Abstract
Flowering time, the major regulatory transition of plant sequential development, is modulated by multiple endogenous and environmental factors. By phenotypic profiling of 80 early flowering mutants of Arabidopsis, we examine how mutational reduction of floral repression is associated with changes in phenotypic plasticity and stability. Flowering time measurements in mutants reveal deviations from the linear relationship between the number of leaves and number of days to bolting described for natural accessions and late flowering mutants. The deviations correspond to relative early bolting and relative late bolting phenotypes. Only a minority of mutants presents no detectable phenotypic variation. Mutants are characterized by a broad release of morphological pleiotropy under short days, with leaf characters being most variable. They also exhibit changes in phenotypic plasticity across environments for florigenic-related responses, including the reaction to light and dark, photoperiodic behavior, and Suc sensitivity. Morphological pleiotropy and plasticity modifications are differentially distributed among mutants, resulting in a large diversity of multiple phenotypic changes. The pleiotropic effects observed may indicate that floral repression defects are linked to global developmental perturbations. This first, to our knowledge, extensive characterization of phenotypic variation in early flowering mutants correlates with the reports that most factors recruited in floral repression at the molecular genetic level correspond to ubiquitous regulators. We discuss the importance of functional ubiquity for floral repression with respect to robustness and flexibility of network biological systems.
Mutational analyses have proved very useful to identify gene functions (Bouché and Bouchez, 2001; Alonso et al., 2003). In turn, the realization that gene functions are involved in reticulate networks of interactions contributed to the emergence of systems biology that is based on exhaustive, simultaneous biological descriptions (Katagiri, 2003). Network systems reveal emergent properties that cannot be predicted from the properties of isolated constituents but are specific of the interactive whole. In particular, the intricacy and flexibility of complex interactions indicates that gene functions are not only primary causal agents of specific processes but can also be recruited directly or indirectly in different processes of a system (Duboule and Wilkins, 1998; Greenspan, 2001). This functional versatility suggests that mutant phenotypes reflect not only specific functional effects but also distortions of wild-type network systems. Canalization or robustness, the capacity of network biological systems to buffer a wide variety of perturbations (Waddington, 1942; Rutherford, 2000; Debat and David, 2001; Siegal and Bergman, 2002) can explain why numerous silent mutations are uncovered in insertion mutagenesis analyses (Bouché and Bouchez, 2001). But mutants are usually less canalized than wild types, and most mutations can affect the expression of numerous unrelated genes and reveal multiple phenotypic variations (Waddington, 1942; Duboule and Wilkins, 1998; Rutherford, 2000; Wagner, 2000; Finnegan, 2001; Greenspan, 2001; Featherstone and Broadie, 2002; Bergman and Siegal, 2003). The degree of pleiotropy and distribution of phenotypic variation in mutants should thus help to characterize robustness and flexibility of network biological systems.
Both robustness and flexibility are crucial properties of network systems that allow developmental stability while enhancing coordinate, orderly dynamic variation necessary for growth and adjustment to the environment (Greenspan, 2001). Phenotypic plasticity, the mode of phenotypic variation that leads to a predictable and homogeneous subset of possible phenotypes for a given genotype in heterogeneous environments —also called its norm of reaction —is especially striking in plants due to their extended and sequential mode of development (Via et al., 1995; Pigliucci, 1996; Sultan, 2000; Debat and David, 2001). Flowering time, a major modulator of plant sequential development (Kuittinen et al., 1997), is influenced by a large number of environmental cues, including light quality and intensity, photoperiod, temperature, and nutrient availability (Garner and Allard, 1920; Bernier, 1988; Millar, 1999; Battey, 2000; Samach and Coupland, 2000). Physiological, biochemical, and genetic studies have shown that this plasticity relies on the adjustment between floral activation and repression processes and involves mobile florigenic and anti-florigenic signals (Lang et al., 1977; Bernier, 1988; Weller et al., 1997; Périlleux and Bernier, 2002). In Arabidopsis, the characterization of a large number of flowering time mutants and loci has led to models of the genetic regulation of flowering time (Koornneef et al., 1991; Mouradov et al., 2002; Périlleux and Bernier, 2002; Simpson and Dean, 2002). These models emphasize the interplay of multiple floral activation pathways while floral repression is still poorly integrated (Pouteau, 2001; Mouradov et al., 2002; Périlleux and Bernier, 2002; Simpson and Dean, 2002).
Mutant lines have been mostly used by developmental biologists for molecular genetic analyses, but the impact of mutations on phenotypic plasticity and pleiotropy has not been well studied (Bagnall, 1993; van Tienderen et al., 1996). Phenotypic plasticity has been mainly studied in the field of ecology and evolution and indirectly addressed in plant breeding in terms of genotype-environment interaction, by using natural accessions and recombinant inbred lines (RILs; Karlsson et al., 1993; Zhang and Lechowicz, 1994; Clarke et al., 1995; Stratton, 1998; Maloof et al., 2001). However, heterochronic mutants exhibiting shifts in developmental timing can prove powerful tools to investigate developmental networks at the organism level (Wiltshire et al., 1994; Diggle, 1999). In this work, we used an extensive mutational analysis to investigate plant phenotypic stability and plasticity at a whole organism level based on phenotypic profiling of early flowering mutants in Arabidopsis. We report the diverse pleiotropic effects associated with reduced floral repression, including variation in morphology and phenotypic plasticity, and discuss the possible role and specificity of floral repression in ontogenetic regulation of flowering time.
RESULTS
Altered Rates of Progression to Flowering
In an initial screen for early flowering mutants under short days (SD), 81 mutants in Ws and Col-0 ecotypes were selected for further analysis (Pouteau et al., 2001; see “Materials and Methods”). Flowering time was measured by using two indicators: the bolting date and the number of nodes bearing leaves. A wide range of variation is observed in the mutant population under SD in controlled growth cabinets (Fig. 1). While Ws produces about 34 rosette leaves and bolts after 7 weeks, flowering time varies between 5 and 28 rosette leaves and 3 to 7 weeks in eav T-DNA insertion mutants. The rate of progression to flowering (RPF) defined as the ratio between the number of nodes bearing leaves and the number of days to bolting is reduced in the 80 eav mutants compared to Ws and Col-0 wild types. No simple linear relation, i.e. relative RPF, can be found between wild types and the mutants (Fig. 1). Since temporal analyses in Arabidopsis indicated that the relative RPF is linear in different genotypes (Koornneef et al., 1991; Bagnall, 1993; Karlsson et al., 1993; Clarke et al., 1995; Kuittinen et al., 1997; Stratton, 1998), we used Ws and Col-0 RPF measurements to calculate a wild-type relative RPF and compared the mutants to this wild-type relative RPF. While a large proportion of the mutants appear to fit in the wild-type relative RPF, about one third of them can be classified as relative early bolting or relative late bolting genotypes (Fig. 1). This suggests that the two indicators of flowering time are not simply surrogates of each other but correspond to specific temporal components that can be uncoupled. In contrast to all eav mutants, ebv1 only bolts early but produces the same number of leaves as the wild type and is characterized by an increased RPF. Preliminary analysis suggests that this is due to acceleration of the rate of leaf initiation (data not shown), a phenotype also reported for the amp1 mutant under long days (LD; Chaudhury et al., 1993).
Figure 1.
Variation in the RPF in SD. Relation between the number of rosette leaves and the number of days to bolting for eav1 to eav61 T-DNA insertion mutants (blue diamonds), eav62 to eav80 EMS mutants (yellow diamonds), ebv1 (green square), and the Ws (red circles) and Col-0 (black circles) ecotypes. Independent repeats are presented (2 on average for eav1 to eav61 and 12 for Ws). sds are shown for Ws and Col-0 only for the sake of clarity. Thin line: linear regression for the population of T-DNA insertion mutants. Dotted red line: linear regression calculated with Ws and Col-0 (out of scale: average 66.2 rosette leaves and 61.3 d to bolting) scores. Area between black dotted lines: range of variation of wild-type relative RPF.
Diversity in Morphological Pleiotropy
To recover the widest range of phenotypic effects associated with early flowering, the only phenotypic selection applied in the initial screen, apart from flowering time, was based on fertility and uniformity of transmission. Strikingly, the mutants exhibit a large range of morphological modifications, such as changes in plant architecture, modified leaf morphology, or altered pigmentation (Fig. 2). An example of a highly pleiotropic phenotype was described for the lhp1-1 and lhp1-2 mutants, including dramatic changes in plant architecture and leaf morphology, as well as dwarfism (Gaudin et al., 2001).
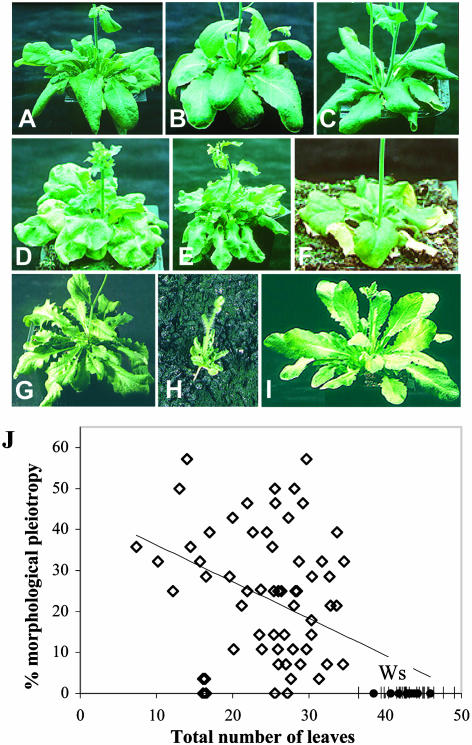
Figure 2.
Morphological pleiotropy of early flowering mutants in SD. A, Ws, rosette and base of the floral stem showing the first cauline leaf. B, eav2. C, eav6. D, eav43. E, eav1. F, eav15. G, eav49. H, eav40. I, ebv1. J, Variation of morphological pleiotropy (proportion of modified parameters in 28 variable morphological parameters) with flowering time in T-DNA insertion mutants (diamonds) and Ws (black circles). sds are shown for 12 Ws independent repeats. Thin line: linear regression.
The range of morphological variation in the mutants under SD was investigated by a phenomenological survey at different developmental stages. We identified 28 variable morphological parameters ranging from vegetative features (for example elongation of petiole and hypocotyl) to floral and inflorescence features (Fig. 3). Only four mutants have no macroscopic morphological modification, and the level of morphological pleiotropy per mutant (estimated as the percentage of modified morphological parameters) is 24.5% on average. A large diversity is observed in the distribution of the variation among mutants (Fig. 3A). Each level of morphological variation (0–16 modifications per mutant) is equally represented in the population of mutants, suggesting that the range of possible multiple morphological changes is not limited.
Figure 3.
Variability of morphological pleiotropy in early flowering mutants under SD. A, Distribution of morphological changes in eav1 to eav61 T-DNA insertion mutants. Empty and filled boxes indicate, respectively, the absence and presence of macroscopically detectable morphological changes. B, Frequency of changes in 28 morphological parameters. Elongation of hypocotyl (1) and petiole (2). Rosette size (3), raising (4), and pigmentation (5). Rosette leaf shape (6), serration (7), surface (8), and trichomes (9). Cauline leaf size (10), shape (11), serration (12), and surface (13). Flower size (14) and number (15). Perianth organ shape (16) and reproductive organ shape (17). Silique size (18), shape (19), and fertility (20). Floral stem elongation (21), length of first internode (22), length of other internodes (23), thickness (24), and appendage arrangement (25). Coflorescence elongation (26), number of secondary inflorescence (27), and secondary inflorescence elongation (28).
The 28 variable parameters are characterized by contrasting frequencies of variation in the mutant population. The variation level is highest for rosette pigmentation and rosette leaf shape (more than 60% of the mutants) and lowest for floral features, silique shape, and elongation of secondary inflorescences (less than 10% of the mutants; Fig. 3B). Changes in rosette leaf shape and pigmentation may be respectively associated with modifications in the leaf heteroblastic transition due to reduction in the number of leaves or temporal changes and perturbations either in photosynthesis and growth capacity or in light perception and signaling. The distribution of the level of morphological pleiotropy with flowering time also shows a wide range of variation among mutants (Fig. 2J). Most of the earliest mutants (less than 20 leaves) are highly pleiotropic while moderately early mutants (between 20 and 30 leaves) exhibit more variable levels of pleiotropy.
Genetic Diversity
Gene tagging analysis indicates that morphological pleiotropy and early flowering are genetically linked in the T-DNA insertion mutants. The average frequency of tagging by a T-DNA expressing kanamycin resistance is 31%. This suggests that each T1 parental line potentially bears three or four mutations. Yet, analyses of F2 and T2 populations for 52 T-DNA insertion mutants reveal no phenotypic segregation of the mutant features. This suggests that only one locus is affected in these mutants. The frequency of linkage is higher in T2 lines having two independent T-DNA insertion loci (55%) than in T2 lines having one single T-DNA insertion locus (28%). Multiple insertion may reflect a higher efficiency and lower rate of abortion of the T-DNA insertion process. Indeed, part of the nontagged mutants are probably due to point mutations caused by aborted insertion. In total 16 mutants show linkage between early flowering and a T-DNA insertion, and for 12 of them the genetic distance is less than 1%.
Despite potentially different underlying mechanisms, no significant difference is observed in the phenotypic behavior of semidominant (11) and recessive (49) T-DNA insertion mutants. Based on morphological similarities, alleles could only be detected for five complementation groups, each of them comprising no more than two or three alleles. Comparison between alleles reveals no significant difference in their phenotypic variation. Over 100 random crosses in a sample population of 24 mutants all resulted in complementation. This suggests that in the absence of morphological similarities allelism is rare and that many mutants in the collection are probably unique members of their corresponding complementation group. The large phenotypic diversity in the collection is thus reflected by a low level of genetic redundancy. Because of this lack of redundancy, it is likely that the collection is not saturated and that the number of loci associated with floral repression is high.
Variation in Phenotypic Plasticity
To further characterize the phenotypic pleiotropy of early flowering mutants, their norms of reaction were assayed in response to environmental constraints having a florigenic influence. Two levels of the regulation of flowering time by photoperiod were investigated (Millar, 1999; Samach and Coupland, 2000). The input level corresponding to light/dark perception by photoreceptors and signaling was indirectly approached by measuring hypocotyl elongation, a classic example of developmental plasticity (Gendreau et al., 1997). The output level corresponding to the emission of rhythms by the circadian clock was analyzed through directly assessing flowering time in response to photoperiod (Samach and Coupland, 2000). Hormonal and metabolic signaling of flowering involves GAs and Suc (Bernier, 1988; Périlleux and Bernier, 2002). This was indirectly addressed by measuring reactions to paclobutrazol (an inhibitor of GA biosynthesis) and to a high concentration of Suc, two conditions that have proved successful for the isolation of signaling mutants (Bethke and Russell, 1998; Gibson, 2000).
Comparison between the two genetic backgrounds shows that Ws is more sensitive to paclobutrazol than Col-0. Ws germination is fully inhibited at a concentration of 3 × 10−5 m while Col-0 still exhibits residual resistance at a concentration of 3 × 10−4 m (1.65% ± 1.23%). Mutants in the two genetic backgrounds also show a differential response to paclobutrazol. No resistance is detected for 61 mutants in the Ws background, while 3 out of 13 mutants in the Col-0 background exhibit a significant decrease in sensitivity (data not shown). The different flexibility of the response to paclobutrazol in the two ecotypes may indicate that mutational effects on phenotypic variation are potentially bound to parental limitations. This may be tentatively explained by the fact that Ws is genetically constrained by a defect in phytochrome D and is an early flowering mutant itself (Auckerman et al., 1997). For the three other environmental factors analyzed, we observed similar levels of phenotypic plasticity and a similar range of mutant variation in both ecotypes and only 20% of the mutant population have no or little changes in phenotypic plasticity.
Norms of Reaction to Photoperiod
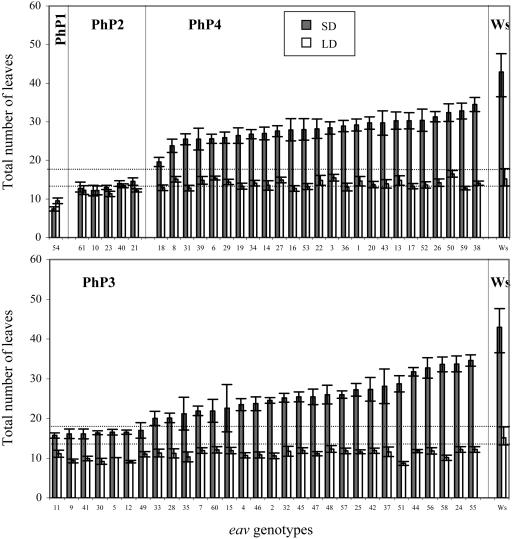
Arabidopsis is a quantitative LD species that flowers later in SD than in LD. By comparing flowering time in LD and SD in the T-DNA insertion mutants and Ws, we identified four classes of response to photoperiod: class PhP1, flowering earlier in SD than in LD; class PhP2, photoperiod-insensitive early flowering; class PhP3, early flowering under both SD and LD; and class PhP4, early flowering specifically under SD (Fig. 4). All ethyl methanesulfonate (EMS) mutants fall into classes PhP3 and PhP4 (data not shown). A photoperiod sensitivity index (PSI) was determined as the ratio between the total number of leaves in SD and the total number of leaves in LD. Although a majority of mutants have a PSI lower than the wild type (between 0.8 and 2.2 compared to 2.8 for Ws), for about 20% of them (mostly in class PhP3) a wild-type PSI is observed.
Figure 4.
Response to photoperiod. Flowering time under SD (shaded bars) and LD (white bars) in eav T-DNA insertion mutants (numbered from 1–61) grouped in four different classes based on t test scores: PhP1 (later in LD than in SD), PhP2 (insensitive to photoperiod), PhP3 (early in SD and in LD), PhP4 (early specifically in SD). The Ws control is shown on both rows (average of 12 and 9 repeats in SD and LD, respectively). sds are indicated.
Norms of Reaction to Light and Dark
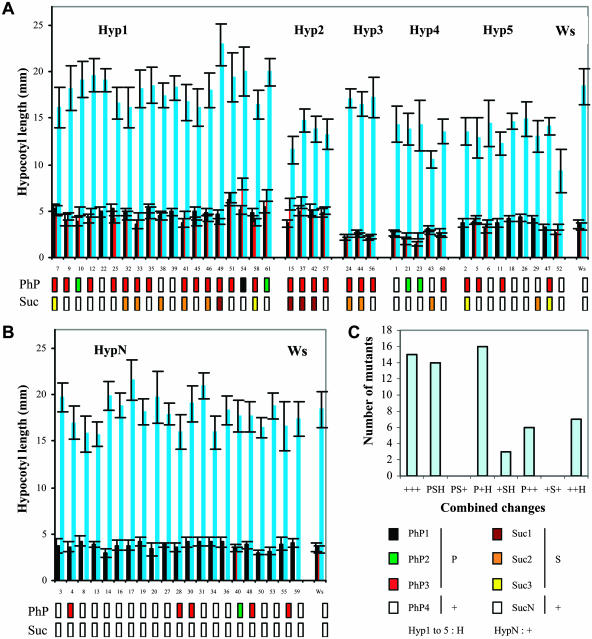
Hypocotyl elongation was determined in classical dark and LD light conditions and, for a number of mutants, in SD light condition. In Ws and Col-0 wild types, hypocotyl elongation is repressed in the light and is strongly activated in the dark (Gendreau et al., 1997). For Ws, no significant difference is observed in the two light conditions, but the kinetics of hypocotyl elongation is slower in SD than in LD (data not shown). For about two-thirds of the T-DNA insertion mutants, hypocotyl elongation is significantly different from the wild type in at least one of the conditions tested. However, only seven mutants are affected in both light and dark conditions, and six mutants exhibit changes only in SD but not in LD. Six classes of response to light and dark based on hypocotyl elongation can be identified: class Hyp1, long in light; class Hyp2, long in light and short in dark; class Hyp3, short in light; class Hyp4, short in light and in dark; class Hyp5, short in dark; and class HypN, similar to the wild type (Fig. 5). Similar results were obtained in EMS mutants: among 10 mutants analyzed, only one is similar to the wild type and two are affected in both light and dark conditions (data not shown).
Figure 5.
Norms of reaction to light and dark, photoperiod, and Suc. A–B, Hypocotyl elongation in LD (black bars), in SD (red bars) and in the dark (blue bars) in eav T-DNA insertion mutants (numbered from 1–61) and in Ws (average of 17 repeats in LD, 8 in SD, and 12 in the dark). Error bars correspond to sds. A, Classes of mutants differing from Ws in the light (Hyp1, Hyp3) or in the dark (Hyp5), or both (Hyp2, Hyp4) based on t test scores. B, Mutants similar to Ws in the light and in the dark (HypN). Corresponding reactions to photoperiod (classes PhP1 to 4) or to 6% Suc (classes Suc1 to N) are indicated. C, Combined changes in photoperiod response (P), Suc sensitivity (S), and hypocotyl elongation (H), or absence of changes (+).
Norms of Reaction to Suc
For Ws and Col-0, the presence of 6% Suc delays germination and early development but subsequent development is little affected. The mutant behavior on 0% Suc is not significantly different from the wild type. We identified four classes of sensitivity to 6% Suc in the T-DNA insertion mutants (Fig. 5; see “Materials and Methods”). Class Suc1 exhibits a high level of early growth arrest (up to 64%) so that only a small proportion of seedlings develop into adult plants, a marked accumulation of anthocyanin and epinasty. For class Suc2, early growth arrest is limited and a majority of seedlings develop into adult plants characterized by a high level of shoot and leaf distortion and anthocyanin accumulation. Finally, class Suc3 shows only weak or variable germination or developmental defects, and class SucN is similar to the wild type.
Multiple Changes in Phenotypic Plasticity
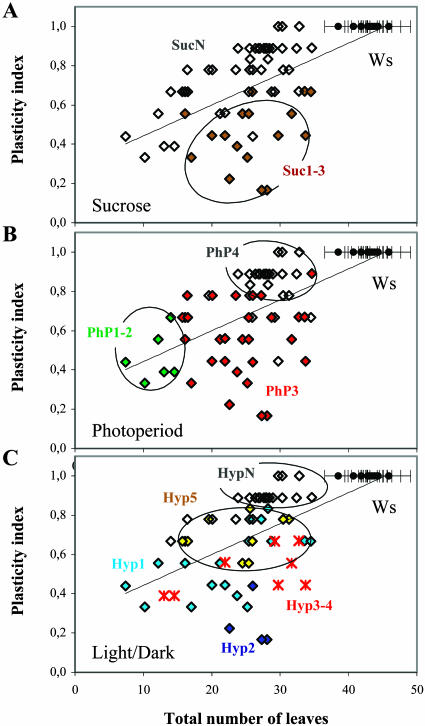
The mutants are characterized by a large diversity in the distribution of the different norms of reaction (Fig. 5, A and B). Multiple changes in response to photoperiod (P), Suc hypersensitivity (S), and hypocotyl elongation (H) are more frequent than single changes (Fig. 5C). To estimate the distribution of phenotypic plasticity with flowering time, P, S, and H phenotypic indices were calculated and used to determine a global plasticity index Pi (0.66 on average in the mutant population). The indices appear differentially distributed in the different categories of mutants (Table I; Fig. 6). Despite a wide range of variation among the mutants, Pi tends to be lower with earlier flowering time, and some categories of mutants localize to different sectors of this distribution.
Table I.
P, S, and H phenotypic indices in the plasticity classes
| Class | Number of Mutants | P Index | S Index | H Index |
|---|---|---|---|---|
| PhP1-2 | 6 | 3.00 ± 0.00 | 0.00 ± 0.00 | 1.83 ± 1.13 |
| PhP3 | 30 | 1.77 ± 0.43 | 0.90 ± 1.13 | 1.28 ± 0.82 |
| PhP4 | 25 | 0.84 ± 0.37 | 0.16 ± 0.47 | 0.48 ± 0.70 |
| Suc1-3 | 17 | 1.59 ± 0.62 | 1.82 ± 0.81 | 1.56 ± 0.58 |
| SucN | 44 | 1.48 ± 0.82 | 0.00 ± 0.00 | 0.80 ± 0.94 |
| Hyp1–4 | 31 | 1.77 ± 0.76 | 0.90 ± 1.14 | 1.71 ± 0.68 |
| Hyp5 | 9 | 1.44 ± 0.53 | 0.33 ± 0.50 | 0.94 ± 0.17 |
| HypN | 21 | 1.14 ± 1.14 | 0.00 ± 0.00 | 0.00 ± 0.00 |
Figure 6.
Variation of phenotypic plasticity with flowering time in eav1 to eav61 T-DNA insertion mutants (diamonds) and Ws (black circles), showing the tendency of the different norms of reaction. A, Response to 6% Suc, classes Suc1 to 3 (brown diamonds), class SucN (white diamonds). B, Response to photoperiod, classes PhP1 to 2 (green diamonds), class PhP3 (red diamonds), class PhP4 (white diamonds). C, Hypocotyl elongation in response to light and dark, class Hyp1 (blue diamonds), class Hyp2 (indigo diamonds), classes Hyp3 to 4 (red crosses), class Hyp5 (yellow diamonds), class HypN (white diamonds). Plasticity index (Pi; see “Materials and Methods”). Thin line: linear regression. sds are shown for 12 Ws independent repeats.
In most cases, S is associated with P and H together, suggesting that it reflects global developmental perturbation (Fig. 5C). Accordingly, classes Suc1-3 comprising the relative late bolting subpopulation are characterized by a low Pi. Because of their moderately early flowering time, they account for the main deviation from a putative linear relation between flowering time and plasticity in the mutant population (Fig. 6A). S is most often combined with class PhP3 and classes Hyp1 to Hyp4 (14 out of 17 mutants in both cases). The H index decreases from class Suc1 to SucN, supporting the possibility of a positive correlation between H and S. In contrast, the P index is not significantly different in the different Suc classes, suggesting that the correlation between P and S is indirect (Table I; data not shown).
P and H are combined in 50% of the mutants (Fig. 5C). A positive correlation between P and H is suggested by the fact that the H index decreases from class PhP1 to PhP4 and that the P index is higher in classes Hyp1 to Hyp4 and intermediate in class Hyp5 compared to class HypN (Table I). The variation in P and H indices is reflected in the distribution of Pi variation with flowering time (Fig. 6, B and C). Classes PhP1-2 and class PhP4 correspond to two separate clusters comprising, respectively, the earliest mutants with a low Pi and most of the least precocious mutants with highest Pi. In contrast, class PhP3 and most Hyp classes do not localize to a specific sector. However, most mutants in classes Hyp1 to Hyp4 have a low Pi, whereas class Hyp5 exhibits an intermediate Pi and is clustered near class HypN characterized by the highest Pi.
According to a mechanistic understanding of photoperiodism, P and H alterations can reveal a reduced capacity to integrate environmental cues at the input level, including light photoreception and signaling, or at the oscillator or output levels due to clock dysfunction (Samach and Coupland, 2000). Indeed, defects in clock functions such as ELF3 have been identified among these mutants (Hicks et al., 2001; I. Carré, V. Gaudin, and S. Pouteau, unpublished data). Specific light quality, such as red or blue light, may be needed for the detection of more subtle changes in mutants with a P phenotype but regular hypocotyl elongation in white light (classes Hyp5 and HypN). But other developmental defects are also involved. For example, reduction in hypocotyl elongation in lhp1-1 and lhp1-2 (eav21 and eav23), two Hyp3 mutants, correlates with reduced cell size and dwarfism (Gaudin et al., 2001), and Hyp2 mutants that are also hypersensitive to Suc may be defective in sugar metabolism or signaling. Finally, the Hyp5 phenotype also points to the possible contribution of photomorphogenesis in the dark or dark-signaling to the regulation of flowering time. In addition, Hyp5, like HypN, is mostly independent from P and S phenotypes and is associated with moderate levels of morphological pleiotropy and a wild-type relative RPF.
DISCUSSION
Based on the first, to our knowledge, extensive characterization of early flowering mutants in Arabidopsis, our work provides new perspectives on the regulation of floral repression in relation to phenotypic variation. First, early flowering mutants exhibit modifications in the coordination of two temporal components of plant ontogeny: an exogenous component corresponding to environmental timing measured by the date of flowering, and an endogenous component related to organismic timing estimated by the number of leaves. The uncoupling of these two components results in relative early or late bolting phenotypes. In contrast, most previous analyses conducted with late flowering mutants or natural accessions and RILs indicated a strong correlation between the date of flowering and the number of leaves (Koornneef et al., 1991; Bagnall, 1993; Karlsson et al., 1993; Clarke et al., 1995; Kuittinen et al., 1997; Stratton, 1998). The difference in the behavior of early and late flowering mutants may be explained by the fact that the latter had been selected for a lack of pleiotropic effects (M. Koornneef, personal communication). A comparison of the early flowering mutants with late flowering mutants not submitted to a phenotypic selective bias would thus be interesting. Because the relative late bolting phenotypes are most affected, it is possible that uncoupling of the two temporal dimensions in late flowering mutants would also be associated with profound disruptions of developmental stability. However, the high level of phenotypic variation in early flowering mutants is probably not coincidental with the type of selection applied but reflects endogenous constraints imposed by advanced ontogenetic phase change (see below). In addition, the uncoupling of endogenous and exogenous temporal components in relative early bolting phenotypes could reflect a slower rate of leaf initiation at early stages (Groot and Meicenheimer, 2000). Additional growth rate measurements would be needed to discriminate between different possible modes of variation in flowering time (Wiltshire et al., 1994; Diggle, 1999).
Second, early flowering mutants exhibit extensive phenotypic variation. This is characterized by a release of morphological pleiotropy under SD and a decrease in phenotypic plasticity across environments resulting in multiple variation profiles. Among the 28 variable morphological parameters identified, leaf characters are most often affected. The lower level of variation observed for floral features may be coincidental with the selection for fertility applied during the mutant screen. But this may also reflect their higher robustness against variation due to their more determinate character. Conversely, vegetative features may be more variable because of their higher flexibility as reflected by heteroblasty, i.e. the gradual change in leaf morphology exhibited by regular ontogenetic sequences (Diggle, 1999). Within a large diversity of alterations in response to light and dark, photoperiod, and Suc, hypersensitivity to Suc coincides with more global developmental defects possibly due to osmotic, metabolic, or signaling changes. Accordingly, many sugar response mutants previously described are also pleiotropic and show altered phytohormone response, metabolism, or osmotolerance (Gibson, 2000). In contrast, most mutants with altered reactions to the dark, unlike those affected in the light, show only moderate developmental defects. This coincides with the finding that in natural populations, hypocotyl elongation in the light but not in the dark correlates with latitude and supports the notion that phenotypic plasticity profiles have different adaptive implications (Via et al., 1995; Maloof et al., 2001). The multiple variation profiles observed may indicate that the functional integrity of developmental network systems is compromised in early flowering mutants. This may prove instrumental to analyze how patterns of phenotypic covariation commonly observed for many morphological and life history traits (Armbruster and Schwaergerle, 1996) are conserved or disrupted by mutations imposing reduced floral repression.
Third, the high level of pleiotropy in early flowering mutants emphasizes the importance of functional ubiquity for the interpretation of floral repression at the molecular genetic level. Indeed, floral repression seems to have recruited more ubiquitous factors than floral activation, including members of the MADS-box gene superfamily that act through complex multimeric protein interactions and epigenetic regulators (Goodrich et al., 1997; Michaels and Amasino, 1999; Hartmann et al., 2000; Finnegan, 2001; Gaudin et al., 2001; Gendall et al., 2001; Kinoshita et al., 2001; Pouteau, 2001; Scortecci et al., 2001; Yoshida et al., 2001; Mouradov et al., 2002; Noh and Amasino, 2003; Piñeiro et al., 2003; Ratcliffe et al., 2003; Sung et al., 2003). In addition to their intricate, interactive mode of action, most of these factors are also characterized by ubiquitous RNA expression. Among the floral repression-defective mutants analyzed in this work, 12 proved to be tagged by a T-DNA insertion. Genetic or molecular characterization of five mutants revealed defects in ubiquitous functions, including chromatin remodeling and circadian clock regulation such as LHP1 and ELF3 (Gaudin et al., 2001; Pouteau et al., 2001; I. Carré, V. Gaudin, and S. Pouteau, unpublished data). In contrast, the seemingly more limited pleiotropy level in floral-activation-defective mutants coincides with the finding that many floral activators are involved in redundant pathways and correspond to more specific transcription factors (Mouradov et al., 2002; Simpson and Dean, 2002). These contrasting features suggest that floral activation and repression operate through different biological processes. While redundancy, in addition to interaction between unrelated genes, is often thought to be a common mode of canalization (Rutherford, 2000; Wagner, 2000; Featherstone and Broadie, 2002), functional ubiquity offers a global basis for different processes to interrelate with each other and undergo coordinated changes without compromising developmental network stability. Accordingly, loci associated with changes in floral activation and repression may contribute respectively to developmental canalization and flexibility.
By integrating a growing number and complexity of regulatory factors, the current genetic models have substantially contributed to advancing our understanding of flowering time in Arabidopsis. More recently attempts have been made to include some repressors in regulatory networks, for example FLC and EFS (Michaels and Amasino, 1999; Sheldon et al., 1999; Soppe et al., 1999; Mouradov et al., 2002). Despite these efforts, Arabidopsis models are still overall dominated by a floral activation rationale. One reason may be that analyses and interpretations are mostly conducted on a deterministic linear basis that is suited for simple chains of reaction but not for complex reticulate networks and ubiquitous processes. By showing differential relationships between flowering time and multiple variation profiles in early flowering mutants, our results point to the need to develop nonlinear dynamic approaches to integrate floral repression and account for functional ubiquity (Pigliucci, 1996; Amzallag, 2001). Further multidimensional analyses will be needed to analyze the biological significance of the typology presented in this work and the possible reticulate correlations between multiple morphological and plastic features and flowering time.
MATERIALS AND METHODS
Plant Material
T-DNA insertion lines of Wassilewskija (Ws) ecotype were obtained from the Versailles collection, INRA, France (Bechtold et al., 1993). EMS mutagenized lines of Columbia (Col-0) ecotype were provided by Catherine Bellini, INRA, Versailles, France. A total of 7,653 T-DNA insertion lines and 384 T2 EMS mutagenized lines were grown in SD, and plants that flowered early were selected and selfed. The progenies were sown on soil in individual pots and grown in growth chamber under SD of 8 h light and 16 h dark under mixed white and incandescent light at 20°C and 70% relative humidity. Progenies that showed uniform early flowering were selected, backcrossed, and outcrossed by standard genetic techniques (Pouteau et al., 2001). The 62 T-DNA insertion mutants and 19 EMS mutants obtained were named eav1 to eav80 for early flowering from versailles and ebv1 for early bolting from versailles. The eav21 and eav23 mutants have been recently renamed respectively lhp1-1 and lhp1-2 since the two alleles proved to be altered in LHP1, a homolog of heterochromatin protein 1 (HP1), a member of the chromo domain protein family in Drosophila melanogaster (Gaudin et al., 2001).
Growth Conditions for Flowering Time Assays
Mutant seeds were sown on soil (Stender A240, Blumenerdenwerk Stender, Schermbeck, Germany) and grown in Sanyo Gallenkamp SGC660 growth cabinets at 20 ± 0.2°C and 70% ± 2% relative humidity. The soil was kept moist by application of nutrient solution three times a week. The light was provided by mixed fluorescent and incandescent tubes and the photon flux density (PFD) measured at soil level was 230 ± 20 μE m−2 s−1 and 2 ± 0.2 μE m−2 s−1, respectively. SD corresponded to 8 h light and 16 h dark, and LD conditions consisted of 16 h light and 8 h dark. Developmental uniformity was obtained by selecting the 10 most uniform plants on average about 12 d after sowing, bringing the plant density to one plant per pot, and rotating the trays three times a week. Bolting time was measured as the number of days from sowing to the first elongation of the floral stem at 0.1 cm height. The number of true leaves produced by the apical meristem was recorded on bolted plants. No major variation was observed in two to four independent repeats (12 repeats in SD and 9 in LD for Ws).
Evaluation of the Morphological Pleiotropy under SD
Repeated characterizations of mutant phenotypes under SD in successive generations obtained from the initial screen and in genetic analyses were further investigated in an exhaustive survey. Four individuals per genotype were selected for developmental uniformity and grown in parallel in the same growth room in SD. Aerial morphological features (vegetative, floral, inflorescence, and plant architecture) were examined after 4 and 10 weeks. Apart from floral features that were observed under a dissection microscope, all parameters were macroscopically recorded. Size or quantity parameters were semiquantified by recording the corresponding ranges of variation for each genotype compared to the wild type. The other parameters such as fertility or leaf surface wrinkling were visually assessed on a qualitative basis.
Growth Conditions in Vitro
Mutant seeds were sterilized and sown on petri dishes containing a standard nutrient medium as described by Santoni et al. (1994). Uniform germination was obtained by allowing the seeds to imbibe at 4°C for 48 h and exposing the plates to white light at 200 μE m−2 s−1 for 4 h before transfer to a growth chamber. Standard growth conditions corresponded to LD of 16 h fluorescent light at a PFD of 200 μE m−2 s−1 and 20°C and 8 h dark at 15°C. For dark growth conditions, the plates were placed in opaque bags under LD conditions. SD conditions consisted of an 8-h light period at 20°C and a 16-h night at 15°C.
Phenotypic Plasticity Assays in Vitro
For hypocotyl length measurements, seedlings were grown on standard nutrient medium without Suc during 10 d for LD and dark conditions according to Gendreau et al. (1997) and during 15 d for SD conditions (D. Lefebvre, S. Pouteau, unpublished data). Seedling spreading, camera recording, and image analysis with Optimas 6.1 software (Bioscan, Imasys, Suresnes, France) was as described by Gendreau et al. (1997). An average of 40 uniform seedlings was analyzed per genotype and per repeat. No major variation was observed in two to four independent repeats (17 repeats in LD, 8 in SD, and 12 in the dark for the wild type).
For Suc sensitivity measurements, seedlings were grown on standard nutrient medium without Suc or with 6% (w/v) Suc. Germination and early development were recorded at different times during a period of 3 to 17 d of growth. Early development was divided into three stages: root emergence (stage 1); shoot emergence (stage 2); and cotyledon expansion and greening (stage 3). Later developmental stages, recorded as stage 3, were examined for morphological features in the initial population and in a subpopulation transferred to fresh medium after 10 to 15 d of growth. The sensitivity of the mutants to 6% Suc was determined by measuring the following parameters: inhibition of germination, early growth arrest (at stage 1 or stage 2), developmental delay (later transition from stage 1 to stage 2 and stage 3), morphological features including shoot or leaf distortion, anthocyanin accumulation, epinasty, and root phenotype. An average of 80 to 100 seeds was analyzed per genotype and per repeat. A consistent behavior was observed in two to four independent repeats performed for 34 mutants comprising all mutants exhibiting sensitivity to 6% Suc (10 repeats for the wild type).
For paclobutrazol resistance assays, an average of 80 to 100 seeds was sown per genotype and per repeat on a 0.7% agar medium containing paclobutrazol (Sopra s.a., Zeneca Agrochemicals, Vélizy-Villacoublay, France) and the percentage of germination was scored after 7 to 10 d in LD. Mutant seeds in Ws and Col-0 backgrounds were assayed at a concentration of 10−5 to 3 × 10−5 m and 3 × 10−4 m paclobutrazol, respectively. Ws and Col-0 responses were assayed in five and four independent repeats, respectively.
Evaluation of Phenotypic and Plasticity Indices
The PSI was determined as the ratio between the total number of leaves in SD and the total number of leaves in LD.
To estimate the deviations of the mutants from the corresponding wild-type plasticity response to photoperiod, Suc, and light/dark, P, S, and H phenotypic indices were calculated. Each index was based on three criteria analyzed: (P1) early in LD, (P2) reduced PSI, (P3) photoperiod insensitive (PSI approximately 1); (S1) morphological modifications and/or developmental delay, (S2) moderate inhibition of development (10%–30% early growth arrest), (S3) strong inhibition of development (50%–65% early growth arrest); and (H1) modification in LD, (H2) modification in SD, (H3) modification in the dark. Each criterion was given a value of 0 or 1 depending on the absence or presence of a significant difference with the wild type based on t test scores.
The P, S, and H phenotypic indices were used to calculate a Pi plasticity index: Pi = [9 − (P + S + H)]/9.
Acknowledgments
We thank Hervé Ferry, Jean-Marie Pollien, and Joël Talbotec for technical assistance with plant culture and glasshouse work and Benoît Lacroix, Virginie Léon, and Cécile Marchenay for their help as trainee students. We are thankful to Nicole Bechtold, Georges Pelletier, and Roger Voisin for providing T-DNA insertion lines of the Versailles collection and to Catherine Bellini for the gift of EMS mutagenized lines. We are grateful to Yves Chupeau for his support and to Nissim Amzallag, Sylvie Dinant, Jean-Louis Durand, Herman Höfte, Jan Traas, Hervé Vaucheret, and Johannes Wirz for comments on this manuscript.
This work, V.F., and F.P. were supported by the European Union (grant no. BI04–CT97–2340). G.Z. received a Chinese Government fellowship.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.104.039453.
References
- Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen H, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk R (2003) Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301: 653–657 [DOI] [PubMed] [Google Scholar]
- Amzallag GN (2001) Data analysis in plant physiology: are we missing the reality? Plant Cell Env 24: 881–890 [Google Scholar]
- Armbruster WS, Schwaergerle KE (1996) Causes of covariation of phenotypic traits among populations. J Evol Biol 9: 261–276 [Google Scholar]
- Auckerman MJ, Hirschfeld M, Wester L, Weaver M, Clack T, Amasino RM, Sharrock RA (1997) A deletion in the PHYD gene of the Arabidopsis Wassilewskija ecotype defines a role for phytochrome D in red/far-red light sensing. Plant Cell 9: 1317–1326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagnall DJ (1993) Light quality and vernalization interact in controlling late flowering in Arabidopsis ecotypes and mutants. Ann Bot (Lond) 71: 75–83 [Google Scholar]
- Battey NH (2000) Aspects of seasonality. J Exp Bot 51: 1769–1780 [DOI] [PubMed] [Google Scholar]
- Bechtold N, Ellis J, Pelletier G (1993) In planta Agrobacterium mediated gene transfer by infiltration of adult Arabidopsis thaliana plants. CR Aca Sci Ser III Sci Vie 316: 1194–1199 [Google Scholar]
- Bergman A, Siegal ML (2003) Evolutionary capacitance as a general feature of complex gene networks. Nature 424: 549–552 [DOI] [PubMed] [Google Scholar]
- Bernier G (1988) The control of floral evocation and morphogenesis. Annu Rev Plant Physiol Plant Mol Biol 39: 175–219 [Google Scholar]
- Bethke PC, Russell LJ (1998) Gibberellin signaling. Curr Opin Plant Biol 1: 440–446 [DOI] [PubMed] [Google Scholar]
- Bouché N, Bouchez D (2001) Arabidopsis gene knockout: phenotypes wanted. Curr Opin Plant Biol 4: 111–117 [DOI] [PubMed] [Google Scholar]
- Chaudhury AM, Letham S, Craig S, Dennis ES (1993) amp1 - a mutant with high cytokinin levels and altered embryonic pattern, faster vegetative growth, constitutive photomorphogenesis and precocious flowering. Plant J 4: 907–916 [Google Scholar]
- Clarke JH, Mithen R, Brown JKM, Dean C (1995) QTL analysis of flowering time in Arabidopsis thaliana. Mol Gen Genet 248: 278–286 [DOI] [PubMed] [Google Scholar]
- Debat V, David P (2001) Mapping phenotypes: canalization, plasticity and developmental stability. Trends Ecol Evol 16: 555–561 [Google Scholar]
- Diggle CS (1999) Heteroblasty and the evolution of flowering phenologies. Int J Plant Sci 160: S123–S134 [DOI] [PubMed] [Google Scholar]
- Duboule D, Wilkins AS (1998) The evolution of ‘bricolage’. Trends Genet 14: 54–59 [DOI] [PubMed] [Google Scholar]
- Featherstone DE, Broadie K (2002) Wrestling with pleiotropy: genomic and topological analysis of the yeast gene expression network. Bioessays 24: 267–274 [DOI] [PubMed] [Google Scholar]
- Finnegan EJ (2001) Is plant gene expression regulated globally? Trends Genet 17: 361–365 [DOI] [PubMed] [Google Scholar]
- Garner WW, Allard HA (1920) Effect on the relative length of day and night and other factors of the environment on growth and reproduction in plants. J Agric Res 18: 553–606 [Google Scholar]
- Gaudin V, Libault M, Pouteau S, Juul T, Zhao G, Lefebvre D, Grandjean O (2001) Mutations in LIKE HETEROCHROMATIN PROTEIN 1 affect flowering time and plant architecture in Arabidopsis. Development 128: 4847–4848 [DOI] [PubMed] [Google Scholar]
- Gendall AR, Levy YY, Wilson A, Dean C (2001) The VERNALIZATION 2 gene mediates the epigenetic regulation of vernalization in Arabidopsis. Cell 107: 525–535 [DOI] [PubMed] [Google Scholar]
- Gendreau E, Traas J, Desnos T, Grandjean O, Caboche M, Höfte H (1997) Cellular basis of hypocotyl growth in Arabidopsis thaliana. Plant Physiol 114: 295–305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson SI (2000) Plant sugar-response pathways. Part of a complex regulatory web. Plant Physiol 124: 1532–1539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodrich J, Puangsomlee P, Martin M, Long D, Meyerowitz EM, Coupland G (1997) A polycomb group gene regulates homeotic gene expression in Arabidopsis. Nature 386: 44–51 [DOI] [PubMed] [Google Scholar]
- Greenspan RJ (2001) The flexible genome. Nat Rev Genet 2: 383–387 [DOI] [PubMed] [Google Scholar]
- Groot EP, Meicenheimer RD (2000) Short-day-grown Arabidopsis thaliana satisfies the assumptions of the plastochron index as a time variable in development. Int J Plant Sci 161: 749–756 [Google Scholar]
- Hartmann U, Höhmann S, Nettesheim K, Wisman E, Saedler H, Huijser P (2000) Molecular cloning of SVP: a negative regulator of the floral transition in Arabidopsis. Plant J 21: 351–360 [DOI] [PubMed] [Google Scholar]
- Hicks KA, Albertson TM, Wagner DR (2001) EARLY FLOWERING3 encodes a novel protein that regulates circadian clock function and flowering in Arabidopsis. Plant Cell 13: 1281–1292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlsson BH, Sills GR, Nienhuis J (1993) Effects of photoperiod and vernalization on the number of leaves at flowering in 32 Arabidopsis thaliana (Brassicaceae) ecotypes. Am J Bot 80: 646–648 [Google Scholar]
- Katagiri F (2003) Attacking complex problems with the power of systems biology. Plant Physiol 132: 417–419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinoshita T, Harada JJ, Goldberg RB, Fischer RL (2001) Polycomb repression of flowering during early plant development. Proc Natl Acad Sci USA 98: 14154–14161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koornneef M, Hanhart CJ, van der Veen JH (1991) A genetic and physiological analysis of late flowering mutants in Arabidopsis thaliana. Mol Gen Genet 229: 57–66 [DOI] [PubMed] [Google Scholar]
- Kuittinen H, Sillanpää MJ, Savolainen O (1997) Genetic basis of adaptation: flowering time in Arabidopsis thaliana. Theor Appl Genet 95: 573–583 [Google Scholar]
- Lang A, Chailakhyan MK, Frolova IA (1977) Promotion and inhibition of flower formation in a dayneutral plant in grafts with a short-day plant and long-day plant. Proc Natl Acad Sci USA 74: 2412–2416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maloof JN, Borevitz JO, Dabi T, Lutes J, Nehring RB, Redfern JL, Trainer GT, Wilson JM, Asami T, Berry CC, Weigel D, Chory J (2001) Natural variation in light sensitivity of Arabidopsis. Nat Genet 29: 441–446 [DOI] [PubMed] [Google Scholar]
- Michaels SD, Amasino RM (1999) FLOWERING LOCUS C encodes a novel MADS domain protein that acts as a repressor of flowering. Plant Cell 11: 949–956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millar AJ (1999) Biological clocks in Arabidopsis thaliana. New Phytol 14: 175–197 [DOI] [PubMed] [Google Scholar]
- Mouradov A, Cremer F, Coupland G (2002) Control of flowering time: interacting pathways as a basis for diversity. Plant Cell 14: S111–S130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noh YS, Amasino RM (2003) PIE, an ISWI family gene, is required for FLC activation and floral repression in Arabidopsis. Plant Cell 15: 1671–1682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Périlleux C, Bernier G (2002) The control of flowering: do genetical and physiological approaches converge? In SD O'Neill, JA Roberts eds, Plant Reproduction, Annual Plant Reviews, Vol 6. Sheffield Academic Press, Sheffield, England, pp 1–32
- Pigliucci M (1996) How organisms respond to environmental changes: from phenotypes to molecules (and vice versa). Trends Ecol Evol 11: 168–173 [DOI] [PubMed] [Google Scholar]
- Piñeiro M, Gómez-Mena C, Schaffer R, Martínez-Zapater JM, Coupland G (2003) EARLY BOLTING IN SHORT DAYS is related to chromatin remodeling factors and regulates flowering in Arabidopsis by repressing FT. Plant Cell 15: 1552–1562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pouteau S (2001) Conceptual context of floral repression in Arabidopsis. Flowering Newsl 31: 12–18 [Google Scholar]
- Pouteau S, Gaudin V, Ferret V, Lefebvre D, Libault M, Prunus F, Sabar M, Zhao G (2001) Analysis of the floral repression process in Arabidopsis. Flowering Newsl 32: 3–9 [Google Scholar]
- Ratcliffe OJ, Kumimoto RW, Wong BJ, Riechmann JL (2003) Analysis of the Arabidopsis MADS AFFECTING FLOWERING gene family: MAF2 prevents vernalization by short periods of cold. Plant Cell 15: 1159–1169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutherford SL (2000) From genotype to phenotype: buffering mechanisms and the storage of genetic information. Bioessays 22: 1095–1105 [DOI] [PubMed] [Google Scholar]
- Samach A, Coupland G (2000) Time measurement and the control of flowering in plants. Bioessays 22: 38–47 [DOI] [PubMed] [Google Scholar]
- Santoni V, Bellini C, Caboche M (1994) Use of two-dimensional protein-pattern analysis for the characterization of Arabidopsis thaliana mutants. Planta 192: 557–566 [Google Scholar]
- Scortecci KC, Michaels SD, Amasino RM (2001) Identification of a MADS-box gene, FLOWERING LOCUS M, that represses flowering. Plant J 26: 229–236 [DOI] [PubMed] [Google Scholar]
- Sheldon CC, Burn JE, Perez PP, Metzger J, Edwards JA, Peacock WJ, Dennis ES (1999) The FLF MADS box gene: a repressor of flowering in Arabidopsis regulated by vernalisation and methylation. Plant Cell 11: 445–458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegal ML, Bergman A (2002) Waddington's canalization revisited: developmental stability and evolution. Proc Natl Acad Sci USA 99: 10528–10532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson GG, Dean C (2002) Arabidopsis, the Rosetta stone of flowering time? Science 196: 285–289 [DOI] [PubMed] [Google Scholar]
- Soppe WJJ, Bentsink L, Koornneef M (1999) The early-flowering mutant efs is involved in the autonomous promotion pathway of Arabidopsis thaliana. Development 126: 4763–4770 [DOI] [PubMed] [Google Scholar]
- Stratton DA (1998) Reaction norm functions and QTL-environment interactions for flowering time in Arabidopsis. Heredity 81: 144–155 [DOI] [PubMed] [Google Scholar]
- Sultan SE (2000) Phenotypic plasticity for plant development, function, and life history. Trends Plant Sci 5: 537–542 [DOI] [PubMed] [Google Scholar]
- Sung ZR, Chen L, Moon YH, Lertpiriyapong K (2003) Mechanisms of floral repression in Arabidopsis. Curr Opin Plant Biol 6: 29–35 [DOI] [PubMed] [Google Scholar]
- van Tienderen PH, Hammad I, Zwaal FC (1996) Pleiotropic effects of flowering time gene in the annual crucifer Arabidopsis thaliana (Brassicaceae). Am J Bot 83: 169–174 [Google Scholar]
- Via S, Gomulkiewicz R, De Jong G, Scheiner SM, Schlichting CD, van Tienderen PH (1995) Adaptive phenotypic plasticity: consensus and controversy. Trends Ecol Evol 5: 212–217 [DOI] [PubMed] [Google Scholar]
- Waddington CH (1942) Canalization of development and the inheritance of acquired characters. Nature 150: 563–565 [Google Scholar]
- Wagner A (2000) Robustness against mutations in genetic networks of yeast. Nat Genet 24: 355–361 [DOI] [PubMed] [Google Scholar]
- Weller JL, Reid JB, Taylor SA, Murfet IC (1997) The genetic control of flowering in pea. Trends Plant Sci 2: 412–418 [Google Scholar]
- Wiltshire RJE, Murfet IC, Reid JB (1994) The genetic control of heterochrony: evidence from developmental mutants of Pisum sativum L. J Evol Biol 7: 447–465 [Google Scholar]
- Yoshida N, Yanai Y, Chen L, Kato Y, Hiratsuka J, Miwa T, Sung ZR, Takahashi S (2001) EMBRYONIC FLOWER2, a novel polycomb group protein homolog, mediates shoot development and flowering in Arabidopsis. Plant Cell 13: 2471–2481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Lechowicz MJ (1994) Correlation between time of flowering and phenotypic plasticity in Arabidopsis thaliana (Brassicaceae). Am J Bot 81: 1336–1342 [Google Scholar]