Abstract
To understand how differentiation and growth may be coordinated during development, we have studied the action of the CINCINNATA (CIN) gene of Antirrhinum. We show that in addition to affecting leaf lamina growth, CIN affects epidermal cell differentiation and growth of petal lobes. Strong alleles of cin give smaller petal lobes with flat instead of conical cells, correlating with lobe-specific expression of CIN in the wild type. Moreover, conical cells at the leaf margins are replaced by flatter cells, indicating that CIN has a role in cell differentiation of leaves as well as petals. A weak semidominant cin allele affects cell types mainly in the petal but does not affect leaf development, indicating these two effects can be separated. Expression of CIN correlates with expression of cell division markers, suggesting that CIN may influence petal growth, directly or indirectly, through effects on cell proliferation. For both leaves and petals, CIN affects growth and differentiation of the more distal and broadly extended domains (leaf lamina and petal lobe). However, while CIN promotes growth in petals, it promotes growth arrest in leaves, possibly because of different patterns of growth control in these systems.
Development involves coordination of two interconnected processes: growth and cellular differentiation. The genetic control of each of these processes has been studied extensively in plants. For example, genes affecting leaf growth or epidermal cell fate have been isolated and analyzed (Masucci et al., 1996; Tsuge et al., 1996; Gu et al., 1998; Kim et al., 1998; Mizukami and Fischer, 2000; Kim et al., 2002). However, the way that growth and differentiation are coupled through the action of genes has been less well studied. To help address this, we have investigated the effects of the CINCINNATA (CIN) gene on growth and cell differentiation in Antirrhinum.
CIN encodes a TCP transcription factor that promotes growth arrest, particularly in leaf margins (Nath et al., 2003). In cin mutants, leaves are larger and have an undulating edge due to excessive growth in marginal regions. In addition to these effects on leaf growth, cin mutants also show alterations in petal shape and cell types. This suggests that the CIN transcription factor may have targets involved in both growth and differentiation pathways. To determine how these pathways may be affected, we have analyzed the role of CIN in petal growth and differentiation.
Flowers of Antirrhinum have five petals, which are united in their proximal region to form a corolla tube. The distal region of each petal forms a lobe, which itself can be subdivided into a more proximal region, termed the lip, and a more distal region (Fig. 1A; Keck et al., 2003). The flowers exhibit dorsoventral asymmetry and comprise two dorsal petals, two lateral petals, and one ventral petal. The lateral and ventral petal lobes form a platform for bees to land on and pry open the flower. A variety of cell types can be recognized in the flower, forming characteristic patterns along the proximodistal and dorsoventral axes (Keck et al., 2003). Several genes encoding transcription factors have been identified that influence these patterns. The CYCLOIDEA (CYC), DICHOTOMA (DICH), and DIVARICATA (DIV) genes encode TCP or MYB proteins that specify differences along the dorsoventral axis (Luo et al., 1996, 1999; Galego and Almeida, 2002). MIXTA (MIX) encodes a MYB-related protein that promotes conical cell differentiation in the lobes of the petal epidermis (Noda et al., 1994), while LIP genes encode APETALA2-like proteins that most strongly affect development in the lip region of the flower (Keck et al., 2003). However, none of these mutants have clear effects on leaf development.
Figure 1.
Phenotype of wild-type and cin mutant flowers. Flowers of the wild type and cin mutants shown in side (left) or face view (right). Wild-type Stock 98 (A), cin-756 (B), cin-628 (C), wild-type Sippe 50 (D), and cincin (E) are shown. The lobe and lip regions are indicated on the face view of wild-type Stock 98 (A).
Here, we describe the effects of CIN on growth and differentiation. A weak semidominant cin allele carries a duplication of the CIN locus, most likely leading to posttranscriptional silencing of CIN. This allele affects cell types mainly in the lip region of the petal but does not affect leaf development. All strong alleles of cin have major disruptions in the CIN gene and give smaller petal lobes with flat instead of conical cells, correlating with lobe-specific expression of CIN in the wild type. In addition, conical cells normally observed at the margins of leaves are flatter than usual, indicating that CIN has a role in promoting conical cell development in leaves as well as petals. Expression of CIN correlates with expression of cell division markers, suggesting that CIN may influence petal growth through effects on cell proliferation. However, while CIN promotes growth in petals, it promotes growth arrest in leaves, possibly because of different patterns of growth control in these systems.
RESULTS
Alleles of cincinnata
To understand the role of CIN in petal development, we first characterized a range of cin alleles. Two alleles with indistinguishable mutant phenotypes, cin-755 and cin-756, had previously been shown to carry deletions that extended into the CIN coding region (Nath et al., 2003). In addition to affecting leaf shape, these putative null alleles had a clear effect on the flower: the petal lobes were smaller, slightly curled, pale, and had altered cell types compared with their progenitor (Fig. 1, A and B).
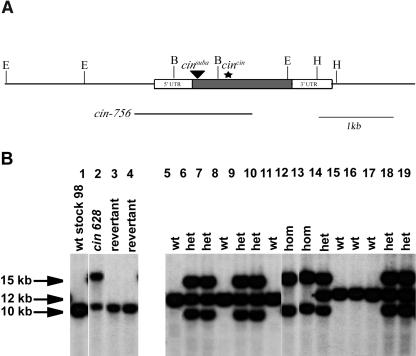
A broadly similar effect on both leaf and floral morphology was seen when comparing the classical allele, cincin (Fig. 1E), with their progenitor (Fig. 1D). Nevertheless, there are some differences in detail. In particular, the degree to which the ventral and lateral petals bend at the tube-lobe boundary is reduced in the classical mutant. The cincin phenotype is very similar to another mutant, subcrispa (Stubbe, 1966). An allelism test showed subcrispa is allelic to cincin and was renamed cinsuba. Genetic analysis indicated that many of the differences in appearance of the cincin and cinsuba alleles compared with cin-756 were a consequence of their distinct genetic backgrounds (the classical cincin and cinsuba alleles were produced in the Sippe 50 background, while the deletion alleles were produced in the John Innes Stock 98 background). Cloning and sequencing of cincin showed that it had a 10-bp insertion 438 bp downstream of the start codon, resulting in a frameshift and a stop codon 444 bp downstream of the start (Fig. 2A). The predicted protein would be truncated at the C terminus by 256 amino acids compared to the wild type CIN protein and is expected to be inactive. PCR analysis on cinsuba DNA revealed an insertion of a 4-kb DNA fragment in the CIN locus (Fig. 2A). Sequencing of the insertion site showed that the insertion was 63 bp downstream of the start codon. The insertion of this fragment disrupts the open reading frame (ORF) and is likely to disrupt CIN function.
Figure 2.
Structure of cin alleles. A, Map of the cin locus. The rectangle represents the transcribed region of the CIN gene with the ORF shaded in gray. The thick black line represents the region deleted in cin-756. The triangle represents the insertion in cinsuba. The star shows the position of the introduced stop codon in cincin. E, B, and H are positions of restriction sites for the enzymes EcoRI, BamHI, and HindIII, respectively. B, DNA blots of wild-type, cin-628, cin-628 homozygous revertant, and an F2 population segregating for cin-628 probed with CIN. DNA was digested with HindIII. Lane 1 contains DNA from wild-type Stock 98 (the background in which the cin-628 allele is maintained). Lane 2 contains DNA from cin-628. Lanes 3 and 4 contain DNA from two independent cin-628 homozygous revertants. Lanes 5 to 19 contain DNA from an F2 population segregating for cin-628; the phenotypes of the plants are wild type (wt), heterozygous mutant (het), and homozygous mutant (hom).
Another allele, cin-628, was also analyzed. Unlike other cin alleles, cin-628 was semidominant and had no clear effect on leaf development. The floral phenotype was different from the cin deletions: The effect of the mutation was primarily on the cell types within the adaxial epidermis of the petal lips (Fig. 1C). In cin-628/CIN heterozygotes, the effect on cell types was weaker than in cin-628/cin-628 homozygotes and confined to more proximal regions of the petal lip. Crosses of cin-628 to cincin gave a strong floral mutant phenotype in the F1, consistent with their allelism.
The cin-628 allele arose from a transposon mutagenesis screen. Southern blots of HindIII digests probed with CIN ORF revealed that in addition to the approximately 10-kb band seen in the wild-type progenitor, cin-628 had a novel band at approximately 15 kb of a similar intensity to the wild-type band (Fig. 2B, lanes 1 and 2). This indicated that cin-628 may carry a transposon-induced duplication of the CIN locus similar to a previously identified mutation of NIVEA in Antirrhinum (Bollmann et al., 1991). To determine whether the additional 15-kb band in cin-628 was responsible for the phenotype, stable homozygous revertants of cin-628 were analyzed (the revertants were obtained by screening progeny of cin-628 plants grown at 15°C, a temperature that favors transposon movement and rearrangements). The 15-kb band was absent in the revertants, although the 10-kb band was still present, indicating that the 15-kb band had been responsible for the cin-628 mutation (Fig. 2B, lanes 3 and 4). PCR analysis indicated that the 15-kb band carried a copy of the Tam3 transposon approximately 3.5 kb upstream of the ATG, indicating that the duplication was associated with a transposition event. To determine whether the 15-kb band was linked to the original 10-kb band, cin-628 was crossed to a wild-type line of a different genetic background (JI-7) with a distinct band of approximately 12 kb in HindIII digests. Analysis of the resulting F2 showed that the approximately 15-kb and approximately 10-kb bands present in cin-628 were linked and cosegregated with the mutant phenotype (Fig. 2B, lanes 5–19).
One explanation for these results is that cin-628 arose by a CIN duplication that led to repression of gene activity, possibly through posttranscriptional gene silencing (PTGS; Kusaba et al., 2003). PTGS would also account for the semidominance of cin-628. Similar transposon-induced semidominant alleles carrying duplications have been described for the NIVEA locus of Antirrhinum and have subsequently been shown to contain small RNAs diagnostic of gene silencing (A. Hamilton and D. Baulcombe, personal communication; Bollmann et al., 1991).
Cell Types in cin Alleles
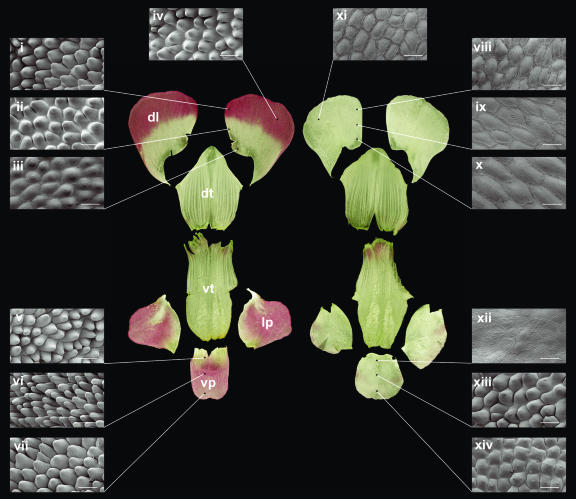
To investigate the effect of CIN on cell types, adaxial epidermal cells of petals from one of the deletion alleles, cin-756, and the wild type were analyzed by scanning electron microscopy (SEM). Four regions of the dorsal petal lobe and three of the ventral petal lobe were recorded (Fig. 3). Cells of the cin-756 were flatter than those of the wild type for all regions analyzed. Instead of conical cells typical of wild-type petal lobes, the cin mutant had nearly flat or weakly conical cells. The effect on conical cell formation was strongest in the lip region of the ventral petal lobe (Fig. 3, compare regions v and xii). Measurements of cell sizes showed that cells were also about 30% larger in area in cin mutants for three out of four regions of the dorsal lobes when compared to the wild type (Fig. 3, regions i–iv and viii–xi; Table I). Although, as these cells are flattened in the mutant, it is possible that the cell volume is not affected.
Figure 3.
SEM analysis of petals from the wild type compared to cin-756. Petals were dissected and flattened to illustrate the various areas used for comparisons of the wild type (i–vii) and cin-756 (viii–xiv). Scale bar = 50 μm. dl, dorsal lobes; dt, dorsal tube; lp, lateral lobes; vp, ventral lobes; vt, ventral/lateral tube.
Table I.
Cell sizes in dorsal petals of cin-756 and the wild type
| Region i/viii | Region ii/ix | Region iii/x | Region iv/xi | |
|---|---|---|---|---|
| cin-756 | 2,022.2 ± 299.8 | 3,027.8 ± 746.4 | 4,785.2 ± 492.1 | 2,731 ± 347.1 |
| Wild type | 1,897.7 ± 93 | 2,024.5 ± 77.1 | 2,731.5 ± 882.1 | 1,784.7 ± 82.2 |
Average size of petal conical cells (μm2) from dorsal petals of cin-756 and the wild type. The sizes were obtained from different regions corresponding to Figure 3.
The cin-628 allele had a slightly different effect on petal cell types. A mixture of flat cells and hair cells (Fig. 4, arrow) was produced in the lip region of the ventral and lateral petals instead of conical cells. The morphology of these hair cells was similar to that of wild-type hair cells from the adaxial surface of the petal tube (Fig. 4, arrowhead). The distal regions of the petal lobes in cin-628 did form conical cells indistinguishable from those found in wild-type petals, consistent with cin-628 being a weak rather than null allele.
Figure 4.
Adaxial epidermis surface of lateral petal in cin-628. Hair cells are produced on the lateral and ventral petal lips in cin-628 (arrow), resembling those normally found in the petal tube (arrowhead).
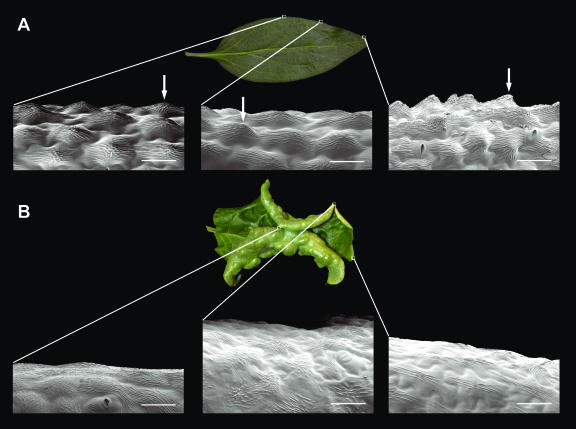
Conical cells are also found along the abaxial-adaxial (dorsal-ventral) boundary (margin) of wild-type Antirrhinum leaves (Fig. 5A). To investigate whether these cells might also be affected by CIN, three regions along the leaf margin in the wild type and cin-756 allele were compared. The conical cells found in the wild type were replaced by flatter cells in cin-756 (Fig. 5B). Thus, CIN has an effect on conical cell development in both leaves and petals.
Figure 5.
SEM analysis of leaf margins of the wild type compared to cin-756. Different areas along the proximal-distal axis of the leaf were compared in the wild type (A) and cin-756 (B). The arrows indicate conical cells found at the leaf margin in the wild type. Scale bar = 50 μm.
Expression of CIN in Wild-Type Floral Tissue
The specific effect of CIN on petal lobe development might reflect restricted expression of CIN or factors that interact with CIN. To distinguish these possibilities, CIN expression in developing petals was studied by RNA in situ hybridization on longitudinal sections of floral buds. Antirrhinum floral development has been divided into 15 stages (Carpenter et al., 1995; Vincent and Coen, 2004). CIN expression in buds was first detected at stage 7, about halfway through floral development. Expression was in the distal regions of the growing petals (corresponding to the developing petal lobes) on both the adaxial and abaxial sides of the petal (Fig. 6A). Some signal was also observed in the distal region of the developing stamens.
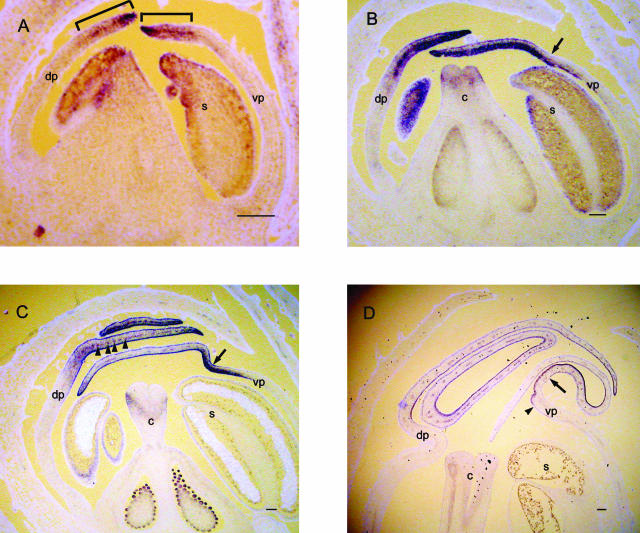
Figure 6.
Expression of CIN in wild-type Sippe 50 floral buds. Longitudinal sections of different stages were probed with digoxigenin-labeled CIN antisense RNA. Staining of RNA is dark blue, and tissue is counterstained with calcofluor (pale blue). In each case the dorsal side of the bud is on the left of the photograph. At stage 7, expression is found in the distal region of the developing petals (bracketed in A). At stage 9, expression is observed throughout the distal portion of the petals (B). At stage 10, expression is observed mainly in the epidermal layers of the distal region of the petal (C). At stage 11, expression is found only in the epidermal layers and vasculature of the lobes of all petals (D). Arrow indicates the position of the ventral furrow. s, stamen; c, carpel; vp, ventral petal; dp, dorsal petal. Scale bars = 100 μm.
At stages 8 to 9, when the petal lobes cover the internal whorls, expression of CIN was observed in the lobes (Fig. 6B). Expression was stronger in, though not exclusive to, the adaxial side of each lobe. A slight kink could be observed at this stage in the ventral petal (Fig. 6B, arrow), corresponding to a furrow that forms between the tube and lobe. This region will give rise to the lip of the lower petals (Keck et al., 2003; Vincent and Coen, 2004). The expression domain of CIN included the region where the ventral furrow was forming.
By stage 10, CIN expression was still in the petal lobes (Fig. 6C). Expression was now concentrated in the epidermal cell layers on both the adaxial and abaxial sides, although the expression was stronger in the adaxial epidermis. The CIN expression pattern included the kink in the ventral petal. Expression was also observed in regularly spaced groups of cells within the petal (Fig. 6C, arrowheads), most likely corresponding to the vascular tissue. At stages 11 to 12, when conical cells begin to form on the lobe, CIN expression was expressed strongly in the adaxial and abaxial epidermal layers of the lobe (Fig. 6D). CIN expression in the lobe stopped at a specific point on the ventral epidermis corresponding to the boundary between the tube and the lobe.
CIN was expressed in both the cin-628 and cincin mutants (data not shown). CIN expression in petals of cin-628 was reduced compared to wild type, although the pattern seemed unaltered (data not shown). The lowered expression in cin-628 is consistent with the hypothesis mentioned above that the effects of this allele may involve PTGS (Van der Krol et al., 1990). Early expression was also reduced in cincin, although later expression was nearer to wild-type levels. Expression of CIN was not detected in mutants carrying the deletion allele, cin-756.
Effect of CIN on Petal Growth
In addition to the effect on cell types, petal lobes of cin-756 mutants were slightly smaller than those of the wild type (Fig. 2). Measurements on several flowers showed that the dorsal petal lobes cin-756 were about 86% of the area of those in the wild type (297 ± 15 mm2 compared to 348 ± 23 mm2). Similarly, cincin dorsal petals were about 62% of the area of their wild-type progenitor (68 ± 8 mm2 compared to 109 ± 8 mm2). The reduction in overall size contrasts with the observed increase in cell size in several regions of the petal lobes, suggesting that cin mutants had fewer cells in the dorsal petal lobes.
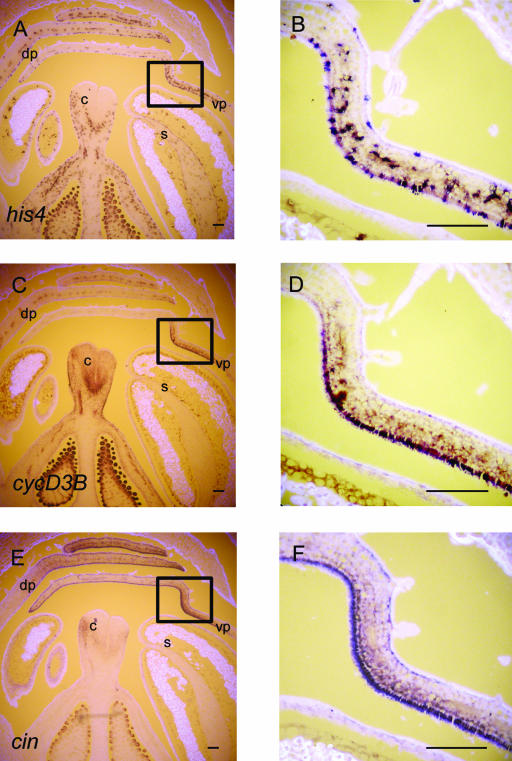
To examine how CIN might be affecting growth of petals, expression of cell-cycle markers HISTONE4 (HIS4) and CYCLIND3B (CYCD3B) was analyzed in wild-type and cin mutant buds in the same genetic background. HIS4 is expressed only in the S phase of the cell cycle, and only a proportion of the cells give a strong signal in proliferating tissue. CYCD3B, a key regulator of the cell cycle in plants, is not restricted to one phase of the cell cycle and is expressed throughout proliferating tissue (Fobert et al., 1994; Riou-Khamlichi et al., 1999; Gaudin et al., 2000).
In stage 9 buds, the spotty pattern of HIS4 expression was observed throughout the floral bud (Fig. 7, A and B). Spots of expression were most frequent in the region corresponding to the ventral furrow of the petal (Fig. 7B). Similarly, strongest CYCD3B expression was observed in the ventral furrow of the petals (Fig. 7, C and D). This indicates a high level of cell division activity and correlates with the highest level of CIN expression in this region (Figs. 7, E and F, and 6C).
Figure 7.
Expression of HIS4, CYCD3B, and CIN in stage 9 wild-type Sippe 50 floral buds. Consecutive longitudinal sections were probed with digoxigenin-labeled HIS4, CYCD3B, and CIN antisense RNA and viewed as in Figure 6. HIS4 expression is observed throughout the floral bud (A) but is more concentrated at the ventral furrow (shown enlarged in B). Strong CYCD3B expression is also found at the ventral furrow (C) and is concentrated on the adaxial side (shown enlarged in D). CIN expression is observed in the epidermal layers in the distal region of the petals (E), with stronger expression in the adaxial epidermal cell layer (shown enlarged in F). Rectangles indicate regions shown as enlargements. s, stamen; c, carpel; vp, ventral petal; dp, dorsal petal. Scale bars = 100 μm.
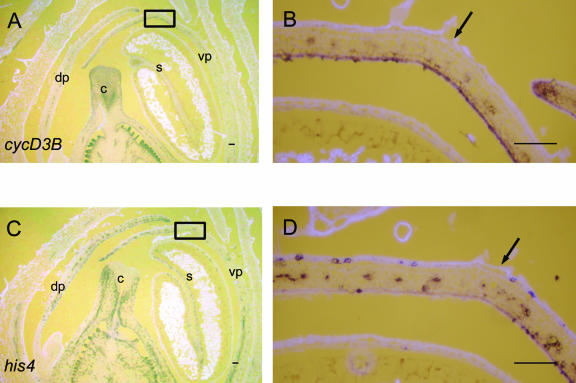
Expression analysis in the cincin allele showed that CYCD3B expression was present in the region corresponding to the ventral furrow, although the prominence of the furrow was reduced (Fig. 8, A and B). The expression was mainly in the epidermal cell layer and was less intense than that in the wild type. Similarly, although HIS4 was expressed throughout floral buds of cincin, the strong expression associated with the ventral furrow in the wild type was absent (Fig. 8, C and D).
Figure 8.
Expression of HIS4 and CYCD3B in stage 9 cincin floral buds. Consecutive longitudinal sections were probed with digoxigenin-labeled HIS4 and CYC3B antisense RNA and viewed as in Figure 6. Expression of CYCD3B is observed in the petals (A) but is not greatly enhanced in the region where the ventral furrow normally forms (shown enlarged in B). Similarly, the frequency of spots of HIS4 expression (C) is not greatly enhanced in the ventral furrow region (shown enlarged in D). Arrows indicate the normal position of the ventral furrow and rectangles indicate regions chosen for enlargements. s, stamen; c, carpel; vp, ventral petal; dp, dorsal petal. Scale bars = 100 μm.
DISCUSSION
We show that CIN affects cell types and growth of petal lobes in Antirrhinum, correlating with specific expression of CIN in these regions (no expression is detected in the petal tube region). There are parallels between these effects on petal development and the effect of CIN on leaves. Petal lobes can be compared to the leaf lamina, as both are broadly extended and positioned distally, while the petal tube can be compared to the petiole, as both are highly polarized and positioned proximally. Thus, for both petals and leaves, CIN plays a role in the development of the more distal and broadly extended regions of the organ. The parallel is further supported by the observation that some of the cell types in the leaf lamina are altered in cin mutants in a comparable way to cell-type alterations in the lobes. However, despite these broad parallels, many of the detailed effects of CIN on petals and leaves are different.
Mature petal lobes of cin null mutant alleles have flat or nearly flat cells instead of the conical cells found in wild-type adaxial epidermis. Conical cells normally begin to form at late stages of floral bud development (Glover et al., 1998; Vincent and Coen, 2004) coinciding with a stage when CIN transcripts are present in the adaxial epidermis of the petal lobe. It is possible that CIN promotes conical cell formation through interactions with MIX, a gene encoding a MYB transcription factor that is also expressed in the adaxial epidermis (Noda et al., 1994); as with cin-756, mix mutants have flat cells instead of conical cells. However, the effect of MIX is more restricted than that of CIN, as organ shape and size are not affected in mix mutants. Moreover, cin mutants have altered conical cell development at the leaf margins while mix mutants did not. As CIN encodes a TCP transcription factor, one possibility is that it influences conical cell development in petals by directly switching on MIX specifically in petal lobes. However, some MIX expression is still detected by RT-PCR in cin-756 mutant petals, indicating that the interaction may be more indirect (our unpublished data). Interactions between TCP and MYB transcription factors have also been described for the control of dorsoventral asymmetry in Antirrhinum flowers. Two TCP transcription factors, CYC and DICH, are known to interact with the MYB transcription factor DIV to establish domains along the dorsal-ventral axis of the flower (Galego and Almeida, 2002). CIN may interact similarly with the family of MIX proteins to establish some of the distinctions within the petal.
The effect of CIN on cell types varies for different regions of the petal lobes. Two results indicate that the lip region of the ventral petal lobe (i.e. the region nearest to the corolla tube) may be more sensitive to the loss of CIN. Firstly, in null cin alleles, the cells in this region are more severely affected (they are completely flat). Secondly, the weak cin-628 allele mainly affects cell types in this region. In this case, hair cells similar to those normally found inside the corolla tube are formed instead of conical cells. This suggests that one role of CIN may be to repress tube cell types while promoting the formation of conical cells. Other explanations are possible; for example, the hair cells in cin-628 may be the result of extended growth of conical cells (Glover et al., 1998). In addition, the difference between effects on ventral and dorsal petals indicates that the effects of CIN may depend on interactions with genes like CYC and DIV controlling dorsoventral asymmetry.
CIN promotes growth in the petal lobes; mature lobes of the cin mutants are smaller than the wild type. The effect on growth depends on genetic background (cin-756 has a smaller effect on petal size than cincin in their respective backgrounds). The promotion of growth by CIN involves extra cell divisions; cin mutants have fewer cells in the lobes. This correlates with reduced expression of cell cycle and histone genes in the developing lip region. CIN expression is strongest in the developing lip, suggesting that it may function in a region-specific manner to promote growth. Such regional specific effects on growth have also been described for other TCP transcription factors, such as CYC and DICH (Luo et al., 1996, 1999; Galego and Almeida, 2002).
In both leaves and petals, CIN acts preferentially in particular regions, having its greatest effects in the petal lip and leaf margins. However, the effect of CIN on overall growth seems to be opposite for petals and leaves. In petals, CIN promotes growth, while in leaves CIN promotes arrest of growth and cell division (cin mutants have larger leaves). This difference may reflect a different mode of action for CIN in petals and leaves or different patterns of growth control in these systems.
MATERIAL AND METHODS
Plant Materials and Growth Conditions
The insertion and deletion alleles of CIN were isolated in the John Innes Stock 98 background (Carpenter et al., 1987), which was used as the standard wild type for comparisons with the mutant. Both the classical mutants, cincinnata and subcrispa, originally came from the Gatersleben Collection (Hammer et al., 1990) and have been described by Stubbe (Stubbe, 1966). Plants were grown in a glasshouse, unless otherwise stated. For Tam3 mutagenesis, plants were grown in a growth cabinet at 15°C. For growth measurements, plants were grown at 25°C in growth cabinet with continuous light.
DNA Analysis
DNA extraction was conducted as described previously (Luo et al., 1996). PCR was carried out in a programmable thermal controller (PTC-100; MJ Research, Waltham, MA). Reactions were made in 10 mm Tris-HCl, pH 8.3; 50 mm KCl; 1.5 mm MgCl2; 0.01% (w/v) gelatin; 0.2 mm each of dATP, dCTP, dGTP, and dTTP; and 0.002% (w/v) Tween 20. Sixty nanograms of DNA and 2 units Taq polymerase were added per 50-μL reaction. Samples were heated to 95°C before addition of polymerase to minimize primer dimerization. Reactions were overlaid with 20 μL of mineral oil and 35 cycles of 30 s at 95°C, 2 min at 55°C, and 40 s at 72°C were performed. Samples were then heated to 72°C for 10 min before being stored at 4°C. Two primers were used to identify the cincin allele: (1) GTA AGG TTG AAT GGC AGG AAC and (2) CCT TCT TTA TCA ACC AAT CAA C. The two primers used to clone the cinsuba allele were (3) ATC CTC ACC ACC ATC ACC AT and primer 2. PCR products were electrophoresed in 0.8% (w/v) agarose gels, purified by Wizard columns (Promega, Madison, WI), and then sequenced with the ABI automatic sequencer (Perkin Elmer, Foster City, CA) using oligos B and D. DNA blots were conducted as described previously (Coen et al., 1990). The probe for Southern hybridization was generated using pJAM193.
Microscopy
Samples for SEM were prepared on plastic replicas as described earlier (Carpenter et al., 1995). Young leaves were dissected from the plants, and a plastic mold and cast were taken (Green and Linstead, 1990). The cast was gold coated, and micrographs were taken using a CT1500 HF microscope (Oxford Instruments, Concord, MA).
In Situ Hybridization
The methods used for tissue preparation, digoxigenin-labeling of RNA probes, and in situ hybridization were as described previously (Coen et al., 1990). The probe used to detect the CIN transcript was a 1048-bp fragment from the cDNA clone, covering the entire ORF. For HIS4, the probe consisted of the entire cDNA (Fobert et al., 1994). For CYCD3B, a 3′-terminal fragment of the cDNA lacking the poly(A) tail was used (Gaudin et al., 2000).
Petal Measurements
Petals were dissected under a microscope, flattened between glass plates, and photographs taken with a Nikon Coolpix 995 digital camera (Tokyo), either fitted on a microscope or a tripod. Quantitative parameters such as area, perimeter, length, and width were measured in the digital images using programs written in Matlab (The MathWorks, Natick, MA; program written by A.G. Rolland-Lagan). The average epidermal cell size was determined by counting the number of cells in SEM of 0.1 to 1 square mm area of the leaf surface, then dividing the area by the number of cells.
Sequence data from this article have been deposited with the EMBL/GenBank data libraries under accession number AY205603.
Acknowledgments
We thank Anne-Gaelle Rolland-Lagan for the Matlab program. In addition, we thank Annabel Whibley for helpful comments on the manuscript.
Present address: Department of Microbiology and Cell Biology, Indian Institute of Science, Bangalore 560 012, India.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.103.036368.
References
- Bollmann J, Carpenter R, Coen ES (1991) Allelic interactions at the nivea locus of Antirrhinum. Plant Cell 3: 1327–1336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpenter R, Copsey L, Vincent C, Doyle S, Magrath R, Coen E (1995) Control of flower development and phyllotaxy by meristem identity genes in Antirrhinum. Plant Cell 7: 2001–2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpenter R, Martin C, Coen ES (1987) Comparison of genetic behaviour of the transposable element Tam3 at two unlinked pigment loci in Antirrhinum majus. Mol Gen Genet 207: 82–89 [Google Scholar]
- Coen ES, Romero JM, Doyle S, Elliott R, Murphy G, Carpenter R (1990) floricaula: a homeotic gene required for flower development in Antirrhinum majus. Cell 63: 1311–1322 [DOI] [PubMed] [Google Scholar]
- Fobert PR, Coen ES, Murphy GJ, Doonan JH (1994) Patterns of cell division revealed by transcriptional regulation of genes during the cell cycle in plants. EMBO J 13: 616–624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galego L, Almeida J (2002) Role of DIVARICATA in the control of dorsoventral asymmetry in Antirrhinum flowers. Genes Dev 16: 880–891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaudin V, Lunness PA, Fobert PR, Towers M, Riou-Khamlichi C, Murray JA, Coen E, Doonan JH (2000) The expression of D-cyclin genes defines distinct developmental zones in snapdragon apical meristems and is locally regulated by the Cycloidea gene. Plant Physiol 122: 1137–1148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glover BJ, Perez-Rodriguez M, Martin C (1998) Development of several epidermal cell types can be specified by the same MYB-related plant transcription factor. Development 125: 3497–3508 [DOI] [PubMed] [Google Scholar]
- Green PB, Linstead P (1990) A procedure for SEM of complex shoot structures applied to the inflorescence of snapdragon (Antirrhinum). Protoplasma 158: 33–38 [Google Scholar]
- Gu Q, Ferrandiz C, Yanofsky MF, Martienssen R (1998) The FRUITFULL MADS-box gene mediates cell differentiation during Arabidopsis fruit development. Development 125: 1509–1517 [DOI] [PubMed] [Google Scholar]
- Hammer K, Knüpffer S, Knüpffer H (1990) Das Gaterslebener Antirrhinum-sortiment. Kulturpflanze 38: 91–117 [Google Scholar]
- Keck E, McSteen P, Carpenter R, Coen E (2003) Separation of genetic functions controlling organ identity in flowers. EMBO J 22: 1058–1066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim GT, Shoda K, Tsuge T, Cho KH, Uchimiya H, Yokoyama R, Nishitani K, Tsukaya H (2002) The ANGUSTIFOLIA gene of Arabidopsis, a plant CtBP gene, regulates leaf-cell expansion, the arrangement of cortical microtubules in leaf cells and expression of a gene involved in cell-wall formation. EMBO J 21: 1267–1279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim GT, Tsukaya H, Uchimiya H (1998) The ROTUNDIFOLIA3 gene of Arabidopsis thaliana encodes a new member of the cytochrome P-450 family that is required for the regulated polar elongation of leaf cells. Genes Dev 12: 2381–2391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusaba M, Miyahara K, Iida S, Fukuoka H, Takano T, Sassa H, Nishimura M, Nishio T (2003) Low glutelin content1: a dominant mutation that suppresses the glutelin multigene family via RNA silencing in rice. Plant Cell 15: 1455–1467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo D, Carpenter R, Copsey L, Vincent C, Clark J, Coen E (1999) Control of organ asymmetry in flowers of Antirrhinum. Cell 99: 367–376 [DOI] [PubMed] [Google Scholar]
- Luo D, Carpenter R, Vincent C, Copsey L, Coen E (1996) Origin of floral asymmetry in Antirrhinum. Nature 383: 794–799 [DOI] [PubMed] [Google Scholar]
- Masucci JD, Rerie WG, Foreman DR, Zhang M, Galway ME, Marks MD, Schiefelbein JW (1996) The homeobox gene GLABRA2 is required for position-dependent cell differentiation in the root epidermis of Arabidopsis thaliana. Development 122: 1253–1260 [DOI] [PubMed] [Google Scholar]
- Mizukami Y, Fischer RL (2000) Plant organ size control: AINTEGUMENTA regulates growth and cell numbers during organogenesis. Proc Natl Acad Sci USA 97: 942–947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nath U, Crawford BC, Carpenter R, Coen E (2003) Genetic control of surface curvature. Science 299: 1404–1407 [DOI] [PubMed] [Google Scholar]
- Noda K, Glover BJ, Linstead P, Martin C (1994) Flower colour intensity depends on specialized cell shape controlled by a Myb-related transcription factor. Nature 369: 661–664 [DOI] [PubMed] [Google Scholar]
- Riou-Khamlichi C, Huntley R, Jacqmard A, Murray JA (1999) Cytokinin activation of Arabidopsis cell division through a D-type cyclin. Science 283: 1541–1544 [DOI] [PubMed] [Google Scholar]
- Stubbe H (1966) Genetik und Zytologie von Antirrhinum L. sect. Antirrhinum. Veb Gustav Fischer Verlag, Jena, Germany
- Tsuge T, Tsukaya H, Uchimiya H (1996) Two independent and polarized processes of cell elongation regulate leaf blade expansion in Arabidopsis thaliana (L.) Heynh. Development 122: 1589–1600 [DOI] [PubMed] [Google Scholar]
- Van der Krol AR, Mur LA, Beld M, Mol JN, Stuitje AR (1990) Flavonoid genes in petunia: Addition of a limited number of gene copies may lead to a suppression of gene expression. Plant Cell 2: 291–299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vincent C, Coen ES (2004) A temporal and morphological framework for flower development in Antirrhinum majus. Can J Bot (in press)