Vleet & Jeffries (2009) and Young et al. (2010)]. In the first strategy, expression of heterologous genes, typically from Sc. stipitis, coding for xylose reductase (XR) and xylitol dehydrogenase (XDH) allowed xylose fermentation (Tantirungkij et al., 1993). However, the resulting strains were suboptimal because XR and XDH require different co-factors (NADPH vs. NAD+), causing a cellular redox imbalance that in turn leads to the secretion of xylitol and a low ethanol yield. Protein engineering, experimental evolution, and fine-tuning of gene expression have been used to further improve these strains. While some strains show significant improvements, yields are often still too low for successful commercial applications (reviewed by Matsushika et al., 2009).
A second strategy to obtain xylose-fermenting S. cerevisiae strains exploits a metabolic route mainly present in prokaryotes. A heterologous gene coding for xylose isomerase is expressed in S. cerevisiae, allowing a one-step conversion from xylose to D-xylulose, which can be readily utilized by S. cerevisiae, albeit at low rate (Kuyper et al., 2003; van Maris et al., 2007; Madhavan et al., 2009). This strategy circumvented the redox imbalance problem of the first strategy, but xylose consumption rates turned out to be lower (Madhavan et al., 2009). To further improve these strains, additional metabolic and evolutionary engineering strategies were used, again with varying levels of success (Matsushika et al., 2009).
In addition to targeted metabolic engineering strategies to further optimize xylose fermentation, other techniques have also been used. Interestingly, these techniques combine aspects of non-GMO techniques (where the best-performing organisms are selected from a large pool of natural or man-made variants) with genetic modification. For instance, Liu et al. (2008) adopted global transcription machinery engineering (gTME; see section on ‘Novel techniques’ below) to select mutants on medium with xylose as the only carbon source and were able to identify an isolate with improved xylose fermentation. Ni et al. (2007) used an insertional transposon mutagenesis screen (see section ‘Novel techniques’) to identify gene deletions that could enhance growth on xylose in an engineered strain.
Although these strategies successfully expanded the substrate range of S. cerevisiae, the resulting strains were rarely directly applicable in an industrial setting because the yields and/or fermentation efficiency and/or the stress resistance of the resulting strains were too low. In addition to optimized xylose metabolism, efficient biofuel strains must also be capable of dealing with general (osmotic pressure, ethanol toxicity) and specific (inhibitors formed during lignocellulose pretreatment) stress factors during the industrial fermentation. Equipping an established industrial biofuel strain, instead of a laboratory strain, with xylose metabolism therefore seems a promising strategy (Demeke et al., 2013).
Ethanol yield
One of the key parameters for industrial application of a biofuel production strain is yield. Therefore, multiple genetic modification approaches aim at improved substrate utilization and decreasing byproduct formation (Nissen et al., 2000a, b, 2001; Kong et al., 2006, 2007; Zhang et al., 2007).
Stuck or sluggish fermentations are the most commonly encountered reasons for the loss of biofuel yields, especially in very high-gravity conditions. The main causes are shown to be the high osmotic pressure in the first phase of the fermentation, and (even more importantly) the gradually increasing ethanol stress during the fermentation (Gibson et al., 2007; Puligundla et al., 2011). In order to tackle this issue, researchers aim at increasing the overall stress tolerance, thereby often increasing yield and/or productivity (see sections ‘Mass mating & Genome Shuffling’ and ‘Enhancing cellular stress tolerance’).
Byproduct formation can also significantly lower the yield of biofuel fermentations. The most abundant byproduct of S. cerevisiae fermentations is glycerol, which is formed both as an osmolyte to counteract osmotic stress in the first phase of the fermentation (Hohmann, 2002) and to convert excess NADH back to NAD+ to maintain the redox balance under anaerobic conditions (van Dijken & Scheffers, 1986). Typically, glycerol accounts for up to 5% of carbon flux (Oura, 1977). Various metabolic engineering strategies have succeeded in reducing glycerol formation, thereby increasing ethanol yield. However, a decrease in growth or fermentation rate (Björkqvist et al., 1997; Nissen et al., 2000b; Hubmann et al., 2011) or decreased stress tolerance (Pagliardini et al., 2013) often hampers industrial applicability of the resulting strains.
Production of novel compounds
The extensive use and knowledge of the production host S. cerevisiae has led to commercial interest in using it to produce advanced biofuels. Because S. cerevisiae does not naturally produce most of the advanced biofuels, which include butanol and biodiesel, multiple research groups started to explore metabolic engineering strategies to expand its product range (see for review de Jong et al., 2012). For instance, a S. cerevisiae strain capable of producing fatty acid ethyl esters was created by targeted deletions of genes involved in producing storage lipids and heterologous expression of a bacterial acyltransferase (Kalscheuer et al., 2004). Multiple studies have explored engineering yeast to produce butanol (e.g. Avalos et al., 2013). Whereas yeast engineering for advanced biofuels seems promising, more research is needed to allow broad industrial implementation.
Cellular stress tolerance
During industrial fermentations, yeast cells are confronted with many different stress factors, including heat or cold stress, high osmotic pressure, and high ethanol levels (Gibson et al., 2007; Puligundla et al., 2011). With some notable exceptions, targeted metabolic engineering strategies have yielded only limited success in increasing stress tolerance (Larsson et al., 2001; Gorsich et al., 2006; Petersson et al., 2006). One of the most important reasons for this is the fact that stress tolerance is a complex, polygenic trait. A complete picture of the genes and molecular mechanisms underlying these traits is often lacking, which complicates targeted modification. In addition to classical non-GMO techniques, including mutagenesis and experimental evolution, several GMO-based approaches recently succeeded in increasing phenotypic variation in an unbiased fashion, followed by the identification of mutants with increased stress resistance. For instance, Alper et al. (2006) used gTME to increase osmotolerance and ethanol tolerance, whereas Park et al. (2003) could significantly improve thermotolerance, osmotolerance, or ketoconazole resistance by screening the mutants expressing artificial transcription factors (see section on ‘Novel Techniques’ below).
Genetic modification of nonconventional yeasts
Although S. cerevisiae and closely related species are by far the major producers of biotechnological products worldwide, nonconventional yeasts have been utilized as industrial organisms for a variety of applications (Johnson, 2013). Below, a selected overview of the recent progress in genetic modification of key nonconventional yeasts is given.
Scheffersomyces (Pichia) stipitis
Scheffersomyces (Pichia) stipitis is an ascomycetous yeast extensively studied for its capacity to ferment xylose to ethanol, L-lactic acid, and other products (Johnson, 2013). Despite the capacity of Sc. stipitis to ferment xylose to ethanol at nearly maximum yield with very limited byproduct formation, few metabolic engineering studies targeting this yeast species have been published (Van Vleet & Jeffries, 2009). The main reasons are its respiratory (Crabtree-negative) lifestyle, low ethanol tolerance, and its slow sugar consumption rate compared to S. cerevisiae (Agbogbo & Coward-Kelly, 2008). Therefore, biotechnological applications involving Sc. stipitis are at this point mostly limited to the transfer of its genes to S. cerevisiae to introduce the ability to ferment pentose sugars. However, the sequencing of the full genome (Jeffries et al., 2007) and the development of more efficient transformation systems (e.g. plasmid vectors and a loxP/Cre recombination system) and drug resistance markers (Laplaza et al., 2006) might create possibilities for genetic modification of industrially applicable Sc. stipitis strains. Indeed, recent introduction of the L-lactate dehydrogenase gene from Lactobacillus helveticus into Sc. stipitis allowed it to efficiently produce L-lactate from xylose-containing medium and illustrates its biotechnological potential (Ilmen et al., 2007).
Yarrowia (Candida) lipolytica
Yarrowia (Candida) lipolytica is an ascomycetous yeast that is the teleomorph (spore-forming form) of C. lipolytica. With its fully sequenced genome (Dujon et al., 2004), it is one of the most extensively studied nonconventional yeasts. It is currently used as a model for the study of protein secretion, peroxisome biogenesis, dimorphism, and degradation of hydrophobic substrates (Fickers et al., 2005). One of its most remarkable features is that it can efficiently use hydrophobic substrates, such as n-alkanes, fatty acids, and oils, as sole carbon source (Fickers et al., 2005; Beopoulos et al., 2009), and that it can accumulate lipids to levels exceeding 50% of cell dry weight (Beopoulos et al., 2009). Yarrowia species are mostly known for the production of ‘single-cell proteins’, but they are currently also used in the production of organic acids (mainly citric acid), aromas (e.g. β-decalactone, 2-phenylethanol), polyols (such as mannitol and erythritol), starters for the food industry, bio-oil, biotransformation of steroids, or waste treatment (Attfield & Bell, 2003; Fickers et al., 2005; Beopoulos et al., 2009; Guo et al., 2012; Celinska et al., 2013; Liu et al., 2013). Additionally, they are important for the production of certain fermented foods such as blue cheese, where they contribute to the sensorial quality (Gkatzionis et al., 2013). Because of its nonpathogenic nature, Y. lipolytica has received a GRAS (generally regarded as safe) status for these processes (Attfield & Bell, 2003), which facilitates further applications of this yeast in food industry.
Although the first publications on genetic recombination in Y. lipolytica date back to the early 1970s (Bassel et al., 1971), the more recently developed molecular and genetic tools to modify Y. lipolytica genomes (reviewed by, e.g., Madzak et al., 2004; Nicaud, 2012) and the availability of its complete genome sequence led to promising recent advances in genetic modification of this yeast. For example, higher production of α-ketoglutarate was established by introducing the acetyl-CoA synthetase gene (ACS1) from S. cerevisiae and the ATP-citrate lyase gene (ACL) from Mus musculus (Zhou et al., 2012), or by using a gene dose-dependent overexpression of genes encoding NADP+-dependent isocitrate dehydrogenase (IDP1) and pyruvate carboxylase (PYC1; Yovkova et al., 2014). Production of β-decalactone, a high-value aromatic compound, was promoted by deleting POX3 and overexpressing POX2, two genes encoding acyl-CoA oxidases with different substrate specificities (Guo et al., 2012). By introducing codon-optimized genes crtB and crtI of Pantoea ananatis and overexpressing native genes GGS1 and HMG1, a metabolic pathway for lycopene production in the otherwise non-carotenoid-producing Y. lipolytica could be established (Matthäus et al., 2013). Because lipid production of Y. lipolytica can be enhanced by the modification of several genes involved in the lipid metabolism (Beopoulos et al., 2008; Dulermo & Nicaud, 2011; Tai & Stephanopoulos, 2013; Wang et al., 2013), additional fine-tuning of the lycopene production pathway could be achieved by deleting POX1-6 (thereby cutting short the peroxisomal β-oxidation) and GUT2 (thereby preventing reduction of the glycerol-3-phosphate pool). This increased the lipid accumulation (and consequently the lycopene yield) significantly and led to the production of 16 mg g−1 (dry weight) in fed-batch cultures, the highest yield reported so far for eukaryotic hosts (Matthäus et al., 2013). Recently, a metabolic engineering approach for the production of ricinoleic acid in Y. lipolytica was described (Beopoulos et al., 2014), describing a yield of 60 mg g−1 (dry weight), the most efficient production of ricinoleic acid to date.
Kluyveromyces lactis and Kluyveromyces marxianus
Kluyveromyces lactis and the closely related K. marxianus have been studied for decades and have a well-established track record of safe use in various food industry applications. The genomes of both species are sequenced (Dujon et al., 2004), and several genetic techniques have been developed for K. marxianus (e.g. plasmid vectors; Pecota et al., 2007; Rocha et al., 2010; Wang et al., 2013; or protocols for the integration of linear DNA; Nonklang et al., 2008) and K. lactis (reviewed by van Ooyen et al., 2006) in the past 20 years. Historically, Kluyveromyces is best known for its production of the bovine milk-clotting enzyme chymosin. This protein was the first heterologous enzyme originating from a higher eukaryote that was produced at low cost in a microorganism. Nowadays, Kluyveromyces is used in the production of many heterologous proteins, such as lactase and interleukin 1-b (van Ooyen et al., 2006).
Other industrially relevant traits of Kluyveromyces are its ability to utilize lactose as a primary carbon source (which has applications in the dairy and biofuel industry), and high production of lactate. For this latter application, the central carbon flux of K. lactis was diverged from the production of ethanol to enhance lactate production. This was achieved by introducing a heterologous L-lactate dehydrogenase gene (LDH) and deleting the unique pyruvate decarboxylase gene KlPDC1 and/or the pyruvate dehydrogenase (PDH) E1 subunit gene (Porro et al., 1999; Bianchi et al., 2001). Kluyveromyces lactis was recently also proposed a host for L-ascorbic acid (vitamin C) production (Rosa et al., 2013). In the metabolically engineered strains, GDP-mannose 3,5-epimerase (GME), GDP-L-galactose phosphorylase (VTC2), and L-galactose-1-phosphate phosphatase (VTC4) from A. thaliana were introduced.
Brettanomyces (Dekkera) bruxellensis
Brettanomyces (teleomorph: Dekkera) is an ascomycetous yeast important in the production of beverages and biofuel. Interestingly, the role of Brettanomyces in the food industry is very ambiguous. Brettanomyces species, with B. bruxellensis as the most important representative, are generally reported as spoilage yeasts. Their off-flavor production in wine, beer, and cider results in huge economic losses. However, more and more authors report that in some cases these yeasts may add positive effects and complexity to, for example, red wines. Additionally, in specialty beers, sour beers, fruit beers, and one Belgian Trappist ale beer, the presence of Brettanomyces is required to obtain the characteristic and complex ‘Brett flavor’, which is (among others) described as clovy, spicy, floral, and/or smoky (Licker et al., 1999). Although still very premature, the growing interest from industry, the multiple particular phenotypes, the recent (independently performed) sequencing of three complete genomes, the development of an efficient transformation protocol, and the subsequent first transformation of B. bruxellensis (Miklenic et al., 2013) are all hinting toward an upcoming field of Brettanomyces engineering. However, more research on efficient and easy-to-use vector systems is still needed.
Komagataella (Pichia) pastoris
The ascomycetous yeast Komagataella (P.) pastoris is mostly known as the prime yeast species for recombinant protein production for both research and industrial purposes. Additionally, it is also a commonly used model organism for methanol assimilation and peroxisomal biogenesis.
Komagataella pastoris possesses several characteristics that make it an excellent host for heterologous protein production: its ability to grow to high cell densities, to produce heterologous proteins at very high levels, and to efficiently secrete them and the availability of straightforward procedures used for genetic transformation (although with rather low efficiency). Nevertheless, the amount of efficient genetic engineering tools available was, until recently, rather disproportionate to its industrial potential. The publication of the full genome in 2009 (De Schutter et al., 2009) provided the information needed to create an extensive toolbox of Ko. pastoris expression vectors, which is now commercially available (reviewed by, e.g., Bollok et al., 2009; Logez et al., 2012).
Being a methylotrophic yeast, Ko. pastoris can utilize methanol as a sole carbon and energy source. This provides a methanol-inducible transgene expression system, where the target protein gene is usually put under control of the strongly inducible promoter of the alcohol oxidase 1 (AOX1) gene (Cereghino & Cregg, 2000). Komagataella pastoris has already been used to produce over 500 different heterologous proteins (see Johnson, 2013; reviewed by Cereghino & Cregg, 2000; Macauley-Patrick et al., 2005). For example, the efficient biosynthesis of high-value carotenoids, such as lycopene [1.141 μg g−1 (dry weight)] and β-carotene [339 μg g−1 (dry weight)], was recently described in Ko. pastoris (Araya-Garay et al., 2012). Three carotenogenic enzymes were expressed for the production of lycopene, geranylgeranyl diphosphate synthase (crtE), phytoene synthase (crtB), and phytoene desaturase (crtI) from Erwinia uredovora. To convert lycopene into β-carotene, another gene encoding a lycopene β-cyclase (crtL) from Ficus carica was additionally expressed. Another example is the production of laccase (4.85 mg L−1) by Ko. pastoris by expressing the lac4 gene of Pleurotus sajor-caju (Soden et al., 2002). Additionally, several studies describe the genetic modification of the glycosylation pathway in Ko. pastoris, enabling these strains to produce complex, mammalian- and human-type N-glycans (e.g. Vervecken et al., 2004), such as functional recombinant erythropoietin (Hamilton et al., 2006).
Schizosaccharomyces pombe
Schizosaccharomyces pombe, also called ‘fission yeast’, is mostly known as a model organism for both molecular and cell biology. For example, the mechanisms of signal transduction and cell cycle regulation in eukaryotic cells (research on the latter resulted in 2001 in a ‘Nobel Prize in Physiology or Medicine’ for Sir Paul Nurse) were elucidated using Sc. pombe (Giga-Hama & Kumagai, 1999). The genome of Sc. pombe, published in 2002, was the second unicellular eukaryotic genome to be fully sequenced and the sixth eukaryotic organism overall (Wood et al., 2002).
The potential of this well-characterized yeast species as an expression tool for heterologous proteins has already been known for a long time (reviewed, e.g., by Takegawa et al., 2009). Because of the intensive use of Sc. pombe in both fundamental and applied research, numerous papers describe the use of (integration-type) plasmids for molecular manipulation (reviewed by Siam et al., 2004; more recent studies by Erler et al., 2006; Chino et al., 2010; Terazawa et al., 2011; Verma & Singh, 2012). Interestingly, notwithstanding the vast phylogenetic distance separating the two species, transformation protocols from S. cerevisiae can usually be used in Sc. pombe (Okazaki et al., 1990).
Schizosaccharomyces pombe has been used as a host for the production of many different heterologous proteins. For example, the introduction of the E. coli B phytase-encoding gene (appA) in Sc. pombe led to the expression of a secreted, glycosylated phytase, intended for use in animal feed (Ciofalo et al., 2003). Another industrially relevant example is the efficient production of vanillin (65 mg L−1), the main constituent of vanilla flavor and worldwide one of the most important flavor compounds (Hansen et al., 2009). By introducing three heterologous genes (a 3-dehydroshikimate dehydratase from Podospora pauciseta, an aromatic carboxylic acid reductase from Nocardia sp., and a human O-methyltransferase), Holic et al. (2012) established a new biosynthetic pathway with glucose as the primary metabolite. By knocking out the Sc. pombe alcohol dehydrogenase ADH6, the reduction of vanillin to vanillyl alcohol was blocked. In 2012, the introduction of a Claviceps purpurea oleate Δ12-hydroxylase gene (FAH12) led to the production at high concentrations (137 mg mL−1) of ricinoleic acid (Holic et al., 2012). Furthermore, the physiology of Sc. pombe (such as its cell cycle, chromosome structure, and RNA splicing) often shows a high similarity to more complex eukaryotic cells and can therefore be used to express heterologous genes from higher eukaryotes (Giga-Hama et al., 1994; Ikeda et al., 2004). However, for heterologous expression of most high eukaryotic genes, Ko. pastoris is still considered the best expression system (cf. supra).
Novel techniques and future perspectives
Genetic modifications as well as non-GMO approaches such as hybridization, mutagenesis, and directed evolution approaches have contributed to the expansion of strains (potentially) useful for industrial purposes. However, as discussed above, each of these techniques has its shortcomings and limitations. In short, non-GMO techniques are often limited to phenotypes that allow efficient selection, and they often involve a risk of improving one property at the expense of others. GMO techniques, on the other hand, require in-depth knowledge of the underlying genetics and are in many cases not well received by consumers. The ever-growing knowledge of yeast physiology and the continuously expanding biotechnological toolbox now allow more adventurous techniques using recombinant DNA technologies to emerge, which target (and sometimes overcome) some of the drawbacks mentioned above. Interestingly, several of these novel techniques combine aspects of both classic non-GMO techniques and genetic modification. Here, we highlight some of these techniques that could either be an alternative or complementary to these well-established approaches (Fig.5). It should be noted though that most of these techniques still need to prove their usefulness to generate commercially applicable yeast strains.
Figure 5.
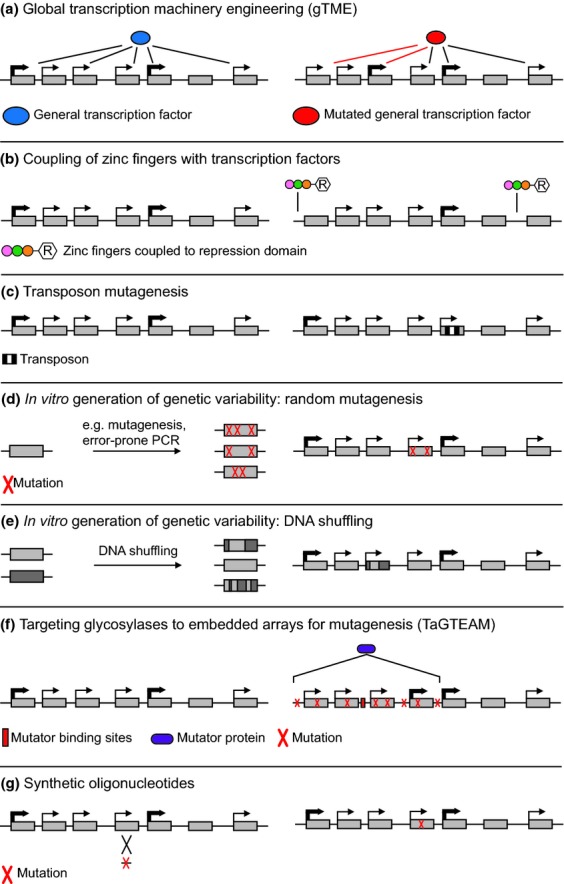
Novel techniques for genetic modification of industrial strains. Schematic overview of selected techniques to generate phenotypic diversity in (industrial) S. cerevisiae strains. The wild-type situation is always shown on the left, and the altered situation on the right. The thickness of the arrows indicates the transcription level of the genes. Global techniques (a–c) include global transcription machinery engineering (a), which exploits genome-wide transcriptional re-wiring generated by a mutated general transcription factor; artificial zinc finger transcription factors (b), creating altered transcription profiles resulting from transcription factors with a wide variety of specificity, for instance by coupling randomized zinc fingers to a repression domain; and transposon mutagenesis (c), which knocks out genes in a random fashion. Targeted techniques comprise in vitro generation of genetic variability using random mutagenesis (d) or DNA shuffling (e), TaGTEAM (f), which can drastically elevate the local mutation rate by targeting a mutator protein (=DNA glycosylase + DNA-binding protein) to a specific genomic region, and the use of oligonucleotide-based approaches like synthetic oligonucleotides (g) to introduce specific gene deletions or mutations. See text for more details.
gTME
Because complex phenotypes are determined by a large number of genes, Alper et al. (2006) hypothesized that huge phenotypic variation could be realized and exploited by (randomly) reprogramming transcription followed by selection of those variants that show improved properties. To this end, they developed a method termed ‘global transcription machinery engineering’ (gTME), which is based on creating a library of randomly mutated versions of a global transcription factor or associated factor. This library is then introduced in the strain of interest, creating a pool of mutants with different mutations in the gene encoding the transcription factor, followed by a screening (as discussed in the section on ‘Selection of phenotypically improved cells’) to identify mutants showing substantial phenotypic improvement. As a proof-of-principle, they created a mutant library of the TATA-binding protein which was transformed into a laboratory yeast strain. Interestingly, strains could be recovered showing increased glucose and ethanol tolerance and improved fermentation capacity (Alper et al., 2006). A later study pointed out that this phenotypic improvement could be explained by fixing a defect in leucine uptake and/or utilization present in the laboratory strain and could only be reproduced in medium with low amounts of leucine (Baerends et al., 2009). Therefore, other studies are needed to confirm the usefulness of this approach to obtain industrial yeasts with increased stress tolerance.
Coupling of zinc fingers with transcription factors
Another approach exploiting global transcriptional variation uses combinatorial libraries of zinc finger-containing proteins (Park et al., 2003). Many zinc fingers are highly specific DNA-binding structural motifs, stabilized by a zinc atom, present in many transcriptional regulators. Each zinc finger binds a specific DNA sequence of three base pairs, and the exact number and configuration of zinc fingers of a transcription factor determines its specificity. In practice, a large library of artificial transcription factors consisting of random combinations of three or four zinc fingers coupled to either an activation or repression domain is generated. Next, plasmids harboring the genes coding for these proteins are transformed in the target strain, after which a screening for the phenotype of interest takes place to select superior variants. When these artificially generated transcription factors were transformed into yeast, the authors were able to obtain cells with increased thermotolerance, osmotolerance, or ketoconazole resistance (Park et al., 2003).
Transposon mutagenesis
Transposon mutagenesis is a technique to introduce gene knockouts in a random fashion to generate variability in a yeast population. In brief, drug resistance cassettes are randomly inserted into the genome by transposition, after which the obtained collection of mutants is screened for superior variants (Hamer et al., 2001). When a mutant of interest is found after screening, a PCR-based sequencing reaction can pinpoint the targeted genomic region. This is an important advantage compared to classical mutagenesis, where it is difficult to map the genetic changes underlying an improved phenotype. Using this technique, researchers were able to identify genes involved in thermo- and ethanol tolerance (Kim et al., 2011).
In vitro generation of genetic variability: random mutagenesis
Although in vivo random mutagenesis (see ‘Generation of artificial diversity’) is a well-established and often powerful technique, the simplicity and more ‘targeted’ characteristics of in vitro mutagenesis of genetic material makes it often the preferred approach to generate variability. To this end, several different techniques have been developed, such as several InDel mutagenesis techniques and sequence saturation mutagenesis (SeSaM). This is particularly useful when the active site of an enzyme is known, but the most optimal combinations of amino acids not. For instance, Young et al. (2014) identified a conserved motif present in transporters that prefer xylose over glucose, and by applying SeSaM on the variable residues in this motif, they could modify primary hexose transporters into xylose transporters. However, the most widespread is the use of error-prone PCR (epPCR; Nair & Zhao, 2010). Typically, the gene of interest, or alternatively promoter regions or even whole genomes, is subjected to PCR amplification with an elevated error rate (by the lack of exonuclease activity and/or nonideal reaction conditions), followed by transformation of the fragments into the microorganism of interest and a screening to recover mutants that display phenotypic improvement. For instance, Runquist et al. (2010) applied epPCR to the cofactor-binding region of the Sc. stipitis gene XYL1, one of the enzymes needed for S. cerevisiae to consume xylose, and transformed this library into S. cerevisiae. After repeated rounds of selection in xylose medium, a strain with increased ethanol productivity could be isolated and two causative mutations in the mutagenized region were identified. Similarly, the epPCR-mediated mutation of the xylose isomerase gene (xylA) from Piromyces sp. and subsequent transformation of the library into S. cerevisiae yielded the identification of significantly improved enzyme variants (Lee et al., 2012). Recently, two reports on the directed evolution by epPCR of multiple genes in the cellobiose utilization pathway (β-glucosidase and the cellodextrin transporter) for efficient biofuel production were published (Eriksen et al., 2013; Yuan & Zhao, 2013). EpPCR can also be applied on promoters (e.g. the promoter of TEF1 (S. cerevisiae) or GAP (Ko. pastoris), cf. supra) or the whole genome. This latter approach was, for example, used to improve ethanol tolerance of S. cerevisiae (Luhe et al., 2011).
Apart from being directly used as a gene improvement method, epPCR of a general transcription factor is the first step in global transcription machinery engineering (cf. supra).
In vitro generation of genetic variability: DNA shuffling
Another way of generating artificial genetic diversity in a protein or pathway in vitro is by DNA shuffling (Stemmer, 1994). In contrast to diversification with random mutagenesis, such as methods using epPCR, DNA shuffling exploits the recombination between genes. The technique was initially proposed in 1994 and often referred to as ‘sexual PCR’. Its original setup involves three steps. First, a heterogeneous pool of closely related genes (or mutagenized copies or different alleles of the same gene) is enzymatically digested by DNase I to generate smaller fragments of DNA. Next, a primer-free PCR allows the small fragments to cross-prime each other for replication, in order to create longer fragments, resulting in hybrid DNA strands with genetic material from multiple parents. Ultimately, the recombined genes are amplified by PCR using specifically designed primers that only target full-length genes, which can subsequently be screened (Bacher et al., 2002).
Today, numerous variants of classical DNA shuffling have been described. However interesting, the numerous techniques developed to improve protein function by gene recombination are only briefly discussed below, but are discussed extensively elsewhere (Nair & Zhao, 2010). For example, ‘restriction enzyme-based shuffling’ (Kikuchi et al., 1999), ‘staggered extension progress’ (StEP; Zhao et al., 1998), ‘random priming recombination’ (Shao et al., 1998), and ‘random chimeragenesis on transient templates’ (RACHITT; Coco et al., 2001) all rely on the same general principle as ‘sexual PCR’, yet with some adaptations to overcome specific drawbacks. Alternatively, oligonucleotide-based approaches, such as ‘synthetic shuffling’, can be applied when the sequence of the parental genes is known and when they have adequate sequence identity (Ness et al., 2002). In this setup, degenerate oligonucleotides are used to construct functional libraries. All these techniques described above rely (much like ‘sexual PCR’ itself) on homologous recombination. Other methods, such as ‘incremental truncation for the creation of hybrid enzymes’ (ITCHY; Ostermeier et al., 1999), ‘sequence homology-independent protein recombination’ (SHIPREC; Sieber et al., 2001), and ‘Golden gate shuffling’ (Engler et al., 2009), use nonhomologous recombination for shuffling.
Saccharomyces cerevisiae is often used as the in vivo model to screen the created library, or sometimes even to further improve diversity by additional DNA shuffling (an approach called ‘combinatorial libraries enhanced by recombination in yeast’, CLERY; Abecassis et al., 2000), but direct use of DNA shuffling for the improvement of industrial yeast strains has not been described so far.
TaGTEAM
In many cases, it is clear which genes are responsible for a certain property, but it is less clear which exact mutations would result in improvements. In this case, it can be valuable to specifically increase the mutation rate in a targeted region of the genome. To this end, Finney-Manchester & Maheshri (2013) developed a method termed ‘targeting glycosylases to embedded arrays for mutagenesis (TaGTEAM)’. In short, a DNA glycosylase is fused to a DNA-binding protein, and the corresponding binding sites are inserted close to the region of interest. As a result, the mutation rate will locally be elevated more than 800-fold. A possible application of this technology is to move a set of genes (for instance, encoding a biochemical pathway or genes involved in a type of stress resistance) into the genomic region showing elevated mutation rates, followed by evolutionary engineering. As such, chances of developing improved mutants are expected to increase, while minimizing the risk of changing other strain properties.
Synthetic oligonucleotides
Various oligonucleotide-based approaches have been developed to introduce genetic alterations without leaving any traces. Moerschell et al. (1988) developed a technique in which transformation of yeasts with synthetic oligonucleotides as short as 20 nucleotides can be employed for site-specific mutagenesis. However, because no selectable markers are involved, this kind of genetic alteration is only applicable to selectable phenotypes. Alternatively, several methods make use of a selectable and/or counter-selectable marker in order to introduce desired genetic changes. Erdeniz et al. (1997) described cloning-free PCR-based allele replacement methods in which either alternative alleles or de novo mutations can be introduced. Through a series of PCRs of both the allele to be inserted and a marker gene (for instance, URA3) and co-transformation of these fragments, the original allele will (after integration) be replaced by a duplication of the novel gene with a URA3 gene in between these two copies. After selection for direct repeat recombinants (pop-out of the marker), only a single copy of the new allele remains. Similarly, the delitto perfetto approach is a two-step cloning-free technique to introduce site-specific genetic alterations or specific deletions up to 200 base pairs. The first step comprises the insertion of a ‘CORE’ (counter-selectable reporter) cassette in the region of interest. In the second step, integrative recombinant oligonucleotides, harboring homologous sequences flanking the CORE cassette are introduced, which mediate removal of the CORE cassette while simultaneously inserting the mutation or deletion (Storici et al., 2001).
Discussion – The new GM techniques bridge the gap between classic breeding and genetic modification
Since its first successful application in 1973, recombinant DNA technology has proven to harbor an enormous potential for strain improvement. While initial approaches only focused on single genes, methodologies such as metabolic engineering and inverse metabolic engineering target complete pathways, resulting in more efficient phenotypic improvements. Moreover, recent technological advances allow other methods, such as TaGTEAM and gTME, to generate ‘semi-random’ diversity, where an enormous number of different mutations can be introduced in a specific genomic region, thereby limiting the risk of off-target deleterious mutations that result in crippled strains. Therefore, these approaches can provide a valuable alternative for (or can be complementary to) the ‘non-GMO’ strain improvement approaches, such as mutagenesis, hybridization, or evolutionary engineering, discussed in ‘Discussion – pros and cons of exploiting natural and artificial diversity’.
However, the major disadvantage of these techniques remains the classification of the resulting strains as GMOs, impeding some industrial applications. Interestingly, strains engineered via ‘self-cloning’ do not have a GMO status in some countries, for example Japan (Akada, 2002). Self-cloning is defined as genetic modification where no DNA from another species is introduced into the genome. Therefore, the use of self-cloning principles may help to obtain formal approval to use the strain for production purposes.
Conclusion
Both traditional and novel yeast-based industries can benefit from using superior yeasts that yield increased production efficiency and/or product quality. Significant progress has already been made by exploring the existing natural diversity, by creating artificial diversity and combining beneficial traits of different strains using hybridization, as well as by targeting specific traits through genetic modification. However, the new knowledge and technologies summarized above show that there are still enormous opportunities to obtain superior industrial yeasts. However, important challenges also still remain.
Firstly, the currently used industrial strains represent only the tip of the proverbial iceberg of the actual genetic diversity present in nature. More extensive genotyping and phenotyping of both natural isolates and nonconventional yeasts will help to identify strains and species with novel and/or improved industrially important properties.
Secondly, although the genetics and physiology of laboratory yeast strains are well characterized, our knowledge about industrial strains is lagging behind. The complex genetic make-up of these strains (which are often polyploid, alloploid, and/or aneuploid) has hampered genetic studies and strain improvement strategies. Importantly, recent studies report the sequencing of industrial strains in their natural ploidy, increasing insight into their genetic architecture.
Thirdly, many industrially relevant phenotypes have a complex genetic base and depend on a large number of genes, scattered throughout the genome. Dissecting these complex traits at the genetic level is currently one of the biggest challenges in molecular biology. The advent of next-generation sequencing technologies allows for fast generation of full-genome sequences at a relatively low cost. Together with large-scale phenotyping approaches, this enables researchers to start mapping the causative mutations for complex industrially relevant traits, such as ethanol tolerance. These approaches not only generate insight into complex phenotypes, but the obtained knowledge can also be used to directly modify strains by inserting only the causative mutation(s). Additionally, novel techniques such as genome shuffling use the existing diversity to a much greater extent and can enhance complex phenotypes much more rapidly compared to more classical approaches of strain improvement. An in-depth study of hybrids generated through this approach could potentially yield insight into the genetics underlying improved phenotypes as well.
Fourthly, it is clear that genetic modification can be a valuable approach to enhance phenotypes for which the causative mutations are known. In spite of this, legislation issues and negative consumer's perception of the use of GMOs in the food chain often hamper industrial application and success of such GM strains. It remains to be seen whether (and how) regulations and public perception will change in the future.
The prospects to obtain superior industrial yeasts are extremely bright. Yeasts offer unique advantages for strain improvement: they combine sexual and asexual life cycles, they can be cultivated in high numbers, and genetic transformation is often easy. Moreover, the examples discussed in this review illustrate that most strain improvement strategies are not mutually exclusive, but can be combined to create even more variation. Additionally, by combining different techniques, the drawbacks of a specific technique can be circumvented. For example, combining random mutagenesis with multiple rounds of genome shuffling is a promising approach to both recombine useful mutations and lose deleterious mutations. Moreover, further development and optimization of current approaches and newly emerging technologies such as next-generation sequencing, combined with a better understanding of complex phenotypes and nonconventional yeasts, pave the way to obtain yeasts that are even better adapted to their industrial tasks.
Acknowledgments
The authors thank all Verstrepen laboratory members for their help and suggestions. Research in the laboratory of K.J.V. is supported by Barry-Callebaut, ERC Starting Grant 241426, VIB, EMBO YIP program, FWO, and IWT. We specifically acknowledge the FWO for the postdoctoral fellowship of K.V.
Authors' contribution
J.S. and T.S. contributed equally to this work.
Supporting Information
Additional Supporting Information may be found in the online version of this article:
Table S1. Research papers targeting strain improvement by hybridization.
Table S2. Research papers targeting strain improvement by genetic modification.
Data S1. Reference list for supplemental files.
References
- Aarnio T, Suikho M. Kauppinen V. Isolation of acetic acid tolerant baker's yeast variants in a turbidostat. Appl Biochem Biotechnol. 1991;27:55–63. [Google Scholar]
- Abecassis V, Pompon D. Truan G. High efficiency family shuffling based on multi-step PCR and in vivo DNA recombination in yeast: statistical and functional analysis of a combinatorial library between human cytochrome P450 1A1 and 1A2. Nucleic Acids Res. 2000;28:E88. doi: 10.1093/nar/28.20.e88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adamo GM, Brocca S, Passolunghi S, Salvato B. Lotti M. Laboratory evolution of copper tolerant yeast strains. Microb Cell Fact. 2012;11:1. doi: 10.1186/1475-2859-11-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agbogbo FK. Coward-Kelly G. Cellulosic ethanol production using the naturally occurring xylose-fermenting yeast, Pichia stipitis. Biotechnol Lett. 2008;30:1515–1524. doi: 10.1007/s10529-008-9728-z. [DOI] [PubMed] [Google Scholar]
- Akada R. Genetically modified industrial yeast ready for application. J Biosci Bioeng. 2002;94:536–544. doi: 10.1016/s1389-1723(02)80192-x. [DOI] [PubMed] [Google Scholar]
- Almario MP, Reyes LH. Kao KC. Evolutionary engineering of Saccharomyces cerevisiae for enhanced tolerance to hydrolysates of lignocellulosic biomass. Biotechnol Bioeng. 2013;110:2616–2623. doi: 10.1002/bit.24938. [DOI] [PubMed] [Google Scholar]
- Alper H. Stephanopoulos G. Engineering for biofuels: exploiting innate microbial capacity or importing biosynthetic potential? Nat Rev Microbiol. 2009;7:715–723. doi: 10.1038/nrmicro2186. [DOI] [PubMed] [Google Scholar]
- Alper H, Fischer C, Nevoigt E. Stephanopoulos G. Tuning genetic control through promoter engineering. P Natl Acad Sci USA. 2005;102:12678–12683. doi: 10.1073/pnas.0504604102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alper H, Moxley J, Nevoigt E, Fink GR. Stephanopoulos G. Engineering yeast transcription machinery for improved ethanol tolerance and production. Science. 2006;314:1565–1568. doi: 10.1126/science.1131969. [DOI] [PubMed] [Google Scholar]
- Angelov AI, Karadjov GI. Roshkova ZG. Strains selection of baker's yeast with improved technological properties. Food Res Int. 1996;29:235–239. [Google Scholar]
- Araya-Garay JM, Feijoo-Siota L, Rosa-dos-Santos F, Veiga-Crespo P. Villa TG. Construction of new Pichia pastoris X-33 strains for production of lycopene and beta-carotene. Appl Microbiol Biotechnol. 2012;93:2483–2492. doi: 10.1007/s00253-011-3764-7. [DOI] [PubMed] [Google Scholar]
- Attfield PV. Bell PJL. Genetics and classical genetic manipulations of industrial yeasts. In: De Winde JH, editor. Functional Genetics of Industrial Yeasts. Berlin: Springer; 2003. pp. 17–56. , Vol. 1 (, ed) &. [Google Scholar]
- Avalos JL, Fink GR. Stephanopoulos G. Compartmentalization of metabolic pathways in yeast mitochondria improves the production of branched-chain alcohols. Nat Biotechnol. 2013;31:335–341. doi: 10.1038/nbt.2509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bacher JM, Reiss BD. Ellington AD. Anticipatory evolution and DNA shuffling. Genome Biol. 2002;3:1021. doi: 10.1186/gb-2002-3-8-reviews1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baerends RJ, Qiu J-L, Rasmussen S, Nielsen HB. Brandt A. Impaired uptake and/or utilization of leucine by Saccharomyces cerevisiae is suppressed by the SPT15-300 allele of the TATA-binding protein gene. Appl Environ Microbiol. 2009;75:6055–6061. doi: 10.1128/AEM.00989-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey JE. Toward a science of metabolic engineering. Science. 1991;252:1668–1675. doi: 10.1126/science.2047876. [DOI] [PubMed] [Google Scholar]
- Bailey JE, Sburlati A, Hatzimanikatis V, Lee K, Renner WA. Tsai PS. Inverse metabolic engineering: a strategy for directed genetic engineering of useful phenotypes. Biotechnol Bioeng. 2002;79:568–579. doi: 10.1002/bit.10441. [DOI] [PubMed] [Google Scholar]
- Bajwa PK, Pinel D, Martin VJ, Trevors JT. Lee H. Strain improvement of the pentose-fermenting yeast Pichia stipitis by genome shuffling. J Microbiol Methods. 2010;81:179–186. doi: 10.1016/j.mimet.2010.03.009. [DOI] [PubMed] [Google Scholar]
- Balakumar S, Arasaratnam V. Balasubramaniam K. Isolation and improvement of a thermotolerant Saccharomyces cerevisiae strain. World J Microbiol Biotechnol. 2001;17:739–746. [Google Scholar]
- Barre P, Vezinhet F, Dequin S. Blondin B. Genetic improvement of wine yeasts. In: Fleet GH, editor. Wine Microbiology and Biotechnology. New York, NY: Taylor & Francis; 1993. pp. 265–287. [Google Scholar]
- Bassel J, Warfel J. Mortimer R. Complementation and genetic recombination in Candida lipolytica. J Bacteriol. 1971;108:609–611. doi: 10.1128/jb.108.1.609-611.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basso LC, de Amorim HV, de Oliveira AJ. Lopes ML. Yeast selection for fuel ethanol production in Brazil. FEMS Yeast Res. 2008;8:1155–1163. doi: 10.1111/j.1567-1364.2008.00428.x. [DOI] [PubMed] [Google Scholar]
- Basso LC, Oliveira AJ, Orelli VFDM, Campos AA, Gallo CR. Amorim HV. Dominância das leveduras contaminantes sobre as linhagens industriais avaliada pela técnica da cariotipagem. Anais Congresso Nacional da STAB. 1993;5:246–250. [Google Scholar]
- Becker JV, Armstrong GO, van der Merwe MJ, Lambrechts MG, Vivier MA. Pretorius IS. Metabolic engineering of Saccharomyces cerevisiae for the synthesis of the wine-related antioxidant resveratrol. FEMS Yeast Res. 2003;4:79–85. doi: 10.1016/S1567-1356(03)00157-0. [DOI] [PubMed] [Google Scholar]
- Bell PJ, Deere D, Shen J, Chapman B, Bissinger PH, Attfield PV. Veal DA. A flow cytometric method for rapid selection of novel industrial yeast hybrids. Appl Environ Microbiol. 1998;64:1669–1672. doi: 10.1128/aem.64.5.1669-1672.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellon JR, Schmid F, Capone DL, Dunn BL. Chambers PJ. Introducing a new breed of wine yeast: interspecific hybridisation between a commercial Saccharomyces cerevisiae wine yeast and Saccharomyces mikatae. PLoS One. 2013;8:e62053. doi: 10.1371/journal.pone.0062053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellon JR, Eglinton JM, Siebert TE, Pollnitz AP, Rose L, de Barros Lopes M. Chambers PJ. Newly generated interspecific wine yeast hybrids introduce flavour and aroma diversity to wines. Appl Microbiol Biotechnol. 2011;91:603–612. doi: 10.1007/s00253-011-3294-3. [DOI] [PubMed] [Google Scholar]
- Benjaphokee S, Hasegawa D, Yokota D, Asvarak T, Auesukaree C, Sugiyama M, Kaneko Y, Boonchird C. Harashima S. Highly efficient bioethanol production by a Saccharomyces cerevisiae strain with multiple stress tolerance to high temperature, acid and ethanol. N Biotechnol. 2012;29:379–386. doi: 10.1016/j.nbt.2011.07.002. [DOI] [PubMed] [Google Scholar]
- Beopoulos A, Cescut J, Haddouche R, Uribelarrea JL, Molina-Jouve C. Nicaud JM. Yarrowia lipolytica as a model for bio-oil production. Prog Lipid Res. 2009;48:375–387. doi: 10.1016/j.plipres.2009.08.005. [DOI] [PubMed] [Google Scholar]
- Beopoulos A, Verbeke J, Bordes F, Guicherd M, Bressy M, Marty A. Nicaud JM. Metabolic engineering for ricinoleic acid production in the oleaginous yeast Yarrowia lipolytica. Appl Microbiol Biotechnol. 2014;98:251–262. doi: 10.1007/s00253-013-5295-x. [DOI] [PubMed] [Google Scholar]
- Beopoulos A, Mrozova Z, Thevenieau F, Le Dall MT, Hapala I, Papanikolaou S, Chardot T. Nicaud JM. Control of lipid accumulation in the yeast Yarrowia lipolytica. Appl Environ Microbiol. 2008;74:7779–7789. doi: 10.1128/AEM.01412-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bianchi MM, Brambilla L, Protani F, Liu CL, Lievense J. Porro D. Efficient homolactic fermentation by Kluyveromyces lactis strains defective in pyruvate utilization and transformed with the heterologous LDH gene. Appl Environ Microbiol. 2001;67:5621–5625. doi: 10.1128/AEM.67.12.5621-5625.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Björkqvist S, Ansell R, Adler L. Liden G. Physiological response to anaerobicity of glycerol-3-phosphate dehydrogenase mutants of Saccharomyces cerevisiae. Appl Environ Microbiol. 1997;63:128–132. doi: 10.1128/aem.63.1.128-132.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blasco L, Veiga-Crespo P, Vinas M. Villa TG. A new disruption vector (pDHO) to obtain heterothallic strains from both Saccharomyces cerevisiae and Saccharomyces pastorianus. Int Microbiol. 2011;14:201–206. doi: 10.2436/20.1501.01.149. [DOI] [PubMed] [Google Scholar]
- Blieck L, Toye G, Dumortier F, Verstrepen KJ, Delvaux FR, Thevelein JM. Van Dijck P. Isolation and characterization of brewer's yeast variants with improved fermentation performance under high-gravity conditions. Appl Environ Microbiol. 2007;73:815–824. doi: 10.1128/AEM.02109-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blomqvist J, Eberhard T, Schnurer J. Passoth V. Fermentation characteristics of Dekkera bruxellensis strains. Appl Microbiol Biotechnol. 2010;87:1487–1497. doi: 10.1007/s00253-010-2619-y. [DOI] [PubMed] [Google Scholar]
- Boeke JD, Trueheart J, Natsoulis G. Fink GR. 5-Fluoroorotic acid as a selective agent in yeast molecular genetics. Methods Enzymol. 1987;154:164–175. doi: 10.1016/0076-6879(87)54076-9. [DOI] [PubMed] [Google Scholar]
- Bollok M, Resina D, Valero F. Ferrer P. Recent patents on the Pichia pastoris expression system: expanding the toolbox for recombinant protein production. Recent Pat Biotechnol. 2009;3:192–201. doi: 10.2174/187220809789389126. [DOI] [PubMed] [Google Scholar]
- Boone C, Sdicu AM, Wagner J, Degre R, Sanchez C. Bussey H. Integration of the yeast K1 killer toxin gene into the genome of marked wine yeasts and its effect on vinification. Am J Enol Vitic. 1990;41:37–42. [Google Scholar]
- Borneman AR, Pretorius IS. Chambers PJ. Comparative genomics: a revolutionary tool for wine yeast strain development. Curr Opin Biotechnol. 2013;24:192–199. doi: 10.1016/j.copbio.2012.08.006. [DOI] [PubMed] [Google Scholar]
- Borneman AR, Desany BA, Riches D, Affourtit JP, Forgan AH, Pretorius IS, Egholm M. Chambers PJ. Whole-genome comparison reveals novel genetic elements that characterize the genome of industrial strains of Saccharomyces cerevisiae. PLoS Genet. 2011;7:e1001287. doi: 10.1371/journal.pgen.1001287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brat D, Weber C, Lorenzen W, Bode HB. Boles E. Cytosolic re-localization and optimization of valine synthesis and catabolism enables inseased isobutanol production with the yeast Saccharomyces cerevisiae. Biotechnol Biofuels. 2012;5:65. doi: 10.1186/1754-6834-5-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brehm-Stecher BF. Johnson EA. Isolation of carotenoid hyperproducing mutants of Xanthophyllomyces dendrorhousPhaffia rhodozyma) by flow cytometry and cell sorting. In: Barredo JL, editor. Microbial Carotenoids From Fungi. New York, NY: Humana Press; 2012. pp. 207–217. [DOI] [PubMed] [Google Scholar]
- Bro C, Knudsen S, Regenberg B, Olsson L. Nielsen J. Improvement of galactose uptake in Saccharomyces cerevisiae through overexpression of phosphoglucomutase: example of transcript analysis as a tool in inverse metabolic engineering. Appl Environ Microbiol. 2005;71:6465–6472. doi: 10.1128/AEM.71.11.6465-6472.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown CA, Murray AW. Verstrepen KJ. Rapid expansion and functional divergence of subtelomeric gene families in yeasts. Curr Biol. 2010;20:895–903. doi: 10.1016/j.cub.2010.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown SW. Oliver SG. Isolation of ethanol-tolerant mutants of yeast by continuous selection. Eur J Appl Microbiol Biotechnol. 1982;16:119–122. [Google Scholar]
- Bryson V. Szybalski W. Microbial selection. Science. 1952;116:45–51. [PubMed] [Google Scholar]
- Buckling A, Craig Maclean R, Brockhurst MA. Colegrave N. The Beagle in a bottle. Nature. 2009;457:824–829. doi: 10.1038/nature07892. [DOI] [PubMed] [Google Scholar]
- Butler PR, Brown M. Oliver SG. Improvement of antibiotic titers from Streptomyces bacteria by interactive continuous selection. Biotechnol Bioeng. 1996;49:185–196. doi: 10.1002/(SICI)1097-0290(19960120)49:2<185::AID-BIT7>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- Cadière A, Ortiz-Julien A, Camarasa C. Dequin S. Evolutionary engineered Saccharomyces cerevisiae wine yeast strains with increased in vivo flux through the pentose phosphate pathway. Metab Eng. 2011;13:263–271. doi: 10.1016/j.ymben.2011.01.008. [DOI] [PubMed] [Google Scholar]
- Cadière A, Aguera E, Caille S, Ortiz-Julien A. Dequin S. Pilot-scale evaluation the enological traits of a novel, aromatic wine yeast strain obtained by adaptive evolution. Food Microbiol. 2012;32:332–337. doi: 10.1016/j.fm.2012.07.010. [DOI] [PubMed] [Google Scholar]
- Cakar ZP, Seker UO, Tamerler C, Sonderegger M. Sauer U. Evolutionary engineering of multiple-stress resistant Saccharomyces cerevisiae. FEMS Yeast Res. 2005;5:569–578. doi: 10.1016/j.femsyr.2004.10.010. [DOI] [PubMed] [Google Scholar]
- Cakar ZP, Alkim C, Turanli B, Tokman N, Akman S, Sarikaya M, Tamerler C, Benbadis L. Francois JM. Isolation of cobalt hyper-resistant mutants of Saccharomyces cerevisiae by in vivo evolutionary engineering approach. J Biotechnol. 2009;143:130–138. doi: 10.1016/j.jbiotec.2009.06.024. [DOI] [PubMed] [Google Scholar]
- Cao X, Hou L, Lu M. Wang C. Improvement of soy-sauce flavour by genome shuffling in Candida versatilis to improve salt stress resistance. Int J Food Sci Technol. 2010;45:17–22. [Google Scholar]
- Cao X, Song Q, Wang C. Hou L. Genome shuffling of Hansenula anomala to improve flavour formation of soy sauce. World J Microbiol Biotechnol. 2012;28:1857–1862. doi: 10.1007/s11274-010-0477-5. [DOI] [PubMed] [Google Scholar]
- Cao X, Hou L, Lu M, Wang C. Zeng B. Genome shuffling of Zygosaccharomyces rouxii to accelerate and enhance the flavour formation of soy sauce. J Sci Food Agric. 2009;90:281–285. doi: 10.1002/jsfa.3810. [DOI] [PubMed] [Google Scholar]
- Capece A, Romaniello R, Siesto G, Pietrafesa R, Massari C, Poeta C. Romano P. Selection of indigenous Saccharomyces cerevisiae strains for Nero d'Avola wine and evaluation of selected starter implantation in pilot fermentation. Int J Food Microbiol. 2010;144:187–192. doi: 10.1016/j.ijfoodmicro.2010.09.009. [DOI] [PubMed] [Google Scholar]
- Carrau JL, Dillon AJP, Serafini LA. Pazqual MS. 1994. & L-malic acid degrading yeast for wine making. (United States Patent 5, 774.)
- Carstens M, Vivier MA. Pretorius IS. The Saccharomyces cerevisiae chitinase, encoded by the CTS1-2 gene, confers antifungal activity against Botrytis cinerea to transgenic tobacco. Transgenic Res. 2003;12:497–508. doi: 10.1023/a:1024220023057. [DOI] [PubMed] [Google Scholar]
- Celinska E, Kubiak P, Bialas W, Dziadas M. Grajek W. Yarrowia lipolytica: the novel and promising 2-phenylethanol producer. J Ind Microbiol Biotechnol. 2013;40:389–392. doi: 10.1007/s10295-013-1240-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cereghino JL. Cregg JM. Heterologous protein expression in the methylotrophic yeast Pichia pastoris. FEMS Microbiol Rev. 2000;24:45–66. doi: 10.1111/j.1574-6976.2000.tb00532.x. [DOI] [PubMed] [Google Scholar]
- Chambers PJ. Pretorius IS. Fermenting knowledge: the history of winemaking, science and yeast research. EMBO Rep. 2010;11:914–920. doi: 10.1038/embor.2010.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambers PJ, Bellon JR, Schmidt SA, Varela C. Pretorius IS. Non-genetic engineering approaches for isolating and generating novel yeasts for industrial applications. In: Satyanarayana T, editor; Kunze G, editor. Yeast Biotechnology: Diversity and applications. Dordrecht, Netherlands: Springer; 2009. pp. 433–457. [Google Scholar]
- Chino A, Watanabe K. Moriya H. Plasmid construction using recombination activity in the fission yeast Schizosaccharomyces pombe. PLoS One. 2010;5:7. doi: 10.1371/journal.pone.0009652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi GW, Um HJ, Kang HW, Kim Y, Kim M. Kim YH. Bioethanol production by a flocculent hybrid, CHFY0321 obtained by protoplast fusion between Saccharomyces cerevisiae and Saccharomyces bayanus. Biomass Bioenergy. 2010;34:1232–1242. [Google Scholar]
- Christiaens JF, Van Mulders SE, Duitama J, Brown CA, Ghequire MG, De Meester L, Michiels J, Wenseleers T, Voordeckers K. Verstrepen KJ. Functional divergence of gene duplicates through ectopic recombination. EMBO Rep. 2012;13:1145–1151. doi: 10.1038/embor.2012.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christianson TW, Sikorski RS, Dante M, Shero JH. Hieter P. Multifunctional yeast high-copy-number shuttle vectors. Gene. 1992;110:119–122. doi: 10.1016/0378-1119(92)90454-w. [DOI] [PubMed] [Google Scholar]
- Ciofalo V, Barton N, Kretz K, Baird J, Cook M. Shanahan D. Safety evaluation of a phytase, expressed in Schizosaccharomyces pombe, intended for use in animal feed. Regul Toxicol Pharmacol. 2003;37:286–292. doi: 10.1016/s0273-2300(03)00005-9. [DOI] [PubMed] [Google Scholar]
- Clarke L. Carbon J. Isolation of a yeast centromere and construction of functional small circular chromosomes. Nature. 1980;287:504–509. doi: 10.1038/287504a0. [DOI] [PubMed] [Google Scholar]
- Coco WM, Levinson WE, Crist MJ, Hektor HJ, Darzins A, Pienkos PT, Squires CH. Monticello DJ. DNA shuffling method for generating highly recombined genes and evolved enzymes. Nat Biotechnol. 2001;19:354–359. doi: 10.1038/86744. [DOI] [PubMed] [Google Scholar]
- Codon AC, Gasent-Ramirez JM. Benitez T. Factors which affect the frequency of sporulation and tetrad formation in Saccharomyces cerevisiae baker's yeasts. Appl Environ Microbiol. 1995;61:1677. doi: 10.1128/aem.61.4.1677-1677b.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coloretti F, Zambonelli C. Tini V. Characterization of flocculent Saccharomyces interspecific hybrids for the production of sparkling wines. Food Microbiol. 2006;23:672–676. doi: 10.1016/j.fm.2005.11.002. [DOI] [PubMed] [Google Scholar]
- Comitini F, Gobbi M, Domizio P, Romani C, Lencioni L, Mannazzu I. Ciani M. Selected non-Saccharomyces wine yeasts in controlled multistarter fermentations with Saccharomyces cerevisiae. Food Microbiol. 2011;28:873–882. doi: 10.1016/j.fm.2010.12.001. [DOI] [PubMed] [Google Scholar]
- Conde J. Fink GR. A mutant of Saccharomyces cerevisiae defective for nuclear fusion. P Natl Acad Sci USA. 1976;73:3651–3655. doi: 10.1073/pnas.73.10.3651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordente AG, Heinrich A, Pretorius IS. Swiegers JH. Isolation of sulfite reductase variants of a commercial wine yeast with significantly reduced hydrogen sulfide production. FEMS Yeast Res. 2009;9:446–459. doi: 10.1111/j.1567-1364.2009.00489.x. [DOI] [PubMed] [Google Scholar]
- Cordier H, Mendes F, Vasconcelos I. Francois JM. A metabolic and genomic study of engineered Saccharomyces cerevisiae strains for high glycerol production. Metab Eng. 2007;9:364–378. doi: 10.1016/j.ymben.2007.03.002. [DOI] [PubMed] [Google Scholar]
- Coulon J, Husnik JI, Inglis DL, van der Merwe GK, Lonvaud A, Erasmus DJ. van Vuuren HJJ. Metabolic engineering of Saccharomyces cerevisiae to minimize the production of ethyl carbamate in wine. Am J Enol Vitic. 2006;57:113–124. [Google Scholar]
- Crook N. Alper HS. Classical strain improvement. In: Patnaik R, editor. Engineering Complex Phenotypes in Industrial Strains. Hoboken, NJ: John Wiley & Sons, Inc; 2012. pp. 1–33. [Google Scholar]
- Curran KA, Karim AS, Gupta A. Alper HS. Use of expression-enhancing terminators in Saccharomyces cerevisiae to increase mRNA half-life and improve gene expression control for metabolic engineering applications. Metab Eng. 2013;19:88–97. doi: 10.1016/j.ymben.2013.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- da Silva-Filho EA, Brito dos Santos SK, Resende Ado M, de Morais JO, de Morais MA., Jr Ardaillon Simoes D. Yeast population dynamics of industrial fuel-ethanol fermentation process assessed by PCR-fingerprinting. Antonie Van Leeuwenhoek. 2005;88:13–23. doi: 10.1007/s10482-004-7283-8. [DOI] [PubMed] [Google Scholar]
- Da Silva NA. Srikrishnan S. Introduction and expression of genes for metabolic engineering applications in Saccharomyces cerevisiae. FEMS Yeast Res. 2012;12:197–214. doi: 10.1111/j.1567-1364.2011.00769.x. [DOI] [PubMed] [Google Scholar]
- Dahabieh MS, Husnik JI. Van Vuuren HJ. Functional enhancement of sake yeast strains to minimize the production of ethyl carbamate in sake wine. J Appl Microbiol. 2010;109:963–973. doi: 10.1111/j.1365-2672.2010.04723.x. [DOI] [PubMed] [Google Scholar]
- de Barros Lopes M, Bellon JR, Shirley NJ. Ganter PF. Evidence for multiple interspecific hybridization in Saccharomyces sensu stricto species. FEMS Yeast Res. 2002;1:323–331. doi: 10.1111/j.1567-1364.2002.tb00051.x. [DOI] [PubMed] [Google Scholar]
- de Jong B, Siewers V. Nielsen J. Systems biology of yeast: enabling technology for development of cell factories for production of advanced biofuels. Curr Opin Biotechnol. 2012;23:624–630. doi: 10.1016/j.copbio.2011.11.021. [DOI] [PubMed] [Google Scholar]
- De Schutter K, Lin YC, Tiels P, Van Hecke A, Glinka S, Weber-Lehmann J, Rouze P, de Peer YV. Callewaert N. Genome sequence of the recombinant protein production host Pichia pastoris. Nat Biotechnol. 2009;27:561–566. doi: 10.1038/nbt.1544. [DOI] [PubMed] [Google Scholar]
- de Souza AP, Vicente Mde A, Klein RC, et al. Strategies to select yeast starters cultures for production of flavor compounds in cachaca fermentations. Antonie Van Leeuwenhoek. 2012;101:379–392. doi: 10.1007/s10482-011-9643-5. [DOI] [PubMed] [Google Scholar]
- de Ullivarri MF, Mendoza LM, Raya RR. Farias ME. Killer phenotype of indigenous yeasts isolated from Argentinian wine cellars and their potential starter cultures for winemaking. Biotechnol Lett. 2011;33:2177–2183. doi: 10.1007/s10529-011-0674-9. [DOI] [PubMed] [Google Scholar]
- Della-Bianca BE, Basso TO, Stambuk BU, Basso LC. Gombert AK. What do we know about the yeast strains from the Brazilian fuel ethanol industry? Appl Microbiol Biotechnol. 2012;97:979–991. doi: 10.1007/s00253-012-4631-x. [DOI] [PubMed] [Google Scholar]
- Demain AL. History of industrial biotechnology. In: Soetaert W, editor; Vandamme EJ, editor. Industrial Biotechnology: Sustainable Growth and Economic Success. Weinheim, Germany: Wiley-VCH Verlag GmbH & Co. KGaA; 2010. pp. 17–77. [Google Scholar]
- Demeke MM, Dietz H, Li Y, et al. Development of a D-xylose fermenting and inhibitor tolerant industrial Saccharomyces cerevisiae strain with high performance in lignocellulose hydrolysates using metabolic and evolutionary engineering. Biotechnol Biofuels. 2013;6:89. doi: 10.1186/1754-6834-6-89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dequin S. The potential of genetic engineering for improving brewing, wine-making and baking yeasts. Appl Microbiol Biotechnol. 2001;56:577–588. doi: 10.1007/s002530100700. [DOI] [PubMed] [Google Scholar]
- Dinh TN, Nagahisa K, Hirasawa T, Furusawa C. Shimizu H. Adaptation of Saccharomyces cerevisiae cells to high ethanol concentration and changes in fatty acid composition of membrane and cell size. PLoS One. 2008;3:e2623. doi: 10.1371/journal.pone.0002623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drake JW. A constant rate of spontaneous mutation in DNA-based microbes. P Natl Acad Sci USA. 1991;88:7160–7164. doi: 10.1073/pnas.88.16.7160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du J, Yuan Y, Si T, Lian J. Zhao H. Customized optimization of metabolic pathways by combinatorial transcriptional engineering. Nucleic Acids Res. 2012;40:e142. doi: 10.1093/nar/gks549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duenas-Sanchez R, Codon AC, Rincon AM. Benitez T. Increased biomass production of industrial bakers' yeasts by overexpression of HAP4 gene. Int J Food Microbiol. 2010;143:150–160. doi: 10.1016/j.ijfoodmicro.2010.08.009. [DOI] [PubMed] [Google Scholar]
- Dujon B, Sherman D, Fischer G, et al. Genome evolution in yeasts. Nature. 2004;430:35–44. doi: 10.1038/nature02579. [DOI] [PubMed] [Google Scholar]
- Dulermo T. Nicaud JM. Involvement of the G3P shuttle and beta-oxidation pathway in the control of TAG synthesis and lipid accumulation in Yarrowia lipolytica. Metab Eng. 2011;13:482–491. doi: 10.1016/j.ymben.2011.05.002. [DOI] [PubMed] [Google Scholar]
- Dunham MJ, Badrane H, Ferea T, Adams J, Brown PO, Rosenzweig F. Botstein D. Characteristic genome rearrangements in experimental evolution of Saccharomyces cerevisiae. P Natl Acad Sci USA. 2002;99:16144–16149. doi: 10.1073/pnas.242624799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn B. Sherlock G. Reconstruction of the genome origins and evolution of the hybrid lager yeast Saccharomyces pastorianus. Genome Res. 2008;18:1610–1623. doi: 10.1101/gr.076075.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duong CT, Strack L, Futschik M, Katou Y, Nakao Y, Fujimura T, Shirahige K, Kodama Y. Nevoigt E. Identification of Sc-type ILV6 as a target to reduce diacetyl formation in lager brewers' yeast. Metab Eng. 2011;13:638–647. doi: 10.1016/j.ymben.2011.07.005. [DOI] [PubMed] [Google Scholar]
- Dutcher SK. Internuclear transfer of genetic information in kar1-1/KAR1 heterokaryons in Saccharomyces cerevisiae. Mol Cell Biol. 1981;1:245–253. doi: 10.1128/mcb.1.3.245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dykhuizen DE. Hartl DL. Selection in chemostats. Microbiol Rev. 1983;47:150–168. doi: 10.1128/mr.47.2.150-168.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dymond JS, Richardson SM, Coombes CE, et al. Synthetic chromosome arms function in yeast and generate phenotypic diversity by design. Nature. 2011;477:471–476. doi: 10.1038/nature10403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekberg J, Rautio J, Mattinen L, Vidgren V, Londesborough J. Gibson BR. Adaptive evolution of the lager brewing yeast Saccharomyces pastorianus for improved growth under hyperosmotic conditions and its influence on fermentation performance. FEMS Yeast Res. 2013;13:335–349. doi: 10.1111/1567-1364.12038. [DOI] [PubMed] [Google Scholar]
- Elena SF. Lenski RE. Evolution experiments with microorganisms: the dynamics and genetic bases of adaptation. Nat Rev Genet. 2003;4:457–469. doi: 10.1038/nrg1088. [DOI] [PubMed] [Google Scholar]
- Ellegren H. Microsatellites: simple sequences with complex evolution. Nat Rev Genet. 2004;5:435–445. doi: 10.1038/nrg1348. [DOI] [PubMed] [Google Scholar]
- Engler C, Gruetzner R, Kandzia R. Marillonnet S. Golden gate shuffling: a one-pot DNA shuffling method based on type IIs restriction enzymes. PLoS One. 2009;4:e5553. doi: 10.1371/journal.pone.0005553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erdeniz N, Mortensen UH. Rothstein R. Cloning-free PCR-based allele replacement methods. Genome Res. 1997;7:1174–1183. doi: 10.1101/gr.7.12.1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eriksen DT, Hsieh PCH, Lynn P. Zhao H. Directed evolution of a cellobiose utilization pathway in Saccharomyces cerevisiae by simultaneously engineering multiple proteins. Microb Cell Fact. 2013;12:61. doi: 10.1186/1475-2859-12-61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erler A, Maresca M, Fu J. Stewart AF. Recombineering reagents for improved inducible expression and selection marker re-use in Schizosaccharomyces pombe. Yeast. 2006;23:813–823. doi: 10.1002/yea.1396. [DOI] [PubMed] [Google Scholar]
- Eschenbruch R, Cresswell KJ, Fisher BM. Thornton RJ. Selective hybridization of pure culture wine yeasts. Eur J Appl Microbiol. 1982;14:155–158. [Google Scholar]
- Farahnak F, Seki T, Ryu DD. Ogrydziak D. Construction of lactose-assimilating and high-ethanol-producing yeasts by protoplast fusion. Appl Environ Microbiol. 1986;51:362–367. doi: 10.1128/aem.51.2.362-367.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fay JC. Benavides JA. Evidence for domesticated and wild populations of Saccharomyces cerevisiae. PLoS Genet. 2005;1:66–71. doi: 10.1371/journal.pgen.0010005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fickers P, Benetti PH, Wache Y, Marty A, Mauersberger S, Smit MS. Nicaud JM. Hydrophobic substrate utilisation by the yeast Yarrowia lipolytica, and its potential applications. FEMS Yeast Res. 2005;5:527–543. doi: 10.1016/j.femsyr.2004.09.004. [DOI] [PubMed] [Google Scholar]
- Finney-Manchester SP. Maheshri N. Harnessing mutagenic homologous recombination for targeted mutagenesis in vivo by TaGTEAM. Nucleic Acids Res. 2013;41:e99. doi: 10.1093/nar/gkt150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuda K, Watanabe M, Asano K, Ouchi K. Takasawa S. Isolation and genetic study of p-fluoro-dl-phenylalanine-resistant mutants overproducing β-phenethyl-alcohol in Saccharomyces cerevisiae. Curr Genet. 1991;20:449–452. doi: 10.1007/BF00334770. [DOI] [PubMed] [Google Scholar]
- Fung E, Wong WW, Suen JK, Bulter T, Lee SG. Liao JC. A synthetic gene-metabolic oscillator. Nature. 2005;435:118–122. doi: 10.1038/nature03508. [DOI] [PubMed] [Google Scholar]
- Futcher B. Carbon J. Toxic effects of excess cloned centromeres. Mol Cell Biol. 1986;6:2213–2222. doi: 10.1128/mcb.6.6.2213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galeote V, Novo M, Salema-Oom M, Brion C, Valerio E, Goncalves P. Dequin S. FSY1, a horizontally transferred gene in the Saccharomyces cerevisiae EC1118 wine yeast strain, encodes a high-affinity fructose/H+ symporter. Microbiology. 2010;156:3754–3761. doi: 10.1099/mic.0.041673-0. [DOI] [PubMed] [Google Scholar]
- Garcia Sanchez R, Solodovnikova N. Wendland J. Breeding of lager yeast with Saccharomyces cerevisiae improves stress resistance and fermentation performance. Yeast. 2012;29:343–355. doi: 10.1002/yea.2914. [DOI] [PubMed] [Google Scholar]
- Gemayel R, Vinces MD, Legendre M. Verstrepen KJ. Variable tandem repeats accelerate evolution of coding and regulatory sequences. Annu Rev Genet. 2010;44:445–477. doi: 10.1146/annurev-genet-072610-155046. [DOI] [PubMed] [Google Scholar]
- Georgieva B. Rothstein R. kar-mediated plasmid transfer between yeast strains: alternative to traditional transformation methods. Methods Enzymol. 2002;350:278–289. doi: 10.1016/s0076-6879(02)50969-1. [DOI] [PubMed] [Google Scholar]
- Gibson BR, Lawrence SJ, Leclaire JP, Powell CD. Smart KA. Yeast responses to stresses associated with industrial brewery handling. FEMS Microbiol Rev. 2007;31:535–569. doi: 10.1111/j.1574-6976.2007.00076.x. [DOI] [PubMed] [Google Scholar]
- Gibson DG, Benders GA, Axelrod KC, Zaveri J, Algire MA, Moodie M, Montague MG, Venter JC, Smith HO. Hutchison CA., 3rd One-step assembly in yeast of 25 overlapping DNA fragments to form a complete synthetic Mycoplasma genitalium genome. P Natl Acad Sci USA. 2008;105:20404–20409. doi: 10.1073/pnas.0811011106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gietz RD. Woods RA. Genetic transformation of yeast. Biotechniques. 2001;30:816–820. doi: 10.2144/01304rv02. &, 822–826, 828 passim. [DOI] [PubMed] [Google Scholar]
- Giga-Hama Y. Kumagai H. Expression system for foreign genes using the fission yeast Schizosaccharomyces pombe. Biotechnol Appl Biochem. 1999;30:235–244. [PubMed] [Google Scholar]
- Giga-Hama Y, Tohda H, Okada H, Owada MK, Okayama H. Kumagai H. High-level expression of human lipocortin-I in the fission yeast Schizosaccharomyces pombe using a novel expression vector. Nat Biotechnol. 1994;12:400–404. doi: 10.1038/nbt0494-400. [DOI] [PubMed] [Google Scholar]
- Giudici P, Solieri L, Pulvirenti AM. Cassanelli S. Strategies and perspectives for genetic improvement of wine yeasts. Appl Microbiol Biotechnol. 2005;66:622–628. doi: 10.1007/s00253-004-1784-2. [DOI] [PubMed] [Google Scholar]
- Gjermansen C. Sigsgaard P. Construction of a hybrid brewing strain of Saccharomyces carlsbergensis by mating of meiotic segregants. Carlsberg Res Commun. 1981;46:1–11. [Google Scholar]
- Gkatzionis K, Hewson L, Hollowood T, Hort J, Dodd CER. Linforth RST. Effect of Yarrowia lipolytica on blue cheese odour development: flash profile sensory evaluation of microbiological models and cheeses. Int Dairy J. 2013;30:8–13. [Google Scholar]
- Gomez-Pastor R, Perez-Torrado R, Cabiscol E, Ros J. Matallana E. Engineered Trx2p industrial yeast strain protects glycolysis and fermentation proteins from oxidative carbonylation during biomass propagation. Microb Cell Fact. 2012;11:4. doi: 10.1186/1475-2859-11-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong J, Zheng H, Wu Z, Chen T. Zhao X. Genome shuffling: progress and applications for phenotype improvement. Biotechnol Adv. 2009;27:996–1005. doi: 10.1016/j.biotechadv.2009.05.016. [DOI] [PubMed] [Google Scholar]
- Gorsich SW, Dien BS, Nichols NN, Slininger PJ, Liu ZL. Skory CD. Tolerance to furfural-induced stress is associated with pentose phosphate pathway genes ZWF1 GND1 RPE1, and TKL1 in Saccharomyces cerevisiae. Appl Microbiol Biotechnol. 2006;71:339–349. doi: 10.1007/s00253-005-0142-3. [DOI] [PubMed] [Google Scholar]
- Gresham D, Desai MM, Tucker CM, Jenq HT, Pai DA, Ward A, DeSevo CG, Botstein D. Dunham MJ. The repertoire and dynamics of evolutionary adaptations to controlled nutrient-limited environments in yeast. PLoS Genet. 2008;4:e1000303. doi: 10.1371/journal.pgen.1000303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guillaume C, Delobel P, Sablayrolles JM. Blondin B. Molecular basis of fructose utilization by the wine yeast Saccharomyces cerevisiae: a mutated HXT3 allele enhances fructose fermentation. Appl Environ Microbiol. 2007;73:2432–2439. doi: 10.1128/AEM.02269-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunge N. Nakatomi Y. Genetic mechanisms of rare matings of the yeast Saccharomyces cerevisiae heterozygous for mating type. Genetics. 1972;70:41–58. doi: 10.1093/genetics/70.1.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunge N. Sakaguchi K. Intergeneric transfer of deoxyribonucleic acid killer plasmids, pGKl1 and pGKl2, from Kluyveromyces lactis into Saccharomyces cerevisiae by cell fusion. J Bacteriol. 1981;147:155–160. doi: 10.1128/jb.147.1.155-160.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y, Song H, Wang Z. Ding Y. Expression of POX2 gene and disruption of POX3 genes in the industrial Yarrowia lipolytica on the gamma-decalactone production. Microbiol Res. 2012;167:246–252. doi: 10.1016/j.micres.2011.10.003. [DOI] [PubMed] [Google Scholar]
- Hall C, Brachat S. Dietrich FS. Contribution of horizontal gene transfer to the evolution of Saccharomyces cerevisiae. Eukaryot Cell. 2005;4:1102–1115. doi: 10.1128/EC.4.6.1102-1115.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamer L, DeZwaan TM, Montenegro-Chamorro MV, Frank SA. Hamer JE. Recent advances in large-scale transposon mutagenesis. Curr Opin Chem Biol. 2001;5:67–73. doi: 10.1016/s1367-5931(00)00162-9. [DOI] [PubMed] [Google Scholar]
- Hamilton SR, Davidson RC, Sethuraman N, et al. Humanization of yeast to produce complex terminally sialylated glycoproteins. Science. 2006;313:1441–1443. doi: 10.1126/science.1130256. [DOI] [PubMed] [Google Scholar]
- Hammond JRM. Genetically-modified brewing yeasts for the 21st century. Progress to date. Yeast. 1995;11:1613–1627. doi: 10.1002/yea.320111606. [DOI] [PubMed] [Google Scholar]
- Hammond JRM. Yeast genetics. In: Priest FG, Campbell I, editors. Brewing Microbiology. New York, NY: Springer; 1996. pp. 43–82. [Google Scholar]
- Hammond JRM. Yeast Genetics. In: Priest FG, Campbell I, editors. Brewing Microbiology. New York, NY: Kluwer Academic/Plenum Publishers; 2003. pp. 67–112. &, eds), Vol. 3 ( [Google Scholar]
- Hammond JRM. Eckersley KW. Fermentation properties of brewing yeast with killer character. J Inst Brew. 1984;90:167–177. [Google Scholar]
- Hansen EH, Moller BL, Kock GR, Buenner CM, Kristensen C, Jensen OR, Okkels FT, Olsen CE, Motawia MS. Hansen J. De novo biosynthesis of vanillin in fission yeast (Schizosaccharomyces pombe) and baker's yeast (Saccharomyces cerevisiae. Appl Environ Microbiol. 2009;75:2765–2774. doi: 10.1128/AEM.02681-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hara S, Iimmura Y, Oyama H, Kozeki T, Kitano K. Otsuka K. The breeding of cryophilic killer wine yeasts. Agric Biol Chem. 1981;45:1327–1334. [Google Scholar]
- Hashimoto S, Ogura M, Aritomi K, Hoshida H, Nishizawa Y. Akada R. Isolation of auxotrophic mutants of diploid industrial yeast strains after UV mutagenesis. Appl Environ Microbiol. 2005;71:312–319. doi: 10.1128/AEM.71.1.312-319.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heluane H, Spencer JF, Spencer DM, De Figueroa L. Callieri DAS. Characterization of hybrids obtained by protoplast fusion, between Pachysolen tannophilus and Saccharomyces cerevisiae. Appl Microbiol Biotechnol. 1993;40:98–100. [Google Scholar]
- Higgins VJ, Bell PJ, Dawes IW. Attfield PV. Generation of a novel Saccharomyces cerevisiae strain that exhibits strong maltose utilization and hyperosmotic resistance using nonrecombinant techniques. Appl Environ Microbiol. 2001;67:4346–4348. doi: 10.1128/AEM.67.9.4346-4348.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgins VJ, Braidwood M, Bell P, Bissinger P, Dawes IW. Attfield PV. Genetic evidence that high noninduced maltase and maltose permease activities, governed by MALx3-encoded transcriptional regulators, determine efficiency of gas production by baker's yeast in unsugared dough. Appl Environ Microbiol. 1999;65:680–685. doi: 10.1128/aem.65.2.680-685.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinnen A, Hicks JB. Fink GR. Transformation of yeast. P Natl Acad Sci USA. 1978;75:1929–1933. doi: 10.1073/pnas.75.4.1929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirata D, Aoki S, Watanabe K, Tsukioka M. Suzuki T. Stable overproduction of isoamyl alcohol by Saccharomyces cerevisiae with chromosome-integrated multicopy LEU4 genes. Biosci Biotechnol Biochem. 1992;56:1682–1683. [Google Scholar]
- Hockney RC. Freeman RF. Gratuitous catabolite repression by glucosamine of maltose utilization in Saccharomyces cerevisiae. J Gen Microbiol. 1980;121:479–482. [Google Scholar]
- Hohmann S. Osmotic stress signaling and osmoadaptation in yeasts. Microbiol Mol Biol Rev. 2002;66:300–372. doi: 10.1128/MMBR.66.2.300-372.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holic R, Yazawa H, Kumagai H. Uemura H. Engineered high content of ricinoleic acid in fission yeast Schizosaccharomyces pombe. Appl Microbiol Biotechnol. 2012;95:179–187. doi: 10.1007/s00253-012-3959-6. [DOI] [PubMed] [Google Scholar]
- Hou J, Tyo KE, Liu Z, Petranovic D. Nielsen J. Metabolic engineering of recombinant protein secretion by Saccharomyces cerevisiae. FEMS Yeast Res. 2012;12:491–510. doi: 10.1111/j.1567-1364.2012.00810.x. [DOI] [PubMed] [Google Scholar]
- Hou L. Novel methods of genome shuffling in Saccharomyces cerevisiae. Biotechnol Lett. 2009;31:671–677. doi: 10.1007/s10529-009-9916-5. [DOI] [PubMed] [Google Scholar]
- Hou L. Improved production of ethanol by novel genome shuffling in Saccharomyces cerevisiae. Appl Biochem Biotechnol. 2010;160:1084–1093. doi: 10.1007/s12010-009-8552-9. [DOI] [PubMed] [Google Scholar]
- Hubmann G, Guillouet S. Nevoigt E. Gpd1 and Gpd2 fine-tuning for sustainable reduction of glycerol formation in Saccharomyces cerevisiae. Appl Environ Microbiol. 2011;77:5857–5867. doi: 10.1128/AEM.05338-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes SR, Gibbons WR, Bang SS, et al. Random UV-C mutagenesis of Scheffersomyces (formerly Pichiastipitis NRRL Y-7124 to improve anaerobic growth on lignocellulosic sugars. J Ind Microbiol Biotechnol. 2011;39:163–173. doi: 10.1007/s10295-011-1012-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Husnik JI, Volschenk H, Bauer J, Colavizza D, Luo Z. van Vuuren HJ. Metabolic engineering of malolactic wine yeast. Metab Eng. 2006;8:315–323. doi: 10.1016/j.ymben.2006.02.003. [DOI] [PubMed] [Google Scholar]
- Huuskonen A, Markkula T, Vidgren V, Lima L, Mulder L, Geurts W, Walsh M. Londesborough J. Selection from industrial lager yeast strains of variants with improved fermentation performance in very-high-gravity worts. Appl Environ Microbiol. 2010;76:1563–1573. doi: 10.1128/AEM.03153-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ichikawa E, Kawato S, Abe Y, Imayasu S, Yoshida K, Inahashi M. Akiyama Y. 2002. & High-flavor component productive yeast. Vol. JP2002253211 A, IPC A21D000802; A21D001300; C12G000302; C12N000116; C12N000118; C12N000116; C12R0001865; C12G000302; C12R0001865; C12N000118; C12R0001865 (Japan BSo)
- Ikeda S, Nikaido K, Araki K, Yoshitake A, Kumagai H. Isoai A. Production of recombinant human lysosomal acid lipase in Schizosaccharomyces pombe: development of a fed-batch fermentation and purification process. J Biosci Bioeng. 2004;98:366–373. doi: 10.1016/S1389-1723(04)00297-X. [DOI] [PubMed] [Google Scholar]
- Ilmen M, Koivuranta K, Ruohonen L, Suominen P. Penttila M. Efficient production of L-lactic acid from xylose by Pichia stipitis. Appl Environ Microbiol. 2007;73:117–123. doi: 10.1128/AEM.01311-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishida-Fujii K, Goto S, Sugiyama H, Takagi Y, Saiki T. Takagi M. Breeding of flocculent industrial alcohol yeast strains by self-cloning of the flocculation gene FLO1 and repeated-batch fermentation by transformants. J Gen Appl Microbiol. 1998;44:347–353. doi: 10.2323/jgam.44.347. [DOI] [PubMed] [Google Scholar]
- Itakura K, Hirose T, Crea R, Riggs AD, Heyneker HL, Bolivar F. Boyer HW. Expression in Escherichia coli of a chemically synthesized gene for the hormone somatostatin. Science. 1977;198:1056–1063. doi: 10.1126/science.412251. [DOI] [PubMed] [Google Scholar]
- Ito H, Fukuda Y, Murata K. Kimura A. Transformation of intact yeast cells treated with alkali cations. J Bacteriol. 1983;153:163–168. doi: 10.1128/jb.153.1.163-168.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang M, Cai L, Udeani GO, et al. Cancer chemopreventive activity of resveratrol, a natural product derived from grapes. Science. 1997;275:218–220. doi: 10.1126/science.275.5297.218. [DOI] [PubMed] [Google Scholar]
- Javadekar VS, SivaRaman H. Gokhale DV. Industrial yeast strain improvement: construction of a highly flocculent yeast with a killer character by protoplast fusion. J Ind Microbiol. 1995;15:94–102. doi: 10.1007/BF01569806. [DOI] [PubMed] [Google Scholar]
- Jeffries TW, Grigoriev IV, Grimwood J, et al. Genome sequence of the lignocellulose-bioconverting and xylose-fermenting yeast Pichia stipitis. Nat Biotechnol. 2007;25:319–326. doi: 10.1038/nbt1290. [DOI] [PubMed] [Google Scholar]
- Jensen PR. Hammer K. Artificial promoters for metabolic optimization. Biotechnol Bioeng. 1998;58:191–195. [PubMed] [Google Scholar]
- Jeppsson M, Johansson B, Jensen PR, Hahn-Hagerdal B. Gorwa-Grauslund MF. The level of glucose-6-phosphate dehydrogenase activity strongly influences xylose fermentation and inhibitor sensitivity in recombinant Saccharomyces cerevisiae strains. Yeast. 2003;20:1263–1272. doi: 10.1002/yea.1043. [DOI] [PubMed] [Google Scholar]
- Jin YS, Ni H, Laplaza JM. Jeffries TW. Optimal growth and ethanol production from xylose by recombinant Saccharomyces cerevisiae require moderate D-xylulokinase activity. Appl Environ Microbiol. 2003;69:495–503. doi: 10.1128/AEM.69.1.495-503.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jingping G, Hongbing S, Gang S, Hongzhi L. Wenxiang P. A genome shuffling-generated Saccharomyces cerevisiae isolate that ferments xylose and glucose to produce high levels of ethanol. J Ind Microbiol Biotechnol. 2012;39:777–787. doi: 10.1007/s10295-011-1076-7. [DOI] [PubMed] [Google Scholar]
- Johnson EA. Biotechnology of non-Saccharomyces yeasts-the ascomycetes. Appl Microbiol Biotechnol. 2013;97:503–517. doi: 10.1007/s00253-012-4497-y. [DOI] [PubMed] [Google Scholar]
- Jones RM, Russell I. Stewart GG. The use of catabolite derepression as a means of improving the fermentation rate of brewing yeast strains. J Am Soc Brew Chem. 1986;44:161. [Google Scholar]
- Jorde LB. Wooding SP. Genetic variation, classification and ‘race'. Nat Genet. 2004;36:S28–S33. doi: 10.1038/ng1435. [DOI] [PubMed] [Google Scholar]
- Kaino T, Tateiwa T, Mizukami-Murata S, Shima J. Takagi H. Self-cloning baker's yeasts that accumulate proline enhance freeze tolerance in doughs. Appl Environ Microbiol. 2008;74:5845–5849. doi: 10.1128/AEM.00998-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalscheuer R, Luftmann H. Steinbuchel A. Synthesis of novel lipids in Saccharomyces cerevisiae by heterologous expression of an unspecific bacterial acyltransferase. Appl Environ Microbiol. 2004;70:7119–7125. doi: 10.1128/AEM.70.12.7119-7125.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katz Ezov T, Chang S-L, Frenkel Z, Segre AV, Bahalul M, Murray AW, Leu J, Korol A. Kashi Y. Heterothallism in Saccharomyces cerevisiae isolates from nature: effect of HO locus on the mode of reproduction. Mol Ecol. 2010;19:121–131. doi: 10.1111/j.1365-294X.2009.04436.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kavanagh K. Whittaker PA. Application of protoplast fusion to the nonconventional yeast. Enzyme Microb Technol. 1996;18:45–51. [Google Scholar]
- Kielland-Brandt M, Nilsson-Tillgren T, Gjermansen C, Holmberg S. Pedersen M. Genetics of brewing yeasts. In: Rose A, Wheals A, Harrison J, editors. The Yeasts. 2nd edn. London: Academic Press; 1995. pp. 223–254. &, eds) &, Vol. 6 ( [Google Scholar]
- Kikuchi M, Ohnishi K. Harayama S. Novel family shuffling methods for the in vitro evolution of enzymes. Gene. 1999;236:159–167. doi: 10.1016/s0378-1119(99)00240-1. [DOI] [PubMed] [Google Scholar]
- Kim HS, Kim NR, Yang J. Choi W. Identification of novel genes responsible for ethanol and/or thermotolerance by transposon mutagenesis in Saccharomyces cerevisiae. Appl Microbiol Biotechnol. 2011;91:1159–1172. doi: 10.1007/s00253-011-3298-z. [DOI] [PubMed] [Google Scholar]
- Kim J, Alizadeh P, Harding T, Hefner-Gravink A. Klionsky DJ. Disruption of the yeast ATH1 gene confers better survival after dehydration, freezing, and ethanol shock: potential commercial applications. Appl Environ Microbiol. 1996;62:1563–1569. doi: 10.1128/aem.62.5.1563-1569.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim TG. Kim K. The construction of a stable starch-fermenting yeast strain using genetic engineering and rare-mating. Appl Biochem Biotechnol. 1996;59:39–51. doi: 10.1007/BF02787856. [DOI] [PubMed] [Google Scholar]
- Kishida M, Muguruma T, Sakanaka K, Katsuragi T. Sakai T. Chromosomal deletion or rearrangement in chimeric hybrids of Saccharomycopsis fibuligera and Saccharomyces diastaticus obtained by cell fusion. J Ferment Bioeng. 1996;81:281–285. [Google Scholar]
- Kishimoto M. Fermentation characteristics of hybrids between the cryophilic wine yeast Saccharomyces bayanus and the mesophilic wine yeast Saccharomyces cerevisiae. J Ferment Bioeng. 1994;77:432–435. [Google Scholar]
- Kitagaki H. Kitamoto K. Breeding research on sake yeasts in Japan: history, recent technological advances, and future perspectives. Annu Rev Food Sci Technol. 2013;4:215–235. doi: 10.1146/annurev-food-030212-182545. [DOI] [PubMed] [Google Scholar]
- Kitamoto K, Oda K, Gomi K. Takahashi K. Genetic engineering of a sake yeast producing no urea by successive disruption of arginase gene. Appl Environ Microbiol. 1991;57:301–306. doi: 10.1128/aem.57.1.301-306.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein CJ, Olsson L, Ronnow B, Mikkelsen JD. Nielsen J. Alleviation of glucose repression of maltose metabolism by MIG1 disruption in Saccharomyces cerevisiae. Appl Environ Microbiol. 1996;62:4441–4449. doi: 10.1128/aem.62.12.4441-4449.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong QX, Zhang AL, Cao LM. Chen X. Over-expressing GLT1 in a gpd2Delta mutant of Saccharomyces cerevisiae to improve ethanol production. Appl Microbiol Biotechnol. 2007;75:1361–1366. doi: 10.1007/s00253-007-0948-2. [DOI] [PubMed] [Google Scholar]
- Kong QX, Gu JG, Cao LM, Zhang AL, Chen X. Zhao XM. Improved production of ethanol by deleting FPS1 and over-expressing GLT1 in Saccharomyces cerevisiae. Biotechnol Lett. 2006;28:2033–2038. doi: 10.1007/s10529-006-9185-5. [DOI] [PubMed] [Google Scholar]
- Krishnamoorthy RN, Vijila K. Kumutha K. Intergeneric protoplast fusion of yeast for high ethanol production from cheese industry waste – Whey. J Yeast Fungal Res. 2010;1:81–87. [Google Scholar]
- Kumari R. Pramanik K. Improvement of multiple stress tolerance in yeast strain by sequential mutagenesis for enhanced bioethanol production. J Biosci Bioeng. 2012;114:622–629. doi: 10.1016/j.jbiosc.2012.07.007. [DOI] [PubMed] [Google Scholar]
- Kunicka-Styczynska A. Rajkowska K. Physiological and genetic stability of hybrids of industrial wine yeasts Saccharomyces sensu stricto complex. J Appl Microbiol. 2011;110:1538–1549. doi: 10.1111/j.1365-2672.2011.05009.x. [DOI] [PubMed] [Google Scholar]
- Kutyna DR, Varela C, Henschke PA, Chambers PJ. Stanley GA. Microbiological approaches to lowering ethanol concentration in wine. Trends Food Sci Technol. 2010;21:293–302. [Google Scholar]
- Kutyna DR, Varela C, Stanley GA, Borneman AR, Henschke PA. Chambers PJ. Adaptive evolution of Saccharomyces cerevisiae to generate strains with enhanced glycerol production. Appl Microbiol Biotechnol. 2012;93:1175–1184. doi: 10.1007/s00253-011-3622-7. [DOI] [PubMed] [Google Scholar]
- Kuyper M, Harhangi HR, Stave AK, Winkler AA, Jetten MSM, de Laat WTAM, den Ridder JJJ, Op den Camp HJM, van Dijken JP. Pronk JT. High-level functional expression of a fungal xylose isomerase: the key to efficient ethanolic fermentation of xylose by Saccharomyces cerevisiae. FEMS Yeast Res. 2003;4:69–78. doi: 10.1016/S1567-1356(03)00141-7. [DOI] [PubMed] [Google Scholar]
- Lang GI. Murray AW. Estimating the per-base-pair mutation rate in the yeast Saccharomyces cerevisiae. Genetics. 2008;178:67–82. doi: 10.1534/genetics.107.071506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laplaza JM, Torres BR, Jin YS. Jeffries TW. Sh ble and Cre adapted for functional genomics and metabolic engineering of Pichia stipitis. Enzyme Microb Technol. 2006;38:741–747. [Google Scholar]
- Larsson S, Nilvebrant NO. Jonsson LJ. Effect of overexpression of Saccharomyces cerevisiae Pad1p on the resistance to phenylacrylic acids and lignocellulose hydrolysates under aerobic and oxygen-limited conditions. Appl Microbiol Biotechnol. 2001;57:167–174. doi: 10.1007/s002530100742. [DOI] [PubMed] [Google Scholar]
- Lee FW. Da Silva NA. Improved efficiency and stability of multiple cloned gene insertions at the delta sequences of Saccharomyces cerevisiae. Appl Microbiol Biotechnol. 1997;48:339–345. doi: 10.1007/s002530051059. [DOI] [PubMed] [Google Scholar]
- Lee S-M, Jellison T. Alper HS. Directed evolution of xylose isomerase for improved xylose catabolism and fermentation in the yeast Saccharomyces cerevisiae. Appl Environ Microbiol. 2012;78:5708–5716. doi: 10.1128/AEM.01419-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legras JL, Merdinoglu D, Cornuet JM. Karst F. Bread, beer and wine: Saccharomyces cerevisiae diversity reflects human history. Mol Ecol. 2007;16:2091–2102. doi: 10.1111/j.1365-294X.2007.03266.x. [DOI] [PubMed] [Google Scholar]
- Levskaya A, Chevalier AA, Tabor JJ, et al. Synthetic biology: engineering Escherichia coli to see light. Nature. 2005;438:441–442. doi: 10.1038/nature04405. [DOI] [PubMed] [Google Scholar]
- Li A, Liu Z, Li Q, Yu L, Wang D. Deng X. Construction and characterization of bidirectional expression vectors in Saccharomyces cerevisiae. FEMS Yeast Res. 2008;8:6–9. doi: 10.1111/j.1567-1364.2007.00335.x. [DOI] [PubMed] [Google Scholar]
- Libkind D, Hittinger CT, Valerio E, Goncalves C, Dover J, Johnston M, Goncalves P. Sampaio JP. Microbe domestication and the identification of the wild genetic stock of lager-brewing yeast. P Natl Acad Sci USA. 2011;108:14539–14544. doi: 10.1073/pnas.1105430108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Licker JL, Acree TE. Henick-Kling T. What is “Brett” (Brettanomyces) Flavor? In: Waterhouse AL, Ebeler SE, editors. Chemistry of Wine Flavour. Washington, DC: American Chemical Society; 1999. pp. 96–115. [Google Scholar]
- Lilly M, Lambrechts MG. Pretorius IS. Effect of increased yeast alcohol acetyltransferase activity on flavor profiles of wine and distillates. Appl Environ Microbiol. 2000;66:744–753. doi: 10.1128/aem.66.2.744-753.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lilly M, Bauer FF, Lambrechts MG, Swiegers JH, Cozzolino D. Pretorius IS. The effect of increased yeast alcohol acetyltransferase and esterase activity on the flavour profiles of wine and distillates. Yeast. 2006;23:641–659. doi: 10.1002/yea.1382. [DOI] [PubMed] [Google Scholar]
- Lin G, Bultman J, Johnson EA. Fell JW. Genetic manipulation of Xanthophyllomyces dendrorhous and Phaffia rhodozyma. Methods Mol Biol. 2012;898:235–249. doi: 10.1007/978-1-61779-918-1_16. [DOI] [PubMed] [Google Scholar]
- Lindegren CC. Chromosome maps of Saccharomyces. Hereditas. 1949;35:338–355. [Google Scholar]
- Lindegren CC. Lindegren G. A new method for hybridizing yeast. P Natl Acad Sci USA. 1943;29:306–308. doi: 10.1073/pnas.29.10.306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lippman ZB. Zamir D. Heterosis: revisiting the magic. Trends Genet. 2007;23:60–66. doi: 10.1016/j.tig.2006.12.006. [DOI] [PubMed] [Google Scholar]
- Liti G. Louis EJ. Advances in quantitative trait analysis in yeast. PLoS Genet. 2012;8:e1002912. doi: 10.1371/journal.pgen.1002912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liti G, Carter DM, Moses AM, et al. Population genomics of domestic and wild yeasts. Nature. 2009;458:337–341. doi: 10.1038/nature07743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Yan M, Lai C, Xu L. Ouyang P. gTME for improved xylose fermentation of Saccharomyces cerevisiae. Appl Biochem Biotechnol. 2008;160:574–582. doi: 10.1007/s12010-008-8431-9. [DOI] [PubMed] [Google Scholar]
- Liu J, Ding W, Zhang G. Wang J. Improving ethanol fermentation performance of Saccharomyces cerevisiae in very high-gravity fermentation through chemical mutagenesis and meiotic recombination. Appl Microbiol Biotechnol. 2011;91:1239–1246. doi: 10.1007/s00253-011-3404-2. [DOI] [PubMed] [Google Scholar]
- Liu L, Redden H. Alper HS. Frontiers of yeast metabolic engineering: diversifying beyond ethanol and Saccharomyces. Curr Opin Biotechnol. 2013;24:1023–1030. doi: 10.1016/j.copbio.2013.03.005. [DOI] [PubMed] [Google Scholar]
- Logez C, Alkhalfioui F, Byrne B. Wagner R. Preparation of Pichia pastoris expression plasmids. Methods Mol Biol. 2012;866:25–40. doi: 10.1007/978-1-61779-770-5_3. [DOI] [PubMed] [Google Scholar]
- Lopes CA, Rodriguez ME, Sangorrin M, Querol A. Caballero AC. Patagonian wines: implantation of an indigenous strain of Saccharomyces cerevisiae in fermentations conducted in traditional and modern cellars. J Ind Microbiol Biotechnol. 2007;34:139–149. doi: 10.1007/s10295-006-0178-0. [DOI] [PubMed] [Google Scholar]
- Lopes TS, Klootwijk J, Veenstra AE, van der Aar PC, van Heerikhuizen H, Raue HA. Planta RJ. High-copy-number integration into the ribosomal DNA of Saccharomyces cerevisiae: a new vector for high-level expression. Gene. 1989;79:199–206. doi: 10.1016/0378-1119(89)90202-3. [DOI] [PubMed] [Google Scholar]
- Loray MA, Spencer JF, Spencer DM. De Figueroa L. Hybrids obtained by protoplast fusion with a salt-tolerant yeast. J Ind Microbiol. 1995;14:508–513. doi: 10.1007/BF01573966. [DOI] [PubMed] [Google Scholar]
- Lu C. Jeffries T. Shuffling of promoters for multiple genes to optimize xylose fermentation in an engineered Saccharomyces cerevisiae strain. Appl Environ Microbiol. 2007;73:6072–6077. doi: 10.1128/AEM.00955-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Y, Cheng Y-F, He X-P, Guo X-N. Zhang B-R. Improvement of robustness and ethanol production of ethanologenic Saccharomyces cerevisiae under co-stress of heat and inhibitors. J Ind Microbiol. 2012;39:73–80. doi: 10.1007/s10295-011-1001-0. [DOI] [PubMed] [Google Scholar]
- Lucca ME, Spencer JF. de Figueroa LI. Glycerol and arabitol production by an intergeneric hybrid, PB2, obtained by protoplast fusion between Saccharomyces cerevisiae and Torulaspora delbrueckii. Appl Microbiol Biotechnol. 2002;59:472–476. doi: 10.1007/s00253-002-1025-5. [DOI] [PubMed] [Google Scholar]
- Lucca ME, Loray MA, de Figueroa LIC. Callieri DA. Characterisation of osmotolerant hybrids obtained by fusion between protoplasts of Saccharomyces cerevisiae and heat treated protoplasts of Torulaspora delbrueckii. Biotechnol Lett. 1999;21:343–348. [Google Scholar]
- Luhe AL, Tan L, Wu J. Zhao H. Increase of ethanol tolerance of Saccharomyces cerevisiae by error-prone whole genome amplification. Biotechnol Lett. 2011;33:1007–1011. doi: 10.1007/s10529-011-0518-7. [DOI] [PubMed] [Google Scholar]
- Ma K, Wakisaka M, Sakai K. Shirai Y. Flocculation characteristics of an isolated mutant flocculent Saccharomyces cerevisiae strain and its application for fuel ethanol production from kitchen refuse. Bioresour Technol. 2009;100:2289–2292. doi: 10.1016/j.biortech.2008.11.010. [DOI] [PubMed] [Google Scholar]
- Macauley-Patrick S, Fazenda ML, McNeil B. Harvey LM. Heterologous protein production using the Pichia pastoris expression system. Yeast. 2005;22:249–270. doi: 10.1002/yea.1208. [DOI] [PubMed] [Google Scholar]
- Maclean CJ. Greig D. Prezygotic reproductive isolation between Saccharomyces cerevisiae and Saccharomyces paradoxus. BMC Evol Biol. 2008;8:1. doi: 10.1186/1471-2148-8-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madhavan A, Tamalampudi S, Srivastava A, Fukuda H, Bisaria V. Kondo A. Alcoholic fermentation of xylose and mixed sugars using recombinant Saccharomyces cerevisiae engineered for xylose utilization. Appl Microbiol Biotechnol. 2009;82:1037–1047. doi: 10.1007/s00253-008-1818-2. [DOI] [PubMed] [Google Scholar]
- Madzak C, Gaillardin C. Beckerich JM. Heterologous protein expression and secretion in the non-conventional yeast Yarrowia lipolytica: a review. J Biotechnol. 2004;109:63–81. doi: 10.1016/j.jbiotec.2003.10.027. [DOI] [PubMed] [Google Scholar]
- Manivasakam P, Weber SC, McElver J. Schiestl RH. Micro-homology mediated PCR targeting in Saccharomyces cerevisiae. Nucleic Acids Res. 1995;23:2799–2800. doi: 10.1093/nar/23.14.2799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mannazzu I, Clementi F. Ciani M. Strategies and criteria for the isolation and selection of autochthonous starters. In: Ciani M, editor. Biodiversity and Biotechnology of Wine Yeasts. Trivandrum, India: Research Signpost; 2002. pp. 19–34. [Google Scholar]
- Marin D, Jimenez A. Fernandez Lobato M. Construction of an efficient amylolytic industrial yeast strain containing DNA exclusively derived from yeast. FEMS Microbiol Lett. 2001;201:249–253. doi: 10.1016/s0378-1097(01)00259-2. [DOI] [PubMed] [Google Scholar]
- van Maris AJA, Winkler AA, Kuyper M. Development of efficient xylose fermentation in Saccharomyces cerevisiae: xylose isomerase as a key component. In: Olsson L, et al., editors. Biofuels. Heidelberg, Germany: Springer; 2007. pp. 179–204. [DOI] [PubMed] [Google Scholar]
- Marullo P, Bely M, Masneuf-Pomarede I, Pons M, Aigle M. Dubourdieu D. Breeding strategies for combining fermentative qualities and reducing off-flavor production in a wine yeast model. FEMS Yeast Res. 2006;6:268–279. doi: 10.1111/j.1567-1364.2006.00034.x. [DOI] [PubMed] [Google Scholar]
- Marullo P, Mansour C, Dufour M, Albertin W, Sicard D, Bely M. Dubourdieu D. Genetic improvement of thermo-tolerance in wine Saccharomyces cerevisiae strains by a backcross approach. FEMS Yeast Res. 2009;9:1148–1160. doi: 10.1111/j.1567-1364.2009.00550.x. [DOI] [PubMed] [Google Scholar]
- Matsuoka M, Uchida K. Aiba S. Cytoplasmic transfer of oligomycin resistance during protoplast fusion of Saccharomycopsis lipolytica. J Bacteriol. 1982;152:530–533. doi: 10.1128/jb.152.1.530-533.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsushika A, Inoue H, Kodaki T. Sawayama S. Ethanol production from xylose in engineered Saccharomyces cerevisiae strains: current state and perspectives. Appl Microbiol Biotechnol. 2009;84:37–53. doi: 10.1007/s00253-009-2101-x. [DOI] [PubMed] [Google Scholar]
- Matthäus F, Ketelhot M, Gatter M. Barth G. Production of lycopene in the non-carotenoid producing yeast Yarrowia lipolytica. Applied and Environmental Microbiology. 2013;80:1660–1669. doi: 10.1128/AEM.03167-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miklenic M, Stafa A, Bajic A, Zunar B, Lisnic B. Svetec IK. Genetic transformation of the yeast DekkeraBrettanomyces bruxellensis with non-homologous DNA. J Microbiol Biotechnol. 2013;23:674–680. doi: 10.4014/jmb.1211.11047. [DOI] [PubMed] [Google Scholar]
- Miller CA, 3rd, Martinat MA. Hyman LE. Assessment of aryl hydrocarbon receptor complex interactions using pBEVY plasmids: expression vectors with bi-directional promoters for use in Saccharomyces cerevisiae. Nucleic Acids Res. 1998;26:3577–3583. doi: 10.1093/nar/26.15.3577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mobini-Dehkordi M, Nahvi I, Zarkesh-Esfahani H, Ghaedi K, Tavassoli M. Akada R. Isolation of a novel mutant strain of Saccharomyces cerevisiae by an ethyl methane sulfonate-induced mutagenesis approach as a high producer of bioethanol. J Biosci Bioeng. 2008;105:403–408. doi: 10.1263/jbb.105.403. [DOI] [PubMed] [Google Scholar]
- Moerschell RP, Tsunasawa S. Sherman F. Transformation of yeast with synthetic oligonucleotides. P Natl Acad Sci USA. 1988;85:524–528. doi: 10.1073/pnas.85.2.524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molzahn SW. A new approach to the application of genetics to brewing yeast. J Am Soc Brew Chem. 1977;35:54. [Google Scholar]
- Morales L. Dujon B. Evolutionary role of interspecies hybridization and genetic exchanges in yeasts. Microbiol Mol Biol Rev. 2012;76:721–739. doi: 10.1128/MMBR.00022-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan AJ. Yeast strain improvement by protoplast fusion and transformation. Experientia Suppl. 1983;46:155–166. doi: 10.1007/978-3-0348-6776-4_20. [DOI] [PubMed] [Google Scholar]
- Mortimer RK. Evolution and variation of the yeast (Saccharomyces) genome. Genome Res. 2000;10:403–409. doi: 10.1101/gr.10.4.403. [DOI] [PubMed] [Google Scholar]
- Mortimer RK. Johnston JR. Genealogy of principal strains of the yeast genetic stock center. Genetics. 1986;113:35–43. doi: 10.1093/genetics/113.1.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortimer RK, Romano P, Suzzi G. Polsinelli M. Genome renewal: a new phenomenon revealed from a genetic study of 43 strains of Saccharomyces cerevisiae derived from natural fermentation of grape musts. Yeast. 1994;10:1543–1552. doi: 10.1002/yea.320101203. [DOI] [PubMed] [Google Scholar]
- Murphy HA. Zeyl CW. Prezygotic isolation between Saccharomyces cerevisiae and Saccharomyces paradoxus through differences in mating speed and germination timing. Evolution. 2012;66:1196–1209. doi: 10.1111/j.1558-5646.2011.01516.x. [DOI] [PubMed] [Google Scholar]
- Murray AW. Szostak JW. Construction of artificial chromosomes in yeast. Nature. 1983;305:189–193. doi: 10.1038/305189a0. [DOI] [PubMed] [Google Scholar]
- Nair NU. Zhao H. Improving protein functions by directed evolution. In: Smolke C, editor. The Metabolic Pathway Engineering Handbook: Tools and Applications. Boca Raton, FL: Taylor & Francis Group; 2010. pp. 2.1–2.35. , Vol. 1 (, ed) &. [Google Scholar]
- Nakagawa S. Ouchi K. Construction from a single parent of baker's yeast strains with high freeze tolerance and fermentative activity in both lean and sweet doughs. Appl Environ Microbiol. 1994;60:3499–3502. doi: 10.1128/aem.60.10.3499-3502.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakazawa N, Okawa K, Sato T, Enei H. Harashima S. Mass mating method in combination with G418- and aureobasidin A-resistance markers for efficient selection of hybrids from homothallic strains in Saccharomyces cerevisiae. J Biosci Bioeng. 1999;88:468–471. doi: 10.1016/s1389-1723(00)87660-4. [DOI] [PubMed] [Google Scholar]
- Ness JE, Kim S, Gottman A, Pak R, Krebber A, Borchert TV, Govindarajan S, Mundorff EC. Minshull J. Synthetic shuffling expands functional protein diversity by allowing amino acids to recombine independently. Nat Biotechnol. 2002;20:1251–1255. doi: 10.1038/nbt754. [DOI] [PubMed] [Google Scholar]
- Nevoigt E. Progress in metabolic engineering of Saccharomyces cerevisiae. Microbiol Mol Biol Rev. 2008;72:379–412. doi: 10.1128/MMBR.00025-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nevoigt E, Kohnke J, Fischer CR, Alper H, Stahl U. Stephanopoulos G. Engineering of promoter replacement cassettes for fine-tuning of gene expression in Saccharomyces cerevisiae. Appl Environ Microbiol. 2006;72:5266–5273. doi: 10.1128/AEM.00530-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni H, Laplaza JM. Jeffries TW. Transposon mutagenesis to improve the growth of recombinant Saccharomyces cerevisiae on D-xylose. Appl Environ Microbiol. 2007;73:2061–2066. doi: 10.1128/AEM.02564-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicaud JM. Yarrowia lipolytica. Yeast. 2012;29:409–418. doi: 10.1002/yea.2921. [DOI] [PubMed] [Google Scholar]
- Nilsson-Tillgren T, Petersen J, Holmberg S. Kielland-Brandt M. Transfer of chromosome III during kar mediated cytoduction in yeast. Carlsberg Res Commun. 1980;45:113–117. [Google Scholar]
- Nilsson-Tillgren T, Gjermansen C, Kielland-Brandt MC, Petersen JGL. Holmberg S. Genetic differences between Saccharomyces carlbergensis and S. cerevisiae: analysis of chromosome III by single chromosome transfer. Carlsberg Res Commun. 1981;46:65–76. [Google Scholar]
- Nissen TL, Kielland-Brandt MC, Nielsen J. Villadsen J. Optimization of ethanol production in Saccharomyces cerevisiae by metabolic engineering of the ammonium assimilation. Metab Eng. 2000a;2:69–77. doi: 10.1006/mben.1999.0140. [DOI] [PubMed] [Google Scholar]
- Nissen TL, Hamann CW, Kielland-Brandt MC, Nielsen J. Villadsen J. Anaerobic and aerobic batch cultivations of Saccharomyces cerevisiae mutants impaired in glycerol synthesis. Yeast. 2000b;16:463–474. doi: 10.1002/(SICI)1097-0061(20000330)16:5<463::AID-YEA535>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- Nissen TL, Anderlund M, Nielsen J, Villadsen J. Kielland-Brandt MC. Expression of a cytoplasmic transhydrogenase in Saccharomyces cerevisiae results in formation of 2-oxoglutarate due to depletion of the NADPH pool. Yeast. 2001;18:19–32. doi: 10.1002/1097-0061(200101)18:1<19::AID-YEA650>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- Nonklang S, Abdel-Banat BM, Cha-aim K, Moonjai N, Hoshida H, Limtong S, Yamada M. Akada R. High-temperature ethanol fermentation and transformation with linear DNA in the thermotolerant yeast Kluyveromyces marxianus DMKU3-1042. Appl Environ Microbiol. 2008;74:7514–7521. doi: 10.1128/AEM.01854-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novo M, Bigey F, Beyne E, et al. Eukaryote-to-eukaryote gene transfer events revealed by the genome sequence of the wine yeast Saccharomyces cerevisiae EC1118. P Natl Acad Sci USA. 2009;106:16333–16338. doi: 10.1073/pnas.0904673106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oda Y. Ouchi K. Hybridization of bakers' yeast by the rare-mating method to improve leavening ability in dough. Enzyme Microb Technol. 1990;12:989–993. [Google Scholar]
- Okazaki K, Okazaki N, Kume K, Jinno S, Tanaka K. Okayama H. High-frequency transformation method and library transducing vectors for cloning mammalian cDNAs by trans-complementation of Schizosaccharomyces pombe. Nucleic Acids Res. 1990;18:6485–6489. doi: 10.1093/nar/18.22.6485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omura F, Fujita A, Miyajima K. Fukui N. Engineering of yeast Put4 permease and its application to lager yeast for efficient proline assimilation. Biosci Biotechnol Biochem. 2005;69:1162–1171. doi: 10.1271/bbb.69.1162. [DOI] [PubMed] [Google Scholar]
- Ostergaard S, Olsson L. Nielsen J. Metabolic engineering of Saccharomyces cerevisiae. Microbiol Mol Biol Rev. 2000;64:34–50. doi: 10.1128/mmbr.64.1.34-50.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostermeier M, Shim JH. Benkovic SJ. A combinatorial approach to hybrid enzymes independent of DNA homology. Nat Biotechnol. 1999;17:1205–1209. doi: 10.1038/70754. [DOI] [PubMed] [Google Scholar]
- Ouchi K, Wickner RB, Toh-E A. Akiyama H. Breeding of killer yeasts for sake brewing by cytoduction. J Ferment Technol. 1979;57:483–487. [Google Scholar]
- Oud B, van Maris AJ, Daran JM. Pronk JT. Genome-wide analytical approaches for reverse metabolic engineering of industrially relevant phenotypes in yeast. FEMS Yeast Res. 2012;12:183–196. doi: 10.1111/j.1567-1364.2011.00776.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oura E. Reaction products of yeast fermentations. Process Biochem. 1977;12:19–21. [Google Scholar]
- Pagliardini J, Hubmann G, Alfenore S, Nevoigt E, Bideaux C. Guillouet SE. The metabolic costs of improving ethanol yield by reducing glycerol formation capacity under anaerobic conditions in Saccharomyces cerevisiae. Microb Cell Fact. 2013;12:29. doi: 10.1186/1475-2859-12-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panadero J, Hernandez-Lopez MJ, Prieto JA. Randez-Gil F. Overexpression of the calcineurin target CRZ1 provides freeze tolerance and enhances the fermentative capacity of baker's yeast. Appl Environ Microbiol. 2007;73:4824–4831. doi: 10.1128/AEM.02651-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pang ZW, Liang JJ, Qin XJ, Wang JR, Feng JX. Huang RB. Multiple induced mutagenesis for improvement of ethanol production by Kluyveromyces marxianus. Biotechnol Lett. 2010;32:1847–1851. doi: 10.1007/s10529-010-0384-8. [DOI] [PubMed] [Google Scholar]
- Paquin C. Adams J. Frequency of fixation of adaptive mutations is higher in evolving diploid than haploid yeast populations. Nature. 1983a;302:495–500. doi: 10.1038/302495a0. [DOI] [PubMed] [Google Scholar]
- Paquin CE. Adams J. Relative fitness can decrease in evolving asexual populations of S. cerevisiae. Nature. 1983b;306:368–370. doi: 10.1038/306368a0. [DOI] [PubMed] [Google Scholar]
- Park KS, Lee DK, Lee H, Lee Y, Jang YS, Kim YH, Yang HY, Lee SI, Seol W. Kim JS. Phenotypic alteration of eukaryotic cells using randomized libraries of artificial transcription factors. Nat Biotechnol. 2003;21:1208–1214. doi: 10.1038/nbt868. [DOI] [PubMed] [Google Scholar]
- Passoth V, Blomqvist J. Schnurer J. Dekkera bruxellensis and Lactobacillus vini form a stable ethanol-producing consortium in a commercial alcohol production process. Appl Environ Microbiol. 2007;73:4354–4356. doi: 10.1128/AEM.00437-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peberdy JF. Protoplast fusion – a tool for genetic manipulation and breeding in industrial microorganisms. Enzyme Microb Technol. 1980;2:23–29. [Google Scholar]
- Pecota DC, Rajgarhia V. Da Silva NA. Sequential gene integration for the engineering of Kluyveromyces marxianus. J Biotechnol. 2007;127:408–416. doi: 10.1016/j.jbiotec.2006.07.031. [DOI] [PubMed] [Google Scholar]
- Pellegrini L, Cardinali G. Martini A. Selection of Saccharomyces cerevisiae strains able to ferment at supraoptimal temperatures. Annali di microbiologia ed enzimologia. 1999;49:55–66. [Google Scholar]
- Perez-Torrado R, Gimeno-Alcaniz JV. Matallana E. Wine yeast strains engineered for glycogen overproduction display enhanced viability under glucose deprivation conditions. Appl Environ Microbiol. 2002;68:3339–3344. doi: 10.1128/AEM.68.7.3339-3344.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Torrado R, Panadero J, Hernandez-Lopez MJ, Prieto JA. Randez-Gil F. Global expression studies in baker's yeast reveal target genes for the improvement of industrially-relevant traits: the cases of CAF16 and ORC2. Microb Cell Fact. 2010;9:56. doi: 10.1186/1475-2859-9-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Traves L, Lopes CA, Barrio E. Querol A. Evaluation of different genetic procedures for the generation of artificial hybrids in Saccharomyces genus for winemaking. Int J Food Microbiol. 2012;156:102–111. doi: 10.1016/j.ijfoodmicro.2012.03.008. [DOI] [PubMed] [Google Scholar]
- Perry C. Meaden P. Properties of a genetically-engineered dextrin-fermenting strain of brewers' yeast. J Inst Brew. 1988;94:64–67. [Google Scholar]
- Petersson A, Almeida JR, Modig T, Karhumaa K, Hahn-Hagerdal B, Gorwa-Grauslund MF. Liden G. A 5-hydroxymethyl furfural reducing enzyme encoded by the Saccharomyces cerevisiae ADH6 gene conveys HMF tolerance. Yeast. 2006;23:455–464. doi: 10.1002/yea.1370. [DOI] [PubMed] [Google Scholar]
- Pina A, Calderon IL. Benitez T. Intergeneric hybrids of Saccharomyces cerevisiae and Zygosaccharomyces fermentati obtained by protoplast fusion. Appl Environ Microbiol. 1986;51:995–1003. doi: 10.1128/aem.51.5.995-1003.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinel D, D'Aoust F, del Cardayre SB, Bajwa PK, Lee H. Martin VJ. Saccharomyces cerevisiae genome shuffling through recursive population mating leads to improved tolerance to spent sulfite liquor. Appl Environ Microbiol. 2011;77:4736–4743. doi: 10.1128/AEM.02769-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piskur J, Rozpedowska E, Polakova S, Merico A. Compagno C. How did Saccharomyces evolve to become a good brewer? Trends Genet. 2006;22:183–186. doi: 10.1016/j.tig.2006.02.002. [DOI] [PubMed] [Google Scholar]
- Porro D, Bianchi MM, Brambilla L, et al. Replacement of a metabolic pathway for large-scale production of lactic acid from engineered yeasts. Appl Environ Microbiol. 1999;65:4211–4215. doi: 10.1128/aem.65.9.4211-4215.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powell CD, Quain DE. Smart KA. The impact of sedimentation on cone yeast heterogeneity. J Am Soc Brew Chem. 2004;62:8–17. [Google Scholar]
- Pretorius IS. Tailoring wine yeast for the new millennium: novel approaches to the ancient art of winemaking. Yeast. 2000;16:675–729. doi: 10.1002/1097-0061(20000615)16:8<675::AID-YEA585>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- Pretorius IS. Bauer FF. Meeting the consumer challenge through genetically customized wine-yeast strains. Trends Biotechnol. 2002;20:426–432. doi: 10.1016/s0167-7799(02)02049-8. [DOI] [PubMed] [Google Scholar]
- Puligundla P, Smogrovicova D, Obulam V. Ko S. Very high gravity (VHG) ethanolic brewing and fermentation: a research update. J Ind Microbiol Biotechnol. 2011;38:1133–1144. doi: 10.1007/s10295-011-0999-3. [DOI] [PubMed] [Google Scholar]
- Putrament A. Baranowska H. Mutagenic action of hydroxylamine and methoxyamine on yeast. I. Hydroxylamine. Mol Gen Genet. 1973;122:61–72. doi: 10.1007/BF00337974. [DOI] [PubMed] [Google Scholar]
- Putrament A, Baranowska H, Ejchart A. Prazmo W. Manganese mutagenesis in yeast. Methods Cell Biol. 1978;20:25–34. doi: 10.1016/s0091-679x(08)62006-3. [DOI] [PubMed] [Google Scholar]
- Qin X, Qian J, Yao G, Zhuang Y, Zhang S. Chu J. GAP promoter library for fine-tuning of gene expression in Pichia pastoris. Appl Environ Microbiol. 2011;77:3600–3608. doi: 10.1128/AEM.02843-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randezgil F, Prieto JA, Murcia A. Sanz P. Construction of bakers' yeast strains that secrete Aspergillus oryzae alpha-amylase and their use in bread-making. J Cereal Sci. 1995;21:185–193. [Google Scholar]
- Ribeiro O, Magalhaes F, Aguiar TQ, Wiebe MG, Penttila M. Domingues L. Random and direct mutagenesis to enhance protein secretion in Ashbya gossypii. Bioengineered. 2013;4:322–331. doi: 10.4161/bioe.24653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richard MS, Van Broock MRG. Figueroa LIC. Restoration of respiratory competence in the yeast Candida utilis after somatic fusion with Saccharomyces cerevisiae. Curr Microbiol. 1987;16:109–112. [Google Scholar]
- Rico J, Pardo E. Orejas M. Enhanced production of a plant monoterpene by overexpression of the 3-hydroxy-3-methylglutaryl coenzyme A reductase catalytic domain in Saccharomyces cerevisiae. Appl Environ Microbiol. 2010;76:6449–6454. doi: 10.1128/AEM.02987-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ro DK, Paradise EM, Ouellet M, et al. Production of the antimalarial drug precursor artemisinic acid in engineered yeast. Nature. 2006;440:940–943. doi: 10.1038/nature04640. [DOI] [PubMed] [Google Scholar]
- Rocha SN, Abrahao-Neto J, Cerdan ME, Gonzalez-Siso MI. Gombert AK. Heterologous expression of glucose oxidase in the yeast Kluyveromyces marxianus. Microb Cell Fact. 2010;9:4. doi: 10.1186/1475-2859-9-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romano P, Soli MG. Suzzi G. Procedure for mutagenizing spores of Saccharomyces cerevisiae. J Bacteriol. 1983;156:907–908. doi: 10.1128/jb.156.2.907-908.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romano P, Soli MG, Suzzi G, Grazia L. Zambonelli C. Improvement of a wine Saccharomyces cerevisiae strain by a breeding program. Appl Environ Microbiol. 1985;50:1064–1067. doi: 10.1128/aem.50.4.1064-1067.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romanos MA, Scorer CA. Clare JJ. Foreign gene expression in yeast: a review. Yeast. 1992;8:423–488. doi: 10.1002/yea.320080602. [DOI] [PubMed] [Google Scholar]
- Rosa JC, Colombo LT, Alvim MC, Avonce N, Van Dijck P. Passos FM. Metabolic engineering of Kluyveromyces lactis for L-ascorbic acid (vitamin C) biosynthesis. Microb Cell Fact. 2013;12:59. doi: 10.1186/1475-2859-12-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossouw D, Naes T. Bauer FF. Linking gene regulation and the exo-metabolome: a comparative transcriptomics approach to identify genes that impact on the production of volatile aroma compounds in yeast. BMC Genomics. 2008;9:530. doi: 10.1186/1471-2164-9-530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rous CV, Snow R. Kunkee RE. Reduction of higher alcohols by fermentation with a leucine-auxotrophic mutant of wine yeast. J Inst Brew. 1983;89:274–278. [Google Scholar]
- Rowlands R. Fungal technology. In: Smith J, Berry DR, Kristiansen B, editors. The Filamentous Fungi. London: Edward Arnold; 1983. pp. 346–372. &, eds), Vol. 4 ( [Google Scholar]
- Rowlands RT. Industrial strain improvement: mutagenesis and random screening procedures. Enzyme Microb Technol. 1984;6:3–10. [Google Scholar]
- Rubio-Texeira M, Van Zeebroeck G, Voordeckers K. Thevelein JM. Saccharomyces cerevisiae plasma membrane nutrient sensors and their role in PKA signaling. FEMS Yeast Res. 2010;10:134–149. doi: 10.1111/j.1567-1364.2009.00587.x. [DOI] [PubMed] [Google Scholar]
- Ruderfer DM, Pratt SC, Seidel HS. Kruglyak L. Population genomic analysis of outcrossing and recombination in yeast. Nat Genet. 2006;38:1077–1081. doi: 10.1038/ng1859. [DOI] [PubMed] [Google Scholar]
- Runquist D, Hahn-Hägerdal B. Bettiga M. Increased ethanol productivity in xylose-utilizing Saccharomyces cerevisiae via a randomly mutagenized xylose reductase. Appl Environ Microbiol. 2010;76:7796–7802. doi: 10.1128/AEM.01505-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell I, Hancock IF. Stewart GG. Construction of dextrin fermentative yeast strains that do not produce phenolic off-flavors in beer. J Am Soc Brew Chem. 1983;41:45–51. [Google Scholar]
- Saerens SM, Duong CT. Nevoigt E. Genetic improvement of brewer's yeast: current state, perspectives and limits. Appl Microbiol Biotechnol. 2010;86:1195–1212. doi: 10.1007/s00253-010-2486-6. [DOI] [PubMed] [Google Scholar]
- Saifitdinova AF, Nizhnikov AA, Lada AG, Rubel AA, Magomedova ZM, Ignatova VV, Inge-Vechtomov SG. Galkin AP. [NSI (+)]: a novel non-Mendelian nonsense suppressor determinant in Saccharomyces cerevisiae. Curr Genet. 2010;56:467–478. doi: 10.1007/s00294-010-0314-2. [DOI] [PubMed] [Google Scholar]
- Sakai A, Shimizu Y. Hishinuma F. Integration of heterologous genes into the chromosome of Saccharomyces cerevisiae using a delta sequence of yeast retrotransposon Ty. Appl Microbiol Biotechnol. 1990;33:302–306. doi: 10.1007/BF00164526. [DOI] [PubMed] [Google Scholar]
- Sakanaka K, Yan W, Kishida M. Sakai T. Breeding a fermentative yeast at high temperature using protoplast fusion. J Ferment Bioeng. 1996;81:104–108. [Google Scholar]
- Salmon JM. Barre P. Improvement of nitrogen assimilation and fermentation kinetics under enological conditions by derepression of alternative nitrogen-assimilatory pathways in an industrial Saccharomyces cerevisiae strain. Appl Environ Microbiol. 1998;64:3831–3837. doi: 10.1128/aem.64.10.3831-3837.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez S, Cea A. Flores M. Selective enrichment of aromatic amino acid auxotrophs in Hansenula polymorpha. Appl Environ Microbiol. 1978;35:228–230. doi: 10.1128/aem.35.2.228-230.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos CN. Stephanopoulos G. Combinatorial engineering of microbes for optimizing cellular phenotype. Curr Opin Chem Biol. 2008;12:168–176. doi: 10.1016/j.cbpa.2008.01.017. [DOI] [PubMed] [Google Scholar]
- Sato M, Kishimoto M, Watari J. Takashio M. Breeding of brewer's yeast by hybridization between a top-fermenting yeast Saccharomyces cerevisiae and a cryophilic yeast Saccharomyces bayanus. J Biosci Bioeng. 2002;93:509–511. doi: 10.1016/s1389-1723(02)80101-3. [DOI] [PubMed] [Google Scholar]
- Sauer U. Evolutionary engineering of industrially important microbial phenotypes. Adv Biochem Eng Biotechnol. 2001;73:129–169. doi: 10.1007/3-540-45300-8_7. [DOI] [PubMed] [Google Scholar]
- Scacco A, Oliva D, Di Maio S, Polizzotto G, Genna G, Tripodi G, Lanza CM. Verzera A. Indigenous Saccharomyces cerevisiae strains and their influence on the quality of Cataratto, Inzolia and Grillo white wines. Food Res Int. 2012;46:1–9. [Google Scholar]
- Scalcinati G, Otero JM, Van Vleet JR, Jeffries TW, Olsson L. Nielsen J. Evolutionary engineering of Saccharomyces cerevisiae for efficient aerobic xylose consumption. FEMS Yeast Res. 2012;12:582–597. doi: 10.1111/j.1567-1364.2012.00808.x. [DOI] [PubMed] [Google Scholar]
- Schacherer J, Shapiro JA, Ruderfer DM. Kruglyak L. Comprehensive polymorphism survey elucidates population structure of Saccharomyces cerevisiae. Nature. 2009;458:342–345. doi: 10.1038/nature07670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitt MJ. Breinig F. The viral killer system in yeast: from molecular biology to application. FEMS Microbiol Rev. 2002;26:257–276. doi: 10.1111/j.1574-6976.2002.tb00614.x. [DOI] [PubMed] [Google Scholar]
- Schoeman H, Vivier MA, Du Toit M, Dicks LM. Pretorius IS. The development of bactericidal yeast strains by expressing the Pediococcus acidilactici pediocin gene (pedA) in Saccharomyces cerevisiae. Yeast. 1999;15:647–656. doi: 10.1002/(SICI)1097-0061(19990615)15:8<647::AID-YEA409>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- Seki T, Choi EH. Ryu D. Construction of killer wine yeast strain. Appl Environ Microbiol. 1985;49:1211–1215. doi: 10.1128/aem.49.5.1211-1215.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serrano L. Synthetic biology: promises and challenges. Mol Syst Biol. 2007;3:158. doi: 10.1038/msb4100202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao Z, Zhao H, Giver L. Arnold FH. Random-priming in vitro recombination: an effective tool for directed evolution. Nucleic Acids Res. 1998;26:681–683. doi: 10.1093/nar/26.2.681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Y, Chen X, Peng B, Chen L, Hou J. Bao X. An efficient xylose-fermenting recombinant Saccharomyces cerevisiae strain obtained through adapted evolution and its global transcription profile. Appl Microbiol Biotechnol. 2012;96:1079–1091. doi: 10.1007/s00253-012-4418-0. [DOI] [PubMed] [Google Scholar]
- Shi DJ, Wang CL. Wang KM. Genome shuffling to improve thermotolerance, ethanol tolerance and ethanol productivity of Saccharomyces cerevisiae. J Ind Microbiol Biotechnol. 2009;36:139–147. doi: 10.1007/s10295-008-0481-z. [DOI] [PubMed] [Google Scholar]
- Siam R, Dolan WP. Forsburg SL. Choosing and using Schizosaccharomyces pombe plasmids. Methods. 2004;33:189–198. doi: 10.1016/j.ymeth.2003.11.013. [DOI] [PubMed] [Google Scholar]
- Sicard D. Legras JL. Bread, beer and wine: yeast domestication in the Saccharomyces sensu stricto complex. C R Biol. 2011;334:229–236. doi: 10.1016/j.crvi.2010.12.016. [DOI] [PubMed] [Google Scholar]
- Sieber V, Martinez CA. Arnold FH. Libraries of hybrid proteins from distantly related sequences. Nat Biotechnol. 2001;19:456–460. doi: 10.1038/88129. [DOI] [PubMed] [Google Scholar]
- Sipiczki M. Interspecies hybridization and recombination in Saccharomyces wine yeasts. FEMS Yeast Res. 2008;8:996–1007. doi: 10.1111/j.1567-1364.2008.00369.x. [DOI] [PubMed] [Google Scholar]
- Smit A, Cordero Otero RR, Lambrechts MG, Pretorius IS. Van Rensburg P. Enhancing volatile phenol concentrations in wine by expressing various phenolic acid decarboxylase genes in Saccharomyces cerevisiae. J Agric Food Chem. 2003;51:4909–4915. doi: 10.1021/jf026224d. [DOI] [PubMed] [Google Scholar]
- Snow R. An enrichment method for auxotrophic yeast mutants using the antibiotic ‘nystatin’. Nature. 1966;211:206–207. doi: 10.1038/211206a0. [DOI] [PubMed] [Google Scholar]
- Soden DM, O'Callaghan J. Dobson ADW. Molecular cloning of a laccase isozyme gene from Pleurotus sajor-caju and expression in the heterologous Pichia pastoris host. Microbiology. 2002;148:4003–4014. doi: 10.1099/00221287-148-12-4003. [DOI] [PubMed] [Google Scholar]
- Sonderegger M. Sauer U. Evolutionary engineering of Saccharomyces cerevisiae for anaerobic growth on xylose. Appl Environ Microbiol. 2003;69:1990–1998. doi: 10.1128/AEM.69.4.1990-1998.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spencer F, Hugerat Y, Simchen G, Hurko O, Connelly C. Hieter P. Yeast kar1 mutants provide an effective method for YAC transfer to new hosts. Genomics. 1994;22:118–126. doi: 10.1006/geno.1994.1352. [DOI] [PubMed] [Google Scholar]
- Spencer JF, Bizeau C, Reynolds N. Spencer DM. The use of mitochondrial mutants in hybridization of industrial yeast strains. Curr Genet. 1985;9:649–652. [Google Scholar]
- Spencer JF, Spencer DM, de Figueroa L, Nougues JM. Heluane H. Transfer of genes for utilization of starch (sta2) and melibiose (mel) to industrial strains of Saccharomyces cerevisiae by single-chromosome transfer, using a kar1 mutant as vector. Appl Microbiol Biotechnol. 1992;37:230–234. doi: 10.1007/BF00178176. [DOI] [PubMed] [Google Scholar]
- Stanley D, Fraser S, Chambers PJ, Rogers P. Stanley GA. Generation and characterisation of stable ethanol-tolerant mutants of Saccharomyces cerevisiae. J Ind Microbiol Biotechnol. 2010;37:139–149. doi: 10.1007/s10295-009-0655-3. [DOI] [PubMed] [Google Scholar]
- Steensels J, Snoek T, Meersman E, et al. Selecting and generating superior yeasts for the brewing industry. Cerevisia. 2012;37:63–67. [Google Scholar]
- Steinmetz LM, Sinha H, Richards DR, Spiegelman JI, Oefner PJ, McCusker JH. Davis RW. Dissecting the architecture of a quantitative trait locus in yeast. Nature. 2002;416:326–330. doi: 10.1038/416326a. [DOI] [PubMed] [Google Scholar]
- Stemmer WP. Rapid evolution of a protein in vitro by DNA shuffling. Nature. 1994;370:389–391. doi: 10.1038/370389a0. [DOI] [PubMed] [Google Scholar]
- Stephanopoulos G. Synthetic biology and metabolic engineering. ACS Synth Biol. 2012;1:514–525. doi: 10.1021/sb300094q. [DOI] [PubMed] [Google Scholar]
- Stephanopoulos G. Vallino JJ. Network rigidity and metabolic engineering in metabolite overproduction. Science. 1991;252:1675–1681. doi: 10.1126/science.1904627. [DOI] [PubMed] [Google Scholar]
- Steyn AJ. Pretorius IS. Co-expression of a Saccharomyces diastaticus glucoamylase-encoding gene and a Bacillus amyloliquefaciens alpha-amylase-encoding gene in Saccharomyces cerevisiae. Gene. 1991;100:85–93. doi: 10.1016/0378-1119(91)90353-d. [DOI] [PubMed] [Google Scholar]
- Storici F, Lewis LK. Resnick MA. In vivo site-directed mutagenesis using oligonucleotides. Nat Biotechnol. 2001;19:773–776. doi: 10.1038/90837. [DOI] [PubMed] [Google Scholar]
- Struhl K. The new yeast genetics. Nature. 1983;305:391–397. doi: 10.1038/305391a0. [DOI] [PubMed] [Google Scholar]
- Sydor T, Schaffer S. Boles E. Considerable increase in resveratrol production by recombinant industrial yeast strains with use of rich medium. Appl Environ Microbiol. 2010;76:3361–3363. doi: 10.1128/AEM.02796-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tai M. Stephanopoulos G. Engineering the push and pull of lipid biosynthesis in oleaginous yeast Yarrowia lipolytica for biofuel production. Metab Eng. 2013;15:1–9. doi: 10.1016/j.ymben.2012.08.007. [DOI] [PubMed] [Google Scholar]
- Takegawa K, Tohda H, Sasaki M, Idiris A, Ohashi T, Mukaiyama H, Giga-Hama Y. Kumagai H. Production of heterologous proteins using the fission-yeast (Schizosaccharomyces pombe) expression system. Biotechnol Appl Biochem. 2009;53:227–235. doi: 10.1042/BA20090048. [DOI] [PubMed] [Google Scholar]
- Tantirungkij M, Nakashima N, Seki T. Yoshida T. Construction of xylose-assimilating Saccharomyces cerevisiae. J Ferment Bioeng. 1993;75:83–88. [Google Scholar]
- Tao X, Zheng D, Liu T, Wang P, Zhao W, Zhu M, Jiang X, Zhao Y. Wu X. A novel strategy to construct yeast Saccharomyces cerevisiae strains for very high gravity fermentation. PLoS One. 2012;7:e31235. doi: 10.1371/journal.pone.0031235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taya M, Honda H. Kobayashi T. Lactose-utilizing hybrid strain derived from Saccharomyces cerevisiae and Kluyveromyces lactis by protoplast fusion. Agric Biol Chem. 1984;48:2239–2243. [Google Scholar]
- Terazawa Y, Wakiyama M. Yokoyama S. pCMV-Leu2/pUCA-Neo, a vector set for screening Schizosaccharomyces pombe transformants expressing heterologous proteins. Anal Biochem. 2011;414:306–308. doi: 10.1016/j.ab.2011.03.034. [DOI] [PubMed] [Google Scholar]
- Teunissen A, Dumortier F, Gorwa MF, Bauer J, Tanghe A, Loiez A, Smet P, Van Dijck P. Thevelein JM. Isolation and characterization of a freeze-tolerant diploid derivative of an industrial baker's yeast strain and its use in frozen doughs. Appl Environ Microbiol. 2002;68:4780–4787. doi: 10.1128/AEM.68.10.4780-4787.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomson JM, Gaucher EA, Burgan MF, De Kee DW, Li T, Aris JP. Benner SA. Resurrecting ancestral alcohol dehydrogenases from yeast. Nat Genet. 2005;37:630–635. doi: 10.1038/ng1553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timberlake WE, Frizzell MA, Richards KD. Gardner RC. A new yeast genetic resource for analysis and breeding. Yeast. 2011;28:63–80. doi: 10.1002/yea.1821. [DOI] [PubMed] [Google Scholar]
- Tosi E, Azzolini M, Guzzo F. Zapparoli G. Evidence of different fermentation behaviours of two indigenous strains of Saccharomyces cerevisiae and Saccharomyces uvarum isolated from Amarone wine. J Appl Microbiol. 2009;107:210–218. doi: 10.1111/j.1365-2672.2009.04196.x. [DOI] [PubMed] [Google Scholar]
- Tristezza M, Vetrano C, Bleve G, Grieco F, Tufariello M, Quarta A, Mita G. Spano G. Autochthonous fermentation starters for the industrial production of Negroamaro wines. J Ind Microbiol Biotechnol. 2012;39:81–92. doi: 10.1007/s10295-011-1002-z. [DOI] [PubMed] [Google Scholar]
- Tsai IJ, Bensasson D, Burt A. Koufopanou V. Population genomics of the wild yeast Saccharomyces paradoxus: quantifying the life cycle. P Natl Acad Sci USA. 2008;105:4957–4962. doi: 10.1073/pnas.0707314105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tubb RS, Searle BA, Goodey AR. Brown AJP. Proc 18th Congr Eur Brew Conv. 1981. pp. 487–496. & Rare mating and transformation for construction of novel brewing yeasts.
- Tyo KEJ. 2008. Forward and inverse metabolic engineering strategies for improving polyhydroxybutyrate production. PhD Thesis, Boston Massachusetts Institute of Technology.
- Urano N, Sahara H. Koshino S. Electrofusion of brewers' yeast protoplasts and enrichment of the fusants using a flow cytometer. Enzyme Microb Technol. 1993;15:959–964. [Google Scholar]
- van Dijken JP. Scheffers WA. Redox balances in the metabolism of sugars by yeasts. FEMS Microbiol Lett. 1986;32:199–224. [Google Scholar]
- van Ooyen AJ, Dekker P, Huang M, Olsthoorn MM, Jacobs DI, Colussi PA. Taron CH. Heterologous protein production in the yeast Kluyveromyces lactis. FEMS Yeast Res. 2006;6:381–392. doi: 10.1111/j.1567-1364.2006.00049.x. [DOI] [PubMed] [Google Scholar]
- Van Vleet JH. Jeffries TW. Yeast metabolic engineering for hemicellulosic ethanol production. Curr Opin Biotechnol. 2009;20:300–306. doi: 10.1016/j.copbio.2009.06.001. [DOI] [PubMed] [Google Scholar]
- van Zyl WH, Lodolo EJ. Gericke M. Conversion of homothallic yeast to heterothallism through HO gene disruption. Curr Genet. 1993;23:290–294. doi: 10.1007/BF00310889. [DOI] [PubMed] [Google Scholar]
- Verma HK. Singh J. New multi-purpose high copy number vector with greater mitotic stability for diverse applications in fission yeast Schizosaccharomyces pombe. Plasmid. 2012;68:186–194. doi: 10.1016/j.plasmid.2012.07.001. [DOI] [PubMed] [Google Scholar]
- Verstrepen KJ, Derdelinckx G, Verachtert H. Delvaux FR. Yeast flocculation: what brewers should know. Appl Microbiol Biotechnol. 2003a;61:197–205. doi: 10.1007/s00253-002-1200-8. [DOI] [PubMed] [Google Scholar]
- Verstrepen KJ, Jansen A, Lewitter F. Fink GR. Intragenic tandem repeats generate functional variability. Nat Genet. 2005;37:986–990. doi: 10.1038/ng1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verstrepen KJ, Derdelinckx G, Delvaux FR, Winderickx J, Thevelein JM, Bauer FF. Pretorius IS. Late fermentation expression of FLO1 in Saccharomyces cerevisiae. J Am Soc Brew Chem. 2001;59:69–76. [Google Scholar]
- Verstrepen KJ, Derdelinckx G, Dufour JP, Winderickx J, Thevelein JM, Pretorius IS. Delvaux FR. Flavor-active esters: adding fruitiness to beer. J Biosci Bioeng. 2003b;96:110–118. [PubMed] [Google Scholar]
- Vervecken W, Kaigorodov V, Callewaert N, Geysens S, De Vusser K. Contreras R. In vivo synthesis of mammalian-like, hybrid-type N-glycans in Pichia pastoris. Appl Environ Microbiol. 2004;70:2639–2646. doi: 10.1128/AEM.70.5.2639-2646.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verwaal R, Wang J, Meijnen J-P, Visser H, Sandmann G, van den Berg JA. van Ooyen AJ. High-level production of beta-carotene in Saccharomyces cerevisiae by successive transformation with carotenogenic genes from Xanthophyllomyces dendrorhous. Appl Environ Microbiol. 2007;73:4342–4350. doi: 10.1128/AEM.02759-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vicente MA, Fietto LG, Castro IM, dos Santos AN, Coutrim MX. Brandao RL. Isolation of Saccharomyces cerevisiae strains producing higher levels of flavoring compounds for production of “cachaca” the Brazilian sugarcane spirit. Int J Food Microbiol. 2006;108:51–59. doi: 10.1016/j.ijfoodmicro.2005.10.018. [DOI] [PubMed] [Google Scholar]
- Victor MY. Bhatia SK. Pathway engineering strategies for production of beneficial carotenoids in microbial hosts. Biotechnol Lett. 2012;34:1405–1414. doi: 10.1007/s10529-012-0921-8. [DOI] [PubMed] [Google Scholar]
- Voordeckers K, Brown CA, Vanneste K, van der Zande E, Voet A, Maere S. Verstrepen KJ. Reconstruction of ancestral metabolic enzymes reveals molecular mechanisms underlying evolutionary innovation through gene duplication. PLoS Biol. 2013;10:e1001446. doi: 10.1371/journal.pbio.1001446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker M, Vystavelova A, Pedler S, Eglinton J. Jiranek V. PCR-based gene disruption and recombinatory marker excision to produce modified industrial Saccharomyces cerevisiae without added sequences. J Microbiol Methods. 2005;63:193–204. doi: 10.1016/j.mimet.2005.03.015. [DOI] [PubMed] [Google Scholar]
- Walton EF, Carter BL. Pringle JR. An enrichment method for temperature-sensitive and auxotrophic mutants of yeast. Mol Gen Genet. 1979;171:111–114. [Google Scholar]
- Wang H. Hou L. Genome shuffling to improve fermentation properties of top-fermenting yeast by the improvement of stress tolerance. Food Sci Biotechnol. 2010;19:145–150. [Google Scholar]
- Wang PM, Zheng DQ, Liu TZ, Tao XL, Feng MG, Min H, Jiang XH. Wu XC. The combination of glycerol metabolic engineering and drug resistance marker-aided genome shuffling to improve very-high-gravity fermentation performances of industrial Saccharomyces cerevisiae. Bioresour Technol. 2012a;108:203–210. doi: 10.1016/j.biortech.2011.12.147. [DOI] [PubMed] [Google Scholar]
- Wang QM, Liu WQ, Liti G, Wang SA. Bai FY. Surprisingly diverged populations of Saccharomyces cerevisiae in natural environments remote from human activity. Mol Ecol. 2012b;21:5404–5417. doi: 10.1111/j.1365-294X.2012.05732.x. [DOI] [PubMed] [Google Scholar]
- Wang Y, Halls C, Zhang J, Matsuno M, Zhang Y. Yu O. Stepwise increase of resveratrol biosynthesis in yeast Saccharomyces cerevisiae by metabolic engineering. Metab Eng. 2011;13:455–463. doi: 10.1016/j.ymben.2011.04.005. [DOI] [PubMed] [Google Scholar]
- Wang ZP, Xu HM, Wang GY, Chi Z. Chi ZM. Disruption of the MIG1 gene enhances lipid biosynthesis in the oleaginous yeast Yarrowia lipolytica ACA-DC 50109. Biochim Biophys Acta. 2013;1831:675–682. doi: 10.1016/j.bbalip.2012.12.010. [DOI] [PubMed] [Google Scholar]
- Warringer J, Zorgo E, Cubillos FA, et al. Trait variation in yeast is defined by population history. PLoS Genet. 2011;7:e1002111. doi: 10.1371/journal.pgen.1002111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe T, Watanabe I, Yamamoto M, Ando A. Nakamura T. A UV-induced mutant of Pichia stipitis with increased ethanol production from xylose and selection of a spontaneous mutant with increased ethanol tolerance. Bioresour Technol. 2010;102:1844–1848. doi: 10.1016/j.biortech.2010.09.087. [DOI] [PubMed] [Google Scholar]
- Wei P, Li Z, He P, Lin Y. Jiang N. Genome shuffling in the ethanologenic yeast Candida krusei to improve acetic acid tolerance. Biotechnol Appl Biochem. 2008;49:113–120. doi: 10.1042/BA20070072. [DOI] [PubMed] [Google Scholar]
- Wickner RB, Kryndushkin D, Shewmaker F, McGlinchey R. Edskes HK. Study of amyloids using yeast. Methods Mol Biol. 2012;849:321–346. doi: 10.1007/978-1-61779-551-0_22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiedemann B. Boles E. Codon-optimized bacterial genes improve L-arabinose fermentation in recombinant Saccharomyces cerevisiae. Appl Environ Microbiol. 2008;74:2043–2050. doi: 10.1128/AEM.02395-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wisselink HW, Toirkens MJ, Wu Q, Pronk JT. van Maris AJ. Novel evolutionary engineering approach for accelerated utilization of glucose, xylose, and arabinose mixtures by engineered Saccharomyces cerevisiae strains. Appl Environ Microbiol. 2009;75:907–914. doi: 10.1128/AEM.02268-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood V, Gwilliam R, Rajandream MA, et al. The genome sequence of Schizosaccharomyces pombe. Nature. 2002;415:871–880. doi: 10.1038/nature724. [DOI] [PubMed] [Google Scholar]
- Woolston BM, Edgar S. Stephanopoulos G. Metabolic engineering: past and future. Annu Rev Chem Biomol Eng. 2013;4:259–288. doi: 10.1146/annurev-chembioeng-061312-103312. [DOI] [PubMed] [Google Scholar]
- Yamano S, Ishii T, Nakagawa M, Ikenaga H. Misawa N. Metabolic engineering for production of beta-carotene and lycopene in Saccharomyces cerevisiae. Biosci Biotechnol Biochem. 1994;58:1112–1114. doi: 10.1271/bbb.58.1112. [DOI] [PubMed] [Google Scholar]
- Yamazaki T. Nonomura H. Inherent G418-resistance in hybridization of industrial yeasts. J Ferment Bioeng. 1994;77:202–204. [Google Scholar]
- Yoshida S, Imoto J, Minato T, et al. Development of bottom-fermenting Saccharomyces strains that produce high SO2 levels, using integrated metabolome and transcriptome analysis. Appl Environ Microbiol. 2008;74:2787–2796. doi: 10.1128/AEM.01781-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshiuchi K, Watanabe M. Nishimura A. Breeding of a non-urea producing sake yeast with killer character using a kar1-1 mutant as a killer donor. J Ind Microbiol Biotechnol. 2000;24:203–209. [Google Scholar]
- Young E, Lee SM. Alper H. Optimizing pentose utilization in yeast: the need for novel tools and approaches. Biotechnol Biofuels. 2010;3:24. doi: 10.1186/1754-6834-3-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young EM, Tong A, Bui H, Spofford C. Alper HS. Rewiring yeast sugar transporter preference through modifying a conserved protein motif. P Natl Acad Sci USA. 2014;111:131–136. doi: 10.1073/pnas.1311970111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young TW. The properties and brewing performance of brewing yeasts possessing killer character. J Am Soc Brew Chem. 1983;41:1–4. [Google Scholar]
- Yovkova V, Otto C, Aurich A, Mauersberger S. Barth G. Engineering the alpha-ketoglutarate overproduction from raw glycerol by overexpression of the genes encoding NADP-dependent isocitrate dehydrogenase and pyruvate carboxylase in Yarrowia lipolytica. Appl Microbiol Biotechnol. 2014;98:2003–2013. doi: 10.1007/s00253-013-5369-9. [DOI] [PubMed] [Google Scholar]
- Yuan YB. Zhao HM. Directed evolution of a highly efficient cellobiose utilizing pathway in an industrial Saccharomyces cerevisiae strain. Biotechnol Bioeng. 2013;110:2874–2881. doi: 10.1002/bit.24946. [DOI] [PubMed] [Google Scholar]
- Zagorc T, Maraz A, Cadez N, Jemec KP, Peter G, Resnik M, Nemanic J. Raspor P. Indigenous wine killer yeasts and their application as a starter culture in wine fermentation. Food Microbiol. 2001;18:441–451. [Google Scholar]
- Zambonelli C, Passarelli P, Rainieri S, Bertolini L, Giudici P. Castellari L. Technological properties and temperature response of interspecific Saccharomyces hybrids. J Sci Food Agric. 1997;74:7–12. [Google Scholar]
- Zaret KS. Sherman F. a-Aminoadipate as a primary nitrogen source for Saccharomyces cerevisiae mutants. J Bacteriol. 1985;162:579–583. doi: 10.1128/jb.162.2.579-583.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang A, Kong Q, Cao L. Chen X. Effect of FPS1 deletion on the fermentation properties of Saccharomyces cerevisiae. Lett Appl Microbiol. 2007;44:212–217. doi: 10.1111/j.1472-765X.2006.02041.x. [DOI] [PubMed] [Google Scholar]
- Zhang CY, Liu YL, Qi YN, Zhang JW, Dai LH, Lin X. Xiao DG. Increased esters and decreased higher alcohols production by engineered brewer's yeast strains. Eur Food Res Technol. 2013;236:1009–1014. [Google Scholar]
- Zhang W. Geng A. Improved ethanol production by a xylose-fermenting recombinant yeast strain constructed through a modified genome shuffling method. Biotechnol Biofuels. 2012;5:46. doi: 10.1186/1754-6834-5-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang YX, Perry K, Vinci VA, Powell K, Stemmer WP. del Cardayre SB. Genome shuffling leads to rapid phenotypic improvement in bacteria. Nature. 2002;415:644–646. doi: 10.1038/415644a. [DOI] [PubMed] [Google Scholar]
- Zhao H, Giver L, Shao Z, Affholter JA. Arnold FH. Molecular evolution by staggered extension process (StEP) in vitro recombination. Nat Biotechnol. 1998;16:258–261. doi: 10.1038/nbt0398-258. [DOI] [PubMed] [Google Scholar]
- Zheng DQ, Wu XC, Wang PM, Chi XQ, Tao XL, Li P, Jiang XH. Zhao YH. Drug resistance marker-aided genome shuffling to improve acetic acid tolerance in Saccharomyces cerevisiae. J Ind Microbiol Biotechnol. 2011a;38:415–422. doi: 10.1007/s10295-010-0784-8. [DOI] [PubMed] [Google Scholar]
- Zheng DQ, Wu XC, Tao XL, Wang PM, Li P, Chi XQ, Li YD, Yan QF. Zhao YH. Screening and construction of Saccharomyces cerevisiae strains with improved multi-tolerance and bioethanol fermentation performance. Bioresour Technol. 2011b;102:3020–3027. doi: 10.1016/j.biortech.2010.09.122. [DOI] [PubMed] [Google Scholar]
- Zheng DQ, Chen J, Zhang K, Gao K-H, Li O, Wang PM, Zhang XY, Du FG, Sun PY. Qu AM. Genomic structural variations contribute to trait improvement during whole-genome shuffling of yeast. Appl Microbiol Biotechnol. 2013a;98:1–12. doi: 10.1007/s00253-013-5423-7. [DOI] [PubMed] [Google Scholar]
- Zheng DQ, Zhang K, Gao K, Liu Z, Zhang X, Li O, Sun J, Zhang X, Du F. Sun P. Construction of novel Saccharomyces cerevisiae strains for bioethanol active dry yeast (ADY) production. PLoS One. 2013b;8:e85022. doi: 10.1371/journal.pone.0085022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J, Yin X, Madzak C, Du G. Chen J. Enhanced alpha-ketoglutarate production in Yarrowia lipolytica WSH-Z06 by alteration of the acetyl-CoA metabolism. J Biotechnol. 2012;161:257–264. doi: 10.1016/j.jbiotec.2012.05.025. [DOI] [PubMed] [Google Scholar]
- Zörgö E, Gjuvsland A, Cubillos FA, Louis EJ, Liti G, Blomberg A, Omholt SW. Warringer J. Life history shapes trait heredity by accumulation of loss-of-function alleles in yeast. Mol Biol Evol. 2012;29:1781–1789. doi: 10.1093/molbev/mss019. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Research papers targeting strain improvement by hybridization.
Table S2. Research papers targeting strain improvement by genetic modification.
Data S1. Reference list for supplemental files.