Abstract
Phosphorus (P) acquisition and partitioning are essential for plant homeostasis. P is available for plant uptake when in its inorganic form (H2PO4−, or Pi), but Pi is often limiting in soils. Plants secrete acid phosphatases (APases) into the apoplastic space, which may be important for obtaining Pi from organic P sources; however, the relative importance of these enzymes for plant P nutrition has yet to be determined. We demonstrate that the root-associated APase pool is increased in Arabidopsis when Pi is limiting and document five APase isoforms secreted from Arabidopsis roots. Previously, we presented the identification of the phosphatase under-producer (pup) mutants, which have decreased in vivo root APase staining when grown under low P conditions. Here, we present the characterization of one of these, pup3, and further studies with pup1. pup3 has 49%, 38%, and 37% less specific APase activity in exudates, roots, and shoots, respectively. Root-associated APase activity is decreased by 16% in pup1 and 25% in pup3, regardless of P treatment. Two APase activity isoforms are reduced in pup3 exudates, and root and shoot isoforms are also affected. One of the two exudate isoforms is recognized by a polyclonal antibody raised to an Arabidopsis purple APase recombinant protein (AtPAP12); however, AtPAP12 transcript levels are unaffected in the mutant. The pup3 mutation was mapped to 68.4 ± 6.0 centimorgans on chromosome 5. Although P concentrations were not altered in pup1 and pup3 tissues when grown in nutrient solution in which Pi was the sole source of P, the mutants had 10% (pup1) and 17% (pup3) lower shoot P concentrations when grown in a peat-vermiculite mix in which the majority of the total P was present as organic P. Therefore, the pup defects, which include secreted APases, are functionally important for plant P nutrition.
Phosphorus (P) deficiency is a major limitation to plant growth (Marschner, 1995; Vance et al., 2003). P is taken up by plants in its inorganic form, Pi or orthophosphate (H2PO4−). Although a macronutrient for plants, the availability of soil Pi is often below that of micronutrients because Pi is immobilized within soil organic complexes, clay complexes, and precipitated salts (for review, see Marschner, 1995). Sparingly available soil Pi can be ameliorated with Pi fertilizers, and applied fertilizers represent more than 80% of world Pi use (Steen, 1998). Modern agriculture relies on crops that provide maximum yields with these fertilizers; however, world resources of extractable Pi are limited, nonrenewable, and increasingly ecologically hazardous to obtain (Steen, 1998). As extractable world Pi stores become depleted, agriculture will be forced to adjust to a lack of Pi fertilizer while continuing to feed an expanding population. Alternatively, the organic P (Po) component of agricultural soil is abundant, representing up to 50% of total soil P (for review, see Vance et al., 2003). Plants may mobilize Po by secreting enzymes into the rhizosphere, including acid phosphatases (APases), ribonucleases, and deoxyribonucleases (for review, see Abel et al., 2002; Vance et al., 2003). Developing plants with enhanced abilities to access the Po component of soils is important to decrease agriculture's reliance on Pi fertilizers. Toward that goal, our group focused on the potential of plant-derived secreted APases to increase Po availability for plant nutrition.
APases may be active against a wide array of organic molecules present in soil Po. These enzymes are nonspecific orthophosphoricmonoester phosphohydrolases (EC 3.1.3.2), cleaving Pi from ester linkage sites. Secreted plant phosphatases preserve >50% activity over a broad pH range (4.0–7.6), maintain >80% activity over a broad temperature range (22°C–48°C), and are stable at temperatures as high as 60°C (LeBansky et al., 1991; Li and Tadano, 1996), making them ideal candidates for active soil enzymes. While most soil enzymes are typically short-lived, APases can be immobilized on or within soil clays and humates that preserve their activity (Burns, 1986). Soil APases are important for Pi acquisition by plant roots (Marschner, 1995), and plants can use Po as a sole source of Pi nutrition in sterile cultures (for review, see Abel et al., 2002). Therefore, plant-derived secreted APases have the potential to facilitate breakdown of soil Po to Pi for plant uptake.
The significance of plant-derived secreted APases to plant P nutrition needs to be determined because microbes and fungi within the rhizosphere also contribute to the overall pool of soil phosphatases (for review, see Richardson, 2001). The model plant Arabidopsis is convenient for studying this problem because, unlike many crop plants, it does not form mycorrhizal associations. To test whether plant-derived secreted APases are important for P nutrition, we identified Arabidopsis mutants defective in phosphatase secretion, the phosphatase under-producer (pup) mutants. Biochemical characterization of the pup1 mutant, as described previously, showed that pup1 is missing a low P-inducible APase isoform and lacks low P-inducible APase staining along its roots but is otherwise not defective in P deficiency responses (Trull and Deikman, 1998).
APases are abundant in Arabidopsis and are represented by at least four gene families. A recent survey of the annotated Arabidopsis genome identified sequences for one His APase, four phosphatidic APases, 10 vegetative storage protein APases, and 29 purple APases (PAPs; Li et al., 2002). Twenty PAP cDNAs were detected by PCR amplification of suspension cell culture cDNAs, and a nomenclature for Arabidopsis PAPs was established based on predicted amino acid sequences. Full-length genes from three APases have been cloned from Arabidopsis: a homolog to mammalian type-5 purple APases known previously as AtACP5 (AtPAP17; del Pozo et al., 1999; Li et al., 2002), a homolog to the kidney bean purple acid phosphatase-1 known previously as PAP1 (AtPAP12; GenBank accession no. U48448; Li et al., 2002), and a homolog to the soybean vegetative storage proteins (AtVSP1; Berger et al., 1995). Transcript accumulation of AtPAP17 is induced in both roots and shoots by low P conditions as well as abscisic acid, peroxide, and senescence (del Pozo et al., 1999). AtVSP transcripts accumulate to high levels in flower parts but are also expressed at lower levels throughout the rest of the plant. Methyl jasmonate and Suc up-regulate transcripts, while high Pi levels depress AtVSP expression (Berger et al., 1995). AtPAP12 is induced by low P conditions (Haran et al., 2000; Li et al., 2002) and has a signal peptide sufficient for secretion of a marker protein from roots into the surrounding medium (Haran et al., 2000). A secreted APase from Arabidopsis exudates has recently been purified and characterized (Coello, 2002). This APase hydrolyzes a variety of Po compounds with kinetic properties similar to other APases studied, including tartrate resistance, inhibition by Mo, Pi, F, and V, and activation with Ca. The protein and DNA sequences of this secreted enzyme have not yet been determined. Since APases are such a large group containing multiple gene families, identification of the secreted isoforms for further functional analysis is important for evaluating their importance to plant P nutrition.
Two Arabidopsis mutants have been described that are defective in P starvation responses, pho3 (Zakhleniuk et al., 2001) and phr1 (Rubio et al., 2001). The Phr1 gene has been cloned, and it is a transcription factor that likely mediates many low P responses, including the up-regulation of high-affinity transporters and ribonucleases as well as APases (Rubio et al., 2001). By contrast, the pup mutants are defective in APase secretion but otherwise respond normally to P deficiency, making them good candidates to test whether plant-derived APases are important for plant P relations. If plant-derived APases are important for P nutrition, then mutants with decreased APase activity may have altered P accumulation. In this study, we present the characterization of a second pup mutant, pup3. We show that both pup1 and pup3 have reduced rhizospheric APase activity and that the pup3 mutation results in lower APase activity in all tissues tested. When provided Pi in solution, pup mutants accumulate P normally, but when grown in a substrate containing Po, the mutants accumulate less shoot P, demonstrating that the pup defects impact plant P nutrition.
RESULTS
Phosphatase In Vivo Activity Staining Is Reduced in pup3 Roots
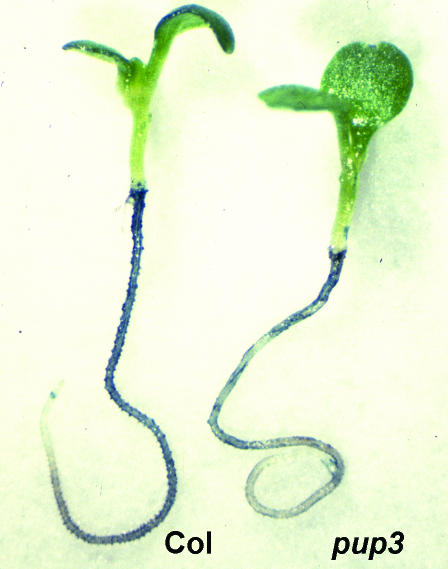
The pup mutants were identified by screening for reduced secreted APase activity when seedlings were grown in −P medium containing the APase stain 5-bromo-4-chloro-3-indolyl phosphate (BCIP; Trull and Deikman, 1998). Under these conditions, pup3 exhibits less intense staining than the Columbia (Col) ecotype control (Fig. 1). The pup3 phenotype is much less dramatic when compared to that of pup1, which completely lacks staining for APase activity with this substrate (Trull and Deikman, 1998).
Figure 1.
In vivo APase staining of pup3 roots under −P conditions. When cleaved by APases, the substrate BCIP forms a blue precipitate on the root surface. The pup3 mutant was identified by reduced in vivo APase staining of 5-dpg seedlings when grown with minimal Pi.
Specific APase Activity Is Reduced in pup3 But Not pup1
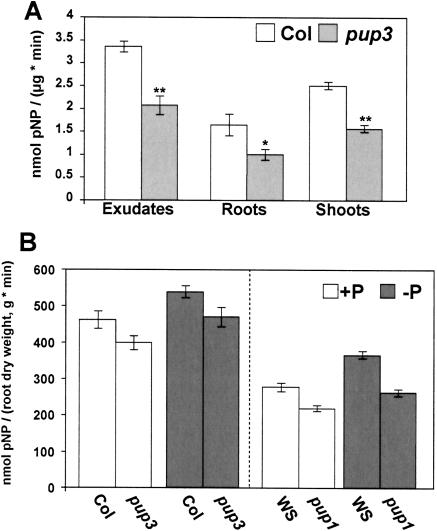
As a first step toward characterization of the pup3 defect, specific APase activity measurements were carried out on concentrated root exudates and protein extracts from root and shoot tissues (Fig. 2A). Relative to Col control plants, the pup3 mutant had 49%, 38%, and 37% less specific APase activity in exudates, roots, and shoots, respectively. pup1 exudates showed no change in specific APase activity (data not shown), and APase activity in tissue extracts was shown previously to be normal (Trull and Deikman, 1998).
Figure 2.
APase activity is decreased in the pup mutants. A, Specific APase activity. The hydrolysis of pNPP at 25°C over 10 min was measured for 0.5 μg of protein (n = 12 for exudates, n = 6 for roots, and n = 6 for shoots). For exudates, each protein sample (n) was the concentrated proteins released into liquid culture media over 2 d from 12 plants, and for roots and shoots each protein sample (n) was extracted from pooled tissues representing at least 20 plants. Error bars are se. Statistical differences between pup3 and Col for each type of extract were determined with Student's t test (*, P < 0.05; **, P < 0.001). B, Root-associated APase activity. Intact root systems were assayed for APase activity by hydrolysis of pNPP and corrected for root dry weight. One representative replication is shown with n ≥ 8 for each treatment. Each sample (n) is the activity of three plants grown together in liquid culture. Error bars are se. Statistical analyses were performed with ANOVA (Table I).
The Rhizospheric APase Pool Is Reduced in pup Mutants
An in vivo root-associated APase activity assay was developed to quantify the reduction in APase activity staining and determine whether rhizospheric APase is affected in the pup mutants (Fig. 2B; Table I). The root-associated APases measured in this assay (and discussed throughout the text) consist of those bound to cell walls at the root surface, those located within the apoplastic space of the root, and those secreted from the root during the course of the assay. Similar assays have been performed on whole root systems of tomato (Boutin et al., 1981), rye, buckwheat, clover, and wheat (McLachlan, 1980a) using an assay buffer containing high levels of sodium acetate. We found that this buffer yielded high levels of APase activity but was inappropriate for the delicate Arabidopsis root system, producing sporadic results with obvious tissue wilting. Instead, we altered the growth medium to serve as the assay buffer by increasing its buffering capacity with MES and balancing it osmotically with Suc.
Table I.
Statistical analyses of the root-associated APase activity experiments
| Variable | Components of Variance | Error df | F Value |
|---|---|---|---|
| Root-associated APase | |||
| pup3 versus Col | Genotype | 68 | 20.52* |
| P treatment | 40.05* | ||
| Genotype × P treatment | 0.78 | ||
| pup1 versus WS | Genotype | 64 | 28.73* |
| P treatment | 41.15* | ||
| Genotype × P treatment | 0.58 | ||
| Root P concentration | |||
| pup3 versus Col | Genotype | 61 | 0.10 |
| P treatment | 116.47* | ||
| Genotype × P treatment | 0.11 | ||
| pup1 versus WS | Genotype | 62 | 0.17 |
| P treatment | 18.87* | ||
| Genotype × P treatment | 0.89 | ||
| Shoot P concentration | |||
| pup3 versus Col | Genotype | 63 | 0.28 |
| P treatment | 158.90* | ||
| Genotype × P treatment | 0.02 | ||
| pup1 versus WS | Genotype | 64 | 0.82 |
| P treatment | 30.28* | ||
| Genotype × P treatment | 0.17 |
ANOVA was carried out on root-associated APase activity measurements (Fig. 2B) as well as the P concentrations of tissues used in these experiments. Replication effects were not significant for root-associated APase activity after transformation (measurements were expressed as a percentage of that replication's average +P control). Replication effects were insignificant for the P concentration data sets without transformation and as such were eliminated as components of variance. df, degrees of freedom.
P < 0.0001.
Using this method, we observed that Arabidopsis, like other plants studied with this technique, increases its root-associated APase activity in response to low P conditions (Fig. 2B). A 2 d −P treatment imposed on adult tissues previously grown under sufficient P conditions increased root-associated APase activity by 22% in Col and 25% in Wassilewskija (WS; Fig. 2B; Table I, P < 0.0001). The pup mutants responded to the −P treatment with the same relative magnitude as their controls (no genotype × P treatment interaction; Table I). Taking both P treatments together, root-associated APase activity was reduced by 16% in pup3 and by 25% in pup1 (Table I, P < 0.0001). These decreases in root-associated APase activity indicate that the pup mutants are defective in the rhizospheric APase pool.
P concentrations were measured from tissues used in the root-associated APase activity assays (Table I). Although the −P treatment dramatically lowered P tissue concentrations by approximately 25% in roots and more than 30% in shoots (data not shown), mutant plants did not differ from their controls in P accumulation under these conditions (no genotype × P treatment interaction; Table I). This result rules out the possibility that the pup mutants' decreased root-associated APase activity is an indirect result of increased Pi accumulation and subsequent repression of APase activity.
pup3 Is Defective in the Activity of Some APase Activity Isoforms
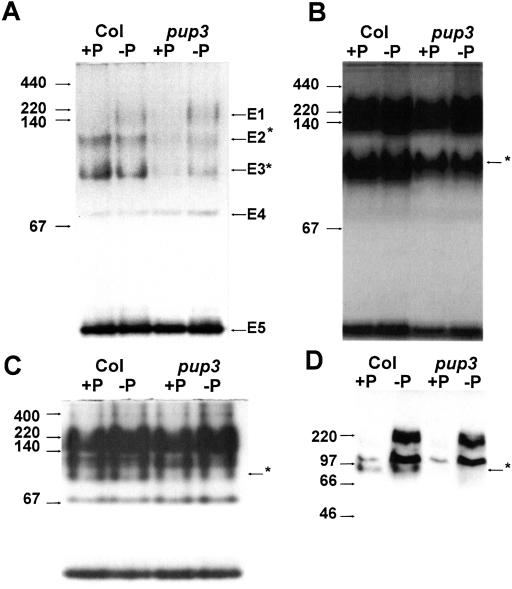
APase isoform analysis was carried out to identify the nature of the decreased activity in the pup mutants (Fig. 3). Concentrated root exudates (Fig. 3A), shoot protein extracts (Fig. 3B), and root protein extracts (Fig. 3C) were run on native PAGE and stained for APase activity. Heavy and heterogeneous glycosylation (Stahl et al., 1994) of these proteins likely caused the bands to migrate nonuniformly in the gels, giving a smeared appearance. Five major APase activity isoforms were detected in Arabidopsis exudates. Molecular weights cannot be estimated on a native gel because SDS was not present to linearize and uniformly charge the proteins, so APase isoforms are denoted as E1, E2, etc. The slowest-running secreted APase isoform, E1, was −P inducible, while the other four isoforms were not affected by P deprivation. Two bands of constitutive APase activity, E2 and E3, were reduced in pup3 exudates. Reductions in APase activity isoforms were also observed in pup3 shoots (Fig. 3B) and roots (Fig. 3C), but these differences were less obvious because multiple APase isoforms migrated together and nonuniformly under the native conditions necessary to retain their maximum activity. It is possible that multiple isoforms may also be affected in pup3 roots and shoots but could not be detected either because they did not retain activity throughout extraction and electrophoresis or because they comigrated with one of the many other APase isoforms found in these tissues.
Figure 3.
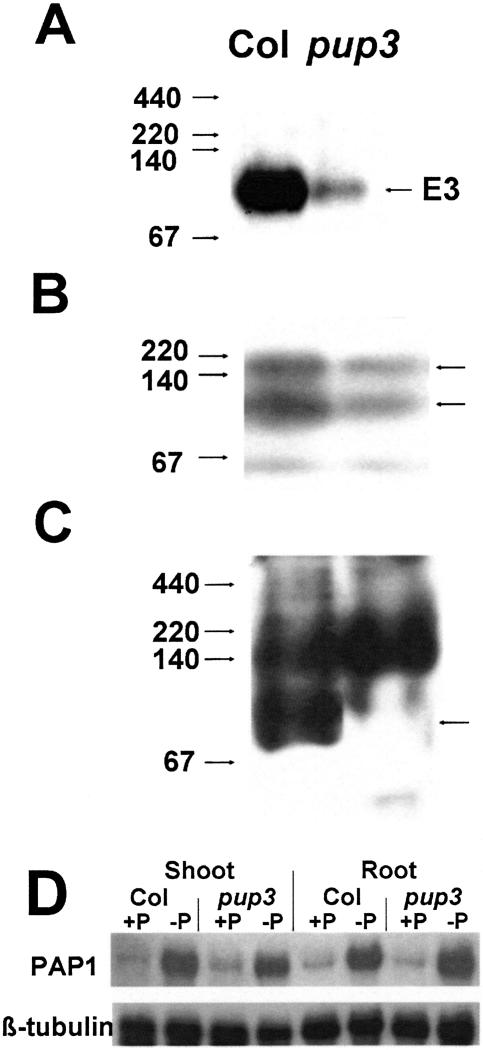
A subset of APase activity isoforms is reduced in pup3. A–C, Proteins were run under native discontinuous PAGE conditions and stained for APase activity using Fast Black K and β-naphthyl acid phosphate. Markers are native protein electrophoresis markers (left), and APase isoforms are noted on the right. Isoforms designated with an asterisk have decreased activity in pup3 exudates. Samples were 7.5 μg of concentrated root exudate proteins (A), 28 μg of shoot proteins (B), and 10 μg of root proteins (C). D, Shoot extracts run under SDS-PAGE but otherwise native conditions and subsequently stained for APase activity also show a reduced pup3 isoform. Markers for D are Rainbow high Mr markers (Amersham Pharmacia Biotech).
When extracts were run on SDS-PAGE under nonreducing and otherwise native conditions, pup3 shoot proteins were missing a 90-kD APase isoform (Fig. 3D). This is different from the 160-kD isoform shown previously to be missing in pup1 under these conditions (Trull and Deikman, 1998). APase isoforms from pup1 exudates were not different from controls (data not shown).
AtPAP12 Is One of the APase Isoforms Defective in pup3
Protein and transcript accumulation of a low P-regulated PAP (AtPAP12) was carried out to analyze the effects of the pup mutations on this secreted APase. In exudates, one APase isoform, E3, was recognized by the AtPAP12 antibody (Fig. 4A). The decreased APase activity of E3 in pup3 extracts (Fig. 3A) correlated with reduced reactivity of E3 with the AtPAP12 antibody. The other isoform with reduced pup3 APase activity, E2, was not recognized by the AtPAP12 antibody.
Figure 4.
AtPAP12 protein but not transcript accumulation is reduced in pup3. Concentrated exudates (A; 7 μg), shoot proteins (B; 7.5 μg), and root proteins (C; 10 μg), were run under native discontinuous PAGE conditions, blotted to a nitrocellulose membrane, and hybridized with the αAtPAP12 antibody. Markers are native protein electrophoresis markers (left). Arrows to the right indicate proteins with reduced reactivity in pup3. D, Total RNA (9.25 μg/lane) isolated from 16-dpg plants grown on a continuous treatment of either high or low P was probed sequentially with an EST corresponding to AtPAP12 or β-tubulin (loading control).
The AtPAP12 antibody reacted with three isoforms in Col shoots (Fig. 4B) and two isoforms in Col roots (Fig. 4C). The multiple isoforms recognized in extracts from these tissues may have been due to AtPAP12 dimerization (Schenk et al., 2000), cross-reactivity with other PAPs (this antibody cross-reacts with a PAP from Spirodela oligorrhiza; Nakazato et al., 1998), or cross-reaction with carbohydrates from other glycoproteins (Miller et al., 2001). As in exudates, these patterns were altered in the pup3 mutant. The AtPAP12 antibody had reduced reactivity with the upper two isoforms in pup3 shoot extracts (Fig. 4B). Of these two bands, the lower one corresponds to the APase isoform with decreased activity in pup3 shoot extracts (Fig. 3B). The AtPAP12 antibody did not recognize the lower isoform in pup3 root extracts (Fig. 4C), and this isoform migrated at the same location as the isoform that had reduced APase activity in pup3 (Fig. 3C).
Accumulation of AtPAP12 transcripts was analyzed to determine if the pup3 defects occur at this level of regulation (Fig. 4D). The expressed sequence tag (EST) used as a probe is the last 550 bp of the 3′ end of the AtPAP12 cDNA, and only one band was recognized on the blot. As shown previously by two other groups (Haran et al., 2000; Li et al., 2002), AtPAP12 transcript accumulation was increased under −P conditions (Fig. 4D). However, pup3 did not differ from control tissues in its accumulation of AtPAP12 mRNA (Fig. 4D). Therefore, the pup3 mutation affects AtPAP12 protein but not transcript levels. This may mean that PUP3 is involved in regulating the stability of AtPAP12 proteins, and possibly other AtPAPs as well, because there is at least one other isoform with reduced APase activity in pup3 exudates that is not recognized by the AtPAP12 antibody.
Proteins from the pup1 mutant were also tested for differences with regards to immunoreactivity with the AtPAP12 antibody and AtPAP12 transcript accumulation, and they were not different from controls (data not shown).
P Accumulation Is Altered in pup Mutants When Organic P Is Present
Although the pup mutants accumulated normal P concentrations when grown in a nutrient solution in which P was supplied entirely as Pi (Table I), the pup mutants exhibited altered P relations when P was supplied as Po within a soil substrate (Fig. 5; Table II). The peat-vermiculite soil mix used in this experiment had 2.67 ± 0.14 μg P/g soil; however, only 0.88 ± 0.02 μg P/g soil was available as Pi. Therefore, the majority of P in this substrate was Po. Plants were also watered with a weak fertilizer solution (Somerville and Ogren, 1982) either containing Pi (+Pi fertilizer) or lacking it (−Pi fertilizer).
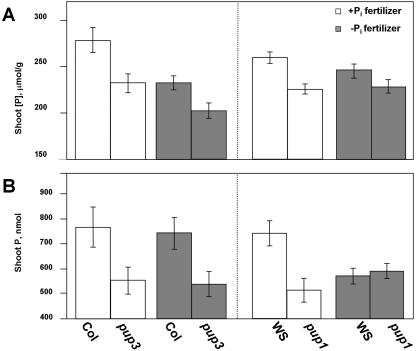
Figure 5.
pup mutants have decreased shoot P concentration and total P accumulation when grown in Po soil. pup mutants and their respective control plants were grown in a peat-vermiculite soil mix for 3 weeks or until the first sign of the primary reproductive inflorescence, then shoots were harvested for P determinations. Plants were watered with complete fertilizer solution (+Pi fertilizer) or one that lacked the Pi component (−Pi fertilizer). Shoot P concentration (A) is expressed on a per gram dry weight basis, and shoot P accumulation (B) is the total amount of P present in an individual shoot. Each data point represents n ≥ 18 (pooled data from two replications of n ≥ 9), and error bars are se. Statistical analyses were performed with ANOVA (Table II).
Table II.
Statistical analyses of shoot P concentration and total P accumulation between the pup mutants and their respective controls
| Variable | Components of Variance | Error df | F Value |
|---|---|---|---|
| P concentration | |||
| pup3 versus Col | Genotype | 69 | 14.3*** |
| Pi fertilizer treatment | 14.1*** | ||
| Genotype × Pi treatment | 0.7 | ||
| pup1 versus WS | Genotype | 75 | 14.0*** |
| P treatment | 0.7 | ||
| Genotype × P treatment | 0.2 | ||
| Total P accumulation | |||
| pup3 versus Col | Genotype | 73 | 10.6** |
| Pi fertilizer | 0.1 | ||
| Genotype × Pi treatment | <0.1 | ||
| pup1 versus WS | Genotype | 79 | 5.8* |
| Pi fertilizer | 1.2 | ||
| Genotype × Pi treatment | 8.2** |
ANOVAs were performed with pooled replications. df, degrees of freedom.
P < 0.05;
P < 0.01;
P < 0.001.
Shoot P concentrations were lower for both the pup3 and pup1 mutants when compared to their controls (Fig. 5A; Table II). In pup3, this decrease was 17% lower with the +Pi fertilizer treatment and 13% lower for the −Pi fertilizer treatment. For pup1, this decrease was not as pronounced but still significant at 13% for the +Pi fertilizer treatment and 7% for the −Pi fertilizer treatment.
Total P accumulation was also altered in pup shoots (Fig. 5B; Table II). For pup3 the decrease was 28%/+Pi fertilizer and 27%/−Pi fertilizer, while pup1 accumulated 31% less P in the +Pi fertilizer treatment but the same amount of P in the −Pi fertilizer treatment.
The fertilizer treatment in this study highlights differences between the two ecotypes in their response to applied Pi. While the +Pi fertilizer treatment increased shoot P concentrations in the Col ecotype (Fig. 5A; Table II), the total amount of P accumulated in shoots remained the same (Fig. 5B; Table II). The WS ecotype exhibited the opposite strategy: Shoot P concentrations remained constant with applied Pi fertilizer (Fig. 5A; Table II), while the total amount of P accumulated increased by 30% (Fig. 5B). These results agree with other findings that different Arabidopsis ecotypes demonstrate growth plasticity with regards to P uptake and accumulation (Krannitz et al., 1991). The pup1 but not the pup3 mutant was altered in its response to Pi fertilizer. pup3 was not different from its Col control in this respect (no genotype × Pi fertilizer treatment interaction; Table II): Shoot P concentrations were increased, while total P accumulation remained constant. However, the pup1 mutant did not increase its shoot P accumulation when the +Pi fertilizer was supplied (Fig. 5B; Table II, creating the genotype × Pi fertilizer treatment interaction). Still, both mutants had decreased P concentrations and total P accumulation when compared to their controls at both fertilizer treatments, demonstrating that even the +Pi fertilizer treatment could not compensate for the decreased amount of Pi available to pup roots as a consequence of their reduced ability to mobilize Pi from Po.
Genetic Analysis
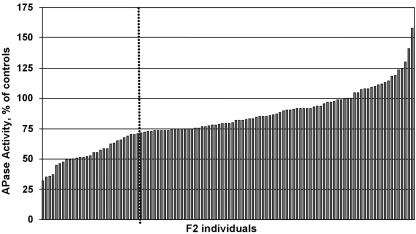
A mapping population to localize the pup3 mutation to a chromosomal region within the Arabidopsis genome was developed using an outcross to the WS ecotype. Since APase activity staining was not a reliable method to determine the pup3 phenotype, the genotype of F2 progeny was determined based on reduced APase activity from crude shoot extracts (Fig. 6). Of the 110 plants assayed, roughly one-quarter had 70% of the crude shoot APase activity of their control parents, which is comparable to the assays with purified pup3 shoot proteins (Fig. 2A). The dispersion pattern of this crude assay implies that pup3 is either a recessive or codominant mutation (Fig. 6). Plants within the lower quartile were confirmed for pup3 isoform phenotypes by native PAGE before codominant amplified polymorphic sequences (CAPS) mapping (Table III). Three markers on chromosome 5 (NIT4, RBCS-B, and ASB2) are linked to the pup3 mutation, with RBCS-B demonstrating the tightest linkage to pup3. Based on the directionality from the other two markers, the pup3 mutation is located at 68.4 ± 6.0 cM on chromosome 5. This position places the pup3 mutation in a different location from both the pup1 mutation and AtPAP12. pup1 was mapped previously between 34.0 cM and 54.8 cM on chromosome 2 (Trull and Deikman, 1998), and AtPAP12 is located on BAC T22013 from chromosome 2.
Figure 6.
APase activity of the pup3 mapping population. Crude shoot proteins were isolated from 110 F2 plants from an outcross to the WS ecotype. APase was assayed and corrected for fresh weight, then compared to the average for Col and WS controls. The dashed line marks the lower quadrant of 29 plants.
TABLE III.
Genetic location of the pup3 mutation
| Marker | Marker Location | n | Segregation Ratio, Col/Col:Col/WS:WS/WS | Distance, cM from Mutation |
|---|---|---|---|---|
| m246 | 11cM, chr 2 | 22 | 7:9:6 | |
| GPA1 | 49 cM, chr 2 | 31 | 8:18:5 | |
| Bgl1 | 75 cM, chr 3 | 25 | 6:15:4 | |
| g4539 | 55 cM, chr 4 | 24 | 5:16:3 | |
| ASA1 | 15, chr 5 | 25 | 8:14:3 | |
| NIT4 | 45, chr 5 | 32 | 16:15:1** | 29.6 ± 10.9 |
| RBCS-B | 80.8, chr 5 | 33 | 25:8:0** | 12.4 ± 6.0 |
| ASB2 | 115, chr 5 | 26 | 10:14:2* | 42.6 ± 17.9 |
CAPS mapping results are shown for markers throughout the Arabidopsis genome. n is the number of F2 plants scored from an outcross to another Arabidopsis ecotype (WS). Significant deviation from the expected segregation ratio of Col/Col:Col/WS:WS/WS (1:2:1; P < 0.1) indicates that the marker is linked to the mutation. chr, chromosome.
P < 0.1;
P < 0.01 (chi square).
DISCUSSION
Plants alter the phosphatase activity of soils. Within the rhizosphere, phosphatase activity is increased and correlates with a zone of Po depletion around plant roots. Roots, microbes, and mycorrhizal fungi all contribute to rhizospheric phosphatase activity. Free-living soil microbes concentrate around root systems because lysed plant cells and root secretions are sources of carbon and nutrients (Marschner, 1995). While microbes secrete phosphatases that liberate Pi for plants, they also compete with plants for that resource. Mycorrhizal fungi are also a significant source of soil phosphatases, and they can be major providers of Pi to plants when available P is limiting (Miyasaka and Habte, 2001). However, not all plants form mycorrhizal associations, and in nonmycorrhizal roots (such as Arabidopsis), APase activity is associated with the root surface and rhizosphere (McLachlan, 1980a; Dodd et al., 1987). Plants grown in solution culture mobilize a variety of Po substrates to fulfill their complete P nutritional requirements when deprived of Pi, so plant-derived phosphohydrolases have the capacity to provide for P growth demands (Richardson et al., 2001; Abel et al., 2002). The work presented in this study addresses the importance of plant-derived APases in plant P nutrition using two Arabidopsis mutants that are defective in rhizospheric APase activity.
Previously, the importance of secreted plant APases to plant P nutrition has only been implied. Increased expression of APases under low P conditions is part of an overall P stress response that functions to increase P availability to the plant and is analogous to the microbial PHO regulon (Abel et al., 2002). Proteoid roots are specialized structures whose secretions, including relatively large amounts of APases, increase P adsorption from soils (Vance et al., 2003). When plant-derived APase secretion was compared between efficient and inefficient P genotypes in maize, the results correlated positively (Guame et al., 2001). However, P efficiency is conferred by multiple traits even between recombinant inbred lines of the same species (Yan et al., 2001), which may explain why another study found no significant effect of root-associated APase activity between efficient and inefficient white clover genotypes (Hunter and McManus, 1999). Conversely, a negative correlation between root-associated APase activity and P efficiency was found in a comparison between wheat genotypes domesticated in the low P soils of Australia and their wild progenitors (McLachlan, 1980b). McLachlan reasoned that the domesticated wheat was more successful at obtaining low levels of P and therefore was not inducing low P-responsive APases. The pup mutants are good experimental tools for evaluating the relative importance of plant-derived APases because they are only altered from their controls in a single locus, eliminating the genetic variation caused between and among species that may exhibit different strategies for obtaining P.
We establish that Arabidopsis, like other plants previously studied (McLachlan, 1980a; Boutin et al., 1981), increases its root-associated APase pool in response to P deprivation (Fig. 2B). This pool of APases includes apoplastic and cell wall-bound enzymes that would have access to the soil solution and therefore may be important for P acquisition. A major APase isoform has recently been purified from Arabidopsis exudates (Coello, 2002), and we add that at least four more APases are secreted from Arabidopsis roots. The activity of one of these isoforms is increased in response to external P levels (Fig. 3A). Since overexpression of substrate-specific phosphohydrolases such as apyrase (Thomas et al., 1999) and phytase (Richardson et al., 2001) throughout the plant leads to increased ability to take up Pi from those substrates, the overexpression of these secreted APases may lead to an enhanced ability to take up P from a variety of Po sources. Further analysis of these secreted APases, including their purification, protein sequencing, and genomic sequencing, may be helpful for determining their relative importance to plant P nutrition.
We also present evidence that plant-derived APase activity can be altered by a single mutation, pup3. The pup3 mutant has reduced APase activity as demonstrated by in vivo staining (Fig. 1), root-associated assay (Fig. 2B), and specific activity (Fig. 2A). The amount of reduction in pup3 APase activity, ranging from 25% to 49% (depending on the localization of the assay within the plant), was unexpected because there are more than 20 actively transcribed APase genes within the Arabidopsis genome (Li et al., 2002). Two secreted APase isoforms are among the APases affected in pup3 (Fig. 3A). However, only one of these isoforms is recognized by the AtPAP12 antibody (Fig. 3B), and AtPAP12 transcripts are not altered in the mutant (Fig. 4D). These facts imply that PUP3 may posttranscriptionally modify at least AtPAP12 and one other APase.
The pup3 mutation maps to chromosome 5 within the Arabidopsis genome between At5g29584 and At5g36210, an area encompassing 2.7 Mb of sequence. This region includes AtPAP26, which is closely related to AtPAP12 (Li et al., 2002), but most genes in this region are annotated as either pseudogenes or genes of unknown function. The AtPAP26 protein is 57% identical and 73% similar to the AtPAP12 protein and of all the AtPAPs is the second most related sequence. AtPAP26 cDNAs have been isolated (Li et al., 2002), so it is an active gene. If pup3 is defective in the AtPAP26 structural gene, then AtPAP26 would have to affect the accumulation of AtPAP12. This phenomenon may be possible if APase enzymes stabilized each other, but this has not been reported for APases despite extensive isozyme and protein purification studies. pup3 is distinct from another mutant reported with decreased APase activity, pho3 (Zakhleniuk et al., 2001). Although genetic mapping was not reported for pho3, it is defective in various P deficiency responses. By contrast, our findings show that pup3 responds normally to low P conditions by inducing P-sensitive root-associated APase activity, APase isoforms, and transcripts (Figs. 2B, 3, A and B, and 4D).
The pup1 mutant was described previously (Trull and Deikman, 1998), and we add to its characterization here. In the earlier work, pup1 was shown to be missing an isoform in root and shoot extracts but only when separated in gel systems that preserve disulfide bonds (Trull and Deikman, 1998). Therefore, PUP1 may be required for dimerization of an APase. PAPs dimerize through disulfide bonds, and disruption of these bonds can lead to decreased activity and stability of the protein (Cashikar and Rao, 1996). Previous work also showed that pup1 lacks APase activity staining along the root surface; however, root and shoot protein extracts have normal specific APase activity (Trull and Deikman, 1998). We add that pup1 has reduced root-associated phosphatase activity (Fig. 2B) but that the pup1 isoform is not detected in exudates. One explanation for this data is that PUP1 activity may be restricted to the apoplastic space. The PUP1 isoform may be bound to cell walls, perhaps similar to the glycosylphosphatidylinositol-anchored phosphatase from S. oligorrhiza (Nishikoori et al., 2001). Ultimately, the cloning of the pup mutations will add to our understanding of this important enzyme family.
Although the rhizospheric APase pool is affected by the pup mutations, APase activity in other parts of the plant may be important as well. The effects of PUP3 are not limited to APases secreted into the rhizosphere because the APase activity pool and APase activity isoforms are affected in both root and shoot tissues. Similarly, the PUP1 isoform is present in shoots as well as roots (Trull and Deikman, 1998). Other groups have also reported that cell wall-bound or secreted APases are not exclusive to plant roots but occur in shoots as well. For example, the AtPAP12 gene has a signal peptide that causes a marker protein to be secreted from roots into the surrounding medium, but studies with its promoter have shown that this gene is transcribed first in shoots in response to P deficiency (Haran et al., 2000). Similarly, a cell wall-localized APase has been isolated from leaf-petiole cell suspension cultures from Brassica nigra (Duff et al., 1991), and in soybean APase activity has been localized to shoot vascular tissues and the lower epidermis of leaves (Staswick et al., 1994). In plants, the role for APases throughout the organism in P partitioning has not been established, but the situation may be analogous to the roles for these enzymes in animal systems. For example, PAPs in mice regulate calcium phosphate deposition and mobilization and are essential for proper osteoclastic bone turnover (Hayman et al., 1996). However, not all shoot APases are important for P recycling and regulation; for instance, the segregation of a major leaf APase in common bean recombinant inbred lines did not correlate with increased P use efficiency (Yan et al., 2001). Further uptake and partitioning studies with the pup mutants may define the role of these enzymes in P cycling throughout the plant.
The pup mutants demonstrate that secreted and/or cell wall-bound APases are important for P nutrition. When grown in a soil mix containing Po as a major source of P, pup1 and pup3 have lower shoot P concentrations (Fig. 5). However, P concentrations are not decreased when Pi is the sole source of P (Table I). Therefore, the pup1 and pup3 phenotypes are functionally important when Po can be utilized as a P source. This implies that proteins affected by the pup mutants are important for obtaining and/or maintaining shoot P concentrations when grown in soil. Even when Pi is added to the soil mix, the pup mutants cannot concentrate as much Pi in their shoots as their wild-type counterparts. The hydrolysis of Po into Pi adds to the total amount of Pi available for uptake, and in soils Pi uptake is a diffusion-limited process resulting in a zone of Pi depletion around plant roots (Marschner, 1995). Therefore, decreased hydrolysis of Po by the pup mutants may decrease the total amount of Pi available, regardless of the amount of Pi added to the system by fertilization. The peat used as a soil substrate in this study, though naturally occurring and high in Po, is not necessarily analogous to agricultural soils with established microbial populations. Therefore, further field studies with these mutants will reveal whether plant-secreted APases are important for plant P nutrition in the context of these microbial populations that also compete for Po.
MATERIALS AND METHODS
Plant Materials, In Vivo Activity Staining, and Growth Conditions
Identification of the pup mutants was based on decreased staining with the APase substrate BCIP or XP (Sigma, St. Louis; Trull and Deikman, 1998). The pup1 mutant originated from Arabidopsis T-DNA insertion lines in the WS ecotype background but was unlinked to an insertion, while the pup3 mutant was identified from ethyl methanesulfonate mutagenized Arabidopsis seeds from the Col ecotype background (Trull and Deikman, 1998). pup3 was backcrossed at least three generations prior to this study to remove unrelated ethyl methanesulfonate mutations.
For the in vivo APase staining, seedlings were grown to 5 d postgermination (dpg) in solid medium (0.5× modified Hoagland salts [Johnson et al., 1957], 0.2% Pi-purified phytagel, 1% Suc, and 0.05% MES, pH 5.7) either +P (1 mm) or −P (0 mm) containing 0.008% BCIP (Trull and Deikman, 1998). When Pi was reduced or eliminated from the growth medium, Pi salts were replaced with appropriate sulfate salts, such that the conjugate cation remained constant. Unless otherwise stated, plants were grown in a growth chamber under the following conditions: 22°C, ambient relative humidity, white light at 100 μmol m−2 s−1, and 16-h-light/8-h-dark photoperiod.
Root-Associated Phosphatase Activity Assays
Seedlings were grown to 7 dpg on +P solid medium (0.5× modified Hoagland salts [Johnson et al., 1957], 0.2% phytagel, 1% Suc, and 0.05% MES, pH 5.7). Plants of uniform size were moved to 10 mL of liquid medium (0.5× modified Hoagland salts with 0.5 mm Pi, 3% Suc, and 2.6 mm MES, pH 5.7), three plants per 150-mL flask. Flasks were shaken at 150 rpm in the dark. After 10 d, plants were rinsed in −P medium, transferred to fresh medium (either +P or −P), and allowed to grow for an additional 48 h.
For the assay, plants were removed from the shaking flasks, briefly rinsed in −P medium, and transferred in a time-dependent manner to rocking Magenta boxes containing 10 mL of reaction buffer (−P medium with 19.1 mm MES, 1.3% Suc, and 5 mm p-nitrophenol phosphate, or pNPP). Reactions proceeded for 30 to 50 min at approximately 22°C, then 185 μL of the reaction buffer was removed to 832 μL of 1 n NaOH. p-Nitrophenol (pNP) accumulation was read as A410 and converted to nanomoles by plotting values against a pNP standard curve generated with assay reagents. Roots and shoots were dissected, transferred to preweighed aluminum foil envelopes, and dried for 2 d at 65°C. Root-associated phosphatase activity was calculated as nanomoles pNP liberated by the root system per min per root dry weight (grams).
Because the two mutants were generated from different ecotype backgrounds, separate experiments were performed with pup1 and pup3 with their respective controls. Completely randomized designs were used, and each of these experiments was replicated at least twice.
Tissue Protein Extraction
Plants used as material for protein extractions were grown as described (Muchhal et al., 1996) with the following exceptions. Seeds were plated on 3 × 3-cm, 300-μm mesh nylon filters (Spectra/Mesh; Spectrum Laboratories, Los Angeles) at a density of nine seeds per filter on 0.1× Murashige and Skoog medium, 1% Suc, and 1% agar. After a 2-d 4°C cold treatment, plants were grown under continuous light until 7 dpg, then the nylon mesh filters were transferred to floating membrane rafts (LifeRafts; Sigma) over 100 mL of liquid 0.5× modified Hoagland medium with 1% Suc. Plants were grown for another 7 d, transferred to fresh +P (1 mm) or −P (0 mm) 0.5× modified Hoagland medium with 1% Suc, and grown for an additional 5 d.
For the protein extraction, roots and shoots were ground in liquid N, then ice-cold extraction buffer (0.1 m K-acetate, pH 5.5, 20 mm CaCl2, 2 mm EDTA, and 0.1 mm phenylmethylsulfonyl fluoride) added to 4 mL/g tissue. Polyvinylpolypyrrolidone was added at 60 mg/g tissue, and the samples were gently agitated at 4°C for 1 h. Samples were centrifuged at 27,000g and 4°C for 30 min, the supernatant removed to a fresh tube, and glycerol added to 20% (v/v). Proteins were quantified with the Bradford method using Coomassie Plus protein assay reagent (Pierce, Rockford, IL) and stored at −80°C. Proteins were isolated from three separate plantings.
Exudate Protein Concentration
Growth medium from the 2-d induction period of the root-associated phosphatase study was passed through a 0.2-μm filter to remove debris, then concentrated >200× with Centriplus-10 centrifugal concentrators (Millipore, Bedford, MA) at 4°C according to manufacturer's instructions. Growth media from four separate flasks were bulked together per exudate sample (therefore, each sample, n, consisted of exudates from 12 plants). Buffer exchange was carried out with protein extraction buffer during the concentration, and glycerol added to 20% (v/v) after concentration was complete. Exuded proteins were stored at −80°C. Exudates were collected from three separate experiments.
Specific APase Activity
Extracted proteins were dialyzed overnight at 4°C in dialysis tubing (6,000–8,000 Mr cutoff; Spectra/Por; Spectrum Laboratories, Los Angeles) against protein extraction buffer with 20% glycerol to remove Pi and other possible APase inhibitors. To measure specific APase activity, 0.5 μg of protein (<40 μL) was added to 300 μL of prewarmed 10 mm pNPP in 50 mm sodium acetate, pH 5.5. Reactions proceeded for 10 min at 25°C, were stopped with 600 μL of 1 n NaOH, and were quantified as for the root-associated APase assay. Samples from each of three protein harvests were assayed simultaneously.
APase Activity Isoforms
Protein electrophoresis was carried out on 5% stacking/10% resolving (w/v) native acrylamide gels at 30 to 60 V and 4°C. Equal amounts of concentrated exuded proteins (7.5 μg), roots (10 μg), or shoots (28 μg) were loaded for comparison between samples. APase staining was carried out with Fast Black K salt and β-naphthyl acid phosphate as described (Trull et al., 1997). High Mr native electrophoresis markers (Amersham Pharmacia Biotech, Piscataway, NJ) were used to estimate isoform migration. Proteins run under SDS-PAGE but otherwise native conditions and subsequently stained for APase activity were isolated and run as described previously (Trull and Deikman, 1998).
αAtPAP12 Immunodetection
αAtPAP12 polyclonal antibody, generated from recombinant Arabidopsis purple acid phosphatase (AtPAP12) gene product (GenBank accession no. U48448), was obtained from Dr. Thomas McKnight (Department of Biology, Texas A&M University). Equal amounts of exuded protein (7 μg) were separated by native PAGE on a Bio-Rad MiniGel apparatus (Hercules, CA) and blotted to a polyvinylidene difluoride membrane with a semidry electroblotter (Panther; Owl Scientific, Woburn, MA). Detection of the antibody/antigen interaction was carried out with the ECL western-blotting analysis system (Amersham Pharmacia Biotech) using a 1:2,500 dilution of αAtPAP12 1° antibody and a 1:1,000 dilution of peroxidase-linked anti-rabbit 2° antibody. Washes and detection were conducted according to manufacturer's instructions using 1× Tris-buffered saline plus Tween 20 (20 mm Tris base, 137 mm NaCl, and 0.1% Tween 20, pH 7.6) as the wash buffer.
RNA Accumulation
Plants for RNA isolation were grown vertically until 16 dpg on either +P (1.2 mm) or −P (9 μm) 0.5× modified Hoagland medium with P-purified phytagel (Trull et al., 1997). Total RNA was isolated using chloroform/phenol/isoamyl alcohol with subsequent LiCl precipitations. Equal amounts of total RNA (9.25 μg) were run on a formaldehyde agarose gel, blotted, hybridized, and washed using standard conditions. Probes were radioactively labeled with the Random Primer DNA labeling system using [32P]dATP (Gibco-BRL, Carlsbad, CA), then run over a Sephadex G-50 spin column to remove unincorporated radioactivity. Blots were hybridized sequentially with 100 ng of a 32P-labeled EST corresponding to AtPAP12 (EST 155C5; Arabidopsis Biological Resource Center, Ohio State University), then 100 ng of 32P-labeled soybean β-1 tubulin cDNA (Guiltinan et al., 1987) was hybridized to the blot as a loading control after stripping the AtPAP12 signal. The AtPAP12 EST 155C5 was excised from the lambda-Ziplox vector (Invitrogen, Carlsbad, CA) using NotI and SalI restriction enzymes, gel purified, and confirmed by sequencing. This experiment was carried out three times with identical results.
Plant Growth Conditions in Peat-Vermiculite Soil Mix
Seeds were planted in 6-cm diameter pots in a standard 55% to 65% sphagnum peat moss/perlite mix (Sunshine Aggregate Plus Mix 4; SunGro, Bellevue, WA). The soil mix contained no additional fertilizer. Prepared pots were stored at 4°C for 2 d and placed in the growth chamber under standard conditions (above). Plants were thinned to a density of four plants per pot at 7 dpg and thereafter fertilized twice weekly by subirrigation with either +P or −P fertilizer (Somerville and Ogren, 1982). No additional watering was needed. Shoots were harvested at 3 weeks postgermination (or at the first sign of the primary reproductive inflorescence) and dried as in the root-associated APase assay. At least nine shoots of each genotype per P fertilizer treatment were harvested in each experiment, and the experiment was replicated fully twice.
P Determinations
Total P determinations were carried out on dried tissues using the molybdate method (Murphy and Riley, 1962). For available soil Pi, 0.3 g of soil was suspended in 10 mL of deionized water and the eluate removed for P determination. To determine the total amount of P in the soil, 0.1 g was converted to ash by baking in a 490°C oven for 12 h prior to P determination. Soil tests were repeated three times each.
Statistical Analysis
Statistical analysis was performed with StatView version 5.0.1 (SAS Institute, Cary, NC). The cutoff was P > 0.1 for values termed not significant.
Genetic Mapping
A pup3 mapping population was generated by outcross to the WS ecotype. F2 mutant segregants were identified first by their decreased APase activity in crude shoot protein extracts. Crude proteins were isolated by grinding individual small leaves from 14 dpg seedlings (grown in peat-vermiculite soil mix) in 500 μL of 50 mm Tris, pH. 7.5, pelleting debris by centrifugation (14,000g, 4°C, 5 min), and collecting the supernatant. APase activity measurements were carried out on these extracts by adding 200 μL of extract to 2 mL of 2.5 mm pNPP in 50 mm sodium acetate, pH 4.8, and the reaction carried out at 37°C for 30 min. Reactions were stopped with the addition of 4 mL of 1 n NaOH. A420 was read with the Brinkmann PC-800 colorimeter (Westbury, NY). APase activity was calculated as A420 per shoot fresh weight per minute and expressed as a percentage of the average between the two control genotypes. Candidate pup3 protein samples with <75% APase activity compared to Col and WS control samples were quantified using the Bradford method (Bradford, 1976) and run on native activity gels to confirm the pup3 phenotype by a slight reduction in a major APase isoform when compared to control extracts (data not shown).
The pup3 mutation was mapped using CAPS as markers (Konieczny and Ausubel, 1993). Chi square analysis was used to determine marker linkage, and the Kosambi method was used to calculate genetic distances from linked markers along a chromosome (Koornneef and Stam, 1992).
Acknowledgments
We thank Dr. Thomas McKnight for use of the AtPAP12 antibody. Discussions with Drs. Jack Schultz, Eva Pell, Seogchang Kang, and Kathleen Brown were also very helpful.
This work was supported by the National Science Foundation (Plant Responses to the Environment Research Training Grant no. 9413204 to John C. Schultz and Eva J. Pell at Penn State University) and the U.S. Department of Agriculture (grant no. 95–37100–1567 to M.C.T., J.P.L., M.J.G., and J.D.).
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.103.036459.
References
- Abel S, Ticconi CA, Delatorre CA (2002) Phosphate sensing in higher plants. Physiol Plant 115: 1–8 [DOI] [PubMed] [Google Scholar]
- Berger S, Bell E, Sadka A, Mullet JE (1995) Arabidopsis thaliana Atvsp is homologous to soybean VspA and VspB, genes encoding vegetative storage protein acid phosphatases, and is regulated similarly by methyl jasmonate, wounding, sugars, light and phosphate. Plant Mol Biol 27: 933–942 [DOI] [PubMed] [Google Scholar]
- Boutin J, Provot M, Roux L (1981) Effect of cycloheximide and renewal of phosphorus supply on surface acid phosphatase activity of phosphorus deficient tomato roots. Physiol Plant 51: 353–360 [Google Scholar]
- Bradford MM (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72: 248–254 [DOI] [PubMed] [Google Scholar]
- Burns R (1986) Interactions of enzymes with soil mineral and organic colloids. In PM Huang, M Schnitzer, eds, Interaction of Soil Minerals with Natural Organics and Microbes, Vol 17. Soil Science Society of America, Madison, WI, pp 429–451
- Cashikar AG, Rao NM (1996) Role of the intersubunit disulfide bond in the unfolding pathway of dimeric red kidney bean purple acid phosphatase. Biochim Biophys Acta 1296: 76–84 [DOI] [PubMed] [Google Scholar]
- Coello P (2002) Purification and characterization of secreted acid phosphatase in phosphorus-deficient Arabidopsis thaliana. Physiol Plant 116: 293–298 [Google Scholar]
- del Pozo JC, Allona I, Rubio V, Leyva A, de la Pena A, Aragoncillo C, Paz-Ares J (1999) A type 5 acid phosphatase gene from Arabidopsis thaliana is induced by phosphate starvation and by some other types of phosphate mobilising/oxidative stress conditions. Plant J 19: 579–589 [DOI] [PubMed] [Google Scholar]
- Dodd J, Burton C, Burns R, Jeffries P (1987) Phosphatase activity associated with the roots and the rhizosphere of plants infected with vesicular-arbuscular mycorrhizal fungi. New Phytol 107: 163–172 [Google Scholar]
- Duff SM, Lefebvre DD, Plaxton WC (1991) Purification, characterization, and subcellular localization of an acid phosphatase from black mustard cell-suspension cultures: comparison with phosphoenolpyruvate phosphatase. Arch Biochem Biophys 286: 226–232 [DOI] [PubMed] [Google Scholar]
- Guame A, Machler F, De Leon C, Narro L, Frossard E (2001) Low-P tolerance by maize (Zea mays L.) genotypes: significance of root growth, and organic acids and acid phosphatase root exudation. Plant Soil 228: 253–264 [Google Scholar]
- Guiltinan MJ, Velten J, Bustos MM, Cyr RJ, Schell J, Fosket DE (1987) The expression of a chimeric soybean beta-tubulin gene in tobacco. Mol Gen Genet 207: 324–328 [Google Scholar]
- Haran S, Logendra S, Seskar M, Bratanova M, Raskin I (2000) Characterization of Arabidopsis acid phosphatase promoter and regulation of acid phosphatase expression. Plant Physiol 124: 615–626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayman AR, Jones SJ, Boyde A, Foster D, Colledge WH, Carlton MB, Evans MJ, Cox TM (1996) Mice lacking tartrate-resistant acid phosphatase (Acp 5) have disrupted endochondral ossification and mild osteoporosis. Development 122: 3151–3162 [DOI] [PubMed] [Google Scholar]
- Hunter DA, McManus MT (1999) Comparison of acid phosphatases in two genotypes of white clover with different responses to applied phosphate. J Plant Nutr 22: 679–692 [Google Scholar]
- Johnson C, Stout P, Broyer T, Carlton A (1957) Comparative chlorine requirements of different plant species. Plant Soil 8: 337–353 [Google Scholar]
- Konieczny A, Ausubel FM (1993) A procedure for mapping Arabidopsis mutations using co-dominant ecotype-specific PCR-based markers. Plant J 4: 403–410 [DOI] [PubMed] [Google Scholar]
- Koornneef M, Stam P (1992) Genetic analysis. In C Koncz, NH Chua, J Schell, ed, Methods in Arabidopsis Research. World Scientific, River Edge, NJ, pp 81–99
- Krannitz PG, Aarssen LW, Lefebvre DD (1991) Relationships between physiological and morphological attributes related to phosphate uptake in 25 genotypes of Arabidopsis thaliana. Plant Soil 133: 169–175 [Google Scholar]
- LeBansky B, McKnight T, Griffing L (1991) Purification and characterization of a secreted purple phosphatase from soybean suspension cells. Plant Physiol 99: 391–395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li D, Zhu H, Liu K, Liu X, Leggewie G, Udvardi M, Wang D (2002) Purple acid phosphatases of Arabidopsis thaliana: comparative analysis and differential regulation by phosphate deprivation. J Biol Chem 277: 27772–27781 [DOI] [PubMed] [Google Scholar]
- Li M, Tadano T (1996) Comparison of characteristics of acid phosphatases secreted from roots of lupin and tomato. Soil Sci Plant Nutr 42: 753–763 [Google Scholar]
- Marschner H (1995) Mineral Nutrition of Plants. Academic Press, New York
- McLachlan KD (1980. a) Acid phosphatase activity of intact roots and phosphorus nutrition in plants. I. Assay conditions and phosphatase activity. Aust J Agric Res 31: 429–440 [Google Scholar]
- McLachlan KD (1980. b) Acid phosphatase activity of intact roots and phosphorus nutrition in plants. II. Variations among wheat roots. Aust J Agric Res 31: 441–448 [Google Scholar]
- Miller SS, Liu J, Allan DL, Menzhuber CJ, Fedorova M, Vance CP (2001) Molecular control of acid phosphatase secretion into the rhizosphere of proteoid roots from phosphorus-stressed white lupin. Plant Physiol 127: 594–606 [PMC free article] [PubMed] [Google Scholar]
- Miyasaka SC, Habte M (2001) Plant mechanisms and mycorrhizal symbioses to increase phosphorus uptake efficiency. Commun Soil Sci Plant Anal 32: 1101–1147 [Google Scholar]
- Muchhal U, Pardo J, Raghothama K (1996) Phosphate transporters from the higher plant Arabidopsis thaliana. Proc Natl Acad Sci USA 93: 10519–10523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy J, Riley JP (1962) A modified single method for the determination of phosphate in neutral waters. Anal Chim Acta 27: 31–36 [Google Scholar]
- Nakazato H, Okamoto T, Nishikoori M, Washio K, Morita N, Haraguchi K, Thompson GA, Jr., Okuyama H (1998) The glycosylphosphatidylinositol-anchored phosphatase from Spirodela oligorrhiza is a purple acid phosphatase. Plant Physiol 118: 1015–1020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishikoori M, Washio K, Hase A, Morita N, Okuyama H (2001) Cloning and characterization of cDNA of the GPI-anchored purple acid phosphatase and its root tissue distribution in Spirodela oligorrhiza. Physiol Plant 113: 241–248 [DOI] [PubMed] [Google Scholar]
- Richardson AE (2001) Prospects for using soil microorganisms to improve the acquisition of phosphorus by plants. Aust J Plant Physiol 28: 897–906 [Google Scholar]
- Richardson AE, Hadobas PA, Hayes JE (2001) Extracellular secretion of Aspergillus phytase from Arabidopsis roots enables plants to obtain phosphorus from phytate. Plant J 25: 641–649 [DOI] [PubMed] [Google Scholar]
- Rubio V, Linhares F, Solano R, Martin AC, Iglesias J, Leyva A, Paz-Ares J (2001) A conserved MYB transcription factor involved in phosphate starvation signaling both in vascular plants and in unicellular algae. Genes Dev 15: 2122–2133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schenk G, Guddat LW, Ge Y, Carrington LE, Hume DA, Hamilton S, de Jersey J (2000) Identification of mammalian-like purple acid phosphatases in a wide range of plants. Gene 250: 117–125 [DOI] [PubMed] [Google Scholar]
- Somerville CR, Ogren WL (1982) Mutants of the cruciferous plant Arabidopsis thaliana lacking glycine decarboxylase activity. Biochem J 202: 373–380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stahl B, Klabunde T, Witzel H, Krebs B, Steup M, Karas M, Hillenkamp F (1994) The oligosaccharides of the Fe(III)-Zn(II) purple acid phosphatase of the red kidney bean. Eur J Biochem 220: 321–330 [DOI] [PubMed] [Google Scholar]
- Staswick PE, Papa C, Huang JF, Rhee Y (1994) Purification of the major soybean leaf acid phosphatase that is increased by seed-pod removal. Plant Physiol 104: 49–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steen I (1998) Phosphorus availability in the 21st century. Phosphorus and Potassium 217: 25–31 [Google Scholar]
- Thomas C, Sun Y, Naus K, Lloyd A, Roux S (1999) Apyrase functions in plant phosphate nutrition and mobilizes phosphate from extracellular ATP. Plant Physiol 119: 543–551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trull MC, Deikman J (1998) An Arabidopsis mutant missing one acid phosphatase isoform. Planta 206: 544–550 [DOI] [PubMed] [Google Scholar]
- Trull MC, Guiltinan MJ, Lynch JP, Deikman J (1997) The responses of wild-type and ABA mutant Arabidopsis thaliana plants to phosphorus starvation. Plant Cell Environ 20: 85–92 [Google Scholar]
- Vance CP, Uhde-Stone C, Allan DL (2003) Phosphorus acquisition and use: critical adaptations by plants for securing a nonrenewable resource. New Phytol 157: 423–427 [DOI] [PubMed] [Google Scholar]
- Yan X, Liao H, Trull MC, Beebe SE, Lynch JP (2001) Induction of a major leaf acid phosphatase does not confer adaptation to low phosphorus availability in common bean. Plant Physiol 125: 1901–1911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zakhleniuk OV, Raines CA, Lloyd JC (2001) pho3: a phosphorus-deficient mutant of Arabidopsis thaliana (L.) Heynh. Planta 212: 529–534 [DOI] [PubMed] [Google Scholar]