Graphical abstract
Keywords: Natural products, Biotransformation, Microbial biocatalysts, Pharmaceutical products
Abstract
Natural products are structurally and biologically interesting metabolites, but they have been isolated in minute amounts. The syntheses of such natural products help in obtaining them in bulk amounts. The recognition of microbial biotransformation as important manufacturing tool has increased in chemical and pharmaceutical industries. In recent years, microbial transformation is increasing significantly from limited interest into highly active area in green chemistry including preparation of pharmaceutical products. This is the first review published on the usage of microbial biocatalysts for some natural product classes and natural product drugs.
Introduction
Natural product compounds are structurally and biologically interesting metabolites. Compounds isolated are often available in minute amounts. Thus, synthesis of natural products also provides a powerful means in solving supply problems in clinical trials and marketing of the drug for obtaining natural products in bulk amounts. If the structure is complex, it is often an impossible task to isolate enough of the natural products for clinical trials [1–3].
The recognition of biotransformation as important manufacturing tool has increased within chemical and pharmaceutical industries in recent years. Biocatalysts can simplify, or in some instances even enable, the production process of complex chemicals and drug intermediates. They can add stereospecificity to the process, eliminating the need for complicated separation and purification steps. The ability of biocatalysts to selectively produce useful products under relatively mild conditions compared to its chemical catalyst counterpart make biocatalysts an interesting and powerful addition. Recent advances in technology have markedly increased the ability of industry to discover new biocatalysts and optimize their performance. These advances are coming at a time when both the chemical and pharmaceutical industries are facing increasing pressure to produce more effective natural products and to make them more efficiently [2]. In this report, we discuss the advances in technology for microbial transformation of natural product compounds.
Microbial transformation
Biocatalysis scope of study involving microbial transformation is increasing significantly from limited interest into highly active area in chemistry today including preparation of pharmaceutical products. Biotransformation can be clarified as the specific modification of a definite compound to a distinct product with structural similarity, by the use of biological catalysts including microorganisms like fungi [4]. The biological catalyst can be described as an enzyme, or a whole, inactivated microorganism that contains an enzyme or several enzymes produced in it. Bioconversion is another term related to microbial transformation for this study in particular. There is only slight difference between a biotransformation and a bioconversion. A bioconversion utilizes the catalytic activity of living organisms and hence can involve several chemical reaction steps. A living microorganism will be continuously producing enzymes and hence bioconversions often involve enzymes which are quite unstable for used substrates. The properties of biotransformations and bioconversions are very similar and in many cases the terms are cited as interchangeable [5].
On the other hand, fermentation, science under zymology utilizes microorganisms, yeast was known to turn sugar into alcohol since 1857 by the French chemist, Louis Pasteur. The biotransformation processes have advantages overcome some of the inherent problems and examples of some commercially successful processes [6]. To utilize from this processes, biocatalysis research have been suggested for the nation’s rich natural resources mainly with the endophytes available.
Biotransformation processes are far more diverse than therapeutic protein production processes [7]. There are many microorganism strains and enzymes required to exploit the selective biotransformation potential for the bioconversion of a myriad of different substances into the desired products especially new optically active main pharmaceutical ingredients. Timeline compressions in the development cycle of pharmaceuticals, in combination with a missing broad strain and enzyme choice, result in the fact that biotransformation typically represents the second generation process choice in the manufacturing of a small molecule pharmaceutical. Novel biocatalysts are needed first and foremost especially oxidoreductases and lyases for biotransformation.
Biotransformation is also known to comply with the green chemistry strategy today. Green chemistry is a term used for sustainable chemical industrial manufacturing processes toward achieving minimal waste production and energy consumption [8]. Thus, biosynthesis and biotransformation are assumed to play a key role in green chemistry in the years to come.
Advantages of microbial transformation
Many benefits can be obtained through microbial transformations studies. The process required in microbial transformation may most probably have the ability to operate at near neutral pH, ambient temperatures and atmospheric pressures [6]. In contrast, chemistry often requires extremes of these conditions which are not exactly environmentally friendly and industrially undesired. Furthermore, extreme pH, temperature and pressure may provide harmful effects toward personnel operating the harsh procedures and may also affect community surrounding the areas.
More importantly biocatalysts are highly reaction specific, enantiomer-specific and regio-specific [6]. This is mainly and directly referring to the chemical structure of a compound one may want to obtain specifically. Many versatile microorganisms can be utilized to carry out extremely specific conversions using substrates of low cost [9].
The basic chemistry reactions include addition reactions, elimination reactions, substitution reactions, pericyclic reactions, rearrangement reactions and redox reactions. The steps may be lengthy and more tedious at times as chemical substances are easily disturbed by the humid environment in tropical areas for instance. Humid tropical climates over here are recorded by hot, wet climates, with average temperatures of 18 °C or higher and an average rainfall of 203 cm or more.
Microorganisms have great potential for inducing many alternatives of innovative and improvised enzyme systems which are capable for converting unfamiliar substrates. Therefore, many studies can be performed to a greater extend regarding different endophyte species toward chemical alterations of molecules and compounds of interest. The genome of a novel thermophilic fungal species can be assessed to provide with gene sequences that encode for thermotolerant enzymes, which are more stable to variations of reaction temperature.
Not as the name may suggest physically, microbes are creature incredibly small for the naked eyes to see but carry major roles today in pharmaceutical industry one way or another. Microorganisms are capable of producing unique enzymes which are stable toward heat, alkali and acids. One of the studies done was regarding hyperthermophilic archaeon Pyrobaculum calidifontis VA1 which produced a thermostable esterase [10,11].
Their small size has by far the largest surface-to-volume ratio in comparison with some living organisms. Thus, this allows them to maximize their metabolic rates because of a high exchange of molecules and metabolites through their surface. With the right cultivation conditions, microorganisms grow exponentially [7].
Microorganisms are capable to produce great variety of enzymes in a short period of time as a result of its natural characteristic to multiply. It is also possible to obtain and cultivate microorganisms that can survive under extreme environments such as low or high temperatures and/or acidic or alkali conditions. Microbial transformation can make feasible reactions that are not likely to be carried out by traditional synthetic procedures. Also, endophytes may produce natural, biodegradable compounds.
Disadvantages and challenges of microbial transformation
Overcoming the existence of well-developed Organic Syntheses technology however is an inherent challenge to biotransformation processes to grow and be frequently applied. Often there is no financial incentive for implementing a new process when old technology is known and investment in plants have been paid for [12].
Technology used to enhance biotransformation and bioconversion processes may include immobilization techniques, genetic engineering and the use of enzymes that cope with organic solvents [6]. Examples of enzyme engineering are protein engineering and crosslinked enzyme crystals. Expertise and equipments along with updated knowledge that is evolving and increasing in the process of microbial transformation are handy if acquired for useful novel compounds to be obtained.
The use of biocatalysts to carry out biotransformed products is often difficult as it involves the challenges of reactant or product toxicity or inhibition, high dilution and the use of pH and temperature labile biocatalysts. However, biological and process solutions do exist to solve some of these problems and methods to compare strategies and techniques for biotransformation operation are being developed [4,13].
Besides that, if the substrate used is toxic, it can kill the microorganism hindering any biotransformation to occur. On the other hand, if the microorganism uses the substrate as an energy source, none of the product desired is likely to be recovered. Time restriction and missing broad strain or enzyme choice cause biotransformation typically represents the second generation process choice in the manufacturing of a small molecule pharmaceutical [7].
Due to involvement of complex biological systems, very low chemical yields are obtained. Enzymes are very specific and therefore the chances of getting high probability of transformation is normally less and slow compared to chemical transformation. Improvement is highly encouraged for the efficiency of microbial transformation to perform incomparably or better industrially in a large scale with greater potential.
Microbial transformation of terpene compounds
Monoterpenes
The microbial transformations by Glomerella cingulata of two saturated acyclic monoterpenoids, tetrahydrogeraniol (1) and tetrahydrolavandulol (3), were investigated by Nankai et al. [14]. Both compounds were hydroxylated regioselectively at the isopropyl group. Tetrahydrogeraniol was transformed to hydroxycitronellol (2), while tetrahydrolavandulol was transformed to 5-hydroxytetrahydrolavandulol (4) (Fig. 1) [14].
Fig. 1.
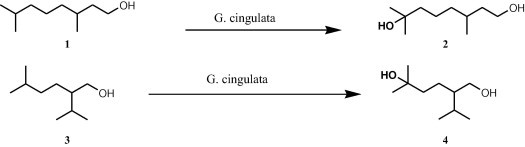
Microbial transformations by Glomerella cingulata of two saturated acyclic monoterpenoids.
The cyclic monoterpene ketone (−)-carvone (5) was metabolized by the plant pathogenic fungus Absidia glauca. After 4 days of incubation, the diol 10-hydroxy-(+)-neodihydrocarveol (8) was formed via (+)-trans-dihydrocarvone (6) and (+)-neodihydrocarveol (7) in 4 days (Fig. 2) [15].
Fig. 2.
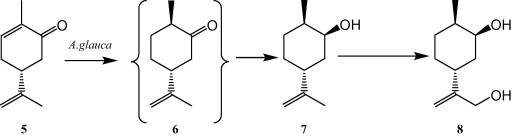
The biotransformation of (−)-carvone (5) and its metabolism to the diol 10-hydroxy-(+)-neodihydrocarveol (8) via (+)-trans-dihydrocarvone (6) and (+)-neodihydrocarveol (7) in 4 days.
Sesquiterpenes
Microbial and chemical transformation studies of the marine sesquiterpene phenols (S)-(+)-curcuphenol (9) and (S)-(+)-curcudiol (18), isolated from the Jamaican sponge Didiscus oxeata, were accomplished. Preparative-scale fermentation of sesquiterpenoid 9 with Kluyveromyces marxianus var. lactis (ATCC 2628) has resulted in the isolation of six new metabolites: (S)-(+)-15-hydroxycurcuphenol (10), (S)-(+)-12-hydroxycurcuphenol (11), (S)-(+)-12,15-dihydroxycurcuphenol (12), (S)-(+)-15-hydroxycurcuphenol-12-al (13), (S)-(+)-12-carboxy-10,11-dihydrocurcuphenol (19), and (S)-(+)-12-hydroxy-10,11-dihydrocurcuphenol (20). Fourteen-days incubation of 9 with Aspergillus alliaceus (NRRL 315) afforded the new compounds (S)-(+)-10β-hydroxycurcudiol (21), (S)-(+)-curcudiol-10-one (22), and (S)-(+)-4-[1-(2-hydroxy-4-methyl)phenyl)]pentanoicacid (25). Rhizopus arrhizus (ATCC 11145) and Rhodotorula glutinus (ATCC 15125) afforded (S)-curcuphenol-1(R)-D-glucopyranoside (14) and (S)-curcudiol-1(R)-D-glucopyranoside (23) when incubated for 6 and 8 days with 9 and 18, respectively.
Reaction of 9 with NaNO2 and HCl afforded (S)-(+)-4-nitrocurcuphenol (15) and (S)-(+)-2-nitrocurcuphenol (16) in a 2:1 ratio. Acylation of 9 and 18 with isonicotinoyl chloride afforded the expected esters (S)-(+)-curcuphenol-1-O-isonicotinate (17) and (S)-(+)-curcudiol-1-O-isonicotinate (24), respectively (Fig. 3A and B) [16].
Fig. 3.
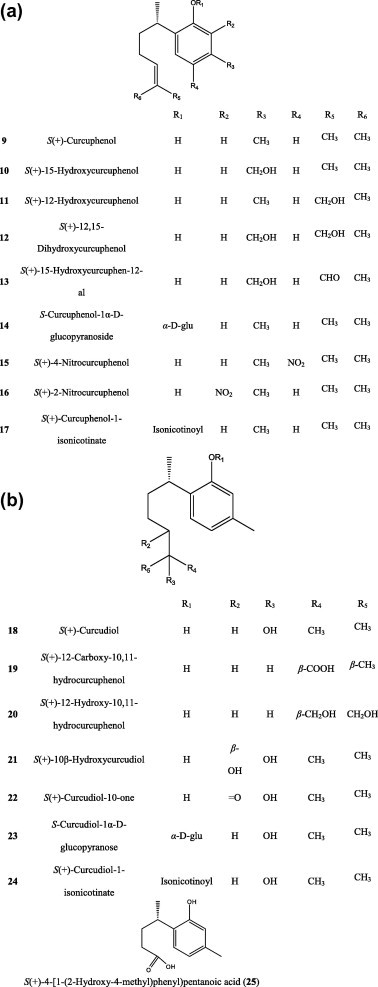
Microbial transformation of sesquiterpe phenols.
Incubations of the fungi Cunninghamella echinulata and Rhizopus oryzae with the sesquiterpene lactones (+)-costunolide (26), (+)-cnicin (27), (+)-salonitenolide (28), (−)-dehydrocostuslactone (30), (−)-lychnopholide (38), and (−)-eremantholide C (41) were performed. Incubation of 26 with C. echinulata afforded Δ 11(13)-dihydrogenation and Δ1(10)-epoxidation products (29, 33–35). C. echinulata also hydrolyzed the side chain of 27, and transformed 30 into (+)-11R,13-dihydrodehydrocostuslactone (31), a new natural product. R. oryzae converted 30 into both Δ11(13)-dihydrogenation and Δ10(14)-epoxidation products (32 and 37). Both fungi transformed 38 into (−)-16-(1-methyl-1-propenyl)eremantholanolide (42), providing experimental evidence for the biosynthesis of the eremantholide hemiketal unit. Formation of 33–35 can be explained by enzymatic epoxidation of 26 to 1β,10α-epoxicostunolide (36), and subsequent electrophilic opening of the epoxide with concomitant rearrangement to the eudesmanolide skeleton, as presumably occurs in plant biogenesis of 1β-hydroxyeudesmanolides. Reaction of 38 with Sodium borohydride (NaBH4) gave the alcohol product 40, and treatment with Bu3–SnH only causes isomerization of the lateral chain, leading to 39. Compounds 28 and 41 were not metabolized by either fungus under the test conditions (Fig. 4) [17].
Fig. 4.
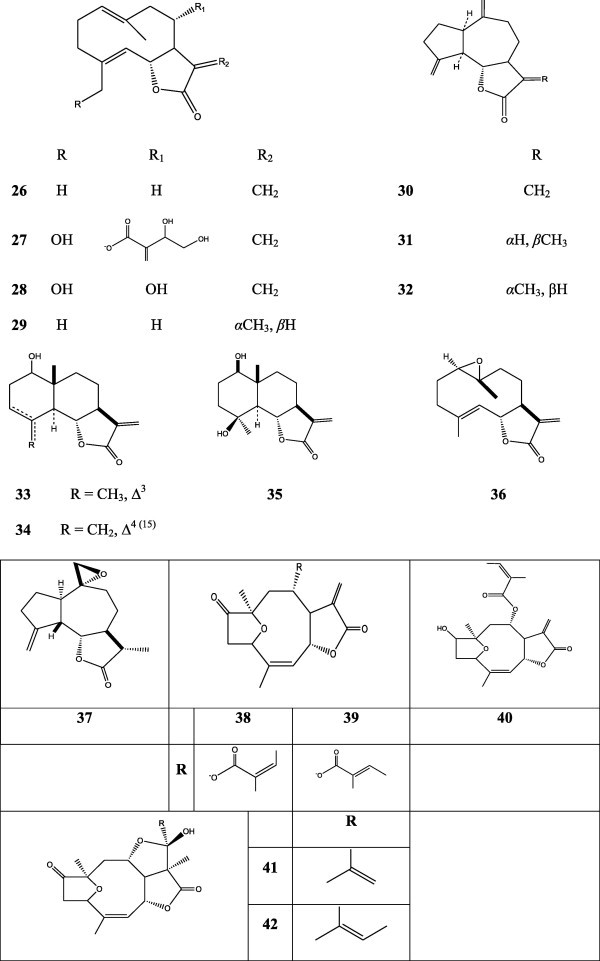
Cunninghamella echinulata and Rhizopus oryzae transformation of sesquiterpene lactones.
Biotransformation studies conducted on (+)-(S)-ar-turmerone (43) and (+)-(S)-dihydro-ar-turmerone (44) by the fungus Aspergillus niger have revealed that 43 was metabolized to give four oxidized metabolites, (+)-(7S)-hydroxydehydro-ar-todomatuic acid (45), (+)-(7S,10E)-12-hydroxydehydro-ar-todomatuic acid (46), (+)-(7S,10E)-7,12-dihydroxydehydroar-todomatuic acid (47), and (+)-(7S)-15-carboxy-9,13-epoxy-7-hydroxy-9,13-dehydro-ar-curcumene (48), and (+)-(S)-dihydro-ar-turmerone (44) was metabolized to (+)-7,11-dihydroxy-ar-todomatuic acid (49) (Fig. 5) [18]. The absolute configurations of 45 at the C-7 position were established to be S after conversion into tetrahydro-2-(4-carbomethoxyphenyl)-2,6,6-trimethyl-4H-pyran-4-one (50).
Fig. 5.
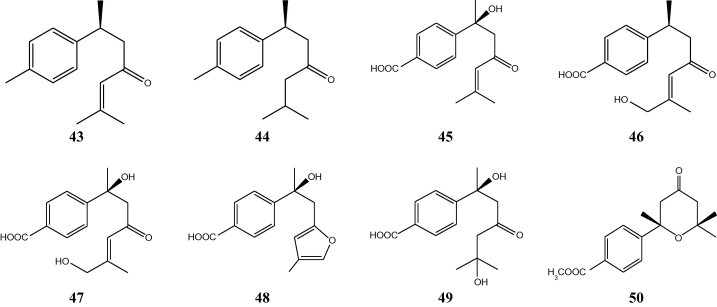
Transformation of (+)-(S)-ar-turmerone (43) and (+)-(S)-dihydro-ar-turmerone (44) by the fungus Aspergillus niger.
Diterpenes
Microbial transformation of 13R,14R,15-trihydroxylabd-7-ene (54) and 13R,14R,15-trihydroxylabd-8(17)-ene (55) by the fungus Debaryomyces hansenii gave 13R,14R,15-trihydroxy-6-oxolabd-8-ene (51) and 7α,13R,14R,15-tetrahydroxy-labd-8(17)-ene (53), respectively. While, microbial transformation of 54 by A. niger afforded 3β,13R,14R,15-tetrahydroxy-labd-7-ene (52), and 13R,14R,15-trihydroxylabd-8,17-ene (56) gave 53 and 3R,14R,15-3-oxotetrahydroxy-labd-7-ene (54) (Fig. 6) [19].
Fig. 6.
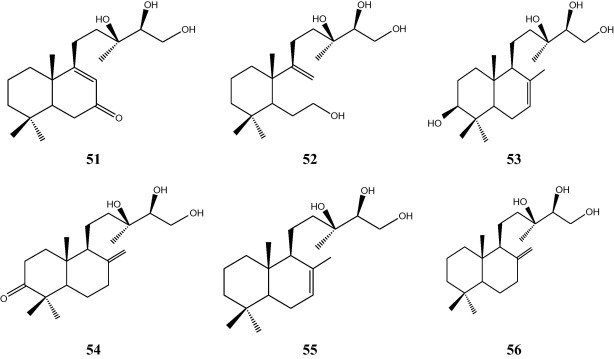
Transformation of 13R,14R,15-trihydroxylabd-7-ene (54) and 13R,14R,15-trihydroxylabd-8(17)-ene (55) by the fungus Debaryomyces hansenii.
The microbiological transformation of candidiol (15α,18-dihydroxy-ent-kaur-16-ene, 57) by Mucor plumbeus led to 3β,15α,18-trihydroxy-ent-kaur-16-ene, 6a,15a,18-trihydroxy-ent-kaur-16-ene (61), 3β,15α,18-trihydroxy-entkaur-16-ene (58), 3α,15α,18-trihydroxy-entkaur-16-ene (59), 11β,15α,18-trihydroxy-ent-kaur-16-ene (62) and 15α,17,18-trihydroxy-11β,16β-epoxy-ent-kaurane (83), while the incubation of 15α,19-dihydroxy-ent-kaur-16-ene (69) gave 9β,15α,19-trihydroxy-ent-kaur-16-ene (80), 3α,15α,19-trihydroxy-ent-kaur-16-ene (70), 11β,15α,19-trihydroxy-ent-kaur-16-ene (74), 6α,15α,19-trihydroxy-ent-kaur-16-ene (61), 15α,17,19-trihydroxy-11β,16β-epoxy-ent-kaurane (82), 19-(β-D-glucopyranosyl)-15α-hydroxy-ent-kaur-16-ene (76) and 19-(β-D-glucopyranosyl)-15-oxo-ent-kaur-16-ene (78). An interesting rearrangement in dilute acid medium of 9β,15α,19-trihydroxy-ent-kaur-16-ene (80) into 16-oxo-19-hydroxy-ent-abiet 8(9),15-diene (84). The possible mechanism of formation of this 8,15-seco-entkaurene diterpene is shown in Fig. 7b, a compound of this type, named hebeiabinin A (85) (Fig. 7A and B) [20]. The following compounds 60, 63, 66–68, 71, 73, 75, 77, 79, and 81, were acetylated products to decrease polarity of its original compounds. Compound 65 suggested to be an artifact formed during the isolation procedure from the true biotransformed metabolite 64.
Fig. 7.
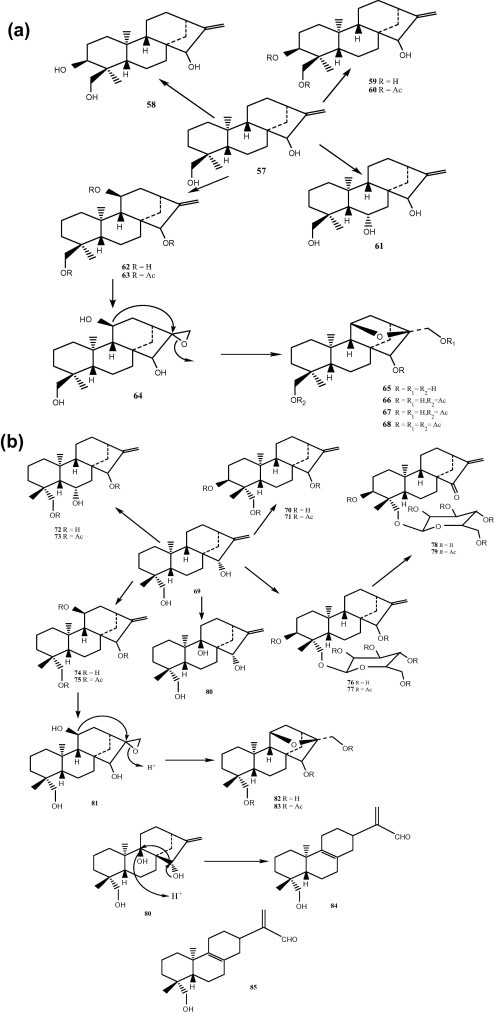
Transformation of candidiol (15α,18-dihydroxy-ent-kaur-16-ene) by Mucor plumbeus.
Triterpenes
Two new metabolites, 15α,16α-dihydroxy-3,4-secocycloarta-4 (28), 17 (20), 17 (E), 24 (E)-triene-3,26-dioic acid (87) and 16α, 20α-dihydroxy-18 (13 → 17β) abeo-3,4-secocycloarta-4 (28), 12 (13), 24 (Z)-triene-3,26-dioic acid (88) were isolated and identified from the co-cultures of nigranoic acid (86) and Trichoderma sp. JY-1. Compound 87 was found to possess an unusual 17(20), 17 (E)-ene structure and compound 88 featured an unprecedented 18(13 → 17β)-abeo-secocyloarta skeleton (Fig. 8) [21].
Fig. 8.
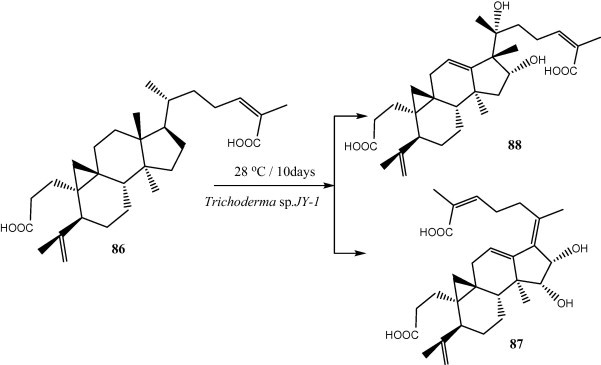
Novel metabolites isolated from cultures of Trichoderma sp. JY-1 grown in the presence of nigranoic acid.
Microbial transformation of 4-olean-type pentacyclic triterpenes (OPTs), 3-oxo oleanolic acid (89), oleanolic acid (93), and esculentoside A (97) was studied. After the screening of 12 strains of microbes, preparative biotransformation by two strains of Streptomyces griseus ATCC 13273 and Aspergillus ochraceus CICC 40330 resulted in the isolation of 10 metabolites (90–92, 94–101). The microbial catalyzed high efficient regio-selective methyl oxidation and glycosylation were discovered, which could be provided as an alternative method to expand the structural diversity of OPTs (Fig. 9) [22].
Fig. 9.
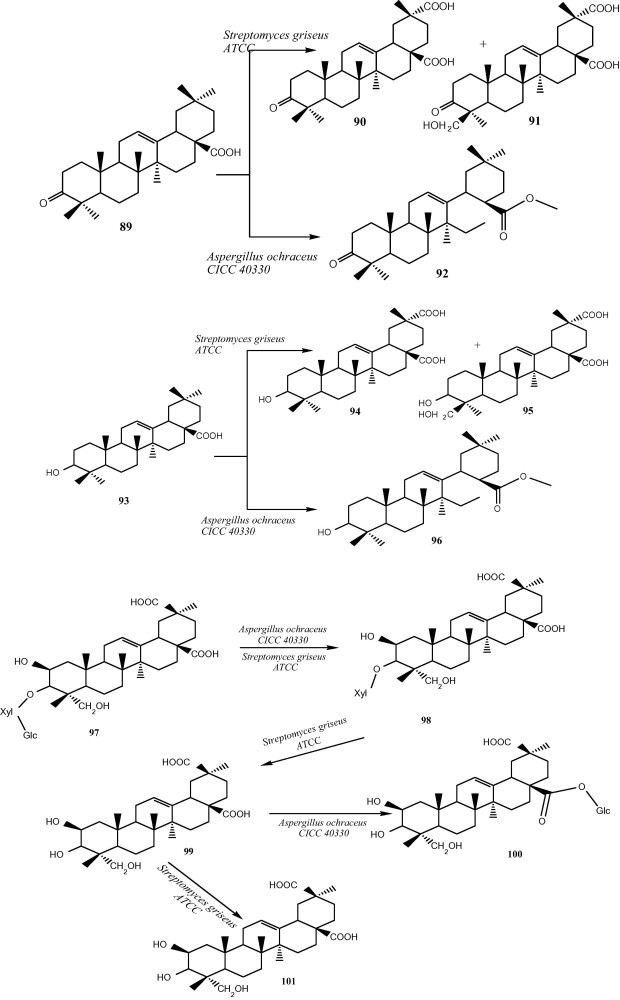
Microbial transformation of 4 olean-type pentacyclic triterpenes (OPTs).
Steroids
Microbial transformation of diosgenin (3β-hydroxy-5-spirostene) (102) using white-rot fungus Coriolus versicolor afforded four previously unreported polyhydroxylated steroids, 25(R)-spirost-5-en-3β,7α,15α,21-tetraol (106), 25(R)-spirost-5-en-3β,7β,12β,21-tetrol (107), (25R)-spirost-5-en-3β,7α,12β,21-tetraol (108), and (25R)-spirost-5-en-3β,7β,11α,21-tetraol (109), along with three known congeners, 25(R)-spirost-5-en-3β,7β-diol (103), 25(R)-spirost-5-en-3β,7β,21-triol (104), and 25(R)-spirost-5-en-3β,7β,12β-triol (105) (Fig. 10) [23].
Fig. 10.
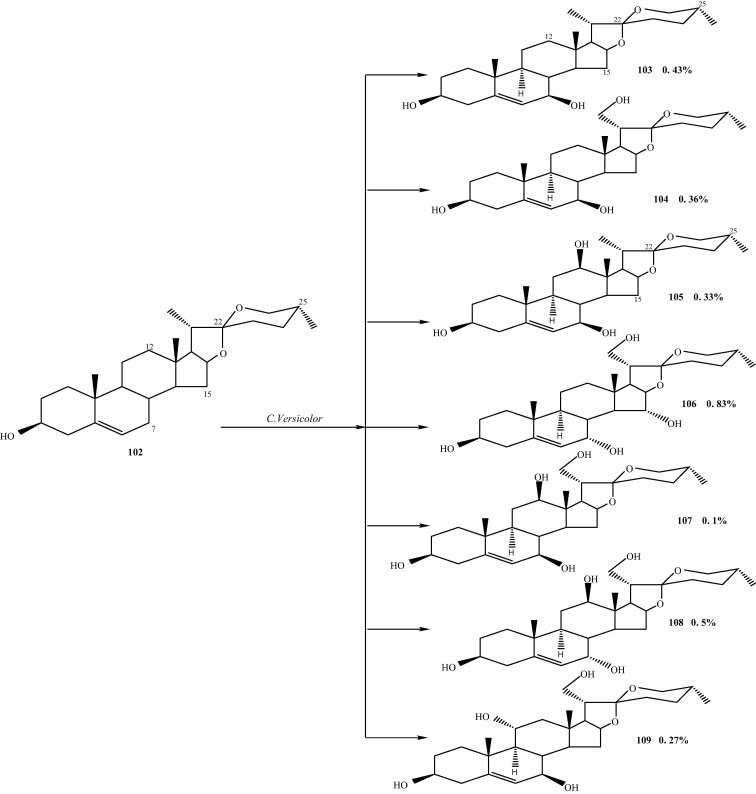
Microbialtransformation of diosgenin (3β-hydroxy-5-spirostene) (102) using white-rot fungus Coriolus versicolor.
Microbial transformation of bioactive natural products
Biotransformation of artemisinin
Artemisinin, a sesquiterpene lactone has an endoperoxide bridge, which was isolated from the Chinese herbal plant, Artemisia annua L. in 1972 [24]. Because of its high therapeutic values in treating malaria, tremendous efforts have been made toward structure modification and analogue synthesis with the aim of developing more potent antimalarial agents with in vivo stability since it was discovered. The structural modifications usually took place at the lactone moiety of artemisinin (110) due mainly to the difficulty of introducing functionalities on the ring systems by conventional chemical methods. Transformation of artemisinin (110) with S. griseus ATCC13273, affording artemisitone-9 (113), 9α-hydroxy-artemisinin, 9β-hydroxy-artemisinin and 3α-hydroxy-deoxyartemisinin (111) [25] (Fig. 11). Metabolites of 9α-hydroxy-artemisinin (114) and 9β-hydroxy-artemisinin (112) were further oxidized to give artemisitone-9 (113). A pathway for the production of artemisitone-9 from artemisinin by S. griseus ATCC 13273 was proposed as well. In the case of using fungi for biotransformation, artemisinin was converted 10β-hydroxy-artemisinin and 3α-hydroxy-deoxyartemisinin by C. echinulata AS 3.3400 and A. niger AS 3.795 [26], as well as biotransformation products of artemisinin by Penicillium chrysogenum ATCC 9480 [27]. The product 10β-hydroxy-artemisinin was obtained in 67% yield when artemisinin was treated with ferrous sulfate in acetonitrile/water [28]. Furthermore, other oxidative and analogues at different positions products of artemisinin have been reported as well. These include deoxyartemisinin, 3α-hydroxy-artemisinin, 10 hydroxy-artemisinin and 9β-hydroxy-11α-artemisinin [27–30].
Fig. 11.
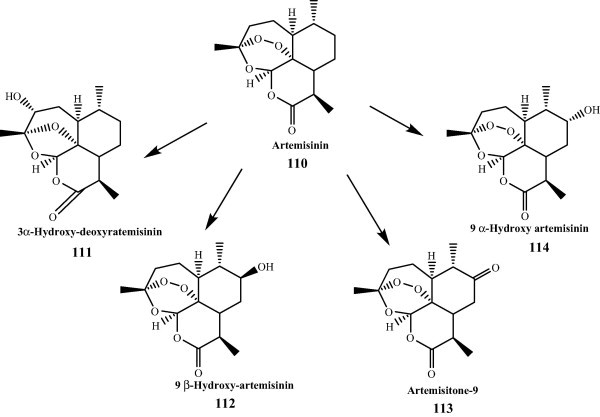
Biotransformation of Artemisinin.
Biotransformation of Taxol and its analogues
Paclitaxel (Taxol®), first isolated from the bark of Taxus brevifolia, is one of the most effective anticancer agents from natural sources. It has been widely used for the treatment of ovarian, breast and lung cancers. In order to reduce side effects and increasing oral bioavailability, more than 500 microorganisms were screened for their ability to achieve useful biotransformation of taxol derivative (cephalomannine) (115). Taxol/cephalomannine (115) was biotransformated by Streptomyces sp. MA 7065, yielding 10-hydroxyacetyl-10-deacetyltaxol (116), 3′-(4-hydroxyphenyl)-3′-dephenyltaxol (117) and 4″-Hydroxycephalomannine (118) [31] (Fig. 12).
Fig. 12.
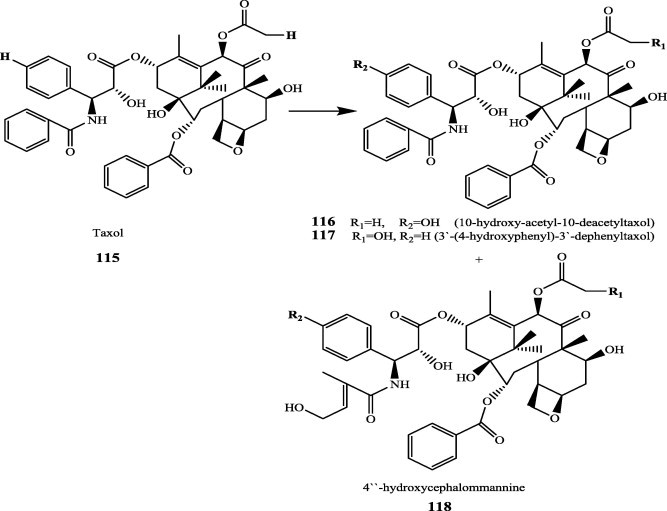
Biotransformation of Paclitaxel and its analogues.
Biotransformation of panaxosides (ginsenosides)
Panaxosides or ginsenosides are the main effective constituents isolated from the traditional Chinese herb ginseng, the roots of Panax ginseng C.A. Meyer. It has been found that the intestinal bacterial metabolites of ginsenosides are responsible for the major pharmacological activities of ginseng roots [32,33]. Investigation on antitumor activities of 20(S)-protopanaxatriol showed that it did not directly inhibit tumor growth in vivo, but that it stimulated splenic NK cells to become cytotoxic to tumor cells [34]. There have been a number of reports on the biotransformation of ginsenosides (119). The ginsenosides 20(S)-Protopanaxatriol (119) was transformed by a fungus Mucor spinosus AS 3.3450, yielding various novel compounds including 12-oxo-15α-hydroxyl-20(S)-protopanaxatriol (128), 27-hydroxyl-20(S)-protopanaxatriol (123), 12-oxo-26-hydroxyl-20(S)-protopanaxatriol (129), 12-oxo-27-hydroxyl-20(S)-protopanaxatriol (125), 12-oxo-23β-hydroxyl-20(S)-protopanaxatriol (126), 20S,24R-epoxy-dammaran-3β,6α,25-triol-12-one (124), 29-hydroxyl-20(S)-protopanaxatriol (121), 12-oxo-11β-hydroxyl-20(S)-protopanaxatriol (127), 28-hydroxyl-20(S)-protopanaxatriol (122) and 12-oxo-20(S)-protopanaxatriol (120) (Fig. 13). MTT assay indicated that eight metabolites had more potent inhibitory effects against HL-60 cell line than the parent compound [35,36]. Transformation of ginsenoside Rg3 by Myrothecium verrucaria furnished the rare ginsenoside Rh2 [37], a more potential molecule than ginsenoside Rg3. Ginsenoside Rh2 can also prepared by enzymatic hydrolysis of ginsenoside Rg3 using β-glucosidase or cell-free extract of Fusarium proliferatum ECU2042 [38,39]. Ginsenoside Rh1 was obtained through transforming ginsenoside Rg1 by A. niger AS 3.1858 or Absidia coerulea AS 3.3538 [40]. A gypenoside-α-(1→6)-L-rhamnosidase isolate from Absidia sp.90 can hydrolyze the β-(1→6)-L-rhamnoside at C-20 position of gypenoside-5 into ginsenoside Rd [41]. A novel β-glucosidase (G-II) from Cladosporium fulvum was also reported. This glucosidase could specifically cleave the β-D-glucosidic linkage at the C-20 position of ginsenoside Rb1 to produce ginsenoside Rd, and did not hydrolyze the other β-D-glucosidic linkages in protopanaxadiol-type ginsenosides [42]. Pythium irregular was used to convert 20(S)-protopanaxadiol ginsenosides such as Rb1, Rb2, Rc, Rd and gypenoside XVII. Nearly all of the 20(S)-protopanaxadiol ginsenosides were metabolized into the minor ginsenoside F2 [43]. Notably, Rb1, the major ginsenoside, was converted to 20(S)-ginsenoside Rg3 by Microbacterium sp. GS514 [44].
Fig. 13.
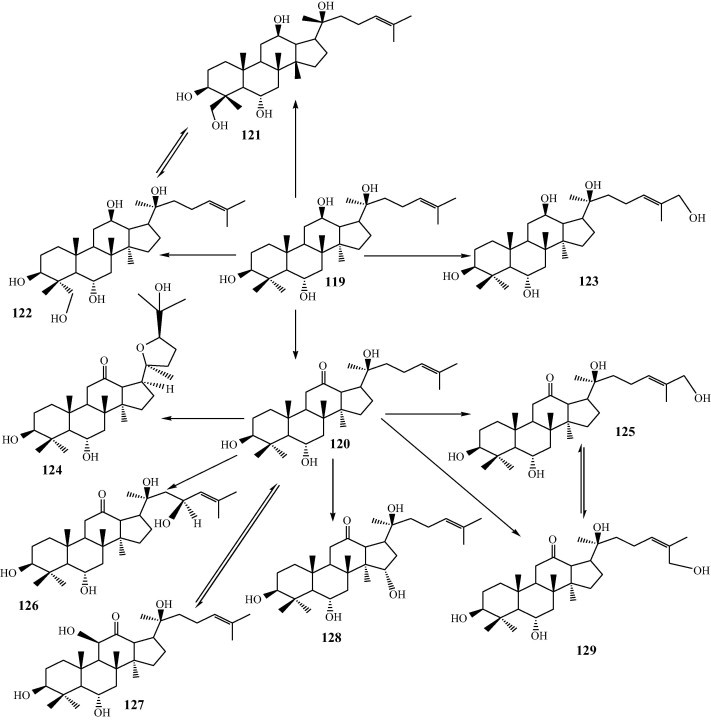
A proposed biotransformation pathway of 20(S)-protopanaxatriol by M. spinosus As 3.3450.
Biotransformation of opiate alkaloid
The biotransformation of alkaloids by microbes and plants was recently reviewed by Rathbone et al. [45,46], in which they provide a summary of the progress of alkaloid biotransformations from mid-1980s to 2002. It is difficult to modify structures of alkaloids because of the complex polycyclic nature of these compounds. Biotransformation offers a versatile tool for structural modification of alkaloids in addition to known chemical methods. Evodiamine is one of the major active alkaloids in Evodia rutaecarpa, a traditional chinese medicine, which has been widely used in China for over two thousand years. Biotransformation of evodiamine (130) by Penicillium janthinellum AS 3.510 resulted in two metabolites, 10-hydroxyevodiamine (131) and 11-hydroxyevodiamine (132) [47–49] (Fig. 14). The microorganisms and rats used similar metabolic pathway for evodiamine. Many fungi have shown to possess enzymes of catalyzing N- and O-demethylation of alkaloids. For example, Mucor piriformis selectively N-demethylated thebaine [48], and Streptomyces and C. echinulata strains N-demethylated a indole alkaloid lergotrile [50], while various Cunninghamella and Fusarium strains N-demethylated codeine [51].
Fig. 14.
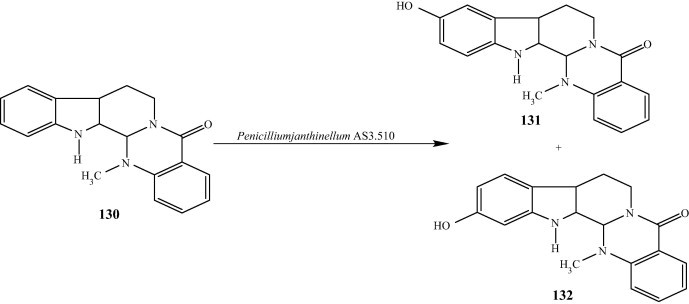
Biotransformation of evodiamine by Penicillium janthinellum AS 3.510.
Moreover, biotransformation of a thebaine derivative using the filamentous fungus C. echinulata NRRL1384 was reported. The thebaine analog was converted to a mixture of N-demethylated and N,O-demethylated products [52].
Biotransformation of bufalin
The biotransformation of steroid compounds by microbes was reviewed by Fernandes in 2003 [53], but considerable progress has been made since then. Many steroids from natural sources, such as bufadienolides, possess significant anticancer activities. Thus, biotransformation has played a role in generating new and more active derivatives. Microbial hydrolysis can achieve very high yield. For instance, cinobufagin (133) and resibufogenin (135) could be completely metabolized by Alternaria alternata AS 3.4578 to generate their 12β-hydroxylated products in greater than 90% yield within 8 h [54]. A. alternata could also convert 3-epi-desacetylcinobufagin into 3-epi-12β-hydroxyl desacetylcinobufagin (137) as the major product (70%). In addition, four dehydrogenated products, 3-keto-resibufogenin (136), 3-keto-cinobufagin (134), 3-ketodeacetylcinobufagin (138) and deacetylcinobufagin (137), were obtained from the biotransformation of resibufogenin and cinobufagin by Pseudomonas aeruginosa AS 1.860 [55] (Fig. 15). However, the biotransformation of resibufogenin by Mucor polymorphosporus AS 3.3443 produced 22 different metabolic products with low yields [54].
Fig. 15.
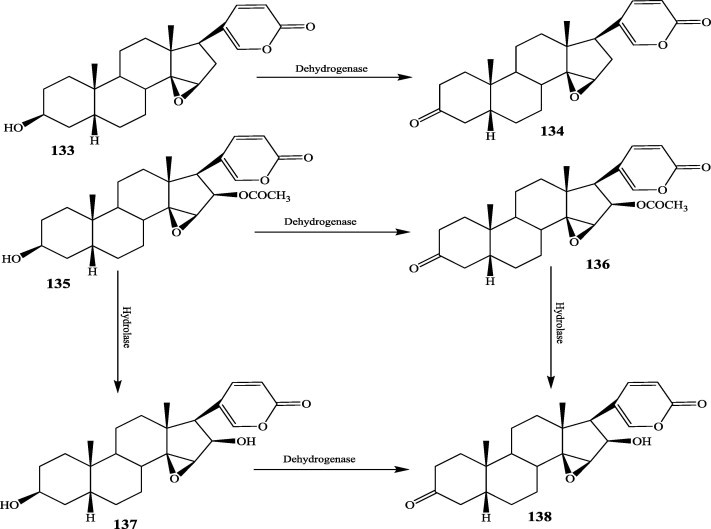
Biotransformation pathways of resibufogenin and cinobufagin by Pseudomonas aeruginosa.
Biotransformation of resibufogenin, cinobufagin, and bufalin by Nocardia sp. NRRL 5646 was also reported [56]. Resibufogenin was converted to 3-acetyl-resibufogenin and 3-acetyl 15β-hydroxyl bufotalin, which showed significantly increased cytotoxic activity than the substrate, while cinobufagin and bufalin were converted to 3-acetyl cinobufagin and 3-acetyl bufalin respectively, in which the biotransformation reaction showed great regio-selectivity on bufadienolides. On the other hand, when Cunninghamella elegans was employed for the biotransformation of cinobufagin, 5 metabolites including 12α-hydroxybufagin, 11α-hydroxybufagin, 12β-hydroxydesacetylcinobufagin, 3-oxo-12α-hydroxybufagin and 12β-hydroxybufagin were produced [57].
Conclusions
Microbial transformation has been studied for centuries. This phenomenon allows for the modification of a compound through an environmentally friendly approach. Microorganisms are capable of producing a great variety of enzymes in a short period of time as a result of a high rate of cell multiplication. In this sense, a reasonable number of compounds of various biological interests can be obtained by microorganisms-driven transformations of natural products.
Conflict of interest
The authors have declared no conflict of interest.
Compliance with Ethics Requirements
This article does not contain any studies with human or animal subjects.
Biographies

Mohamed-Elamir F. Hegazy, Associate Professor in Chemistry of Medicinal plant Department, National Research Center, who has two Ph.D. degrees: A Ph.D. degree from Hiroshima University, Japan, and a Ph.D. degree from Elminia University, Egypt. Dr. Hegazy is working in the field of natural products chemistry and biotransformation of natural compounds with cultured plant cells from ten years ago and he had a strong experience in the isolation, purification and identification of natural compounds form medicinal plants and marine organisms using high technique for identification (1D and 2D NMR analysis).

Tarik A. Mohamed, Researcher in National Research Centre, Egypt. His research interest focused on Chemical Constituents of Medicinal Plants and Marine Organisms, Extraction, Isolation and Purification of Natural Bioactive Compounds, Structural Elucidation of Natural Products by Modern Techniques of Spectroscopic Analysis, MS, HRMS, 1D and 2D NMR and X-ray analysis, Biological Activities of Natural Products against different common diseases and Biotransformation for Natural Compounds.

Abd El-Samid I. El-Shamy, Researcher in National Research Centre, Egypt. His research experiences are focused on isolation, identification of phenanthrenes, flavonoids, sterols, terpenes, coumarines, volatile oils, ceramides from medicinal plants and marines by different isolation and identification methods. Synthesis of derivatives of natural products. Bioactive assay in vivo and in vitro of natural products such as hepatoprotective, anticancer, antimicrobial and antiulcer.

Abou-El-Hamd H. Mohamed, Professor of Natural products chemistry. He is a specialist in natural products isolation and purification of natural product compounds by using different technique (Column chromatography, TLC, HPLC) Identification of naturally isolated pure compounds by using 1D and 2D NMR analysis II – Biotransformation and biocatalysis with Plant cell tissue culture; Biotransformation of organic and natural compounds; Enzyme purification Bioassay.

Usama A. Mahalel, Associate Professor of plant taxonomy. His research interest, include Medicinal plant and natural products chemistry.

Eman H. Reda, a master student, has an experience in isolation and purification of the active constituents from medicinal plants using modern techniques.

Alaa M. Shaheen, a master student, has an experience in isolation and purification of the active constituents from medicinal plants using the modern techniques.

Wafaa A. Tawfik, Assoc. Prof. of Phytochemistry. She has experience in Phytochemical screening of medicinal plants, isolation and identification of the active constituents by using the modern physiochemical techniques, isolating colors and flavors from natural resources, Extraction of oils from plants, interpretation of spectral data with special emphasis to NMR analysis.

Abdelaaty A. Shahat, Professor of Phytochemistry. He is a specialist in Phytochemical evaluation of the Medicinal Plants as flavonoids, alkaloids, coumarine, terpens, proanthocyanidins, phenolic compounds, lignans, organic acids, etc. Isolation and Identification of the active constituents of the medicinal plants using chromatographic techniques {column, Thin Layer, and Paper chromatography, High Performance Liquid Chromatography (HPLC)}. Bioassay guided isolation-pharmacological and biological evaluation of medicinal plant used in tradition medicine. For example antiviral, antibacterial, antioxidant, anticomplement modulation, anti-osteoporosis, anti-obesity, anti-anemia, etc.

Khalid A. Shams, Professor of Phytochemistry, he has experience in isolation and identification of the active constituents from medicinal plants, by using the modern physiochemical techniques. He is familiar with extraction techniques such as Microwave-Assisted Extraction (MAE), Ultrasonic-Assisted Extraction (UAE), Accelerated Solvent Extraction (ASE) and supercritical fluid extraction (SFE).

Nahla S. Abdel-Azim, Professor of Phytochemistry. She has experience in isolation and purification of the active constituents from medicinal plants by using the modern techniques, interpretation of spectral data with special emphasis to NMR analysis, pharmacological screening of medicinal plants & Extraction of medicinal plants using innovative, green and friendly environmental extraction techniques such as Microwave-Assisted Extraction (MAE), Ultrasonic-Assisted Extraction (UAE), Accelerated Solvent Extraction (ASE) and supercritical fluid extraction (SFE).

Faiza M. Hammouda, Professor of Phytochemistry Awarded “State Recognition Prize in the realm of Advanced Technological Sciences (Basic Sciences)” 2003.Co-author of 160 published research papers in national and international journals. Supervised 55 Ph.D. and 42 M. Sc. Thesis, Co-author in 10 books. Principle Investigator of more than 20 national and international projects (FDA, UNDP, GEF, IUCN). Member in Scientific Ethics Council in AST.
Footnotes
Peer review under responsibility of Cairo University.
References
- 1.Roussis V., Wu Z., Fenical W., Stobel S.A., Van Duyne D.G., Clardy J. New antiinflammatory pseudopterosins from the marine octacoral Pseudopterogorgia elisabethae. J Org Chem. 1990;55:4916–4922. [Google Scholar]
- 2.Lilies G. Gambling on marine biotechnology. Bioscience. 1996;46:250–253. [Google Scholar]
- 3.Frydman A., Weisshaus O., Huhman D.V., Sumner L.W., Bar-Peled M., Lewinsohn E. Metabolic engineering of plant cells for biotransformation of hesperedin into neohesperidin, a substrate for production of the low-calorie sweetener and flavor enhancer NHDC. J Agric Food Chem. 2005;53(25):9708–9712. doi: 10.1021/jf051509m. [DOI] [PubMed] [Google Scholar]
- 4.Lilly M.D. Advances in biotransformation processes. Chem Eng Sci. 1994;49(2):151–159. [Google Scholar]
- 5.Walker J.M., Cox M. 2nd ed. ACS Professional Reference Book, ACS; USA: 1995. The language of biotechnology – a dictionary of terms. [Google Scholar]
- 6.Collins A.M., Kennedy M.J. Biotransformations and bioconversions in New Zealand: past endeavours and future potential. Austral Biotechnol. 1999;9(2):86–94. [Google Scholar]
- 7.Leresche J.E., Meyer H.P. Chemocatalysis and biocatalysis (biotransformation): some thoughts of a chemist and a biotechnologist. Org Proc Res Dev. 2006;10:572–580. [Google Scholar]
- 8.Tang F.H., Zhao Y.J., Tang A.K. Presence of ectoparasitic trichodinids (Ciliophora, Oligohymenophorea, Peritrichida) on the gills of cultured freshwater fish, Carassius auratus in Chongqing, China, with the description of a new species of the genus Trichodina. Acta Zootaxon Sin. 2005;30:35–40. [Google Scholar]
- 9.Rozenbaum H.F., Patitucci M.L., Antunes O.A.C., Pereira N. Production of aromas and fragrances through microbial oxidation of monoterpenes. Brazil J Chem Eng. 2006;23(3):273–279. [Google Scholar]
- 10.Gershwin L. Nematocysts of the Cubozoa. Zootaxa. 2006;1232:1–57. [Google Scholar]
- 11.Hotta Y., Ezaki S., Atomi H., Imanaka T. Extremely stable and versatile carboxylesterase from a hyperthermophilic. Appl Environ Microbiol. 2002;68(8):3925–3931. doi: 10.1128/AEM.68.8.3925-3931.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Faber K., Franssen M.C.R. Prospects for the increased application of biocatalysts in organic transformations. Trends Biotechnol. 1993;11:461–470. doi: 10.1016/0167-7799(93)90079-O. [DOI] [PubMed] [Google Scholar]
- 13.Woodley J.M., Lilly M.D. Biotransformation reactor selection and operation. In: Cabral J.M.S., Best D., Boross L., Tramper J., editors. Applied biocatalysis. Harwood Academic, Chur; 1994. [Google Scholar]
- 14.Nankai H., Miyazawa M., Kameoka H. Hydroxylation of 2 saturated acyclic monoterpenoids, tetrahydrogeraniol and tetrahydrolavandulol, by the plant–pathogenic fungus glomerella-cingulata. J Nat Prod. 1997;60(3):287–289. [Google Scholar]
- 15.Demirci F., Noma Y., Kirimer N., Baser K., Huesnue C. Microbial transformation of (−)-carvone. J Biosci. 2004;59(5/6):389–392. [PubMed] [Google Scholar]
- 16.El Sayed K.A., Yousaf M., Hamann M.T., Avery M.A., Kelly M., Wipf P. Microbial and chemical transformation studies of the bioactive marine sesquiterpenes (S)-(+)-curcuphenol and -curcudiol isolated from a deep reef collection of the Jamaican sponge Didiscus oxeata. J Nat Prod. 2002;65(11):1547–1553. doi: 10.1021/np020213x. [DOI] [PubMed] [Google Scholar]
- 17.Barrero A.F., Oltra J.E., Raslan D.S., Saude D.A. Microbial transformation of sesquiterpene lactones by the fungi Cunninghamella echinulata and Rhizopus oryzae. J Nat Prod. 1999;62(5):726–729. doi: 10.1021/np980520w. [DOI] [PubMed] [Google Scholar]
- 18.Fujiwara M., Marumoto S., Yagi N., Miyazawa M. Biotransformation of turmerones by Aspergillus niger. J Nat Prod. 2011;74(1):86–89. doi: 10.1021/np100416v. [DOI] [PubMed] [Google Scholar]
- 19.Haridy M.S.A., Ahmed A.A., Doe M. Microbiological transformation of two labdane diterpenes, the main constituents of Madia species, by two fungi. Phytochemistry. 2006;67(14):1455–1459. doi: 10.1016/j.phytochem.2006.05.036. [DOI] [PubMed] [Google Scholar]
- 20.Fraga B.M., de Alfonso I., Gonzalez-Vallejo V., Guillermo R. Microbial transformation of two 15a-hydroxy-ent-kaur-16-ene diterpenes by Mucor plumbeus. Tetrahedron. 2010;66(1):227–234. [Google Scholar]
- 21.Yang Y., Sun R., Song H., Xu Y., Yang P., Yang D. Microbial transformation of the triterpene nigranoic acid in Trichoderma sp. Phytochem Lett. 2012;5(1):123–127. [Google Scholar]
- 22.Zhu Y., Qian L., Zhang J., Liu J., Yu B. New approaches to the structural modification of olean-type pentacyclic triterpenes via microbial oxidation and glycosylation. Tetrahedron. 2011;67(23):4206–4211. [Google Scholar]
- 23.Wu G., Gao J., Shi X., Zhang Q., Wei S., Ding K. Microbial transformations of diosgenin by the white-rot basidiomycete Coriolus versicolor. J Nat Prod. 2011;74(10):2095–2101. doi: 10.1021/np2003484. [DOI] [PubMed] [Google Scholar]
- 24.Klayman D.L. Qinghaosu (artemisinin): an antimalarial drug from China. Science. 1985;228:1049–1055. doi: 10.1126/science.3887571. [DOI] [PubMed] [Google Scholar]
- 25.Liu J.H., Chen Y.G., Yu B.Y., Chen Y.J. A novel ketone derivative of artemisinin biotransformed by Streptomyces griseus ATCC 13273. Bioorg Med Chem Lett. 2006;16:1909–1912. doi: 10.1016/j.bmcl.2005.12.076. [DOI] [PubMed] [Google Scholar]
- 26.Zhan J., Guo H., Dai J., Zhang Y., Guo D. Microbial transformation of artemisinin by Cunninghamella echinulata and Aspergillus niger. Tetrahedron Lett. 2002;43:4519–4521. [Google Scholar]
- 27.Lee I.S., Elsohly H.N., Croom E.M., Hufford C.D. Microbial metabolism studies of the antimalarial sesquiterpene artemisinin. J Nat Prod. 1989;52:337–341. doi: 10.1021/np50062a020. [DOI] [PubMed] [Google Scholar]
- 28.Wu W.M., Wu Y., Wu Y.L., Yao Z.J., Zhou C.M., Li Y. Unified mechanistic framework for the Fe(II)-induced cleavage of qinghaosu and derivatives/analogues. The first spintrapping evidence for the previously postulated secondary C-4 radical. J Am Chem Soc. 1998;120:3316–3325. [Google Scholar]
- 29.Tatineni R., Doddapaneni K.K., Dalavayi S., Kulkarni S.M., Mangamoori L.N. Microbacterium trichotecenolyticum enzyme mediated transformation of arteannuin B to artemisinin. Process Biochem. 2006;41:2464–2467. [Google Scholar]
- 30.Parshikov I.A., Muraleedharan K.M., Avery M.A., Williamson J.S. Transformation of artemisinin by Cunninghamella elegans. Appl Microbiol Biotechnol. 2004;64:782–786. doi: 10.1007/s00253-003-1524-z. [DOI] [PubMed] [Google Scholar]
- 31.Chen T.S., Li X., Bollag D., Liu Y., Chang C. Biotransformation of taxol. Tetrahedron Lett. 2001;42:3787–3789. [Google Scholar]
- 32.Bae E.A., Han M.J., Choo M.K., Park S.Y., Kim D.H. Metabolism of 20(S)- and 20(R)-ginsenoside Rg3 by human intestinal bacteria and its relation to in vitro biological activities. Biol Pharm Bull. 2002;25:58–63. doi: 10.1248/bpb.25.58. [DOI] [PubMed] [Google Scholar]
- 33.Liu Y., Li W., Li P., Deng M.C., Yang S.L., Yang L. The inhibitory effect of intestinal bacterial metabolite of ginsenosides on CYP3A activity. Biol Pharm Bull. 2004;27:1555–1560. doi: 10.1248/bpb.27.1555. [DOI] [PubMed] [Google Scholar]
- 34.Hasegawa H., Suzuki R., Nagaoka T., Tezuka Y., Kadota S., Saiki I. Prevention of growth and metastasis of murine melanoma through enhanced natural-killer cytotoxicity by fatty acid-conjugate of protopanaxatriol. Biol Pharm Bull. 2002;25:861–866. doi: 10.1248/bpb.25.861. [DOI] [PubMed] [Google Scholar]
- 35.Zhang J., Guo H., Tian T., Liu P., Li N., Zhou J. Biotransformation of 20(S)-protopanaxatriol by Mucor spinosus and the cytotoxic structure activity relationships of the transformed products. Phytochemistry. 2007;68:2523–2530. doi: 10.1016/j.phytochem.2007.05.028. [DOI] [PubMed] [Google Scholar]
- 36.Tian Y., Guo H., Han J., Guo D. Microbial transformation of 20(S)-protopanaxatriol by Mucor spinosus. J Nat Prod. 2005;68:678–680. doi: 10.1021/np049688+. [DOI] [PubMed] [Google Scholar]
- 37.Wu X., Wang Y., Zhao W., Zhang Y. Fungal biotransformation of ginse-noside Rg3. Acta Microbiol Sin. 2008;48:1181–1185. [PubMed] [Google Scholar]
- 38.Su J.H., Xu J.H., Lu W.Y., Lin G.Q. Enzymatic transformation of ginsenoside Rg 3 to Rh 2 using newly isolated Fusarium proliferatum ECU2042. J Mol Catal B Enzym. 2006;38:113–118. [Google Scholar]
- 39.Su J.H., Xu J.H., Yu H.L., He Y.C., Lu W.Y., Lin G.Q. Fusarium proliferatum ECU2042 that converts ginsenoside Rg 3 into Rh 2. J Mol Catal B Enzym. 2009;57:278–283. [Google Scholar]
- 40.Dong A.L., Cui Y., Cuo H. J Chin Pharm Sci. 2001;10:115–118. [Google Scholar]
- 41.Yu H., Liu H., Zhang C., Tan D., Lu M., Jin F. Purification and characterization of gypenoside-alpha-L-rhamnosidase hydrolyzing gypenoside-5 into ginsenoside Rd. Process Biochem. 2004;39:861–867. [Google Scholar]
- 42.Zhao X., Gao L., Wang J., Bi H., Gao J., Du X., Zhou Y., Tai G. A novel ginsenoside Rb1-hydrolyzing b-D-glucosidase from Cladosporium fulvum. Process Biochem. 2009;44:612–618. [Google Scholar]
- 43.Yousef L.F., Bernards M.A. In vitro metabolism of ginsenosides by the ginseng root pathogen Pythium irregulare. Phytochemistry. 2006;67:1740–1749. doi: 10.1016/j.phytochem.2005.06.030. [DOI] [PubMed] [Google Scholar]
- 44.Cheng L.Q., Na J.R., Bang M.H., Kim M.K., Yang D.C. Conversion of major ginsenoside Rb1 to 20(S)-ginsenoside Rg3 by Microbacterium sp. GS514. Phytochemistry. 2008;69:218–224. doi: 10.1016/j.phytochem.2007.06.035. [DOI] [PubMed] [Google Scholar]
- 45.Rathbone D.A., Lister D.L., Bruce N.C. Biotransformation of alkaloids. Alkaloids Chem Biol. 2001;57:1–74. doi: 10.1016/s0099-9598(01)57002-9. [DOI] [PubMed] [Google Scholar]
- 46.Rathbone D.A., Bruce N.C. Microbial transformation of alkaloids. Curr Opin Microbiol. 2002;5:274–281. doi: 10.1016/s1369-5274(02)00317-x. [DOI] [PubMed] [Google Scholar]
- 47.Madyastha K.M., Sridhar G.R. A novel pathway for the metabolism of caffeine by a mixed culture consortium. Biochem Biophys Res. 1998;249:178–181. doi: 10.1006/bbrc.1998.9102. [DOI] [PubMed] [Google Scholar]
- 48.Madyastha K.M., Reddy G.V.B. Mucor piriformis, an efficient N-dealkylating reagent for thebaine and its N-variants. J Chem Soc Perkin Trans. 1994;1:911–912. [Google Scholar]
- 49.Li L., Liu R., Ye M., Hu X., Wang Q., Bi K. Microbial metabolism of evodiamine by Penicillium janthinellum and its application for metabolite identification in rat urine. Enzyme Microb Technol. 2006;39:561–567. [Google Scholar]
- 50.Davis P.J., Glade J.C., Clark A.M., Smith R.V. N-demethylation of lergotrile by Streptomyces platensis. Appl Environ Microbiol. 1979;38:891–893. doi: 10.1128/aem.38.5.891-893.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gibson M., Spoer G.J., Parfitt R.T., Sewell G.J. Enzyme-mediated N-demethylation of codeine. Enzyme Microb Technol. 1984;6:471–475. [Google Scholar]
- 52.Abel A.M., Carnell A.J., Davis J.A., Paylor M. The synthesis of buprenorphine intermediates by regioselective microbial N- and O-demethylation reactions using Cunninghamella echinulata. Enzyme Microb Technol. 2003;33:743–748. [Google Scholar]
- 53.Fernandes P., Cruz A., Angelova B., Pinheiro H.M., Cabral J.M.S. Microbial conversion of steroid compounds: recent developments. Enzyme Microb Technol. 2003;32:688–705. [Google Scholar]
- 54.Ye M., Han J., An D., Tu G., Guo G. New cytotoxic bufadienolides from the biotransformation of resibufogenin by Mucor polymorphosporus. Tetrahedron. 2005;61:8947–8955. [Google Scholar]
- 55.Zhan J., Liu W., Guo H., Zhang Y., Guo D. Selective dehydrogenation of resibufogenin and cinobufagin at 3-OH by Pseudomonas aeruginosa. Enzyme Microb Technol. 2003;33:29–32. [Google Scholar]
- 56.Zhang J., Sun Y., Liu J.H., Yu B.Y., Xu Q. Microbial transformation of three bufadienolides by Nocardia sp. and some insight for the cytotoxic structure–activity relationship (SAR) Bioorg Med Chem Lett. 2007;17:6062–6065. doi: 10.1016/j.bmcl.2007.09.065. [DOI] [PubMed] [Google Scholar]
- 57.Qiao L., Zhou Y.Z.H., Qi X.L., Lin L.H., Chen H.C.H., Pang L.Y. Biotransformation of cinobufagin by Cunninghamella elegans. J Antibiot. 2007;60(4):261–264. doi: 10.1038/ja.2007.32. [DOI] [PubMed] [Google Scholar]



