Abstract
Malignant gliomas, such as glioblastoma multiforme (GBM), present some of the greatest challenges in the management of cancer patients worldwide. Even with aggressive surgical resections and recent advances in radiotherapy and chemotherapy, the prognosis for GBM patients remains dismal and quality of life is poor. Although new molecular pathways crucial to the biology and invasive ability of GBM are coming to light, translation of basic science achievements into clinical practice is slow. Optimal management requires a multidisciplinary approach and knowledge of potential complications arising from both disease and treatment. To help illustrate “where we are going” with GBM, we here include a detailed depiction of the molecular alterations underlying this fatal disease, as well as intensive research over the past two decades that has led to considerable advances in the understanding of basic GBM biology, pathogenesis and therapeutic approaches.
Keywords: Glioblastoma, targeted therapies, temozolomide (TMZ)
Introduction
The term ‘glioma’ encompasses all tumors thought to be of glial cell origin, including astrocytoma grades I, II (astrocytoma), III (anaplastic astrocytoma) and IV (glioblastoma multiforme or GBM), oligodendrogliomas, ependymomas, and mixed gliomas (1,2). GBMs are the most common primary malignant brain tumors (2) and account for 12% to 15% of all intracranial tumors and 50% to 60% of astrocytic tumors with an annual incidence of 5.26 per 100,000 population or 17,000 new diagnoses per year (3). Although uncommon, GBMs are exceedingly lethal with the worst prognosis of any brain tumor and a 5-year survival rate of only 5% (4). They are extremely proliferative and invasive, display highly aberrant vascularization and are resistant to traditional radiotherapy and chemotherapy regimens, making them difficult to remove completely with the usual standard of care (4).
The concept of molecular subtypes of GBM is an old one, beginning with the distinction of primary (also known as de novo) and secondary GBM (5). Genome-wide expression studies in GBM have revealed four transcriptional subclasses with distinct molecular characteristics, though molecular abnormalities, such as PTEN loss, may be common to all (2,6). The classical GMB displays chromosome 7 amplifications, chromosome 10 deletions, epidermal growth factor receptor (EGFR) amplification, EGFR mutations and Ink4a/ARF locus deletion (6). Mesenchymal GBM displays a high frequency of neurofibromatosis type 1 (NF1) mutation/deletion and high expression of CHI3L1, MET and genes involved in the tumor necrosis factor (TNF) and nuclear factor of κ-light polypeptide gene enhancer in B-cells (NFκB) pathways (7). Proneural GBM is one of the best studied subclass, characterized by alterations of platelet derived growth factor receptor a (PDGFRa) and bears the glioma-CpG island methylator phenotype (GCIMP), usually associated with mutations in isocitrate dehydrogenase (IDH) 1 and 2 (8). It shares gene expression features with lower-grade gliomas and secondary glioblastomas (8). Further refinement of the proneural class has distinguished tumors with the glioma-GCIMP phenotype from GCIMP negative tumors, which are usually IDH wild-type (9). Finally, in neural GBM particular molecular abnormalities remain unidentified (6,7). While gene expression studies have led to the concept of GBM subclasses, collective accumulation of transcriptional data suggest the distinction between classes may not be so rigid and can also lead to mosaicism or even class switching, possibly under the influence of the tumor microenvironment (10).
In this review we discuss GBM molecular biology in depth with the aim to stimulate the development of new therapeutic strategies that can improve patient outcomes. We also provide an update of the standard GBM therapeutic approach.
Standard therapy for GBM patients
The standard treatment for GBM patients is surgery, followed by radiotherapy with concurrent temozolomide (TMZ) and six monthly cycles of adjuvant TMZ (11). Use of the DNA alkylating agent TMZ concomitantly with radiotherapy is supported by a randomized phase III study that found addition of TMZ increased median survival to 15 months compared to 12 months with radiotherapy alone (11). The 2-year survival rate was 27% versus 10%, respectively (11). The other agent approved by the US Food and Drug Administration (FDA) for first-line treatment of GMB is biodegradable polymers containing the alkylating agent carmustine, implanted into the tumor bed following resection (12). Although a phase III trial suggested a survival benefit, the study has methodological problems: toxicity is severe, and a direct comparison of standard chemoradiotherapy with TMZ is lacking (12).
Since surgery remains the first and most important treatment modality for patients suffering from brain tumors, steps to improve many aspects of surgical care are of great relevance. Santagata et al. presented a technical innovation that permits rapid molecular characterization of tissue samples at the time of surgery (13). The tumor metabolite 2-hydroxyglutarate (2-HG), generated from IDH1 mutant gliomas, can be monitored during surgery with ambient mass spectrometry techniques and may quickly provide crucial information (13). 2-HG-expressing central nervous system tumors are nearly always gliomas, carry IDH1 or IDH2 mutations and have more favorable outcomes (13). Monitoring 2-HG with intraoperative mass spectrometry could conceivably become routinely used for surgery of primary brain tumors, first to classify the tumor and then, if 2-HG is present, to guide optimal resection (13). In tumors lacking 2-HG, surgical guidance would require monitoring lipid signatures or other metabolites (13).
Bevacizumab, a humanized vascular endothelial growth factor (VEGF) monoclonal antibody that targets angiogenesis, has been examined in first-line GBM treatment, added to chemoradiotherapy with TMZ. In the study of Gilbert et al., first-line use of bevacizumab prolonged progression-free survival (PFS) but did not reach the prespecified improvement target and did not improve overall survival (OS) in patients with newly diagnosed GBM (14). Also, in a second study from Chinot et al., addition of bevacizumab to radiotherapy and TMZ improved PFS but the rate of adverse events was higher with bevacizumab compared to placebo and there was no improvement in OS (15). Other antiangiogenetic agents have also been explored. αvβ3 and αvβ5 integrins are thought to be key mediators of crosstalk between tumour cells and the brain microenvironment in GBM and are overexpressed in tumour cells and vasculature (16,17). However a study with cilengitide, a selective inhibitor of αvβ3 and αvβ5 integrins, with standard TMZ chemoradiotherapy in GBM patients was negative and the drug was shelved (18).
After first-line treatment, almost all GBM patients experience disease progression after a median PFS of 7 to 10 months (3). Apart from surgical resection, which may be considered for mass effect relief, and updating histology and molecular characteristics of the tumor, salvage chemotherapy options include bevacizumab, TMZ rechallenge, and other alkylating agents, such as nitrosoureas (carmustine and lomustine) and carboplatin (3). Several phase II studies have reported high responses and 6-month PFS with bevacizumab (19-21). The value of bevacizumab in management of progressive GBMs in clinical practice is almost universally accepted because of evident symptom relief and steroid-sparing effects (4). Bevacizumab has been approved for treatment of recurrent GBM in several countries (though not in the EU), on the basis of findings from two prospective phase II trials that showed radiological response rates of 30% or more and PFS and OS times superior to those of historical controls (20,21). Such findings, in combination with the results of phase III trials in newly diagnosed GBM, mean that the OS benefit of bevacizumab treatment remains unclear. No active combination partner for bevacizumab has been identified in the setting of progressive disease (4). The BELOB study is the first randomized phase II trial to examine the role of bevacizumab in treatment of recurrent GBM and includes a bevacizumab free control group (22). The combination of bevacizumab and the synthetic alkylating agent lomustine met prespecified criteria for assessment in a phase III trial by the EORTC, but the results in the bevacizumab-alone group do not justify further studies of this treatment (4,22). Finally, another anti-VEGF agent is cediranib, an orally pan-VEGFR TKI (23). The efficacy of cediranib has been tested as monotherapy and in combination with lomustine versus lomustine alone in patients with recurrent GBM. The study did not meet its primary end point of PFS prolongation with cediranib alone or in combination with lomustine compared to lomustine monotherapy (24).
Immunotherapy has recently been incorporated into general clinical practice with the approval of ipilimumab for metastatic melanoma in 2011, and even before with the approval of sipuleucel-T for castrate-resistant prostate cancer in 2010 (25,26). Immunotherapy for GBM has afforded valuable insights but failed to generate comparable clinical results with other tumors. An approach tailored to the unique aspects of glioma biology in order to establish the role of immunotherapy in GBM is needed. EGFRvIII has been considered an ideal target for antitumor immunotherapy. In a phase II clinical trial, the efficacy of rindopepimut EGFRvIII-targeted peptide vaccine was assessed in newly diagnosed EGFRvIII positive GBM patients (27). Rindopepimut is a 14-amino acid peptide (PEPvIII) conjugated to the foreign antigen keyhole limpet hemocyanin (KLH) (27). Vaccinating patients was safe, induced specific immunity against EGFRvIII and was associated with elimination of EGFRvIII-expressing cells at recurrence. PEPvIII improved PFS and OS in this population and warrants further investigation in a phase III, randomized trial (27). Rindopepimut plus granulocyte and macrophage colony stimulating factor has also been evaluated in the ACT III phase II trial; 65 patients with newly diagnosed GBM were included and the reported 21 months median OS compared favorably with historic controls (28). A multiple antigen vaccine targeting MAGE-1, HER-2, AIM-2, TRP-2, gp100, and IL-13Rα2 has been evaluated in a phase II multicenter randomized trial which found a statistically significant difference of two months in terms of PFS (29). Immune checkpoint blockade, through targeting cytotoxic T lymphocyte antigen-4 (CTLA-4) or programmed cell death-1 (PD-1) signaling, requires more detailed understanding of the role of these pathways, both within the central nervous system and peripherally in this patient population (28).
DNA repair and resistance to TMZ
TMZ is an alkylating agent applied to malignant glioma, including GBM. TMZ causes cytotoxicity by spontaneously converting to the reactive methylating agent 5-(3-methyltriazen-1-yl) imidazole-4-carboxamide (MTIC), which then degrades to the methyldiazonium cation (16). The methyldiazonium cation reacts with DNA to form methyl adducts, such as N3-methyladenine, N7-methyl-guanine, and O6-methyl-guanine, resulting in DNA strand breaks which, if not repaired by RAD51-driven homologous recombination (HR), lead to cell-cycle arrest and delayed cell death (30,31).
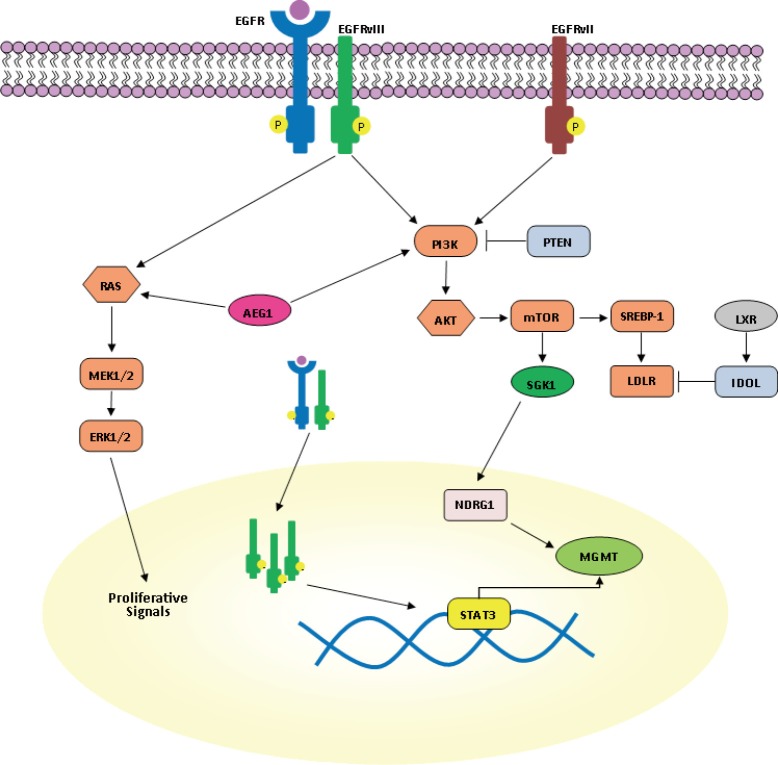
Resistance to TMZ can be caused through removal of methylation from O6 position of guanine by O6-methylguanine methyltransferase (MGMT) (32). GBM patients with MGMT promoter methylation and inhibition of MGMT expression were reported to have an improved 2-year survival with TMZ treatment together with irradiation (33). Therefore, therapeutic agents which suppress MGMT expression are highly desirable for TMZ resistant patients. In the study of Kohsaka et al., enhanced phosphorylation of STAT3 was observed in MGMT overexpressing GBM cells, such as KMG4, U138 and LN308, and STAT3 was found to be necessary for post-transcriptional elevation of MGMT (Figure 1) (34). In the same study, increased levels of both MGMT and pSTAT3 were observed in recurrent tumor compared to the primary GBM of identical patients, suggesting that STAT3 can be a therapeutic target for TMZ resistance in GBM (34). Interestingly, common therapeutic measures for GMB patients, like irradiation, corticosteroids and chronic exposure to alkylating agents, induce expression of mTOR and target N-myc downstream regulated gene 1 (NDRG1), a key determinant of resistance to alkylating chemotherapy which binds and stabilizes MGMT (Figure 1) (35). Recently, it was found that in GBM patients, MGMT promoter methylation in tumor tissue is not more predictive for response to alkylating chemotherapy in patients who received concomitant corticosteroids (35). mTORC2 transcriptionally and post-transcriptionally regulates NDRG1 through the serum glucocorticoid-induced protein kinase 1 (SGK1), making the mTORC2/SGK1/NDRG1 pathway a target for future preclinical and clinical research into GBM therapy resistance (35). However, the therapeutic activity of temsirolimus, an intravenous mTOR inhibitor, in patients with newly diagnosed GBM with an unmethylated MGMT promoter was found to be too low (36).
Figure 1.
The EGFR pathway in glioblastoma. EGFR, epidermal growth factor receptor.
MGMT expression is not the only DNA repair mechanism associated with TMZ resistance. Mismatch repair (MMR) plays a crucial role in TMZ-induced cytotoxicity. The MMR pathway removes thymidine but is unable to repair the original O6-methyl-guanine lesion. Repeated unsuccessful MMR attempts to repair DNA lead to double-strand breaks, replication arrest and cell death (37). A deficiency in the MMR pathway would result in failure to recognize and repair at this position, resulting in continued DNA replication past the O6-methyl-guanine block and thereby producing no double-strand breaks, no cell death and, ultimately, resistance to TMZ (38).
Another mechanism of TMZ resistance involves the base excision repair (BER) pathway which consists of several DNA repair proteins that cooperate on removal of damaged or inappropriate DNA bases, such as N7-methyl-guanine (39). Once these methyl groups are removed, the cells survive, meaning that high levels of BER proteins can contribute to TMZ resistance (40,41). Poly (ADP-ribose) polymerase (PARP) cooperates with the BER system to ensure genomic stability by repairing single-stranded DNA breaks. If PARP activity is inhibited, these single-stranded DNA breaks become double-strand breaks, leading to induction of cell death (42,43). Therefore, PARP inhibitors may enhance TMZ induced cytotoxicity. Recent in vitro and in vivo experiments reported that NEO212, a conjugate of TMZ to perillyl alcohol, can be cytotoxic to several types of TMZ resistant glioma cells, resulting from mechanisms other than MGMT expression (44). NEO212 functions similar to temozolamide but is unique since it induces endoplasmic reticulum ER stress and inhibits autophagy (44). When tested on three human TMZ resistant glioma cell lines, due to high levels of MGMT, MMR deficiencies, or overexpression of BER proteins, NEO212-induced cell death independent of the mechanisms of resistance (44).
Although the modern definition of primary GBM is a group of tumors lacking IDH mutations, it is worth mentioning in passing the role of IDH1 mutations. These mutations are detected in more than 70% of grade II and III gliomas and in secondary GBMs (45) and cause distinct alterations in lipid metabolism that can be detected noninvasively by 31P magnetic resonance spectroscopic imaging (46). A higher rate of response to up-front TMZ has been reported in IDH1 mutant low-grade glioma patients relative to that noted in patients with histologically identical IDH1 tumors, implying that mutant IDH1 contributes to TMZ sensitivity (47). However, the contribution of IDH mutations to drug sensitivity is complex. A recent study showed that expression of mutant IDH1 confers TMZ resistance rather than sensitivity (48). This resistance is not a direct effect of mutant IDH1 expression but mutant IDH1 drives a unique set of transformative events that indirectly enhance HR and facilitate repair of TMZ-induced DNA damage and TMZ resistance, suggesting that inhibitors of HR may be a viable means to enhance TMZ response in IDH1-mutant glioma (48).
The EGFR pathway in GBM-targeting EGFR
Although increased expression of other receptor tyrosine kinases (RTK) such as PDGFRa and VEGFR contribute to growth of GBM through the activation of RAS/ERK- or phosphatidylinositide-3 kinase (PI3K)/AKT-dependent signaling pathways, EGFR gene amplification and mutation are the most striking abnormalities detected in about 40-50% of patients with GBM and are usually found in the classical GBM subtype (34). Indeed, a specific EGFR mutant (EGFR type III, EGFRvIII) is detected in about 50% of tumors with EGFR amplification (49). EGFRvIII is generated from a deletion in exons 2-7 of the EGFR gene which results in an in-frame deletion of 267 amino acids in the extracellular domain of EGFR (49). Therefore, it is unable to bind ligand and signals constitutively. However, in contrast to EGFR mutants in lung cancer, treatment with EGFR tyrosine kinase inhibitors (TKIs) appears to be less successful in GBM compared with lung cancer, possibly secondary to altered kinetics of inhibitor binding or sensitivity of EGFRvIII (50,51). At the preclinical level, the second generation EGFR TKI dacomitinib is effective in EGFR amplified cells with or without EGFRvIII (52). A phase II clinical trial with dacomitinib is ongoing in patients with recurrent GBM with amplification of EGFR (NCT01520870) (52).
Recently, it has been shown that wild type EGFR is required for the oncogenic effect of EGFRvIII (Figure 1) (53). Dimerization of EGFRvIII is important for its activation and EGFRvIII may homo-or heterodimerize with EGFR (53). There is a feed-forward loop: EGFRvIII is activated when wild type EGFR is co-expressed. Wild type EGFR in turn is activated by heparin-binding epidermal growth factor (HB-EGF)-like growth factor, induced by EGFRvIII (53). The resultant increased activation of EGFRvIII leads to increased transactivation of multiple RTK families such as Met and EphA2 that may mediate EGFRvIII oncogenicity. An EGFR antibody [528] that blocks ligand binding to wild type EGFR inhibits EGFRvIII mediated tumor growth in xenograft studies and blocks this positive feedback loop (54).
An EGFR-EGFRvIII-STAT3 signaling axis in a subset of GBMs that co-amplify EGFR and EGFRvIII within individual tumor cells has been identified (55). Fan et al. highlighted the cooperation between EGFR and EGFRvIII in transformation in vivo, showing that EGF treatment of cells expressing both EGFR and EGFRvIII resulted in phosphorylation of both kinases. Indeed, EGFR promotes unidirectional EGFRvIII signaling in GBM cells (55). It is intriguing that enhanced STAT3 signaling is selectively affected by EGFR/EGFRvIII crosstalk, whereas signaling through PI3K and MAPK is less prominently affected. EGFR phosphorylation of EGFRvIII leads to nuclear transport of EGFRvIII and enhanced formation of a complex between EGFRvIII and STAT3 in the nucleus (55). These data suggest that EGFR and EGFRvIII coordinate to drive enhanced and prolonged STAT3 activity in the nucleus (Figure 1). It is still possible, however, that very high levels of EGFR could subvert this role even in the absence of EGFRvIII (55). Therefore, targeting EGFR in conjunction with STAT3 signaling may be a therapeutic strategy for patients with EGFRvIII-positive GBM.
However, the extent of clonal diversity of EGFR mutants in GBM and their functional and therapeutic properties require further characterization. The study from Francis et al. demonstrated the presence of multiple EGFR variants in a single GBM tumor, highlighting the intratumoral heterogeneity of GBM conferred by plasticity of EGFR amplicons (56). EGFRvII is generated by deletion of exons 2-7 of the EGFR gene and can be present in 9% of cases with focally amplified (56). Constitutive expression of EGFRvII results in downstream activation of AKT signaling, consistent with that of EGFRvIII, but not with enhanced ERK activation. It is possible that EGFRvII activates an alternative pathway such as STAT3 and poses a more direct means of inducing transcriptional changes; it is interesting to note that EGFRvII enhanced sensitivity to EGFR TKIs (56,57) (Figure 1).
EGFR amplification or activating mutations as well as loss of PTEN keep the PI3K signaling hyperactivated in nearly 90% of GBMs (58). PI3K promotes tumor growth and survival, including through sterol regulatory element-binding protein 1 (SREBP-1)-dependent lipogenesis (59). Guo et al., were able to identify in GBM cell lines, xenograft models, and clinical samples an EGFRvIII-activated, PI3K/SREBP-1-dependent tumor survival pathway through the low-density lipoprotein receptor (LDLR) (59). GW3965, a liver X receptor (LXR) agonist, activates the inducible degrader of LDLR (IDOL), inhibits LDLR and promotes tumor cell death in an in vivo GBM model (Figure 1). Therefore, GW3965 may have a role in the treatment of GBM patients (59).
Other molecular pathways involved in GBM pathogenesis
NFκB is a transcription factor activated by the EGFR pathway. Aberrant constitutive activation of NFκB has been observed in glioblastoma (60). NFκB functions as an oncogenic driver in many human cancers and is an attractive target for cancer prevention or treatment. However, as NFκB also regulates normal immune response, targeting it can produce immunotoxicity. A number of NFκB-targeting drugs have been tested in clinical trials, sometimes with disappointing results (61).
The encoding nuclear factor of κ-light polypeptide gene enhancer in B-cells inhibitor-α (NKFBIA) is an inhibitor of NFκB which abrogates signaling in the NFκB and EGFR pathways and has a suppression role in GBM tumors (62). NFKBIA dosage or expression is independently associated with survival of GBM patients and plays an important role as a determinant of GBM behavior, including response to TMZ. Bredel et al. reported that NFKBIA deletion and EGFR amplification are mutually exclusive but have a similar effect in GBM pathogenesis (62). A 2-gene model based on expression of NFKBIA and MGMT was strongly associated with the clinical course of the disease (62).
Interestingly, a relationship between NKκB and IL-6 SphK1/S1P/S1PR1 axis has been identified as a nexus between NFκB and STAT3 (63). Sphingosine-1-phosphate (S1P) induced by sphingosine kinase 1 (SphK1) is essential for production of the multifunctional NFκB regulated cytokine IL-6 with persistent activation of the transcription factor STAT3 and consequent upregulation of the S1P receptor, S1PR1 (63). The prodrug FTY720 decreased SphK1 and S1PR1 expression and eliminated the NFκB/IL-6/STAT3 amplification cascade and development of colitis-associated cancer in mice (63,64). FTY270 blocks SphK1 and S1PR1. It was previously demonstrated that SphK1, an intracellular S1P, plays a direct role in TNFα signaling and the canonical NFκB activation pathway, important in inflammatory, anti-apoptotic and immune processes (65). SphK1 mRNA is increased in several tumors, including brain, and has been correlated with a multidrug resistance phenotype (66).
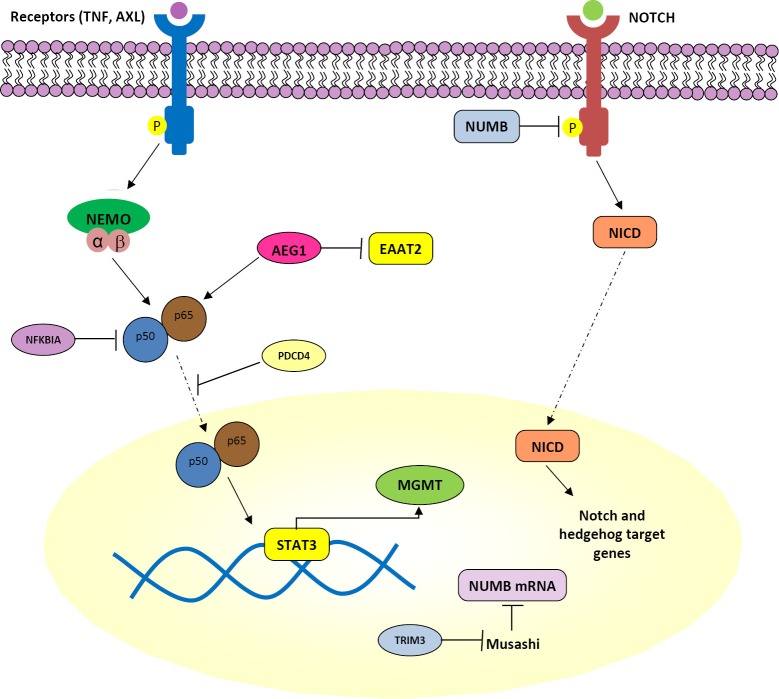
PDCD4 is a tumor suppressor gene that inhibits translation in an mRNA-selective manner by interacting with translation initiation factor eIF4A and inhibiting its RNA helicase activity (67). The effect of PDCD4 on NFκB-dependent transcriptional activity in GBM was recently demonstrated (68). PDCD4 expression inhibits NFκB transcriptional activation in a p65-dependent manner (68) (Figure 2) through interaction of the PDCD4 protein with p65 to inhibit its nuclear localization (68). NFκB target genes matrix metalloproteinase-9 (MMP-9) and VEGF—overexpressed in GBM tissues which also have overactivated NFκB—are regulated by PDCD4 (68,69). Since novel ways to target NFκB are being sought, stabilizing or mimicking PDCD4, or its binding to p65 to sequester p65 in the cytoplasm, may be useful alternatives.
Figure 2.
The NFκB and other signaling pathways in glioblastoma.
Astrocyte elevated gene-1 (AEG1) was first cloned in 2002 as a human immunodeficiency virus (HIV)-1- and TNF-α-inducible gene in primary human fetal astrocytes. Since then, it has become clear that it plays a key role in the carcinogenic process in diverse organs (70). AEG1 is overexpressed in several types of human cancers, including more than 90% of brain tumors (71). As a target of RAS, AEG1 activates multiple oncogenic signaling pathways including PI3K-AKT, mitogen-activated protein kinase (MAPK/ERK), Wnt, and NFκB which are involved in regulation of proliferation, invasion, chemoresistance, angiogenesis, and metastasis (70,72) (Figures 1,2). Enhanced expression of AEG1 increases binding of the transcriptional activator p50/p65 complex of NFκB, corresponding with degradation of NFKBIA, nuclear translocation of p65 and induction of NFκB downstream genes (73). Additionally, a strong negative correlation between expression of AEG1 and the excitatory amino acid transporter 2 (EAAT2) has been described (74). EAAT2 repression causes a reduction of glutamate uptake by glial cells, resulting in induction of neuronal cell death which contributes to glioma-induced neurodegeneration, a hallmark of this fatal tumor (74). In view of the effects of AEG1 in the context of GBM, this gene provides a viable target not only for limiting direct pathogenesis of brain tumors but also reducing indirect toxicity to neurons promoted by defects in glutamate transport (74). Increased insights into the detailed molecular mechanisms of AEG-1 action will facilitate development of improved therapies for GBM and methods to ameliorating its pathogenesis.
The role of AXL overexpression in astrocytoma cell migration and invasion has been reported (75). Keating et al. demonstrated abnormal expression of Mer and AXL RTKs in astrocytoma cell lines and primary patient samples, and the role of AXL in cell survival, proliferation, and migration (76). The cancer-promoting characteristics of the TAM family of RTK make them attractive therapeutic targets, while inhibition introduces minimal increased therapeutic toxicity. Targeted inhibition of either Mer or AXL in astrocytoma, through antibodies and small-molecule kinase inhibitors, may have clinical benefit and could enhance the efficacy of currently used chemotherapeutics (76). On the other hand, AXL has been identified as a novel target of enhancer of zeste homolog 2 (EZH2), adding further evidence to the role of the molecular network influenced by EZH2 in sustaining malignant tumor phenotype (77). Specifically, EZH2 expression—involved in epigenetically regulating gene transcription programs during development and cellular differentiationv—is found in human gliomas with AXL overexpression (77). Histone deacetylase inhibition results in depletion of EZH2 (78). Ott et al., demonstrated that inhibition of HDAC suppress AXL transcription by transcriptional control of EZH2, indicating that EZH2 is under transcriptional control of HDAC in GBM (77). It is worth mentioning that EZH2 has a dual role: besides histone methylation-dependent transcriptional silencing, it can contribute to GBM stemlike cell self-renewal and GBM malignancy through STAT3 activation (79). EZH2 phosphorylation by AKT is critical for this EZH2-STAT3 interaction (79). Therefore EZH2 is a promising therapeutic target in GBM.
As with intrahepatic cholangiocarcinoma, transcript of fused-in-glioblastoma (FIG)—which encodes a protein associated with the Golgi apparatus and plays a role in its function—has been reported as a 5’ fusion partner to the RTK ROS in the U118MG GBM cell line (80,81). Constitutive activation of FIG-ROS requires localization to the Golgi apparatus and FIG sequences deletion eliminates the transformation capacity of FIG-ROS. Thus, the role of ROS as a therapeutic target in GBM should be further explored. Another oncogenic chromosomal translocation described in almost 3% of GBM patients is the in-frame fusing of the tyrosine kinase coding domains of fibroblast growth factor receptor (FGFR) genes (FGFR1 or FGFR3) to transforming acidic coiled-coil (TACC) coding domains of TACC1 or TACC3. In in vivo models, oral administration of an FGFR inhibitor prolongs survival of mice harboring intracranial FGFR3-TACC3-initiated glioma, meaning FGFR-TACC fusions could potentially identify a subset of GBM patients who could benefit from targeted FGFR kinase inhibition (82).
Zhu et al. reported novel insights into the mechanisms that can transform membrane protein tyrosine phosphatase receptor U (PTPRU) from a tumor suppressor to a tumor promoting agent in GBMs (83). PTPRU belongs to the R2B subfamily of receptor protein tyrosine phosphatases (RPTPs) which are involved in cell adhesion and function as tumor suppressors primarily through dephosphorylation of β-catenin (84). While expression of full-length PTPRU protein is low in glioma cells, a number of non-full-length PTPRU isoforms are highly expressed and promote glioma progression. PTPRU knockdown-induced inhibition of glioma cell motility is mediated through β-catenin signaling inactivation and ubiquitin ligase-mediated degradation of focal adhesion proteins (83).
Tripartite motif (TRIM) proteins belong to the family of E3 ubiquitin ligases that have a TRIM containing RING finger domain, one or two zinc-binding B-box domains and coiled-coil domains (85). These proteins regulate cellular processes such as proliferation, apoptosis and transcriptional regulation (85). TRIM3 was first identified and characterized as a brain-enriched RING finger protein with its gene localized to chromosome 11p15.5 (86). TRIM3 loss, through deletion or DNA methylation, is detected in approximately 25% of GBMs, as well as in lower-grade gliomas (87). TRIM3 deletion is highly associated with the proneural transcriptional class of GBM, which is enriched for genes that regulate neural developmental and proliferation (8). TRIM3 expression is related to suppressed expression of c-Myc and stem cell markers like Nestin, Nanog, and Musashi, antagonizing stem-like behavior (87). Musashi is the human homologue of Drosophila Musashi protein. In Droshophila it regulates precursors of neurons and sensory bristles, and flies with gene mutations have double-shafted bristles; the gene was named after the samurai Miyamoto Musashi who fought with two swords (88). In vertebrates, Musashi1, and the closely related Musashi2 protein, bind consensus motifs in mRNAs to inhibit their transcription. Known targets include Numb, a membrane-bound protein that has a role in determining binary cell fates during development by inhibiting the intracellular domain of Notch (NICD) and suppressing Notch and Hedgehog signaling (88-90) (Figure 2). Therefore, TRIM3 regulates proliferation and differentiation through the Musashi/Numb/Notch/Hedgehog pathway, as well as c-Myc, and its loss increases the glioma stem-cell population by disrupting asymmetric cell division and cellular differentiation (87).
As for most solid tumors, treatments for GBM that inhibit individual mutant proteins related to cancer maintenance or its blood supply are ineffective. Kitambi et al. screened a relatively small set of compounds looking for agents that alter the size or shape of GBM cells and identified a compound they termed Vacquinol-1 (91,92). Vacquinol-1 kills cancer cells not through apoptosis or autophagy but through a mechanism the researchers named “methuosis”, from the Greek “methuo”, meaning to drink to intoxication (91). Vacquinol-1 and MOMIPP, a compound also identified by this group, induce rapid endocytic-like activity in GBM cells, leading to formation of massive numbers of empty, variably sized, intracellular vacuoles, each bound by a single membrane. The cell membrane then ruptures, resulting in death (91,93). These results identify a vulnerability to massive vacuolization that can be targeted by small molecules and point to the possibility of exploiting this process in the design of anticancer therapies.
Do extracranial metastases occur in GBM patients?
It has long been unclear whether glioma cells can pass through the blood brain barrier and grow in the secondary organ microenvironment, or whether they simply do not have time to grow elsewhere due to deadly rapid growth of the primary tumor (94). Suppression of extracranial growth of GBM cells by the immune system, the absence of lymphatic channels in the central nervous system, or the inability of GBM cells to invade and loosen connective tissue in the extracranial space are additional reasons for the low incidence of extracranial metastasis (94). However, the dogma that glioma cells survive only in the brain is challenged by the findings of Müller et al. (95) who detected circulating tumor cells (CTCs) in peripheral blood of 20.6% GBM patients, most frequently in those with EGFR gene amplification in the corresponding tumor tissues compared to patients with non-EGFR-amplified tumor (95). High numbers of CTCs may identify patients as long-term survivors and could be used to monitor response to therapy or disease progression (96). Thus, this group’s discovery has the potential to impact on diagnosis, prognosis and treatment of GBM patients (96).
Conclusions
GBM, the most common primary brain tumor, has few available therapies providing significant improvement in survival. It has been demonstrated that molecular heterogeneity among GBMs is prominent, and pathological diagnosis cannot always predict tumor clinical behavior. As a result of increasing knowledge and molecular classification of GBM into a number of subgroups, with the resulting application to clinical practice, we are now better informed of the reasons for differing outcomes. This understanding, along with the potential for individualized patient-directed therapy, will continue to evolve in this lethal disease. A number of prognostic or predictive biomarkers have been identified which may contribute to clinical management of GBM (see Table 1). Although the clinical implications of these alterations remain to be determined in prospective studies, a growing number of candidate biomarkers have been investigated. To date, MGMT promoter methylation and IDH1 and 2 mutations have proven clinical applications to GBM prognosis and treatment. The usefulness of other molecular alterations in predicting outcome or guiding decisions about disease management remains to be clinically validated. Given that recent medical treatment strategies have been moving toward individualized therapy, with many targeted drugs investigated, the identification of reliable molecular biomarkers in GBM will be of considerable therapeutic importance.
Table 1. Activated pathways in GBM; biomarkers and potential therapeutic opportunities.
| Biomarker | Pathway activated | Prognostic significance | Predictive significance | Ref | |
|---|---|---|---|---|---|
| MGMT | |||||
| MGMT methylation | Defects in DNA repair mechanisms | Better survival | Better response to TMZ | (32,33) | |
| mTOR2/SGK1/NDRG1 activation | MGMT stabilization | – | NDRG1 expression more predictive for response to TMZ than MGMT | (35) | |
| IDH | |||||
| IDH mutations | Affect DNA repair mechanisms | Better survival | Better response to TMZ; under debate | (46-48) | |
| EGFR | |||||
| EGFRvIII mutation | AKT, ERK and STAT3 | – | EGFR TKIs not successful in GBM. Interesting results with dacomitinib | (49-52) | |
| HB-EGF/EGFR wild type/EGFRvIII crosstalk | Prolonged STAT3 activity in the nucleus | – | EGFR antibody [528] that blocks binding of ligand to wild type EGFR. Targeting EGFR in conjunction with STAT3 | (53,55) | |
| EGFRvII mutation | Activation of AKT and STAT3 | – | Enhanced sensitivity to EGFR TKIs | (56,57) | |
| mTOR/SREBP-1/LDLR pathway | Tumor biosynthesis | – | Sensitivity to GW3965 (LXR agonist) | (59) | |
| NFκB | |||||
| NFκB activation | Expression of genes involved in inflammation, immune response, proliferation, and apoptosis | – | Disappointing results with NFκB-targeting drugs | (60,61) | |
| NFKBIA deletion | Mutually exclusive with EGFR amplification. NFκB suppression gene | NFKBIA/MGMT: 2-gene model associated with prognosis | Restoration of NFKBIA expression: sensitivity to chemotherapy | (62) | |
| IL-6/SphK1/S1P/S1PR1axis | NFκB and STAT3 activation | – | FTY270: SphK1 and S1PR1inhibitor | (63) | |
| PDCD4downregulation | Inhibits NFκB-dependent transcriptional activity | – | Potential alternative target for NFκB inhibition | (68) | |
| AEG1 | |||||
| AEG1 overexpression | PI3K-AKT, MAPK/ERK, Wnt, and NFκB activation. EAAT2 inhibition | – | Potential target for GBM therapy | (70-72,74) | |
| Other biomarkers | |||||
| AXL/Mer overexpression | – | – | AXL or Mer inhibitors | (75,76) | |
| EZH2 overexpression | AXL overexpression, | – | HDAC inhibitors | (77-79) | |
| STAT3 activation | |||||
| FIG-ROS fusion | – | – | ROS as a therapeutic target | (80,81) | |
| FGFR1-TACC1 and FGFR3-TACC3 fusions | – | – | FGFR inhibitors | (82) | |
| PTPRU (non-full-length isoforms) | β-catenin signaling activation | – | – | (83) | |
| TRIM3 loss | Musashi/Numb/NICD/Notch/Hedgehog pathway c-Myc expression | – | – | (87) | |
TMZ, temozolomide; EGFR, epidermal growth factor receptor; MGMT, methylguanine methyltransferase; IDH, isocitrate dehydrogenase; AEG1, astrocyte elevated gene-1; GBM, glioblastoma multiforme; TKIs, tyrosine kinase inhibitors; LXR, liver X receptor; FGFR, fibroblast growth factor receptor.
Acknowledgements
Disclosure: The authors declare no conflict of interest.
References
- 1.Schwartzbaum JA, Fisher JL, Aldape KD, et al. Epidemiology and molecular pathology of glioma. Nat Clin Pract Neurol 2006;2:494-503; quiz 491 p following 516. [DOI] [PubMed]
- 2.Rosell R, de Las Penas R, Balana C, et al. Translational research in glioblastoma multiforme: molecular criteria for patient selection. Future Oncol 2008;4:219-28. [DOI] [PubMed] [Google Scholar]
- 3.Omuro A, DeAngelis LM. Glioblastoma and other malignant gliomas: a clinical review. JAMA 2013;310:1842-50. [DOI] [PubMed] [Google Scholar]
- 4.Weller M, van den Bent M, Hopkins K, et al. EANO guideline for the diagnosis and treatment of anaplastic gliomas and glioblastoma. Lancet Oncol 2014;15:e395-403. [DOI] [PubMed] [Google Scholar]
- 5.Scherer HJ. A Critical Review: The Pathology of Cerebral Gliomas. J Neurol Psychiatry 1940;3:147-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Theeler BJ, Yung WK, Fuller GN, et al. Moving toward molecular classification of diffuse gliomas in adults. Neurology 2012;79:1917-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bredel M, Scholtens DM, Harsh GR, et al. A network model of a cooperative genetic landscape in brain tumors. JAMA 2009;302:261-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Verhaak RG, Hoadley KA, Purdom E, et al. Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell 2010;17:98-110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ozawa T, Riester M, Cheng YK, et al. Most Human Non-GCIMP Glioblastoma Subtypes Evolve from a Common Proneural-like Precursor Glioma. Cancer Cell 2014;26:288-300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bhat KP, Balasubramaniyan V, Vaillant B, et al. Mesenchymal differentiation mediated by NF-kappaB promotes radiation resistance in glioblastoma. Cancer Cell 2013;24:331-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stupp R, Mason WP, van den Bent MJ, et al. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N Engl J Med 2005;352:987-96. [DOI] [PubMed] [Google Scholar]
- 12.Westphal M, Hilt DC, Bortey E, et al. A phase 3 trial of local chemotherapy with biodegradable carmustine (BCNU) wafers (Gliadel wafers) in patients with primary malignant glioma. Neuro Oncol 2003;5:79-88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Santagata S, Eberlin LS, Norton I, et al. Intraoperative mass spectrometry mapping of an onco-metabolite to guide brain tumor surgery. Proc Natl Acad Sci U S A 2014;111:11121-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gilbert MR, Dignam JJ, Armstrong TS, et al. A randomized trial of bevacizumab for newly diagnosed glioblastoma. N Engl J Med 2014;370:699-708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chinot OL, Wick W, Mason W, et al. Bevacizumab plus radiotherapy-temozolomide for newly diagnosed glioblastoma. N Engl J Med 2014;370:709-22. [DOI] [PubMed] [Google Scholar]
- 16.Schnell O, Krebs B, Wagner E, et al. Expression of integrin alphavbeta3 in gliomas correlates with tumor grade and is not restricted to tumor vasculature. Brain Pathol 2008;18:378-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Roth P, Silginer M, Goodman SL, et al. Integrin control of the transforming growth factor-beta pathway in glioblastoma. Brain 2013;136:564-76. [DOI] [PubMed] [Google Scholar]
- 18.Stupp R, Hegi ME, Gorlia T, et al. Cilengitide combined with standard treatment for patients with newly diagnosed glioblastoma with methylated MGMT promoter (CENTRIC EORTC 26071-22072 study): a multicentre, randomised, open-label, phase 3 trial. Lancet Oncol 2014;15:1100-8. [DOI] [PubMed] [Google Scholar]
- 19.Vredenburgh JJ, Desjardins A, Herndon JE, 2nd, et al. Phase II trial of bevacizumab and irinotecan in recurrent malignant glioma. Clin Cancer Res 2007;13:1253-9. [DOI] [PubMed] [Google Scholar]
- 20.Kreisl TN, Kim L, Moore K, et al. Phase II trial of single-agent bevacizumab followed by bevacizumab plus irinotecan at tumor progression in recurrent glioblastoma. J Clin Oncol 2009;27:740-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Friedman HS, Prados MD, Wen PY, et al. Bevacizumab alone and in combination with irinotecan in recurrent glioblastoma. J Clin Oncol 2009;27:4733-40. [DOI] [PubMed] [Google Scholar]
- 22.Taal W, Oosterkamp HM, Walenkamp AM, et al. Single-agent bevacizumab or lomustine versus a combination of bevacizumab plus lomustine in patients with recurrent glioblastoma (BELOB trial): a randomised controlled phase 2 trial. Lancet Oncol 2014;15:943-53. [DOI] [PubMed] [Google Scholar]
- 23.Wedge SR, Kendrew J, Hennequin LF, et al. AZD2171: a highly potent, orally bioavailable, vascular endothelial growth factor receptor-2 tyrosine kinase inhibitor for the treatment of cancer. Cancer Res 2005;65:4389-400. [DOI] [PubMed] [Google Scholar]
- 24.Batchelor TT, Mulholland P, Neyns B, et al. Phase III randomized trial comparing the efficacy of cediranib as monotherapy, and in combination with lomustine, versus lomustine alone in patients with recurrent glioblastoma. J Clin Oncol 2013;31:3212-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lipson EJ, Drake CG. Ipilimumab: An Anti-CTLA-4 Antibody for Metastatic Melanoma. Clin Cancer Res 2011;17:6958-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Drake CG. Prostate cancer as a model for tumour immunotherapy. Nat Rev Immunol 2010;10:580-93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sampson JH, Heimberger AB, Archer GE, et al. Immunologic escape after prolonged progression-free survival with epidermal growth factor receptor variant III peptide vaccination in patients with newly diagnosed glioblastoma. J Clin Oncol 2010;28:4722-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jackson CM, Lim M, Drake CG. Immunotherapy for brain cancer: recent progress and future promise. Clin Cancer Res 2014;20:3651-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wen PY, Reardon DA, Phuphanich S, et al. A randomized, double-blind, placebo-controlled phase 2 trial of dendritic cell (DC) vaccination with ICT-107 in newly diagnosed glioblastoma (GBM) patients. J Clin Oncol 2014;32:abstr 2005. [DOI] [PMC free article] [PubMed]
- 30.Friedman HS, Kerby T, Calvert H. Temozolomide and treatment of malignant glioma. Clin Cancer Res 2000;6:2585-97. [PubMed] [Google Scholar]
- 31.Darkes MJ, Plosker GL. Pneumococcal conjugate vaccine (Prevnar; PNCRM7): a review of its use in the prevention of Streptococcus pneumoniae infection. Paediatr Drugs 2002;4:609-30. [DOI] [PubMed] [Google Scholar]
- 32.Kokkinakis DM, Bocangel DB, Schold SC, et al. Thresholds of O6-alkylguanine-DNA alkyltransferase which confer significant resistance of human glial tumor xenografts to treatment with 1,3-bis(2-chloroethyl)-1-nitrosourea or temozolomide. Clin Cancer Res 2001;7:421-8. [PubMed] [Google Scholar]
- 33.Hegi ME, Diserens AC, Gorlia T, et al. MGMT gene silencing and benefit from temozolomide in glioblastoma. N Engl J Med 2005;352:997-1003. [DOI] [PubMed] [Google Scholar]
- 34.Kohsaka S, Wang L, Yachi K, et al. STAT3 inhibition overcomes temozolomide resistance in glioblastoma by downregulating MGMT expression. Mol Cancer Ther 2012;11:1289-99. [DOI] [PubMed] [Google Scholar]
- 35.Weiler M, Blaes J, Pusch S, et al. mTOR target NDRG1 confers MGMT-dependent resistance to alkylating chemotherapy. Proc Natl Acad Sci U S A 2014;111:409-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wick W, Gorlia T, Van Den Bent MJ, et al. Radiation therapy and concurrent plus adjuvant temsirolimus (CCI-779) versus chemoirradiation with temozolomide in newly diagnosed glioblastoma without methylation of the MGMT gene promoter. J Clin Oncol 2014;32:abstr 2003.
- 37.Fu D, Calvo JA, Samson LD. Balancing repair and tolerance of DNA damage caused by alkylating agents. Nat Rev Cancer 2012;12:104-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cahill DP, Levine KK, Betensky RA, et al. Loss of the mismatch repair protein MSH6 in human glioblastomas is associated with tumor progression during temozolomide treatment. Clin Cancer Res 2007;13:2038-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang J, Stevens MF, Laughton CA, et al. Acquired resistance to temozolomide in glioma cell lines: molecular mechanisms and potential translational applications. Oncology 2010;78:103-14. [DOI] [PubMed] [Google Scholar]
- 40.Curtin NJ, Wang LZ, Yiakouvaki A, et al. Novel poly(ADP-ribose) polymerase-1 inhibitor, AG14361, restores sensitivity to temozolomide in mismatch repair-deficient cells. Clin Cancer Res 2004;10:881-9. [DOI] [PubMed] [Google Scholar]
- 41.Trivedi RN, Almeida KH, Fornsaglio JL, et al. The role of base excision repair in the sensitivity and resistance to temozolomide-mediated cell death. Cancer Res 2005;65:6394-400. [DOI] [PubMed] [Google Scholar]
- 42.Johannessen TC, Bjerkvig R. Molecular mechanisms of temozolomide resistance in glioblastoma multiforme. Expert Rev Anticancer Ther 2012;12:635-42. [DOI] [PubMed] [Google Scholar]
- 43.Tang JB, Goellner EM, Wang XH, et al. Bioenergetic metabolites regulate base excision repair-dependent cell death in response to DNA damage. Mol Cancer Res 2010;8:67-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cho HY, Wang W, Jhaveri N, et al. NEO212, Temozolomide Conjugated to Perillyl Alcohol, Is a Novel Drug for Effective Treatment of a Broad Range of Temozolomide-Resistant Gliomas. Mol Cancer Ther 2014;13:2004-17. [DOI] [PubMed] [Google Scholar]
- 45.Ohgaki H, Kleihues P.The definition of primary and secondary glioblastoma. Clin Cancer Res 2013;19:764-72. [DOI] [PubMed] [Google Scholar]
- 46.Esmaeili M, Hamans BC, Navis AC, et al. IDH1 R132H Mutation Generates a Distinct Phospholipid Metabolite Profile in Glioma. Cancer Res 2014;74:4898-907. [DOI] [PubMed] [Google Scholar]
- 47.Houillier C, Wang X, Kaloshi G, et al. IDH1 or IDH2 mutations predict longer survival and response to temozolomide in low-grade gliomas. Neurology 2010;75:1560-6. [DOI] [PubMed] [Google Scholar]
- 48.Ohba S, Mukherjee J, See WL, et al. Mutant IDH1-Driven Cellular Transformation Increases RAD51-Mediated Homologous Recombination and Temozolomide Resistance. Cancer Res 2014;74:4836-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hatanpaa KJ, Burma S, Zhao D, et al. Epidermal growth factor receptor in glioma: signal transduction, neuropathology, imaging, and radioresistance. Neoplasia 2010;12:675-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Barkovich KJ, Hariono S, Garske AL, et al. Kinetics of inhibitor cycling underlie therapeutic disparities between EGFR-driven lung and brain cancers. Cancer Discov 2012;2:450-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Vivanco I, Robins HI, Rohle D, et al. Differential sensitivity of glioma- versus lung cancer-specific EGFR mutations to EGFR kinase inhibitors. Cancer Discov 2012;2:458-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sepúlveda JM, Zahonero C, Hernandez-Lain A, et al. Targeting EGFR in glioblastoma: Preclinical testing of dacomitinib. J Clin Oncol 2014;32:abstr e13015.
- 53.Li L, Chakraborty S, Yang CR, et al. An EGFR wild type-EGFRvIII-HB-EGF feed-forward loop regulates the activation of EGFRvIII. Oncogene 2014;33:4253-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Johns TG, Perera RM, Vernes SC, et al. The efficacy of epidermal growth factor receptor-specific antibodies against glioma xenografts is influenced by receptor levels, activation status, and heterodimerization. Clin Cancer Res 2007;13:1911-25. [DOI] [PubMed] [Google Scholar]
- 55.Fan QW, Cheng CK, Gustafson WC, et al. EGFR phosphorylates tumor-derived EGFRvIII driving STAT3/5 and progression in glioblastoma. Cancer Cell 2013;24:438-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Francis JM, Zhang CZ, Maire CL, et al. EGFR Variant Heterogeneity in Glioblastoma Resolved through Single-Nucleus Sequencing. Cancer Discov 2014;4:956-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gini B, Mischel PS. Greater Than the Sum of Its Parts: Single-Nucleus Sequencing Identifies Convergent Evolution of Independent EGFR Mutants in GBM. Cancer Discov 2014;4:876-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chakravarti A, Zhai G, Suzuki Y, et al. The prognostic significance of phosphatidylinositol 3-kinase pathway activation in human gliomas. J Clin Oncol 2004;22:1926-33. [DOI] [PubMed] [Google Scholar]
- 59.Guo D, Reinitz F, Youssef M, et al. An LXR Agonist Promotes Glioblastoma Cell Death through Inhibition of an EGFR/AKT/SREBP-1/LDLR-Dependent Pathway. Cancer Discov 2011;1:442-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bredel M, Bredel C, Juric D, et al. Tumor necrosis factor-alpha-induced protein 3 as a putative regulator of nuclear factor-kappaB-mediated resistance to O6-alkylating agents in human glioblastomas. J Clin Oncol 2006;24:274-87. [DOI] [PubMed] [Google Scholar]
- 61.Staudt LM. Oncogenic activation of NF-kappaB. Cold Spring Harb Perspect Biol 2010;2:a000109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bredel M, Scholtens DM, Yadav AK, et al. NFKBIA deletion in glioblastomas. N Engl J Med 2011;364:627-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Liang J, Nagahashi M, Kim EY, et al. Sphingosine-1-phosphate links persistent STAT3 activation, chronic intestinal inflammation, and development of colitis-associated cancer. Cancer Cell 2013;23:107-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Pyne NJ, Pyne S. Sphingosine 1-phosphate and cancer. Nat Rev Cancer 2010;10:489-503. [DOI] [PubMed] [Google Scholar]
- 65.Alvarez SE, Harikumar KB, Hait NC, et al. Sphingosine-1-phosphate is a missing cofactor for the E3 ubiquitin ligase TRAF2. Nature 2010;465:1084-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Pilorget A, Demeule M, Barakat S, et al. Modulation of P-glycoprotein function by sphingosine kinase-1 in brain endothelial cells. J Neurochem 2007;100:1203-10. [DOI] [PubMed] [Google Scholar]
- 67.Yang HS, Cho MH, Zakowicz H, et al. A novel function of the MA-3 domains in transformation and translation suppressor Pdcd4 is essential for its binding to eukaryotic translation initiation factor 4A. Mol Cell Biol 2004;24:3894-906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hwang SK, Baker AR, Young MR, et al. Tumor suppressor PDCD4 inhibits NF-kappaB-dependent transcription in human glioblastoma cells by direct interaction with p65. Carcinogenesis 2014;35:1469-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zanotto-Filho A, Braganhol E, Schroder R, et al. NFkappaB inhibitors induce cell death in glioblastomas. Biochem Pharmacol 2011;81:412-24. [DOI] [PubMed] [Google Scholar]
- 70.Sarkar D, Emdad L, Lee SG, et al. Astrocyte elevated gene-1: far more than just a gene regulated in astrocytes. Cancer Res 2009;69:8529-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Emdad L, Sarkar D, Lee SG, et al. Astrocyte elevated gene-1: a novel target for human glioma therapy. Mol Cancer Ther 2010;9:79-88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lee SG, Su ZZ, Emdad L, et al. Astrocyte elevated gene-1 (AEG-1) is a target gene of oncogenic Ha-ras requiring phosphatidylinositol 3-kinase and c-Myc. Proc Natl Acad Sci U S A 2006;103:17390-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Emdad L, Lee SG, Su ZZ, et al. Astrocyte elevated gene-1 (AEG-1) functions as an oncogene and regulates angiogenesis. Proc Natl Acad Sci U S A 2009;106:21300-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Lee SG, Kim K, Kegelman TP, et al. Oncogene AEG-1 promotes glioma-induced neurodegeneration by increasing glutamate excitotoxicity. Cancer Res 2011;71:6514-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Vajkoczy P, Knyazev P, Kunkel A, et al. Dominant-negative inhibition of the Axl receptor tyrosine kinase suppresses brain tumor cell growth and invasion and prolongs survival. Proc Natl Acad Sci U S A 2006;103:5799-804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Keating AK, Kim GK, Jones AE, et al. Inhibition of Mer and Axl receptor tyrosine kinases in astrocytoma cells leads to increased apoptosis and improved chemosensitivity. Mol Cancer Ther 2010;9:1298-307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ott M, Litzenburger UM, Sahm F, et al. Promotion of glioblastoma cell motility by enhancer of zeste homolog 2 (EZH2) is mediated by AXL receptor kinase. PloS One 2012;7:e47663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Orzan F, Pellegatta S, Poliani PL, et al. Enhancer of Zeste 2 (EZH2) is up-regulated in malignant gliomas and in glioma stem-like cells. Neuropathol Appl Neurobiol 2011;37:381-94. [DOI] [PubMed] [Google Scholar]
- 79.Kim E, Kim M, Woo DH, et al. Phosphorylation of EZH2 activates STAT3 signaling via STAT3 methylation and promotes tumorigenicity of glioblastoma stem-like cells. Cancer Cell 2013;23:839-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Charest A, Kheifets V, Park J, et al. Oncogenic targeting of an activated tyrosine kinase to the Golgi apparatus in a glioblastoma. Proc Natl Acad Sci U S A 2003;100:916-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Saborowski A, Saborowski M, Davare MA, et al. Mouse model of intrahepatic cholangiocarcinoma validates FIG-ROS as a potent fusion oncogene and therapeutic target. Proc Natl Acad Sci U S A 2013;110:19513-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Singh D, Chan JM, Zoppoli P, et al. Transforming fusions of FGFR and TACC genes in human glioblastoma. Science 2012;337:1231-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Zhu Z, Liu Y, Li K, et al. Protein tyrosine phosphatase receptor U (PTPRU) is required for glioma growth and motility. Carcinogenesis 2014;35:1901-10. [DOI] [PubMed] [Google Scholar]
- 84.Nikolaienko RM, Agyekum B, Bouyain S. Receptor protein tyrosine phosphatases and cancer: new insights from structural biology. Cell Adh Migr 2012;6:356-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Hatakeyama S.TRIM proteins and cancer. Nature reviews. Cancer 2011;11:792-804. [DOI] [PubMed] [Google Scholar]
- 86.El-Husseini AE, Fretier P, Vincent SR. Cloning and characterization of a gene (RNF22) encoding a novel brain expressed ring finger protein (BERP) that maps to human chromosome 11p15.5. Genomics 2001;71:363-7. [DOI] [PubMed] [Google Scholar]
- 87.Chen G, Kong J, Tucker-Burden C, et al. Human Brat Ortholog TRIM3 Is a Tumor Suppressor That Regulates Asymmetric Cell Division in Glioblastoma. Cancer Res 2014;74:4536-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Moore MA. A cancer fate in the hands of a samurai. Nat Med 2010;16:963-5. [DOI] [PubMed] [Google Scholar]
- 89.Nishimoto Y, Okano H.New insight into cancer therapeutics: induction of differentiation by regulating the Musashi/Numb/Notch pathway. Cell Res 2010;20:1083-5. [DOI] [PubMed] [Google Scholar]
- 90.Muto J, Imai T, Ogawa D, et al. RNA-binding protein Musashi1 modulates glioma cell growth through the post-transcriptional regulation of Notch and PI3 kinase/Akt signaling pathways. PloS One 2012;7:e33431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Kitambi SS, Toledo EM, Usoskin D, et al. Vulnerability of glioblastoma cells to catastrophic vacuolization and death induced by a small molecule. Cell 2014;157:313-28. [DOI] [PubMed] [Google Scholar]
- 92.Gilbertson RJ. Driving glioblastoma to drink. Cell 2014;157:289-90. [DOI] [PubMed] [Google Scholar]
- 93.Robinson MW, Overmeyer JH, Young AM, et al. Synthesis and evaluation of indole-based chalcones as inducers of methuosis, a novel type of nonapoptotic cell death. J Med Chem 2012;55:1940-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Fonkem E, Lun M, Wong ET. Rare phenomenon of extracranial metastasis of glioblastoma. J Clin Oncol 2011;29:4594-5. [DOI] [PubMed] [Google Scholar]
- 95.Müller C, Holtschmidt J, Auer M, et al. Hematogenous dissemination of glioblastoma multiforme. Sci Transl Med 2014;6:247ra101. [DOI] [PubMed]
- 96.Perryman L, Erler JT. Brain cancer spreads. Sci Transl Med 2014;6:247fs228. [DOI] [PubMed]