Abstract
The abscisic aldehyde oxidase 3 (AAO3) gene product of Arabidopsis catalyzes the final step in abscisic acid (ABA) biosynthesis. An aao3-1 mutant in a Landsberg erecta genetic background exhibited a wilty phenotype in rosette leaves, whereas seed dormancy was not affected (Seo et al., 2000a). Therefore, it was speculated that a different aldehyde oxidase would be the major contributor to ABA biosynthesis in seeds (Seo et al., 2000a). Through a screening based on germination under high-salt concentration, we isolated two mutants in a Columbia genetic background, initially named sre2-1 and sre2-2 (for salt resistant). Complementation tests with different ABA-deficient mutants indicated that sre2-1 and sre2-2 mutants were allelic to aao3-1, and therefore they were renamed as aao3-2 and aao3-3, respectively. Indeed, molecular characterization of the aao3-2 mutant revealed a T-DNA insertional mutation that abolished the transcription of AAO3 gene, while sequence analysis of AAO3 in aao3-3 mutant revealed a deletion of three nucleotides and several missense mutations. Physiological characterization of aao3-2 and aao3-3 mutants revealed a wilty phenotype and osmotolerance in germination assays. In contrast to aao3-1, both aao3-2 and aao3-3 mutants showed a reduced dormancy. Accordingly, ABA levels were reduced in dry seeds and rosette leaves of both aao3-2 and aao3-3. Taken together, these results indicate that AAO3 gene product plays a major role in seed ABA biosynthesis.
Abscisic acid (ABA) plays a major role in seed development, adaptive plant responses to water deprivation, and sugar sensing (Cheng et al., 2002; Finkelstein et al., 2002). The level of ABA increases in plants during seed development and under environmental stresses, particularly drought and salinity (Finkelstein and Rock, 2002). During seed development, ABA levels rise at the end of embryogenesis, are maximal during mid development when storage reserves are accumulated, and then decline during desiccation (Karssen et al., 1983). During embryogenesis and seed formation, ABA is implicated in the control of many events such as embryo morphogenesis, storage protein synthesis, desiccation tolerance, as well as the onset and maintenance of dormancy (Bentsink and Koornneef, 2002). Recently, the importance of ABA to prevent seed germination and to promote postgermination developmental arrest under unfavorable water conditions has been genetically documented (Lopez-Molina et al., 2001; Gonzalez-Guzman et al., 2002). Later on, the increase of ABA levels in response to salinity and drought stress is a crucial adaptation to cope with these stresses, which has a wide impact on regulation of gene expression (Hoth et al., 2002; Seki et al., 2002).
Recently, all the major genes encoding the enzymes that catalyze the different steps of ABA biosynthesis have been identified and the biosynthetic pathway mostly elucidated (Schwartz et al., 2003; Xiong and Zhu, 2003). Thus, the biosynthesis of ABA can be traced from the epoxidation of zeaxanthin by zeaxanthin epoxidase, which leads to all-trans-violaxanthin (Marin et al., 1996). Next, this compound is converted in 9-cis-violaxanthin and 9-cis-neoxanthin through an uncharacterized mechanism. Nine-cis-epoxycarotenoids suffer an oxidative cleavage catalyzed by nine-cis-epoxicarotenoid dioxigenase (NCED), leading to production of xanthoxin (Schwartz et al., 1997). Then, xanthoxin is converted to abscisic aldehyde by a short-chain alcohol dehydrogenase, which is a NAD-dependent enzyme (Cheng et al., 2002; Gonzalez-Guzman et al., 2002). Finally, abscisic aldehyde is oxidized to ABA by AAO3 (abscisic aldehyde oxidase; Seo et al., 2000a), which requires a molybdenum cofactor (MoCo) that is synthesized by the MoCo sulfurase ABA3 (Bittner et al., 2001; Xiong et al., 2001).
While the main features of the pathway of ABA biosynthesis have been elucidated during the last years, the regulatory mechanisms of the biosynthetic genes remain largely unknown at the molecular level. Additionally, some aspects of ABA biosynthesis still are not well characterized. For instance, the formation of the 9-cis isomers that are cleaved by NCED has not been clearly established (Schwartz et al., 2003). Another point that deserves further research is the biosynthesis of ABA in seeds. Thus, while it has been demonstrated that AAO3 is responsible for ABA biosynthesis in leaves, according to the phenotypic characterization of the aao3-1 allele, it has been suggested that a different aldehyde oxidase catalyzes the last step of ABA biosynthesis in seeds (Seo et al., 2000a). For instance, aao3-1 seeds (in a Landsberg erecta [Ler] genetic background) do not show a clear change in dormancy (Seo et al., 2000a), whereas other known ABA-deficient mutants have much less dormant seeds. Additionally, Seo et al. (2000b) failed to detect AAO3 gene expression in siliques and dry seeds.
Our understanding of the pathway of ABA biosynthesis (and by extension of ABA functions) has been greatly aided by the identification and characterization of ABA-deficient mutants. These mutants have been identified on the basis of their wilty phenotype or seed germination characteristics (Koornneef et al., 1982; Leon-Kloosterziel et al., 1996). Particularly, mutants able to germinate and carry out early growth in medium containing a high-NaCl concentration (sre mutants for salt resistant) were found to be allelic to ABA-deficient mutants (Gonzalez-Guzman et al., 2002). Thus, the sre1 mutant was found to be allelic to aba2-1 mutant (Gonzalez-Guzman et al., 2002). In this work, we report the characterization of two new sre mutants in a Columbia (Col) genetic background, sre2-1 and sre2-2, which were found to be allelic to aao3-1. These new alleles of AAO3, in addition to a wilty phenotype, show reduced dormancy, paclobutrazol- and osmotic stress-resistant germination, as well as reduced ABA levels in seeds, indicating that the AAO3 gene product plays an important role in ABA biosynthesis in seeds.
RESULTS
Identification and Physiological Characterization of New AAO3 Alleles
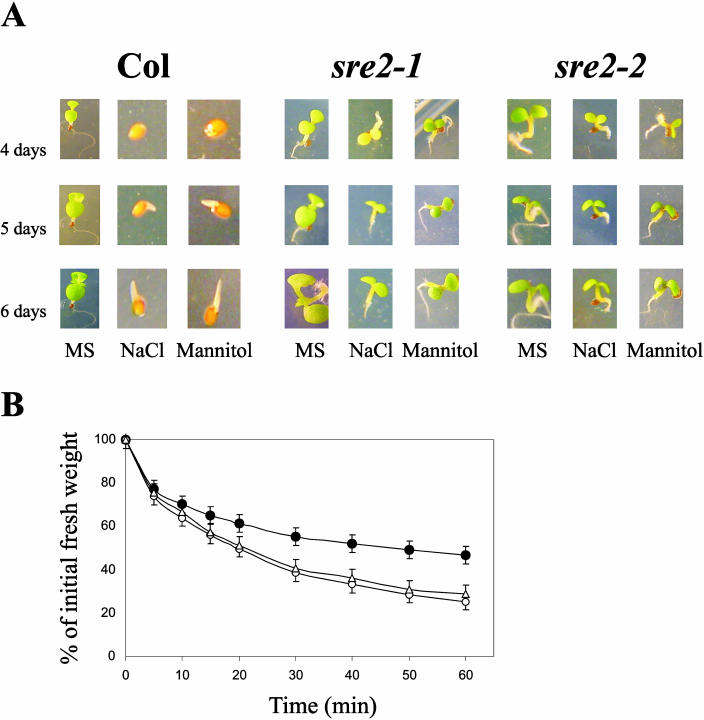
During the course of a screening for mutants able to germinate and develop green cotyledons under inhibitory concentrations of NaCl, we identified four complementation groups, which were named sre1 to sre4, in a germination assay (Gonzalez-Guzman et al., 2002). The sre2 mutant was able to bypass the developmental arrest induced by high-osmoticum, that is, 200 mm NaCl or 400 mm mannitol (Fig. 1A). Two allelic mutants (in a Col genetic background), sre2-1 and sre2-2, were identified for this locus (Table I). Genetic analysis indicated that the mutant phenotype was caused by a single recessive mutation (Table I). ABA promotes the inhibition of both seed germination and early seedling development under low-water-potential conditions. Therefore, screenings for mutants able to bypass the ABA-mediated blockage of germination and development under osmotic stress usually result in the identification of ABA-deficient mutants (Leon-Kloosterziel et al., 1996; Quesada et al., 2000; Gonzalez-Guzman et al., 2002). Indeed, complementation analyses of both sre2-1 and sre2-2 showed that they were allelic to aao3-1 (Table I), which is the only mutant allele described currently for the AAO3 gene. Therefore, we renamed sre2-1 and sre2-2 mutants as aao3-2 and aao3-3, respectively. The aao3-1 allele was identified in a mutant showing a mild wilty phenotype under greenhouse conditions. Transpiration assays of aao3-2 and aao3-3 mutants also indicated enhanced water loss as compared to wild type (Fig. 1B).
Figure 1.
Phenotype of sre2-1 and sre2-2 mutants. A, Seed germination in the presence of NaCl and mannitol. Wild-type Col ecotype (left), sre2-1 (middle), and sre2-2 (right) seeds were germinated on either MS medium (MS) or medium supplemented with either 200 mm NaCl or 400 mm mannitol. The picture was taken at 4, 5, and 6 d after sowing. B, Enhanced transpiration rate of sre2-1 (aao3-2) and sre2-2 (aao3-3) plants compared to wild type. Percentage of initial fresh weight was measured in detached rosette leaves of either Col (black circles), sre2-1 (white circles) or sre2-2 (white triangles) plants.
Table I.
Complementation tests of Arabidopsis sre2, aba, and aao3 mutants
| Cross (Female × Male) | Generation | Total Seeds Sown | Germinated | χ2 |
|---|---|---|---|---|
| sre2-1/sre2-1 × sre2-2/sre2-2 | F1 | 154 | 135 | |
| sre2-1/sre2-1 × Col | F2 | 1,377 | 304 | 6.26 |
| sre2-2/sre2-2 × Col | F2 | 1,444 | 359 | 0.015 |
| sre2-1/sre2-1 × aba1-1/aba1-1 | F1 | 107 | 0 | |
| sre2-1/sre2-1 × aba2-1/aba2-1 | F1 | 112 | 0 | |
| sre2-1/sre2-1 × aba3-1/aba3-1 | F1 | 113 | 0 | |
| sre2-1/sre2-1 × aao3-1/aao3-1 | F1 | 89 | 74 | |
| sre2-2/sre2-2 × aao3-1/aao3-1 | F1 | 85 | 71 |
Complementation tests were done by analyzing intercrosses among sre2, aba, and aao3 homozygous mutants. The first individual of each cross acted as female. F1 and F2 seeds were scored for germination in 150 mm NaCl 5 d after sowing.
As both aao3-2 and aao3-3 were isolated in a screening based on seed germination under high-osmoticum, we presumed that they could have reduced ABA levels in seeds. To experimentally support this hypothesis, we measured ABA levels both in dry seeds and seeds imbibed and incubated for 24 h in 200 mm NaCl (Table II). ABA levels in dry seeds of aao3-2 and aao3-3 were approximately 35% of wild type. Interestingly, upon seed imbibition and incubation in 200 mm NaCl, ABA levels were still approximately 3-fold lower in aao3-2 and aao3-3 than in wild type. In addition to the phenotypes observed in seeds of aao3-2 and aao3-3 mutants, we also noticed a wilty phenotype under low humidity conditions (Fig. 1B). Therefore, we also measured ABA levels in rosette leaves of both aao3-2 and aao3-3 under unstressed or water-stress conditions (Table II). Rosette leaves of aao3-2 and aao3-3 mutants contained less ABA than wild type, and ABA levels upon water stress were notably lower in the mutants compared to wild type.
Table II.
ABA content (ng ABA/g dry weight)
| Dry Seeds | 200 mm NaCl | Turgid Leaves | Wilted Leaves | |
|---|---|---|---|---|
| Col | 151 ± 15 | 250 ± 20 | 103 ± 13 | 811 ± 76 |
| aao3-2 | 52 ± 9 | 83 ± 10 | 30 ± 3 | 112 ± 15 |
| aao3-3 | 57 ± 10 | 91 ± 12 | 35 ± 5 | 162 ± 16 |
ABA levels were measured in dry seeds, imbibed seeds treated with 200 mm NaCl for 24 h, and turgid or wilted rosette leaves of wild-type, aao3-2 and aao3-3 plants.
Molecular Characterization of the aao3-2 and aao3-3 Alleles
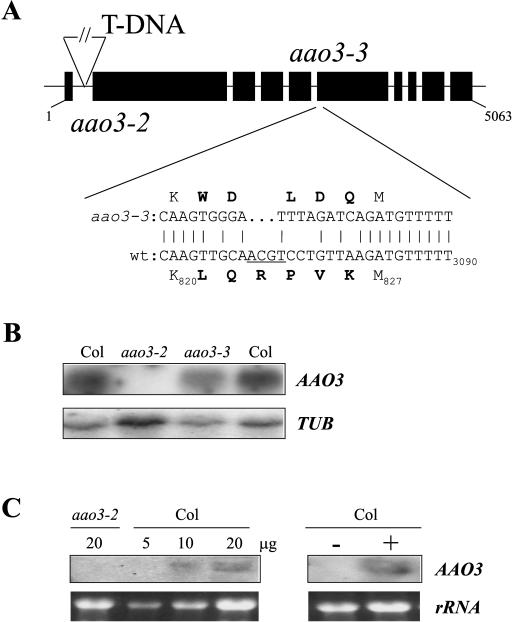
The aao3-2 and aao3-3 mutants were isolated from a seed population mutagenized with T-DNA. Therefore, we analyzed whether the mutant phenotypes were linked to a T-DNA insertion. Homozygous aao3-2 plants were crossed to Col wild-type plants. From the segregating F2 generation, homozygous aao3-2 individuals were selected and scored for the presence of the T-DNA by Southern blot analysis. The analysis of 88 F2 chromosomes revealed cosegregation of the salt-resistant phenotype and the presence of the T-DNA (data not shown). Plant T-DNA flanking sequences were isolated by plasmid rescue, and sequence analysis revealed that the T-DNA was inserted at nucleotide 282 of AAO3 (Fig. 2A). The T-DNA insertion in the aao3-2 allele abolished the expression of the AAO3 gene, as detected by northern blot analysis (Fig. 2B).
Figure 2.
Molecular characterization of the aao3-2 and aao3-3 alleles. Northern blot analysis of AAO3 mRNA. A, Structure of the AAO3 gene and mutations in the aao3-2 and aao3-3 alleles. The position of the T-DNA insertion in aao3-2 is indicated. The three-nucleotide deletion and missense mutations present in aao3-3, as well as the predicted amino acid substitutions, are shown. A TaiI restriction site is underlined in the wild-type nucleotide sequence. Amino acid numbering refers to the initial Met residue. Nucleotide numbering refers to the ATG start codon. B, Northern blot analysis of AAO3 gene expression in Col wild type, aao3-2 and aao3-3 mutants. Each track of the blot contained approximately 10 μg total RNA extracted from rosette leaves. RNA analysis was performed as described previously (Gonzalez-Guzman et al., 2002). RNA loading of the gel was quantified by hybridization with a tubulin probe (TUB). C, Northern blot analysis of AAO3 gene expression in siliques (left) and seeds (right). For siliques, increasing amounts (5, 10, and 20 μg) of total RNA from Col wild type and 20 μg total RNA from aao3-2 mutant were loaded in the blot. For seeds, approximately 20 μg of total RNA were loaded. Total RNA was extracted from seeds that were imbibed for 60 h and then mock-treated (−) or incubated with 400 mm mannitol for 24 h (+). RNA loading of the gel was visualized by ethidium bromide staining.
A similar analysis to the one described above failed to show cosegregation between the salt-resistant phenotype of aao3-3 mutant and a T-DNA insertion. Sequence analysis of the AAO3 gene in aao3-3 revealed a complex mutation that affects a gene stretch from nucleotides 3,066 to 3,080, including deletion of three nucleotides and resulting in several missense mutations (Fig. 2A). This mutation leads to loss of a TaiI restriction site, and, consequently, a CAPS marker was developed based on this DNA polymorphism. Analysis of F2 chromosomes of homozygous aao3-3 individuals revealed cosegregation of the three-nucleotide deletion observed in the AAO3 gene with the ABA-deficient phenotype (data not shown). Contrary to the aao3-2 mutation, the aao3-3 mutation does not appreciably affect the level of AAO3 mRNA (Fig. 2B).
The AAO3 Gene Plays an Important Role in ABA Biosynthesis Both in Leaves and Seeds
The role of AAO3 in ABA biosynthesis had been restricted to leaves on the basis that seed dormancy of aao3-1 mutant was unaffected and its ABA levels in mature dry seeds were only reduced by 40% compared to the wild type (Seo et al., 2000a). Additionally, gene expression studies failed to detect the AAO3 transcript both in siliques and dry seeds from a Col ecotype (Seo et al., 2000b). As both aao3-2 and aao3-3 were isolated in a seed germination screening, we reasoned that AAO3 gene product must contribute to ABA biosynthesis in seeds. Indeed, ABA levels in dry seeds or seeds imbibed and treated with 200 mm NaCl of both aao3-2 and aao3-3 were approximately one-third of wild type (Table II). We have reexamined the expression level of AAO3 in siliques using increasing amounts of total RNA. Between 10 and 20 μg of total RNA from siliques was required to detect AAO3 by northern blot analysis (Fig. 2C). Using only 5 μg, we could not detect the transcript (in agreement with Seo et al., 2000b). Likewise, in dry seeds we were not able to detect AAO3 mRNA (data not shown); however, upon imbibition and osmotic stress treatment, we could detect it (Fig. 2C).
To further investigate the role of AAO3 in seeds, we compared the phenotype of all three aao3 alleles in seed germination assays. The aao3-1 mutant, as well as aao3-2 and aao3-3 mutants, showed an osmotolerant phenotype in seed germination (Table III). However, whereas aao3-2 and aao3-3 mutants were able to germinate under 10 μm paclobutrazol, aao3-1 was not (Table III). Additionally, aao3-2 and aao3-3 mutants showed a reduced dormancy, whereas aao3-1 showed a dormancy similar to wild type (Table III). These data reveal notable phenotypic differences among aao3-1 and the new aao3 alleles and suggest that, at least in a Col genetic background, the AAO3 gene product plays a role in seed ABA biosynthesis.
Table III.
Seed germination and early seedling growth of wild type, aao3-2, aao3-3, and aao3-1 mutants in different media
| 150 mm NaCl | 200 mm NaCl | 400 mm Mannitol | 10 μm Paclobutrazol | Dormancy | |
|---|---|---|---|---|---|
| Col | 1.0 ± 2.8 | 0.0 ± 0 | 0.0 ± 0 | 0.7 ± 1.2 | 13.3 ± 3.9 |
| aao3-2 | 82.3 ± 12.3 | 61.3 ± 16.5 | 84.4 ± 10 | 76.1 ± 19.1 | 67.9 ± 15.0 |
| aao3-3 | 88.6 ± 7.7 | 82.8 ± 5.3 | 91.7 ± 7.1 | 86.7 ± 15.1 | 73.0 ± 18.6 |
| aao3-1 | 50.7 ± 6.4 | 28.4 ± 2 | 91.1 ± 11 | 3.0 ± 4.2 | 9.4 ± 3.7 |
| Ler | 6.6 ± 0.7 | 0 ± 0 | 0.8 ± 1.1 | 0 ± 0 | 5.6 ± 7.5 |
Wild-type (Col or Ler background) and mutant seeds were sowed on MS agar plates supplemented either with 150 mm NaCl, 200 mm NaCl, 400 mm mannitol, or 10 μm paclobutrazol. Approximately 200 seeds were sowed and scored 5 (NaCl) or 8 (mannitol and paclobutrazol) d later. Percentage of seeds that germinated and developed green cotyledons in the different media was scored, and sd values were calculated from three independent experiments. Germination in MS medium was between 95% and 100%. A dormancy assay was performed with freshly harvested seeds (data from last column) by scoring the germination percentage after 5 d in the absence of stratification.
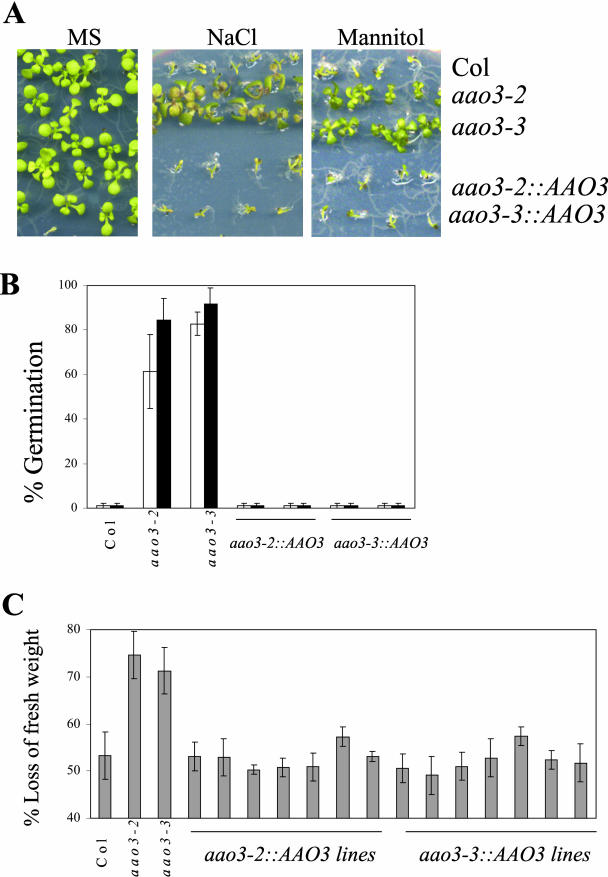
The demonstration that, for the aao3-2 and aao3-3 independent mutant alleles, a heritable change in phenotype is associated with a mutation in AAO3 suggests that this gene is responsible for the phenotype being studied. Therefore, introduction of a wild-type AAO3 allele in aao3-2 and aao3-3 should complement the phenotype observed in the mutants. The AAO3 gene driven by its own promoter region (ProAAO3-AAO3 construct; Seo et al., 2000a) was introduced into aao3-2 and aao3-3 mutants by Agrobacterium tumefaciens-mediated transformation. Transformants were selected for kanamycin resistance, and a T3 population homozygous for the transgene was obtained. Experiments of seed germination and early seedling growth under high-osmoticum, as well as measures of leaf transpiration, indicated that the phenotype of aao3-2 and aao3-3 mutants was complemented upon introduction of a wild-type AAO3 transgene (Fig. 3). These results confirm that in a Col genetic background, the AAO3 gene plays an important role in ABA biosynthesis both in leaves and seeds.
Figure 3.
Complementation of aao3-2 and aao3-3 mutants by AAO3 gene. A, Seed germination and early seedling growth in the presence of NaCl or mannitol. Wild-type (Col), aao3-2, aao3-3, and T3 homozygous progeny of the mutants transformed with the binary vector pPZP211 containing a ProAAO3-AAO3 insert. Seeds were germinated on MS medium (MS) or medium supplemented with either 200 mm NaCl or 400 mm mannitol. The picture was taken at 10 d after sowing. B, Percentage of seeds that germinated and developed green cotyledons under osmotic stress for wild-type, aao3-2, aao3-3, and representative transformed lines (T3 progeny) of aao3-2 and aao3-3. Seeds were sowed on MS agar plates supplemented either with 200 mm NaCl (white bars) or 400 mm mannitol (black bars). Approximately 200 seeds were sowed and scored 5 (NaCl) or 8 (mannitol) d later. Germination in MS medium was between 95% and 100%. C, The enhanced transpiration rate of both aao3-2 and aao3-3 plants is complemented by a wild-type AAO3 transgene. Loss of fresh weight was measured in detached rosette leaves of wild type, aao3-2, aao3-3, and T1 transformed lines of aao3-2 and aao3-3. Four leaves per individual were excised, and fresh weight was determined at ambient conditions (25°C and approximately 40% relative humidity) after 60 min. The percentage of fresh weight lost is indicated.
DISCUSSION
The availability of ABA-deficient mutants has allowed the substantiation of the role of ABA in different physiological processes. Additionally, these mutants have been very useful to clone the genes that encode the ABA biosynthetic enzymes. Currently, most of these genes have been cloned in Arabidopsis and other plant species, and only the step involving the conversion of all-trans-violaxanthin to 9-cis-violaxanthin or 9-cis-neoxanthin remains to be characterized (Schwartz et al., 2003). The pioneering biochemical studies based on 18O2-labeling experiments, together with the characterization of ABA-deficient mutants and subsequent cloning of the corresponding loci, have contributed to our current understanding of ABA biosynthesis.
The last step of ABA biosynthesis is the oxidation of ABA-aldehyde to ABA, which requires the activity of both AAO3 and ABA3 enzymes (Seo et al., 2000a; Xiong et al., 2001). ABA3 is a MoCo sulfurase required for aldehyde oxydase function in ABA biosynthesis (Bittner et al., 2001; Xiong et al., 2001). The sulfide form of MoCo is a cofactor of AAO3, and the AAO3 holoenzyme catalyzes the oxidation of the 1-aldehyde group of abscisic aldehyde to the 1-carboxylic acid group, generating ABA (Seo et al., 2000b). The Arabidopsis aldehyde oxidase family comprises four genes: AAO1, AAO2, AAO3, and AAO4 (Seo et al., 2000b). Aldehyde oxidase assays indicated that AAO1, AAO2, and AAO4 had almost no activity using abscisic aldehyde as a substrate (Seo et al., 1998, 2000b). Instead, AAO3 oxidized efficiently abscisic aldehyde to ABA (Seo et al., 2000b). Additionally, the isolation and characterization of an ABA-deficient mutant (in a Ler genetic background) that mapped to the AAO3 gene provided evidence that this gene is involved in the last step of ABA biosynthesis. Accordingly, aao3-1 mutant showed a reduced ABA level in rosette leaves (20% of wild type in wilted rosettes) and increased transpiration rate compared to wild type. However, aao3-1 exhibited wild-type dormancy, and ABA levels in dry seeds were reduced only by 40% with respect to the wild type (Seo et al., 2000a). Therefore, the major function of AAO3 was supposed to be the catalysis of the final step of ABA biosynthesis in leaves, but not in seeds (Seo et al., 2000a).
In this work, we report the isolation and characterization of two new alleles of AAO3, aao3-2, and aao3-3, that in contrast to aao3-1, showed a reduced dormancy and a significant reduction of ABA levels in dry seeds or seeds submitted to salt stress (Tables II and III). As aao3-1, both aao3-2 and aao3-3 showed a wilty phenotype and reduced ABA levels in rosette leaves. Molecular characterization of aao3-2 revealed a T-DNA insertional mutation that abolished mRNA expression of AAO3, representing therefore a null allele of AAO3. In the case of aao3-3, a complex mutation was found that is predicted to result in loss of an amino acid residue as well as several amino acid substitutions (L821QRPVK826→WDLDQ). The amino acid stretch affected by the mutation lies between the first MoCo binding site (residues 797-803) and a predicted substrate binding site (residues 878-886; Sekimoto et al., 1998). Additionally, the PROSITE program predicts a nucleotide-binding site located between residues 796 and 826, which would be affected by the mutation. Taken together, these data indicate that the aao3-3 mutation might have a severe effect on AAO3 enzyme activity. Indeed, the phenotypes of aao3-3 were quite similar to that of aao3-2, and ABA content was reduced at similar levels in both mutants.
According to Seo et al. (2000b), AAO3 gene expression was detected in 8-d-old seedlings, roots, rosette leaves, stems, and flowers; however, AAO3 mRNA was not detected in dry seeds or siliques. The phenotypes of the aao3-2 and aao3-3 mutants prompted us to reanalyze the expression of AAO3 in siliques and seeds. As a result, AAO3 transcript was detected in siliques and seeds imbibed and submitted to osmotic stress (Fig. 2C). We had to use at least between 10 and 20 μg total RNA to detect a weak AAO3 gene expression in siliques. In agreement with Seo et al. (2000b), we could not detect AAO3 gene expression in dry seeds. However, upon seed imbibition and submission to osmotic stress, expression of the AAO3 mRNA was induced (Fig. 2C). This result and the increased ABA levels measured in wild-type seeds treated with 200 mm NaCl suggest that ABA biosynthesis takes place in the seed upon osmotic stress. In agreement with that idea, ABA-deficient mutants behave as salt resistant in germination assays (Leon-Kloosterziel et al., 1996; Quesada et al., 2000; Gonzalez-Guzman et al., 2002; this work).
Physiological characterization of both aao3-2 and aao3-3 reveals notable differences with respect to aao3-1. Although aao3-1, aao3-2, and aao3-3 seeds exhibit salt and osmotic stress resistance, only aao3-2 and aao3-3 seeds show a reduced dormancy and paclobutrazol-resistant germination (Table III). These data, together with the reduced ABA levels in seeds of aao3-2 and aao3-3, clearly support a role for the AAO3 gene product in seed ABA biosynthesis. The differential features of aao3-2 and aao3-3 with respect to aao3-1 might be attributed to, at least, two reasons. First, the aao3-1 mutation might be leaky to some extent. This mutation is a single bp substitution found at the end of the ninth intron of the AAO3 gene, which results in incorrect splicing of the primary AAO3 transcript (Seo et al., 2000a). However, shorter transcripts with only a six-nucleotide deletion still occur in aao3-1 (Seo et al., 2000a). Therefore, a residual activity below the detection limit of the activity staining assay employed cannot be excluded. Second, the aao3-1 mutation is present in a Ler background, whereas aao3-2 and aao3-3 mutations are in a Col background. The erecta mutant has a lesion in a Leu-rich repeat receptor-like kinase that affects development of aerial plant organs (Torii et al., 1996) and also leads to additional unexpected phenotypes (Godiard et al., 2003). Indeed, seeds of Ler ecotype are more sensitive to inhibition of germination by low exogenous concentrations of ABA than Col ecotype (data not shown). Therefore, the threshold of ABA concentration required to show reduced dormancy could be different in a Ler than in a Col genetic background. In any case, the identification of genetic lesions in the AAO3 locus that lead to an ABA-deficient phenotype in seeds of Col genotype strongly suggests that this gene plays a major role in seed ABA biosynthesis.
MATERIALS AND METHODS
Plant Material
Arabidopsis plants (ecotype Columbia) were routinely grown under greenhouse conditions in pots containing a 1:3 perlite-soil mixture. For in vitro culture, seeds were surface-sterilized by treatment with 70% ethanol for 20 min, followed by commercial bleach (2.5%) containing 0.05% Triton X-100 for 10 min, and, finally, four washes with sterile distilled water. Stratification of the seeds was conducted during 3 d at 4°C. Afterward, seeds were sowed on Murashige and Skoog (MS) plates (Murashige and Skoog, 1962) containing solid medium composed of MS basal salts and 1% Suc, solidified with 1% agar and pH adjusted to 5.7 with KOH before autoclaving. Different concentrations of NaCl and mannitol were made by adding appropriate amounts of reagents to the basal medium. Plates were sealed and incubated in a controlled environment growth chamber at 22°C under a 16-h light, 8-h dark photoperiod at 80 to 100 μE m−2 s−1.
Screening Conditions
T-DNA lines were constructed in the D. Weigel laboratory (Salk Institute, La Jolla, CA) using the pSKI15 vector. Approximately 17,000 lines, stock numbers N21995 and N21991, were provided by the Arabidopsis Biological Resource Center (Ohio State University, Columbus). The distributed T-DNA pools are T4 seeds. Approximately 2 × 105 seeds were screened at high seed density (50 petri plates of 14-cm diameter containing approximately 4,000 seeds per plate) on MS medium (plus 1% Suc) containing 200 mm NaCl. Seeds were considered to be salt-resistant only after they produced fully green expanded cotyledons. Selected salt-resistant candidates (T4) were grown in soil to obtain the T5 progeny for further studies. The T5 progeny of the candidates was retested at low seed density (up to 200 seeds per 9-cm-diameter petri plate) under 150 to 200 mm NaCl.
Genetic Analysis of sre2 Mutants
Backcrosses of sre2 mutants to the wild type, intercrosses among sre2 mutants as well as those of sre2 with aba mutants were performed by transferring pollen to the stigmas of emasculated flowers. F1 and F2 seeds were scored for germination in 150 to 200 mm NaCl. In order to map the sre2-2 (aao3-3) locus, homozygous sre2-2 plants (in a Col background) were crossed to wild-type plants of the Ler background. From the segregating F2 generation, homozygous sre2-2 individuals were selected, and DNA was individually extracted. Mapping of the sre2-2 locus was carried out by testing linkage with simple sequence length polymorphism (SSLP) markers (Bell and Ecker, 1994; Lukowitz et al., 2000). The analysis of 44 F2 chromosomes revealed linkage of the sre2-2 locus and the nga1126 marker (which is only 150 kb away from AAO3).
A CAPS molecular marker (Konieczny and Ausubel, 1993) was developed based on the three-nucleotide deletion present in the AAO3 gene of sre2-2. This mutation destroys a TaiI restriction site, and therefore TaiI digestion of a 1-kb AAO3 DNA fragment amplified using the primers F2672 and R3661 (see below) reveals polymorphic bands in sre2-2 compared to wild type. Thus, TaiI digestion of AAO3 DNA amplified from wild type leads to four DNA fragments (154, 186, 228, and 395 bp). Instead, TaiI digestion of AAO3 DNA amplified from F2 sre2-2 individuals leads to three DNA fragments (228, 340, and 395 bp).
Germination Assays
Seeds were plated on solid medium composed of MS basal salts, 1% Suc, and 150 to 200 mm NaCl or 400 mm mannitol. After the indicated days of incubation, the percentage of seeds that had germinated and developed fully green expanded cotyledons was determined. To measure paclobutrazol sensitivity, seeds were plated on medium containing 10 μm paclobutrazol, and germination was determined after 7 d.
ABA Extraction and Determination
Dry seeds, seeds imbibed for 60 h and incubated for 24 h in 200 mm NaCl, as well as rosette leaves of unstressed or drought-stressed plants (unwatered for a week) were ground to a fine powder with mortar and pestle under liquid nitrogen. Duplicate samples (50 mg dry weight each) were extracted with 5 mL 80% acetone containing 100 mg/L 2,6-ditert-butyl-methyl phenol (BHT) and 500 mg/L citric acid, for 16 h at 4°C in the dark. The extracts were further homogenized in a polytron homogenizer at maximum speed for 1 min and centrifuged at 3,000g for 5 min. A 1-mL aliquot of the extract was evaporated in a vacuum centrifuge. The sample was resuspended in 100 μL Tris saline buffer (TBS, 50 mm Tris, 1 mm MgCl2, 150 mm NaCl, pH 7.8) and analyzed directly or diluted with TBS in order to fit the ABA content of the extracts within the linear range of the ABA standard curve of the assay. Quantitative analysis of ABA was performed by the indirect ELISA method, using the Phytodetek ABA kit (Agdia, Elkhart, IN).
Molecular Characterization of aao3-2 and aao3-3 Alleles
Plasmid rescue was used to isolate plant DNA flanking sequences of the T-DNA insertion in aao3-2. To this end, 5 μg genomic DNA from a aao3-2 homozygous plant was digested with BamHI (left border rescue). The reaction mixture was extracted once with an equal volume of phenol:chloroform:isoamylalcohol (25:24:1, v/v), once with chloroform:isoamylalcohol (24:1, v/v), and then ethanol precipitated. The DNA was ligated in a 100-μL reaction, and the ligation mixture was precipitated with ethanol. Ligated DNA was introduced by electroporation into the Escherichia coli XL-1-Blue MRF′ strain (Stratagene, La Jolla, CA). The transformed colonies contained two classes of plasmid. The most common class contained a plasmid whose restriction pattern corresponded to a direct repeat of T-DNA. The second class was represented by the plasmid PR9B. Sequence analysis of the PR9B plasmid revealed that the T-DNA insertion in the aao3-2 mutant is located at nucleotide 282 of AAO3.
In order to identify the mutation occurring in aao3-3, oligonucleotides (see below) were designed according to the AAO3 gene sequence (At2g27150), and overlapping fragments encompassing the entire gene were PCR-amplified from aao3-3. The amplified products were sequenced on both strands. To avoid errors caused by PCR, three independent PCR samples were mixed and batch sequenced. The following oligonucleotides were used:
F5: 5′AAATCTAACCTTATAATTGG
R1010: 5′ATGTTATGAAGCTCAGCCAC
F900: 5′CAAAGACCATCTTGTAACAT
R1880: 5′ATCAAGACTACATATTCTAT
F1800: 5′CACCTTGCACTCGGAATATA
R2790: 5′GAAGTACTGTGACCCTAGCC
F2672: 5′GCTGAGCGAAAGATAATCTCC
R3661: 5′GGTACTCGAGAAATCCCTCTC
F3550: 5′GATCCTGATGAATATACACTGCC
R4544: 5′CAGTAGTGTACTCTTCCATCATG
F4360: 5′TCCGATATTATTTATGACTGTGG
R5139: 5′TGTAACTTAGCAGCAACGAGAG
Upon request, all novel materials described in this publication will be made available in a timely manner for noncommercial research purposes, subject to the requisite permission from any third-party owners of all or parts of the material. Obtaining any permissions will be the responsibility of the requestor.
Acknowledgments
We thank M. Seo and T. Koshiba (Plant Science Center, RIKEN, Yokohama, Japan) for providing seeds of the aao3-1 mutant and the ProAAO3-AAO3 construct. We also thank the Arabidopsis Biological Resource Center (Ohio State University, Columbus)/Nottingham Arabidopsis Stock Centre (University of Nottingham, Loughborough, UK) for providing seed stocks and the group of Lorenzo Zacarías (Instituto de Agroquimica y Tecnologia de Alimentos, Valencia, Spain) for invaluable help in determining ABA content. M.G.G. was supported by a Ministerio de Educacion y Cultura fellowship.
This work was supported by the Ministerio de Ciencia y Tecnologia (grant no. BIO2002–03090) and FEDER.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.103.036590.
References
- Bell CJ, Ecker JR (1994) Assignment of 30 microsatellite loci to the linkage map of Arabidopsis. Genomics 19: 137–144 [DOI] [PubMed] [Google Scholar]
- Bentsink L, Koornneef M (2002) Seed dormancy and germination. In CR Somerville, EM Meyerowitz, eds, The Arabidopsis Book. American Society of Plant Biologists, Rockville, MD, pp 1–17. http://www.aspb.org/publications/Arabidopsis
- Bittner F, Oreb M, Mendel RR (2001) ABA3 is a molybdenum cofactor sulfurase required for activation of aldehyde oxidase and xanthine dehydrogenase in Arabidopsis thaliana. J Biol Chem 276: 40381–40384 [DOI] [PubMed] [Google Scholar]
- Cheng WH, Endo A, Zhou L, Penney J, Chen HC, Arroyo A, Leon P, Nambara E, Asami T, Seo M, et al. (2002) A unique short-chain dehydrogenase/reductase in Arabidopsis glucose signaling and abscisic acid biosynthesis and functions. Plant Cell 14: 2723–2743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkelstein RR, Gampala SS, Rock CD (2002) Abscisic acid signaling in seeds and seedlings. Plant Cell 14(Suppl): S15–S45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkelstein RR, Rock CD (2002) Abscisic acid biosynthesis and response. In CR Somerville, EM Meyerowitz, eds, The Arabidopsis Book. American Society of Plant Biologists, Rockville, MD, pp 1–52. http://www.aspb.org/publications/Arabidopsis [DOI] [PMC free article] [PubMed]
- Godiard L, Sauviac L, Torii KU, Grenon O, Mangin B, Grimsley NH, Marco Y (2003) ERECTA, an LRR receptor-like kinase protein controlling development pleiotropically affects resistance to bacterial wilt. Plant J 36: 353–365 [DOI] [PubMed] [Google Scholar]
- Gonzalez-Guzman M, Apostolova N, Belles JM, Barrero JM, Piqueras P, Ponce MR, Micol JL, Serrano R, Rodriguez PL (2002) The short-chain alcohol dehydrogenase ABA2 catalyzes the conversion of xanthoxin to abscisic aldehyde. Plant Cell 14: 1833–1846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoth S, Morgante M, Sanchez JP, Hanafey MK, Tingey SV, Chua NH (2002) Genome-wide gene expression profiling in Arabidopsis thaliana reveals new targets of abscisic acid and largely impaired gene regulation in the abi1-1 mutant. J Cell Sci 115: 4891–4900 [DOI] [PubMed] [Google Scholar]
- Karssen CM, Brinkhorst-van der Swan DLC, Breekland AE, Koornneef M (1983) Induction of dormancy during seed development by endogenous abscisic acid: studies on abscisic acid deficient genotypes of Arabidopsis thaliana (L.) Heynh. Planta 157: 158–165 [DOI] [PubMed] [Google Scholar]
- Konieczny A, Ausubel FM (1993) A procedure for mapping Arabidopsis mutations using co-dominant ecotype-specific PCR-based markers. Plant J 4: 403–410 [DOI] [PubMed] [Google Scholar]
- Koornneef M, Jorna ML, Brinkhorst-Van der Swan DLC, Karssen CM (1982) The isolation of abscisic acid (ABA) deficient mutants by selection of induced revertants in non-germinating gibberellin-sensitive lines of Arabidopsis thaliana. Theor Appl Genet 61: 385–393 [DOI] [PubMed] [Google Scholar]
- Leon-Kloosterziel KM, Gil MA, Ruijs GJ, Jacobsen SE, Olszewski NE, Schwartz SH, Zeevaart JA, Koornneef M (1996) Isolation and characterization of abscisic acid-deficient Arabidopsis mutants at two new loci. Plant J 10: 655–661 [DOI] [PubMed] [Google Scholar]
- Lopez-Molina L, Mongrand S, Chua NH (2001) A postgermination developmental arrest checkpoint is mediated by abscisic acid and requires the ABI5 transcription factor in Arabidopsis. Proc Natl Acad Sci USA 98: 4782–4787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukowitz W, Gillmor CS, Scheible WR (2000) Positional cloning in Arabidopsis. Why it feels good to have a genome initiative working for you. Plant Physiol 123: 795–805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marin E, Nussaume L, Quesada A, Gonneau M, Sotta B, Hugueney P, Frey A, Marion-Poll A (1996) Molecular identification of zeaxanthin epoxidase of Nicotiana plumbaginifolia, a gene involved in abscisic acid biosynthesis and corresponding to the ABA locus of Arabidopsis thaliana. EMBO J 15: 2331–2342 [PMC free article] [PubMed] [Google Scholar]
- Murashige T, Skoog F (1962) A revised medium for rapid growth and bioassays with tobacco tissue culture. Physiol Plant 15: 473–497 [Google Scholar]
- Quesada V, Ponce MR, Micol JL (2000) Genetic analysis of salt-tolerant mutants in Arabidopsis thaliana. Genetics 154: 421–436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz SH, Qin X, Zeevaart JA (2003) Elucidation of the indirect pathway of abscisic acid biosynthesis by mutants, genes, and enzymes. Plant Physiol 131: 1591–1601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz SH, Tan BC, Gage DA, Zeevaart JA, McCarty DR (1997) Specific oxidative cleavage of carotenoids by VP14 of maize. Science 276: 1872–1874 [DOI] [PubMed] [Google Scholar]
- Seki M, Ishida J, Narusaka M, Fujita M, Nanjo T, Umezawa T, Kamiya A, Nakajima M, Enju A, Sakurai T, et al. (2002) Monitoring the expression pattern of around 7,000 Arabidopsis genes under ABA treatments using a full-length cDNA microarray. Funct Integr Genomics 2: 282–291 [DOI] [PubMed] [Google Scholar]
- Sekimoto H, Seo M, Kawakami N, Komano T, Desloire S, Liotenberg S, Marion-Poll A, Caboche M, Kamiya Y, Koshiba T (1998) Molecular cloning and characterization of aldehyde oxidases in Arabidopsis thaliana. Plant Cell Physiol 39: 433–442 [DOI] [PubMed] [Google Scholar]
- Seo M, Akaba S, Oritani T, Delarue M, Bellini C, Caboche M, Koshiba T (1998) Higher activity of an aldehyde oxidase in the auxin-overproducing superroot1 mutant of Arabidopsis thaliana. Plant Physiol 116: 687–693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo M, Koiwai H, Akaba S, Komano T, Oritani T, Kamiya Y, Koshiba T (2000. b) Abscisic aldehyde oxidase in leaves of Arabidopsis thaliana. Plant J 23: 481–488 [DOI] [PubMed] [Google Scholar]
- Seo M, Peeters AJ, Koiwai H, Oritani T, Marion-Poll A, Zeevaart JA, Koornneef M, Kamiya Y, Koshiba T (2000. a) The Arabidopsis aldehyde oxidase 3 (AAO3) gene product catalyses the final step in abscisic acid biosynthesis in leaves. Proc Natl Acad Sci USA 97: 12908–12913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torii KU, Mitsukawa N, Oosumi T, Matsuura Y, Yokoyama R, Whittier RF, Komeda Y (1996) The Arabidopsis ERECTA gene encodes a putative receptor protein kinase with extracellular leucine-rich repeats. Plant Cell 8: 735–746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong L, Ishitani M, Lee H, Zhu JK (2001) The Arabidopsis los5/aba3 locus encodes a molybdenum cofactor sulfurase and modulates cold stress and osmotic stress-responsive gene expression. Plant Cell 13: 2063–2083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong L, Zhu JK (2003) Regulation of abscisic acid biosynthesis. Plant Physiol 133: 29–36 [DOI] [PMC free article] [PubMed] [Google Scholar]