Abstract
The resurrection plant Craterostigma plantagineum has the ability to survive complete dehydration. In an attempt to further understand desiccation tolerance in this plant, the CpMYB10 transcription factor gene was functionally characterized. CpMYB10 is rapidly induced by dehydration and abscisic acid (ABA) treatments in leaves and roots, but no expression was detected in fully hydrated tissues. Electrophoretic mobility shift assay experiments showed binding of rCpMYB10 to specific mybRE elements within the LEA Cp11-24 and CpMYB10 promoters. Localization of CpMYB10 transcript by in situ reverse transcription-PCR reactions showed expression in vascular tissues, parenchyma, and epidermis both in leaves and roots in response to ABA. Transgenic Arabidopsis plants transformed with CpMYB10 promoter fused to GUS gene showed reporter expression under ABA and stress conditions in several organs. Overexpression of CpMYB10 cDNA in Arabidopsis led to desiccation and salt tolerance of transgenics lines. Interestingly, it was found that plants overexpressing CpMYB10 exhibited Glc-insensitive and ABA hypersensitive phenotypes. Therefore, our results indicate that CpMYB10 in Arabidopsis is mediating stress tolerance and altering ABA and Glc signaling responses.
Diurnal and seasonal environmental fluctuations as well as extreme conditions have been a major selective pressure for plant evolution. Plants are sessile organisms that cannot move to escape from adverse environmental cues, thus complex metabolic and anatomical adaptations have been developed to cope with abiotic stresses. Availability of water is probably the most limiting factor for crop productivity and yield, compromising economical output and human food supply. Therefore, there is a strong need to understand plant adaptation mechanisms against adverse environmental conditions to improve stress tolerance.
Plant stress responses involve the expression of a plethora of genes with an adaptive role. Among the products of these genes are enzymes catalyzing the synthesis of osmoprotectants or antioxidants, late-embryogenesis abundant (LEA) proteins, chaperones and heat shock proteins, lipid desaturases, water channels, and ion transporters, representing some of the best characterized examples (Ingram and Bartels, 1996). Abscisic acid (ABA) plays a major role in transducing stress responses (Knight and Knight, 2001). Rapid stress responses are in most cases ABA-independent, and there is growing evidence that ABA-dependent and independent pathways cross-talk (Shinozaki and Yamaguchi-Shinozaki, 2000). Signal transduction components include protein kinases such as calcium-dependent protein kinases (CDPK) and mitogen-activated protein (MAP) kinases, G-proteins, phosphatase 2C, and second messengers such as Ca2+ and phosphoinositides. A phospholipase D raises its activity minutes after dehydration (Frank et al., 2000). Also, within 1 min after osmotic shock, inositol 1,4,5-P3, a breakdown product of phospholipase C, dramatically increases its concentration, and rapid changes in cytosolic free Ca2+ concentrations are triggered during this process (DeWald et al., 2001). It has been claimed that nitric oxide is involved in ABA-induced response to stomatal closure and requires cGMP and cADPR (Neill et al., 2002). An Arabidopsis transmembrane His kinase functions as an osmosensor in a yeast mutant, suggesting a similar role in plants (Urao et al., 1999).
Several plant model systems have been used to study responses to water deficit, according to the severity of the stress. Upon a mild water deficit, plants reduce water loss by closing stomata, retain water by osmotic adjustment, and increase water uptake. These responses have been thoroughly studied in Arabidopsis and other mesophytes (Tabaeizadeh, 1998). A different situation occurs in the so-called resurrection plants that exhibit protoplasmic desiccation tolerance. These organisms withstand long periods with air of 0% (v/v) relative humidity, reviving a few hours after exposure to water. The best characterized example is Craterostigma plantagineum, a South African plant living on rocks in shallow soil (Gaff, 1971). An important question is whether the biochemical and molecular mechanisms to cope with dehydration stress that are present in Arabidopsis, crops, or seeds are also in Craterostigma. Although some common molecular components have been found in all these plants, in mature seeds or Craterostigma some differences with other systems have been uncovered, such as the presence of large concentrations of sugars (Bartels and Salamini, 2001). The C8 sugar octulose is very abundant in fully hydrated leaves of Craterostigma, but as soon as desiccation proceeds its concentration drastically drops down and concomitantly Suc reaches high levels. Disaccharides such as Suc or trehalose have been shown to protect enzymes and membrane structures under the dehydrated state and are abundant in anhydrobiotic organisms (Crowe et al., 1998; Hoekstra et al., 2001).
Another potential difference between Craterostigma and Arabidopsis might be the pattern of gene regulation coordinated by transcriptional activators and their tissue-specificity. Genetic and molecular approaches have identified transcription factors that modulate gene expression in response to abiotic stress and ABA. The transcription activators DREB1A, DREB2A, and CBF1, involved in ABA-independent stress response, bind to the consensus dehydration-responsive element (DRE) TACCGACAT, which is present in promoter regions of genes induced by osmotic, saline, and cold stresses (Stockinger et al., 1997; Liu et al., 1998). Genes responsive to ABA usually contain ABA-responsive elements consisting of the (C/T)ACGTGGC consensus sequence and are transactivated by bZIP transcription factors (Choi et al., 2000). Three classes of transcription factors have been characterized in Craterostigma: a heat shock transcription factor (Bockel et al., 1998), two members of the homeodomain Leu zipper family (Frank et al., 1998), and three MYB genes (Iturriaga et al., 1996). Here we show that Arabidopsis transgenic lines overexpressing the heterologous MYB transcription factor gene CpMYB10 are stress tolerant, Glc-insensitive, and ABA hypersensitive. The expression pattern of CpMYB10 in Craterostigma suggests a key role in desiccation tolerance.
RESULTS
CpMYB10 Gene Is Induced by Desiccation and ABA and Is Under Repression Control in Unstressed Conditions
We have previously reported the cloning of a cDNA and two genomic MYB genes from Craterostigma (Iturriaga et al., 1996). The CpMYB7 cDNA and CpMYB5 and CpMYB10 genomic clones share between them 96% to 98% identity and have the canonical MYB DNA-binding domain with the R2R3-type structure that is predominant in plants (Stracke et al., 2001). In Arabidopsis, the closest homolog to these MYB genes from Craterostigma is AtMYB2, which is induced by ABA, dehydration, and salt stresses (Urao et al., 1993). Although the DNA-binding domain of CpMYB10 and AtMYB2 shares 92% similarity, the 230-amino acid C terminus of CpMYB10 is different from the corresponding AtMYB2 region, except for a stretch of 31 amino acids that shows 87% similarity (Iturriaga et al., 1996).
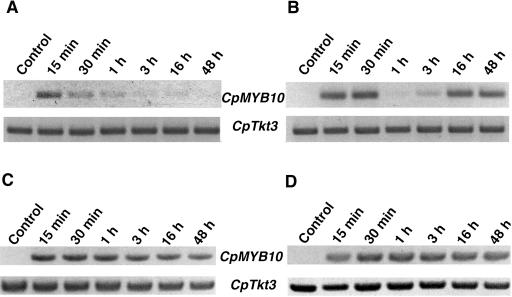
To test whether CpMYB10 is expressed in leaves and roots, a reverse transcription coupled to PCR (RT-PCR) analysis was used. Specific oligonucleotides for the 5′and 3′ unstranslated region of CpMYB10 gene were used in this expression analysis. To confirm specific amplification of CpMYB10 cDNA, the amplification product was cloned and sequenced. As shown in Figure 1, A and B, CpMYB10 is induced upon desiccation in leaves and roots, respectively. No expression could be detected in unstressed organs. Since detached leaves were used for these experiments, the absence of expression also suggests that CpMYB10 is not induced by wounding. Both in leaves and roots the CpMYB10 transcript is detected at significant levels 15 min after dehydration suggesting that its expression began earlier. In leaves, CpMYB10 reaches its maximum level at about 15 min, sharply declining thereafter and is absent 48 h after stress treatment began (Fig. 1A). The CpTkt3 gene that encodes a transketolase (Bernacchia et al., 1995) was used as a control for constitutive expression.
Figure 1.
Expression of CpMYB10 is regulated by drought and ABA. A, Total RNA was isolated from dehydrated Craterostigma leaves at the indicated times and the expression of CpMYB10 was determined by coupled RT and PCR. The PCR product of Cptkt3 gene was included as a cDNA loading control. B, RNA from dehydrated roots. C, RNA from leaves treated with 100 μm ABA. D, RNA from roots treated with 100 μm ABA. Specific primers were used to amplify CpMYB10 and Cptkt3 gene transcripts. The lengths of the PCR products are 1,117 and 733 bp, respectively. The linear phase of the exponential PCR reaction was corroborated for each gene (data not shown). A representative experiment from three biologically independent experiments is shown.
In desiccated roots, CpMYB10 has a biphasic expression pattern where a first transcript peak is observed from 15 to 30 min after stress was initiated and drastically declining after 1 h (Fig. 1B). A second burst of CpMYB10 expression was observed 16 h after desiccation treatment at similarly high levels as the first peak, and it is maintained during the 48 h of the experimental time course. To determine if CpMYB10 expression is induced by ABA, fully hydrated plants were treated with 100 μm ABA at same time points as above (Fig. 1, C and D). In contrast to desiccation treatment, ABA switches on CpMYB10 at constant levels both in leaves and roots after 15 min of treatment and continues up through 48 h. These two sets of experiments show that CpMYB10 is induced by desiccation and ABA, although its differential expression pattern upon ABA treatment suggests that the endogenous and exogenous ABA signals are sensed differently during the desiccation treatment.
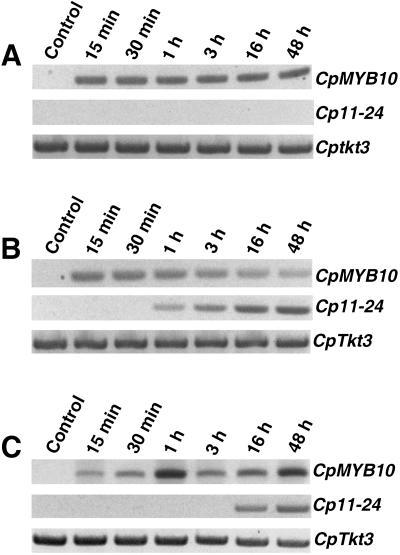
To test whether CpMYB10 expression depends on de novo protein synthesis, fully hydrated Craterostigma leaves were incubated in the presence of the protein synthesis inhibitor cycloheximide (CHX) and/or ABA at different time points. The LEA Cp11-24 gene was used as a control of ABA treatment (Velasco et al., 1998). As shown in Figure 2A, CHX treatment blocked completely Cp11-24 gene expression, suggesting its dependence on de novo protein synthesis. Surprisingly, CpMYB10 expression was not inhibited but rather induced with CHX treatment (Fig. 2A) at relatively higher levels after comparison to its induction by ABA (Fig. 2B). Control experiments showed that ABA induced expression of CpMYB10 was present 15 min after treatment, whereas Cp11-24 was not detected until 1 h after ABA exposure (Fig. 2B). This delay in Cp11-24 expression may be due to the fact that it requires de novo protein synthesis, as suggested by Figure 2A. Incubation of Craterostigma leaves in the presence of both CHX and ABA resulted in higher levels of CpMYB10 expression than in ABA alone, whereas LEA Cp11-24 transcription was restored although only after 16 h of ABA treatment (Fig. 2C). These results showed that CpMYB10 does not require prior protein synthesis for its expression, instead inhibition of translation triggers CpMYB10 transcription, suggesting that somehow it is repressed under unstressed conditions.
Figure 2.
Expression of CpMYB10 in the presence of cycloheximide. A, Total RNA was isolated from Craterostigma leaves treated with 10 μm cycloheximide at the indicated times and the expression of CpMYB10 was determined by coupled RT and PCR. The Craterostigma LEA gene Cp11-24 was included in the experiment as a control of ABA treatment, whereas Cptkt3 was used as a cDNA loading control. B, Craterostigma leaves were treated with 100 μm ABA. C, Craterostigma leaves were treated with 100 μm ABA and 10 μm cycloheximide. Specific primers were used to amplify CpMYB10, Cp11-24, and Cptkt3 gene transcripts. The lengths of the PCR products are 1,117, 629, and 733 bp, respectively. The linear phase of the exponential PCR reaction was corroborated for each gene (data not shown). A representative experiment from three biologically independent experiments is shown.
CpMYB10 Protein Binds to Cp11-24 and CpMYB10 Promoters
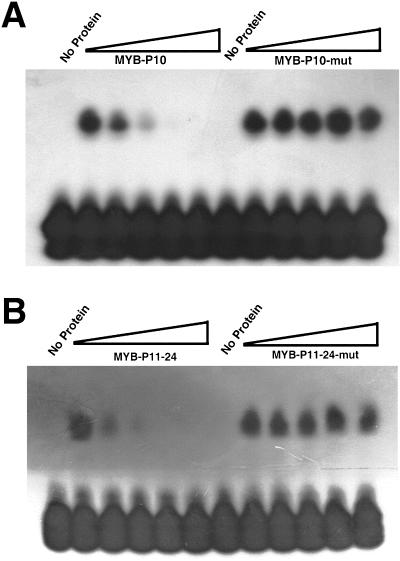
To study the DNA-binding properties of CpMYB10 protein, its cDNA was expressed in Escherichia coli as a fusion protein to a hexapeptide of His residues. The purified recombinant protein was used to perform gel mobility shift assays with double-strand oligonucleotide probes 32P labeled. These probes contained the MYB binding sequence found in the LEA Cp11-24 (Velasco et al., 1998) and CpMYB10 (Iturriaga et al., 1996) gene promoters of Craterostigma. The consensus DNA-binding recognition sequence of several plant MYB genes including AtMYB2 has been defined as T/GAACTG/A (Urao et al., 1993; Higo and Ugawa, 1999). Three TAACTG elements and a GAACTA sequence were found in CpMYB10 and Cp11-24 promoter regions, respectively. Each DNA motif was included in either MYB-P10 or MYB-P11-24 oligonucleotide probes. To analyze the binding affinity of recombinant MYB protein (rCpMYB10) to MYB-P10 and MYB-P11-24 probes, the optimal concentration of required protein was determined (data not shown). Figure 3A shows binding of rMYB10 protein to MYB-P10 probe. To analyze the specificity of the DNA binding activity, a mutated form of MYB-P10 was used (MYB-P10-mut) as well as MYB-P10 as probes (Fig. 3A). The MYB-P10-mut oligonucleotide contained two point mutations in the MYB binding site (TCCCTG instead of TAACTG). In the electrophoretic mobility shift assay (EMSA), the DNA binding activity rMYB10 protein was reduced by the addition of excess unlabeled MYB-P10 but not by that of unlabeled MYB-P10-mut. Thus, rMYB10 protein bound to MYB-P10 but not to MYB-P10-mut. These results indicate that rMYB10 binds sequence specifically to the MYB-P10 oligonucleotide probe and suggests that CpMYB10 protein binds to its own promoter.
Figure 3.
DNA-binding assays of rCpMYB10 protein. A, Purified recombinant rCpMYB10 protein expressed in E. coli was used in EMSA assays. Complementary 20-nucleotide length primers corresponding to the MYB-P10 DNA-binding region were used as 32P-labeled probe. Unlabeled mutant (MYB-P10-mut) and unmodified versions of primers were used to compete radioactive probe. The concentrations used in the competition represented by the triangles were 0, 5, 25, 125, and 625 ng. B, EMSA assays using MYB-P11-24 DNA-binding region as a 32P-labeled probe. Unlabeled mutant (MYB-P11-24-mut) probe was used for competition.
To further characterize the DNA binding properties of rCpMYB10 protein, it was incubated with MYB-P11-24 probe and used to perform an EMSA, as shown in Figure 3B. Unlabeled competitor MYB-P11-24 decreased rCpMYB10 protein binding activity, whereas a mutated form of MYB-P11-24, MYB-P11-24-mut (GCCCTA instead of GAACTA), was unable to compete for protein binding. Therefore, rCpMYB10 protein also binds specifically to MYB-P11-24 probe, suggesting that the LEA 11-24 might be a possible target gene.
Expression of CpMYB10 Is Localized in Discrete Tissues in Craterostigma
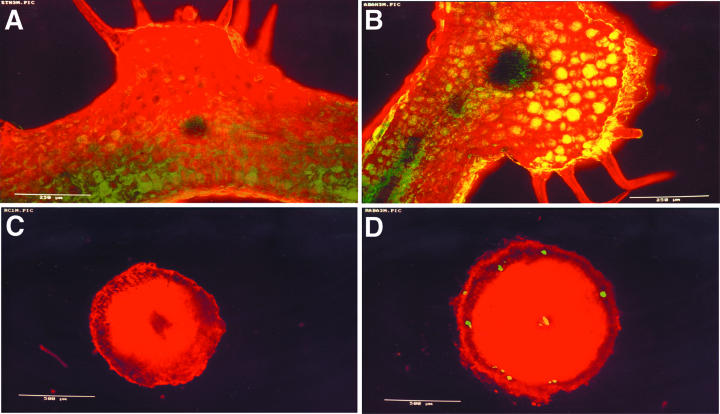
The expression pattern of CpMYB10 was further investigated by in situ RT-PCR analysis in Craterostigma. Leaf and root tissue sections obtained from control or 100 μm ABA pretreated plants were processed for in situ RT-PCR reactions, which yields high-resolution specific mRNA amplification signals in plant tissues (Xoconostle-Cázares et al., 1999). Red chlorophyll autofluorescence was superimposed with the green signal produced by the in situ RT-PCR reaction, leading to a yellow fluorescence (Fig. 4). In untreated leaves, CpMYB10 amplification products were visible at low levels in epidermis, in some parenchyma cells, and in the vascular bundle central region (Fig. 4A). In contrast, PCR products in ABA treated leaves were detected at higher levels in epidermis, palisade, and spongy parenchyma, and vascular bundle and undetectable in trichoma (Fig. 4B). ABA treated roots showed a more discrete pattern of CpMYB10 expression, limited to vascular cylinder and some isolated cortex cells (Fig. 4D). No fluorescent PCR products were detected in untreated roots (Fig. 4C). It was not possible to perform in situ RT-PCR assays from dehydrated plants since tissue sections were difficult to handle.
Figure 4.
Tissue localization of CpMYB10 expression in Craterostigma. A, Confocal laser scanning microscope images of transverse sections from fully hydrated Craterostigma leaves or B, treated with 100 μm ABA. C, Tissue sections of fully hydrated Craterostigma roots or D, treated with 100 μm ABA. The sections were processed for in situ coupled RT and PCR using specific CpMYB10 primers. Overlaps of green and red fluorescence from the same field of view are presented. Bars = 500 μm. A representative experiment from at least three biologically independent experiments is shown.
CpMYB10 Promoter Is Regulated in Transgenic Arabidopsis
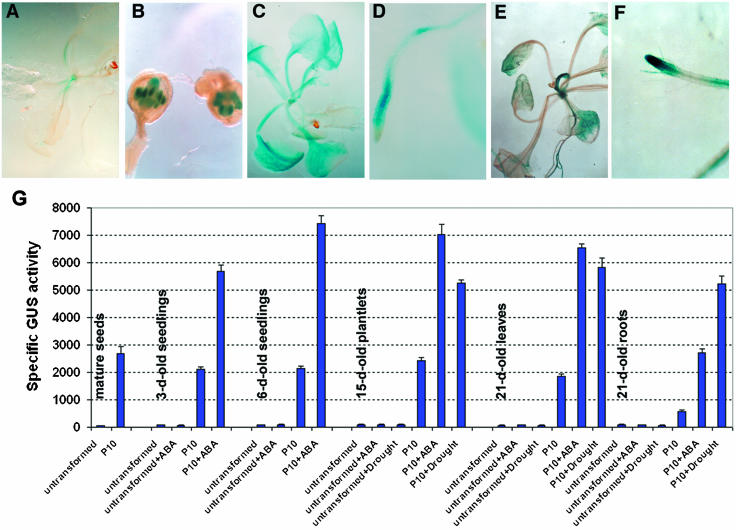
More detailed expression patterns of CpMYB10 under stress and ABA treatments were determined by histochemical β-glucoronidase (GUS) staining of Arabidopsis transgenic plants that harbored a CpMYB10 promoter-GUS fusion construct. Three independent T3 homozygous (9.2, 11.1, and 13.5) lines with similar performance in response to ABA and dehydration were analyzed. In unstressed transgenic plants, GUS activity was detected in apical shoot meristem at low levels (Fig. 5A) and in pollen grains (Fig. 5B). After subjecting transgenic plants to 100 μm ABA treatment, a strong staining was observed in all vegetative tissues (Fig. 5C), although staining was stronger in the subapical region of roots and no staining was visible in root tip (Fig. 5D). Dehydration treatment led to a more localized staining, mainly in leaf vascular tissues and conspicuously strong in apical shoot meristem, emerging leaves, and roots (Fig. 5E). In contrast to ABA treatment, dehydration led to a stronger GUS activity in the root tip (Fig. 5F). These results strongly suggest that in transgenic Arabidopsis the CpMYB10 promoter is regulated by ABA and dehydration as in Craterostigma.
Figure 5.
Expression pattern of CpMYB10 in transgenic Arabidopsis. Three-week-old plants of a representative transgenic Arabidopsis line (9.2) carrying a 1.5-kb CpMYB10 promoter-GUS construct were processed for histochemical GUS staining (A–F). β-Glucoronidase activity is visualized by the blue color. A, Unstressed plants. B, Anthers from unstressed plants. C, Plants treated with 100 μm ABA. D, Roots from plants treated with 100 μm ABA. E, Dehydrated plants. F, Roots from dehydrated plants. G, Quantification of β-Glucoronidase activity in mature seeds, and different developmental stages of untreated or 100-μm ABA-treated seedlings for 16 h or seedlings subjected to dehydration. Treated and untreated untransformed plants were included as negative controls. The results are means of GUS activities from three independent experiments with a repetition in each experiment. Specific GUS activities are expressed as pmol of 4-methylunbelliferone per mg of total protein per min.
To have a quantitative analysis of CpMYB10 promoter strength in Arabidopsis, GUS activity was determined by fluorimetric assays. Transgenic plants 3, 6, 15, and 21 d after germination were dehydrated, treated with 100 μm ABA, or unstressed to measure GUS activity. No significant GUS activities were detected in untransformed Arabidopsis plants (dehydrated, ABA treated, or untreated) at the developmental stages analyzed. GUS activity in unstressed transgenic plants exhibited similarly low levels at different ages, basal expression was around 10% to 30% of that observed for ABA or dehydrated plants of 3 to 21 d old. This basal β-glucoronidase activity is consistent with the histochemical GUS staining described above (Fig. 5A). As shown in Figure 5G, ABA treatment induced the highest levels of GUS activity, reaching a peak in 6-d-old plants. In 15-d-old plants, ABA induced GUS activity was 25% greater than that observed in dehydration stress. The maximum level of GUS activity induced by dehydration was seen in the leaves of 21-d-old plants. Only in roots were levels caused by dehydration greater than those of ABA treatment (Fig. 5G).
Ectopic Expression of CpMYB10 Gene Confers Stress Tolerance in Arabidopsis
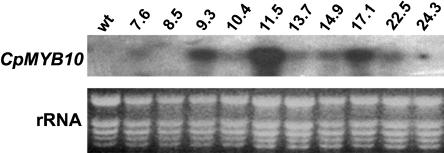
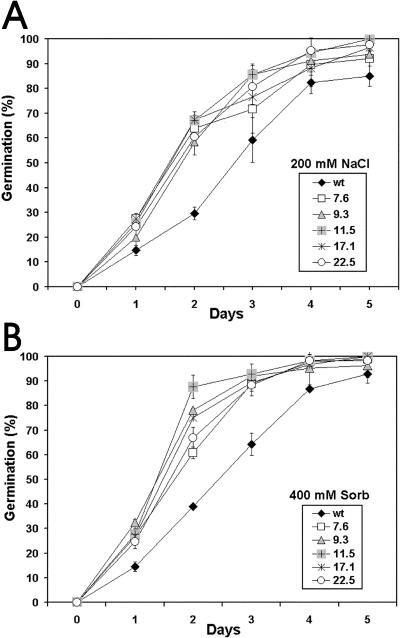
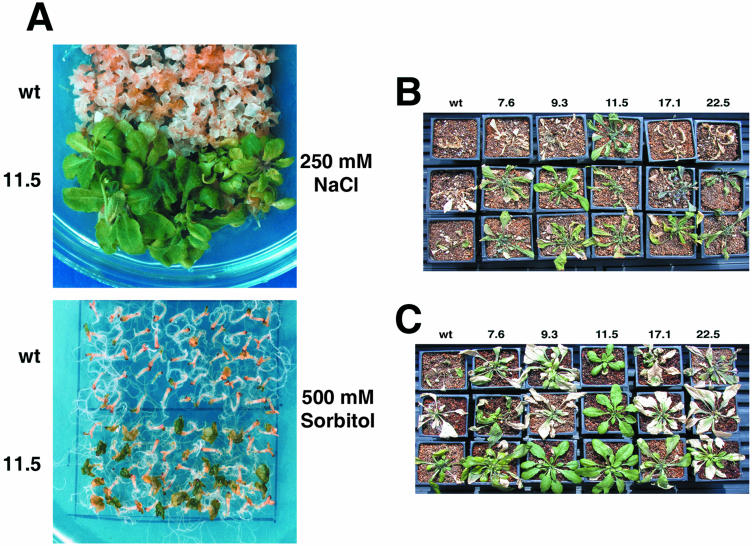
To investigate the in vivo function of CpMYB10, its cDNA was overexpressed in Arabidopsis using the 35S promoter. Thirty independent T2 lines, named 35S-CpMYB10, were recovered and checked by RT-PCR for CpMYB10 expression and 10 T3 homozygous lines were corroborated by RNA gel blot (Fig. 6). Eight T3 homozygous lines showing transgene expression were used for stress tolerance tests. Representative lines 11.5, 17.1, 9.3, 22.5, and 7.6 with decreasing levels of CpMYB10 transcript were further analyzed. Comparison of 35S-CpMYB10 lines with wild-type plants showed no morphological alterations or growth retardation except for a bulky root system in transgenic lines that was clearly visible in 3-week- old plants (data not shown). The germination rate of wild-type and 35S-CpMYB10 lines was assayed in Murashige and Skoog (MS) (1962) media containing different concentrations of osmoticum compounds. Germination was defined as radicule emergence. The sharpest differences were observed in media containing 200 mm NaCl or 400 mm sorbitol (Fig. 7). After 2 d in 200 mm NaCl, 60% to 70% of 5 selected 35S-CpMYB10 lines had already germinated compared to 30% of wild-type plants (Fig. 7A). In media containing 400 mm sorbitol, 90% of 35S-CpMYB10 lines germinated in contrast to only 40% of wild-type seedlings (Fig. 7B). After 5 d, close to 100% and around 90% of transgenic and wild-type seedlings, respectively, germinated in both treatments. All lines germinated at the same rate in the absence of osmoticum compounds. These results showed a faster rate of germination of Arabidopsis lines overexpressing CpMYB10 gene in osmotic stress conditions. In a further experiment, transgenic seeds were germinated media and grown for 4 weeks in high osmoticum media. As shown in Figure 8A, plants from the representative 11.5 line grew normally in 250 mm NaCl, whereas wild-type plants became chlorotic and died. In 500 mm sorbitol, transgenic line 11.5 grew poorly but better than wild-type plants (Fig. 8A).
Figure 6.
Analysis of CpMYB10 expression in transgenic Arabidopsis. Northern blot from Col wild- type and several transgenic lines (line number on top of the figure) grown for 15 d on MS medium. Each lane contains 10 μg of total RNA. The complete 1.1-kb CpMYB10 cDNA was used as a probe. On bottom part of the figure rRNAs are shown as a loading control.
Figure 7.
Kinetics of germination of 35S-CpMYB10 lines. A, Growth of transgenic plants on MS media added with 200 mm NaCl. B, Growth of transgenic plants on MS media added with 400 mm sorbitol. The figure represents the average of 3 replicates of 100 seeds from wild-type and 5 independent transgenic (T3 generation) homozygous lines. Error bar represents sds. Germination was defined as complete protrusion of the radicule.
Figure 8.
Salt and drought tolerance of 35S-CpMYB10 plants. A, Wild-type and 11.5 transgenic seeds were germinated on MS media containing 250 mm NaCl or 500 mm sorbitol during 4 weeks. B, Wild- type and five independent (T3 generation) homozygous lines expressing the 35S CpMYB10 transcript were withheld from water for 14 d and then rewatered before being photographed. C, Wild-type and five independent 35S-CpMYB10 lines were stressed with increasing concentrations of NaCl (50, 100, 150, 200, and 250 mm). Three plants per line are shown. See “Materials and Methods” section for details.
To asses if the overexpression of the CpMYB10 gene conferred stress tolerance, drought and salt tolerance tests in adult plants grown in soil were performed. Plants from five selected 35S lines were grown for 4 weeks under fully watered conditions followed by 2 weeks of water deprivation. As shown in Figure 8B, most 35S-CpMYB10 lines recovered water deprivation after rewatering, whereas wild-type plants did not survive this treatment. To test for salt-stress tolerance, transgenic plants overexpressing CpMYB10 were grown with increasing concentrations of salt up to 250 mm NaCl. Figure 8C shows that transgenic plants grew well, whereas wild-type plants are wilted and chlorotic. Plants from transgenic lines under both treatments continued normal growth and set viable seeds. These results suggest that the overexpression of CpMYB10 gene in Arabidopsis could be up-regulating genes involved in stress tolerance.
Altered Response to Glc and ABA in 35S-CpMYB10 Plants
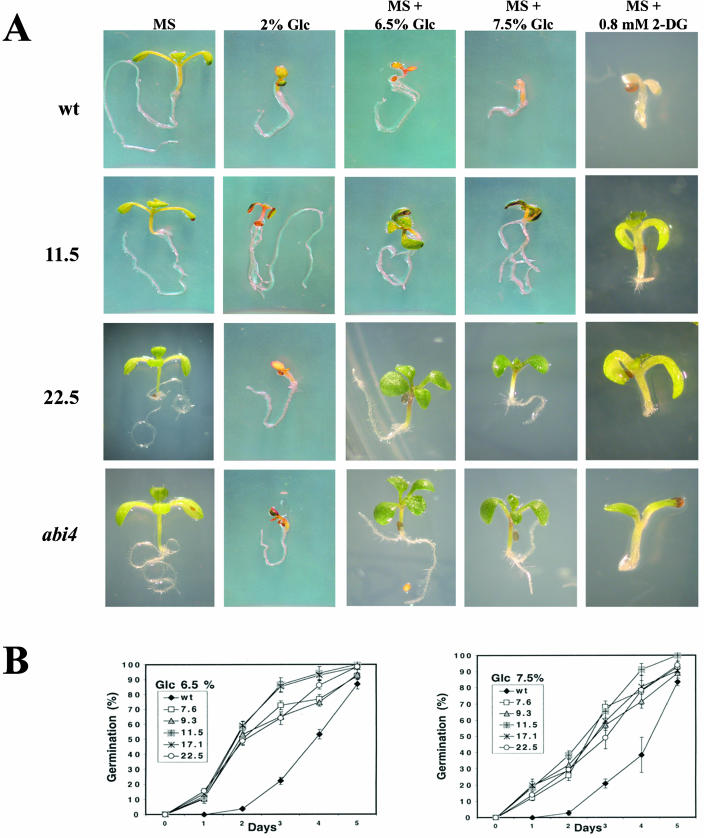
Several reports have documented that mutations in ABA-insensitive-4 (ABI4) transcription factor gene led to a Glc-insensitive phenotype (Arenas-Huertero et al., 2000) and displayed salt and osmotic stress tolerance during germination (Quesada et al., 2000), thus suggesting a cross-talk between signaling pathways for stress tolerance, ABA, and Glc. In an effort to further characterize our transgenic plants overexpressing CpMYB10 and test their germination in response to Glc, seeds from 7.6, 9.3, 11.5, 17.1 and 22.5 CpMYB10 transgenic lines were germinated in MS media supplemented with various concentrations of Glc and allowed to grow for 10 d. In Figure 9A it can be observed that MS medium supplemented with 6.5% or 7.5% Glc, or 2% Glc without MS salts, have a detrimental effect in development of wild-type seedlings, whereas no effect could be detected in the representative 11.5 transgenic line that showed cotyledon expansion and greening. A low concentration of Glc without MS salts was tested to discard a possible effect of nitrate on germination since it is known to antagonize Glc responses (Moore et al., 2003). Thus, germination of CpMYB10 transgenic lines on 2% Glc without MS supports the observation that it is caused by Glc and it is not an osmotic effect. This Glc-insensitive phenomenon was further characterized by comparing the germination rate on different Glc concentrations, of wild-type and five selected 35S-CpMYB10 transgenic lines, as shown in Figure 9B. The largest differences in germination rate were observed after 2 d on media containing 6.5% Glc, where more than 50% of transgenic lines initiated root emergence and elongation, whereas less than 5% of wild-type seedlings had germinated on the same conditions. On 7.5% Glc, germination rate of transgenic lines after 2 d was roughly 30%, whereas the germination rate in wild-type seedlings was near 5%. All these results indicated that overexpression of CpMYB10 gene confers Glc insensitivity in Arabidopsis.
Figure 9.
Glc sensitivity of 35S-CpMYB10 plants. A, Transgenic lines overexpressing CpMYB10 gene were germinated on MS salts added with 6.5 or 7.5% Glc, or 2% Glc without MS, or 0.8 mm 2-DG, or MS medium as a control, and growth for 10 d. Representative lines 11.5 and 22.5 are shown in comparison to wild type and abi4 mutant. B, Germination of 35S-CpMYB10 transgenic lines in MS with 6.5% or 7.5% Glc. Error bar represents sds. The figure represents the average of 3 or more replicates of 100 seeds. Germination was defined as complete protrusion of the radicule.
Glc analogs have been used to discriminate the sugar signaling pathway involved in specific responses such as germination, cotyledon expansion, and greening as well as gene expression. 2-deoxy-Glc (2-DG) has been shown to trigger a potent sugar response even at very low concentrations compared to Glc. For example, 2-DG strongly represses photosynthetic gene expression as well as photosynthetic efficiency (Jang and Sheen, 1994; Van Oosten et al., 1997) in an HXK mediated manner. To get insight into the sugar signaling pathway altered in CpMYB10 overexpressing lines, we examined greening and expansion of cotyledons, known to be inhibited by a low 2-DG concentration (0.8 mm; Jang et al., 1997). As in the Glc assay, the 35S-CpMYB10 lines were 2-DG hyposensitive and appeared green and had more expanded cotyledons compared to wild-type plants (Fig. 9A). The rest of the transgenic lines expressing CpMYB10 showed similar phenotypes, although at different degrees, most likely due to variations in transgene expression. Additionally, the ABI4 gene, involved in sugar and ABA signaling (Arenas-Huertero et al., 2000), has been shown to participate in the HXK-dependent sugar signaling (Pego et al., 1999; Van Oosten et al., 1997); thus we included the abi4 mutant in our sugar bioassays. Interestingly, this mutant also showed 2-DG hyposensitivity in this assay, confirming its participation in this sugar signaling pathway. Together, these data suggest that the CpMYB10 overexpressing lines are affected at least in part in the hexose phosphorylation-dependent sugar signaling, and also indicate that Glc resistance of these lines can be separated from their osmotic resistance.
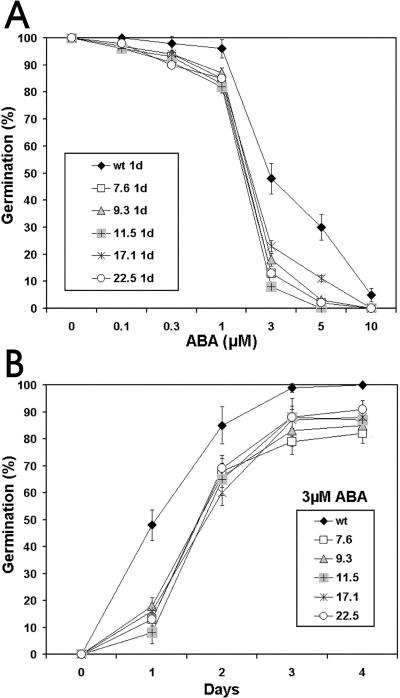
As mentioned above, there is strong evidence for a cross-talk of Glc and ABA signaling pathways (Arenas-Huertero et al., 2000; Huijser et al., 2000; Laby et al., 2000). Therefore, we decided to investigate the ABA sensitivity of CpMYB10 transgenic lines. As shown in Figure 10A, an ABA dose response curve in germination was conducted, which showed that 3 μm ABA was sufficient to inhibit 35S-CpMYB10 lines germination efficiency to 20% or less, whereas wild-type plants retained 50% germination under the same conditions. The line 11.5 showed 10% of germination rate at 3 μm ABA. Additional evidence was gathered by a germination kinetics experiment using 3 μm ABA, where selected lines of CpMYB10 seeds had only about 10% germination after 1 d. In contrast, 50% germination was observed in wild-type seeds, as shown in Figure 10B. These results indicated that overexpression of CpMYB10 gene in Arabidopsis conferred ABA hypersensitivity.
Figure 10.
ABA sensitivity of 35S-CpMYB10 plants. A, ABA dose response in germination on MS media suplemented with 0.1, 0.3, 1, 3, 5, or 10 μm ABA. B, Kinetic of germination of the CpMYB10 transgenic lines on MS containing 3 μm ABA. The data correspond to the average of 3 different experiments each containing 50 seeds per data point. Five independent transgenic (T3 generation) homozygous lines were used in the analysis. Germination was defined as complete protrusion of the radicule.
CpMYB10 Regulates Stress Related Genes in Arabidopsis
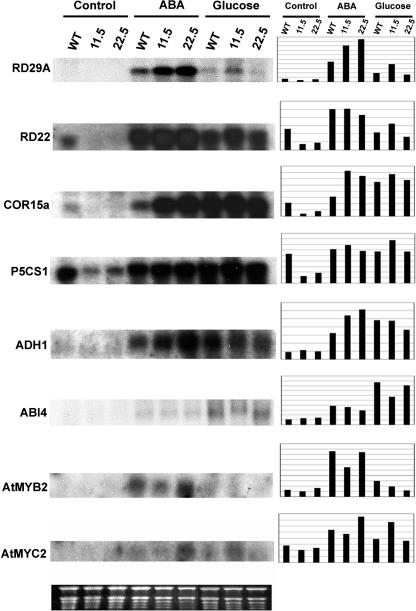
So far, we have shown that overexpression of CpMYB10 confers osmotic stress tolerance, Glc insensitivity, and ABA hypersensitive phenotypes in Arabidopsis. Since CpMYB10 is a transcription factor, it might be modulating the expression of genes involved in these processes. To test this hypothesis, gene expression analysis of possible regulated genes in 11.5 and 22.5 transgenic lines was carried out by RNA gel blot (Fig. 11). We selected the following genes for expression analysis: AtEM6 (Vicient et al., 2000) and ERD10 (Kiyosue et al., 1994), which correspond to LEA group I and group II genes, respectively; dehydration- and ABA-responsive genes RD29A, RD29B, and RD22 (Yamaguchi-Shinozaki and Shinozaki, 1993, 1994); cold-responsive gene COR15a (Baker et al., 1994), Δ1-pyrroline-5-carboxylate synthetase gene P5CS1 (Yoshiba et al., 1999), alcohol dehydrogenase gene ADH1 (de Bruxelles et al., 1996), and transcription factor genes AtMYB2 (Urao et al., 1993), AtMYC2 (Abe et al., 1997), DREB1A, and DREB2A (Liu et al., 1998) as they are involved in stress response; hexokinase gene HXK2 (Jang et al., 1997) and Suc synthase gene SUS1 (Martin et al., 1993) for their role in sugar sensing and metabolism, respectively; and ABA-insensitive transcription factor genes ABI4 (Finkelstein et al., 1998) and ABI5 (Finkelstein and Lynch, 2000) and zeaxanthine epoxidase gene ZEP1 (Arabidopsis Genome Initiative, 2000) involved in ABA signaling and biosynthesis. Two-week-old wild-type, 11.5, and 22.5 transgenic lines were treated for 16 h with 7.5% Glc, 100 μm ABA, or untreated before RNA-gel blot analysis. It can be observed in Figure 11 that RD29A, COR15a, and ADH1 genes are overexpressed in transgenic lines upon ABA treatment, whereas without treatment RD22, COR15a, and P5CS1 are repressed in both 11.5 and 22.5 transgenics after comparison to wild-type plants. Under Glc treatment, RD29A, RD22, P5CS1, and AtMYC2 transcripts were up-regulated only in 11.5 transgenic line. In the case of ABI4 no differences in gene expression could be observed after comparison with wild-type plants except for a lower level of induction upon Glc treatment in 11.5 line. Also, the AtMYB2 gene was partially repressed upon ABA treatment in 11.5 transgenic plants. None of the other analyzed genes (AtEM6, ERD10, RD29B, SUS1, HXK2, ABI5, ZEP1, DREB1A, and DREB2A) were up- or down-regulated under the described conditions in both 11.5 and 22.5 transgenic lines (data not shown).
Figure 11.
Expression of CpMYB10 target gene mRNAs in 35S-CpMYB10 plants. RNA gel blotting was conducted to measure the amount of RD29A, RD22, COR15A, P5CS1, ADH1, ABI4, AtMYB2, and AtMYC2 mRNA in transgenic Arabidopsis plants. Each lane was loaded with 10 μg of total RNA from 2-week-old plants grown in normal MS medium and transferred to MS plates containing the indicated compound 16 h before collecting tissue. Graphics show the densitometric quantification of shown bands after normalization against loaded RNA according to Image 1.61 software from NIH. Ethidium bromide-stained rRNAs were used as a loading control (bottom section).
DISCUSSION
In this work, we analyzed the function of a MYB transcription factor gene, CpMYB10, from the resurrection plant Craterostigma using different techniques including overexpression in a heterologous background. First of all, the expression pattern of the CpMYB10 gene in Craterostigma was adressed by coupled RT and PCR since RNA blot experiments did not detect gene expression, suggesting that CpMYB10 could be a low-abundance transcript. Here we showed that CpMYB10 gene is induced by desiccation and ABA treatments in leaves and roots a few minutes after treatment began. In leaves, maximum CpMYB10 expression was observed around 15 min after desiccation, and thereafter sharply declined. This early induction and rapid shut off suggests a key role of CpMYB10 in gene activation for stress tolerance. In roots, a biphasic pattern of CpMYB10 expression was observed, raising the question of whether the same or a different set of genes are transactivated at early (30 min) and late (16 h and thereafter) desiccation stages, which physiologically represent quite different water status levels. ABA turns on CpMYB10 in leaves and roots also around 15 min after dehydration, maintaining the gene expression for as long as 48 h, the duration of the experiment. Upon dehydration, within the first 30 min, Craterostigma leaves have a relative water content of 90% and several genes are already being expressed (Bartels et al., 1990). In contrast, 16 h after dehydration the relative water content drops to only 20% and it goes down to 2.5% in plants desiccated for 48 h (Bartels et al., 1990). After 2 d, Craterostigma leaves have lost their chlorophyll, and ultrastructural changes are evident such as convoluted cell walls, shrunk organelles freely suspended in the cytosol, and loss of chloroplast grana (Schneider et al., 1993). Therefore, it is likely that two set of genes for early and late desiccation stages are transactivated by CpMYB10 in Craterostigma.
The expression pattern of CpMYB10 gene was consistent with its role in stress tolerance. Using in situ coupled RT and PCR analysis in Craterostigma tissue sections, CpMYB10 was found in epidermis, palisade, and spongy parenchyma, and vascular bundle of ABA treated leaves, whereas in ABA treated roots CpMYB10 expression was limited to the vascular cylinder and some isolated cortex cells. In untreated leaves, CpMYB10 was faintly expressed in epidermis and in some parenchyma and vascular bundle cells and totally undetectable in roots. Therefore, CpMYB10 is expressed at basal levels in unstressed leaves and shortly after stress its transcript increases dramatically in a translation-independent manner as mentioned above, suggesting a rapid posttranslational activation mechanism.
The promoter analysis of CpMYB10 gene in Arabidopsis further supported its role in stress tolerance. These experiments showed GUS gene induction after dehydration in leaf vascular tissues and conspicuously strong expression in root tip, apical shoot meristem, and emerging leaves. Roots and root cap sense water deficit triggering ABA translocation from roots to shoots (Zhang et al., 1987), thus CpMYB10 gene might play a major role on this process. Treatment with ABA led to a strong GUS staining in all vegetative tissues, although no staining was visible in root tip. Quantification of GUS activity showed that although ABA treatment induced the highest levels of GUS at the whole plant level, in roots it was induced 2-fold higher upon dehydration. Also, CpMYB10 promoter induction by ABA reached its maximum in 6-d-old plants, whereas upon dehydration GUS activity peaked in 21-d-old plants. On the other hand, in unstressed plants GUS activity was detected at low levels in apical shoot meristem and in pollen grains. Expression of CpMYB10 promoter in mature pollen grains probably reflects that this tissue is developmentally programed for desiccation tolerance and hence prone to regulate genes responsive to stress. In addition, since 5′ untranslated region leader sequence was present in the construct, posttranscriptional regulatory events cannot be excluded. Other regions, such as intergenic or 3′ untranslated region, could be also required for regulation.
The use of protein synthesis inhibitors in transcription analysis in animal and plant cells allowed characterization of genes whose induction is stimulated in the absence of protein synthesis as primary response genes and many of them correspond to transcription factor genes (Herschman, 1991; Fujimoto et al., 2000). Those genes whose induction requires protein synthesis are termed secondary response genes. When unstressed tissues are pretreated with cycloheximide (CHX), CpMYB10 was highly expressed but not the LEA encoding-gene Cp11-24, suggesting that the former gene is independent on prior protein synthesis for its expression. The induction of CpMYB10 expression in the presence of CHX suggests that translation of a putative repressor protein that could act on its promoter is inhibited, thus releasing CpMYB10 transcription. Alternatively, it could indicate protection of CpMYB10 mRNA from degradation by inhibition of labile mRNases. Similar experimental criteria using CHX has been widely used to characterize several plant genes as primary responsive to stimuli (Koshiba et al., 1995; Fujimoto et al., 2000; Bouquin et al., 2001; Laskowski et al., 2002).
A role of CpMYB10 as a transcription factor was elucidated after analyzing the DNA binding properties of a recombinant CpMYB10 protein by using EMSA assays. Our results showed that rCpMYB10 protein has DNA binding recognition for two specific MYB motifs, namely TAACTG and GAACTA sequences present in CpMYB10 and Cp11-24 promoter regions, respectively. The TAACTG sequence is also the recognition site of AtMYB2 protein (Urao et al., 1993). Binding of rCpMYB10 protein to Cp11-24 promoter element suggests that a possible target gene of CpMYB10 transcription factor is the Cp11-24 gene which has been detected 30 min after dehydration of Craterostigma leaves (Bartels et al., 1990). The Cp11-24 gene encodes a LEA (late-embryogenesis abundant) protein which is among other proteins involved in stress tolerance (Ingram and Bartels, 1996). rCpMYB10 protein also binds to a MYB responsive element present in its own promoter. The binding of rCpMYB10 to its own promoter could be indicating a possible autoregulation of CpMYB10 gene. This mechanism has been reported in other plant genes (Schwarz-Sommer et al., 1992; Finkelstein and Lynch, 2000), although it cannot be ruled out that another MYB homolog might regulate CpMYB10 promoter. Although CpMYB10 encoded sequence has three potential phosphorylation sites (Iturriaga et al., 1996), apparently no such protein modification is required for DNA binding in vitro.
A major finding of the present study was that overexpression of CpMYB10 in Arabidopsis using the 35S promoter confers both osmotic stress tolerance of transgenic seedlings germinated in tissue culture and desiccation and salt tolerance in adult plants grown in soil. Other reports have shown that overexpression of transcription factor genes improves stress tolerance. Freezing tolerance was shown in Arabidopsis overexpressing CBF1 (Jaglo-Ottosen et al., 1998). Plants overexpressing DREBA1 gene improved their freezing, drought, and salt stress tolerance, although constitutive expression of this gene led to growth retardation, whereas minimal effects were observed when a stress-inducible promoter was used (Kasuga et al., 1999). On the other hand, except for a bulky root system, no other morphological or growth alterations in all transgenic lines overexpressing CpMYB10 compared to wild-type plants were observed, suggesting that CpMYB10 may also play an important role in promoting root growth and differentiation. In contrast, overexpression of AtMYB2 in Arabidopsis showed a dwarf phenotype and no stress tolerance was reported, only coexpression of both AtMYB2 and AtMYC2 conferred moderate stress tolerance (measured in an electrolyte leakage test in the presence of mannitol) and plants exhibited severe growth retardation as well (Abe et al., 2003). In contrast, in this study it was found that overexpression of the CpMYB10 gene alone led to an improved stress tolerance to severe drought and salt conditions in planta.
An important question was to determine which genes are responsible for the stress tolerance phenotype in Arabidopsis overexpressing the CpMYB10 gene. In an effort to find downstream regulated genes by CpMYB10 in Arabidopsis, we analyzed the expression pattern of 17 genes involved in either stress tolerance, sugar sensing and metabolism, or ABA signaling and biosynthesis by RNA gel blot. Only 8 of these genes showed altered gene expression in plants overexpressing CpMYB10. Two genes encoding hydrophilic proteins, RD29A and Cor15A, and the alcohol dehydrogenase ADH1 gene were up-regulated in 35S-CpMYB10 plants upon ABA treatment. Overexpression of DREB1A or CBF1 transcription factor genes in Arabidopsis led to up-regulation of several hydrophilic protein genes including RD29A and COR15a (Jaglo-Ottosen et al., 1998; Kasuga et al., 1999). In contrast, the overexpression of AtMYB2 in Arabidopsis only activates ADH1, although coexpression of both AtMYB2 and AtMYC2 up-regulates several other genes, including RD22, COR6.6, and SUS1 (Abe et al., 2003). We also found that RD22, Cor15a, and P5CS1 are down-regulated in 35S-CpMYB10 plants without stress but not under ABA or Glc treatments, suggesting that CpMYB10 might be acting on these genes as a repressor and activator. A transcriptomics profile would reveal other target genes of CpMYB10 in Arabidopsis. Noteworthy is the presence of multiple plant myb binding sites in the 1-kb upstream sequence of the putative promoter region of all the putative target genes analyzed in this study, as revealed by PLACE computer analysis program (Higo and Ugawa, 1999), supporting a possible transactivation by CpMYB10 upon them.
Taking together these results with the EMSA analysis, it is tempting to speculate whether this dual role is also present in Craterostigma, CpMYB10 acting as a repressor in unstressed tissues and as an activator upon dehydration. For instance, it could be involved in maintaining CpMYB10 gene shut off in unstressed conditions, and could also activate Cp11-24 and other target genes upon dehydration. This dual mechanism could allow a fast down-regulation of target genes responsive to abiotic stress. This shift in activity could well be modulated by a kinase or phosphatase in order to rapidly activate gene response for adapting cells to abiotic stress.
Besides having an essential function in plant metabolism, sugars play a role in regulating developmental and physiological processes such as photosynthesis, photomorphogenesis, flowering, and germination (Smeekens, 2000). The isolation of Arabidopsis mutants exhibiting normal development and greening when germinated on high Glc concentrations has been reported. The molecular identity of some mutants with a similar phenotype corresponded to the ABI4 and ABI5 transcription factors required for ABA signaling, suggesting a link between sugar- and ABA-signaling pathways (Arenas-Huertero et al., 2000; Finkelstein and Lynch, 2000). The molecular mechanisms by which abi4 and abi5 mutants affect sugar and stress responses are currently unknown. In this work it was found that overexpression of CpMYB10 gene in Arabidopsis conferred insensitivity to high Glc concentrations and also ABA hypersensitive response. To correlate these phenotypes with a possible altered expression pattern of well-characterized genes involved in Glc and ABA responses, the transcription of ABI4, ABI5, and HXK2 genes was analyzed. ABI5 and HXK2 transcription pattern was not modified; only ABI4 was partially repressed in 11.5 line but not in 22.5 under Glc treatment. Overexpression of AtMYB2 also conferred ABA hypersensitive response but no Glc response was reported (Abe et al., 2003).
The use of 2-DG analog allows us to suggest that these transgenic lines are affected at least in part in the hexose phosphorylation-dependent sugar signaling, commonly interpreted as the HXK-dependent pathway. These data correlate with the abi4 phenotype observed in presence of this Glc analog, which has been demonstrated to participate in the HXK-dependent sugar signaling (Pego et al., 1999). In fact, sun6 mutant, an abi4 allele, was shown to be less sensitive to photosynthesis inhibition by 2-DG than wild-type plants in adult stages (Van Oosten et al., 1997), further supporting our observations at earlier developmental stages. However, interpretations based on the use of Glc analogs must be taken carefully as no specific studies about their metabolism and effects on different tissues and plant species have been performed (Gibson, 2000). A direct link between the sugar response and stress resistance of the transgenic lines expressing CpMYB10 is still difficult to establish. Data about the molecular bases for both phenotypes in these transgenics remains to be determined in the future. However, osmotolerant phenotypes on sugar response mutants have been reported (Gibson, 2000), which correlates with our results.
In summary, in this study it is shown that a MYB homolog (CpMYB10) from the resurrection plant Craterostigma plantagineum is induced by dehydration and ABA treatments in leaves and roots and in transgenic Arabidopsis as well. The expression of CpMYB10 is induced early and could be activated by CHX treatment alone, suggesting its regulation by unknown and short-lived repressor or RNase. CpMYB10 might regulate its own promoter and transactivate the LEA Cp11-24 gene. We propose that this MYB gene might account in part for the differences between Craterostigma and Arabidopsis regarding drought tolerance. This is substantiated by the ectopic expression of CpMYB10 in Arabidopsis that confers salt and drought tolerance and led to an altered expression of several stress-responsive genes. Additionally, overexpression of CpMYB10 in Arabidopsis led to ABA hypersensitive and Glc insensitive phenotypes by an unknown mechanism. This is the first report of a transgenic plant with improved stress tolerance using a gene from a resurrection plant. Additionally, heterologous expression of CpMYB10 transcription factor gene represents a potential approach to improve stress tolerance in crops avoiding endogenous mechanisms that often cosuppress the transgene of interest.
MATERIALS AND METHODS
Plant Growth Conditions and Stress Treatments
Craterostigma plantagineum Hochst. and Arabidopsis ecotype Columbia (wild type or transgenic) were grown at 24°C/20°C with 16 h light/8 h dark cycle on sterile MS medium supplemented with 2.0% and 1.0% Suc, respectively, and solidified with 0.8% phytoagar. The carbon source was as mentioned, unless another is indicated. To break dormancy, seeds were incubated at 4°C for 4 d before germination. Fully grown Craterostigma plants (before flowering) were used to perform dehydration and ABA treatments. For Craterostigma dehydration experiments the plants were placed on filter paper in a growth chamber under described conditions. For ABA treatment, Craterostigma plants were submerged in a solution containing 100 μm ABA (Sigma-Aldrich, St. Louis). For dehydration treatment of Arabidopsis, 4-week-old plants grown on a 1:1:1 mixture of vermiculite, perlite, and peat moss were used. At week 5, watering was stopped for 2 weeks and then rewatered, and allowed to grow one more week before being photographed. For salt stress treatment in pots, 4-week-old Arabidopsis plants were watered every 4 d with increasing concentrations of NaCl, starting from 50 mm, 100 mm, 150 mm, and 200 mm, and twice with 250 mm. For salt, sugar, ABA, and osmotic stress treatments performed in plates, the wild-type and transgenic Arabidopsis seeds were germinated in MS media containing the indicated salt, Glc, 2-DG analog, or osmotic agent concentration. For treatments in GUS experiments, 3-week-old plants were transferred from standard MS medium to plates containing the indicated compounds.
RT-PCR Expression Analysis
RT-PCR experiments were performed using 5 μg of total RNA extracted from Craterostigma by standard procedures and used for first strand cDNA synthesis with SuperScript II reverse transcriptase (Invitrogen, Gaithersburg, MD) and oligo(dT). PCR program consisted of 25 to 40 cycles of amplification (1 min, 95°C; 30 s, 52°C; and 2 min, 72°C) using Taq polymerase (Roche Diagnostics, Indianapolis) and sequence-specific primers for each gene. The specific oligonucleotides designed to amplify the cDNA corresponding to CpMYB10 have the following sequence: primer CpMYB10-sense 5′-AGGCATCAGCTTTTTCTT-3′, and CpMYB10-antisense 5′-ATGGTACGTCCCTTGATT-3′. The expected 1.1-kb PCR product was cloned and sequenced, and corresponded to the CpMYB10 cDNA after comparison with the corresponding genomic clone (Iturriaga et al., 1996). Specific oligonucleotides to amplify Cp11-24 (forward 5′-GAAGTTCGATGCTAACGA-3′, reverse 5′-TGCTCATCCGCAGCAGCAGC-3′) and Cptkt3 (forward 5′-TGGATGGGAAAAAGCTC-3′, reverse 5′-AAACAACCCTCACTCCC-3′) gene transcripts of Craterostigma yielded 629- and 733-bp fragments, respectively. Each RT-PCR result was confirmed in three biologically independent experiments. Aliquots at different cycles of PCR reactions were analyzed to select for the linearity phase of the exponential PCR reaction. RT-PCR products were resolved in 1× Tris-acetate EDTA, 1% agarose gels stained with ethidium bromide. The figures present the negatives of the fluorescent images.
Purification of Recombinant CpMYB10 Protein
To express a hexahistidine-CpMYB10 fusion protein in bacteria, the CpMYB10 cDNA was amplyfied by PCR using BglII-CPM10.5 (5′-GAAGATCTATGAACCAACAGCAGGTTA-3′) and CPM10.3-KpnI (5′-GGGGTACCTTCGTATATCTAAAAGCAGC-3′) primers and cloned in the pQE30 vector (Quiagen, Valencia, CA) on BamHI and KpnI restriction sites. DNA fusion was sequenced to confirm the in-frame cloning and used to transform BL21 Escherichia coli strain. Protein expression was achieved by adding 1 mm isopropylthio-β-galactoside to bacterial cultures and let grown for 2 h at 37°C. The soluble protein was purified according to manufacturer's instructions by affinity chromatography in Ni-NTA resin (Quiagen) and concentrated by Centricon-10 centrifugal concentrators (Amicon, Billerica, MA), before diluting in 3 mL of binding buffer (15 mm HEPES, 8 mm Tris, 120 mm KCl, 0.14 mm EDTA, 7 mm β-ME, 0.1 mm phenylmethylsulfonyl fluoride, and 10% glycerol). The protein concentration was determined with Bradford reagent (Bio-Rad Laboratories, Hercules, CA) and checked by SDS-PAGE.
EMSA
The EMSA protocol was essentially as previously described by Armstrong et al. (1992). The double-strand complementary oligonucleotides used as probes were radioactive labeled using α-32P dATP and α-32P dCTP and Klenow enzyme (Roche Diagnostics), and diluted to 1 ng/μL containing at least 50,000 cpm. For each reaction, around 100 ng of pure recombinant protein was used, 1.0 μg of salmon sperm DNA, and 1 μL milk powder suspension (20 mg/mL low fat milk powder). The reaction mixture was loaded on a native polyacrylamide gel (4% acrylamide, 0.1% N,N′-methylenbisacrylamide, 0.2× Tris-acetate EDTA, 7% glycerol, 0.06% ammonium persulfate, and 0.06% N,N,N′,N′-tetramethylethylenediamine). The gel was prerun at 100 V for 1 h at room temperature and run at 150 V for 3 to 4 h at 4°C. After electrophoresis, the gel was dried and exposed to x-ray film. The primer sequences used were: MYB-P10 forward 5′-GTGTATATAGTTAACTGAAACTGC-3′, MYB-P10 reverse 5′-GTGTGCAGTTTCAGTTAACTATAT-3′, MYB-P10-mut forward 5′-GTGTATATAGTTCCCTGAAACTGC-3′, MYB-P10-mut reverse 5′-GTGTGCAGTTTCAGGGAACTATAT-3′, MYB11-24 forward 5′-GTGTACAGAGGTGAACTACCGAATC-3′, MYB11-24 reverse 5′-GTGTGATTCGGTAGTTCACCTCTGT-3′, MYB11-24-mut forward 5′-GTGTACAGAGGTGCCCTACGAATC-3′, MYB11-24-mut reverse 5′-GTGTGATTCGGTAGGGCACCTCTGT-3′.
RT-PCR in Situ
Transversal leaf or root hand-cut sections (100–200 μm) of Craterostigma plants pretreated with 100 μm ABA for 1 h were tested for RT-PCR in situ technique (Xoconostle-Cázares et al., 1999) using Oregon Green 488-5-dUTP as fluorescent label (Molecular Probes, Eugene, OR) and GeneAmp EZ rTth RNA PCR kit (Perkin Elmer, San Jose, CA). The primers used in the amplification were the same as described before for CpMYB10 cDNA in the expression analysis section. The fluorescent signal was visualized in a confocal laser scanning microscope.
Plant Transformation
Two binary vector constructs were used to transform Arabidopsis. For the overexpression of CpMYB10, its 1.1-kb cDNA was amplified by RT-PCR with primers CpMYB10-sense and CpMYB10-antisense, cloned in pBluescript SK− (Stratagene, La Jolla, CA) and the DNA sequence was determined before subcloning in pBin19 vector (Bevan, 1984) containing the 0.8-kb 35S-promoter and 0.3-kb NOS polyadenylation site. This construct was introduced by electroporation in Agrobacterium tumefaciens C58C1 strain containing the pGV2260 plasmid. The second construct consisted of the 1.5 kb CpMYB10 promoter region (P10) fused to the uidA (GUS) gene (Jefferson, 1987). The promoter fragment was amplified by PCR using Expand High Fidelity PCR System (Roche) using as a template the genomic CPM10 clone (Iturriaga et al., 1996), and primers MVP.5 (5′-GGGGTACCTATATGGCTAGGATAGGATAC-3′) and MVP.3 (5′-CGAATTCATGTCTGTATTTTCTTCTTCC-3′), cloned in pBluescript SK+ and sequenced. A translational fusion between the first putative ATG of CpMYB10 and the first codon of GUS gene was created before subcloning in pBin19 vector. This construct was introduced by electroporation in A. tumefaciens C58C1 strain. The resulting bacteria were used to transform wild-type Arabidopsis (Col) by in planta vacuum infiltration (Bechtold et al, 1993). Seedlings were grown on MS medium supplemented with 1% Suc and for selection of transgenic plants 50 μg mL−1 kanamycin (Sigma-Aldrich) was added to the medium. One-week-old plantlets were transferred to pots under described conditions until plants formed seeds. To select homozygous lines, T2 generation seeds were analyzed for germination on kanamycin. Only T3 plants with a 3:1 segregation ratio were used. Ten independent homozygous lines for 35S-CpMYB10 and three for P10-GUS construct were isolated.
Distribution of Materials
Upon request, all novel materials described in this publication will be made available in a timely manner for noncommercial research purposes, subject to the requisite permission from any third-party owners of all or parts of the material. Obtaining any permissions will be the responsibility of the requestor.
GUS Histochemical and Fluorimetric Assays
The histochemical staining of GUS activity in transgenic plants was performed as described by Jefferson et al. (1987). Seedlings were fixed in a 0.3 (w/v) formaldehyde solution containing 10 mm MES, pH 7.5, and 0.3 m mannitol for 30 min on ice prior to staining. Stained samples were rinsed extensively in 70% ethanol to remove chlorophyll. The stained plants were analyzed using a bright-field microscopy (Type 104, Nikon, Tokyo). Crude plant extracts for fluorimetric assay of GUS enzyme activity were prepared with an extraction buffer consisting of 50 mm sodium phosphate buffer, pH 7.0, 10 mm EDTA, 0.1% (w/v) Triton X-100, 0.1% (w/v) sodium lauryl sarcosine, and 10 mm β-mercaptoethanol. GUS activity was assayed as described by Jefferson et al. (1987) with 4-methylumbelliferyl-β-d-glucoronide (Sigma-Aldrich) as a substrate.
Gel Blot Analysis
Total RNA was isolated (Ausubel et al., 1989) from seedlings grown as indicated using standard protocols. For northern blots, total RNA was fractionated by electrophoresis in 1.2% (w/v) agarose gels and transferred onto Hybond N+ nylon membrane (Amersham, Buckingamshire, UK). Hybridizations and washes were performed at high stringency conditions according to standard procedures, using 32P-radiolabeled probes. Probes for all the used genes were obtained by PCR using specific primers and cDNAs prepared from ABA-treated Arabidopsis plants. The graphic representation of densitometric quantification of the northern experiments were done using the public domain NIH Image program (developed at the United States National Institutes of Health and available on the Internet at http://rsb.info.nih.gov/nih-image) and normalized in each case by the amount of rRNA in the gel.
Sequence data from this article have been deposited with the EMBL/GenBank data libraries under accession number AF510112 for the CpMYB10 cDNA.
Acknowledgments
We thank Drs. Analilia Arroyo for critical reading of the manuscript, Juan Estevez for his technical advice on Arabidopsis transformation, and Beatriz Xoconostle-Cázares and Roberto Ruiz-Medrano for training on RT-PCR in situ technique. We also thank Xochitl Alvarado for assistance on confocal laser scanning microscope and Paul Gaytán and Eugenio López for oligonucleotide synthesis.
This work was supported in part by CONACYT (grant no. 27703–N [Mexico] to G.I.) and by ICGEB (grant no. CRP/MEX98–01 [Trieste] to G.I.). M.A.V. was supported by a CONACyT PhD fellowship.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.103.034199.
References
- Abe H, Yamaguchi-Shinozaki K, Urao T, Iwasaki T, Shinozaki K (1997) Role of MYC and MYB homologs in drought- and abscisic acid-regulated gene expression. Plant Cell 9: 1859–1868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abe H, Urao T, Ito T, Seki M, Shinozaki K, Yamaguchi-Shinozaki K (2003) Arabidopsis AtMYC (bHLH) and AtMYB2 (MYB) function as transcriptional activators in abscisic acid signaling. Plant Cell 15: 63–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arabidopsis Genome Initiative (2000) Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408: 796–815 [DOI] [PubMed] [Google Scholar]
- Arenas-Huertero F, Arroyo A, Zhou L, Sheen J, León P (2000) Analysis of Arabidopsis glucose insensitive mutants, gin5 and gin6, reveals a central role of the plant hormone ABA in the regulation of plant vegetative development by sugar. Genes Dev 14: 2085–2096 [PMC free article] [PubMed] [Google Scholar]
- Armstrong GA, Weisshaar B, Hahlbrock K (1992) Homodimeric and heterodimeric leucine zipper proteins and nuclear factors from parsley recognize diverse promoter elements with ACGT cores. Plant Cell 4: 525–537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ausubel FM, Brent R, Kingstone RE, Moore DD, Siedman JG, Smith JA, Struhl K (1989) Current Protocols in Molecular Biology. John Wiley & Sons, New York
- Baker SS, Wilhelm KS, Tomashow MF (1994) The 5′-region of Arabidopsis thaliana cor15a has cis-acting elements that confer cold-, drought- and ABA-regulated gene expression. Plant Mol Biol 24: 701–713 [DOI] [PubMed] [Google Scholar]
- Bartels D, Schneider K, Terstappen G, Piatkowski D, Salamini F (1990) Molecular cloning of abscisic acid-modulated genes which are induced during desiccation of the resurrection plant Craterostigma plantagineum. Planta 181: 27–34 [DOI] [PubMed] [Google Scholar]
- Bartels D, Salamini F (2001) Dessication tolerance in the resurrection plant Craterostigma plantagineum. A contribution to the study of drought tolerance at the molecular level. Plant Physiol 127: 1346–1353 [PMC free article] [PubMed] [Google Scholar]
- Bechtold N, Ellis J, Pelletier G (1993) In planta Agrobacterium mediated gene transfer by infiltration of adult Arabidodpsis thaliana plants. C R Acad Sci Paris 316: 1194–1199 [Google Scholar]
- Bernacchia G, Schwall G, Lottspeich F, Salamini F, Bartels D (1995) The transketolase gene family of the resurrection plant Craterostigma plantagineum: differential expression during the rehydration phase. EMBO J 14: 610–618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bevan M (1984) Binary Agrobacterium vectors for plant transformation. Nucleic Acids Res 12: 8711–8721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bockel C, Salamini F, Bartels D (1998) Isolation and characterization of genes expressed during early events of the dehydration process in the resurrection plant Craterostigma plantagineum. J Plant Physiol 152: 158–166 [Google Scholar]
- Bouquin T, Meier C, Foster R, Nielsen ME, Mundy J (2001) Control of specific gene expression by gibberellin and brassinosteroid. Plant Physiol 127: 450–458 [PMC free article] [PubMed] [Google Scholar]
- Choi H, Hong J, Ha J, Kang J, Kim SY (2000) ABFs, a family of ABA-responsive element binding factors. J Biol Chem 275: 1723–1730 [DOI] [PubMed] [Google Scholar]
- Crowe JH, Carpenter JF, Crowe LM (1998) The role of vitrification in anhydrobiosis. Annu Rev Physiol 60: 73–103 [DOI] [PubMed] [Google Scholar]
- de Bruxelles GL, Peacock WJ, Dennis ES, Dolferus R (1996) Abscisic acid induces the alcohol dehydrogenase gene in Arabidopsis. Plant Physiol 111: 381–391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeWald DB, Torabinejad J, Jones CA, Shope JC, Cangelosi AR, Thompson JE, Prestwich GD, Hamma H (2001) Rapid accumulation os phosphatidylinositol 4,5-biphosphate and inositol 1,4,5-triphosphate correlates with calcium mobilization in salt-stressed Arabidopsis. Plant Physiol 126: 759–769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkelstein RR, Wang ML, Lynch TJ, Rao S, Goodman HM (1998) The Arabidopsis abscisic acid response locus ABI4 encodes an APETALA 2 domain protein. Plant Cell 10: 1043–1054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkelstein RR, Lynch TJ (2000) The Arabidopsis abscisic acid response gene ABI5 encodes a basic leucine zipper transcription factor. Plant Cell 12: 599–609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank W, Phillips J, Salamini F, Bartels D (1998) Two dehydration-inducible transcripts from the resurrection plant Craterostigma plantagineum encode interacting homeodomain-leucine zipper proteins. Plant J 15: 413–421 [DOI] [PubMed] [Google Scholar]
- Frank W, Munnik T, Kerkmann K, Salamini F, Bartels D (2000) Water deficit triggers phospholipase D activity in the resurrection plant Craterostigma plantagineum. Plant Cell 12: 111–123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujimoto SY, Ohta M, Usui A, Shinshi H, Ohme-Takagi M (2000) Arabidopsis ethylene-responsive element binding factors act as transcriptional activators or repressors of GCC box-mediated gene expression. Plant Cell 12: 393–404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaff DF (1971) Dessication-tolerant flowering plants in Southern Africa. Science 174: 1033–1034 [DOI] [PubMed] [Google Scholar]
- Gibson SI (2000) Plant sugar-response pathways. Part of a complex regulatory web. Plant Physiol 124: 1532–1539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herschman HR (1991) Primary response genes induced by growth factors and tumor promoters. Annu Rev Biochem 60: 281–319 [DOI] [PubMed] [Google Scholar]
- Higo K, Ugawa Y, Iwamoto M, Korenaga T (1999) Plant cis-acting regulatory DNA elements (PLACE) database: 1999. Nucleic Acids Res 27: 297–300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoekstra FA, Golovina EA, Buitink J (2001) Mechanisms of plant dessication tolerance. Trends Plant Sci 6: 431–438 [DOI] [PubMed] [Google Scholar]
- Huijser C, Kortstee A, Pego J, Weisbeek P, Wisman E, Smeekens S (2000) The Arabidopsis SUCROSE UNCOUPLED-6 gene is identical to ABSCISIC ACID INSENSITIVE-4: involvement of abscisic acid in sugar responses. Plant J 23: 577–585 [DOI] [PubMed] [Google Scholar]
- Ingram J, Bartels D (1996) The molecular basis of dehydration tolerance in plants. Annu Rev Plant Physiol Plant Mol Biol 47: 377–403 [DOI] [PubMed] [Google Scholar]
- Iturriaga G, Leyns L, Villegas A, Gharaibeh R, Salamini F, Bartels D (1996) A family of novel myb-related genes from the resurrection plant Craterostigma plantagineum are specifically expressed in callus and roots in response to ABA or desiccation. Plant Mol Biol 32: 707–716 [DOI] [PubMed] [Google Scholar]
- Jaglo-Ottosen KR, Gilmour SJ, Zarka DG, Schabenberger O, Thomashow MF (1998) Arabidopsis CBF1 overexpression induces COR genes and enhances freezing tolerance. Science 280: 104–106 [DOI] [PubMed] [Google Scholar]
- Jang J-C, Sheen J (1994) Sugar sensing in higher plants. Plant Cell 6: 1665–1679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang J-C, Leon P, Zhou L, Sheen J (1997) Hexokinase as a sugar sensor in higher plants. Plant Cell 9: 5–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jefferson RA (1987) Assaying chimeric genes in plants: the GUS gene fusion system. Plant Mol Biol Rep 5: 387–405 [Google Scholar]
- Jefferson RA, Kavanagh TA, Bevan MW (1987) GUS fusions: β-glucoronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J 6: 3901–3907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasuga M, Liu Q, Miura S, Yamaguchi-Shinozaki K, Shinozaki K (1999) Improving plant drought, salt, and freezing tolerance by gene transfer of a single stress-inducible transcription factor. Nat Biotechnol 17: 287–291 [DOI] [PubMed] [Google Scholar]
- Kiyosue T, Yamaguchi-Shinozaki K, Shinozaki K (1994) Characterization of two cDNAs (ERD10 and ERD14) corresponding to genes that respond rapidly to dehydration stress in Arabidopsis thalina. Plant Cell Physiol 35: 225–231 [PubMed] [Google Scholar]
- Knight H, Knight MR (2001) Abiotic stress signalling pathways: specificity and cross-talk. Trends Plant Sci 6: 262–267 [DOI] [PubMed] [Google Scholar]
- Koshiba T, Ballas N, Wong LM, Theologis A (1995) Transcriptional regulation of PS-IAA4/5 and PS-IAA6 early gene expression by indoleacetic acid and protein synthesis inhibitors in pea (Pisum sativum). J Mol Biol 253: 396–413 [DOI] [PubMed] [Google Scholar]
- Laby RJ, Kincaid S, Kim D, Gibson S (2000) The Arabidopsis sugar-insensitive mutants sis4 and sis5 are defective in abscisic acid synthesis and response. Plant J 23: 587–596 [DOI] [PubMed] [Google Scholar]
- Laskowski M, Dreher KA, Gehring MA, Abel S, Gensler AL, Sussex IM (2002) FQRI, a novel primary axin-response gene, encodes a flavin mononucleotide-binding quinine reductase. Plant Physiol 128: 578–590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q, Kasuga M, Sakuma Y, Abe H, Yamaguchi-Shinozaki K, Shinozaki K (1998) Two transcription factors, DREB1 and DREB2, with an EREBP/AP2 DNA binding domain separate two cellular signal transduction pathways in drought- and low-temperature-responsive gene expression, respectively, in Arabidopsis. Plant Cell 10: 1391–1406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin T, Frommer WB, Salanoubat M, Willmitzer L (1993) Expression of an Arabidopsis sucrose synthase gene indicates a role in metabolization of sucrose both during phloem loading and in sink organs. Plant J 4: 367–377 [DOI] [PubMed] [Google Scholar]
- Moore B, Zhou L, Rolland F, Hall Q, Cheng W-A, Liu Y-X, Hwang I, Jones T, Sheen J (2003) Role of the Arabidopsis glucose sensor HXKI in nutrient, light, and hormonal signaling. Science 300: 332–336 [DOI] [PubMed] [Google Scholar]
- Murashige T, Skoog F (1962) A revised medium for rapid growth and bioassays with tobacco tissue culture. Physiol Plant 15: 473–497 [Google Scholar]
- Neill SJ, Desikan R, Clarke A, Hancock JT (2002) Nitric oxide is a novel component of abscisic acid signaling in stomatal guard cells. Plant Physiol 128: 13–16 [PMC free article] [PubMed] [Google Scholar]
- Pego JV, Weibeek PJ, Smeekens SCM (1999) Mannose inhibits Arabidopsis germination via a hexokinase-mediated step. Plant Physiol 119: 1017–1023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quesada V, Ponce MR, Micol JL (2000) Genetic analysis of salt-tolerant mutants in Arabidopsis thaliana. Genetics 154: 421–436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider K, Wells B, Schmelzer E, Salamini F, Bartels D (1993) Desiccation leads to the rapid accumulation of both cytosolic and chloroplastic proteins in the resurrection plant Craterostigma plantagineum Hochst. Planta 189: 120–131 [Google Scholar]
- Schwarz-Sommer Z, Hue I, Huijser P, Flor PJ, Hansen R, Tetens F, Lönning WE, Saedler H, Sommer H (1992) Characterization of the Antirrhinum floral homeotic MADS-box gene deficiens: evidence for DNA binding and autoregulation of its persistent expression throughout flower development. EMBO J 11: 251–263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinozaki K, Yamaguchi-Shinozaki K (2000) Molecular responses to dehydration and low temperature: differences and cross-talk between two stress signaling pathways. Curr Opin Plant Biol 3: 217–223 [PubMed] [Google Scholar]
- Smeekens S (2000) Sugar-induced siganal transduction in plants. Annu Rev Plant Physiol Plant Mol Biol 51: 49–81 [DOI] [PubMed] [Google Scholar]
- Stockinger EJ, Gilmour SJ, Thomashow MF (1997) Arabidopsis thaliana CBF1 encodes an AP2 domain-containing transcriptional activator that binds to the C-repeat/DRE, a cis-acting DNA regulatory element that stimulates transcription in response to low temperature and water deficit. Proc Natl Acad Sci USA 94: 1035–1040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stracke R, Werber M, Weisshaar B (2001) The R2R3-MYB gene family in Arabidopsis thaliana. Curr Opin Plant Biol 4: 447–456 [DOI] [PubMed] [Google Scholar]
- Tabaeizadeh Z (1998) Drought-induced responses in plant cells. Int Rev Cytol 182: 193–246 [DOI] [PubMed] [Google Scholar]
- Urao T, Yamaguchi-Shinozaki K, Urao S, Shinozaki K (1993) An Arabidopsis myb homolog is induced by dehydration stress and its gene product binds to the conserved MYB recognition sequence. Plant Cell 5: 1529–1539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urao T, Yakubov B, Satoh R, Yamaguchi-Shinozaki K, Seki M, Hirayama T, Shinozaki K (1999) A transmembrane hybrid-type histidine kinase in Arabidopsis functions as an osmosensor. Plant Cell 11: 1743–1754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Oosten JJM, Gerbaut A, Huijser C, Dijkwel PP, Chua N-H, Smeekens SCM (1997) An Arabidopsis mutant showing reduced feedback inhibition of photosynthesis. Plant J 12: 1011–1020 [DOI] [PubMed] [Google Scholar]
- Velasco R, Salamini F, Bartels D (1998) Gene structure and expression analysis of the drought- and abscisic acid-responsive CDeT11-24 gene family from the resurrection plant Craterostigma plantagineum Hochst. Planta 204: 459–471 [DOI] [PubMed] [Google Scholar]
- Vicient CM, Hull G, Guilleminot J, Devic M, Delseny M (2000) Differential expression of the Arabidopsis genes coding for Em-like proteins. J Exp Bot 51: 1211–1220 [PubMed] [Google Scholar]
- Xoconostle-Cázares B, Xiang Y, Ruiz-Medrano R, Wang HL, Yoo BC, McFarland KC, Franceschi VR, Lucas WJ (1999) Plant paralog to viral movement protein that potentiates transport of mRNA into the phloem. Science 283: 94–98 [DOI] [PubMed] [Google Scholar]
- Yamaguchi-Shinozaki K, Shinozaki K (1993) The plant hormone abscisic acid mediates the drought-induced expression but not the seed-specific expression of rd22, a gene responsive to dehydration stress in Arabidopsis thaliana. Mol Gen Genet 238: 17–25 [DOI] [PubMed] [Google Scholar]
- Yamaguchi-Shinozaki K, Shinozaki K (1994) A novel cis-acting element in an Arabidopsis gene is involved in responsiveness to drought, low-temperature, or high-salt stress. Plant Cell 6: 251–264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshiba Y, Nanjo T, Miura S, Yamaguchi-Shinozaki K, Shinozaki K (1999) Stress-responsive and developmental regulation of Δ1-pyrroline-5-carboxylate synthetase 1 (P5CS1) gene expression in Arabidopsis thaliana. Biochem Biophys Res Commun 261: 766–772 [DOI] [PubMed] [Google Scholar]
- Zhang J, Schurr U, Davies WJ (1987) Control of stomatal behaviour by abscisic acid which apparently originates in roots. J Exp Bot 38: 1174–1181 [Google Scholar]