Abstract
Gibberellins (GAs) are involved in regulation of many aspects during plant development. To investigate the impact of altered GA levels on plant growth and metabolism, transgenic tobacco (Nicotiana tabacum) plants have been engineered to express either a GA20-oxidase (AtGA20-ox) or a GA2-oxidase (AtGA2-ox) gene from Arabidopsis under control of the cauliflower mosaic virus 35S promoter. Resulting plants were characterized by elongated or stunted shoot growth, respectively, indicating changes in the content of bioactive GAs. In accordance with the effect on plant growth, biomass production was increased or decreased in AtGA20-ox or AtGA2-ox plants, respectively, and was found to be positively correlated with the rate of photosynthesis as determined at the whole plant level. Differences in dry matter accumulation were most likely due to changes in lignin deposition as indicated by histochemical staining and quantitative measurements. Altered lignification of transgenic plants was paralleled by up- or down-regulation of the expression of lignin biosynthetic genes. Short-term GA3 feeding of excised petioles induced lignin formation in the absence of a transcriptional activation of pathway-specific genes. Thus, short-term GA treatment mediates lignin deposition most likely by polymerization of preformed monomers, whereas long-term effects on lignification involve elevated production of precursors by transcriptional stimulation of the biosynthetic pathway. Interestingly, analysis of stem cross sections revealed a differential effect of GA on the formation of xylem and pith cells. The number of lignified vessels was increased in AtGA20-ox plants pointing to a stimulation of xylem formation while the number of pith cells declined indicating a negative regulation.
Gibberellins (GAs) are plant hormones that contribute to the control of growth and development of higher plants throughout their life cycle. Although a bewildering number of different GA species have been identified, only a few of them are believed to possess biological activity (Hedden and Phillips, 2000). The most obvious function of GAs is to promote stem elongation, which is best illustrated by GA-deficient mutants of Arabidopsis, pea (Pisum sativum), and maize (Zea mays; Hedden and Proebsting, 1999). Furthermore, GAs are involved in the regulation of seed germination, leaf expansion, flowering, and fruit development, and the mediation of environmental signals such as day length. On the cellular level GAs stimulate both cell elongation and cell division (Kende and Zeevaart, 1997).
GAs originate from a branch of the diterpenoid pathway. Their biosynthesis can be divided into three major stages according to the localization and the nature of the enzymes involved (Graebe, 1987; Hedden and Kamiya, 1997; Hedden and Phillips, 2000). GA formation is initiated by the cyclization of geranylgeranyldiphosphate in the plastid. Subsequently, GA12-aldehyde is formed by the action of cytochrome-P450-dependent mono-oxygenases on the exterior of the endoplasmatic reticulum. GA12-aldehyde is the first GA produced and is thus the precursor of all other GA species. They are formed in the cytosol in a complex metabolic network, varying between species, with developmental stage and in a tissue-specific manner. These later steps of the pathway are catalyzed by soluble 2-oxogluterate- dependent dioxygenases. This group comprises enzymes either involved in the formation of the bioactive GA1 and/or GA4 such as GA20-oxidases and GA3-oxidases or involved in their deactivation such as GA2-oxidases. Both GA20-oxidase and GA2-oxidase have been considered as important regulators of the biosynthetic pathway in biochemical and molecular studies (summarized in Hedden and Kamiya, 1997; Hedden and Phillips, 2000).
The recent cloning of most genes coding for enzymes of GA biosynthesis offered the opportunity to modify their expression in transgenic plants to study their function and regulation. Genes encoding the multifunctional GA20-oxidase have been cloned from various species including pumpkin (Cucurbita moschata; Lange et al., 1994), Arabidopsis (Phillips et al., 1995; Xu et al., 1995), pea (Martin et al., 1996), tobacco (Nicotiana tabacum; Tanaka-Ueguchi et al., 1998), potato (Solanum tuberosum; Carrera et al., 1999), and hybrid aspen (Populus tremula × Populus tremuloides; Eriksson and Moritz, 2002). Overexpression of different Arabidopsis GA20-oxidases in transgenic Arabidopsis plants gave rise to elongated hypocotyls of seedlings, increased shoot growth, an early flowering phenotype, and led to a 2- to 3-fold increase in the level of GA4 (Coles et al., 1999). Thus, the overexpression of GA20-oxidase provides an effective tool to increase the level of bioactive GA. Several approaches have been followed to reduce the level of bioactive GAs including antisense expression of GA20-oxidase in Arabidopsis and potato (Coles et al., 1999; Carrera et al., 2000) or the expression of a pumpkin GA20-oxidase, which is an unusual enzyme producing C-20-carboxylic acid GAs rather than the precursors of bioactive GAs (Curtis et al., 2000; reviewed in Hedden and Phillips, 2000). However, these strategies have only been partially successful. Another approach to reduce the endogenous GA level is to increase the rate of its catabolism. This is initiated by GA2-oxidases that inactivate both the bioactive GAs and their immediate precursors. The first cDNA encoding a GA2-oxidase was isolated from runner bean (Phaseolus coccineus) by a functional screening (Thomas et al., 1999). Meanwhile, the sequence information is available from a number of other species such as Arabidopsis (Thomas et al., 1999), pea (Lester et al., 1999; Martin et al., 1999), and rice (Oryza sativa; Sakamoto et al., 2001). Ectopic expression of GA2-oxidase in rice resulted in inhibition of stem growth, small, dark green leaves, and impaired development of the reproductive organs (Sakamoto et al., 2001), whereas expression of the same gene under control of the shoot-specific OsGA3-ox2 promoter induced only semi-dwarfism without any negative effect on flower and grain development (Sakamoto et al., 2003). Since it has been shown that the so-called green revolution genes are involved in GA signaling and biosynthesis (Peng et al., 1999; Spielmeyer et al., 2002) this approach offers an alternative strategy to introduce beneficial traits, such as dwarf architecture into cereal varieties to improve grain yield.
In summary, the results demonstrate that GA levels and, hence, plant growth and development, can be manipulated by genetic engineering. Apart from the promotive effect of GA on plant elongation, Eriksson et al. (2000) reported that expression of Arabidopsis GA20-oxidase in hybrid aspen also improved biomass production. Moreover, the authors could show that increased levels of endogenous GAs led to more and longer xylem fibers, which is a desirable feature of tree-breeding programs. These results were in agreement with earlier GA application experiments showing a stimulation of xylem fiber elongation and cambial activity (Wareing, 1958; Digby and Wareing, 1966; Ridoutt et al., 1996). However, remarkably little is known about how GAs affect photosynthesis, which supplies carbon skeletons and energy for growth (Nagel and Lambers, 2002). The few available reports reveal contradictory results showing that application of GA3 either stimulates (Yuan and Xu, 2001; Ashraf et al., 2002) or reduces the photosynthetic rate (Dijkstra et al., 1990).
The aim of our work was to elucidate the impact of altered GA biosynthesis on plant growth, morphology, and metabolism with special emphasis on the relationship between changed plant growth, biomass production, lignin formation, and photosynthetic capacity. To this end, transgenic tobacco plants have been created expressing either an Arabidopsis GA20-oxidase or GA2-oxidase under the control of the 35S cauliflower mosaic virus (CaMV) promoter. Here, we provide a thorough morphological and anatomical characterization of the transgenic lines and show that GA not only affects plant elongation but seems also to exert different effects on xylem and pith cell formation. Moreover, GA content was found to be positively correlated with biomass accumulation, lignin formation, and the rate of photosynthesis. The short-term effect of GA on lignin biosynthesis was studied by feeding experiments revealing a GA-induced accumulation of lignin without transcriptional activation of pathway-specific genes.
RESULTS
Expression of Both AtGA20-ox and AtGA2-ox Affects Plant Growth and Morphology
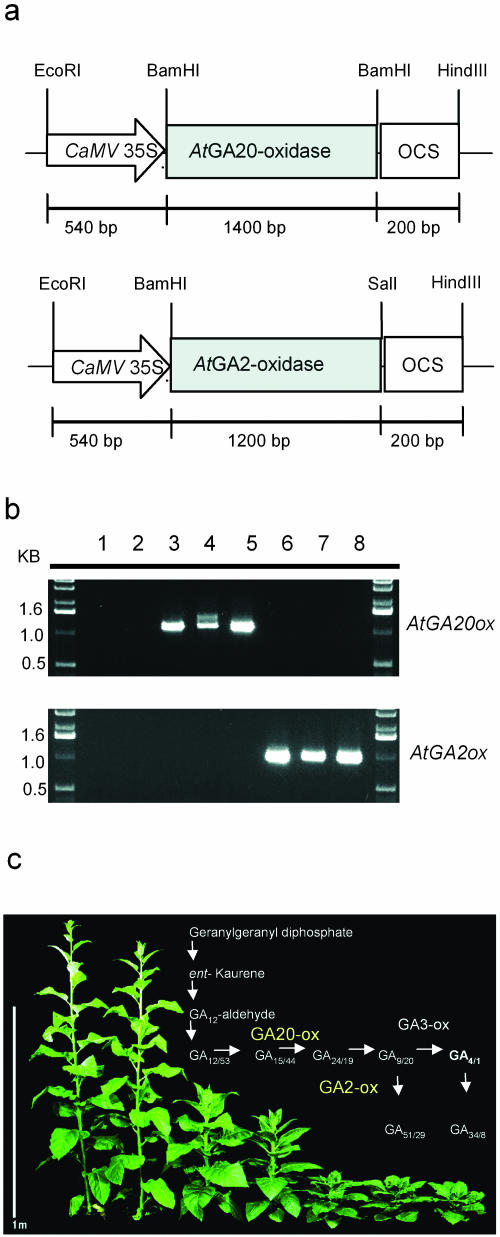
To alter GA levels in transgenic tobacco plants, genomic clones coding either for GA20-oxidase (accession no. X8339; AtGA20-ox) or GA2-oxidase (accession no. AJ123435; AtGA2-ox) from Arabidopsis have been amplified by PCR and inserted between the CaMV 35S promoter and the octopin synthase polyadenylation signal of the pBinAR vector (Höfgen and Willmitzer, 1990; Fig. 1a). These constructs were used to transform tobacco plants using Agrobacterium-mediated gene transfer. About 75 transgenic lines were obtained from each construct. Since the most obvious effect of GA is on plant growth, transgenic plants could be easily prescreened for expression of AtGA20-ox or AtGA2-ox by the occurrence of a slender or a dwarf phenotype, respectively (Fig. 1c). Expression of either transgene was verified by northern-blot analysis in preselected transgenic tobacco lines (data not shown).
Figure 1.
Expression of GA20-oxidase and GA2-oxidase from Arabidopsis in transgenic tobacco plants. a, Schematic structure of AtGA20-ox and AtGA2-ox constructs used to transform tobacco plants. b, RT-PCR of selected transgenic lines using gene-specific primers for either AtGA20-ox or AtGA2-ox as given in “Materials and Methods.” Size of amplified products corresponds to the size of the respective cDNA clones (1,133 bp for AtGA20-ox, 990 bp for AtGA2-ox). Lane 1 to 2, wild type (SNN); lane 3 to 5, independent transgenic lines of AtGA20-ox expressing plants (nos. 15, 10, and 7); and lanes 6 to 8, independent transgenic lines of AtGA2-ox expressing plants (nos. 41, 32, and 2). c, Phenotypic alteration of transgenic tobacco plants caused by the expression of AtGA20-ox or AtGA2-ox after 8 weeks in the greenhouse. From left to right: AtGA20-ox line numbers 15 and 7; wild type; AtGA2-ox line numbers 74 (low expression), 2, 32, and 41. The inset schematically illustrates the GA biosynthetic pathway. GA20-ox catalyzes the conversion from GA12/53 to GA9/20. GA2-oxidase inactivates the bioactive GA4/1 and their precursors GA9/20.
Three highly expressing lines of each construct were chosen (Fig. 1b) and subjected to a thorough morphological characterization (Table I). Homozygous T2 plants were selected by their kanamycin-resistance and the appearance of a homogenous phenotype. Transgenic tobacco plants expressing the AtGA20-ox exhibited elongated shoots, slightly thinner and pale leaves compared to wild-type plants, which was also reflected by a reduced content of chlorophyll per leaf area (Table I, Fig. 1c). These plants tended to flower earlier (S. Biemelt and U. Sonnewald, unpublished data) and had enlarged flowers (Table I). Shoot growth of AtGA2-ox expressing plants was strongly retarded and stem length corresponded to only 16% of that of wild-type plants (Fig. 1c, Table I). Counting the number of leaves revealed that the observed differences in shoot height were primarily due to the different length of internodes. AtGA2-ox tobacco plants possess small, thick, dark green leaves coinciding with an increased chlorophyll content compared to control leaves (Table I). Leaf cross sections showed a thicker spongy parenchyma layer and an extra but incomplete palisade layer compared to wild-type and AtGA20-ox leaves (data not shown). Flowering of these plants was delayed (S. Biemelt and U. Sonnewald, unpublished data) and flowers formed were smaller than wild-type flowers (Table I). Moreover, the number of seed capsules was reduced by 40% to 70% in the dwarf AtGA2ox lines. However, seeds were fertile and developed into seedlings, although with strongly shortened hypocotyls. Treatment of seedling with 10 μm GA3 restored their growth to that of GA3-treated wild-type controls, whereas treatment of GA20-ox seedlings resulted in no further increase of hypocotyl length (data not shown).
Table I.
Phenotypic characteristics of soil-grown transgenic tobacco plants expressing either a GA20-oxidase (AtGA20-ox) or a GA2-oxidase (AtGA2-ox) from Arabidopsis
| Wild Type
|
AtGA20-ox
|
AtGA2-ox
|
|||||
|---|---|---|---|---|---|---|---|
| GA20-7 | GA20-10 | GA20-15 | GA2-2 | GA2-32 | GA2-41 | ||
| Stem height (cm) | 68.6 ± 9.9 | 88.2 ± 6.7 | 86.8 ± 12.5 | 107.6 ± 7.8 | 11.6 ± 0.6 | 11.8 ± 0.2 | 11.3 ± 1.3 |
| Number of leaves | 22.8 ± 0.4 | 24.5 ± 0.5 | 24.2 ± 0.7 | 24.3 ± 1.0 | 24.4 ± 0.8 | 23.2 ± 1.2 | 22.2 ± 1.2 |
| Leaf thickness (mm) | 0.23 ± 0.02 | 0.19 ± 0.02 | 0.19 ± 0.02 | 0.19 ± 0.02 | 0.31 ± 0.02 | 0.33 ± 0.01 | 0.37 ± 0.01 |
| Stem fwt (g) | 73.9 ± 7.0 | 93.9 ± 11.3 | 75.3 ± 9.0 | 93.3 ± 5.7 | 32.1 ± 2.8 | 28.5 ± 3.1 | 24.3 ± 6.3 |
| Stem dry wt (g) | 8.9 ± 2.0 | 15.9 ± 1.9 | 11.5 ± 2.3 | 16.6 ± 1.3 | 3.1 ± 1.0 | 2.5 ± 1.0 | 1.8 ± 0.7 |
| fwt/dwt | 8.3 | 5.9 | 6.5 | 5.6 | 10.7 | 11.4 | 13.5 |
| Stem diameter (mm) | 9.6 ± 0.6 | 9.6 ± 0.6 | 10.2 ± 0.7 | 12.4 ± 0.7 | 15.3 ± 1.2 | 15.8 ± 0.9 | 14.7 ± 1.5 |
| Flower length (mm) | 48.3 ± 1.25 | 53.3 ± 1.25 | 50.3 ± 0.47 | 53.3 ± 0.94 | 34.7 ± 0.47 | 33.0 ± 1.41 | 33.7 ± 0.94 |
| Total chlorophyll (mg m−2) | 416 ± 31 | 335 ± 27 | 394 ± 38 | 330 ± 50 | 556 ± 11 | 588 ± 26 | 678 ± 30 |
Results are given for three independent lines of each construct and represent the mean ±. fwt, fresh weight. dry wt, dry weight.
To assess the impact on biomass production we determined fresh and dry weight of stems of fully mature plants. As shown in Table I there were significant differences in stem biomass between wild type and the two groups of transgenic lines. While the stem fresh weight was increased up to 27% in AtGA20-ox lines, it was reduced by 70% in AtGA2-ox lines. On dry weight basis the differences were even more pronounced, which is reflected by a decreased or increased fresh to dry weight ratio for AtGA20-ox and AtGA2-ox lines, respectively.
Manipulation of GA Biosynthesis Led to an Altered Number of Lignified Vessels
One possible reason for the changed fresh to dry weight ratio could be the altered composition or deposition of structure-forming substances like cellulose and/or lignin. To visualize differences in lignification, cross sections of petioles from different transgenic lines were stained with phloroglycinol-HCl. The histochemical comparison revealed that the number of lignified vessels was higher in the tall AtGA20-ox plants and lower in dwarf AtGA2-ox plants as compared to wild-type controls. In addition, red color appeared more intense in AtGA20-ox plants (data not shown). These initial experiments indicated that altered GA levels not only influence plant growth but also the lignin content.
For more detailed investigations we selected one line of each construct because the general morphological features within one group were essentially the same. In the following experiments selected lines (AtGA20-15; AtGA2-41) and wild-type plants were grown on sand and watered daily with a nutrient solution to standardize growth conditions.
Lignification is a characteristic feature of secondary growth and increases with maturity. Therefore, different parts of stems were analyzed (apical part, eighth internode; middle part, fifth internode; basal part, second internode). The transgenic lines differed substantially in their shoot height but not in their leaf number. Thus, lignin accumulation was compared in stem sections derived from the same internodes of plants at the same age.
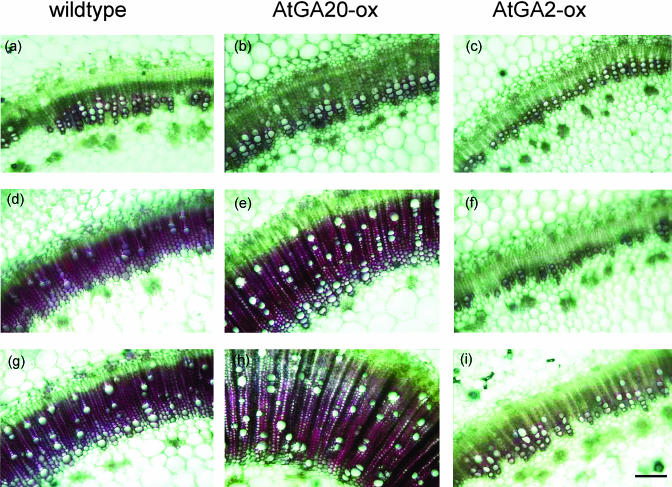
A representative lignin staining pattern is shown in Figure 2. In general, there was a progressive increase in the number of lignified vessels and the amount of lignin deposition from the apex toward the base of the stem. Comparison of cross sections through the upper part of the stems of transgenic and wild-type plants revealed only minor differences because of the low degree of lignification (Fig. 2, a–c), although a slightly less intense staining could be observed in the dwarf AtGA2-ox plants. Differences between transgenics and wild type became clearly evident when cross sections of the fifth and second internodes were inspected (Fig. 2, d–i). Transgenic tobacco plants expressing the AtGA20-ox displayed increased staining of the xylem area compared to the control (Fig. 2, d, f, g, and h). Stems of tall AtGA20-ox plants apparently formed more xylem cells. In contrast, staining intensity was clearly lowered in AtGA2-ox plants indicating a reduced lignin content (Fig. 2, f and i). This seems to be mainly brought about by a decreased number of lignified xylem vessels.
Figure 2.
Histochemical detection of lignin in stem cross sections of transgenic tobacco plants altered in their GA biosynthesis. Stem material from different parts of the axis of wild type, AtGA20-ox number 15, and AtGA2-ox number 41 was dehydrated by increasing ethanol concentrations (plants at 16-leaf stage). Subsequently, cross sections were stained for lignin using phloroglycinol-HCl reagent. Similar results were obtained in independent experiments. a–c, Cross sections through the upper stem part corresponding to eighth internode (counted from bottom to top) of wild type, AtGA20-ox, and AtGA2-ox, respectively. d–f, Cross sections through the middle stem part corresponding to fifth internode of wild type, AtGA20-ox, and AtGA2-ox, respectively. g–i, Cross sections through the lower stem part corresponding to second internode of wild type, AtGA20-ox, and AtGA2-ox, respectively. Bar is equivalent to 200 μm.
The histochemical data could be confirmed by measuring the lignin content using the thioglycolic acid method (Campbell and Ellis, 1992). As shown in Table II the lignin content increased more than 2-fold in stems of AtGA20-ox lines compared to the respective parts of wild-type plants. However, the highest relative increase (3.6-fold) was obtained in the apical part of these plants. In contrast, AtGA2-ox plants deposited only small amounts of lignin and, as already observed histochemically, the differences were more pronounced in the middle and basal parts (Table II).
Table II.
Lignin content in different stem parts of transgenic and wild-type plants
| Stem Part
|
Lignin Content
|
||
|---|---|---|---|
| Wild Type | AtGA20-ox | AtGA2-ox | |
| mg g−1 fwt | |||
| Apical | 1.5 ± 0.5 | 5.4 ± 0.7 | 1.3 ± 0.1 |
| Medial | 20.6 ± 4.7 | 40.6 ± 4.1 | 3.7 ± 0.8 |
| Basal | 32.5 ± 9.1 | 76.8 ± 5.8 | 13.0 ± 1.5 |
Stem material was harvested from sand-grown plants with a total of 16 leaves. Samples were taken from the eighth, fifth, and second internodes (counted from bottom to top of the plants) of wild type, AtGA20-ox number 15, and AtGA2-ox number 41 corresponding to apical, medial, and basal stem parts, respectively. The values represent the mean ± sd of two independent experiments with three samples each.
In summary, manipulation of GA biosynthesis led to an altered biomass accumulation and a changed fresh to dry matter ratio that was most likely caused by a modified lignin deposition.
Unequal Stimulation of Cell Division by GA
In contrast to the thickened xylem the mean stem diameter of AtGA20-ox plants was almost unaltered compared to wild-type stems, whereas AtGA2-ox plants exhibited clearly thicker stems (Table I). Closer inspection of fifth internode cross sections revealed that stems of AtGA2-ox exhibited an enlarged pith area. In stems of AtGA20-ox lines, however, a more extended xylem and a reduced pith area was observed indicating anatomical differences compared to wild type. To examine whether the differences in the core were caused by an altered size and/or number of cells, the number of pith cells was determined. Within a 1-mm distance 7.5 ± 0.8, 5.5 ± 0.6, and 9.3 ± 1.2 (n = 12) were counted for wild-type, AtGA20-ox, and AtGA2-ox plants, respectively. Thus, the size of pith cells was increased in AtGA20-ox and decreased in AtGA2-ox plants. Hence, stems of AtGA2-ox plants were thicker because of an enlarged pith consisting of a higher number of small cells. Taken together, these results indicate that GA apparently promotes cell division in the fascicular cambium, but may negatively control pith cell formation.
Changes in Lignification Were Accompanied by Altered Expression of Biosynthetic Genes
To explore whether changes in lignin content could be correlated to altered gene expression, cDNAs coding for enzymes of lignin biosynthesis were isolated by PCR and transcriptional changes were studied in identical stem parts as used for the determination of lignin content.
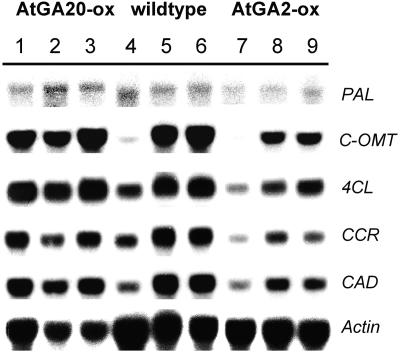
Transcripts coding for Phe ammonia-lyase (Pal) were found to be almost unchanged between the different internodes and also between wild type and the different transgenics (Fig. 3). The enzyme catalyzes the initial step of the phenylpropanoid pathway(s) also feeding into lignin formation. Expression of genes encoding enzymes operating more downstream in the lignin biosynthesis such as catechol O-methyltransferase (COMT), 4-coumarate:CoA ligase (4CL), cinnamoyl:CoA reductase (CCR), and cinnamyl alcohol dehydrogenase (CAD) showed a clear increase with lignin accumulation in wild-type plants (Fig. 3). Compared to wild type, the amounts of these transcripts were clearly reduced in AtGA2-oxidase expressing lines. This effect was most obvious in the middle and basal stem sections coinciding with a significantly decreased lignin formation in these parts. Expression of the lignin biosynthetic genes was uniformly high in the different stem parts of AtGA20-ox plants. Compared to wild type, transcript accumulation was more abundant in the apical part where also the highest difference (3.6-fold) in lignin content was determined. This indicates that transgenic lines with increased GA biosynthesis started to induce lignin formation already at younger developmental stages. However, changes in lignification observed in transgenic lines were associated with an altered expression of pathway-specific genes.
Figure 3.
Effect of altered GA metabolisms on expression of lignin biosynthesis genes. Northern-blot analysis of PAL and transcripts specific for lignin biosynthesis in stems of wild type (lanes 4–6), AtGA20-ox number 15 (lanes 1–3), and AtGA2-ox number 41 (lanes 7–9). Samples were taken from upper (eighth internode, lanes 1, 4, and 7), middle (fifth internode, lanes 2, 5, and 8), and lower stem parts (second internode, lanes 3, 6, and 9) of plants at 16-leaf stage. Twenty micrograms of total RNA were loaded per lane and probed with PAL, COMT, 4CL, CCR, and CAD, respectively. An actin probe was used as a loading control.
Short-Term GA3 Treatment Led to Increased Lignification but Not to a Transcriptional Activation of Lignin-Biosynthetic Genes
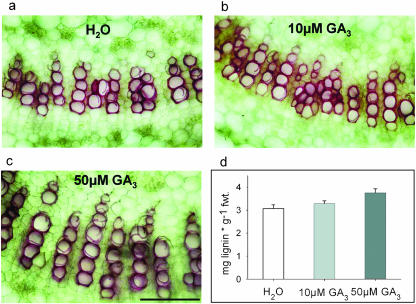
We aimed to elucidate whether lignin formation was transcriptionally induced by GA or whether the observed effects were rather indirect. Therefore, different methods to apply GA3 were tested such as spraying or watering plants. Incubating excised petioles in darkness has been proven to be the most reliable and appropriate system in our hands to study short-term responses. Thus, petioles were incubated in either water, 10 μm GA3, or 50 μm GA3, and expression of 4CL or CCR as marker genes for lignin biosynthesis was analyzed following 6, 18, or 24 h of treatment. Northern-blot analysis did not reveal significant differences in the amount of transcripts (data not shown). Nevertheless, a gradual increase in the degree of lignification could be seen in cross sections of petioles after incubation for 24 h with increasing GA3 concentrations (Fig. 4, a–c). While in control, sections at an average four xylem vessels were stained, this number increased up to seven in petioles treated with 50 μm GA3. Using the quantitative assay a 22% increase in lignin content was measured in petioles treated with 50 μm GA3 as compared to water control (Fig. 4d), which confirms the histological observations.
Figure 4.
Effect of GA3 treatment on lignin content. Petioles of mature tobacco plants were infiltrated with water, 10 μm GA3, or 50 μm GA3, respectively, and kept for 24 h in darkness at room temperature. a–c, Histochemical detection of lignin in cross sections using thioglycol-HCl staining: (a) SNN + water, (b) SNN+ 10 μm GA3, and (c) SNN + 50 μm GA3. Bar corresponds to 200 μm. d, Lignin content in petioles treated with water, 10 μm GA3, or 50 μm GA3. Values represent the mean ± sd of three different experiments (n = 9).
Single Leaf Measurement of Photosynthesis Did Not Reflect Altered Biomass Accumulation of Transgenic Plants
We showed that manipulation of GA biosynthesis led to altered biomass production in transgenic tobacco plants. To evaluate whether this effect was correlated by an enhanced or a reduced photosynthetic capacity, CO2 uptake was measured in leaves of AtGA20-ox, AtGA2-ox, and wild-type plants. For these investigations one line of each group was analyzed using the same experimental set-up described earlier (cultivation of plants on sand). Under these conditions the mean dry weight per plant (n = 24–27) was 3.94 g ± 0.24 g in case of the wild-type plants, 4.74 g ± 0.25 g for AtGA20-ox plants, and 1.93 g ± 0.14 g for the AtGA2-ox plants, respectively.
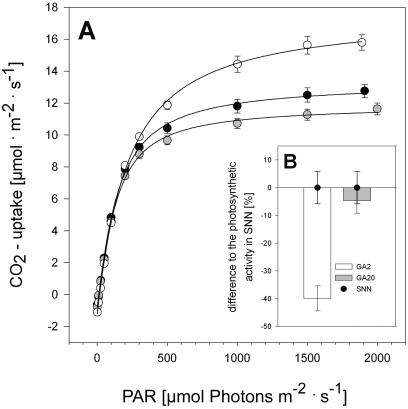
Figure 5A illustrates the effect of actinic light intensity on CO2 uptake of a fully mature leaf of the different lines. The photosynthetic CO2 uptake in leaves of AtGA20-ox plants was slightly decreased compared to the wild type. In contrast, photosynthetic activity was increased in AtGA2-ox plants under nearly saturating illumination. The enhanced photosynthetic activity of AtGA2-ox leaves was more remarkable in older (leaf 8, about 75%) than in younger leaves (leaf 12, about 15%; Table III). The maximum photosynthetic rate of corresponding AtGA20-ox leaves was not significantly altered compared to wild type.
Figure 5.
Single leaf measurements of photosynthesis. A, Light response curves of net photosynthetic rate were monitored in fully mature leaves of wild-type (SNN, black circles) and transgenic plants with altered GA biosynthesis (AtGA2-ox no. 41 [GA2], white circles; AtGA20-ox no. 15 [GA20], gray circles). Each data point is the mean ± se of 18 to 20 independent measurements. B, Photosynthetic activity of whole plants was calculated from 4 to 5 leaves of different age at 200 μmol quanta m−2 s−1 light intensity. Columns represent the difference (in percent) to the mean value of SNN plants (black circle). The mean gas exchange rate of wild-type plants was 0.762 ± 0.044 μmol CO2 s−1/plant. Data shown are the mean ± se of 24 to 27 independent values.
Table III.
Characteristic photosynthetic features of tobacco plants with altered GA biosynthesis
| Leaf 12
|
Leaf 8
|
|||||
|---|---|---|---|---|---|---|
| Wild Type | AtGA20-ox | AtGA2-ox | Wild Type | AtGA20-ox | AtGA2-ox | |
| Maximum CO2 uptake (μmol m−2 s−1) | 15.8 ± 0.5 | 15.4 ± 0.3 | 20.0 ± 1.0 | 7.6 ± 0.3 | 8.0 ± 0.4 | 13.4 ± 0.3 |
| Maximum quantum yield of CO2 fixation (mmol CO2 mol−1 quanta) | 0.058 ± 0.001 | 0.059 ± 0.001 | 0.060 ± 0.001 | 0.053 ± 0.002 | 0.051 ± 0.001 | 0.059 ± 0.001 |
| Fv/Fm | 0.830 ± 0.003 | 0.830 ± 0.002 | 0.841 ± 0.002 | 0.811 ± 0.004 | 0.802 ± 0.004 | 0.838 ± 0.002 |
| Chlorophyll (mg m−2) | 376 ± 9.2 | 332 ± 8 | 628 ± 26 | 308 ± 14 | 252 ± 12 | 522 ± 14 |
Parameters were measured from different leaves of wild-type and transgenic plants expressing AtGA20-ox (no. 15) or AtGA2-ox (no. 41; number of leaves counted from bottom to top, plants at 16-leaf stage). Maximum CO2 uptake and maximum quantum yield of CO2 fixation were calculated from data of light response curves at light saturation or their initial slope, respectively. Each value represents the mean ± se of 18 to 20 or 8 to 11 (in case of data for Fv/Fm and chlorophyll content) independent measurements.
The maximum quantum yield for the CO2 assimilation of leaves can be calculated from the initial slope of the light response curves. The comparison of these parameters revealed a weak but significant increase in the maximum quantum yield in leaves of AtGA2-ox plants (up to 11%), whereas it remained unaffected in AtGA20-ox plants (Table III). Note that the differences between wild-type and AtGA2-ox plants were again most strikingly in the older leaves. This result was in agreement with data obtained for the maximum quantum yield of PS II (Fv/Fm), a chlorophyll fluorescence parameter (Table III). In all leaves measured from AtGA2-ox plants the values for Fv/Fm were clearly higher than in wild-type leaves. The quantum yield of PS II was maintained at a high level in older leaves of dwarf plants, whereas it decreased with increasing leaf age in wild-type and AtGA20-ox plants (data not shown). The tendency that the differences between wild-type and AtGA2-ox lines were more pronounced in older leaves was also apparent for the chlorophyll content (Table III) and might be indicative for a delayed development.
We had expected that the tall AtGA20-ox plants with the highest biomass accumulation would also be characterized by increased net assimilation rates. However, the photosynthetic activity measured from single leaves contradicts this assumption. In an attempt to resolve the contradiction we aimed to assess the photosynthetic capacity of an entire plant. Therefore, CO2 uptake was measured in up to five different leaves of each plant. The light intensity in these experiments was 200 μmol quanta m−2 s−1, an intensity that was reached on average in the greenhouse during the day. The photosynthetic activity of all leaves and finally of whole plants was calculated based on these data and under consideration of the specific leaf area per plant, which was 1,342 ± 57, 1,172 ± 46, and 814 ± 37 cm2 for wild-type, AtGA20-ox, and AtGA2-ox plants, respectively. As shown in Figure 5B, this calculation revealed that the photosynthetic activity of the small AtGA2-ox plants was reduced to 60.1% ± 6.8% of that of wild-type plants. However, by means of this method, no difference between wild-type plants and the tall AtGA20-ox plants was obtained.
Whole Plant Measurement of Photosynthetic Capacity Revealed Changes in Photosynthesis That Were in Agreement with Biomass Production
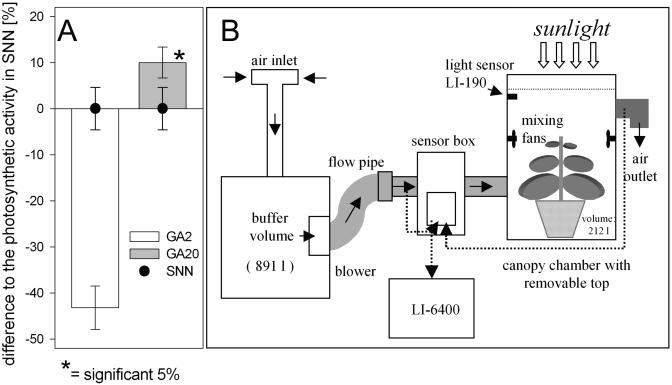
The described procedure ignores some important parameters that influence the photosynthetic capacity of a whole plant such as size of plants, position, or self shading of leaves. Therefore, we used an experimental system (PMK-1), which is schematically depicted in Figure 6B. This experimental set-up allows the measure of the gas exchange of whole plants by analyzing the continuously exchanged gas volume of the canopy chamber. Air is sucked in by a blower during the measurement and flushed through the system. A big case serves as a buffer volume and is connected to the canopy chamber by a flexible tube and a flow pipe. The function of the sensor box containing the sensors and pumps is controlled by the LI-6400. The temperature and the molar fraction of CO2 and H2O are measured inside the flow pipe and inside the tube at the chamber outlet. From these continuously recorded data the CO2 uptake of the whole plant is calculated by the LI-6400. These measurements were conducted in the greenhouse under daylight conditions to exclude artifacts caused by artificial light sources. For comparison it was necessary to calculate the photosynthetic rate for each plant under the same light condition, which was 200 μmol photons m−2 s−1 (see “Materials and Methods”).
Figure 6.
Photosynthetic activities of whole plants measured in the canopy chamber PMK1. A, CO2 uptake rates of AtGA2-ox (GA2) and AtGA20-ox (GA20 expressing tobacco plants are shown as difference, in percent, to the mean value of wild-type plants (SNN). Mean CO2 uptake of wild-type plants was 0.722 ± 0.033 μmol CO2 s−1/plant. Light intensity was 200 μmol quanta m−2 s−1. Each bar is the mean ± se of 24 to 27 independent measurements. B, Schematic illustration of the continuous flow canopy chamber system PMK1 for measuring whole plant gas exchange.
The photosynthetic CO2 uptake of whole wild-type plants measured in the canopy chamber (0.722 ± 0.033 μmol CO2 s−1/ plant) was only slightly lower compared to the activity calculated on the basis of single leaf measurements (0.762 ± 0.044 μmol CO2 s−1/plant). Both methods to determine the photosynthetic capacity of a whole plant revealed a significant reduction in net assimilation of AtGA2-ox plants (Figs. 5B and 6A) correlating with a low amount of dry matter accumulation and the dwarf phenotype. However, in case of the AtGA20-ox plants a clear difference between the methods was apparent. Contrary to the calculation based on single leaf measurements, the whole plant measurement in the canopy chamber indicated a significant increased CO2 uptake in AtGA20-ox plants (10% ± 4.6%). It is worth mentioning that the exact same plants were used for both measurements. Thus, the greater biomass production of the tall AtGA20-ox plants correlates with an enhanced photosynthetic capacity. These results demonstrate the importance of whole plant measurements if plant stature has to be taken in account.
DISCUSSION
Genetic manipulation of GA biosynthesis or degradation has become an alternative approach to the widespread use of chemical regulators to modify plant growth and stature since most of the biosynthetic genes have been identified. There is accumulating evidence that the dioxygenases catalyzing the later steps in the pathway, namely GA20-oxidases and GA2-oxidases, are important regulators of GA biosynthesis using feedback or feedforward mechanisms. Additionally, expression of these genes is under environmental and developmental control.
In order to engineer transgenic plants with increased or decreased GA levels, we cloned both a GA20-oxidase and a GA2-oxidase from Arabidopsis and expressed them constitutively in tobacco plants. The phenotype of the transgenic plants obtained is consistent with phenotypes observed in previous studies using transgenic approaches as well as mutants that have either higher GA levels or are GA-deficient. A GA overproduction phenotype is characterized by longer hypocotyls, increased internode length, pale-green leaves, and early flower induction. These phenotypic effects were attained by overexpression of endogenous GA20-oxidase genes in Arabidopsis (Coles et al., 1999) or potato (Carrera et al., 2000) and also in our study by heterologous expression of an Arabidopsis GA20-oxidase in tobacco. Thus, these transgenic plants behave like control plants that have been treated with GA. The main characteristic of GA-deficient mutants like the Arabidopsis ga1-3 mutant is their dwarf growth and the occurrence of small dark-green leaves (Koornneef and van der Veen, 1980). We obtained tobacco plants resembling the phenotype of GA-deficient mutants by expression of an Arabidopsis GA2-oxidase. The dwarf growth phenotype of GA2-ox lines can be overcome by treatment with GA3 that is indicative for a lower content of bioactive GA. A similar phenotype was caused by expressing a GA2-oxidase in transgenic rice (Sakamoto et al., 2001) and recently in transgenic Arabidopsis and tobacco plants (Schomburg et al., 2003).
Manipulation of GA biosynthesis does not only affect plant growth and morphology but also biomass accumulation. For example, overexpression of an Arabidopsis GA20-oxidase in hybrid aspen resulted in an increased biomass production (Eriksson et al., 2000). In our experiments the relative dry matter accumulation was also enhanced in tall AtGA20-ox, but it was reduced in dwarf AtGA2-ox plants. However, little is known about the effect of GA on photosynthesis and the studies published are contradictory. For example, GA3 treatment of salt-stressed wheat plants resulted in an increased photosynthetic capacity, which was discussed as a major factor for a greater dry matter production (Ashraf et al., 2002). A stimulatory effect of applied GA3 on photosynthesis was also observed by Yuan and Xu (2001) in broad bean and soybean. In another report, GA treatment promoted growth of Plantago major, but decreased the photosynthetic rate (Dijkstra et al., 1990). No significant differences in photosynthetic rates were found between a GA-deficient tomato mutant and wild-type plants (Cramer et al., 1995). Different ways of measuring and calculating photosynthesis as well as the different experimental systems used (application of GA or GA inhibitors, use of mutants) were thought to be the reason for these contradictory results (Nagel and Lambers, 2002). The latter authors investigated the interrelationship between photosynthesis, respiration and growth using tomato GA mutants. For this purpose, CO2 exchange of intact plants was followed over 24 h and used to calculate the carbon budget (Nagel and Lambers, 2002). Although photosynthesis per unit leaf area was unaffected in the GA-deficient mutant, carbon assimilation per unit plant mass was reduced compared to wild type, which was brought about by the lower specific leaf area. The fact that the leaf area is an important factor is in accordance with our results obtained for the dwarf AtGA2-ox expressing tobacco plants. Based on single leaf measurements AtGA2-ox plants exhibited increased rates of CO2 uptake and a higher maximum quantum yield of PS II in comparison to wild-type plants. At first glance this result contradicts the lower biomass accumulation of these plants. However, a reduced CO2 was detected when the photosynthetic capacity of whole plants (based on single leaf measurements) was estimated. This approach considered the total leaf area per plant, which was clearly decreased in the AtGA2-ox line. Thus, less carbon can be fixed in total that in turn means that less carbon is available to support shoot growth.
In contrast to the AtGA2-ox plants, no significant changes in the net assimilation rate or other photosynthetic parameters could be detected in AtGA20-ox plants using both single leaf gas exchange measurements and the calculation of whole plant photosynthetic capacity. This result was in contradiction to the increased biomass production of these plants. However, calculations based on single leaf measurements neglect some important features such as distance and position of leaves to each other. To resolve this problem an experimental set-up was established enabling us to measure the gas exchange of whole plants. Generally, the data obtained in the canopy chamber for wild-type and AtGA2-ox plants were in good agreement with photosynthetic activity calculated on basis of single leaf measurements. In contrast to these results, for AtGA20-ox plants the gas exchange measurements performed in the canopy chamber revealed approximately 10% higher photosynthetic activity compared to wild-type plants. Apparently, the rise in photosynthetic activity was not caused by an increased CO2 assimilation rate per leaf area, but was most likely due to an optimal arrangement of leaves along the enlarged internodes, thereby avoiding self-shading of leaves. This was reflected by approximately 20% higher biomass accumulation of AtGA20-ox plants, which is in strong agreement with the observed changes in photosynthesis.
Biomass determination revealed that the fresh to dry weight ratio was altered in the transgenic lines. Both histochemical and biochemical analysis indicated that these changes can be assigned to differences in lignin deposition. In addition, changes in lignin content were found to be accompanied by altered expression of genes encoding enzymes of the lignin pathway. Thus, our data show that GA promotes plant growth and simultaneously affects lignification. Until now, there is no report showing a transcriptional activation of lignin biosynthesis by GA. In order to investigate whether GA directly activates lignin biosynthesis on the transcriptional level, petioles were infiltrated with GA3. Albeit increased lignin formation could be induced by short-term GA3 treatment, no significant change in the abundance of 4CL and CCR-specific transcripts was detected. An increased polymerization of existing monolignols may account for this short-term response. However, the observed transcriptional activation of lignin-biosynthetic genes in AtGA20-ox expressing plants might be the result of long-term adaptations to altered GA contents. Using microarrays, Israelsson et al. (2003) reported an up-regulation of some genes involved in lignin biosynthesis in the wood-forming tissue of AtGA20-oxidase overexpressing hybrid aspen such as a caffeolyl-coenzyme A methyltransferase, a putative sinapyl alcohol dehydrogenase, and a CAD isoform. Here, the induction of gene expression was apparently also due to long-term exposure to higher GA levels.
Transgenic hybrid aspen expressing an AtGA20-ox had more and longer xylem fibers compared to control plants, suggesting that elevated GA levels stimulate secondary growth (Eriksson et al., 2000). This is in accordance with results obtained by Ridoutt and coworkers (1996). Therein, the treatment of eucalyptus saplings with a GA biosynthesis inhibitor led to reduced fiber length and a decreased number of cambial cells and differentiating fibers. In very early studies a stimulation of cambial cell division and cell expansion by GA was already described, but differentiation and lignification of the resulting cambial derivatives was facilitated by indole-acetic acid (Wareing, 1958; Digby and Wareing, 1966). In our study, we observed an increased or decreased number of xylem vessels in stems of AtGA20-ox and AtGA2-ox plants, respectively. Interestingly, stems of dwarf AtGA2-ox plants were thicker than stems of wild-type plants, whereas the stem diameter of tall AtGA20-ox was almost unaltered. Anatomical characterization of the AtGA2-ox plants revealed that this was mainly caused by an increased number of pith cells, since the number of xylem vessels was clearly reduced. AtGA20-ox lines displayed a thicker (and stronger lignified) xylem tissue and a smaller portion of the pith. Remarkably, the size of the pith cells was increased, but the number was reduced. These data implicate that GA has opposite effects on cell division of cambial and pith cells. Generally, it is assumed that GA promotes both cell division and cell elongation. The underlying mechanisms were thoroughly studied in deepwater rice (Sauter and Kende, 1992; Sauter et al., 1995). GA exerts its control on cell division through transcriptional activation of cyclin-dependent protein kinases and mitotic cyclin genes (Sauter et al., 1995). However, GA is thought to act preferentially in young, meristematic cells (Kende and Zeevaart, 1997). The different orientation of micro fibrils is discussed as one possible reason for that. This may explain the induction of cell division in the fascicular cambium, but not the negative impact on pith cell formation. More detailed studies need to be performed to elucidate the cause for the reverse effect of GA on cell division of specific cell types.
CONCLUSION
GA biosynthesis of tobacco plants was modified by expressing either an AtGA20-ox or AtGA2-ox in order to investigate the effect of altered GA levels on plant growth and its relationship to biomass production and rate of photosynthesis. The phenotypic changes are consistent with those observed in other transgenic approaches using the same enzymes and are indicative for changes in the content of bioactive GAs. Enhanced and reduced stem growth of AtGA20-ox and AtGA2-ox plants, respectively, coincided with higher and lower biomass production and rate of photosynthesis. Increased photosynthetic activity of AtGA20-ox plants was only detectable by gas exchange measurements of entire plants and was most likely due to an optimal arrangement of leaves. This emphasizes the importance of considering plant stature for calculation of photosynthetic capacity of whole plants. Altered biomass accumulation was brought about by modified lignin deposition that was associated with changes in expression of pathway-specific genes. Short-term GA3 feeding experiments caused an increased lignin formation as well as a stimulation of cambial activity. Therefore, transcriptional activation of lignin biosynthesis might be the result of long-term adaptation. Moreover, we demonstrate for the first time, to our knowledge, that GA has a differential influence on cell division of different cell types.
MATERIALS AND METHODS
Plant Material and Growth Conditions
Tobacco plants (Nicotiana tabacum cv Samsun NN) were grown in tissue culture under 16 h light/8 h dark regime (irradiance 150 μmol quanta m−2 s−1) at 50% humidity on Murashige Skoog medium containing 2% (w/v) Suc. For basic characterization, tobacco plants were cultivated in soil in a greenhouse with 16 h supplemental light (approximately 250 μmol quanta photosynthetically active radiation m−2 s−1) followed by 8 h darkness. The temperature regime followed the day/night cycle with 25°C/18°C. Detailed investigations of biomass accumulation and lignification as well as single leaf versus whole plant measurements of photosynthesis were performed with homozygous, kanamycin-selected transgenic plants of the T2 generation. These plants were grown in a greenhouse on sand and were watered daily with a nutrient solution (concentration 25% according to Knop nutrient solution; Schropp, 1951). Pods were covered with black foil to avoid growth of microorganisms.
GA3 Treatments
Petioles from leaves 9 and 10 of plants with a total of 16 leaves (counted from bottom to top) were cut in approximately 2.5-cm long pieces and incubated in petri dishes containing either water, 10 μm GA3, or 50 μm GA3. To ensure equal distribution of the solution within the tissue, the material was vacuum infiltrated for about 3 min. Petri dishes were kept in the dark at room temperature. After 24 h petiole pieces were dried between filter paper and either frozen in liquid nitrogen or dehydrated using increasing ethanol concentrations for histochemical purposes.
Plasmid Construction and Plant Transformation
Standard procedures were performed as in Sambrook et al. (1989). The Arabidopsis GA20-oxidase (AtGA20ox) and GA2-oxidase (AtGA2ox) clones were amplified by PCR using genomic DNA as template. Gene-specific primers for amplification of AtGA20ox were designed from the published sequence (accession no. X83379) and comprised the following sequences (restriction sites are underlined): 5′GA20 primer 5′-AGGATCCATGGCCGTAAGTTTCGTAAC-3′ and 3′GA20 primer 5′-TGGATCCTTAGATGGGTTTGGTGAGCC-3′. For amplification of AtGA2ox, specific primers were derived from the sequence of the AtGA2-ox1 gene (accession no. AJ132435) and were as follows (restriction sites are underlined): 5′GA2 primer 5′-GGATCCATCAATGGCGGTATTGTCTAAACCG-3′ and 3′GA2 primer 5′-GTCGACTCAATTTAGGAGATTTTTTATAGTC-3′. The resulting PCR products were subcloned into the pGEMT vector (Promega, Madison, WI) and sequenced to verify identity of the clones. Subsequently, the fragments were excised using the BamHI (AtGA20-ox1) or BamHI and SalI (AtGA2-ox) restriction sites and inserted into a Bin19-derived binary vector (Höfgen and Willmitzer, 1990) containing the CaMV 35S promoter and octopin synthase polyadenylation signal. Orientation of the AtGA20-ox construct was confirmed by restriction endonuclease digestion. The binary constructs were transformed into Agrobacterium tumefaciens, strain CV58C1, carrying the virulence plasmid pGV2260 (Deblaere et al., 1985). Tobacco plants were transformed by Agrobacterium-mediated gene transfer as described by Rosahl et al. (1987).
RNA Isolation, RT-PCR, and Northern-Blot Analysis
Isolation of total RNA was essentially performed as described in Logemann et al. (1987). For reverse transcription (RT)-PCR, 20 μg of total RNA were treated with 10 units of RNase-free DNase (Roche, Basel) for 45 min at 37°C followed by inhibition of the reaction for 10 min at 65°C. A total of 2.5 μg of DNase-treated RNA were reverse transcribed for 60 min at 37°C in a 25-μL reaction volume containing 1 unit of M-MLV (H−) Reverse Transcriptase (Promega), 20 μm each dNTP, 2 μm oligo(dT)[30] V[G/C/A] primer, and 40 units of RNase Inhibitor (Roche). One microliter of the RT reaction was applied for PCR using the gene-specific primers (5′GA20, 3′GA20, 5′GA2, and 3′GA2) as described above.
For northern-blot analysis 20 to 30 μg of total RNA were separated on 1.5% formaldehyde containing agarose gels and blotted onto nylon membrane (GeneScreen, NEN, Boston) by capillary blotting overnight. The membranes were prehybridized and hybridized at 65°C. Radioactive labeling of cDNA fragments was carried out with [32P]dCTP by means of the High Prime Kit (Roche). After stringent washing, radioactive membranes were exposed to x-ray film (Kodak, Rochester, NY) at −70°C or to Phosphoimager screens (Fuji, Tokyo).
cDNA fragments coding for enzymes of lignin biosynthesis were amplified by PCR from known sequences: CAD (accession no. X62344), Phe ammonia-lyase (PAL; accession no. AB008199), 4CL (accession no. U50845), COMT (accession no. X74452.1), and CCR (accession no. A86534) using specific primers. PCR products were cloned into pCR2.1 vector (Invitrogen, CA) and fragments used as probe for hybridization were excised using the EcoRI restriction sites. Equal amounts of loaded RNA were controlled using an actin probe that was made by PCR using primers sequences described in Romeis et al. (2001).
Histochemical Visualization and Quantitative Determination of Lignin
For histochemical experiments hand-cut transverse sections from the eighth, fifth, or second internodes (counted from bottom to top; plants at approximately 16-leaf stage) were analyzed. Plant material was dehydrated using increasing ethanol concentrations starting from 30% up to 70%. Lignin staining was performed by means of phloroglycinol-HCl reagent and sections were analyzed using a light microscope (Axioscop, Zeitz, Germany). All photographs were taken after 30 to 40 min of staining.
For quantitative analysis internodes of indicated stem parts were frozen in liquid nitrogen. Lignin content was determined spectrophotometrically (280 nm) from about 150 mg fresh weight using thioglycolic acid following the protocol described by Campbell and Ellis (1992). Calculation was done according to a calibration curve prepared with pure lignin (5–25 mg).
Gas Exchange Measurements and Calculation
CO2 uptake rates of single leaves were measured with a portable photosynthesis system LI-6400 (LI-COR, Lincoln, NE). The CO2 concentration of the air entering the leaf chamber was adjusted to 400 μmol mol−1 and the leaf temperature was maintained at 20°C. During the measurements of light response curves of the CO2 gas exchange the photosynthetic photon flux density varied between 0 and 2,000 μmol photons m−2 s−1.
For calculation of the photosynthetic capacity of the whole plant on the basis of single leaf measurements, gas exchange of at least 4 to 5 different leaves of each plant was recorded. Light intensity in these cases was 200 μmol photons m−2 s−1. Using these data, the total photosynthetic capacity of each plant was estimated in relation to the leaf area.
Gas exchange of whole plants was measured directly by means of a canopy chamber PMK-1 (DMP, Fehraltorf, Switzerland; see also Fig. 6A). These measurements were performed in the greenhouse under daylight conditions. Intensity of the illumination was in the range of 100 to 280 μmol photons m−2 s−1 and the CO2 concentration of the inlet gas varied between 365 to 395 μmol mol−1 during the experiments. Volume flow of the inlet gas, light intensity, temperature, and the molar fraction of CO2 and water of the inlet and the outlet gas were recorded continuously by the LI-6400 and the CO2 gas exchange rate of the whole plant was calculated. To compare the different plants the photosynthetic rate for every plant under illumination with an equal light intensity (200 μmol photons m−2 s−1) was determined. For this purpose, light curves were measured for three different representative leaves of each plant directly after recording the gas exchange of the whole plants. The averaged data were fitted by the equation of a nonrectangular hyperbola (Hajirezaei et al., 2002). The resulting value was used to calculate photosynthetic capacity of the whole plant at a light intensity of 200 μmol photons m−2 s−1.
Chlorophyll fluorescence was measured using a PAM-2000 portable fluorometer (Walz, Effeltrich, Germany). For the determination of maximum quantum yield of PS II plants were preincubated for 30 min in the dark. Minimum fluorescence yield (F0) was measured by applying a weak pulsed red light (<1 μmol quanta m−2 s−1). Maximum fluorescence yield (Fm) was determined during a 0.8 s pulse of white light of intensity 3,500 μmol quanta m−2 s−1. Maximum quantum yield of PS II was calculated as (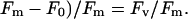
Chlorophyll Determination
Chlorophyll content of leaf discs (0.5 cm2 diameter) was determined in 96% ethanol, as described by Lichtenthaler (1987).
Morphological Parameters
The area of detached leaves was measured using the portable area meter LI-3000 in combination with the transparent belt conveyor accessory LI-3050A (LI-COR). Fresh weight was determined directly after harvest. Dry weight was determined after incubation at 70°C overnight or until no further decrease in weight could be detected.
Sequence data from this article have been deposited with the EMBL/GenBank data libraries under accession numbers X83379, AJ132435, X62344, AB008199, U50845, X74452.1, and A86534.
Acknowledgments
We thank Melanie Ruff for excellent technical assistance, and Andrea Knospe and Sibylle Freist for plant transformation and taking care of tissue-culture plants. We are grateful to Martin Peisker for initial help with gas exchange measurements and helpful comments.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.103.036988.
References
- Ashraf M, Karim F, Rasul E (2002) Interactive effects of gibberellic acid (GA3) and salt stress on growth, ion accumulation and photosynthetic capacity of two spring wheat (Triticum aestivum L.) cultivars differing in salt tolerance. Plant Growth Regul 36: 49–59 [Google Scholar]
- Campbell MM, Ellis BE (1992) Fungal elicitor mediated responses in pine cell cultures I. Induction of phenylpropanoid metabolism. Planta 186: 409–417 [DOI] [PubMed] [Google Scholar]
- Carrera E, Bou J, Garcia-Martinez JL, Prat S (2000) Changes in GA20-oxidase gene expression strongly affect stem length, tuber induction and tuber yield of potato plants. Plant J 22: 247–256 [DOI] [PubMed] [Google Scholar]
- Carrera E, Jackson SD, Prat S (1999) Feedback control and diurnal regulation of gibberellin 20-oxidase transcript levels in potato. Plant Physiol 119: 765–773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coles JP, Phillips AL, Croker SJ, Garcia-Lepe R, Lewis MJ, Hedden P (1999) Modification of gibberellin production and plant development in Arabidopsis by sense and antisense expression of gibberellin 20-oxidase. Plant J 17: 547–556 [DOI] [PubMed] [Google Scholar]
- Cramer MD, Nagel OW, Lips SH, Lambers H (1995) Reduction, assimilation and transport of N in normal and gibberellin-deficient tomato plants. Physiol Plant 95: 347–354 [Google Scholar]
- Curtis I, Ward DA, Thomas SG, Phillips AL, Davey MR, Power JB, Lowe KC, Croker SJ, Lewis MJ, Magness SL, et al. (2000) Induction of dwarfism in transgenic Solanum dulcamara by over-expression of a gibberellin 20-oxidase cDNA from pumpkin. Plant J 23: 329–338 [DOI] [PubMed] [Google Scholar]
- Deblaere R, Bytebier B, DeGreve H, Beboeck F, Schell J, Van Montagu M, Leemanns J (1985) Efficient octopine Ti plasmid-derived vectors for Agrobacterium mediated gene transfer to plants. Nucleic Acids Res 13: 4777–4788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Digby J, Wareing PF (1966) The effect of applied growth hormones on cambial division and the differentiation of the cambial derivates. Ann Bot (Lond) 30: 539–548 [Google Scholar]
- Dijkstra P, Ter Reegen H, Kuiper PJC (1990) Relation between relative growth rate, endogenous gibberellins and the response to applied gibberellic acid for Plantago major. Physiol Plant 79: 629–634 [DOI] [PubMed] [Google Scholar]
- Eriksson ME, Israelsson M, Olsson O, Moritz T (2000) Increased gibberellin biosynthesis in transgenic trees promotes growth, biomass production and xylem fibre length. Nat Biotechnol 18: 784–788 [DOI] [PubMed] [Google Scholar]
- Eriksson ME, Moritz T (2002) Daylength and spatial expression of a gibberellin 20-oxidase isolated from hybrid aspen (Populus tremula L. × P. tremuloides Michx.). Planta 214: 920–930 [DOI] [PubMed] [Google Scholar]
- Graebe JE (1987) Gibberellin biosynthesis and control. Annu Rev Plant Physiol Plant Mol Biol 38: 419–465 [Google Scholar]
- Hajirezaei MR, Peisker M, Tschiersch H, Palatnik JF, Valle EM, Carrillo N, Sonnewald U (2002) Small changes in the activity of chloroplastic NAPD+-dependent ferredoxin oxidoreductase lead to impaired plant growth and restrict photosynthetic activity of transgenic tobacco plants. Plant J 29: 281–293 [DOI] [PubMed] [Google Scholar]
- Hedden P, Kamiya Y (1997) Gibberellin biosynthesis: enzymes, genes their regulation. Annu Rev Plant Physiol Plant Mol Biol 48: 431–460 [DOI] [PubMed] [Google Scholar]
- Hedden P, Phillips AL (2000) Gibberellin metabolism: new insights revealed by the genes. Trends Plant Sci 5: 523–530 [DOI] [PubMed] [Google Scholar]
- Hedden P, Proebsting WM (1999) Genetic analysis of gibberellin biosynthesis. Plant Physiol 119: 365–370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Höfgen R, Willmitzer L (1990) Biochemical and genetic analysis of different patattin isoforms expressed in various organs of potato (Solanum tuberosum ). Plant Sci 66: 221–230 [Google Scholar]
- Israelsson M, Eriksson ME, Hertzberg M, Aspeborg H, Nilsson P, Mortiz T (2003) Changes in gene expression in the wood-forming tissue of transgenic hybrid aspen with increased secondary growth. Plant Mol Biol 52: 893–903 [DOI] [PubMed] [Google Scholar]
- Kende H, Zeevaart JAD (1997) The five “classical” plant hormones. Plant Cell 9: 1197–1210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koornneef M, van der Veen JH (1980) Induction and analysis of gibberellin sensitive mutants in Arabidopsis thaliana (L). Heynh Theor Appl Genet 58: 257–263 [DOI] [PubMed] [Google Scholar]
- Lange T, Hedden P, Graebe JE (1994) Expression cloning of a gibberellin 20-oxidase, a multifunctional enzyme involved in gibberellin biosynthesis. Proc Natl Acad Sci USA 91: 8552–8556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lester DR, Ross JJ, Smith JJ, Elliott RC, Reid JB (1999) Gibberellin 2-oxidation and the SLN gene of Pisum sativum. Plant J 19: 65–73 [DOI] [PubMed] [Google Scholar]
- Lichtenthaler HK (1987) Chlorophylls and carotinoids: pigments of photosynthetic biomembranes. Methods Enzymol 148: 350–382 [Google Scholar]
- Logemann J, Schell J, Willmitzer L (1987) Improved methods for the preparation of RNA from plant tissues. Anal Biochem 163: 16–20 [DOI] [PubMed] [Google Scholar]
- Martin DN, Proebsting WM, Hedden P (1999) The SLENDER gene of pea encodes a gibberellin 2-oxidase. Plant Physiol 121: 775–781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin DN, Proebsting WM, Parks TD, Dopugherty WG, Lange T, Lewis MJ, Gaskin P, Hedden P (1996) Feed-back regulation of gibberellin biosynthesis and gene expression in Pisum sativum L. Planta 200: 159–166 [DOI] [PubMed] [Google Scholar]
- Nagel OW, Lambers H (2002) Changes in the acquisition and partitioning of carbon and nitrogen in the gibberellin-deficient mutants A70 and W335 of tomato (Solanum lycopersicum L.). Plant Cell Environ 25: 883–891 [Google Scholar]
- Peng J, Richards DE, Hartley NM, Murphy GP, Devos KM, Flintham JE, Beales J, Fish LJ, Worland AJ, Pelica F, et al. (1999) Green revolution” genes encode mutant gibberellin response modulators. Nature 400: 256–261 [DOI] [PubMed] [Google Scholar]
- Phillips AL, Ward DA, Uknes S, Appleford NEJ, Lange T, Huttley AK, Gaskin P, Graebe JE, Hedden P (1995) Isolation and expression of three gibberellin 20-oxidases cDNA clones from Arabidopsis. Plant Physiol 108: 1049–1057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ridoutt BG, Pharis RP, Sands R (1996) Fibre length and gibberellins A1 and A20 are decreased in Eucalyptus globulus by acylcyclohexanedione injected into stem. Physiol Plant 96: 559–566 [Google Scholar]
- Romeis T, Ludwig AA, Martin R, Jones JD (2001) Calcium-dependent protein kinases play an essential role in a plant defence response. EMBO J 20: 5556–5567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosahl S, Schmidt R, Schell J, Willmitzer L (1987) Expression of a tuber specific storage protein in transgenic tobacco plants: demonstration of an esterase activity. EMBO J 6: 1155–1159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakamoto T, Kobayashi M, Ihoh H, Tagiri A, Kayano T, Tanaka H, Iwahori S, Matsuoka M (2001) Expression of a gibberellin 2-oxidase gene around the shoot apex is related to phase transition in rice. Plant Physiol 125: 1508–1516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakamoto T, Morinaka Y, Ishiyama K, Kobayashi M, Ihoh H, Kayano T, Iwahori S, Matsuoka M, Tanaka H (2003) Genetic manipulation of gibberellin metabolism in transgenic rice. Nat Biotechnol 21: 909–913 [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T (1989) Molecular Cloning. A Laboratory Manual, Ed 2. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY
- Sauter M, Kende H (1992) Levels of β-glucan and lignin in elongating internodes of deepwater rice. Plant Cell Physiol 33: 1089–1097 [Google Scholar]
- Sauter M, Mekhedov SL, Kende H (1995) Gibberellin promotes histon H1 kinase activity and the expression of cdc2 and cyclin genes during the induction of rapid growth in deepwater rice. Plant J 7: 623–632 [DOI] [PubMed] [Google Scholar]
- Schomburg FM, Bizzell CM, Lee DJ, Zeevart JAD, Amasino RM (2003) Overexpression of a novel class of gibberellin 2-oxidases decreases gibberellin levels and created dwarf plants. Plant Cell 15: 151–163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schropp W (1951) Der Vegetationsversuch. 1. Die Methodik der Wasserkultur höherer Pflanzen. Neuman, Berlin, p 132
- Spielmeyer W, Ellis MH, Chandler P (2002) Semidwarf (sd-1), “green revolution” rice, contains a defective gibberellin 20-oxidase gene. Proc Natl Acad Sci USA 99: 9043–9048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka-Ueguchi M, Itoh H, Oyama N, Koshioka M, Matsuoka M (1998) Over-expression of a tobacco homeobox gene NTH15 decreases the expression of gibberellin biosynthetic gene encoding GA20-oxidase. Plant J 15: 391–400 [DOI] [PubMed] [Google Scholar]
- Thomas SG, Phillips AL, Hedden P (1999) Molecular cloning and functional expression of gibberellin 2-oxidases multifunctional enzymes involved in gibberellin deactivation. Proc Natl Acad Sci USA 96: 4698–4703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wareing PF (1958) Interaction between indole-acetic acid and gibberellic acid in cambial activity. Nature 181: 1744–1745 [DOI] [PubMed] [Google Scholar]
- Xu YL, Li L, Wu K, Peeters AJM, Gage DA, Zeevart JAD (1995) The GA5 locus of Arabidopsis thaliana encodes a multifunctional gibberellin 20-oxidase: molecular cloning and functional expression. Proc Natl Acad Sci USA 92: 6640–6644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan L, Xu DQ (2001) Stimulation effect of gibberellic acid short-term treatment on leaf photosynthesis related to the increase in Rubisco content in broad bean and soybean. Photosynth Res 68: 39–47 [DOI] [PubMed] [Google Scholar]