Abstract
To broaden our understanding of gibberellin (GA) biosynthesis and the mechanism whereby GA homeostasis is maintained in plants, we have investigated the degree to which the enzyme GA 3-oxidase (GA3ox) limits the formation of bioactive GAs in elongating shoots of hybrid aspen (Populus tremula × Populus tremuloides). We describe the cloning of a hybrid aspen GA3ox and its functional characterization, which confirmed that it has 3β-hydroxylation activity and more efficiently converts GA9 to GA4 than GA20 to GA1. To complement previous studies, in which transgenic GA 20-oxidase (GA20ox) overexpressers were found to produce 20-fold higher bioactive GA levels and subsequently grew faster than wild-type plants, we overexpressed an Arabidopsis GA3ox in hybrid aspen. The generated GA3ox overexpresser lines had increased 3β-hydroxylation activity but exhibited no major changes in morphology. The nearly unaltered growth pattern was associated with relatively small changes in GA1 and GA4 levels, although tissue-dependent differences were observed. The absence of increases in bioactive GA levels did not appear to be due to feedback or feed-forward regulation of dioxygenase transcripts, according to semiquantitative reverse transcription polymerase chain reaction analysis of PttGA20ox1, PttGA3ox1, and two putative PttGA2ox genes. We conclude that 20-oxidation is the limiting step, rather than 3β-hydroxylation, in the formation of GA1 and GA4 in elongating shoots of hybrid aspen, and that ectopic GA3ox expression alone cannot increase the flux toward bioactive GAs. Finally, several lines of evidence now suggest that GA4 has a more pivotal role in the tree hybrid aspen than previously believed.
Gibberellins (GAs) form a group of more than 130 tetracyclic diterpenes, some of which are biologically active and act as growth regulators in higher plants. Work on GA-deficient mutants has established that bioactive GAs play an important role in controlling diverse developmental processes such as seed germination, stem elongation, flowering, and fruit ripening (Davies, 1995). The GA biosynthetic pathway has been elucidated and its key components identified (for review, see Hedden and Phillips, 2000; Yamaguchi and Kamiya, 2000; Olszewski et al., 2002). The final steps in the pathway are catalyzed by the soluble 2-oxoglutarate-dependent dioxygenases GA 20-oxidase (GA20ox), GA 3-oxidase (GA3ox), and GA 2-oxidase (GA2ox). The pathway branches at GA12, which can be 13-hydroxylated into GA53, marking the starting points for two parallel routes catalyzed by the above dioxygenases: the early nonhydroxylated and the13-hydroxylated pathways forming the bioactive GAs, GA4 and GA1, respectively. The multifunctional GA20ox removes a carbon by successive oxidation of GA12 to GA9 and GA53 to GA20. However, the final interconversion into the bioactive GA4 or GA1 requires the action of the enzyme GA3ox. The deactivation of the bioactive species is catalyzed by GA2ox, which can also divert GA9 and GA20 away from the route toward active GAs by forming GA51 and GA29, respectively. The genes encoding the three described enzymes have been cloned in several species and have been found to belong to small multigene families. In Arabidopsis there are at least four genes coding for each enzyme (Hedden et al., 2001).
Maintaining the levels of the bioactive GAs in appropriate temporal and spatial patterns, while controlling the responsiveness of each tissue to GAs, are vital processes for the plant during its growth and development. Recent studies in Arabidopsis, rice (Oryza sativa), and barley (Hordeum vulgare) have identified several positive and negative regulators of GA signaling pathways, all involved in regulating GA responsiveness during development (for review, see Olszewski et al., 2002; Gomi and Matsuoka, 2003). In addition, a large number of studies have found evidence that GA biosynthesis is finely modulated via regulation of the GA dioxygenases. Work on GA mutants and plants treated with chemical inhibitors of GA biosynthesis has shown that transcript levels of GA20ox and GA3ox increase, while GA2ox transcripts decrease, in response to lowered amounts of GA (Chiang et al., 1995; Phillips et al., 1995; Thomas et al., 1999). In contrast, plants treated with bioactive GAs depress GA20ox and GA3ox transcription, and increase the transcription of GA2ox (Phillips et al., 1995; Yamaguchi et al., 1998; Thomas et al., 1999). Furthermore, evidence for interactions between GA signaling and biosynthesis has come from studies of GA signaling mutants. For instance, the gai mutant has increased levels of bioactive GAs and GA20ox transcript (Peng et al., 1999). As a result of the feedback and feed-forward regulation, governed by the endogenous levels of GA1 and GA4, the plant can maintain appropriate levels of GAs throughout its growth and development.
In hybrid aspen (Populus tremula × Populus tremuloides), expression of a GA20ox gene under the cauliflower mosaic virus (CaMV) 35S promoter led to a 20-fold increase in bioactive GA levels and subsequently increased growth and taller trees (Eriksson et al., 2000). Accelerated growth as a response to ectopic GA20ox expression and increased levels of GA growth regulator have also been observed in Arabidopsis (Huang et al., 1998; Coles et al., 1999), potato (Solanum tuberosum; Carrera et al., 2000), and tobacco (Nicotiana tabacum; Vidal et al., 2001), implying that this enzyme has a limiting role in regulating the production of bioactive GAs. In contrast, when the genes coding for ent-copalyl diphosphate synthase and ent-kaurene synthase, which are involved in early stages of GA biosynthesis, are overexpressed in Arabidopsis, no significantly increased levels of bioactive GA are observed (Fleet et al., 2003). This implies that key regulation of the flux through the GA biosynthesis pathway may take place downstream of ent-kaurene formation.
To complement the previous transgenic GA20ox overexpresser (OE) findings, we wanted to elucidate the role of GA3ox in controlling GA homeostasis in Populus. In this report, we describe the cloning and characterization of a functional GA3ox isolated from P. tremula × P. tremuloides and results from a study in which we ectopically expressed an Arabidopsis GA3ox in hybrid aspen. Increased levels of GA3ox transcript had relatively minor effects on GA1 and GA4 homeostasis, although tissue-dependent differences were observed. Our results suggest that GA 20-oxidation is much more important as a rate-limiting step than GA 3β-hydroxylation in GA-controlled shoot elongation. Finally, several lines of evidence now suggest that GA4 may have a more central role in the tree hybrid aspen than previously believed.
RESULTS
Isolation and Functional Characterization of a GA 3-Oxidase from Hybrid Aspen
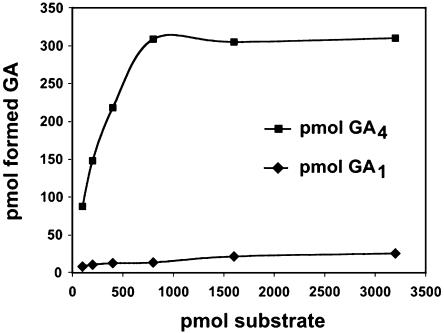
We screened a hybrid aspen cambial cDNA library (Hertzberg and Olsson, 1998) with a PttGA3ox PCR probe generated by reverse transcription (RT)-PCR. Out of 1,000,000 plaque-forming units, two cDNA clones were identified in a low stringency screening. Both clones were identified as partial PttGA3ox fragments, with identical, overlapping sections. The missing 5′ end was subsequently obtained in a RACE experiment. Finally, a full-length cDNA lacking5′- and 3′-untranslated regions was reconstructed by RT-PCR using primers based on the 5′ information and the isolated cambial library clones. This last step provided confirmation that the fragments obtained originated from the same gene. The resulting full-length cDNA was named PttGA3ox1 (GenBank accession no. AY433958). The cDNA contained an open reading frame of 1,122 bp encoding a putative protein of 374 amino acids. This deduced amino acid sequence showed the highest similarity to a GA3ox of Nicotiana sylvestris, NsGA3ox2, with which it shared 71% identity (AF494090). The binding sites of Fe2+ and 2-oxoglutarate in one member of the 2-oxoglutarate-dependent dioxygenase family have been determined by x-ray crystallography (Valegard et al., 1998), and these conserved sites were also found in the deduced amino acid sequence of PttGA3ox1. To test whether PttGA3ox1 encoded a functional GA3ox we subcloned the coding region in the expression vector pGEX-4T-2 to produce an inframe fusion protein and expressed this (and an empty vector construct serving as a control) in the Escherichia coli strain XL1 Blue. Lysates of bacteria, in which recombinant GA3ox protein production had been induced, and controls were used for functional assays. The recombinant PttGA3ox1 protein converted 14C-labeled GA20 and GA9 to GA1 and GA4, respectively, as identified by gas chromatography-mass spectrometry (GC-MS; data not shown), demonstrating that the clone encoded a functional enzyme. No conversion of substrate took place when the vector control was incubated with substrate. We then used unlabeled substrates at various concentrations in a competitive assay and analyzed products by GC/MS-SRM. The recombinant PttGA3ox1 protein was shown to have a higher affinity for GA9 than GA20, and formed GA4 more efficiently than GA1 (Fig. 1).
Figure 1.
Competitive assay for GA 3β-hydroxylation with cell lysates from recombinant E. coli expressing PttGA3ox1 in pGEX-4T-2. Cell lysates were incubated with cofactors and a mixture of equal amounts of the substrates GA9 and GA20 at varying concentrations for 1 h at 20°C, and the corresponding formation of GA4 and GA1 was monitored by GC/MS-SRM using deuterated GAs as internal standards.
Southern blot analysis using the full-length PttGA3ox1 sequence as a probe suggested the occurrence of at least three GA3ox genes in Populus (data not shown). The existence of four Populus GA3ox genes was confirmed by BLAST searches of PttGA3ox1 in the ongoing Populus trichocarpa genome sequencing program (http://genome.jgi-psf.org/poplar0/poplar0.home.html; data not shown). In comparison, GA3ox in Arabidopsis is encoded by four genes, and their functions have been confirmed by heterologous expression in E. coli (Hedden et al., 2001).
Generation of Overexpression Lines
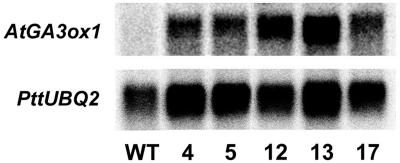
Transgenic hybrid aspen plants expressing the Arabidopsis GA4 gene (AtGA3ox1) under control of the CaMV 35S promoter were obtained by Agrobacterium tumefaciens-mediated transformation. Growth traits of 12 of the resulting transgenic lines and wild type (WT) were characterized. The results were used to select five representative lines (lines 4, 5, 12, 13, and 17) for further studies, after confirming that the transgenic plants were OEs by RNA blot analysis. The selected lines showed high levels of AtGA3ox1 expression, with line 12 having the highest and line 5 the lowest levels (Fig. 2). Southern blot analysis of genomic DNA from the selected lines confirmed that they were independent of each other (data not shown). It also revealed that lines 12 and 13 contained one insert and the other lines two or more inserts.
Figure 2.
Northern analysis of AtGA3ox1 expression in young expanding leaf tissue of WT and 35S-AtGA3ox1 transgenic lines (4, 5, 12, 13, and 17). Twenty micrograms of total RNA was loaded per lane and hybridized under stringent conditions at 65°C to a full-length AtGA3ox1 probe. A ubiquitin-like EST, PttUBQ2, was used as loading control.
No Dramatic Alterations in AtGA3ox OE Morphology
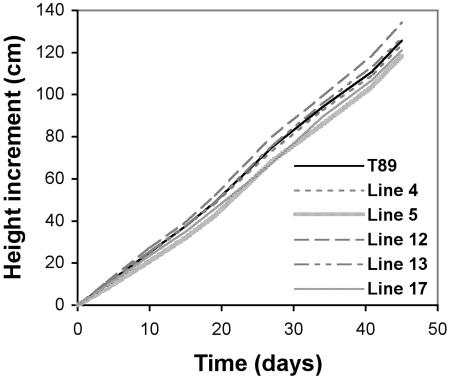
Anticipating that they would display changes in GA-related traits, we characterized the transgenic lines in terms of height increment, final internode lengths, and number of internodes formed. The total growth increment of the transgenic GA3ox OE lines was similar to WT during the period plant growth was monitored (Fig. 3). Line 12 had the highest overall total height growth, line 5 the lowest, while WT and the other lines were intermediate. Thus, ectopic expression of GA3ox resulted in lines that were slightly taller or shorter than WT. All transgenic lines formed fewer internodes than WT (Table I), with line 12 forming the least. The increase in total height increment in line 12, despite forming fewer internodes, was due to its internodes being significantly (approximately 30%) longer than WT (Table I). The results of the growth and developmental studies were consistent with data from the initial growth characterization of the 12 lines (data not shown).
Figure 3.
Relatively minor changes observed in height growth increment of 35S-AtGA3ox1 transgenic plants. Actively growing hybrid aspen plants cultivated in the greenhouse under LD conditions were monitored during a 45-d period. The depicted growth pattern is based on averages per line, n = 9 except for line 17 (n = 5). Student's t test analysis of the significance of differences between each line and WT based on the last data point per genotype showed that line 5 (P < 0.01) and line 17 (P < 0.05) were shorter and line 12 taller (P < 0.01) than WT.
Table I.
Comparison of internode formation between WT and AtGA3ox1 OEs
| Internode Length | Number of Internodes Formed | |
|---|---|---|
| cm | ||
| WT | 2.8 ± 0.036 | 45 ± 0.75 |
| Line 4 | 3.0 ± 0.033* | 41 ± 0.47* |
| Line 5 | 2.9 ± 0.040 | 41 ± 0.65* |
| Line 12 | 3.7 ± 0.045* | 36 ± 0.84* |
| Line 13 | 2.9 ± 0.048 | 43 ± 0.58 |
| Line 17 | 2.9 ± 0.018 | 42 ± 0.49* |
Internode lengths of 80-d-old plants and the number of new internodes formed during a 45-d period were determined in nine plants per genotype except for line 17 (five plants). Data presented are means for each genotype ±se. Differences that are significant at the <0.05 probability level between WT and each of the overexpressing lines for the two traits, according to Student's t test, are marked with asterisks.
Effects of Altered GA Biosynthesis on GA Levels and Expression of PttGA20ox1, PttGA3ox1, and Two Putative PttGA2ox Genes
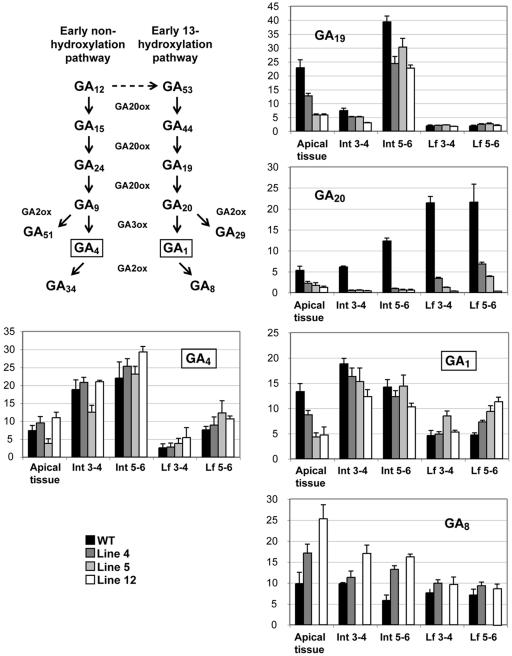
The substrate of GA3ox, GA20, was dramatically reduced in all lines and tissues analyzed, providing strong evidence of successful overexpression at the protein level (Fig. 4). In accordance with this finding, in vivo metabolic studies of 14C-labeled GA20 confirmed that GA3ox enzyme activity was increased in transgenic plants (data not shown). Unexpectedly, however, no marked increase in bioactive GAs was observed. The bioactive GA1 and GA4 levels remained unchanged or increased in leaf tissue compared to WT, and in apical and internode tissues the GA1 levels generally decreased while the GA4 levels slightly increased in lines 4 and 12. In contrast, the levels of the deactivated GA8 increased in apical and internode tissues, while remaining largely unaffected in leaves.
Figure 4.
GA content of apical, internode, and leaf tissue in 35S-AtGA3ox1 transgenic plants and WT. Tissue from nine individuals was pooled per genotype and tissue type. GAs from 200 mg fresh weight of pooled tissue were purified and analyzed by GC/MS-SRM using 2H2-GAs as internal standards. Data presented are the means of three technical replicates of pooled sample ±sd. GA8 levels in line 5 were not measured. Boxed GAs depict bioactive GAs.
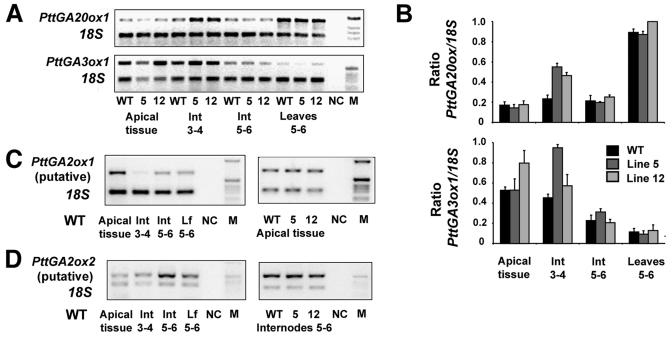
To elucidate whether the transcriptional regulation of GA biosynthesis had been altered in OEs, thus limiting the formation of bioactive GAs, we investigated the expression of genes encoding three key enzymes in GA biosynthesis (PttGA20ox1, the endogenous PttGA3ox1, and two putative PttGA2ox genes) by semiquantitative RT-PCR. The expression of the Arabidopsis homologs has been demonstrated to be feedback and feed-forward regulated by bioactive GAs, respectively (Chiang et al., 1995; Phillips et al., 1995; Xu et al., 1995; Thomas et al., 1999) and previously both PttGA20ox1 and PttGA3ox1 have been shown to be feedback regulated in hybrid aspen (Eriksson and Moritz, 2002; M. Chono, unpublished data). We found that PttGA20ox1 and PttGA3ox1 had not been down-regulated in the OE lines; instead, they had remained unchanged or even become up-regulated as in young elongating stem tissue (Fig. 5). Moreover, the putative GA2ox exhibited distinct expression patterns; PttGA2ox1 was mainly expressed in apical tissue, while PttGA2ox2 was mainly expressed in internode tissue, but neither of the genes was up-regulated due to the ectopic GA3ox expression (Fig. 5; data not shown). Therefore the lack of a large increase in bioactive GAs in the transgenic lines was not caused by any major changes in the expression of the examined GA dioxygenases. More likely, there is a limiting activity of GA20ox in supplying GA9 and GA20 as substrates for GA3ox. As a consequence, ectopic GA3ox will quickly deplete the limiting substrate pool without largely affecting the bioactive GA levels.
Figure 5.
GA dioxygenase expression levels, as determined by semiquantitative RT-PCR in various tissues of WT and the 35S-AtGA3ox lines 5 and 12. Different gene-specific and intron-spanning primer pairs were used together with a primer pair for the internal standard 18S. One cDNA source was used per sample that originated from pooled plant tissues (nine plants per genotype). A, The expression of PttGA20ox1 and PttGA3ox1 in various tissues of WT and line 5 and 12. B, Quantification results of two GA genes were normalized to the internal standard 18S. The average of 3 to 4 independent RT-PCRs were calculated and results plotted; bars depict se. C, The PttGA2ox1 expression was analyzed first in WT and then compared to line 5 and 12 in apical tissue. D, PttGA2ox2 transcript levels were determined in WT and compared to line 5 and 12 in young expanding internodes. Int, internodes; NC, negative control—no cDNA added to the PCR mixture; M, DNA size marker.
GA4 Levels, But Not GA1 Levels, in Expanding Internodes Are Correlated with Final Internode Lengths
The morphological data from our growth characterization had shown that line 12 developed 30% longer internodes than WT, and the GA analysis had revealed slightly elevated levels of GA4 in line 12 internodes (Table I; Fig. 4). Therefore, an additional experiment was conducted in which line 12 was grown together with WT to investigate the influence of GA1 and GA4 on final internode length. For this purpose, actively elongating internodes representing stages at which they were 0% to 20%, 20% to 40%, and 40% to 80% of their final length were harvested from five plants per genotype (Table III), and treated as replicates in a further analysis of GAs. Compared to the previous experiment (Fig. 4), the GA levels were much higher, especially for GA4 (Table II). This could be explained by changes in the growth conditions, which increased growth rates considerably (data not shown), in addition to the use of a new, improved GA extraction method, which extracted some GAs, such as GA4, more efficiently from different tissues of Populus (Fig. 4; Table II). Thus it seems that the initial GA analysis (Fig. 4) underestimated the levels of GA4.
Table III.
Internode lengths of WT and two selected transgenic 35S-AtGA3ox1 lines in three developmental stages of elongation
| Plant Line | Internode A 20%–40% | Internode B 40%–60% | Internode C 60%–80% | Final Internode Length 100% |
|---|---|---|---|---|
| WT | 2.0 ± 0 | 11.2 ± 1.1 | 18.4 ± 0.5 | 32 ± 1.2 |
| Line 5 | 2.0 ± 0 | 8.4 ± 0.6 | 18.6 ± 1.4 | 29 ± 0.9 |
| Line 12 | 2.5 ± 0.2 | 12.4 ± 0.5 | 22.2 ± 0.7 | 39 ± 0.9 |
Data are means from five biological replicates, in mm, ±se.
Table II.
GA contents in elongating internodes of WT hybrid aspen and transgenic 35S-AtGA3ox1 in a complementary experiment
| Genotype | GA53 | GA19 | GA20 | GA1 | GA8 | GA9 | GA4 | GA34 |
|---|---|---|---|---|---|---|---|---|
| WT-a | 24.3 ± 7.0a | 29.8 ± 7.0 | 26.8 ± 7.3 | 22.7 ± 4.3 | 138.0 ± 14.9 | 18.0 ± 3.6 | 199.8 ± 40.7 | 119.7 ± 24.8 |
| WT-b | 1.2 ± 0.2 | 6.1 ± 0.8 | 31.5 ± 3.1 | 14.6 ± 2.7 | 76.0 ± 11.1 | 11.7 ± 0.9 | 14.0 ± 3.6 | 39.3 ± 1.5 |
| WT-c | 3.1 ± 1.2 | 15.1 ± 4.0 | 34.0 ± 2.4 | 30.0 ± 5.2 | 40.3 ± 4.4 | 11.2 ± 1.1 | 7.7 ± 3.4 | 15.4 ± 1.1 |
| Line 12-a | 31.3 ± 19.0 | 15.7 ± 7.7 | ndb | 19.8 ± 10.7 | 212.7 ± 98.5 | nd | 571.5 ± 237.6 | 65.3 ± 33.6 |
| Line 12-b | 0.7 ± 0.1 | 1.3 ± 0.1 | nd | 3.4 ± 0.7 | 49.6 ± 5.2 | nd | 62.9 ± 6.2 | 33.9 ± 1.6 |
| Line 12-c | 0.9 ± 0.2 | 4.2 ± 0.8 | nd | 21.1 ± 2.6 | 35.0 ± 2.2 | nd | 36.0 ± 7.4 | 18.7 ± 2.3 |
GA contents of internodes in developmental stages at which they had reached (a) 20% to 40%, (b) 40% to 60%, and (c) 60% to 80% of their final length are shown (pg mg−1 fresh weight).
Values represent the average of five biological replicates ±se.
nd, Not detectable.
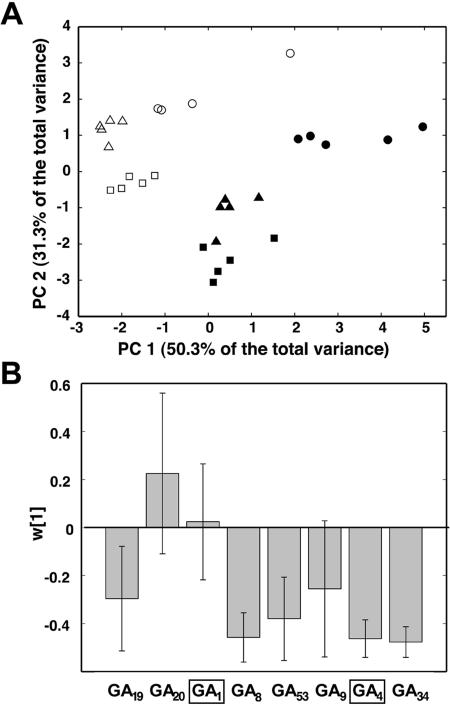
A number of different multivariate statistical tools can be used for displaying data that contain a large number of variables. In Principal Component Analysis (PCA; Wold et al., 1987) the aim is to extract one or more principal components or latent variables to describe the majority of variance in a data set. Although the dataset acquired from the GA analysis does not fulfill the general multivariate requirement that the number of variables should be much larger than the number of samples, PCA can still provide useful visual displays of the data. The PCA-score plot using all GA values as X-matrix shows that the different classes of samples, i.e. the three developmental stages of internode elongation and the two genotypes, are clearly distinct (Fig. 6A). This is because the GA levels differ both between lines and between internodes in various stages of elongation. Partial Least Square to Latent Structures (PLS) is a supervised method in which any underlying covariation between an X-matrix, here GA-levels, and a Y-matrix, here internode length, can be found (Wold et al., 2001). We found a correlation between GA4 levels and internode length for both WT (Fig. 6B) and line 12 (data not shown). The absence of a similar correlation for the bioactive GA1 leads us to suggest that the absolute levels of GA4 in the earlier stages of internode elongation determine the final internode length. The longer internodes of line 12 compared to WT can thus be explained by its consistently higher GA4 levels (Table II).
Figure 6.
A, PCA-score plot from the analysis of GAs in five WT and five line 12 plants. All variables were log-transformed, centered, and scaled to unit variance. Each point in the plot corresponds to a separate sample type, i.e. internode tissue of different developmental stages. Internode A, circles; internode B, triangles; and internode C, boxes. Line 12 is represented by white symbols, and WT is depicted in black. A clear separation can be observed between all samples, suggesting developmental regulation of GA levels. B, PLS-loading plots from the first loading vector (w) in the analysis of GA levels and internode length from internodes at different developmental stages of five WT plants. The height of the bars shows the relative correlation between GAs and internode length. If the confidence interval (calculated with jackknifing; Efron, 1986; Martens and Martens, 2000) does not include 0, a variable is considered significant. Bioactive GAs are boxed, and the plot shows that GA4 levels, and not GA1 levels, are correlated with internode length.
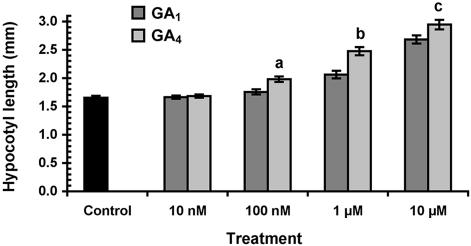
As our results suggested that GA4 levels determine the final length of internodes (Fig. 6B), we performed a sensitivity assay to determine which GA has the strongest effect on P. tremula hypocotyl elongation. There was a significant difference in the response to GA4 as compared to GA1 in our assay. However, the difference was only subtle, providing relatively weak support for a higher responsiveness to GA4 (Fig. 7). As a consequence, it is more likely that the increased internode elongation observed in line 12 is due to the higher concentration of GA4 (Table II).
Figure 7.
Sensitivity of P. tremula seedlings to GA4 and GA1. Hypocotyl length of 2.5-d-old seedlings are presented as an average of 40 seedlings ±se, except for the 10-μm treatment (30 ± se). Letters indicate statistically significant differences, according to Student's t test, in sensitivity to GA4 as compared to GA1; a, P < 0.001; b, P < 0.0001; and c, P < 0.01. At GA4 concentrations greater than 100 nm and GA1 concentrations higher than 1 μm the seedlings grew significantly taller than controls, P < 0.00001. The experiment was repeated once with P. tremuloides seedlings with very similar results.
DISCUSSION
It is well established that GA 3-oxidase catalyzes the conversion of inactive GA species into GAs with biological activity and that it is subject to strict developmental controls in the life cycle of a plant (Lester et al., 1997; Martin et al., 1997; Williams et al., 1998; Itoh et al., 1999; Yamaguchi et al., 2001). As well as cloning a P. tremula × P. tremuloides GA3ox, in this study we investigated transgenic hybrid aspen expressing the AtGA3ox1 under the CaMV 35S promoter, in order to broaden our understanding of the regulation of GA biosynthesis. Because GA 3-oxidase catalyzes the last step in the formation of bioactive GAs, it was tempting to believe that overexpression of the enzyme may increase the flux of GA precursors to bioactive GAs.
We identified a GA3ox from hybrid aspen that possessed the ability to convert GA9 and GA20 into the bioactive GA4 and GA1, respectively, demonstrating that we had cloned a functional GA3ox. Furthermore, the GA4-producing activity of PttGA3ox1 was higher than its GA1-producing activity, suggesting that it may make a more important contribution to the early nonhydroxylated pathway than the early 13-hydroxylation pathway. Data on the expression of PttGA3ox1 during early seedling development in P. tremula shows that its expression is 40% inhibited, via feedback regulation, by the bioactive GA4 at 36 h following germination, according to semiquantitative RT-PCR analysis (M. Chono, unpublished data). The transcript profile of PttGA3ox1 has now been extended to apical tissue and young expanding internodes, sites in which GA3ox expression and GA-induced stem elongation are known to occur (Itoh et al., 1999).
To elucidate the way in which GA homeostasis is maintained and the consequences of altering GA levels in planta, we overexpressed the Arabidopsis GA4 gene AtGA3ox1 in hybrid aspen. The total growth increment remained relatively stable in relation to WT, but the GA3ox OEs developed fewer internodes and the internode length remained the same or increased. The substrate for GA3ox GA20 was dramatically reduced in all the transgenes and tissues (Fig. 5). Nevertheless, the levels of the bioactive GA1 and GA4 did not increase correspondingly, although some tissue-dependent changes occurred.
The transcriptional regulation of the GA biosynthesis is well documented: treatment with bioactive GAs triggers feedback regulation of GA20ox and GA3ox, and feed-forward regulation of GA2ox transcription, enabling GA homeostasis to be maintained (Chiang et al., 1995; Phillips et al., 1995; Xu et al., 1995; Thomas et al., 1999). It could be argued that the absence of an increase in bioactive GA formation may be due to the shortage of GA3ox substrate and/or increased deactivation activity. Scarcity of the substrates GA20 and GA9 may, in turn, be due to inherent limitations on the activity of GA20ox and/or to down-regulation of GA20ox in the transgenics. To seek evidence for alterations in dioxygenase gene regulation, we first determined the transcript levels of PttGA20ox1 and the endogenous PttGA3ox1 in lines 5, 12, and WT. The regulation of both genes was similar in that the transcript levels remained unchanged or increased compared to WT (Fig. 5). In Arabidopsis, GA20ox and GA3ox are both encoded by at least four genes (Hedden et al., 2001), so the possibility that the expression of other gene family members is negatively affected in the transgenic lines cannot be excluded. Because both PttGA20ox1 and PttGA3ox1 have been shown to be feedback regulated by bioactive GAs (Eriksson and Moritz, 2002; M. Chono, unpublished data), it is unlikely that only other family members would respond to altered GA levels. Instead, we conclude that GA20ox and GA3ox are not down-regulated. Similarly, semiquantitative RT-PCR of two putative PttGA2ox genes revealed unaltered transcript profiles in the OEs suggesting that the deactivation of GA1 and GA4 had not been changed. It must be emphasized, however, that changes at the protein or activity levels of the gene family members of GA20ox or GA2ox may also contribute to an altered GA biosynthesis. Without conclusive evidence of increased feedback effects of GA20ox and GA3ox, or feed-forward regulation of GA2ox in the OEs, an alternative explanation for the lack of elevated bioactive GAs must be sought. Unlike the relatively minor phenotype of 35S-GA3ox plants, ectopic GA20ox expression in hybrid aspen elevates the bioactive GA levels 20-fold, leading to highly increased internode elongation (Eriksson et al., 2000). Therefore, we suggest that GA 20-oxidation, relative to GA 3β-hydroxylation, acts as a limiting step in GA biosynthesis controlling shoot elongation. As a consequence, ectopic GA3ox will quickly deplete the limiting substrate pool, explaining the observed GA profile in 35S-GA3ox plants. In accordance with our findings, a recent study has shown that overexpression of ent-copalyl diphosphate synthase and ent-kaurene synthase, which catalyze the first two GA-committed steps, does not result in increased levels of bioactive GAs either (Fleet et al., 2003).
An interesting observation was that line 12 developed 30% longer internodes, causing this line to grow more quickly than WT, although it formed fewer internodes. In order to study if GAs (and if so, which ones) determine final internode length, we sampled internodes from line 12 and WT in three representative stages of internode elongation and determined the levels of GAs in the individual internodes. The PCA-score plot (Fig. 6A) showed that GA levels of each class of internode developmental stage was clearly separated from the others, suggesting that the GA levels are not only different between WT and line 12, but they are also developmentally regulated along the elongating shoot. Furthermore, the results from the PLS analysis showed that GA4 levels, but not GA1 levels, are correlated with internode length (i.e. developmental stage; Fig. 6B). As GAs are considered to act in the subapical parts of the elongating shoot (Olsen et al., 1995; Vogler et al., 2003) it is possible that GA4 is the main bioactive GA involved in Populus internode elongation and ultimately determines their final length, explaining both the 30% longer internodes in line 12 and the correlation between GA4 and developmental stage.
The early nonhydroxylation pathway, in which GA4 is produced, has previously been suggested to be less active in hybrid aspen based on feeding studies to shoot cuttings using radioactively labeled GA12 (Eriksson and Moritz, 2002). However, recombinant PttGA20ox1 was shown to be capable of metabolizing both GA12 and GA53 and therefore able to accept substrates of both pathways. In our study, GA4 levels were found to be higher than GA1 levels, why the focus turned to GA4 as the main bioactive GA in hybrid aspen. We determined the relative sensitivity of P. tremula seedlings to these GAs and found that they were slightly more responsive to GA4 (Fig. 7). It is widely accepted that GA4 is the bioactive GA in Arabidopsis, since it is more sensitive to, and has higher concentrations of, GA4 than GA1 (Xu et al., 1997; Cowling et al., 1998). Although the response to GA4 in Populus is only marginally higher, the results from the functional PttGA3ox1 assay, which revealed a higher formation rate of GA4 than GA1, the higher concentration of GA4, and the correlation between GA4 and final internode length suggest that GA4 plays a pivotal role in hybrid aspen growth and development.
Finally, one of the phenotypic changes observed in the transgenic lines was the lower number of leaves formed, especially in the most strongly divergent transgenic line. The process that governs the rate of leaf formation is initiated in the peripheral zone of the shoot apical meristem and is measured in plastochrons. It is separate from the subsequent cell divisions and cell elongation in the subapical region, which dictate final internode length (Steeves and Sussex, 1989). Further plastochron investigations are now required which can detect regional GA differences in various apical regions, for instance the peripheral zone of the apex where the leaf primordia are established, but at present we are unable to achieve such analytical resolution.
In conclusion, our study has shown that GA 3β-hydroxylation is much less important as a rate-limiting step than GA 20-oxidation in the formation of the GAs that control shoot elongation in hybrid aspen. In addition, by using the most strongly divergent transgenic line and WT we showed that the GA4 levels are correlated with final internode lengths, explaining the increased stem elongation of this OE line. The role of GA4 as the predominant bioactive GA was further supported by its higher concentrations as compared to GA1.
MATERIAL AND METHODS
Cloning of PttGA3ox1
Total RNA was isolated from growing apical buds of hybrid aspen, using the cetyl-trimethyl-ammonium bromide (CTAB) method (Chang et al., 1993). Poly(A+)RNA was then isolated using Dynabeads Oligo(dT)25 resin as described by the manufacturer (Dynal, Oslo, Norway), and first-strand cDNA was synthesized using the first strand cDNA synthesis kit according to the manufacturer's protocol (Amersham-Pharmacia Biotech, Uppsala). A PCR probe was generated using this cDNA as template with degenerate primers based on conserved regions of GA3ox sequences. The 5′ primer was 5′-ATGTGGTMNGARGGNTTYAC-3′ and the 3′ primer was 5′-GTRTGIGSIGCNARNCCCAT-3′. A band of expected size was purified by gel electrophoresis, subcloned, and sequenced. The 0.3-kb insert was radiolabeled and used to probe a hybrid aspen λGT11 cambial cDNA library (Hertzberg and Olsson, 1998) using standard techniques (Sambrook et al., 1989). From the 1,000,000 plaque-forming units that were screened at low stringency, two partial cDNA clones were identified, subcloned, and sequenced. The missing 5′ end was obtained by RACE on a Marathon (CLONTECH Laboratories, Palo Alto, CA) double-stranded cDNA library originating from the cDNA described above, using the gene specific primer 5′-TGGTGAATCCCTCTGACCACATGAGC-3′ and adapter primer 1 supplied by the manufacturers, using their protocol. Finally, the complete cDNA was reconstructed in a PCR experiment with the 5′ primer including an EcoRI site, 5′-CCCTCCAGTAAGAATTCCCATGCCTTCAAG-3′ and the 3′ primer including an XhoI site, 5′-GTGATCACTCGAGTCCTAACCAACTTTTACAC-3′. A Marathon (CLONTECH) cDNA library of growing apical buds was used as a template in the PCR, using the Advantage PCR system (CLONTECH). The product of the expected size was purified by gel electrophoresis, subcloned, sequenced, and the full-length clone was named PttGA3ox1 (GenBank accession no. AY433958).
Heterologous Expression and Enzyme Assays
The entire coding region of PttGA3ox1 was subcloned in frame into the expression vector pGEX-4T-2 (Amersham-Pharmacia Biotech), and both this construct and empty vector controls were subsequently transformed into the Escherichia coli strain XL1 Blue. Heterologous expression was induced, and lysates were prepared as described in Martin et al. (1997). The presence of induced protein in the crude extract was confirmed by SDS-PAGE analysis. The enzyme assay mixtures, consisting of 0.1 m Tris-HCl, 4 mm 2-oxoglutarate, 4 mm ascorbate, 4 mm dithiothreitol, and 0.5 mm FeSO4 added to the lysate (10 μL) in a final volume of 100 μL, were incubated for 1 h with gibberellin substrate at 20°C. The following GAs (purchased from Professor L. Mander, Australian National University, Canberra, Australia) were used as substrates: 14C2-17,17-GA9 (1,900 Bq mol−1), 14C2-17,17-GA20 (1,900 Bq mol−1), 14C2-17,17-GA19 (1,900 Bq mol−1), 14C2-17,17-GA1 (1,370 Bq mol−1), 14C2-17,17-GA4 (1,370 Bq mol−1), GA9, and GA20.
Plant Transformation and Growth Conditions
cDNA corresponding to the Arabidopsis GA4 locus, AtGA3ox1 (supplied by Professor Howard Goodman, Harvard Medical School, Boston), was ligated in sense orientation into the BamHI cloning site of the binary vector pPCV702.kana (Walden et al., 1990) under the control of the CaMV 35S promoter. The construct was transformed into hybrid aspen, Populus tremula × Populus tremuloides Michx. clone T89, and plants were regenerated essentially as previously described (Nilsson et al., 1992). Following an initial characterization of 12 transgenic lines, five representative lines were chosen for further studies. WT control plants did not receive any plant vector but were subjected to simultaneous vegetative propagation in tissue culture as transgenes. Once the plantlets had developed sufficient roots, they were potted in a fertilized peat:perlite mixture (5:1) and grown in a growth chamber under long day (LD) conditions described elsewhere (Eriksson et al., 2000) for 34 d, when the growth characterization began. More detailed analysis of three different stages of internode elongation in WT and line 12 was carried out under greenhouse LD conditions, with an 18/6-h photoperiod, 60% humidity, 20°C, during January and February with supplementary lighting (Osram Powerstar HQI-BT 400 W/D, Osram, Germany) switched on and off when incoming light fell below and increased above 20 W/m−2, respectively, during the photoperiod. In both growth studies, plants were watered daily and fertilized with a complete nutrient solution (SuperbaS, Supra Hydro, Landskrona, Sweden) once a week.
RNA Blot Analysis
Total RNA was extracted from young leaf tissues of transgenic lines and WT plants. The RNA was isolated, prepared for gel blot analysis, and hybridized. The relative RNA amounts were then quantified as previously described (Eriksson et al., 2000). The full-length AtGA3ox1 and a hybrid aspen ubiquitin-like expressed sequence tag (EST; AI165341) were used as probes under high stringency conditions at 65°C (last wash 0.1 × sodium chloride/sodium phosphate/EDTA and 0.1% SDS) to analyze the ectopic GA3ox expression and reference gene expression patterns, respectively.
Growth Measurements, Sampling, and Statistical Analysis
Thirty-five days after potting, the plants were marked at an actively growing internode, approximately 10 cm above the root-stem junction. This was used as a reference point for measuring internode formation and height increment. The first internode was defined as the first one below a leaf at least 1 cm long, and the number of internodes was counted between the first internode and the reference point. Measurements were made approximately every 5 d until the plants reached the age of 80 d. At this point, apical tissues were sampled by dissecting out as many leaf primordia as possible, internodes 3 to 4, internodes 5 to 6, and the corresponding leaves of these internodes. Nine individual plants per genotype were analyzed and subsequently harvested, except for five plants of line 17. After sampling, the plant material was frozen immediately in liquid nitrogen. Tissues from the different individual plants of each line were pooled, ground in liquid nitrogen, and divided for GA analysis and RT-PCR analysis. For the complementary study of WT and line 12 internode elongation, the first 20 internodes were individually measured to establish the final internode length per genotype and plant. Actively elongating internodes representing stages at which they had reached 0% to 20%, 20% to 40%, and 40% to 80% of their final length were harvested from five plants per genotype, and internodes were sampled and processed for GA analysis as separate biological samples. The dataset was analyzed by multivariate statistical tools using SIMCA-P+ 10.0.4.0 software (Umetrics, Umeå, Sweden).
RT-PCR Analysis
First-strand cDNA was synthesized using the first strand cDNA synthesis kit according to the manufacturer's protocol (Amersham-Pharmacia Biotech) from total RNA isolated using the CTAB method (Chang et al., 1993) from the tissues described in “Growth Measurements, Sampling, and Statistical Analysis” (see above). Gene-specific and intron-spanning primer pairs, capable of amplifying a 0.47-kb 3′ end fragment of PttGA20ox1 and a 0.60-kb 3′ end fragment of PttGA3ox1, respectively, were GA20THR Fw (5′-TTAGGCACCGGTCCTCATTGT-3′); GA20THR.Rev (5′-AATAGCAGGCCCCCAAGTGCAT-3′) and 3bhF11 (5′-AAGCTCATGTGGTCAGAGGGATTC-3′); and CN10-1ny (5′-GTGATTGGGCGATAGAGAGGAGG-3′). Primers to amplify GA2ox were designed based on sequences of two putative GA2ox genes generated from a Populus EST sequencing effort: PttGA2ox1 (GenBank accession no. BU835271) and PttGA2ox2 (GenBank accession no. BU877509). Gene-specific and intron-spanning primer pairs, capable of amplifying a 0.73-kb PttGA2ox1 fragment and a 0.58-kb PttGA2ox2 fragment, respectively, were T071g08For (5′-ACTTCTTTGCCAAAACATTCGATGA-3′); T071g08Rev (5′-AGGCTTGATGGGTGTAACCATTTCT-3′) and V035d04For (5′-CAGCTGTGAAGAAAATGGCA-3′); and V035d04Rev (5′-ATCAGCCAATCTGGAGCTGT-3′). The RT-PCR was performed with a gene-specific primer pair and an 18S primer pair as an internal control (QuantumRNA 18S internal standards, Ambion, Austin, TX) according to the manufacturer's instructions. PCR products were analyzed on a 1.5% (w/v) agarose gel containing ethidium bromide and the signal intensities were determined with a Gel Doc 1000 DNA Gel Analysis and Documentation System and Molecular Analyst software (Bio-Rad Laboratories, Hercules, CA). Each experiment was repeated at least three times using the same cDNA source, originating from pooled material of five to nine plants per genotype. For the GA20ox and GA3ox reactions the average mean and se values of relative transcript abundance were calculated.
GA1 and GA4 Treatment of P. tremula Seedlings
Approximately 50 wild P. tremula seeds per treatment were sown on filter paper in petri dishes drenched in water with GA1 or GA4 at the following concentrations: 10 nm, 100 nm, 1 μm, and 10 μm. Controls received no addition of GA. The petri dishes were cold-treated for 5 d at 4°C before being placed under LD greenhouse conditions for 2 d, after which they were harvested in order to analyze the lengths of their hypocotyls. The seedlings were analyzed under a Leica MZFLIII microscope (Leica Microsystems, Wetzlar, Germany) equipped with a digital camera, and the lengths were determined using the program Leica QWin 2.3. Forty seedlings for each treatment, except the 10-μm treatment (30 seedlings), were measured. The experiment was repeated once with P. tremuloides seedlings, and similar results were obtained.
Quantification of GAs
GAs from 200 mg (fresh weight) of tissue were purified and analyzed, essentially as described by Eriksson et al. (2000) by GC/MS-SRM (JMS-MStation 700, JEOL, Tokyo), using 2H2-GAs (Professor L. Mander) as internal standards. For the GA analysis of the internodes in specific developmental stages, the extraction was modified as follows. Plant tissues, between 10 and 50 mg fresh weight, were extracted using an MM 301 Vibration Mill (Retsch, Haan, Germany) at a frequency of 30 Hz s−1 for 3 min after adding 3 mm tungsten carbide beads (Retsch) to each tube to increase the extraction efficiency. After centrifugation in an Eppendorf centrifuge for 10 min at 14,000 rpm, the supernatant was further purified as earlier described (Eriksson et al., 2000).
Sequence data from this article have been deposited with the EMBL/GenBank data libraries under accession numbers AY433958, BU835271, and BU877509.
Acknowledgments
We thank Dr. Maria E. Eriksson for discussions and suggestions and Inga-Britt Carlsson for help with GA analysis.
This work was supported by the Swedish Research Council for Environment, Agricultural Sciences and Spatial Planning; by European Union Strategic Funding; and by the Swedish Research Council.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.104.038935.
References
- Carrera E, Bou J, Garcia-Martinez JL, Prat S (2000) Changes in GA 20-oxidase gene expression strongly affect stem length, tuber induction and tuber yield of potato plants. Plant J 22: 247–256 [DOI] [PubMed] [Google Scholar]
- Chang S, Puryear J, Cairney J (1993) A simple and efficient method for isolating RNA from pine trees. Plant Mol Biol Report 11: 113–116 [Google Scholar]
- Chiang HH, Hwang I, Goodman HM (1995) Isolation of the Arabidopsis GA4locus. Plant Cell 7: 195–201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coles JP, Phillips AL, Croker SJ, Garcia-Lepe R, Lewis MJ, Hedden P (1999) Modification of gibberellin production and plant development in Arabidopsis by sense and antisense expression of gibberellin 20-oxidase genes. Plant J 17: 547–556 [DOI] [PubMed] [Google Scholar]
- Cowling RJ, Kamiya Y, Seto H, Harberd NP (1998) Gibberellin dose-response regulation of GA4gene transcript levels in Arabidopsis. Plant Physiol 117: 1195–1203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies P (1995) Plant hormones: physiology, biochemistry and molecular biology, Ed 2. Kluwer Academic Publishers, Dordrecht, The Netherlands
- Efron B (1986) Discussion: jackknife, bootstrap and other resampling methods in regression analysis. Ann Stat 14: 1301–1304 [Google Scholar]
- Eriksson ME, Israelsson M, Olsson O, Moritz T (2000) Increased gibberellin biosynthesis in transgenic trees promotes growth, biomass production and xylem fiber length. Nat Biotechnol 18: 784–788 [DOI] [PubMed] [Google Scholar]
- Eriksson ME, Moritz T (2002) Daylength and spatial expression of a gibberellin 20-oxidase isolated from hybrid aspen (Populus tremula L. × P. tremuloides Michx.). Planta 214: 920–930 [DOI] [PubMed] [Google Scholar]
- Fleet CM, Yamaguchi S, Hanada A, Kawaide H, David CJ, Kamiya Y, Sun TP (2003) Overexpression of AtCPS and AtKS in Arabidopsis confers increased ent-kaurene production but no increase in bioactive gibberellins. Plant Physiol 132: 830–839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomi K, Matsuoka M (2003) Gibberellin signaling pathway. Curr Opin Plant Biol 6: 489–493 [DOI] [PubMed] [Google Scholar]
- Hedden P, Phillips AL (2000) Gibberellin metabolism: new insights revealed by the genes. Trends Plant Sci 5: 523–530 [DOI] [PubMed] [Google Scholar]
- Hedden P, Phillips AL, Rojas MC, Carrera E, Tudzynski B (2001) Gibberellin biosynthesis in plants and fungi: a case of convergent evolution? J Plant Growth Regul 20: 319–331 [DOI] [PubMed] [Google Scholar]
- Hertzberg M, Olsson O (1998) Molecular characterisation of a novel plant homeobox gene expressed in the maturing xylem zone of Populus tremula × tremuloides. Plant J 16: 285–295 [DOI] [PubMed] [Google Scholar]
- Huang SS, Raman AS, Ream JE, Fujiwara H, Cerny RE, Brown SM (1998) Overexpression of 20-oxidase confers a gibberellin-overproduction phenotype in Arabidopsis. Plant Physiol 118: 773–781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh H, Tanaka-Ueguchi M, Kawaide H, Chen XB, Kamiya Y, Matsuoka M (1999) The gene encoding tobacco gibberellin 3 beta-hydroxylase is expressed at the site of GA action during stem elongation and flower organ development. Plant J 20: 15–24 [DOI] [PubMed] [Google Scholar]
- Lester DR, Ross JJ, Davies PJ, Reid JB (1997) Mendel's stem length gene (Le) encodes a gibberellin 3 beta-hydroxylase. Plant Cell 9: 1435–1443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martens H, Martens M (2000) Modified Jack-knife estimation of parameter uncertainty in bilinear modelling by partial least squares regression (PLSR). Food Qual Pref 11: 5–16 [Google Scholar]
- Martin DN, Proebsting WM, Hedden P (1997) Mendel's dwarfing gene: cDNAs from the Le alleles and function of the expressed proteins. Proc Natl Acad Sci USA 94: 8907–8911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsson O, Aldén T, Sitbon F, Little CHA, Chalupa V, Sandberg G, Olsson O (1992) Spatial pattern of cauliflower mosaic virus 35S promoter-luciferase expression in transgenic hybrid aspen trees monitored by enzymatic assay and non-destructive imaging. Transgenic Res 1: 209–220 [Google Scholar]
- Olsen JE, Junttila O, Moritz T (1995) A localised decrease of GA(1) in shoot tips of Salix pentandra seedlings precedes cessation of shoot elongation under short photoperiod. Physiol Plant 95: 627–632 [Google Scholar]
- Olszewski N, Sun TP, Gubler F (2002) Gibberellin signaling: Biosynthesis, catabolism, and response pathways. Plant Cell 14: S61–S80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng JR, Richards DE, Moritz T, Cano-Delgado A, Harberd NP (1999) Extragenic suppressors of the Arabidopsis gai mutation alter the dose-response relationship of diverse gibberellin responses. Plant Physiol 119: 1199–1207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips AL, Ward DA, Uknes S, Appleford NEJ, Lange T, Huttly AK, Gaskin P, Graebe JE, Hedden P (1995) Isolation and expression of three gibberellin 20-oxidase cDNA clones from Arabidopsis. Plant Physiol 108: 1049–1057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Fritsch E, Maniatis T (1989) Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY
- Steeves TA, Sussex IM (1989) Patterns in Plant Development. Cambridge University Press, Cambridge, UK
- Thomas SG, Phillips AL, Hedden P (1999) Molecular cloning and functional expression of gibberellin 2-oxidases, multifunctional enzymes involved in gibberellin deactivation. Proc Natl Acad Sci USA 96: 4698–4703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valegard K, van Scheltinga ACT, Lloyd MD, Hara T, Ramaswamy S, Perrakis A, Thompson A, Lee HJ, Baldwin JE, Schofield CJ, et al. (1998) Structure of a cephalosporin synthase. Nature 394: 805–809 [DOI] [PubMed] [Google Scholar]
- Vidal AM, Gisbert C, Talon M, Primo-Millo E, Lopez-Diaz I, Garcia-Martinez JL (2001) The ectopic overexpression of a citrus gibberellin 20-oxidase enhances the non-13-hydroxylation pathway of gibberellin biosynthesis and induces an extremely elongated phenotype in tobacco. Physiol Plant 112: 251–260 [DOI] [PubMed] [Google Scholar]
- Walden R, Koncz C, Schell J (1990) The use of gene vectors in plant molecular biology. Methods Mol Cell Biol 1: 175–194 [Google Scholar]
- Williams J, Phillips AL, Gaskin P, Hedden P (1998) Function and substrate specificity of the gibberellin 3 beta-hydroxylase encoded by the Arabidopsis GA4gene. Plant Physiol 117: 559–563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wold S, Esbensen K, Geladi P (1987) Principal component analysis. Chemom Intell Lab Syst 2: 37–52 [Google Scholar]
- Wold S, Sjostrom M, Eriksson L (2001) PLS-regression: a basic tool of chemometrics. Chemom Intell Lab Syst 58: 109–130 [Google Scholar]
- Xu YL, Gage DA, Zeevaart JAD (1997) Gibberellins and stem growth in Arabidopsis thaliana - Effects of photoperiod on expression of the GA4and GA5loci. Plant Physiol 114: 1471–1476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu YL, Li L, Wu KQ, Peeters AJM, Gage DA, Zeevaart JAD (1995) The GA5locus of Arabidopsis thaliana encodes a multifunctional gibberellin 20-oxidase: molecular cloning and functional expression. Proc Natl Acad Sci USA 92: 6640–6644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogler H, Caderas D, Mandel T, Kuhlemeier C (2003) Domains of expansin gene expression define growth regions in the shoot apex of tomato. Plant Mol Biol 53: 267–272 [DOI] [PubMed] [Google Scholar]
- Yamaguchi S, Kamiya Y (2000) Gibberellin biosynthesis: its regulation by endogenous and environmental signals. Plant Cell Physiol 41: 251–257 [DOI] [PubMed] [Google Scholar]
- Yamaguchi S, Kamiya Y, Sun TP (2001) Distinct cell-specific expression patterns of early and late gibberellin biosynthetic genes during Arabidopsis seed germination. Plant J 28: 443–453 [DOI] [PubMed] [Google Scholar]
- Yamaguchi S, Smith MW, Brown RGS, Kamiya Y, Sun TP (1998) Phytochrome regulation and differential expression of gibberellin 3 beta-hydroxylase genes in germinating Arabidopsis seeds. Plant Cell 10: 2115–2126 [DOI] [PMC free article] [PubMed] [Google Scholar]